Summary
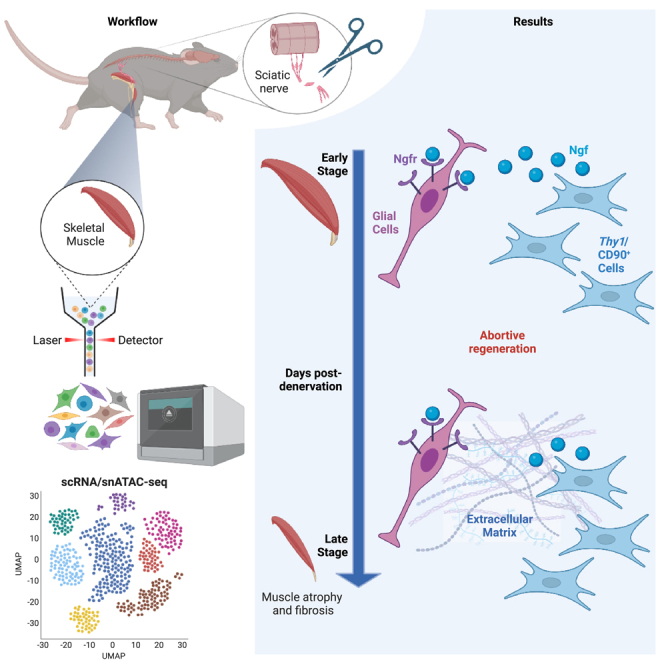
We performed scRNA-seq/snATAC-seq of skeletal muscles post sciatic nerve transection to delineate cell type-specific patterns of gene expression/chromatin accessibility at different time points post-denervation. Unlike myotrauma, denervation selectively activates glial cells and Thy1/CD90-expressing mesenchymal cells. Glial cells expressed Ngf receptor (Ngfr) and were located near neuromuscular junctions (NMJs), close to Thy1/CD90-expressing cells, which provided the main cellular source of NGF post-denervation. Functional communication between these cells was mediated by NGF/NGFR, as either recombinant NGF or co-culture with Thy1/CD90-expressing cells could increase glial cell number ex vivo. Pseudo-time analysis in glial cells revealed an initial bifurcation into processes related to either cellular de-differentiation/commitment to specialized cell types (e.g., Schwann cells), or failure to promote nerve regeneration, leading to extracellular matrix remodeling toward fibrosis. Thus, interactions between denervation-activated Thy1/CD90-expressing and glial cells represent an early abortive process toward NMJs repair, ensued by the conversion of denervated muscles into an environment hostile for NMJ repair.
Subject areas: Natural sciences, biological sciences, Biochemistry, Physiology
Graphical abstract
Highlights
-
•
Selective expansion of NGFRpos glial and CD90pos mesenchymal cells after denervation
-
•
Functional communication between glial and mesenchymal cells via NGFR/NGF
-
•
Recombinant NGF or NGF-expressing cells maintain glial cells survival ex vivo
-
•
Denervation ultimately culminates with muscle fibrosis and failure to repair NMJs
Natural sciences; Biological sciences; Biochemistry; Physiology
Introduction
Skeletal muscle response to homeostatic perturbations involves the coordinated activation of a multitude of muscle-resident cell types that cooperate to restore the original homeostasis.1,2,3 Recent single-cell-based transcriptional analyses have revealed the identity of muscle-resident cells and their dynamic heterogeneity in response to acute or chronic perturbations of skeletal muscle homeostasis.4,5,6,7,8,9,10,11,12,13,14,15,16 These studies have established that homeostatic perturbation of skeletal muscles by physical injury is highly influenced by the inflammatory infiltrate, which transiently provides regulatory signals to other muscle-resident cells, including fibro-adipogenic progenitors (FAPs) and other interstitial cell types. The interplay between signals released by these cells ultimately promotes muscle stem cells (MuSCs)-mediated regeneration. Lack of complete resolution of inflammation and/or dysregulated interactions between muscle-resident cells have been implicated in the pathogenesis of muscular diseases, including muscular dystrophies.4,5,12,16,17,18,19,20
By contrast, perturbation of muscle homeostasis by denervation is accompanied by a milder inflammatory response, activation of FAP-derived pro-atrophic signals and negligible activation of MuSCs-mediated regeneration.3,21,22 Irreversible denervation by sciatic nerve transection provides an optimal experimental setting for longitudinal single cell-based transcriptional analysis of muscles post-denervation. By capturing cell type-specific transcriptional signatures it is in fact possible to identify denervation-activated muscle-resident cell types and predict their contribution to muscle-derived signals implicated in muscle response to denervation. Previous studies have implicated different cell types, including Schwann cells and MuSCs, in muscle responses to nerve injury, such as NMJ repair and extra-cellular matrix (ECM) remodeling.22,23,24,25,26 However, a systematic, unbiased identification of the muscle-resident cell types activated by denervation, and the activation of related functional networks, has not been performed to date. This gap of knowledge currently prevents our complete understanding of the pathogenesis of denervation-based muscular diseases and the identification of targets for therapeutic interventions toward repairing diseased or injured NMJs.
Here we present a longitudinal analysis of the changes in the transcriptional profiles and chromatin accessibility of muscle-resident cells at the single-cell level, by parallel single-cell RNA sequencing (scRNA-seq) and single nucleus assay for transposase-accessible chromatin sequencing (snATAC-seq), upon acute denervation by sciatic nerve transection.
Our data reveal an abortive attempt to repair NMJs, whereby NGFR and NGF expression in denervation-activated glial cells and mesenchymal cells, respectively, mediate functional interactions between these cell types at early time points post-denervation, followed by a global conversion of denervated muscles into an environment hostile for NMJ repair.
Results
scRNA-seq-mediated identification of denervation-responsive muscle-resident cell types
We used scRNA-seq analysis of whole skeletal muscles subjected to denervation, as compared to unperturbed counterparts, in order to capture gene expression patterns that could identify transcriptional signatures of muscle-resident cell types activated in response to denervation. We performed scRNA-seq of single-cell suspensions prepared from 2 muscles downstream to sciatic nerve—tibialis anterior (TA) and gastrocnemius (GA) muscles—isolated from 3-month-old male mice, either unperturbed (control - CTR) or subjected to denervation by complete sciatic nerve transection (Figure 1A). We selected 3 time points post-denervation (p.d.), based on previous studies reporting on the activation kinetics of muscle-resident cells by the first 48 h post-denervation (day 2 p.d.) and the expansion of FAPs with persistent activation of STAT3-IL6 pathway at later time points (days 5 and 15 p.d.) that eventually leads to myofiber atrophy and fibrosis.21 scRNA-seq datasets were generated from two biological replicates of a pool of TA and GA muscles isolated at our specific time points post-denervation (abbreviated as DEN2d, DEN5d, and DEN15d in figures and tables) or from unperturbed mice (abbreviated as CTR in figures and tables). Fluorescence-activated cell sorting (FACS) was used to isolate single cells and eliminate contaminating cell debris, doublets, and dead cells (Figures S1A–S1D). Single-cell libraries were generated with the 10X Genomics Chromium platform and sequenced on the Illumina Nova-Seq platform. We isolated a total of 48,666 cells across all the experimental conditions, with 44,403 cells analyzed post-filtering, and detected a total of 36,185 genes (average of 6,000 cells per sample) with a read depth between 30k and 50k per sample (Figures S1A–S1D). Seurat package27 was used for most of the data analysis (see STAR Methods section scRNA-seq data preprocessing, dimensionality reduction, and visualization, for details). Data were filtered to exclude cells with higher rate of mitochondrial genes and cell doublets, as predicted with Scrublet28 (Figures S1A–S1D). Principal component analysis (PCA), used for dimensionality reduction, revealed no obvious batch effect between biological replicate samples from the different experimental points (Figures S1E and S1G) or for cells captured at different cell cycle phases (Figures S1F and S1H). Nevertheless, cells from all experimental points were normalized together taking into consideration their mitochondrial gene content, number of detected genes, and cell cycle-related genes.
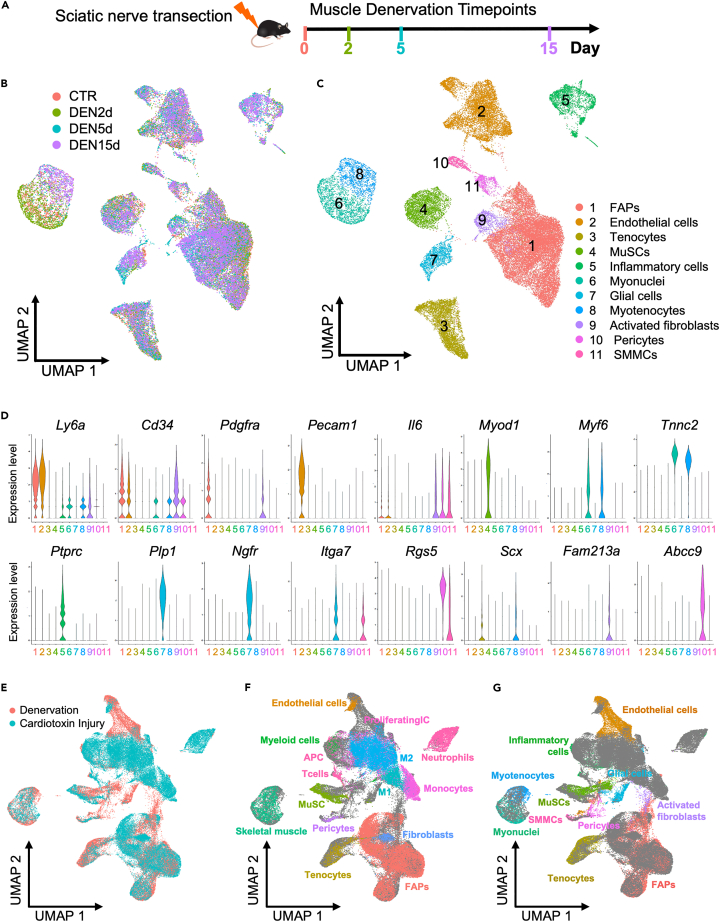
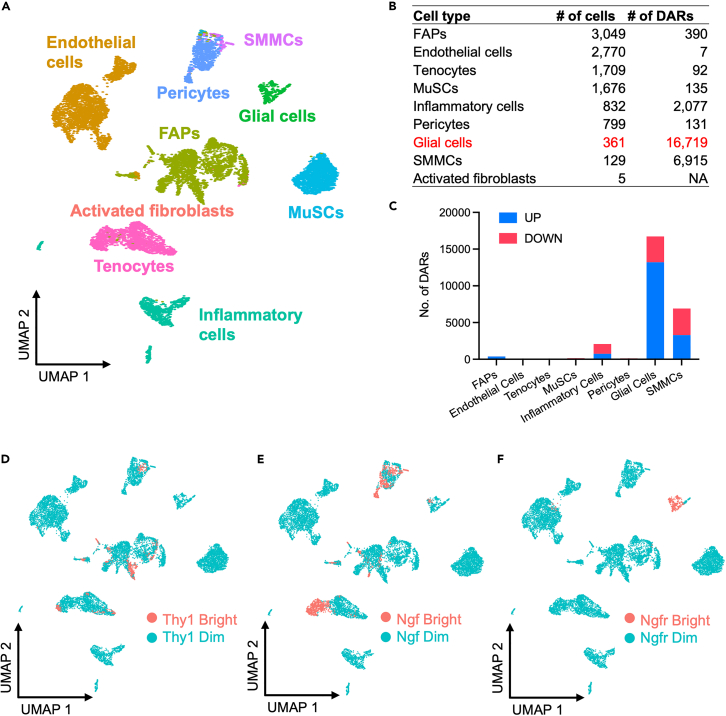
Figure 1.
scRNA-seq-mediated identification of denervation-responsive muscle-resident cell types
(A) Experimental design. Single-cell suspensions were generated from whole TA and GA muscles from non-denervated mice and from three time points following denervation through sciatic nerve transection (namely, 0-, 2-, 5-, or 15-day post-denervation, abbreviated as CTR (Non-DEN), DEN2d, DEN5d, and DEN15d in figures and tables).
(B and C) UMAP embedding of all the single cells comprising the dataset colored either by experimental condition (B) or by meta-cluster (C).
(D) Violin plots of the expression profile of marker genes for each meta-cluster, color legend as in 1C.
(E–G) UMAP embedding of the integration analysis of denervation and injury (Oprescu et al., 2020)10 datasets (E), with Oprescu’s meta-cluster identities (F), or with the meta-cluster identities from our dataset (G)—color legend as in 1C.
Independent unsupervised Shared Nearest Neighbor (SNN) clustering was used to define clusters representing specific cell types and sub-types, and Universal Mani-fold Approximation and Projection (UMAP) was used as a non-linear dimensionality reduction method to represent single-cell transcriptomes from all conditions in the 2D space. These analyses identified 18 clusters of single cells, likely representing sub-populations or cellular states of muscle-resident cell types isolated from unperturbed muscles or at defined time points following denervation. Marker identification analysis found cell type-specific gene expression patterns, which revealed the expression of well-known lineage markers and further reduced single-cell distribution into 11 meta-clusters (Figure 1C). This analysis revealed the identity of FAPs (based on the simultaneous expression of Ly6a, Cd34, and Pdgfra),29,30 as the most represented population of muscle-resident cells (Figures 1C, 1D, and S2). Other major clusters identified additional and well-known muscle-resident cell types, including endothelial cells (based on Pecam1 and Ly6a co-expression),31 pericytes (marked by the co-expression of Abcc9, Pdgfrb, and Kcnj8),32 inflammatory cells (a heterogeneous group of immune cells that share the selective expression of Ptprc - CD45),33 and tenocytes (enriched for the expression of Scx and Pdgfrb)34 (Figures 1C, 1D, and S2). MuSCs were identified as a cluster of heterogeneous cells expressing Pax7, Myod1, and Calcr in various combinations35,36,37 (Figures 1C, 1D, and S2). We also detected a cluster of cells that were identified as myonuclei, likely derived from contamination of our cell preparation. They clustered adjacent to myotenocytes, as they both shared the selective expression of Tnnc2, but differ on the expression of Scx, which was restricted to myotenocytes (Figures 1C, 1D, and S2). Lastly, three additional clusters were assigned to recently identified muscle-resident cell types, such as smooth muscle and mesenchymal cells (SMMCs), activated fibroblasts, and muscle glial cells9,15 (Figures 1C, 1D, and S2). For single cells dispersed between clusters, we could not identify any cell-type specific transcriptional signature or identity markers that could discriminate them from other muscle-resident cells. As these clusters share gene expression with surrounding cell types, we speculate that they could be composed by cells in dynamic transition from one cluster to another, within the spectrum of adjacent cell lineages.
We next performed an integrative analysis of our denervation dataset with a publicly available scRNA-seq dataset of muscle regeneration after cardiotoxin injury,10 in order to cross-verify the cell identity of muscle-resident cell types and to determine their differential transcriptional response to distinct types of perturbations—e.g., denervation vs. myotrauma. This analysis demonstrates a large overlap between the muscle-resident cell types identified after denervation and those observed in previous studies on skeletal muscles undergoing regeneration post-injury (Figures 1E–1G). Of note, unlike regenerating muscles, which exhibit extensive changes in the size of multiple cell clusters following myotrauma,9,10,11,13,14 denervation did not appear to significantly alter the average number of cells in most of the clusters, such as FAPs, endothelial cells, tenocytes, and MuSCs (Figure S3 and Table S1). In this regard, we reasoned that scRNA-seq provides a tool to measure expansions or contractions of specific cell types, when performed at sequential time points following an experimental perturbation. Indeed, the relative percentage of cluster representation is a rough reflection of their relative amount within the whole number of single cells isolated, assuming that there is no bias in cell type-specific extraction preference. With the exception of myonuclei, whose extraction is clearly biased by their anatomical position inside fibers, muscle-resident cell extraction is unlikely to be biased toward any particular cell types, as also indicated by recent studies of whole muscles showing that scRNA-seq analysis could capture quantitative dynamics of muscle-resident cell expansion post-injury that were previously detected based on immunohistochemistry or FACS analyses.9,10,11,13,14 For instance, a dramatic expansion of the inflammatory infiltrate and immune-resident cells occurs in the 48 h immediately after myotrauma, with the inflammatory cells accounting for about 90% of the single cells.10 We argue that this massive expansion of one cell type invariably reduces the capture of other muscle-resident cell types, which are by consequence underrepresented within the 10% of the non-inflammatory cell types analyzed. Figure 2A and Table S1 show that, unlike muscle injury by myotrauma, denervation did not trigger an immediate and massive infiltration of immune cells, which only by day 5 p.d. started to display a modest increase in the number that remained unaltered by day 15 p.d. (Figure S3). As such, denervated muscles did not exhibit any specific bias in the simultaneous analysis of muscle-resident cell types at any time point p.d. Another potential bias in the quantitative interpretation of the longitudinal analysis of perturbed muscles by scRNA-seq could be provided by the massive loss of one or more cell types that account for a large proportion of cells, thereby skewing the representation of the other cell types (Figure 2A and Table S1). Upon denervation, we observed the expected drastic reduction only of the myonuclei, whose number dramatically drops by day 5 p.d., after a paradoxical expansion observed on day 2 p.d. However, we note that myonuclei represent an accidental contamination from myofiber leakage typically occurring during single-cell extraction, as also observed by others.9,10,14,15 Since myonuclei account for less than 5% of cells post-filtering (Figure 2A and Table S1), they do not provide a significant bias. The transient drop of endothelial cell number observed on day 2 p.d., which coincided with a consensual increase in the proportion of FAPs, also did not significantly alter the percentage of cell type distribution (Figures 2A and S3).
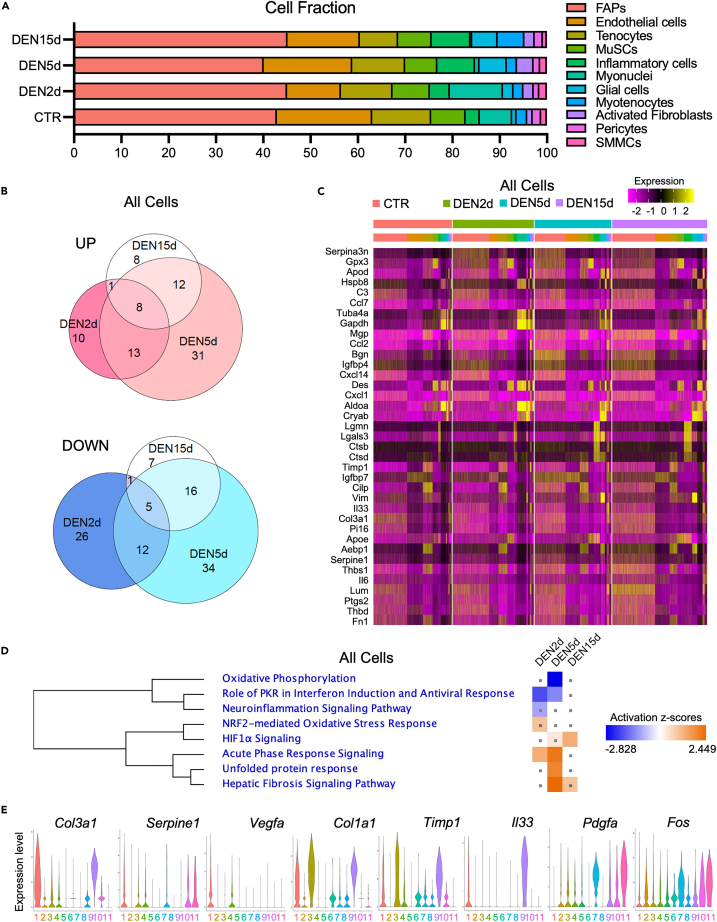
Figure 2.
Dynamic changes in cell number and gene expression in muscle-resident cells following denervation
(A) Barplot representing the proportion of single cells belonging to each meta-cluster for each experimental condition.
(B) Venn diagram of upregulated genes (upper) and downregulated genes (bottom) in all single cells at DEN2d, DEN5d, and DEN15d compared with CTR. DE genes with p-value-adj ≤ 0.01 are counted.
(C) Heatmap of top 20 upregulated genes in all cells at DEN2d, DEN5d, and DEN15d respectively compared with CTR, clustered by condition and meta-cluster identity. Gene expression values visualized on a scale from low (purple) to high (yellow), with black for zero expression value.
(D) IPA comparison of DE genes in all cells at DEN2d, DEN5d, and DEN15d respectively compared with CTR. Pathways significantly enriched in either time point with absolute Z score > 1 were shown. Pathways with p value ≥ 0.05 were indicated with gray dots. Pathways activation status (Z-scores) visualized on a scale from repressed (blue) to activated (orange), with neutral status (Z-Score = 0) set to white.
(E) Violin plots in DEN5d cells of genes involved in hepatic fibrosis signaling pathways.
Our scRNA-seq analysis revealed that, following denervation, the majority of the most represented clusters of muscle-resident cells did not exhibit dramatic alterations in their number; however, two cell types that were underrepresented in unperturbed muscles exhibited a progressive increase in both absolute number and percentage relative to all muscle-resident cells (Figure 2A and Table S1). These putative denervation-activated populations coincided with clusters identified as muscle-resident glial cells22 and a cell population previously defined as injury-activated fibroblasts10,13 (Figure 2A and Table S1). Their patterns of expansion were similar during the first 5 days p.d., with a progressive increase in both percentage and absolute cell number; however, by day 15 p.d. both cell types declined in terms of percentage, despite the observed increase in the absolute number of glial cells (Figures 2A and S3, and Table S1). Progressive and moderate expansion of two cell types that are abundant in unperturbed muscles—myotenocytes and inflammatory cells—was also observed (Figures 2A and S3B, and Table S1). Modest fluctuations in FAPs and endothelial cells, tenocytes, and SMMCs were observed during our time-course post-denervation, while negligible changes in MuSCs number were observed at every time point post-denervation (Figures 2A and S3B, and Table S1).
The dynamic changes in the abundance of each cell type at sequential time points post-denervation are represented by the UMAP embedding shown in Figure S3A, which illustrates the selective and progressive expansion of muscle glial-derived cells and activated fibroblasts in response to denervation (Figure S3A). At the same time, it shows that, within other cell types that did not undergo significant expansions or reductions in number, there were changes in the internal distribution of gene expression, which predict transitions between cell states within the same lineage. Indeed, we found that sub-clusters identified within FAPs, endothelial cells, and inflammatory cells underwent extensive reconfiguration along with the muscle response to denervation (Figure S3A). We also determined the magnitude of transcriptional changes induced by denervation in each cell type, by comparing the number of differentially expressed (DE) genes in all cell types at each p.d. condition against control. Table S2 shows that the number of DE genes was higher in muscle glial-derived cells and activated fibroblasts (Table S2).
Overall, among the muscle-resident cells, glial cells and activated fibroblasts were the cell types that exhibited the most significant changes in abundance and gene expression, indicating that they are the two major denervation-activated muscle-resident cell types.
Global dynamics of transcriptional responses of muscle-resident cells following denervation
Global gene expression analysis revealed that the magnitude of changes in gene expression of muscle-resident cells in response to denervation was much reduced when compared to the response to myotrauma.10 Venn diagram shows that the largest changes in gene expression occurred between days 2 and 5 p.d., while day 15 p.d. shows a reduced extent of DE genes (Figure 2B). Functional analysis by Ingenuity Pathway Analysis (IPA) of DE genes across experimental conditions predicted general trends of repression of oxidative phosphorylation, PKR-mediated viral response, and neuroinflammation within the initial stages of muscle response to denervation—from days 2–5 p.d.—followed by activation of processes related to fibrosis and neo-angiogenesis that were detected at later stages—days 5–15 p.d. (Figures 2C and 2D).
While these data indicate a general biphasic response of skeletal muscles to denervation, eventually leading to a persistent activation of fibrosis-related processes, global analysis of gene expression could be largely biased by the relative amounts of different cell types analyzed. In particular, a global analysis might not be informative on genes activated by denervation in cell types representing a very small percentage of the total muscle-resident cells, such as muscle glial cells and activated fibroblasts. Indeed, when we identified the top marker genes for each cell type across all conditions (Table S3), we found that the top genes of the largest populations were the same genes detected as DE in the heatmap shown in Figure 2C. For instance, top DE genes were enriched in ECM and inflammatory genes, which mostly derived from larger populations, such as FAPs, endothelial/pericytes, and inflammatory cells. Conversely, top marker genes of the denervation-activated populations (muscle glial cells and activated fibroblasts), which represent a small percentage of the whole population of muscle-resident cells, were largely under-represented in the same heatmap (Figure 2C and Table S3). Figure 2E illustrates the relative contribution of multiple cell types, by day 5 p.d., to the expression levels of upregulated genes implicated in the major biological processes activated by denervation, such as ECM remodeling and fibrosis.
This prompted an interest to restrict our focus and further analyze DE genes and changes in chromatin accessibility in the two most activated muscle-resident cell types (glial cells and activated fibroblasts) in response to denervation.
Muscle glial cells exhibit a biphasic activation of gene expression predictive of an abortive pro-regenerative response
Muscle glial cells are one of the least represented cell clusters in unperturbed muscles (Figures 2A and S3A, and Table S1); hence, their response to perturbations that trigger massive activation of larger populations (i.e., myotrauma) could not be entirely appreciated by previous scRNA-seq studies.9,10,13 Our analysis shows that muscle glial cells quickly responded to denervation with a progressive increase in cell number (Figures 2A and S3A, and Table S1) that was accompanied by significant changes in gene expression along with the time points p.d. (Table S2). Interestingly, most of the DE genes were detected at days 2 and 5 p.d., with a strong bias toward upregulated genes (Figure 3A). Figure 3B displays the heatmaps of top DE genes at each experimental time point, showing a general upregulation of genes at days 2 and 5 p.d., followed by a consensual downregulation at day 15 p.d., although some genes, like Igfbp3, Tnc, Vim, and Btc, exhibited a progressive increase from day 2 to day 15 p.d. (Figure 3B). The glial identity of these cells was confirmed by a set of markers (Plp1, Kcna1, Mpz, and Ptn) used in two earlier scRNA-seq datasets for Schwann cell- or glial cell-specific gene expression9,15 (Table S3).
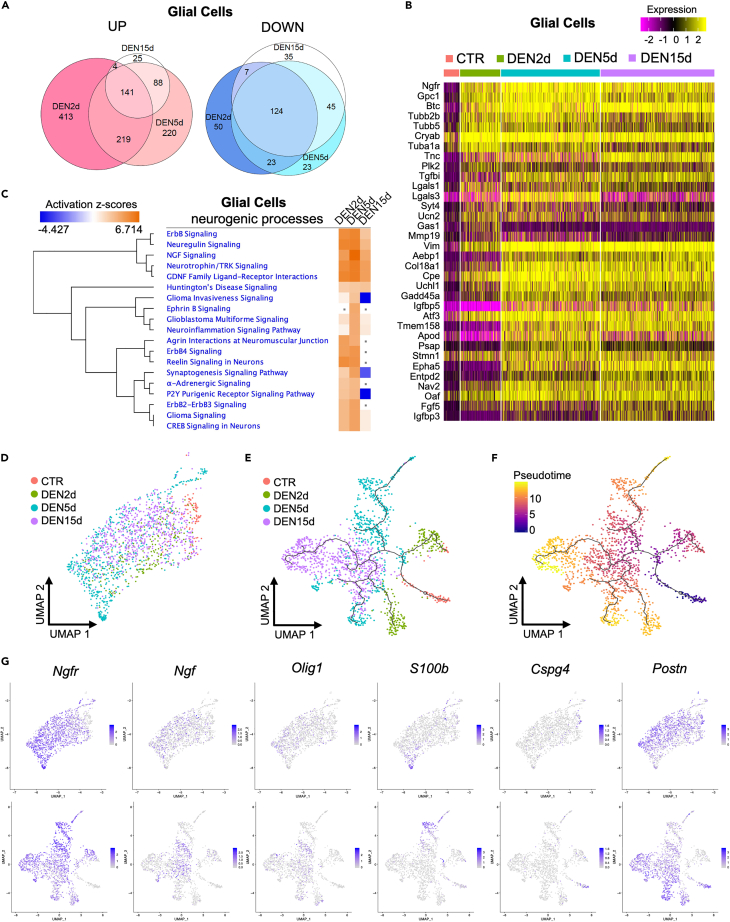
Figure 3.
Response of muscle-resident glial cells to denervation
(A) Venn diagram of upregulated genes (left) and downregulated genes (right) in glial cells at DEN2d, DEN5d, and DEN15d compared with CTR. Genes with p-value-adj ≤ 0.01 are counted.
(B) Heatmap of top 20 upregulated genes in glial cells at DEN2d, DEN5d, and DEN15d respectively compared with CTR. Gene expression values visualized on a scale from low (purple) to high (yellow), with black for zero expression value.
(C) IPA comparison of DE genes in glial cells at DEN2d, DEN5d, and DEN15d respectively compared with CTR. Selected pathways related to neurogenic processes are shown. Pathways with p value ≥ 0.05 were indicated with gray dots. Pathways activation status (Z-scores) visualized on a scale from repressed (blue) to activated (orange), with neutral status (Z-Score = 0) set to white.
(D) UMAP embedding subset from the whole dataset of glial cells colored by time point.
(E) UMAP embedding of glial cells after re-clustering, with superimposition of pseudo-time trajectories from Monocle3. Glial cells are colored by time point.
(F) Same as (E), but glial cells are colored by pseudo-time. Pseudotime visualized on a scale from early (blue) to late (yellow), with intermediate stages in purple and orange.
(G) UMAP embedding of glial cells before (upper) and after (bottom) re-clustering, showing the expression of selected genes associated with different glial cell-derived lineages.
IPA analysis of DE genes across all experimental conditions predicted an activation of multiple neurogenic processes, including glial cell-derived neurotrophic factor (GDNF)- and nerve growth factor (NGF)-mediated signaling, neuregulin signaling, and other processes implicated in nerve repair, such as neuronal survival, synaptogenesis, and axon guidance, between days 2 and 5 p.d. (Figure 3C and Table S3).38 However, these neurogenic processes were no longer found activated by day 15 p.d. (Figure 3C), suggesting that the progression of denervation-activated muscle glial cells toward nerve repair was interrupted from day 5 to day 15 p.d. We further investigated the neurogenic progression of muscle-glial-derived cells following denervation, by pseudo-time analysis, using Monocle3,39 which infers temporal dynamics of gene expression as cells progress through time points. This analysis revealed that muscle glial-derived progenies diverged into two pseudo-temporal trajectories during our experimental conditions, at days 2 and 5 p.d.; however, by day 15 p.d. most of the muscle glial-derived cells collapsed into one common central trajectory (Figures 3D–3F). Module analysis of the DE genes within the pseudo-temporal trajectories could define 8 distinct functional modules (Figure S4A). According to IPA analysis, module 4, which was more represented in the trajectory of cells at day 2 p.d., was enriched in genes implicated in metabolic reprogramming toward oxidative phosphorylation—a process associated with a general commitment to differentiation. Modules 2 and 3, which were enriched in neurogenic processes, such as axonal guidance, or pathways implicated in neurogenesis, including integrin-linked kinase (ILK) signaling and vitamin D /retinoid X receptors (VDR/RXR) activation, were mostly represented in the trajectory of cells at day 5 p.d. (Figures S4A–S4C). However, while module 2 was still represented in the trajectory of cells at day 15 p.d., the same trajectory was dominated by the upregulation of genes implicated in ECM-related processes, including fibrosis and acute phase response signaling (Figures S4A–S4C). The comparison of the initial UMAP embedding of the whole dataset with the UMAP embedding of muscle glial-derived cells used for pseudo-time analysis revealed that the cell trajectories identified by Monocle corresponded to distinct glial cell-derived progenies, whose dynamic gene expression patterns predicted a commitment to specialized cell types in response to denervation (Figures S4D and S4E). Indeed, while the glial cell identity marker Plp1 was invariably expressed by all muscle glial cells in unperturbed muscles as well as at each time point p.d., the majority of neurogenic genes were induced only upon denervation (Figures S5A and S5B). Among them, Ngfr expression was induced in the large majority of glial-derived cells following denervation (Figures 3G and S5A). The identity of Ngfr-expressing cells, as denervation-activated glial cells, was further confirmed by histological analysis of Plp1-tdTomato transgenic mice, generated by crossing transgenic mice expressing a tamoxifen-inducible Cre driven by the mouse Plp1 promoter with a loxP-flanked STOP cassette preventing transcription of a CAG promoter-driven red fluorescent protein variant (tdTomato). While Plp1-tdTomato-positive cells were found in non-denervated mice as NGFR-negative cells, upon denervation all Plp1-tdTomato-positive cells turned positive for NGFR expression (Figure S5C).
Interestingly, Figures 3G and S4G illustrate how Ngfr-expressing cells aligned toward a gradient of increasing Ngfr expression, with higher levels being observed in cells polarized toward the lower tip of the UMAP that was only detected at day 5 p.d. (Figures 3G and S4G). Of note, this lower tip was composed of cells that also expressed presumptive markers of Schwann cells, such as S100b (Figures 3G and S4D). S100b expression also marked a distinct streak of cells in the UMAP (Figure 3G) that corresponded to cell trajectories identified by the pseudo-time analysis at days 2 and 5 p.d. (Figures 3D–3F, S4D, and S4E). Interestingly, by day 5 p.d. a subset of S100b expressing cells were aligned into an outstanding streak that uniformly exhibited a gene expression profile predictive of G2-M phase progression (Figures S4E and S4F), suggesting that they could represent an expansion of Schwann cell progenitors. A recent work has established the identity of a specialized sub-population of Schwann cells within the NMJ, also referred to as perisynaptic Schwann cells (PSCs), which co-express S100b and Cspg4 (NG2)—a gene otherwise expressed in other S100B negative cell types.40 However, our data show that co-expression of S100b and Cspg4 could be only detected in glial cells from unperturbed muscles (Figures 3G, S5A, and S5B). Thus, the streak of proliferating cells that express S100b, but not Cspg4, might represent a gradient of Schwann cell progenitors before their functional specialization into PSCs, similar to what was observed during development.40
The finding that S100b/Cspg4 expressing cells accounted for the large majority of muscle-resident glial cells in unperturbed muscles, but were not detected at any time point p.d., suggests an abortive attempt to repair NMJ in denervated muscles, possibly because of the irreversible nature of denervation by complete sciatic nerve transection. This hints at the possibility that glial cells of unperturbed muscles are mostly composed of Schwann cells, whereas denervation-activated muscle glial cell progenies are more heterogeneous and might contain different populations of cell types in dynamic transition along the time points p.d. (Figure S4G). For instance, within the high Ngfr-expressing cells we identified a subset of cells that also expressed low levels of Ngf in response to denervation, in concomitance with the appearance of Olig1 expression (Figure 3G). These events have been previously associated with Schwann cell de-differentiation, as an early response to nerve injury that typically precedes their expansion and differentiation into Schwann cells toward nerve repair.41 Consistently, these cells were polarized toward a cluster of high Ngfr-expressing cells at day 5 p.d.; however, by day 15 p.d. these cells re-dispersed among the other muscle glial-derived cells (Figures 3G, S4D–S4G, S5A, and S5B). A third distinct group of denervation-activated glial cells was marked by the expression of Postn (Figure 3G), without co-expression of Ngfr, and was enriched in genes implicated in ECM remodeling, including Postn itself, Tnc, and Itgb8 (Figures 3G, S4D, S4E, S5A, and S5B) as well as Serpine1, Col1a1, and Timp1 (data not shown). This subset coincided with the cell trajectories identified by pseudo-time analysis at days 2 and 5 p.d. that were found in the opposite direction of the cell trajectories enriched with Ngfr at the same time points (Figures 3D–3F, S4D, and S4E). This population progressively expanded along with the time points p.d. and represented the majority of muscle glial cells at day 15 p.d. Interestingly, while these three putative sub-populations of muscle glial-derived cells could be clearly separated, as distinct clusters/trajectories of cells during days 2 and 5 p.d., by day 15 p.d. most of the muscle glial-derived cells repositioned into a diffuse and heterogeneous population of Postn-expressing cells (Figures 3G, S4D–S4G, S5A, and S5B).
Overall, the trend of DE genes in muscle glial-derived cells along with our time points p.d. suggests an initial activation of pro-neurogenic processes toward nerve regeneration and recovery of NMJs that eventually fails and is followed by the retention of a heterogeneous population of glial-derived cells expressing genes implicated in ECM remodeling.
Denervation-activated fibroblasts exhibit patterns of gene expression predictive of mesenchymal cell types
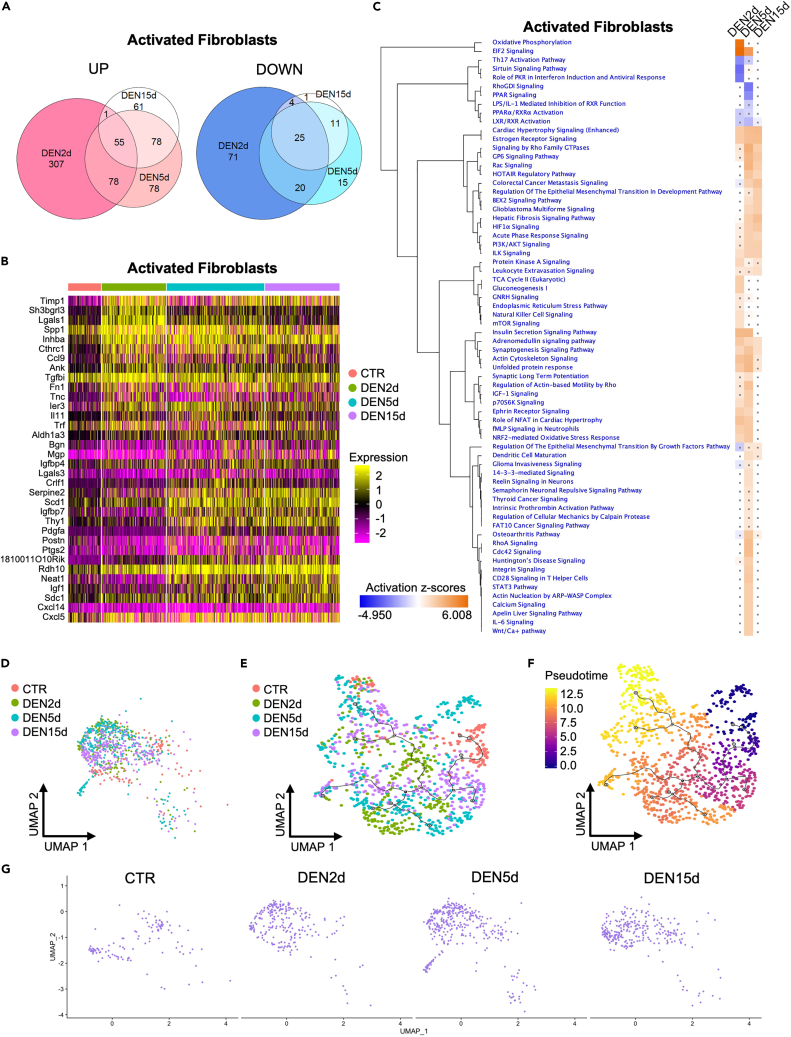
Denervation-activated fibroblasts represent a population of mesenchymal cells that share many gene expression features with FAPs, the most contiguous cell type, as shown by the UMAP embedding (Figure 1C). While both activated fibroblasts and FAPs express Cd34, a cell surface glycoprotein that is generally expressed in hematopoietic and mesenchymal muscle-resident cells,42 lower expression of Pdgfra and Ly6a could discriminate denervation-activated fibroblasts as a cellular cluster independent from FAPs (Figure S2). Moreover, at variance with FAPs, which represent the most abundant fraction of muscle-resident cells, a modest amount of activated fibroblasts (2.2% of the whole dataset) was detected in unperturbed muscles (Figure 2A and Table S1); however, similar to muscle glial cells, activated fibroblasts progressively expanded following denervation (Table S1, Figures 4G and S3A) and exhibited significant changes in gene expression between days 2 and 5 p.d., with a strong bias toward gene upregulation (Table S2; Figures 4A and 4B). IPA analysis of DE genes at each time point p.d. predicted a consensual activation of several signaling pathways implicated in neurogenic processes related to synaptogenesis, axon guidance, and neuronal migration, among others (Figure 4C). Interestingly, similar to muscle glial cells, also in denervation-activated fibroblasts these neurogenic signals were downregulated by day 15 p.d.; in contrast, DE genes implicated in ECM remodeling toward cell proliferation, migration, and fibrosis (such as Serpine 2, Scd1, Igfbp7, and Thy1) continued to be expressed across all time points p.d. (Figure 4B). While pseudo-time analysis could not identify well-defined cellular trajectories in denervation-activated fibroblasts along with the time points p.d. (Figures 4D–4F), comparative analysis with UMAP indicated a general trend to form sub-clusters at days 2 and 5 p.d., followed by a progressive dispersion within the heterogeneous cell population observed by day 15 p.d. (Figures 4D–4G). Overall, the gene expression profile shown by denervation-activated fibroblasts along with the time points p.d. is predictive of a mixture of mesenchymal cell types. Consistently, an integrative analysis of the UMAP of our scRNA-seq from denervated muscle with that of a scRNA-seq from mouse sciatic nerve43 shows that the denervation-activated fibroblasts are adjacent to two distinct populations of muscle mesenchymal cells identified in our dataset and overlap with a cluster of cells contiguous to nerve-associated fibroblasts and non-myelinating Schwann Cells (nmSC) in the sciatic nerve (Figures S6A and S6B). On the other hand, the same analysis confirmed that a fraction of denervation-activated glial cells overlap with myelinating Schwann cells (mySC) detected in the sciatic nerve (Figures S6A and S6B).
Figure 4.
Response of muscle-resident mesenchymal cells to denervation
(A) Venn diagram of upregulated genes (left) and downregulated genes (right) in activated fibroblasts at DEN2d, DEN5d, and DEN15d compared with CTR. Genes with p-value-adj ≤ 0.01 are counted.
(B) Heatmap of top 20 upregulated genes in activated fibroblasts at DEN2d, DEN5d, and DEN15d respectively compared with CTR. Gene expression values visualized on a scale from low (purple) to high (yellow), with black for zero expression value.
(C) IPA comparison of DE genes in activated fibroblasts at DEN2d, DEN5d, and DEN15d respectively compared with CTR. Pathways significantly enriched in either time point with absolute Z score > 1 were shown. Pathways with p value ≥ 0.05 were indicated with gray dots. Pathways activation status (Z-scores) visualized on a scale from repressed (blue) to activated (orange), with neutral status (Z-Score = 0) set to white.
(D) UMAP embedding subset from the whole dataset of activated fibroblasts colored by time point.
(E) UMAP embedding of activated fibroblasts after re-clustering, with superimposition of pseudo-time trajectories from Monocle3. Activated fibroblasts are colored by time point.
(F) Same as (E), but here activated fibroblasts are colored by pseudo-time. Pseudotime visualized on a scale from early (blue) to late (yellow), with intermediate stages in purple and orange.
(G) UMAP embedding of activated fibroblasts before re-clustering at each time point.
snATAC-seq analysis reveals that activated fibroblasts represent a cellular state of CD90-positive mesenchymal cells that express NGF after denervation
To further clarify the identity of denervation-activated cell types in our experimental model, we performed a parallel snATAC-seq, as it captures changes in chromatin accessibility that typically anticipate changes in gene expression. In particular, an integrative analysis of the scRNA-seq and snATAC-seq datasets can identify cell clusters as defined by the presence of cell cluster-specific peaks of chromatin accessibility at promoters of cell clusters’ marker genes. This analysis was performed on datasets generated by parallel scRNA-seq and snATAC-seq of denervated muscles at day 5 p.d. vs. control muscles. Interestingly, coherent patterns of differentially accessible regions (DARs) at promoters of DE genes were detected for most of the cell types identified in Figure 1C, with the notable exception of denervation-activated fibroblasts, which could not be detected by this criterion as a distinct cluster (Figures 5A and 5B), but were rather identified as sporadic cells within the cluster of tenocytes (Figure 5A). The large majority of DARs were observed in glial cells from denervated muscles (Figure 5B). Moreover, DARs in glial cells were univocally biased toward increased chromatin accessibility, again indicating a strong correlation between denervation and extensive chromatin remodeling toward activation of gene expression in these cells (Figure 5C). Conversely, DARs detected in other cell types were equally distributed between increased (UP) and decreased (DOWN) chromatin accessibility (Figure 5C). Of note, we observed a strong bias in the genomic distribution of DARs toward non-promoter elements in glial cells—e.g. introns and intergenic elements, which typically harbor enhancers. (Figure S7A). Regardless of their genomic distribution, glial cell-specific DARs detected at day 5 p.d. were associated with the activation of downstream genes implicated in two major processes, i.e., neuronal regeneration—axon growth/guidance and cell cycle regulation (Figures S7A–S7E). Of note, gene ontology of DARs at gene promoters in glial cells predicted an enrichment in promoter-binding by SOX (SRY-related HMG-box) family of transcription factors (TFs), with the Sox10—a neural TF that has been described to regulate gene expression in glial cells44—ranking as top TF (Figures S8A and S8B, and Table S4). Among the putative downstream targets of Sox10, we detected Ngfr (Table S4), whose expression in glial cells coincides with increased chromatin accessibility at its promoter (Figure S8C).
Figure 5.
Chromatin accessibility at single nucleus level of muscle-resident cells upon denervation
(A) UMAP embedding of snATAC-seq data after integration, colored according to our scRNA-seq data.
(B) Table illustrating the number of nuclei assigned to each muscle-resident cell population and the number of differential accessibility regions (DARs) between DEN5d and CTR.
(C) Histogram of the number of DARs overlapping promoters (red, UP) or non-promoter (enhancer, in blue, DOWN) regions, per cell type.
(D–F) UMAP embedding of snATAC-seq data, showing the predicted gene expression profile for Thy1 (CD90) (D), Ngf (E) ,and Ngfr (F), either dim (blue) or bright (red).
Overall, these data support the conclusion that glial cells detected at day 5 p.d. represent a distinct cluster of denervation-activated cells, in which co-expression of high levels of Sox10 and Ngfr marks a fraction of Schwann progenies. By contrast, denervation-activated fibroblasts are likely representing a cellular state in which different mesenchymal cell types might converge by virtue of similar patterns of gene expression, although they exhibit different patterns of DARs.
In order to better define the identity of denervation-activated fibroblasts, we reasoned that they could derive from different muscle mesenchymal cells that express a common surface marker. To this purpose, we used the cell surface marker CD90 (Thy1), which has been previously associated with muscle mesenchymal cells activated in response to pathological perturbations, including denervation21 and diabetes.45 Indeed, scRNA-seq UMAP shows that CD90 was expressed in tenocytes, FAPs, and SMMCs from non-denervated muscles and was specifically enriched in denervation-activated fibroblasts (Figure S8D). However, the predicted gene expression analysis of snATAC-seq shows that the Thy1 signal was indeed distributed among tenocytes, FAPs, and SMMCs in all experimental conditions (Figure 5D). In contrast, the unique marker of glial cells, Ngfr, exhibited a univocal glial-specific predicted expression pattern (Figure 5F). Of note, we observed an overlap between Thy1 and Ngf signals in the integrative analysis of the scRNA-seq and snATAC-seq (Figures 5D and 5E), suggesting that CD90-positive mesenchymal cells are the major source of NGF. Consistently, Ngf expression was induced by denervation in activated fibroblasts, as well as tenocytes, FAPs, and SMMCs (Figure S8D), consistent with the conclusion that the large majority of CD90-positive mesenchymal cells express Ngf in response to denervation.
Heterotypic communication between glial cells and CD90-positive cells upon denervation is mediated by NGFR/NGF interactions
Overall, the consensual activation pattern of muscle glial and mesenchymal cells suggests that these two denervation-activated muscle-resident cell types might reciprocally exchange neurogenic signals toward the repair of NMJs and possibly nerve regeneration. The interruption of this neurogenic signaling network by day 15 p.d. in both cell types suggests a functional interdependence between muscle glial cells and activated fibroblasts in promoting nerve repair. At the same time, their simultaneous conversion into cellular sources of pro-fibrotic ECM components by day 15 p.d. also suggests that these cell types might cooperate to promote persistent fibrosis in chronically denervated muscles. We performed a global ligand-receptor interactome analysis based on RNA-seq collection of human ligand-receptor pairs from CellPhoneDB,46 and manually integrated the results with those obtained from the IPA upstream regulator analysis of DE genes at different time points, which revealed multiple interactions between various muscle-resident cell types (Figures 6A and 6B).
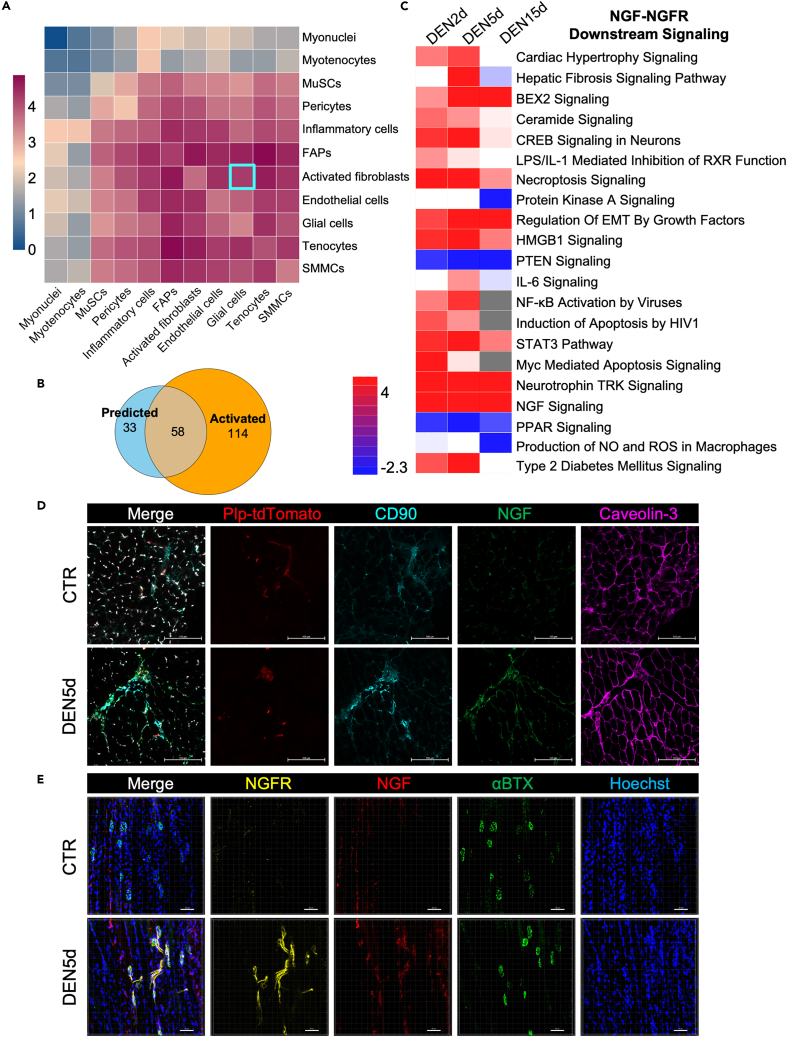
Figure 6.
Ligand-receptor interaction analysis
(A) Heatmap plot from CellPhoneDB showing the total number of interactions (log count) between cell types in our scRNA-seq data. Interaction log count visualized on a scale from low (blue) to high (dark red).
(B) Venn diagram of the intersection between molecular pathways downstream to receptors on glial cells predicted to interact with ligands expressed by activated fibroblasts (Predicted), and molecular pathways predicted to be activated in glial cells based on gene expression (Activated).
(C) Heatmap containing the molecular pathways connected to a specific receptor-ligand pair (only downstream of NGF-NGFR; complete information can be found in Figure S9) for the interactions between glial cells and activated fibroblasts (Interactome). On the right each pathway activation status calculated by Ingenuity Pathway Analysis (IPA; red for activation, blue for repression, and gray or white for activation status info missing) at DEN2d, DEN5d, and DEN15d, respectively compared to the control. EMT: epithelial-mesenchymal transition; NO: nitric oxide; ROS: reactive oxygen species. Pathways activation status (Z-scores) visualized on a scale from repressed (blue) to activated (red), with neutral status (Z-Score = 0) set to white. Pathways not reported in a specific timepoint, with Z-score = N/A, are represented in grey.
(D) Representative images of CD90 (cyan) and NGF (green) immunofluorescence staining of TA sections from Control (CTR, upper) and denervated (DEN5d, lower) Plp1-TdTomato (red) mice. Nuclei were counterstained with DAPI (white). Serial section was stained for Caveolin-3 (magenta). Scale bar: 100μm. N = 4 mice.
(E) Representative z stack images of whole mount staining of NGFR (yellow) and NGF (red) in extensor digitorum longus (EDL) isolated from CTR and DEN5d B6 mice. Acetylcholine receptor (AChR) at post-synaptic membrane of myofiber was stained with αBTX (green): α-Bungarotoxin. Nuclei were counterstained with Hoechst (blue). Scale bar: 50μm. N = 3 mice.
Among them, interactions between glial cells and denervation-activated fibroblasts were identified and included various putative ligand/receptor interactions, largely biased by two main categories: interactions mediated by components of the ECM, which were dominated by collagens; and interactions mediated by secreted soluble factors and their receptors, which were dominated by NGF/NGFR (Figure S9). Moreover, the downstream processes regulated by these two putative types of interactions were almost mutually exclusive (Figure S9), with NGF/NGFR signaling mostly associated with cell survival, proliferation, and neurogenesis (Figures 6C and S9). The expression patterns of Ngfr and Ngf induced by denervation in glial cells and denervation-activated fibroblasts, respectively (Figures S2, S5A, and S8D), predict that these cells might be in close proximity after denervation, in order to favor ligand (NGF)/receptor (NGFR) interactions. We tested this prediction by using two equivalent, parallel mouse models. By using Plp1-tdTomato transgenic mice we could detect tdTomato-positive glial cells surrounded by CD90/NGF-positive cells only following denervation (hereby defined as CD90-positive mesenchymal cells) (Figure 6D). Likewise, in wild-type mice, NGF-positive cells were found adjacent to NGFR-positive glial cells and in close proximity to NMJs, as detected by alpha-bungarotoxin (αBTX) staining, only in denervated muscles (Figure 6E).
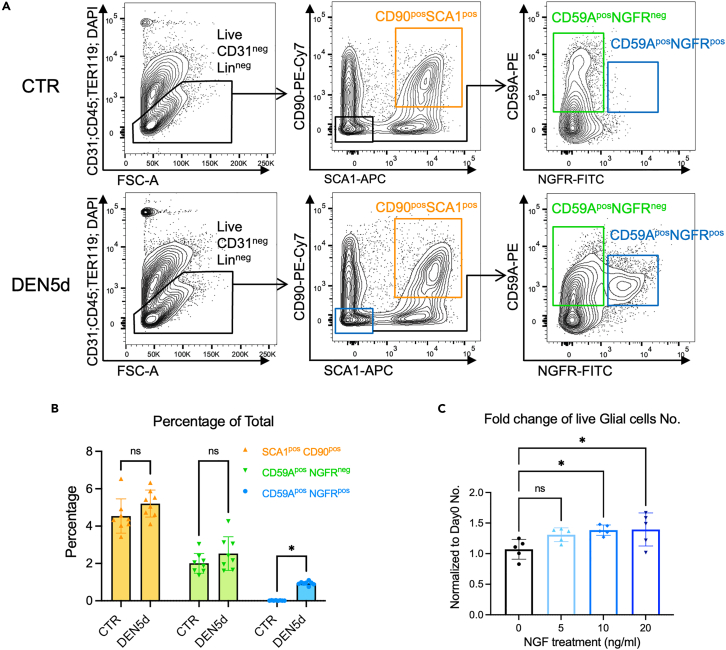
We evaluated whether glial cells and CD90-positive mesenchymal cells could establish functional interactions mediated by NGF/NGFR, following denervation. To this purpose, we established FACS sorting strategies to isolate these cell types from muscles either unperturbed (CTR) or 5 days p.d. (DEN5d). Based on the expression patterns of Thy1 and Ngf in our scRNA-seq data (Figure S8D), we could isolate two distinct Thy1-expressing cell populations among Sca1 positive cells, namely CD90/DPP4 double-positive cells, which are accounted only by FAPs, and CD90pos/DPP4neg cells, which included denervation-activated fibroblasts, tenocytes, and FAPs (Figure S10A). Although only CD90pos/DPP4neg cells exhibited a significant increase in the number after denervation (Figures S10B and S10C), both CD90/DPP4 double-positive and CD90pos/DPP4neg cells showed denervation-induced expression of the mesenchymal marker Col8a1 and Ngf (Figure S10D). Likewise, glial cells were sorted as CD59A/NGFR double-positive cells (denervation-inducible nmSC) and CD59Apos/NGFRneg myelinating cells47 that are also present in unperturbed muscles (Figures 7A and 7B). A global strategy for simultaneous FACS-mediated isolation of glial cells and CD90-positive mesenchymal cells from unperturbed and denervated muscles is illustrated in Figures 7A and 7B. The expression of marker genes in FACS-isolated cells was further validated by real-time qPCR, respectively (Figure S11).
Figure 7.
Recombinant NGF promotes expansion of NGFR-positive glail cells ex vivo
(A) FACS plot showing the sorting strategy for CD90posSCA1pos activated fibroblasts (abbreviated as CD90pos cells), CD59AposNGFRneg cells (abbreviated as CD59Apos cells), and CD59AposNGFRpos Glial cells (abbreviated as NGFRpos cells) simultaneously from control (upper, CTR) and DEN5d (bottom) muscles.
(B) Percentage of 3 populations (gated in A) by FACS at CTR and DEN5d conditions. N = 8 mice.
(C) Sorted NGFRpos glial cells were treated with 0/5/10/20 ng/mL beta-NGF for 2 days in culture. Hoechst and propidium iodide (PI) were added into the medium to help detect total cell number and dead cell number, respectively. Images were captured at day 0 and day 2. Quantified live cell number at day 2 was normalized to that at day 0. N = 5 mice.
In B and C, data are represented as mean ± SD. Statistical significance was analyzed by two-way ANOVA in (B) and one-way ANOVA in (C) with p values shown as ∗p < 0.05 and ns for not significant.
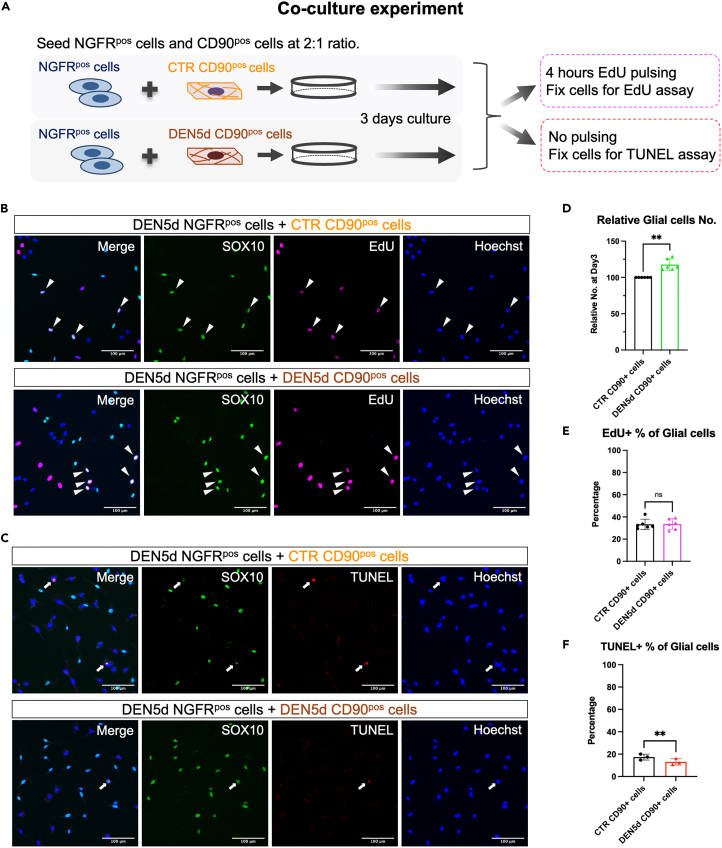
We next determined if CD59A/NGFR double-positive glial cells isolated from denervated muscles could respond to recombinant NGF when cultured ex vivo. Figure 7C shows that 48 h of incubation with recombinant NGF increased the number of CD59A/NGFR double-positive glial cells in a dose-dependent manner (Figure 7C). We next tested whether co-culture with control vs. denervation-activated CD90-positive mesenchymal cells, as a cellular source of endogenous NGF, was sufficient to induce an equivalent increase in the number of CD59A/NGFR double-positive glial cells (Figure 8). In this experiment, co-cultured CD59A/NGFR double-positive glial cells were identified, after 3 days of culture, as SOX10-positive cells by immunofluorescence. In comparison with control CD90-positive mesenchymal cells (expressing low Ngf), CD90-positive mesenchymal cells isolated from denervated muscles (expressing high Ngf) increased the number of co-cultured CD59A/NGFR double-positive glial cells more (Figure 8D). Of note, we could not appreciate significant changes in EdU incorporation in the SOX10-positive glial cells co-cultured with denervation-activated CD90-positive mesenchymal cells (Figures 8B and 8E), indicating that their increase in number was not due to increased cell proliferation. Consistently, we observed reduced cell death of SOX10-positive cells by terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) assay either treated with 10 ng/ml recombinant NGF ex vivo (Figure S12) or co-cultured with DEN5d CD90-positive cells (Figures 8C and 8F). Thus, our data indicate that after denervation, CD90-positive mesenchymal cells support the survival of CD59A/NGFR double-positive glial cells, possibly through the NGF/NGFR-activated downstream signaling.
Figure 8.
Co-culture with NGF-expressing mesenchymal cells promotes expansion of NGFR-positive glial cells ex vivo
(A) Graphic illustration of co-culture experiment with NGFRpos cells and CTR- or DEN5d- CD90pos cells (abbreviated as CTR group or DEN5d group, respectively). Cells were fixed after 3 days culture for EdU assay (EdU was added for 4 h pulsing before fixation) and TUNEL assay, respectively.
(B) Representative cell staining images of SOX10 (green) and EdU (magenta), counterstained with Hoechst (blue). Arrow heads indicate SOX10posEdUpos Glial cells. Scale bar: 100μm.
(C) Representative cell staining images of SOX10 (green) and TUNEL (red), counterstained with Hoechst (blue). Arrows indicate SOX10posTUNELpos Glial cells. Scale bar: 100μm.
(D) SOX10pos Glial cell number per field was quantified in two co-culture groups. By normalization to the CTR group, relative Glial cell number was shown. N = 6 mice.
(E) EdUpos percentage of SOX10pos Glial cells in two co-culture groups. N = 6 mice.
(F) TUNELpos percentage of SOX10pos Glial cells in two co-culture groups. N = 3 mice. Statistical significance was analyzed by paired Student’s t test in (D-F) with p values shown as ∗∗p < 0.01 and ns for not significant.
Discussion
Our longitudinal genome-wide analysis of gene expression and chromatin accessibility at the single-cell level from denervated skeletal muscles provides an unprecedented atlas of the transcriptional response of skeletal muscle-resident cells to acute denervation. This analysis identified muscle-glial cells and CD90-expressing mesenchymal cells as the main muscle-resident cell types activated in response to denervation. We show that, at variance with the dramatic expansion of multiple cell types observed upon acute muscle injury, the transcriptional perturbations following acute denervation are much less extensive, while specifically directed toward the selective activation of two cell types implicated in nerve repair.
Previous studies have implicated muscle-resident interstitial cells located in the proximity of NMJs as potential progenitors of glial cells upon denervation.22,23,24,26 However, analysis of the kinetic of glial cell accumulation48 inferred from our scRNA-seq datasets is not consistent with the expansion of a pool of muscle-resident glial cell progenitors by proliferation, as they double their number every 24 h during the first 2 days p.d. (Table S1), without a corresponding pattern of DNA synthesis and mitotic activity observed in these cells (Figure S4F). Indeed, only a subset of these cells exhibited a gene expression profile predictive of the G2-M phase, most likely accounted by Schwann cell progenitors (Figures S4E and S4F). These proliferating cells might represent a gradient of Schwann cell progenitors before their functional specialization into PSCs, similar to what was observed during development.40 The doubling time of glial cells showed a progressive reduction by the following time points (days 5 and 15 p.d.) with a final plateau of glial cell accumulation observed at day 15 p.d. Thus, it is likely that the large majority of glial cells accumulated post-denervation might derive from motoneuron-associated glial cells released as a consequence of axonal degeneration following the nerve transection. Consistent with this hypothesis, their survival in a highly degenerative environment, such as denervated muscles, is dependent on the trophic action of neurotrophins and the relative engagement of their specific receptors.49
Interestingly, we found that the cellular source of the most abundant intra-muscular ligand of NGFR—the neurotrophin NGF—in denervated muscles is provided by a population of denervation-activated CD90-positive mesenchymal cells, with only minimal, negligible contribution from glial cells. The population of NGF and CD90-expressing cells in denervated muscles included several mesenchymal cell types detected by scRNA-seq, such as the activated fibroblasts and sub-clusters of tenocytes, FAPs, and SMMCs (Figure S8D). However, integration of scRNA-seq and snATAC-seq datasets and gene expression prediction from snATAC-seq indicates that activated fibroblasts indeed represent a cellular cluster of denervation-activated cells, in which converge several mesenchymal cell types that share common transcriptional features, while exhibiting distinct patterns of chromatin accessibility. Consistently, the Ngf promoter exhibits multiple denervation-induced DARs, whose TF-binding motif analysis predicts cell type-specific patterns of NGF transcriptional activation from different types of mesenchymal cells (data not shown). Thus, it is likely that denervation-activated mesenchymal cells might also contain nerve-associated fibroblasts released by degenerating axons. Although lineage tracing experiments are required to conclusively determine the cell origin of denervation-activated glial and mesenchymal cells, nonetheless our data indicate that: (i) these cell types might functionally interact during the first 5 days p.d.; (ii) these interactions occur in the proximity of the NMJ and are mediated by NGF/NGFR signaling; and (iii) NGF/NGFR signaling might promote glial cell survival and could help generating Schwann cells toward nerve and NMJ repair.
Our longitudinal analysis reveals that at late time points (i.e., day 15 p.d.) the pattern of gene expression exhibited by denervation-activated glial and mesenchymal cells is skewed toward the expression of genes implicated in ECM remodeling for fibrosis. This pattern suggests a biphasic activation of specific cell types, with an initial abortive attempt to repair the NMJ that eventually culminates with fibrosis and muscle atrophy—two events invariably associated with complete sciatic nerve transection.21 In this scenario, denervation-activated mesenchymal cells initially produce specific ECM components of the NMJ-associated basal lamina,50 but upon lack of completion of NMJ repair these cells might continue to constitutively produce ECM components ultimately leading to fibrosis. The most reasonable explanation for this biphasic response is that the severe extent of the nerve lesion in our experimental model—complete transection of the sciatic nerve—is anatomically incompatible with the full repair by endogenous regeneration. Conceivably, at variance with nerve lesions that are permissive to repair and muscle re-innervation, upon complete sciatic nerve transection, the lack of signals from dying motor neurons and the loss of coordination between denervation-activated cell types might lead to a persistent pathological reconfiguration of ECM composition.
Overall, our data predict a functional interdependence between denervation-responsive cell types and identify networks implicated in both nerve regeneration and pathogenic events (myofiber atrophy and fibrosis) that could be exploited for interventions toward promoting nerve repair and limiting muscle degeneration and atrophy. Thus, these data will provide an initial atlas of reference for further studies in experimental conditions of reversible nerve damage, in order to capture cellular networks that can be targeted by interventions toward promoting nerve repair. Our data could also guide the selection of genes that discriminate specific cell types or sub-clusters activated by denervation for future strategies of FACS-mediated sorting or genetic models of lineage tracing of specific cell types of interest.
Limitations of the study
A primary limitation of this study relates to the lack of knowledge on the anatomical location of cellular clusters identified by scRNA-seq. Although our immunofluorescence analysis predicted that NGFR-expressing glial cells and NGF-expressing mesenchymal cells might be in close proximity to the NMJs, further spatial omics studies will provide definitive evidence on the anatomical distribution of these and other muscle resident cell types. Another limitation that is intrinsic to most of the currently available single cell/nuclei transcriptome analysis concerns the impossibility to assign lineage derivation to the cell types detected and to determine potential changes of lineage within muscle-resident cells during the time point post-denervation. Finally, while our study predicts several functional interactions between denervation-activated cell types—e.g., glial and mesenchymal cells—these predictions need to be further investigated in term of both their impact in vivo and mechanistic insights.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-Caveolin 3 (Clone 26) mouse antibody | BD Biosciences | Cat# 610421; RRID:AB_397801 |
| Anti-p75NTR (D4B3) XP®Rabbit antibody | Cell Signaling Technology | Cat# 8238; RRID:AB_10839265 |
| Anti-CD90.2 (Thy-1.2) Monoclonal Antibody (30-H12) | Thermo Fisher Scientific | Cat# 16-0903-82; RRID:AB_469034 |
| Anti-NGF Rabbit polyclonal antibody | Abcam | Cat# 6199; RRID:AB_2152414 |
| Anti-mouse/human NGF mouse antibody | BioLegend | Cat# 509701; RRID:AB_315543 |
| Anti-SOX10 antibody | Abcam | Cat# ab227680; RRID:AB_2927464 |
| AffiniPure Fab Fragment Goat Anti-Mouse IgG (H+L) | Jackson ImmunoResearch Labs | Cat# 115-007-003; RRID:AB_2338476 |
| Anti-Rabbit IgG (H+L), Alexa Fluor 647 | Thermo Fisher Scientific | Cat# A21244; RRID:AB_2535812 |
| Anti-Mouse IgG (H+L), Alexa Fluor 568 | Thermo Fisher Scientific | Cat# A10037; RRID:AB_2534013 |
| CD31 Monoclonal Antibody (390), Pacific Blue™ | Thermo Fisher Scientific | Cat# RM5228; RRID:AB_10373114 |
| CD45 Monoclonal Antibody (30-F11), eFluor™ 450 | Thermo Fisher Scientific | Cat# 48-0451-82; RRID:AB_1518806 |
| TER-119 Monoclonal Antibody (TER-119), eFluor™ 45 | Thermo Fisher Scientific | Cat# 48-5921-82; RRID:AB_1518808 |
| CD90.2 (Thy-1.2) Monoclonal Antibody (53-2.1), PE-Cyanine7 | Thermo Fisher Scientific | Cat# 25-0902-81; RRID:AB_469641 |
| CD271 (LNGFR) Antibody, Vio® Bright FITC | Miltenyi Biotec | Cat# 130-110-115; RRID:AB_2656844 |
| APC anti-mouse Ly-6A/E (Sca-1) | BioLegend | Cat# 122512; RRID:AB_756197 |
| PE anti-mouse CD59a | BioLegend | Cat# 143104; RRID:AB_10962439 |
| FITC anti-mouse CD26 (DPP-4) | BioLegend | Cat# 137805; RRID:AB_10662380 |
| Chemicals, peptides, and recombinant proteins | ||
| Collagenase, Type 2 (Collagenase II) | Worthington-Biochem | Cat# LS004177 |
| Dispase®II (neutral protease, grade II) | Sigma-Aldrich | Cat# 4942078001 |
| Goat Serum, New Zealand origin | Thermo Fisher Scientific | Cat# 16210064 |
| Bovine Serum Albumin (BSA) | Rockland Immunochemicals | Cat# NC9418785 |
| α-Bungarotoxin, Alexa Fluor™ 488 conjugate | Thermo Fisher Scientific | Cat# B13422 |
| EMS Glycerol Fluoro Mount w/ Para Phenylenediamine (PPD) | Electron Microscopy Sciences | Cat# 17989-70 |
| Propidium Iodide | Thermo Fisher Scientific | Cat# P3566 |
| DAPI (4',6-Diamidino-2-Phenylindole, Dihydrochloride) | Thermo Fisher Scientific | Cat# D1306 |
| Hoechst 33258, Pentahydrate (bis-Benzimide) | Thermo Fisher Scientific | Cat# H3569 |
| Ham's F-10 Nutrient Mix | Thermo Fisher Scientific | Cat# 11550043 |
| DMEM/F-12 | Gibco | Cat# 11320033 |
| Horse Serum, heat inactivated, New Zealand origin | Thermo Fisher Scientific | Cat# 26050088 |
| Fetal Bovine Serum, U.S. Source | Omega Scientific | Cat# FB-11 |
| L-Glutamine (200 mM) | Thermo Fisher Scientific | Cat# 25030081 |
| TRIzol™ Reagent | Thermo Fisher Scientific | Cat# 15596018 |
| Recombinant Mouse beta-NGF Protein | R&D | Cat# 1156-NG |
| Recombinant Human NRG1-beta 1 | R&D | Cat# 396-HB |
| Penicillin-Streptomycin (10,000 U/mL) | Thermo Fisher Scientific | Cat# 15140122 |
| Critical commercial assays | ||
| Click-iT™ EdU Cell Proliferation Kit for Imaging, Alexa Fluor™ 647 dye | Life Technology | Cat# C10340 |
| Click-iT™ Plus TUNEL Assay Kits for In Situ Apoptosis Detection, Alexa Fluor™ 594 dye | Life Technology | Cat# C10618 |
| High-Capacity cDNA Reverse Transcription Kit | Life Technologies | Cat# 4368814 |
| PowerUp SYBR green Master Mix | Applied Biosystems | Cat# 4368708 |
| Deposited data | ||
| Raw scRNA-seq and snATAC-seq data and processed matrices | This paper | GEO:GSE221736 |
| scRNA-seq processed matrices | Oprescu et al.10 | GEO:GSE138826 |
| scRNA-seq processed matrices | Wolbert et al.43 | GEO:GSE142541 |
| Experimental models: Organisms/strains | ||
| C57BL/6J (B6 mice) | The Jackson Laboratory | Cat# 000664; RRID:IMSR_JAX:000664 |
| B6.Cg-Tg(Plp1-cre/ERT)3Pop/J | The Jackson Laboratory | Cat# 005975; RRID:IMSR_JAX:005975 |
| B6.Cg-Gt(ROSA)26Sortm14(CAG-tdTomato)Hze/J | The Jackson Laboratory | Cat# 007914; RRID:IMSR_JAX:007914 |
| Software and algorithms | ||
| Zen 3.4 (blue edition) | Carl Zeiss Microscopy | N/A |
| Prism 9.2.0 | GraphPad | https://www.graphpad.com |
| ImageJ v1.53 | N/A | https://imagej.net/ij/index.html |
| CellRanger v3.0.0 | 10X Genomics | Release version 3.0.0 Official 10x Genomics Support https://www.10xgenomics.com/software |
| Seurat v3.1.2 | Butler et al. (2018) | Release version 3.1.2 remotes::install_version(package = 'Seurat', version = package_version('3.1.2')) |
| Scrublet v0.2.1 | Wolock et al.28 | Release version 0.2.1 https://github.com/swolock/scrublet |
| Monocle v3.0.2.1 | Cao et al.39 | Release version 3.0.2.1 https://cole-trapnell-lab.github.io/monocle3/ |
| CellPhoneDB v2.0.0 | Efremova et al.46 | Release version 2.0.0 https://github.com/ventolab/CellphoneDB |
| CellRanger-ATAC v2.0.0 | 10X Genomics | Release version 2.0.0 Official 10x Genomics Support https://www.10xgenomics.com/software |
| CellRanger-Arc v2.0.0 | 10X Genomics | Release version 2.0.0 Official 10x Genomics Support https://www.10xgenomics.com/software |
| Ingenuity Pathway Analysis (IPA) v01-16 | QIAGEN | Release version 01-16 https://www.qiagen.com/us/products/discovery-and-translational-research/next-generation-sequencing/informatics-and-data/interpretation-content-databases/ingenuity-pathway-analysis |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to the lead contact, Pier Lorenzo Puri (lpuri@sbpdiscovery.org).
Materials availability
This study did not generate new unique reagents. Requests for mouse strains in this study could be shared by lead contact upon request, Pier Lorenzo Puri (lpuri@sbpdiscovery.org).
Experimental model and study participant details
Mice
All experiments in this study were performed in accordance with protocols approved by the Sanford Burnham Prebys Medical Discovery Institute (SBP) Animal Care and Use Committee (IACUC) and the Italian Ministry of Health, National Institute of Health (IIS) and Sapienza University of Rome. The study is compliant with all relevant ethical regulations regarding animal research. Male mice at 2-5-month-old were used in this study. C57BL/6J mice were provided by the SBP Animal Facility (La Jolla, CA, USA) or purchased from The Jackson Laboratory (JAX:000664). Plp1-tdTomato transgenic mice (Plp1CreERT+/-Tomatofl/-) was generated by crossing B6.Cg-Tg(Plp1-cre/ERT)3Pop/J mice (The Jackson Laboratory, Strain #:005975) with B6.Cg-Gt(ROSA)26Sortm14 (CAG-tdTomato)Hze/J mice (The Jackson Laboratory, Strain #:007914). Genotype was assessed as describe by the seller. Mice were treated for 5 consecutive days with intraperitoneal injection of 100ml tamoxifen (20 mg/ml in corn oil solution). Denervation was performed 7 days after the final tamoxifen injection.
Method details
Muscle denervation
Briefly, male C57BL/6J or Plp1-tdTomato mice at 10-12 weeks old are anesthetized by intraperitoneally injection of Avertin (0.25-0.5 mg/g), the hair is clipped from the surgical site (the right hindquarter; 1mm posterior and parallel to the femur) and the site is prepped with betadine solution. Next, the right sciatic nerve of mice will be exposed and excised for nearly 3mm from a small incision made in the mid-lateral thigh by a scissor, followed with wound closure by tissue adhesive topical application (Vetbond). Buprenorphine (0.05-0.1 mg/kg) is administrated subcutaneously prior to animal regaining consciousness. All instruments are sterilized with glass bead sterilizer at 200°C for 20-30 seconds. Mice at 2 days, 5 days, 15 days post denervation surgery were sacrificed and used in this project.
Muscle processing for scRNA-seq and snATAC-seq
For each time-point post denervation (namely, 2, 5 and 15 days p.d., abbreviated as DEN2d, DEN5d, and DEN15d in all figures and tables) as well as the non-denervated control (abbreviated as CTR in all figures and tables), two male C57BL/6J mice at 3 months of age were used for the two biological replicates of scRNA-seq, respectively. After sciatic nerve transection, we collected two muscles downstream to the sciatic nerve, the tibialis anterior (TA) and gastrocnemius (GA) muscles. Samples were processed as previously described protocol,51 and sorted by fluorescence activated cell sorting (FACS) to remove cell debris, doublets and dead cells. 100,000 live cells/sample were sorted and used for downstream processing with the 10X Genomics 3’ v3 kit for single-cell gene expression. 12,000 cells were used for library preparation, a little over the 10,000 suggested by the 10X protocol, in order to reach a higher capture rate. Libraries were constructed per manufacturer’s instructions and sequenced using Illumina’s NextSeq platform.
scRNA-seq data preprocessing, dimensionality reduction, and visualization
Seurat (v3.1.2) was used for data filtering, normalization, differential marker expression and visualization.27 For scRNA-seq, average number of cells across samples was 6,000 and average read depth was 44,500 reads/cell. Reads were then aligned to the mouse genome mm10/GRCm38. Biological replicates for each time point were initially merged together and filtered for quality control parameters (cells with more than 20% reads mapping to mitochondrial genes, less than 200 or greater than 6,000 features (i.e., genes), and more than 50,000 reads were filtered out, as well as cells with a doublet score, derived from Scrublet, greater than 0.1). Data from different time points were then merged, and Seurat’s SCTransform function was used to normalize and scale the data to minimize batch effects,52 normalizing counts for mitochondrial genes content, number of features and cell cycle phase. Dimensionality reduction was performed through Principal Component Analysis (PCA) on top variable features, with npcs=100. FindNeighbors, FindClusters and UMAP embedding functions were based on the top 20 PCs of the PCA. Clusters were identified through FindClusters with parameters resolution= 0.4, algorithm= 3. UMAP embedding was run with metric= ‘euclidean’.
Differential marker expression and cell type identification
Identified cell clusters were evaluated for marker gene expression to determine cell types through Seurat’s FindAllMarkers function. Meta-clusters were manually annotated based on shared enriched markers expression of genes reported in literature as cell type specific markers. The FindMarkers function was also used to derive, for each condition, cumulative gene expression changes vs the control condition (i.e., non-denervated; Figure 2C), considering only upregulated genes (parameter only.pos=T). Finally, each cell type (meta-cluster) was subset from the Seurat Object and the FindMarkers function was used to derive, for each meta-cluster, differential gene expression per condition (number of genes tested for differential expression: 36,189), where each time point post-denervation was compared to the control condition (i.e., non-denervated; Table S2). In both differential analyses the parameter min.pct was set to 0.25, restricting the analysis to genes (among the initial 36,189) that were detected in at least 25% of cells in either of the two compared conditions.
Cell type-specific analysis
We used Seurat’s subset function to extract cells belonging to each meta-cluster, on which we performed new clustering and dimensionality reduction, keeping the same parameters used for the analysis of the complete dataset. The subset data was used for trajectory inference with Monocle.
Trajectory inference and enriched gene modules analysis
Monocle (v3.0.2.1)39 was used for pseudotime analysis in glial cells and activated fibroblasts and to find enriched gene modules along the pseudotime trajectory of glial cells. Raw data from glial cells and activated fibroblasts as well as their meta-data were used as input for the new_cell_data_set function, to create Monocle objects for trajectory inference. The preprocess_cds function was used to perform PCA analysis and normalize the data, with num_dim = 100, while UMAP embedding was inherited from the original cell population specific Seurat object. Also, gene loadings from the 20 PCs from the cell population specific Seurat object were imported in the Monocle object. For pseudotime inference, the non-denervated time point was chosen as root node.
To identify genes associated to the different cell fates in the glial cells pseudotime trajectory, we calculated differential genes with the graph_test function (with neighbor_graph=”principal_graph”) and selected genes with q-value< 0.000001. These genes were then used to derive gene modules through the find_gene_modules function.
Pathway analysis
Differentially expressed genes (p-value-adjusted < 0.01) in each cell group at DEN2d, DEN5d, DEN15d respectively compared with CTR were subjected to Ingenuity Pathway Analysis (IPA) (QIAGEN, version 01-16) for “Core Analysis” and further “Comparison Analysis” and “Upstream Regulator Analysis”. Selected pathways, significantly enriched in either DEN2d, DEN5d, or DEN15d with absolute z score >1, were shown in figures and supplementary tables. Pathways with p-value >= 0.05 were indicated with gray dots.
snATAC-seq data preprocessing, dimensionality reduction and scRNA-seq integration
Samples for day 5 p.d. and control were processed according to 10X protocols. Data were aligned to the mm10/GRCm38 version of the mouse genome with CellRanger-ATAC (v2.0.0) for the snATAC samples (one biological replicate for each condition) and with CellRanger-Arc (v2.0.0) for the Multiome snATAC samples (one biological replicate for each condition). Doublets presence for each replicate was inferred with Scrublet (v0.2.1)28 within Python (v3.8). Filtered barcode and count matrices produced by CellRanger were used for downstream analyses in R (v4.0.3). Data from both experiments were analyzed with Seurat (v4.0.1) and Signac.53 Cells were initially filtered out if they contained less than 200 features (i.e., ATAC peaks), and features were filtered out if they were detected in less than 10 cells. A second round of filtering was performed with the following thresholds: peak_region_fragments > 3000, peak_region_fragments < 60,000, nCount_peaks < 100,000, nCount_peaks > 1000, pct_reads_in_peaks > 15, blacklist_fraction < 0.05, nucleosome_signal < 2 and TSS.enrichment > 1. Filtered data were subjected to latent semantic indexing to reduce their dimensionality and label transfer anchors were calculated to integrate the snATAC-seq data to our previously generated scRNA-seq data. Gene expression prediction from snATAC-seq data was achieved through the creation of a gene activity matrix with the GeneActivity function.
Interactome analysis
In order to predict cell communication signals between different muscle-resident cell populations, normalized counts were extracted from the Seurat object containing cells from all experimental conditions, and their meta data (e.g. cluster information). Since CellPhoneDB uses a data base of human genes to calculate receptor-ligand interactions, mouse genes were converted to human with biomaRt.54 CellPhoneDB statistical analysis was then run with 10 iterations, and its results were visualized through dot_plot and heatmap_plot functions. Downstream pathway activation/repression resulting from the predicted receptor-ligand interactions for glial cells and fibroblasts was then evaluated through the manual integration of the list of receptors found by CellPhoneDB with the list of Upstream Regulators from IPA and the pathways that we found predicted to be activated or repressed through IPA Pathway analysis.
Integration with scRNA-seq data from injured muscle (Oprescu et al., 2020)10 and sciatic nerve (Wolbert et al., 2020)43
scRNA-seq raw counts were retrieved from GEO (accession numbers are listed in the key resources table), and filtered following the filtering parameters used for our own scRNA-seq data. Integration with our denervation dataset was achieved through an initial merge with our Seurat object, followed by a concurrent normalization with SCTransform, selection of integration features (SelectIntegrationFeatures, with 2000 nfeatures) and anchors (FindIntegrationAnchors, with SCT as normalization method). Finally, dimensionality reduction through PCA and UMAP (with 20 PCs) allowed us to visualize the integrated object, with the cell labels assigned by (Oprescu et al.) or (Wolbert et al.), or from our meta-cluster identities.
Immunofluorescence staining of muscle sections
Isolated TA muscles were fixed in 1% Paraformaldehyde (PFA) for 1 hour and dehydrated in 20 % sucrose solution in PBS at 4°C overnight. Then, muscles were embeded in Tissue-Tek® O.C.T. Compound (Sakura® Finetek, 4583) and frozen in isopentane cooled in liquid nitrogen. Muscle sections at 8μm thick were obtained using a Cryostat and fixed with 4% PFA for 10 minutes. After permeabilizing with 0.5% Triton X-100 in PBS for 15 minutes and blocking with 4% BSA (Millipore Sigma, A7030) in PBS for 1 hour, muscle sections were incubated at 4°C overnight with Caveolin-3 (1:1000), NGFR (p75NTR, 1:600), CD90.2 (Thy-1.2, 1:150), and/or NGF (1:100) primary antibodies diluted in 0.1% Triton-X 100, 4% BSA in PBS. Then muscle sections were incubated for 1 hour with fluorophore-conjugated secondary antibodies (1:1000) and DAPI (1:10000, Thermo Fisher Scientific, D1306) diluted in 0.1% Triton X-100 in PBS. After wash with PBS twice, muscle sections were mounted with glycerol (3:1 in PBS), covered with cover glass, and sealed with nail polish. Images were captured with a Zeiss confocal microscope (LSM 900 with Airyscan2) using a 20x air objective, and then edited using the ImageJ software.
Whole mount immunofluorescence staining
Extensor digitorum longus (EDL) muscles were isolated, separated into 4 pieces according to Uezumi’s paper55 (Uezumi, J Clin Invest. 2021) and fixed in 0.5% PFA for 2 hours at room temperature with gentle shaking, following with 2 times PBS wash, overnight incubation in the blocking solution (5% Goat Serum, 2% BSA, 0.5% Triton X-100 in PBS), and Fab Fragment Goat Anti-Mouse IgG (1:50, Jackson ImmunoResearch Labs., Inc., 115-007-003) in PBS for 1 hour. Then, muscles were incubated for 1 day with NGFR rabbit antibody (1:600) and NGF mouse antibody (BioLegend, 509701, 1:100) diluted in blocking solution. After 3 times wash with 0.5% Triton X-100 in PBS (0.5% PBST, 1 hour per wash), muscles were incubated for 1 day with secondary reagents (Alexa Fluor 488-conjugated alpha-Bungarotoxin (1:600), Hoechst 33258 (1:10000), Alexa Fluor 647-conjugated Goat anti-Rabbit IgG antibody (1:1000), Fluor 568-conjugated Donkey anti-Mouse IgG Antibody (1:1000)) diluted in blocking solution. After 3 times wash with 0.5% PBST and 1 time wash with PBS (1 hour per wash), muscles were moved on a glass slide and mounted using EMS Glycerol Fluoro Mount w/Para Phenylenediamine Anti-fading agent and cover glass. The incubation and wash steps were carried on a rotator at 4°C during the whole mount staining. Images were captured with a Zeiss LSM710 (Carl Zeiss, Jena, Germany) using a 20x air objective and processed using Zeiss Zen Black software. Z-stack images were processed using the Imaris Single Full Suite with ClearView Deconvolution software.
Cell isolation by FACS
GA and TA muscles were dissected and processed as previously described in Liu’s protocol51 (Liu et al., 2015) to isolate cells by BD FACSAriaIIu 16-color high-speed cell sorter. In general, dead cells are excluded by DAPI (1:2000), CD31-CD45-TER119-SCA1+CD90+ cells (abbreviated as CD90+ cells) are sorted as Ngf-expressing mesenchymal cells including activated fibroblasts, CD31-CD45-TER119-SCA1-CD90-CD59A+NGFR- cells (abbreviated as CD59A+ cells) are sorted as non-myelinating Schwann cells, and CD31-CD45-TER119-SCA1-CD90-CD59A+NGFR+ cells (abbreviated as NGFR+ cells) are sorted as Glial cells. Antibodies used are listed in the key resources table.
NGF-treatment experiment
FACS isolated NGFR+ cells from DEN5d muscles were seeded in 384-well plate (VWR, 82051-282) and cultured in Schwann cell medium containing 0, 5, 10, or 20 ng/ml Recombinant Mouse beta-NGF Protein (1156-NG-100) in a 37°C incubator with 5% CO2. Hoechst 33258 (1:10000) and Propidium Iodide (PI, Life Technology, P3566, 1:2000) were added into the medium to help detect total cell number and dead cell number, respectively. Images were captured at Day 0 and Day 2 using an Image Xpress confocal microscope (Molecular Devices) and quantification was performed with Molecular Device software. Quantified live cells number at Day 2 was normalized to that at Day 0. Schwann cell medium: DMEM:F12 (1:1, Gibco, 11320-033) medium containing 10% Fetal Bovine Serum (U.S. Source, FB-11), 1x L-Glutamine (Life, 25030081), 1% penicillin-streptomycin (Life, 15140122), 5μM Forsklin (Sigma, F3917), and 10 ng/ml Recombinant human beta1-Neuregulin (R&D, 396-HB-050). For EdU assay, cells were pulsed with 10nM EdU during the last 4 hours before fixation. For both EdU assay and TUNEL assay, 10 ng/ml Recombinant Mouse beta-NGF Protein was added to cultured NGFR+ cells at Day1. After 2 days culture, cells were fixed using 4% PFA for the indicated assay, respectively. EdU was detected by Click-iT EdU Alexa Fluor 647 Imaging Kit (Life Technology, C10340). TUNEL assay was performed according to the manufacturer's protocol using Click-iT™ Plus TUNEL Assay Kits for In Situ Apoptosis Detection (Life Technology, C10618).
Co-culture experiment
FACS isolated NGFR+ cells and CD90+ cells were seeded in 384-well plate at a 2:1 ratio and cultured in Schwann cell medium for 3 days. EdU (10nM) was added into medium in the last 4 hours before fixation at Day3 using 4% PFA. After wash twice with PBS, cells were permeabilized in 0.5% PBST for 10 min and blocked in 4% BSA PBS for 1 hour at RT. EdU was detected by Click-iT EdU Alexa Fluor 647 Imaging Kit (Life Technology, C10340) and TUNEL assay was performed according to the manufacturer's protocol (Life Technology, C10618), respectively. After EdU assay or TUNEL assay, SOX10 rabbit antibody (ab227680, 1:200) was diluted at 1:200 in antibody solution and added to cells for overnight incubation at 4°C. After 3 times wash with 0.1% PBST, cells were incubated with secondary antibody (A-21206) and Hoechst 33258 for 1 hour at RT. After 3 times wash with 0.1% PBST, cells were kept in PBS at 4°C. Images capture and quantification of the percentages of SOX10-positive or EdU-positive cells were performed by using the MetaXpress, supported by Dr. Alexandre R Colas at SBP.
RNA extraction and real time PCR
Total RNA was extracted from sorted cells using TRIzol Reagent (Life Technologies, 15596018) following the manufacturer’s protocol and retrotranscribed using the High-Capacity cDNA Reverse Transcription Kit (Life Technologies, 4368814). PowerUp SYBR green Master Mix reagents (Applied Biosystems, 4368708), cDNA and primers were used in a 10 μl reaction which was performed in a 384 well plate on the ABI Applied Biosystems 7900HT PCR system. The 2-ΔΔCT method was used for calculation. Primers used in this study are listed in the Table S5.
Quantification and statistical analysis
All statistical analyses were performed using Prism version 9.0 (GraphPad Software, San Diego CA, USA). Data are presented as the mean ± SD. Statistical significance was analyzed by paired Student’s t-test, one-way ANOVA, or two-way ANOVA, as indicated in each figure legend. P values of <0.05 were considered significant (∗∗∗∗p < 0.0001, ∗∗∗p < 0.001, ∗∗p < 0.01, and ∗p < 0.05) and P values ≥ 0.05 were considered not significant (ns).
Acknowledgments
We thank Brian James for scRNA-seq library preparation, Amy Cortez, Benji Portillo, and Yoav Altman for assistance on FACS-mediated isolation of single cells, Alexandre Colas for support of the Image Xpress confocal microscope (Molecular Devices), and David Huhta for assistance in software installation and analysis troubleshooting. We thank Dr. Francesca Malerba for sharing material and suggestions regarding immunofluorescence for NGF. The graphical abstract was created with BioRender.com.
The snATAC-seq data was performed at the Center for Epigenomics at UC San Diego, which is partly funded by the UC San Diego School of Medicine. This publication includes data generated at the UC San Diego IGM Genomics Center utilizing an Illumina NovaSeq 6000 that was purchased with funding from a National Institutes of Health SIG grant (#S10 OD026929). This work was supported by NIH/NIAMS grant (R01AR076247) to P.L.P.; American Heart Association Postdoctoral Fellowship (19POST34450187) to C.N.; Fishman Fund Postdoctoral Fellowship to U.E.; Italian Ministry of Health (GR-2013- 02356592) and “Roche per la Ricerca 2019” grant to L.M.
Author contributions
C.N. performed scRNA-seq and snATAC-seq data analysis and contributed to the experimental design, manuscript writing, and figure preparation. X.W. contributed to data analysis and interpretation, performed the experiments of denervation for scRNA-seq and snATAC-seq analysis, for isolation of muscle-resident cells and their culture or co-culture ex vivo, performed muscle section and whole mount staining, and contributed to manuscript writing and figure preparation; U.E. contributed to the experimental design and scRNA-seq sample preparation; C.D.E. and L.M. contributed to the data interpretation by performing immunofluorescence staining on muscle sections of PLP1-tdTomato mice and sharing unpublished datasets. R.P. contributed to scRNA-seq data generation. P.L.P. designed the experiments, contributed to the data analysis and interpretation, and wrote the manuscript.
Declaration of interests
The authors declare no competing interests.
Inclusion and diversity
We support inclusive, diverse, and equitable conduct of research.
Published: June 13, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2023.107114.
Supplemental information
Data and code availability
-
•
Single-cell RNA-seq and single-nucleus ATAC-seq data have been deposited at GEO and are publicly available as of the date of publication. Accession numbers are listed in the key resources table.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- 1.Farup J., Madaro L., Puri P.L., Mikkelsen U.R. Interactions between muscle stem cells, mesenchymal-derived cells and immune cells in muscle homeostasis, regeneration and disease. Cell Death Dis. 2015;6:e1830. doi: 10.1038/cddis.2015.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Biferali B., Proietti D., Mozzetta C., Madaro L. Fibro-Adipogenic Progenitors Cross-Talk in Skeletal Muscle: The Social Network. Front. Physiol. 2019;10:1074. doi: 10.3389/fphys.2019.01074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wei X., Nicoletti C., Puri P.L. Fibro-Adipogenic Progenitors: Versatile keepers of skeletal muscle homeostasis, beyond the response to myotrauma. Semin. Cell Dev. Biol. 2021;119:23–31. doi: 10.1016/j.semcdb.2021.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Saleh K.K., Xi H., Switzler C., Skuratovsky E., Romero M.A., Chien P., Gibbs D., Gane L., Hicks M.R., Spencer M.J., Pyle A.D. Single cell sequencing maps skeletal muscle cellular diversity as disease severity increases in dystrophic mouse models. iScience. 2022;25:105415. doi: 10.1016/j.isci.2022.105415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Scripture-Adams D.D., Chesmore K.N., Barthélémy F., Wang R.T., Nieves-Rodriguez S., Wang D.W., Mokhonova E.I., Douine E.D., Wan J., Little I., et al. Single nuclei transcriptomics of muscle reveals intra-muscular cell dynamics linked to dystrophin loss and rescue. Commun. Biol. 2022;5:989. doi: 10.1038/s42003-022-03938-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Leinroth A.P., Mirando A.J., Rouse D., Kobayahsi Y., Tata P.R., Rueckert H.E., Liao Y., Long J.T., Chakkalakal J.V., Hilton M.J. Identification of distinct non-myogenic skeletal-muscle-resident mesenchymal cell populations. Cell Rep. 2022;39:110785. doi: 10.1016/j.celrep.2022.110785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lovrić A., Rassolie A., Alam S., Mandić M., Saini A., Altun M., Fernandez-Gonzalo R., Gustafsson T., Rullman E. Single-cell sequencing deconvolutes cellular responses to exercise in human skeletal muscle. Commun. Biol. 2022;5:1121. doi: 10.1038/s42003-022-04088-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cutler A.A., Pawlikowski B., Wheeler J.R., Dalla Betta N., Elston T., O’Rourke R., Jones K., Olwin B.B. The regenerating skeletal muscle niche drives satellite cell return to quiescence. iScience. 2022;25:104444. doi: 10.1016/j.isci.2022.104444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.De Micheli A.J., Laurilliard E.J., Heinke C.L., Ravichandran H., Fraczek P., Soueid-Baumgarten S., De Vlaminck I., Elemento O., Cosgrove B.D. Single-Cell Analysis of the Muscle Stem Cell Hierarchy Identifies Heterotypic Communication Signals Involved in Skeletal Muscle Regeneration. Cell Rep. 2020;30:3583–3595.e5. doi: 10.1016/j.celrep.2020.02.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Oprescu S.N., Yue F., Qiu J., Brito L.F., Kuang S. Temporal Dynamics and Heterogeneity of Cell Populations during Skeletal Muscle Regeneration. iScience. 2020;23:100993. doi: 10.1016/j.isci.2020.100993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Petrany M.J., Swoboda C.O., Sun C., Chetal K., Chen X., Weirauch M.T., Salomonis N., Millay D.P. Single-nucleus RNA-seq identifies transcriptional heterogeneity in multinucleated skeletal myofibers. Nat. Commun. 2020;11:6374. doi: 10.1038/s41467-020-20063-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chemello F., Wang Z., Li H., McAnally J.R., Liu N., Bassel-Duby R., Olson E.N. Degenerative and regenerative pathways underlying Duchenne muscular dystrophy revealed by single-nucleus RNA sequencing. Proc. Natl. Acad. Sci. USA. 2020;117:29691–29701. doi: 10.1073/pnas.2018391117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wheeler J.R., Whitney O.N., Vogler T.O., Nguyen E.D., Pawlikowski B., Lester E., Cutler A., Elston T., Dalla Betta N., Parker K.R., et al. RNA-binding proteins direct myogenic cell fate decisions. Elife. 2022;11:e75844. doi: 10.7554/eLife.75844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dell’Orso S., Juan A.H., Ko K.-D., Naz F., Perovanovic J., Gutierrez-Cruz G., Feng X., Sartorelli V. Single cell analysis of adult mouse skeletal muscle stem cells in homeostatic and regenerative conditions. Development. 2019;146:dev174177. doi: 10.1242/dev.174177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Giordani L., He G.J., Negroni E., Sakai H., Law J.Y.C., Siu M.M., Wan R., Corneau A., Tajbakhsh S., Cheung T.H., Le Grand F. High-Dimensional Single-Cell Cartography Reveals Novel Skeletal Muscle-Resident Cell Populations. Mol. Cell. 2019;74:609–621.e6. doi: 10.1016/j.molcel.2019.02.026. [DOI] [PubMed] [Google Scholar]
- 16.Malecova B., Gatto S., Etxaniz U., Passafaro M., Cortez A., Nicoletti C., Giordani L., Torcinaro A., De Bardi M., Bicciato S., et al. Dynamics of cellular states of fibro-adipogenic progenitors during myogenesis and muscular dystrophy. Nat. Commun. 2018;9:3670. doi: 10.1038/s41467-018-06068-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Morgan J., Partridge T. Skeletal muscle in health and disease. Dis. Model. Mech. 2020;13:dmm042192. doi: 10.1242/dmm.042192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Serrano A.L., Muñoz-Cánoves P. Fibrosis development in early-onset muscular dystrophies: Mechanisms and translational implications. Semin. Cell Dev. Biol. 2017;64:181–190. doi: 10.1016/j.semcdb.2016.09.013. [DOI] [PubMed] [Google Scholar]
- 19.Lemos D.R., Babaeijandaghi F., Low M., Chang C.-K., Lee S.T., Fiore D., Zhang R.-H., Natarajan A., Nedospasov S.A., Rossi F.M.V. Nilotinib reduces muscle fibrosis in chronic muscle injury by promoting TNF-mediated apoptosis of fibro/adipogenic progenitors. Nat. Med. 2015;21:786–794. doi: 10.1038/nm.3869. [DOI] [PubMed] [Google Scholar]
- 20.Judson R.N., Zhang R.-H., Rossi F.M.A. Tissue-resident mesenchymal stem/progenitor cells in skeletal muscle: collaborators or saboteurs? FEBS J. 2013;280:4100–4108. doi: 10.1111/febs.12370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Madaro L., Passafaro M., Sala D., Etxaniz U., Lugarini F., Proietti D., Alfonsi M.V., Nicoletti C., Gatto S., De Bardi M., et al. Denervation-activated STAT3–IL-6 signalling in fibro-adipogenic progenitors promotes myofibres atrophy and fibrosis. Nat. Cell Biol. 2018;20:917–927. doi: 10.1038/s41556-018-0151-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Proietti D., Giordani L., De Bardi M., D’Ercole C., Lozanoska-Ochser B., Amadio S., Volonté C., Marinelli S., Muchir A., Bouché M., et al. Activation of skeletal muscle-resident glial cells upon nerve injury. JCI Insight. 2021;6:e143469. doi: 10.1172/jci.insight.143469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu W., Wei-LaPierre L., Klose A., Dirksen R.T., Chakkalakal J.V. Inducible depletion of adult skeletal muscle stem cells impairs the regeneration of neuromuscular junctions. Elife. 2015;4:e09221. doi: 10.7554/eLife.09221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu W., Klose A., Forman S., Paris N.D., Wei-LaPierre L., Cortés-Lopéz M., Tan A., Flaherty M., Miura P., Dirksen R.T., Chakkalakal J.V. Loss of adult skeletal muscle stem cells drives age-related neuromuscular junction degeneration. Elife. 2017;6:e26464. doi: 10.7554/eLife.26464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu W., Chakkalakal J.V. The Composition, Development, and Regeneration of Neuromuscular Junctions. Curr. Top. Dev. Biol. 2018;126:99–124. doi: 10.1016/bs.ctdb.2017.08.005. [DOI] [PubMed] [Google Scholar]
- 26.Kang H., Tian L., Thompson W. Terminal Schwann cells guide the reinnervation of muscle after nerve injury. J. Neurocytol. 2003;32:975–985. doi: 10.1023/B:NEUR.0000020636.27222.2d. [DOI] [PubMed] [Google Scholar]
- 27.Stuart T., Butler A., Hoffman P., Hafemeister C., Papalexi E., Mauck W.M., Hao Y., Stoeckius M., Smibert P., Satija R. Comprehensive Integration of Single-Cell Data. Cell. 2019;177:1888–1902.e21. doi: 10.1016/j.cell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wolock S.L., Lopez R., Klein A.M. Scrublet: Computational Identification of Cell Doublets in Single-Cell Transcriptomic Data. Cell Syst. 2019;8:281–291.e9. doi: 10.1016/j.cels.2018.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Joe A.W.B., Yi L., Natarajan A., Le Grand F., So L., Wang J., Rudnicki M.A., Rossi F.M.V. Muscle injury activates resident fibro/adipogenic progenitors that facilitate myogenesis. Nat. Cell Biol. 2010;12:153–163. doi: 10.1038/ncb2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Uezumi A., Fukada S.I., Yamamoto N., Takeda S., Tsuchida K. Mesenchymal progenitors distinct from satellite cells contribute to ectopic fat cell formation in skeletal muscle. Nat. Cell Biol. 2010;12:143–152. doi: 10.1038/ncb2014. [DOI] [PubMed] [Google Scholar]
- 31.Albelda S.M., Muller W.A., Buck C.A., Newman P.J. Molecular and cellular properties of PECAM-1 (endoCAM/CD31): a novel vascular cell-cell adhesion molecule. J. Cell Biol. 1991;114:1059–1068. doi: 10.1083/jcb.114.5.1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vanlandewijck M., He L., Mäe M.A., Andrae J., Ando K., Del Gaudio F., Nahar K., Lebouvier T., Laviña B., Gouveia L., et al. A molecular atlas of cell types and zonation in the brain vasculature. Nature. 2018;554:475–480. doi: 10.1038/nature25739. [DOI] [PubMed] [Google Scholar]
- 33.Hermiston M.L., Xu Z., Weiss A. CD45: A Critical Regulator of Signaling Thresholds in Immune Cells. Annu. Rev. Immunol. 2003;21:107–137. doi: 10.1146/annurev.immunol.21.120601.140946. [DOI] [PubMed] [Google Scholar]
- 34.Murchison N.D., Price B.A., Conner D.A., Keene D.R., Olson E.N., Tabin C.J., Schweitzer R. Regulation of tendon differentiation by scleraxis distinguishes force-transmitting tendons from muscle-anchoring tendons. Development. 2007;134:2697–2708. doi: 10.1242/dev.001933. [DOI] [PubMed] [Google Scholar]
- 35.Baghdadi M.B., Castel D., Machado L., Fukada S.-I., Birk D.E., Relaix F., Tajbakhsh S., Mourikis P. Reciprocal signalling by Notch-Collagen V-CALCR retains muscle stem cells in their niche. Nature. 2018;557:714–718. doi: 10.1038/s41586-018-0144-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fukada S.I., Uezumi A., Ikemoto M., Masuda S., Segawa M., Tanimura N., Yamamoto H., Miyagoe-Suzuki Y., Takeda S. Molecular signature of quiescent satellite cells in adult skeletal muscle. Stem Cell. 2007;25:2448–2459. doi: 10.1634/stemcells.2007-0019. [DOI] [PubMed] [Google Scholar]
- 37.Seale P., Sabourin L.A., Girgis-Gabardo A., Mansouri A., Gruss P., Rudnicki M.A. Pax7 is required for the specification of myogenic satellite cells. Cell. 2000;102:777–786. doi: 10.1016/s0092-8674(00)00066-0. [DOI] [PubMed] [Google Scholar]
- 38.Birchmeier C., Bennett D.L.H. In: Current Topics in Developmental Biology Essays on Developmental Biology, Part A. Wassarman P.M., editor. Academic Press; 2016. Chapter Four - Neuregulin/ErbB Signaling in Developmental Myelin Formation and Nerve Repair; pp. 45–64. [DOI] [PubMed] [Google Scholar]
- 39.Cao J., Spielmann M., Qiu X., Huang X., Ibrahim D.M., Hill A.J., Zhang F., Mundlos S., Christiansen L., Steemers F.J., et al. The single-cell transcriptional landscape of mammalian organogenesis. Nature. 2019;566:496–502. doi: 10.1038/s41586-019-0969-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Castro R., Taetzsch T., Vaughan S.K., Godbe K., Chappell J., Settlage R.E., Valdez G. Specific labeling of synaptic schwann cells reveals unique cellular and molecular features. Elife. 2020;9:e56935. doi: 10.7554/eLife.56935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jessen K.R., Arthur-Farraj P. Repair Schwann cell update: Adaptive reprogramming, EMT, and stemness in regenerating nerves. Glia. 2019;67:421–437. doi: 10.1002/glia.23532. [DOI] [PubMed] [Google Scholar]
- 42.Sidney L.E., Branch M.J., Dunphy S.E., Dua H.S., Hopkinson A. Concise review: evidence for CD34 as a common marker for diverse progenitors. Stem Cell. 2014;32:1380–1389. doi: 10.1002/stem.1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wolbert J., Li X., Heming M., Mausberg A.K., Akkermann D., Frydrychowicz C., Fledrich R., Groeneweg L., Schulz C., Stettner M., et al. Redefining the heterogeneity of peripheral nerve cells in health and autoimmunity. Proc. Natl. Acad. Sci. USA. 2020;117:9466–9476. doi: 10.1073/pnas.1912139117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lopez-Anido C., Sun G., Koenning M., Srinivasan R., Hung H.A., Emery B., Keles S., Svaren J. Differential Sox10 genomic occupancy in myelinating glia. Glia. 2015;63:1897–1914. doi: 10.1002/glia.22855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Farup J., Just J., de Paoli F., Lin L., Jensen J.B., Billeskov T., Roman I.S., Cömert C., Møller A.B., Madaro L., et al. Human skeletal muscle CD90+ fibro-adipogenic progenitors are associated with muscle degeneration in type 2 diabetic patients. Cell Metab. 2021;33:2201–2214.e11. doi: 10.1016/j.cmet.2021.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Efremova M., Vento-Tormo M., Teichmann S.A., Vento-Tormo R. CellPhoneDB: inferring cell–cell communication from combined expression of multi-subunit ligand–receptor complexes. Nat. Protoc. 2020;15:1484–1506. doi: 10.1038/s41596-020-0292-x. [DOI] [PubMed] [Google Scholar]
- 47.Skelly D.A., Squiers G.T., McLellan M.A., Bolisetty M.T., Robson P., Rosenthal N.A., Pinto A.R. Single-Cell Transcriptional Profiling Reveals Cellular Diversity and Intercommunication in the Mouse Heart. Cell Rep. 2018;22:600–610. doi: 10.1016/j.celrep.2017.12.072. [DOI] [PubMed] [Google Scholar]
- 48.Lindström H.J.G., Friedman R. Inferring time-dependent population growth rates in cell cultures undergoing adaptation. BMC Bioinf. 2020;21:583. doi: 10.1186/s12859-020-03887-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Richner M., Ulrichsen M., Elmegaard S.L., Dieu R., Pallesen L.T., Vaegter C.B. Peripheral Nerve Injury Modulates Neurotrophin Signaling in the Peripheral and Central Nervous System. Mol. Neurobiol. 2014;50:945–970. doi: 10.1007/s12035-014-8706-9. [DOI] [PubMed] [Google Scholar]
- 50.Singhal N., Martin P.T. Role of extracellular matrix proteins and their receptors in the development of the vertebrate neuromuscular junction. Dev. Neurobiol. 2011;71:982–1005. doi: 10.1002/dneu.20953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu L., Cheung T.H., Charville G.W., Rando T.A. Isolation of skeletal muscle stem cells by fluorescence-activated cell sorting. Nat. Protoc. 2015;10:1612–1624. doi: 10.1038/nprot.2015.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hafemeister C., Satija R. Normalization and variance stabilization of single-cell RNA-seq data using regularized negative binomial regression. Genome Biol. 2019;20:296. doi: 10.1186/s13059-019-1874-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stuart T., Srivastava A., Madad S., Lareau C.A., Satija R. Single-cell chromatin state analysis with Signac. Nat. Methods. 2021;18:1333–1341. doi: 10.1038/s41592-021-01282-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Durinck S., Spellman P.T., Birney E., Huber W. Mapping identifiers for the integration of genomic datasets with the R/Bioconductor package biomaRt. Nat. Protoc. 2009;4:1184–1191. doi: 10.1038/nprot.2009.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Uezumi A., Ikemoto-Uezumi M., Zhou H., Kurosawa T., Yoshimoto Y., Nakatani M., Hitachi K., Yamaguchi H., Wakatsuki S., Araki T., et al. Mesenchymal Bmp3b expression maintains skeletal muscle integrity and decreases in age-related sarcopenia. J. Clin. Invest. 2021;131:e139617. doi: 10.1172/JCI139617. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
Single-cell RNA-seq and single-nucleus ATAC-seq data have been deposited at GEO and are publicly available as of the date of publication. Accession numbers are listed in the key resources table.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.