Abstract
Background
Neisseria gonorrhoeae is a major public health problem due to increasing incidence and antimicrobial resistance. Genetic markers of reduced susceptibility have been identified; the extent to which those are representative of global antimicrobial resistance is unknown. We evaluated the performance of whole-genome sequencing (WGS) used to predict susceptibility to ciprofloxacin and other antimicrobials using a global collection of N. gonorrhoeae isolates.
Methods
Susceptibility testing of common antimicrobials and the recently developed zolifodacin was performed using agar dilution to determine minimum inhibitory concentrations (MICs). We identified resistance alleles at loci known to contribute to antimicrobial resistance in N. gonorrhoeae from WGS data. We tested the ability of each locus to predict antimicrobial susceptibility.
Results
A total of 481 N. gonorrhoeae isolates, collected between 2004 and 2019 and making up 457 unique genomes, were sourced from 5 countries. All isolates with demonstrated susceptibility to ciprofloxacin (MIC ≤0.06 μg/mL) had a wild-type gyrA codon 91. Multilocus approaches were needed to predict susceptibility to other antimicrobials. All isolates were susceptible to zoliflodacin, defined by an MIC ≤0.25 μg/mL.
Conclusions
Single marker prediction can be used to inform ciprofloxacin treatment of N. gonorrhoeae infection. A combination of molecular markers may be needed to determine susceptibility for other antimicrobials.
Keywords: Neisseria gonorrhoeae, antimicrobial susceptibility testing, whole-genome sequencing
A single genetic marker, gyrA codon 91, correctly predicted ciprofloxacin susceptibility in a global collection of Neisseria gonorrhoeae isolates with genome sequences and phenotypic antimicrobial susceptibility data. For other antimicrobials, multiple molecular markers are needed to accurately predict antimicrobial susceptibility.
Neisseria gonorrhoeae infection has become a major threat to public health owing to increasing incidence and antimicrobial resistance [1–5]. Globally, there are >87 million cases of N. gonorrhoeae every year [6]. Resistance has emerged to each antimicrobial used to treat N. gonorrhoeae, and World Health Organization (WHO) guidelines suggest that empiric therapy should be discontinued when the prevalence of antimicrobial resistance reaches 5% of the N. gonorrhoeae population [2, 7]. Because of the emergence and expansion of N. gonorrhoeae lineages resistant to previously recommended therapies, treatment with ceftriaxone (an injectable extended-spectrum cephalosporin) is the currently recommended therapy for N. gonorrhoeae infection in the United States [8]; however, cases of reduced susceptibility have increasingly been documented [3–5, 8–20]. Zoliflodacin is a promising new antimicrobial with a unique mechanism of action for the treatment of N. gonorrhoeae infection in a time where the antimicrobial pipeline is limited. A global phase III clinical trial for of zoliflodacin for the treatment of uncomplicated N. gonorrhoeae infection is underway (ClinicalTrials.gov NCT03959527) [21, 22].
As an alternative to empiric therapy, antibiotic prescribing for gonorrhea could be guided by antimicrobial susceptibility testing (AST). However, there are technical and resource constraints to widespread AST. Antimicrobial susceptibility is measured on a continuous scale using the minimum inhibitory concentration (MIC), and antimicrobial resistance is defined as having greater than a certain threshold MIC. Currently, N. gonorrhoeae AST uses phenotypic methods that require live organisms. Newer methods of AST are urgently needed.
Screening for bacterial genotypes to predict antimicrobial resistance phenotypes may be a valuable method to guide treatment [23–25]. For example, commercialized molecular tests (eg, SpeedDx ResistancePlus GC) [26] targeting gyrA codon 91 have been developed and marketed in Europe and Australia for the simultaneous detection of N. gonorrhoeae infection and prediction of ciprofloxacin susceptibility [27]. In addition, whole-genome sequencing (WGS) has advanced, and the application of this technology might aid in the assessment of drug susceptibility. Genetic markers of reduced susceptibility and resistance in N. gonorrhoeae have been identified for currently available antimicrobials [28–32]. However, those markers may not explain all of the resistance or reduced susceptibility identified through phenotypic methods. In addition, data sets with a global distribution of strains are limited and thus the extent to which those genetic determinants are representative of global antimicrobial resistance is unknown.
In the current study, we collected gonococcal isolates from diverse geographic locations where antimicrobial selection pressures vary and validated WGS for the prediction of antimicrobial susceptibility in N. gonorrhoeae. The primary outcome measures were to compare the performance of WGS and the genotype at previously reported molecular genetic markers in N. gonorrhoeae isolates to predict susceptibility to ciprofloxacin to a culture-based phenotypic method as a nonreference standard. The secondary outcome measures were to assess the performance of WGS with previously reported molecular genetic markers for the detection of susceptibility to ceftriaxone, cefixime, azithromycin, spectinomycin, penicillin, tetracycline, and zoliflodacin in N. gonorrhoeae isolates compared with a culture-based phenotypic method. Additional secondary outcome measures included calculating the fraction of reduced susceptibility that do not have previously reported genetic markers of resistance and describing the genetic relatedness between isolates across geographic areas. The results of this study will contribute to the development of diagnostic tools to guide clinical treatment for N. gonorrhoeae infections.
METHODS
Study Design
This study was designed to evaluate the validity of WGS to predict susceptibility to ciprofloxacin using a global collection of N. gonorrhoeae isolates. Additional aims were to determine the performance of WGS to predict susceptibility to ceftriaxone, cefixime, azithromycin, spectinomycin, penicillin, tetracycline and zoliflodacin in this collection of isolates. N. gonorrhoeae isolates from Canada, the Dominican Republic, Hong Kong, South Africa, and Vietnam were selected and underwent AST by agar dilution, and the isolates’ genomes were sequenced (Table 1). Positive percent agreement (PPA), negative percent agreement (NPA), positive predictive value (PPV), and negative predictive value (NPV) were all calculated for susceptibility to antimicrobials by comparing the reference standard (MIC breakpoint) results (susceptible vs resistant or reduced susceptibility) and the genotype based on the presence of previously reported genetic markers of resistance, as described below.
Table 1.
Number of Neisseria gonorrhoeae Isolates and Sample Collection Years From Each Geographic Site
| Site | Collection Years | Isolates, No. |
|---|---|---|
| Canada | 2004–2017 | 42 |
| South Africa | 2013–2014 | 55 |
| Hong Kong | 2014–2018 | 225 |
| Vietnam | 2019 | 123 |
| Dominican Republic | 2018–2019 | 36 |
| Total | … | 481 |
Study Procedures/Evaluations
Isolates
A total of 481 N. gonorrhoeae isolates were sourced from 5 countries to create the collection analyzed in this study.
Phenotypic AST
The laboratory at the University of Washington performed confirmatory tests on all N. gonorrhoeae isolates, conducted multiple subcultures to obtain pure N. gonorrhoeae colonies from contaminated cultures, and then performed culture-based AST by the Clinical and Laboratory Standards Institute (CLSI)–recommended reference agar dilution methods to determine the MIC [33]. The panel of antimicrobial agents and ranges of concentrations for AST included penicillin (0.008–64.0 µg/mL), tetracycline (0.008–64.0 µg/mL), cefixime (0.001–2.0 µg/mL), ceftriaxone (0.001–2.0 µg/mL), ciprofloxacin (0.001–64.0 µg/mL), azithromycin (0.008–256.0 µg/mL), zoliflodacin (0.001–2.0 µg/mL), and spectinomycin (16.0–256.0 µg/mL).
In addition, β-lactamase production was also determined using the nitrocefin reference test [28]. For quality assurance, each AST run performed included a set of 4 well-characterized reference N. gonorrhoeae strains with known MICs, including American Type Culture Collection 49226 recommended by CLSI and 3 of the whole-genome sequenced 2016 WHO reference strains, WHO L, WHO O, and WHO U [28]. Reference N. gonorrhoeae strain AST results had to be within established MIC range for the AST run to be valid. All measured MICs for each isolate can be found in Supplementary Table 1.
WGS and Assembly
DNA was extracted from bacterial cells grown overnight on GCB-K plates at 37°C with 5% carbon dioxide using the Invitrogen PureLink Genomic DNA Mini Kit. Libraries were prepped and sequenced on the Illumina NextSeq 2000 sequencer at the Microbial Whole Genome Sequencing Center or the Bauer Core Facility at Harvard University. Read quality was assessed using FastQC (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/). Reads were mapped to the NCCP11945 (NC_011035.1) reference genome using BWA-MEM software (version 0.7.17) [34], mapped reads were sorted and indexed with Samtools (version 1.9) [35], duplicates were marked with Picard tools (version 2.20.1) (https://broadinstitute.github.io/picard/), and variants were called using Pilon software (version 1.23) [36]. Variants were called using a minimum mapping quality of 20 and a minimum coverage of 10×.
Reads were also mapped to single copies of the 23S and 16S ribosomal DNA (rDNA) sequences using the same procedure to call variants. The quality and coverage of mapped reads were assessed using Qualimap software, version 2.2.1 [37]. We created pseudogenomes by editing the reference genome with variants supported by ≥90% of mapped reads and replacing ambiguous bases with “N” and deletions with “-”. We performed de novo genome assembly with SPAdes software (version 3.12.0) [38] and filtered contigs <500 nucleotides in length or with <10× coverage. Sequencing reads have been deposited at the National Center for Biotechnology Information (BioProject accession no. PRJNA776899).
Resistance Allele Identification
Using the WGS data, we identified resistance alleles at all loci known to contribute to antimicrobial resistance in N. gonorrhoeae. The full list of molecular markers that modulate antibiotic susceptibility assessed is included in Table 2. The presence of single-nucleotide polymorphisms associated with resistance was determined from variant calls after mapping to the reference genome. If coverage, mapping quality, or allele frequency did not meet the thresholds described above, the genotype was not defined.
Table 2.
Molecular Markers That Modulate Antibiotic Susceptibility
| Antimicrobial | CLSI MIC Breakpoint, µg/mL | Sitesa | |
|---|---|---|---|
| Susceptibility | Resistance | ||
| Penicillin | ≤0.06 | ≥2 | bla TEM, mosaic mtr, mosaic penA, mtr promoter mutations, mtrC loss of function, mtrR loss of function, mtrR 39, mtrR 45, penA 501, penA 513, penA 517, penA 542, penA 543, penA 551 porB 120, porB 121 |
| Tetracycline | ≤0.25 | ≥2 | Mosaic mtr, mtr promoter mutations, mtrR loss of function, mtrC loss of function, mtrR 39, mtrR 45, porB 120, porB 121, rpsJ 57, tetM |
| Ciprofloxacin | ≤0.06 | ≥1 | gyrA 91, gyrA 95, mosaic mtr loci, mtr promoter mutations, mtrC loss of function, mtrR loss of function, mtrR 39, mtrR 45, parC 86, parC 87, parC 91, porB 120, porB 121 |
| Spectinomycin | ≤32 | ≥128 | 16S rDNA 1187, rpsE 24 rpsE del 27, rpsE 82 |
| Azithromycin | ≤1 | … | 23S rRNA 2059, 23S rRNA 2611, duplications in rpIV, ermB, ermC, mosaic mtr, mtr promoter mutations, mtrC loss of function, mtrR loss of function, mtrR 39, mtrR 45, rpID 70 |
| Cefixime | ≤0.25 | … | Mosaic penA, mtr promoter mutations, mtrC loss of function, mtrR loss of function, mtrR 39, mtrR 45, penA 501, penA 513, penA 517, penA 542, penA 543, penA 551, porB 120, porB 121, rpoB 157, rpoB 158, rpoB 201, rpoD 98, rpoD del 92–95 |
| Ceftriaxone | ≤0.25 | … | Mosaic penA, mtr promoter mutations, mtrC loss of function, mtrR loss of function, mtrR 39, mtrR 45, penA 501, penA 513, penA 517, penA 542, penA 543, penA 551, porB 120, porB 121, rpoB 157, rpoB 158, rpoB 201, rpoD 98, rpoD del 92–95 |
| Zoliflodacin | … | … | gyrB 429, gyrB 450 |
Abbreviations: CLSI, Clinical and Laboratory Standards Institute; del, deletion; MIC, minimum inhibitory concentration; rDNA, ribosomal DNA; rRNA, ribosomal RNA.
Numbers indicate the codon analyzed. If a specific codon is not indicated, gene, loss of function allele, or mosaicism presence was used.
Variants in 16S and 23S rDNA were separately called based on mapping to these loci; the copy number of 23S rDNA variants was determined based on allele frequency, as described elsewhere [39]. The penA alleles were typed according to the NGSTAR database (last accessed 2 March 2021) from assemblies. Resistance-associated insertions and deletions were identified from assemblies. For mtrCDE, mtrR, and the mtr promoter, mosaic alleles were defined as those with <95% identity to the wild-type mtr locus from FA1090 (NC_002946.2). When ≥1 of mtrCDE in addition to the promoter region showed evidence of mosaicism, the mtr operon was designated as mosaic [40]. The presence of tetM, blaTEM, and erm genes was determined using blastn software (version 2.9.0) [41] from assemblies using the following accessions as the query: MG874353.1, NG_068038.1, EU048318.1, and M14730.1.
Phylogenetic Analysis
An alignment of pseudogenomes was used for phylogenetic analysis. Gubbins (version 2.4.1) [42] and RAxML (version 8.2.12) [43] software were used for recombination detection and building a recombination-corrected maximum likelihood phylogeny.
Statistical Analysis
We assessed the performance of susceptibility prediction, using genetic markers from WGS data and 4 methods: predicting susceptibility using single markers, predicting susceptibility from the presence of susceptibility-associated allele at all loci, predicting MICs from multiple markers using multivariate linear regression, and predicting MICs using previously described equations [44]. We varied MIC breakpoints for determination of antimicrobial susceptibility: ciprofloxacin (MIC, ≤0.06 and ≤0.5 μg/mL), ceftriaxone (≤0.06, ≤0.125, and ≤0.25 μg/mL), cefixime (≤0.06, ≤0.125, and ≤0.25), azithromycin (≤0.5 and ≤1 μg/mL), spectinomycin (≤32 and ≤64 μg/mL), penicillin (≤0.06 and ≤1 μg/mL), tetracycline (≤0.25 and ≤1 μg/mL), zoliflodacin (≤0.25 and ≤0.5 μg/mL) to calculate measures of validity, PPVs, and NPVs. For each antimicrobial that has a designated CLSI breakpoint, we included that breakpoint in the above (CLSI MIC breakpoints for susceptibility: ciprofloxacin, ≤0.06 μg/mL; ceftriaxone, cefixime, and tetracycline, ≤0.25 μg/mL; azithromycin, ≤1 μg/mL; spectinomycin, ≤32 μg/mL; penicillin, ≤0.06 μg/mL) [45].
We estimated the PPA, NPA, PPV, and NPV of known genetic markers (Table 2) for the detection of susceptibility to each antimicrobial (ciprofloxacin, ceftriaxone, cefixime, azithromycin, spectinomycin, tetracycline, penicillin, and zoliflodacin) compared with phenotypic antimicrobial susceptibility determination by agar dilution (measures were calculated twice using each of 2 MIC breakpoints for susceptibility: ≤0.06 and ≤0.5 μg/mL). Because multiple molecular markers may contribute to antimicrobial resistance, we also conducted multivariate linear regression modeling to predict the MIC for each isolate from a combination of molecular antimicrobial reduced susceptibility determinants for each antimicrobial [44]. We used log2(MIC) as the dependent variable in the linear regression and the genetic markers for resistance (Table 2) as the independent variables in the regression model.
The weights used for each molecular marker were generated from the coefficients from the regression and we report those equations to calculate the predicted MIC. From those we calculated the predicted MIC. The predicted MICs and MICs measured by means of agar dilution AST were plotted, and R2 was calculated for each antimicrobial. In addition, we calculated the PPV to demonstrate the variation that existed in the prediction models.
We also predicted MICs using the multivariate regression equations from Demczuk et al [44], which were trained on a set of isolates distinct from those described here. The performance of those equations was assessed by comparing predicted and measured MICs and calculating the R2 for each antimicrobial. In addition, we calculated the PPV for susceptibility category (susceptible, nonsusceptible, resistant, etc) given the predicted MIC and the categorical cutoffs for each antibiotic.
This research was deemed nonhuman subject research by the institutional review boards at the University of California San Diego, University of Washington, and Harvard T.H. Chan School of Public Health and therefore did not require institutional review board oversight.
RESULTS
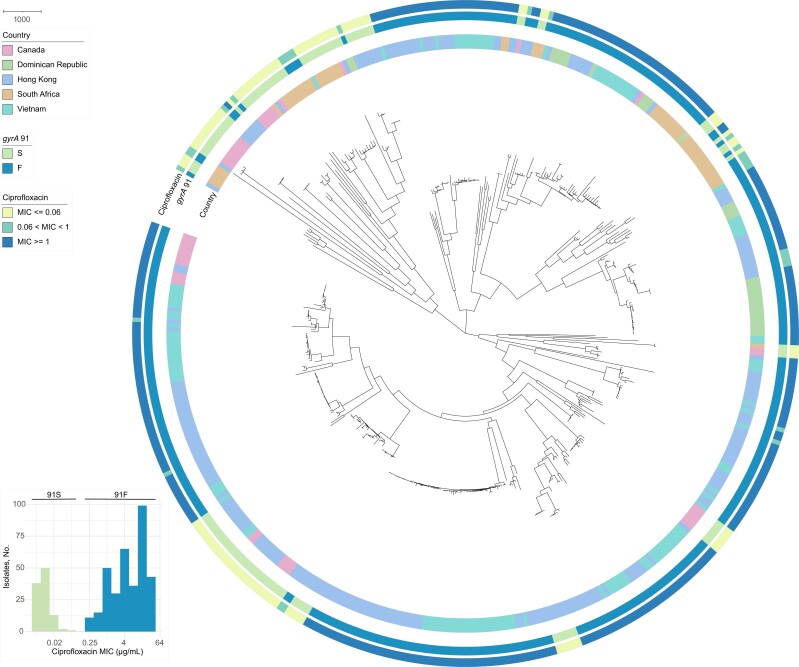
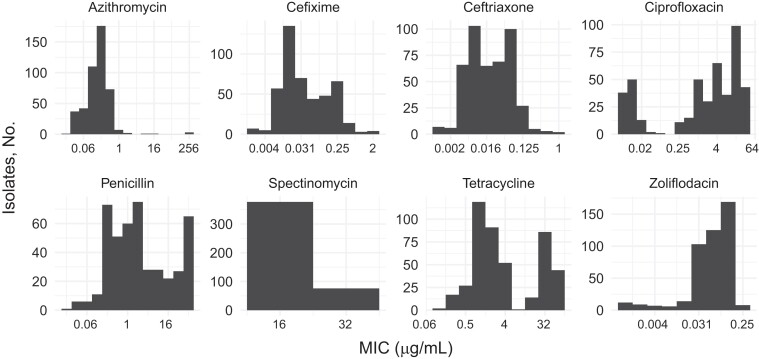
We conducted AST and WGS for 481 N. gonorrhoeae isolates. The sequencing reads from 1 isolate were heavily contaminated, and comparison of single-nucleotide polymorphism distances from pseudogenomes demonstrated that an additional 23 isolates were identical to another isolate in the collection. We removed 1 isolate from the identical pairs based on assembly quality, yielding a final data set of 457 genomes (Table 1). Isolates from each geographic location were diverse and interspersed on the phylogeny (Figure 1). The collection of 457 isolates represented a range of MICs for ciprofloxacin, cefixime, ceftriaxone, ciprofloxacin, penicillin, and tetracycline, with MICs in both the susceptible and resistant or reduced susceptibility ranges (Figure 2). We did not observe any isolates with decreased susceptibility or resistance to spectinomycin or zoliflodacin.
Figure 1.
The gyrA 91S genotype is associated with ciprofloxacin susceptibility across geographic locations. A maximum likelihood whole-genome phylogeny of 457 Neisseria gonorrhoeae isolates mapped to the NCCP11945 (NC_011035.1) reference genome was generated using Gubbins software. Branch lengths represent nonrecombinant substitutions. Inner annotation ring represents the geographic locations of isolates: Canada (pink), Dominican Republic (green), Hong Kong (blue), South Africa (tan), or Vietnam (teal). Middle ring represents gyrA codon 91: serine, S (green) or phenylalanine, F (blue), and outer ring, the ciprofloxacin minimum inhibitory concentration (MIC) category: ≤0.06 μg/mL (yellow), 0.06 to <1 μg/mL (green), or ≥1 μg/mL (blue). Inset depicts the distribution of ciprofloxacin MICs; isolates with the gyrA 91S genotype have ciprofloxacin MICs ranging from 0.008–0.06 μg/mL, and isolates with the gyrA 91F genotype have ciprofloxacin MICs ranging from 0.25 – >32 μg/mL.
Figure 2.
Histograms indicating the azithromycin, cefixime, ceftriaxone, ciprofloxacin, penicillin, spectinomycin, tetracycline, and zoliflodacin minimum inhibitory concentration (MIC) distribution in the collection of Neisseria gonorrhoeae isolates (N = 457).
All isolates with demonstrated susceptibility to ciprofloxacin, defined by an MIC ≤0.06 μg/mL, had a serine (the wild-type allele) encoded at gyrA codon 91 (lower confidence limit for 95% confidence interval [LCL], 97.2%) (Figure 1). The NPA for a phenylalanine (the mutant allele) encoded at gyrA position 91 and reduced susceptibility to ciprofloxacin (MIC, >0.06 μg/mL) was also 100% (LCL, 99.2%). When using a higher MIC breakpoint for ciprofloxacin susceptibility determination (≤0.5 μg/mL), the PPA and NPA for the gyrA 91 genotype were 78.8% (LCL, 72.1%) and 100% (99.1%), respectively. However, the PPVs and NPVs for the gyrA 91 genotype and ciprofloxacin susceptibility determination, defined by that higher breakpoint, were high, at 100% (LCL, 97.2%) and 92.1% (89.3%), respectively. Performance characteristics for antimicrobial susceptibility determination by other single molecular markers are included in Supplementary Table 2.
For ciprofloxacin, all of the assessed molecular markers indicated susceptibility in 7 (6.7%) of the 104 isolates that were susceptible based on phenotypic determination (MIC, ≤0.06 μg/mL). In addition, 8 of 452 (1.8%) ceftriaxone-susceptible, 8 of 438 (1.8%) cefixime-susceptible, and 3 of 450 (0.7%) azithromycin-susceptible isolates had all respective molecular markers indicating susceptibility. Thus, among isolates in which all assessed molecular markers indicated susceptibility, 100% (LCL, 65.2%) were ciprofloxacin susceptible based on the phenotypic determination, 100% (68.8%) were ceftriaxone susceptible based on the phenotypic determination (MIC, ≤0.25 μg/mL), 100% (LCL, 68.8%) were cefixime susceptible based on the phenotypic determination (≤0.25 μg/mL), and 100% (36.8%) were azithromycin susceptible based on the phenotypic determination (≤1 μg/mL) (Supplementary Table 3).
Since multiple loci may contribute to antimicrobial susceptibility and isolates with susceptibility-associated alleles at all molecular markers were rare, we also used regression analysis to generate equations to predict MIC from multiple molecular markers (Supplementary Figure 1). We found some variance in the data such that our predicted MICs did not perfectly match measured MICs (Supplementary Figure 2); however, the PPVs were >90% for each antimicrobial (Supplementary Table 4). In addition, we used the regression equations reported by Demczuk et al [44] to create a second set of predicted MICs that we compared to our measured MICs (Supplementary Figure 3). We found that these equations also performed well with PPVs >95% for ciprofloxacin, ceftriaxone, cefixime and azithromycin susceptibility (Table 3). However, using both sets of equations, we found that some isolates with nonsusceptible MICs were incorrectly predicted to be susceptible for some antimicrobials. Using the equations from Demczuk et al [44], that was 4.2% of isolates for cefixime, 1.1% for ceftriaxone and 0.5% for azithromycin (Table 2 and Supplementary Table 4).
Table 3.
Performance of Genotype-Based Prediction of Susceptibility Calculated Using Previously Published Regression EquationsS Compared to the Phenotypic Susceptibility in a Collection of Neisseria gonorrhoeae Isolates From 5 Countries (N = 457)a
| Antimicrobial | MIC Breakpoint, μg/mL | TP | FP | TN | FN | PPV, % | |
|---|---|---|---|---|---|---|---|
| Est | LCLb | ||||||
| Ciprofloxacin | ≤0.06 | 104 | 0 | 353 | 0 | 100 | 97.2 |
| ≤0.5 | 130 | 57 | 268 | 2 | 69.5 | 63.5 | |
| Cefixime | ≤0.06 | 255 | 104 | 29 | 67 | 71.0 | 66.8 |
| ≤0.125 | 370 | 83 | 1 | 1 | 81.7 | 78.4 | |
| ≤0.25 | 436 | 19 | 0 | 0 | 95.8 | 93.9 | |
| Ceftriaxone | ≤0.06 | 260 | 2 | 33 | 160 | 99.2 | 97.6 |
| ≤0.125 | 366 | 5 | 4 | 80 | 98.7 | 97.2 | |
| ≤0.25 | 449 | 5 | 0 | 1 | 98.9 | 97.7 | |
| Azithromycin | ≤0.5 | 409 | 5 | 9 | 30 | 98.8 | 97.5 |
| ≤1 | 437 | 2 | 5 | 9 | 99.5 | 98.6 | |
| Penicillin | ≤ 0.06 | 0 | 0 | 442 | 13 | … | … |
| ≤1 | 110 | 3 | 241 | 101 | 97.3 | 93.3 | |
| Tetracycline | ≤0.06 | 0 | 0 | 434 | 19 | … | … |
| ≤1 | 19 | 0 | 289 | 145 | 100 | 85.4 | |
Abbreviations: FN, false-negative; FP, false-positive; LCL, lower confidence limit for 95% confidence interval; MIC, minimum inhibitory concentration; TN, true-negative; TP, true-positive.
Susceptibility predictions were based on regression equations from Demczuk et al [44] and were compared with phenotypically measured susceptibility.
LCL based on 1-sided 95% Clopper-Pearson confidence interval.
DISCUSSION
We collected and analyzed N. gonorrhoeae isolates from multiple geographic sites around the world to assess the performance of WGS and bioinformatic analysis of known genetic markers to predict antimicrobial susceptibility. Encouragingly, all isolates in our diverse collection were susceptible to zoliflodacin by an MIC breakpoint of ≤0.25 μg/mL, confirming a large prior in silica study [21]. Our results affirm previous findings and indicate that N. gonorrhoeae diagnostics based on the gyrA genotype will perform well across geographic locations [26, 27]. In all isolates, gyrA 91S predicted ciprofloxacin susceptibility defined by an MIC of ≤0.06 μg/mL. In addition, there was 100% agreement between those with gyrA 91F and nonsusceptibility to ciprofloxacin defined by the CLSI breakpoint of MIC >0.06 μg/mL. The PPV for susceptibility is the most important factor when considering using genetic markers for susceptibility determination, because a high PPV indicates that the genetic marker can reliably be used in clinical contexts to indicate susceptibility (eg, wild-type gyrA codon 91S in the case of ciprofloxacin) and the expectation of successful treatment [46].
Single marker prediction did not perform as well for any of the other antimicrobials we evaluated. Moreover, few isolates had all of the molecular markers associated with susceptibility, and thus it would be rare that testing for susceptible genotypes at all markers would be a useful way to inform therapy selection for N. gonorrhoeae infection. However, equations derived from linear regression performed relatively well in predicting susceptibility to antimicrobials; there were some cases of false-positive susceptible determination for a small percentage of isolates for ceftriaxone, cefixime, and azithromycin (<5% demonstrating susceptibility when the measured MIC is above the CLSI breakpoint for susceptibility). Those isolates for which the regression equation–predicted MIC indicated susceptibility but that were not deemed susceptible by phenotypic characterization may have undescribed genetic variants or more complicated molecular interactions causing contributing to the observed high MICs.
The training data set for genotype-based prediction informs performance on test data sets of isolates obtained from other settings. Using a similar regression-based approach and a training data set of 1280 strains, the majority from Canada, a genotype-based prediction of resistance phenotypes yielded output best for a test data set representing 1095 Canadian strains and somewhat lower correlation among 431 strains from the United States and the United Kingdom [44]. While our data set aimed to reduce the likelihood of bias from restricted geographic sampling, it was neither geographically nor temporally comprehensive. Ongoing monitoring of resistance patterns and emergence of novel determinants of resistance will be critical if molecular AST is introduced [47].
Ceftriaxone is the recommended empiric treatment for N. gonorrhoeae infection in the United States and other parts of the world, and concern about the emergence of resistance underscores the need for strategies to help maintain its clinical utility. In the current study, we provide evidence supporting the use of genotype-based prediction of antibiotic susceptibility to expand treatment options from empiric ceftriaxone to a range of possible tailored therapies. Such an approach could have several benefits. First, reduction of the selective pressures from ceftriaxone use could slow the emergence of resistance [23, 48, 49]. Second, when isolates are known to be susceptible to ≥2 antibiotics, the availability of choice provides treatment flexibility when there are supply chain challenges and in clinical scenarios where the empiric antibiotic option is not ideal (eg, because of allergies or the mode of administration). The linear regression approach used here could also be used more broadly in molecular surveillance programs to identify N. gonorrhoeae strains with unexplained antimicrobial resistance. Continued global surveillance and growing understanding of the genetic basis of resistance, even as new antibiotics are introduced into clinical practice, will be critical for maintaining accuracy and the broad utility of genotype-based diagnostics.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Supplementary Material
Contributor Information
Claire C Bristow, Department of Medicine, University of California San Diego, La Jolla, California, USA.
Tatum D Mortimer, Department of Immunology and Infectious Diseases, Harvard T.H. Chan School of Public Health, Boston, Massachusetts, USA.
Sheldon Morris, Department of Medicine, University of California San Diego, La Jolla, California, USA.
Yonatan H Grad, Department of Immunology and Infectious Diseases, Harvard T.H. Chan School of Public Health, Boston, Massachusetts, USA.
Olusegun O Soge, Departments of Global Health, Allergy and Infectious Disease, Laboratory Medicine and Pathology, University of Washington, Seattle, Washington, USA.
Erika Wakatake, Departments of Global Health, Allergy and Infectious Disease, Laboratory Medicine and Pathology, University of Washington, Seattle, Washington, USA.
Rushlenne Pascual, Departments of Global Health, Allergy and Infectious Disease, Laboratory Medicine and Pathology, University of Washington, Seattle, Washington, USA.
Sara McCurdy Murphy, Social & Scientific Systems, a DLH Holdings Company, Silver Spring, Maryland, USA.
Kyra E Fryling, Department of Immunology and Infectious Diseases, Harvard T.H. Chan School of Public Health, Boston, Massachusetts, USA.
Paul C Adamson, Division of Infectious Diseases at the David Geffen School of Medicine, University of California, Los Angeles, California, USA.
Jo-Anne Dillon, Department of Biochemistry, Microbiology and Immunology, University of Saskatchewan, Saskatoon, Sikkim, Canada.
Nidhi R Parmar, Department of Biochemistry, Microbiology and Immunology, University of Saskatchewan, Saskatoon, Sikkim, Canada.
Hai Ha Long Le, Department of Microbiology, Mycology and Parasitology, National Hospital of Venereology and Dermatology, Hanoi, Vietnam; Department of Clinical Microbiology and Parasitology, Hanoi Medical University, Hanoi, Vietnam.
Hung Van Le, Department of Microbiology, Mycology and Parasitology, National Hospital of Venereology and Dermatology, Hanoi, Vietnam; Department of Dermatology and Venereology, Hanoi Medical University, Hanoi, Vietnam.
Reyna Margarita Ovalles Ureña, Laboratorio Nacional Dr. Defillo, Santo Domingo, Dominican Republic.
Nireshni Mitchev, University of KwaZulu-Natal: Durban, KwaZulu-Natal, Glenwood, Durban, South Africa.
Koleka Mlisana, University of KwaZulu-Natal: Durban, KwaZulu-Natal, Glenwood, Durban, South Africa; National Health Laboratory Service, Johannesburg, South Africa; Centre for the AIDS Programme of Research in South Africa (CAPRISA), Durban, South Africa.
Teodora Wi, World Health Organization, Geneva, Switzerland.
Samuel P Dickson, Pentara Corporation, Salt Lake City, Utah, USA.
Jeffrey D Klausner, Department of Population and Public Health Sciences, Keck School of Medicine of the University of Southern California, Los Angeles, California, USA.
Notes
Acknowledgments. We acknowledge the scientific and programmatic guidance of Peter Wolff, MPA, Leah Vincent, PhD, and Carolyn Deal, PhD, of the Division of Microbiology and Infectious Diseases, National Institutes of Health. We thank the site research, laboratory, and clinic staff for their support in this study and Entasis Therapeutics for its provision of zoliflodacin powder. We also acknowledge the Hong Kong Department of Health for providing isolates for this project.
Disclaimer. The contents of this publication are solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Health.
Financial support. This work was supported by the Division of Microbiology and Infectious Diseases, National Institute of Allergy and Infectious Diseases, National Institutes of Health (support for the STAR Sexually Transmitted Infections Clinical Trials Group and protocol sites [contract HHSN272201300014I/HHSN27200011]), the National Institute of Allergy and Infectious Diseases (grants K01AI136725 to C. C. B. and F32AI145157 to T. D. M.), the National Institute of Mental Health (grant T32 MH080634 to P. C. A.), and the Fogarty International Center (grant K01TW012170 to P. C. A).
References
- 1. Newman L, Rowley J, Vander Hoorn S, et al. Global estimates of the prevalence and incidence of four curable sexually transmitted infections in 2012 based on systematic review and global reporting. PLoS One 2015; 10:e0143304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Barry PM, Klausner JD. The use of cephalosporins for gonorrhea: the impending problem of resistance. Expert Opin Pharmacother 2009; 10:555–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cole MJ, Spiteri G, Jacobsson S, et al. Is the tide turning again for cephalosporin resistance in Neisseria gonorrhoeae in Europe? results from the 2013 European surveillance. BMC Infect Dis 2015; 15:321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lee H, Unemo M, Kim HJ, Seo Y, Lee K, Chong Y. Emergence of decreased susceptibility and resistance to extended-spectrum cephalosporins in Neisseria gonorrhoeae in Korea. J Antimicrob Chemother 2015; 70:2536–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Shimuta K, Watanabe Y, Nakayama S, et al. Emergence and evolution of internationally disseminated cephalosporin-resistant Neisseria gonorrhoeae clones from 1995 to 2005 in Japan. BMC Infect Dis 2015; 15:378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Rowley J, Vander Hoorn S, Korenromp E, et al. Chlamydia, gonorrhoea, trichomoniasis and syphilis: global prevalence and incidence estimates, 2016. Bull World Health Organ 2019; 97:548–62P. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. World Health Organization . Global action plan to control the spread and impact of antimicrobial resistance in Neisseria gonorrhoeae. World Health Organization, 2012. [Google Scholar]
- 8. St Cyr S, Barbee L, Workowski KA, et al. Update to CDC's treatment guidelines for gonococcal infection, 2020. MMWR Morb Mortal Wkly Rep 2020; 69:1911–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Tuddenham S, Ghanem KG. Delaying the widespread emergence of cephalosporin-resistant gonorrhoea: what is the best target? Sex Transm Infect 2015; 91:232–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Allen VG, Mitterni L, Seah C, et al. Neisseria gonorrhoeae treatment failure and susceptibility to cefixime in Toronto, Canada. JAMA 2013; 309:163–70. [DOI] [PubMed] [Google Scholar]
- 11. Allen VG, Seah C, Martin I, Melano RG. Azithromycin resistance is coevolving with reduced susceptibility to cephalosporins in Neisseria gonorrhoeae in Ontario, Canada. Antimicrob Agents Chemother 2014; 58:2528–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gratrix J, Bergman J, Egan C, Drews SJ, Read R, Singh AE. Retrospective review of pharyngeal gonorrhea treatment failures in Alberta, Canada. Sex Transm Dis 2013; 40:877–9. [DOI] [PubMed] [Google Scholar]
- 13. Ison CA, Hussey J, Sankar KN, Evans J, Alexander S. Gonorrhoea treatment failures to cefixime and azithromycin in England, 2010. Euro Surveill 2011; 16:19833. [PubMed] [Google Scholar]
- 14. Martin I, Sawatzky P, Allen V, et al. Emergence and characterization of Neisseria gonorrhoeae isolates with decreased susceptibilities to ceftriaxone and cefixime in Canada: 2001–2010. Sex Transm Dis 2012; 39:316–23. [DOI] [PubMed] [Google Scholar]
- 15. Ohnishi M, Golparian D, Shimuta K, et al. Is Neisseria gonorrhoeae initiating a future era of untreatable gonorrhea? : detailed characterization of the first strain with high-level resistance to ceftriaxone. Antimicrob Agents Chemother 2011; 55:3538–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ohnishi M, Saika T, Hoshina S, et al. Ceftriaxone-resistant Neisseria gonorrhoeae, Japan. Emerg Infect Dis 2011; 17:148–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Unemo M, Golparian D, Nicholas R, Ohnishi M, Gallay A, Sednaoui P. High-level cefixime- and ceftriaxone-resistant Neisseria gonorrhoeae in France: novel penA mosaic allele in a successful international clone causes treatment failure. Antimicrob Agents Chemother 2012; 56:1273–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Unemo M, Golparian D, Stary A, Eigentler A. First Neisseria gonorrhoeae strain with resistance to cefixime causing gonorrhoea treatment failure in Austria, 2011. Euro Surveill 2011; 16:19998. [PubMed] [Google Scholar]
- 19. Unemo M, Golparian D, Syversen G, Vestrheim DF, Moi H. Two cases of verified clinical failures using internationally recommended first-line cefixime for gonorrhoea treatment, Norway, 2010. Euro Surveill 2010; 15:19721. [DOI] [PubMed] [Google Scholar]
- 20. Martin I, Sawatzky P, Liu G, et al. Decline in decreased cephalosporin susceptibility and increase in azithromycin resistance in Neisseria gonorrhoeae, Canada. Emerg Infect Dis 2016; 22:65–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Adamson PC, Lin EY, Ha SM, Klausner JD. Using a public database of Neisseria gonorrhoeae genomes to detect mutations associated with zoliflodacin resistance. J Antimicrob Chemother 2021; 76:2847–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Taylor SN, Marrazzo J, Batteiger BE, et al. Single-dose zoliflodacin (ETX0914) for treatment of urogenital gonorrhea. N Engl J Med 2018; 379:1835–45. [DOI] [PubMed] [Google Scholar]
- 23. Buono SA, Watson TD, Borenstein LA, Klausner JD, Pandori MW, Godwin HA. Stemming the tide of drug-resistant Neisseria gonorrhoeae: the need for an individualized approach to treatment. J Antimicrob Chemother 2015; 70:374–81. [DOI] [PubMed] [Google Scholar]
- 24. Siedner MJ, Pandori M, Castro L, et al. Real-time PCR assay for detection of quinolone-resistant Neisseria gonorrhoeae in urine samples. J Clin Microbiol 2007; 45:1250–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Parmar NR, Singh R, Martin I, et al. Genomic analysis reveals antibiotic-susceptible clones and emerging resistance in Neisseria gonorrhoeae in Saskatchewan, Canada. Antimicrob Agents Chemother 2020; 64:e02514–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ebeyan S, Windsor M, Bordin A, et al. Evaluation of the ResistancePlus GC (beta) assay: a commercial diagnostic test for the direct detection of ciprofloxacin susceptibility or resistance in Neisseria gonorrhoeae. J Antimicrob Chemother 2019; 74:1820–4. [DOI] [PubMed] [Google Scholar]
- 27. Allan-Blitz LT, Klausner JD. Codon 91 gyrase A testing is necessary and sufficient to predict ciprofloxacin susceptibility in Neisseria gonorrhoeae. J Infect Dis 2017; 215:491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Unemo M, Golparian D, Sanchez-Buso L, et al. The novel 2016 WHO Neisseria gonorrhoeae reference strains for global quality assurance of laboratory investigations: phenotypic, genetic and reference genome characterization. J Antimicrob Chemother 2016; 71:3096–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Grad YH, Kirkcaldy RD, Trees D, et al. Genomic epidemiology of Neisseria gonorrhoeae with reduced susceptibility to cefixime in the USA: a retrospective observational study. Lancet Infect Dis 2014; 14:220–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Mortimer TD, Grad YH. Applications of genomics to slow the spread of multidrug-resistant Neisseria gonorrhoeae. Ann N Y Acad Sci 2019; 1435:93–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Unemo M, Nicholas RA. Emergence of multidrug-resistant, extensively drug-resistant and untreatable gonorrhea. Future Microbiol 2012; 7:1401–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Unemo M, Shafer WM. Antimicrobial resistance in Neisseria gonorrhoeae in the 21st century: past, evolution, and future. Clin Microbiol Rev 2014; 27:587–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Clinical and Laboratory Standards Institute . Performance standards for antimicrobial susceptibility testing. 30th ed. CLSI supplement M100. Wayne, PA: Clinical and Laboratory Standards Institute, 2020. [Google Scholar]
- 34. Li H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv [Preprint: not peer reviewed]. 26 May 2013. Available from: https://arxiv.org/abs/1303.3997. [Google Scholar]
- 35. Li H, Handsaker B, Wysoker A, et al. The sequence alignment/map format and SAMtools. Bioinformatics 2009; 25:2078–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Walker BJ, Abeel T, Shea T, et al. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One 2014; 9:e112963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Okonechnikov K, Conesa A, Garcia-Alcalde F. Qualimap 2: advanced multi-sample quality control for high-throughput sequencing data. Bioinformatics 2016; 32:292–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Bankevich A, Nurk S, Antipov D, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 2012; 19:455–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Johnson SR, Grad Y, Abrams AJ, Pettus K, Trees DL. Use of whole-genome sequencing data to analyze 23S rRNA-mediated azithromycin resistance. Int J Antimicrob Agents 2017; 49:252–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wadsworth CB, Arnold BJ, Sater MRA, Grad YH. Azithromycin resistance through interspecific acquisition of an epistasis-dependent efflux pump component and transcriptional regulator in Neisseria gonorrhoeae. mBio 2018; 9:e01419–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Camacho C, Coulouris G, Avagyan V, et al. BLAST+: architecture and applications. BMC Bioinformatics 2009; 10:421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Croucher NJ, Page AJ, Connor TR, et al. Rapid phylogenetic analysis of large samples of recombinant bacterial whole genome sequences using Gubbins. Nucleic Acids Res 2015; 43:e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014; 30:1312–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Demczuk W, Martin I, Sawatzky P, et al. Equations to predict antimicrobial MICs in Neisseria gonorrhoeae using molecular antimicrobial resistance determinants. Antimicrob Agents Chemother 2020; 64:e02005–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Clinical and Laboratory Standards Institute . Performance standards for antimicrobial susceptibility testing. 31st ed. CLSI supplement M100. Wayne, PA: Clinical and Laboratory Standards Institute, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Klausner JD, Bristow CC, Soge OO, et al. Resistance-guided treatment of gonorrhea: a prospective clinical study. Clin Infect Dis 2021; 73:298–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Hicks AL, Kissler SM, Mortimer TD, et al. Targeted surveillance strategies for efficient detection of novel antibiotic resistance variants. Elife 2020; 9:e56367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Tuite AR, Gift TL, Chesson HW, Hsu K, Salomon JA, Grad YH. Impact of rapid susceptibility testing and antibiotic selection strategy on the emergence and spread of antibiotic resistance in gonorrhea. J Infect Dis 2017; 216:1141–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Klausner JD, Kerndt P. Cephalosporin resistance in Neisseria gonorrhoeae infections. JAMA 2013; 309:1989. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.