Abstract
Opportunistic yeast pathogens arose multiple times in the Saccharomycetes class, including the recently emerged, multidrug-resistant (MDR) Candida auris. We show that homologs of a known yeast adhesin family in Candida albicans, the Hyr/Iff-like (Hil) family, are enriched in distinct clades of Candida species as a result of multiple, independent expansions. Following gene duplication, the tandem repeat–rich region in these proteins diverged extremely rapidly and generated large variations in length and β-aggregation potential, both of which are known to directly affect adhesion. The conserved N-terminal effector domain was predicted to adopt a β-helical fold followed by an α-crystallin domain, making it structurally similar to a group of unrelated bacterial adhesins. Evolutionary analyses of the effector domain in C. auris revealed relaxed selective constraint combined with signatures of positive selection, suggesting functional diversification after gene duplication. Lastly, we found the Hil family genes to be enriched at chromosomal ends, which likely contributed to their expansion via ectopic recombination and break-induced replication. Combined, these results suggest that the expansion and diversification of adhesin families generate variation in adhesion and virulence within and between species and are a key step toward the emergence of fungal pathogens.
Keywords: Candida auris, adhesin, gene duplication, convergent evolution, opportunistic yeast pathogen, natural selection
Pathogenic fungi emerged repeatedly and independently. In this study, Smoak and Snyder et al. focus on the budding yeast subphylum, which includes some of the most important opportunistic fungal pathogens, including Candida albicans and Candida auris. They find multiple, repeated expansions of an adhesin family enriched in pathogenic yeast species and show how the sequences rapidly diverged under selection forces. Their results suggest that expansion and diversification of adhesin families are key steps toward the emergence of fungal pathogens.
Introduction
Candida auris, a newly emerged multidrug-resistant (MDR) yeast pathogen, is associated with a high mortality rate [up to 60% in a multicontinent meta-analysis (Lockhart et al. 2017)] and has caused multiple outbreaks across the world (CDC global C. auris cases count, 2021 February 15). As a result, it became the first fungal pathogen to be designated by CDC as an urgent threat (CDC 2019). The emergence of C. auris as a pathogen is part of a bigger evolutionary puzzle: Candida is a polyphyletic genus that contains most of the human yeast pathogens. Phylogenetically, species like Candida albicans, C. auris, and Candida glabrata belong to distinct clades with close relatives that either do not or only rarely infect humans (Fig. 1a). This strongly suggests that the ability to infect humans evolved multiple times in yeasts (Gabaldón et al. 2016). Because many of the newly emerged Candida pathogens are either resistant or can quickly evolve resistance to antifungal drugs (Lamoth et al. 2018; Srivastava et al. 2018), it is urgent to understand how yeast pathogenesis arose and what increases their survival in the host. We reason that any shared genetic changes among independently derived Candida pathogens will reveal key factors for host adaptation.
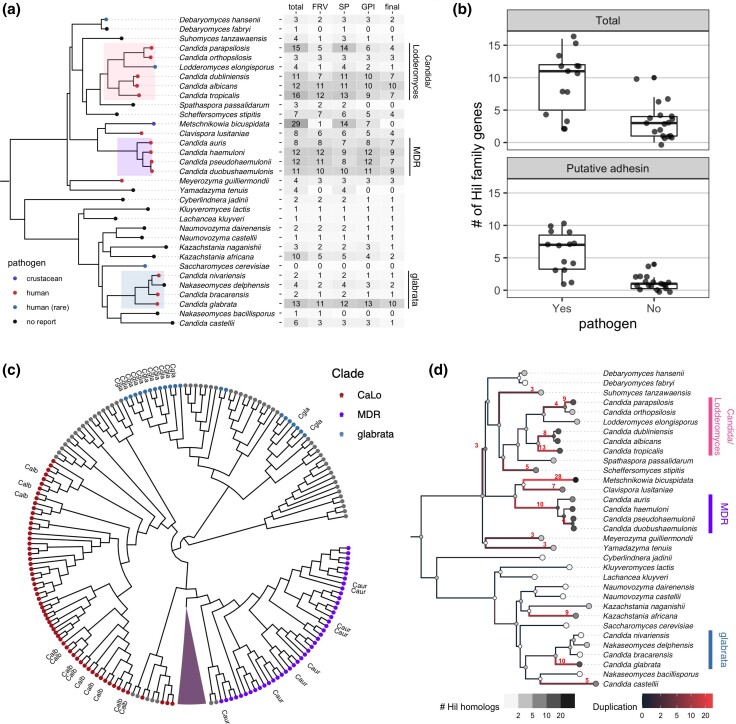
Fig. 1.
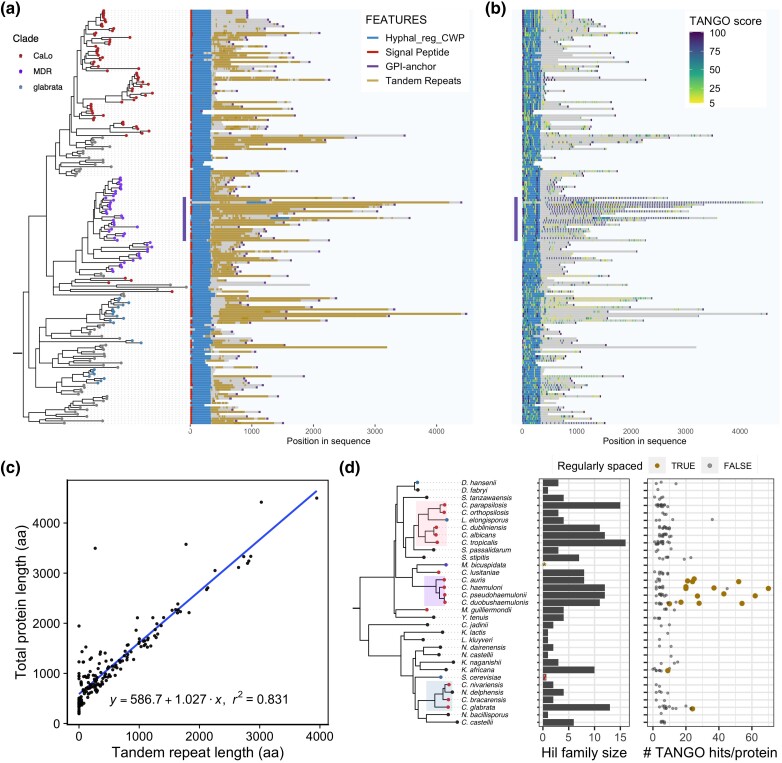
Phylogenetic distribution of the yeast Hil family and its parallel expansion in independently derived pathogenic Candida species. a) Species tree is based on the phylogeny for 332 yeast species from Shen et al. (2018), except for 3 species in the MDR clade other than C. auris, whose phylogenetic relationships are based on Muñoz et al. (2018). The tip colors show the pathogenic status of the species. The highlighted clades are enriched in known human pathogens. In the table, the first column shows the total number of Hil family homologs per species. The number of homologs that pass each of the 3 tests for determining their adhesin status is shown in the next 3 columns. FRV: FungalRV, SP: signal peptide, and GPI: GPI anchor (see Materials and methods for details). The number of homologs passing all 3 tests is shown in the “final” columns. b) Boxplots comparing the number of Hil homologs (upper) or the number of putative adhesins passing all 3 tests (lower) per species between known pathogens and low pathogenic potential species. Individual species numbers are shown as dots on top of the boxplot. Homologs from M. bicuspidata were excluded (see text). Both comparisons are significant at a 0.005 level by either a t-test with unequal variance or Mann–Whitney U test. c) Maximum likelihood tree based on the Hyphal_reg_CWP domain of the Hil family was shown as a cladogram. All 29 homologs in M. bicuspidata formed a single group, which is shown as a triangle. Homologs from the species in the 3 highlighted clades in (A) are colored accordingly. CaLo: Candida/Lodderomyces. Homologs from C. albicans, C. auris, and C. glabrata are labeled as Calb, Caur, and Cgla, respectively. d) Species tree showing the number of inferred duplication events on each branch. The shading of the tip and internal nodes represent the identified and inferred number of Hil homologs, respectively. The branch color shows the inferred number of duplication events, with 3 or more duplications also shown as a number next to the branch.
Gene duplication and the subsequent functional divergence are a major source for the evolution of novel phenotypes (Zhang 2003; Qian and Zhang 2014; Kuang et al. 2016; Eberlein et al. 2017). In a genome comparison of 7 pathogenic Candida species and 9 low pathogenic potential relatives, 3 of the top 6 pathogen-enriched gene families encode glycosylphosphatidylinositol (GPI)–anchored cell wall proteins, namely, Hyr/Iff-like (Hil), Als-like, and Pga30-like (Butler et al. 2009). The first two encode known fungal adhesins (Bailey et al. 1996; Hoyer 2001; Luo et al. 2010). These glycosylated cell wall proteins play key roles in fungal attachment to host endo- and epithelial cells, mediate biofilm formation and iron acquisition, and are well-established virulence factors (Hoyer et al. 2008; de Groot et al. 2013; Lipke 2018). It has been suggested that expansion of the cell wall protein repertoire, particularly adhesins, is a key step toward the evolution of yeast pathogens (Gabaldón et al. 2016). This is supported by a study showing that several adhesin families independently expanded in C. glabrata and close relatives (Gabaldón et al. 2013). Interestingly, studies of pathogenic Escherichia coli found that multiple strains independently acquired genes mediating intestinal adhesion, giving credence to the hypothesis from a different kingdom (Reid et al. 2000).
Despite the importance of adhesins in both the evolution and virulence of Candida pathogens, there is a lack of detailed phylogenetic analysis elucidating their evolutionary history (Hoyer 2001; Linder and Gustafsson 2008; Gabaldón et al. 2013). Even less is known about their sequence divergence and the role of natural selection in their evolution (Xie et al. 2011). In the newly emerged C. auris, individual adhesins have been characterized but there is little information about their evolutionary relationship with homologs in other Candida species and how their sequences diverged (Kean et al. 2018; Singh et al. 2019; Muñoz et al. 2021). In this study, we characterized the detailed evolutionary history of a yeast adhesin family and used C. auris as a focal group to determine how adhesin sequences diverged under various natural selection forces. To choose a candidate adhesin family in C. auris, we compared it with the well-studied C. albicans, which belongs to the same CUG-Ser1 clade. Of the known adhesins in C. albicans, C. auris lacks the Hwp family and has only 3 Als or Als-like proteins compared with 8 Als proteins in C. albicans (Muñoz et al. 2018). By contrast, C. auris has 8 genes with a Hyphal_reg_CWP (PF11765) domain found in the Hyr/Iff family in C. albicans (Muñoz et al. 2021). This family was one of the most highly enriched in pathogenic Candida species relative to nonpathogenic ones (Butler et al. 2009). Transcriptomic studies identified two C. auris HIL genes as being upregulated during biofilm formation and under antifungal treatment (Kean et al. 2018). Interestingly, isolates from the less virulent C. auris Clade II lack 5 of the 8 HIL genes (Muñoz et al. 2021). It is currently not known whether the C. auris HIL genes encode adhesins, how they relate to the C. albicans Hyr/Iff family genes, and how their sequences diverged after duplication.
We show that the Hil family independently expanded multiple times, including in C. auris and C. albicans. Using C. auris as a focal species, we show in detail how sequence features and predicted structures of the effector domain offer support for the hypothesis that its Hil family members encode adhesins, while rates of nonsynonymous to synonymous substitutions reveal varying strengths of selective constraint and positive selection acting on the effector domain during the expansion of the family. The observed pattern of rapid divergence in the repeat-rich central domain was found to be general across the entire family and led to large variations in length and β-aggregation potential both between and within species, likely contributing to phenotypic diversity in adhesion and virulence.
Materials and methods
Software
Versions and sources of the main software used in this study are listed in Table 1.
Table 1.
Software and algorithm list.
Identify Hil family homologs in yeasts and beyond
To identify the Hil family proteins in yeasts and beyond, we used the Hyphal_reg_CWP domain sequence from 3 distantly related Hil homologs as queries, namely, C. albicans Hyr1 (XP_722183.2), C. auris Hil1 (XP_028889033), and C. glabrata CAGL0E06600g (XP_722183.2). We performed BLASTP searches in the RefSeq protein database with an E-value cut-off of 1 × 10−5, with a minimum query coverage of 50%, and with the low-complexity filter on. All hits were from Ascomycota (yeasts), and all but one were from the Saccharomycetes class (budding yeast). A single hit was found in the fission yeast Schizosaccharomyces cryophilus. Using that hit as the query, we searched all fission yeasts in the nr protein database, with a relaxed E-value cut-off of 10−3, and identified no additional hits. We thus excluded that one hit from downstream analyses. To supplement the RefSeq database, which lacks some yeast species such as those in the Nakaseomyces genus, we searched the Genome Resources for Yeast Chromosomes (GRYC, http://gryc.inra.fr/). Using the same criteria, we recovered 16 additional sequences. To allow for gene tree and species tree reconciliation, we excluded 3 species that are not part of the 322 species yeast phylogeny (Shen et al. 2018) and not a member of the MDR clade (Muñoz et al. 2018). Further details, including additional quality control steps taken to ensure that the homolog sequences are accurate and complete, can be found in Supplementary Text 1. In total, we curated a list of 215 Hil family homologs from 32 species.
Gene family enrichment analysis
To determine if the Hil family is enriched in the pathogenic yeasts, we performed 2 analyses. In the first analysis, we divided the species into pathogens vs low pathogenic potential groups and performed a t-test with unequal variance (also known as Welch's test) as well as a nonparametric Mann–Whitney U test to compare the Hil family size in the 2 groups. For both tests, we used either the total size of the family or the number of putative adhesins as the random variable, and the results were consistent. We excluded homologs from Metschnikowia bicuspidata because 10 of its 29 Hil family proteins were annotated as incomplete in the RefSeq protein database and also because as a parasite of freshwater crustaceans, it does not fit into either the human pathogen or the low pathogenic potential group. Saccharomyces cerevisiae was included in the comparison as an example of species with zero members of the Hil family. We chose S. cerevisiae because we could be confident about its lack of a Hil family homolog thanks to its well-assembled and well-annotated genome.
In the second test, we used phylogenetic logistic regression (Ives and Garland 2010) to account for the phylogenetic relatedness between species. We used the “phyloglm” function in the “phylolm” package in R, with {method = “logistic_IG10,” btol = 50, boot = 100}. The species tree, including the topology and branch lengths, were based on the 322 species phylogeny from Shen et al. (2018), supplemented by the phylogenetic relationship for the MDR clade based on Muñoz et al. (2018). The P-values based on phylogenetically specified residual correlations were reported.
Phylogenetic analysis of the Hil family and inference of gene duplications and losses
To infer the evolutionary history of the Hil family, we reconstructed a maximum likelihood tree based on the alignment of the Hyphal_reg_CWP domain. First, we used hmmscan (HmmerWeb version 2.41.2) to identify the location of the Hyphal_reg_CWP domain in each Hil homolog. We used the “envelope boundaries” to define the domain in each sequence and then aligned their amino acid sequences using Clustal Omega with the parameter {–iter = 5}. We then trimmed the alignment using ClipKit with its default smart-gap trimming mode (Steenwyk et al. 2020). RAxML-NG v1.1.0 was run in the MPI mode with the following parameters on the alignment: “raxml-ng-mpi –all –msa INPUT –model LG + G –seed 123 –bs-trees autoMRE.” The resulting tree was corrected using GeneRax, which seeks to maximize the joint likelihood of observing the alignment given the gene family tree (GFT) and observing the GFT given the species phylogeny, using the parameter {–rec-model UndatedDL}. The species tree used is the same as the one used for the phylogenetic logistic regression above. In addition to correcting the GFT, GeneRax also reconciled it with the species tree and inferred duplication and loss event counts on each branch. Tree annotation and visualization were done in R using the treeio and ggtree packages (Yu 2020; Wang et al. 2020).
To infer the phylogenetic tree for the Hil family homologs in various C. auris strains and infer gains and losses within species, we identified orthologs of the HIL genes in representative strains from the 4 major clades of C. auris (B8441, B11220, B11221, B11243) (Muñoz et al. 2018). Orthologs from 2 MDR species, Candida haemulonii and Candida pseudohaemulonis, and from Debaryomyces hansenii were included to help root the tree. The gene tree was constructed as described above. To root the tree, we first inferred a gene tree without the outgroup (D. hansenii) sequences in the alignment. Then, the full alignment with the outgroup sequences along with the gene tree from the first step were provided to RAxML to run the Evolutionary Placement Algorithm (EPA) algorithm (Berger et al. 2011), which identified a unique root location. To reconcile the gene tree with the species tree, we performed maximum likelihood–based gene tree correction using GeneRax (v2.0.1) with the following parameters: {–rec-model UndatedDL}. The species tree was based on Muñoz et al. (2018).
Prediction for fungal adhesins and adhesin-related sequence features
(1) The potential of Hil homologs encoding fungal adhesins was assessed using FungalRV, a support vector machine–based fungal adhesin predictor (Chaudhuri et al. 2011). Proteins passing the recommended cut-off of 0.511 were considered positive. (2) Signal peptide was predicted using the SignalP 6.0 server, with the “organism group” set to Eukarya. The server reported the proteins that had predicted signal peptides. No further filtering was done. (3) GPI anchor was predicted using PredGPI using the general model. Proteins with a false-positive rate of 0.01 or less were considered as containing a GPI anchor. (4) Tandem repeats were identified using XSTREAM with the following parameters: {-i.7 -I.7 -g3 -e2 -L15 -z -Asub.txt -B -O}, where the “sub.txt” was provided by the software package. (5) β-aggregation–prone sequences were predicted using TANGO v2.3.1 with the following parameters: {ct=“N” nt=“N” ph=“7.5” te=“298” io=“0.1” tf=“0” stab=“−10” conc=“1” seq=“SEQ”}. (6) Serine and threonine content in proteins were quantified using “freak” from the EMBOSS suite, with a sliding window of 100 or 50 aa and a step size of 10 aa. To compare with proteome-wide distribution of Ser/Thr frequency, the protein sequences for C. albicans (SC5314), C. glabrata (CBS138), and C. auris (B11221) were downloaded from NCBI Assembly database (IDs in Supplementary Table 7) and the frequency of serine and threonine residues was counted for each protein. (7) O-linked and N-linked glycosylations were predicted using NetOGlyc (v4.0) and NetNGlyc (v1.0) servers.
Structural prediction and visualization for the Hyphal_reg_CWP domain
To perform structural predictions using AlphaFold2, we used the Google Colab notebook (https://colab.research.google.com/github/deepmind/alphafold/blob/main/notebooks/AlphaFold.ipynb) authored by the DeepMind team. This is a reduced version of the full AlphaFold version 2 in that it searches a selected portion of the environmental BFD database and does not use templates. The Amber relaxation step is included, and no other parameters other than the input sequences are required. DALI was used to search for similar structures in the PDB50 database. Model visualization and annotation were done in PyMol v2.5.2. Secondary structure prediction for C. auris Hil1's central domain was performed using PSIPred.
Dotplot
To determine the self-similarity and similarity between the 8 C. auris Hil proteins, we made dotplots using JDotter (Brodie et al. 2004). The window size and contrast settings were labeled in the legends for the respective plots. The self-alignment for C. auris Hil1 tandem repeats was visualized using Jalview v2.11.
Identification of intraspecific tandem repeat copy number variations among C. auris strains
To identify polymorphisms in Hil1–Hil4 in diverse C. auris strains, we downloaded the genome sequences for the following strains from NCBI: Clade I, B11205 and B13916; Clade II, B11220, B12043, and B13463; Clade III, B11221, B12037, B12631, and B17721; and Clade IV, B11245 and B12342 (Supplementary Table 4). The amino acid sequences for Hil1–Hil4 from the strain B8441 were used as the query to search the nucleotide sequences of the above assemblies using TBLASTN, with the following parameters {-db_gencode 12 -evalue 1e-150 -max_hsps 2}. Orthologs in each strain were curated based on the BLAST hits to either the Hyphal_reg_CWP domain alone or the entire protein query. All Clade II strains had no hits for Hil1–Hil4. Several strains in Clades I, III, and IV were found to lack one or more Hil proteins (Supplementary Table 5). But upon further inspection, it was found that they had significant TBLASTN hits for part of the query, e.g. the central domain, and the hits were located at the end of a chromosome, suggesting the possibility of incomplete or misassembled sequences. Further experiments will be needed to determine if those HIL genes are present in those strains.
Estimation of dN/dS ratios and model comparisons
We used “codeml” in PAML (v4.9e) to perform evolutionary inferences on the Hyphal_reg_CWP domain in C. auris. We first used Clustal Omega to align the amino acid sequences for the Hyphal_reg_CWP domain from Hil1–Hil8 from C. auris similar to how we generated the multiple sequence alignment for all Hil proteins. A closely related outgroup (XP_018709340.1 from M. bicuspidata) was included to root the tree. We then generated a coding sequence alignment from the protein alignment using PAL2NAL (Suyama et al. 2006). We used GARD (Kosakovsky Pond et al. 2006) to analyze the coding sequence alignment to detect gene conversion events. The web service of GARD on datamonkey.org was run with the following parameters: {data type: nucleotide, run mode: normal, genetic code: yeast alternative nuclear, site-to-site rate variation: general discrete, rate classes: 3}. Based on the results, we identified 2 putatively nonrecombining partitions, P1 = 1–414 and P2 = 697–981 (the numbers refer to the alignment columns). We then separately analyzed the 2 partitions in PAML. To test hypotheses about positive selection on a subset of the sites on all branches, we compared models M2a vs M1a, M8 vs M7, and M8a vs M8. The first 4 models were specified by {seqtype = 1, CodonFreq = 1, model = 0, NSsites = 0,1,2,7,8, icode = 8, fix_kappa = 0, kappa = 2, fix_omega = 0, omega = 0.4, cleandata = 1}. The model M8a is additionally specified by { seqtype = 1, CodonFreq = 1, model = 0, NSsites = 8, fix_omega = 1 and omega = 1, cleandata = 1}. To test hypotheses for variable dN/dS on different branches (no variation across sites), we used {model = 0 or 1 or 2, NSsites = 0}, with the rest being the same as the site tests. Model = 0 specified the single-ratio model, model = 1 the free-ratio model, and model = 2 the user-defined model. For the user-defined model, we first used estimates from the free-ratio model to designate a set of branches with dN/dS > 10 as the foreground and then tested if their dN/dS was significantly different from the rest of the tree by comparing a 2-ratio model with the single-ratio model. Since the results were significant, we further tested if the foreground dN/dS was significantly greater than 1, by comparing the 2-ratio model to a constrained version of the model where omega was fixed at 1. For branch-site test, we used {model = 2, NSsites = 2, fix_omega = 0, omega = .4} as the alternative model and {model = 2, NSsites = 2, fix_omega = 1, omega = 1} as the null to test for positive selection on a subset of the sites on the foreground branches. Sites under positive selection were identified using the Bayes Empirical Bayes (BEB) procedure, with a posterior probability threshold of 0.99.
Chromosomal locations of Hil family genes
To compare the chromosomal locations of the Hil family genes to the background distribution, we selected 8 species whose genomes were assembled to a chromosomal level and are not within a closely related group, including C. albicans, D. hansenii, Candida orthopsilosis, Kazachstania africana, Kluyveromyces lactis, Naumovozyma dairenensis, C. auris, and C. glabrata (Supplementary Table 7). We did not include some species, e.g. Candida dubliniensis, to minimize statistical dependence due to shared ancestry. The RefSeq assembly for C. auris was included even though it was at a scaffold level because a recent study showed that 7 of its longest scaffolds were chromosome-length, allowing the mapping of the scaffolds to chromosomes (Muñoz et al. 2021, supplementary table 1). To determine the chromosomal locations of the Hil homologs in these 8 species, we used Rentrez v1.2.3 (Winter 2017) in R to retrieve their chromosome ID and coordinates. To calculate the background gene density on each chromosome, we downloaded the feature tables for the 8 assemblies from the NCBI assembly database and calculated the location of each gene as its start coordinate divided by the chromosome length. To compare the chromosomal location of the Hil family genes to the genome background, we divided each chromosome into 5 equal-sized bins based on the physical distance to the nearest chromosomal end. We calculated the proportion of genes residing in each bin for the Hil family or for all protein-coding genes. To determine if the 2 distributions differ significantly from 1 another, we performed a goodness-of-fit test using either a log–likelihood ratio (LLR) test or a chi-squared test, as implemented in the XNomial package in R (Engels 2015). The LLR test P-value was reported.
Results
Phylogenetic distribution of the Hil family and its potential to encode adhesin
The Hyr/Iff family was first identified and characterized in C. albicans (Bailey et al. 1996; Richard and Plaine 2007). The family is defined by its N-terminal hyphally regulated cell wall protein domain (Hyphal_reg_CWP, PF11765), followed by a highly variable central domain rich in tandem repeats (Boisramé et al. 2011). Because the effector domain is more conserved than the repeat region and plays a prominent role in mediating adhesion in known yeast adhesins (Willaert 2018), here, we define the Hil family as the group of evolutionarily related proteins sharing the Hyphal_reg_CWP domain, different from a previous definition based on sequence similarity in either the Hyphal_reg_CWP domain or the repeat region (Butler et al. 2009).
To determine the phylogenetic distribution of the Hil family and its association with the pathogenic potential of species, we performed BLASTP searches using the Hyphal_reg_CWP domain from 3 distantly related Hil homologs as queries (from C. auris, C. albicans, and C. glabrata). We scrutinized the database hits and searched additional assemblies to ensure that their sequences are complete and accurate given the available genome assemblies (Supplementary Text 1). Using the criteria of E-value < 10−5 and query coverage > 50%, we identified a total of 215 proteins containing the Hyphal_reg_CWP domain from 32 species (Fig. 1a, Supplementary Table 1). No credible hits were identified outside the budding yeast subphylum even after a lower E-value cut-off of 10−3 was tested, suggesting that this family is specific to this group (see Materials and Methods). Species with 8 or more Hil family genes fell largely within the multidrug resistant (MDR) and the Candida/Lodderomyces (CaLo) clades, which include C. auris and C. albicans, respectively. Only 3 such species were found outside of the 2 clades: C. glabrata, M. bicuspidata, and K. africana. C. glabrata is a major opportunistic pathogen that is more closely related to S. cerevisiae than to most other Candida species (Dujon et al. 2004; Butler et al. 2009; Gabaldón et al. 2013). M. bicuspidata is part of the CUG-Ser1 clade. While not a pathogen in humans, it is a parasite of freshwater animals (Hall et al. 2010; Jiang et al. 2022). K. africana is not closely related to any known yeast pathogen, and its ecology is poorly understood (Gordon et al. 2011).
We then asked how many of the Hil family genes in each species are likely to encode yeast adhesins. To get an initial estimate, we combined a machine learning tool for predicting fungal adhesins (Chaudhuri et al. 2011) with predictions for the N-terminal signal peptide and C-terminal GPI anchor sequence, 2 features shared by the majority of known fungal adhesins (Lipke 2018). Half of all Hil homologs passed all 3 tests (Fig. 1a). Notably, M. bicuspidata has the largest Hil family among all species, but none of its 29 Hil genes passed all tests. We found most of the identified hits in this species were short relative to the rest of the family (Supplementary Fig. 1), and 10 of the 29 hits were annotated as being incomplete in the RefSeq database. Further analyses with a better assembled genome and functional studies are needed to determine if the Hil family in this species has unique properties and functions.
Independent expansion of the Hil family in multiple pathogenic Candida lineages
Pathogenic yeast species have on average a larger Hil family, and also, more of its members were predicted to encode adhesins than in low pathogenic potential species (Fig. 1b, t-test with unequal variance and Mann–Whitney U test both yielded P < 0.005, one-sided test). This naive comparison does not account for phylogenetic relatedness between species and could result in a false-positive association (Levy et al. 2018; Bradley et al. 2018). To address this, we performed phylogenetic logistic regression, which uses the known phylogeny to specify the residual correlation structure among species with shared ancestry (Ives and Garland 2010). We tested for associations between the pathogen status with either the total number of Hil homologs or the number of putative adhesins in each species. Both tests were significant (P = 0.005 and 0.007, respectively). Together, these results strongly support an enrichment of the Hil family and the putative adhesins therein among the pathogenic yeast species.
Some adhesin families have undergone independent expansions even among closely related species (Gabaldón et al. 2013). This would result in overestimation of the phylogenetic signal in the above analysis. To further characterize the evolutionary history of the Hil family, including among closely related Candida lineages, we reconstructed a species tree–aware maximum likelihood phylogeny for the Hil family based on the Hyphal_reg_CWP domain alignment (Fig. 1c, Supplementary Fig. 2). We found that homologs from the MDR clade and the CaLo clade separated into 2 groups, suggesting that the duplications of the Hil family genes in the 2 clades occurred independently. To better illustrate the history of gene duplications in the Hil family, we reconciled the gene tree with the species tree and mapped the number of duplications onto the species phylogeny (see Materials and Methods). The result showed that the Hil family has independently expanded multiple times, not only between clades but also among closely related species within a clade, such as in C. albicans and C. tropicalis (Fig. 1d).
Sequence features of the C. auris Hil family support their adhesin status
Experiments have demonstrated that Hil family members function as adhesin in C. albicans and more recently for one member in C. glabrata (Bailey et al. 1996; Boisramé et al. 2011; Rosiana et al. 2021; Reithofer et al. 2021). To further evaluate the adhesin function of Hil family proteins, we focused on C. auris, in which Hil family members were implicated in biofilm formation and response to antifungal treatments but still remain poorly characterized (Kean et al. 2018). We named the 8 C. auris Hil family proteins Hil1–Hil8 ordered by their length (Supplementary Table 2). This differs from the literature, which referred to them by their most closely related Hyr/Iff genes in C. albicans (Kean et al. 2018; Jenull et al. 2021; Muñoz et al. 2021). The renaming avoids the incorrect implication of one-to-one orthology between the 2 species (Fig. 1c).
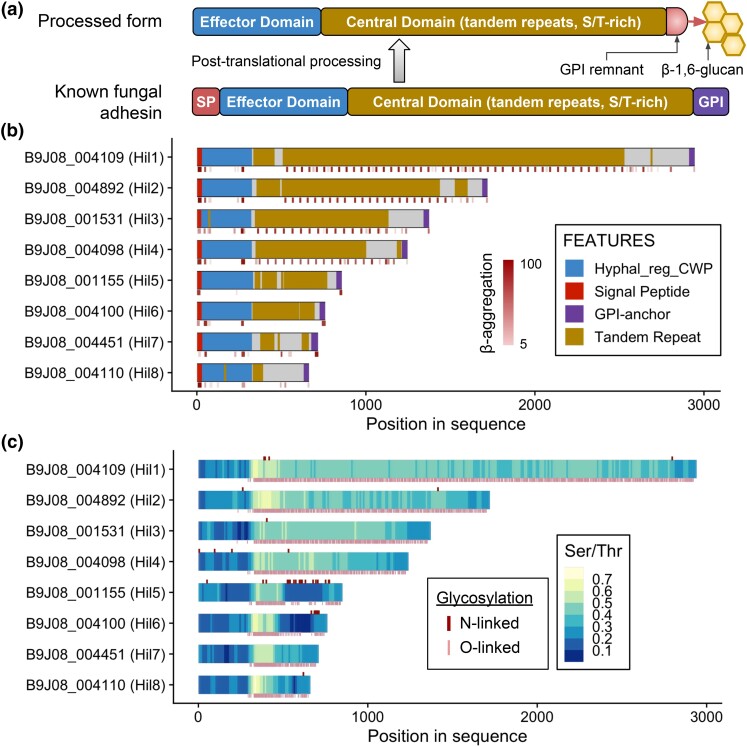
To further assess the adhesin potential for the C. auris Hil family, we compared their domain architecture and sequence features to those typical of known yeast adhesins, including a signal peptide, an effector domain, a Ser/Thr-rich and highly glycosylated central domain with tandem repeats and β-aggregation–prone sequences, and a GPI anchor signal (Fig. 2a) (de Groot et al. 2013; Lipke 2018). All 8 C. auris Hil proteins followed this domain architecture (Fig. 2b). Hil1–4 were additionally characterized by an array of regularly spaced β-aggregation–prone sequences (red ticks below the protein, Fig. 2b). All 8 proteins also had elevated Ser/Thr frequencies in their central domain and were predicted to be heavily O-glycosylated (Fig. 2c). Predicted N-glycosylation was rare except in Hil5 and Hil6 (Fig. 2c). The overall Ser/Thr frequencies in the Hil family proteins were significantly elevated compared with the rest of the proteome (Supplementary Fig. 3). All 8 members were predicted to be fungal adhesins by FungalRV, a support vector machine–based classifier that showed high sensitivity and specificity in 8 pathogenic fungi based on sequence features (Chaudhuri et al. 2011).
Fig. 2.
Domain architecture and adhesin-associated features of the C. auris Hil family. a) Diagram depicting the domain organization of a typical yeast adhesin before and after the posttranslational processing, adapted from de Groot et al. (2013). b) Domain features of the 8 Hil proteins in C. auris (strain B8441). Gene IDs and names are labeled on the left. The short stripes below each diagram are the TANGO-predicted β-aggregation–prone sequences, with the intensity of the color corresponding to the score of the prediction. c) Serine and threonine (Ser/Thr) frequencies in each protein are plotted in 50 aa sliding windows with step size of 10 aa. N-linked and O-linked glycosylation sites were predicted by NetNGlyc 1.0 and NetOGlyc 4.0, respectively, and are shown as short ticks above and below each protein schematic.
Hyphal_reg_CWP domain in the Hil family is predicted to adopt a β-helical fold similar to unrelated bacterial adhesin binding domains
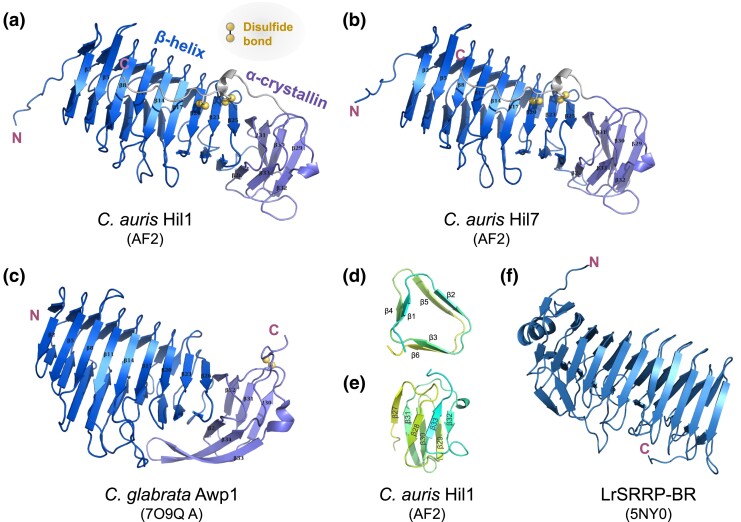
Crystal structures of the effector domain in several yeast adhesin families, including Als, Epa, and Flo, revealed carbohydrate- or peptide-binding activities supporting the proteins’ adhesin functions (Willaert 2018). The structure of the Hyphal_reg_CWP domain in the Hil family in this study has not yet been experimentally determined. However, crystal structures for the effector domains of 2 adhesin-like wall proteins (Awp1 and Awp3b) in C. glabrata, which are distantly related to those in the Hil family, were recently reported, and the predicted structure of 1 of C. glabrata's Hil family members (Awp2) was found to be highly similar to the 2 solved structures (Reithofer et al. 2021). We used AlphaFold2 (Jumper et al. 2021) to predict the structures of the effector domain for 2 C. auris Hil proteins, Hil1 and Hil7 (Fig. 3, a and b). Both resemble the C. glabrata Awp1 effector domain (Fig. 3c), consisting of a right-handed β-helix at the N-terminus followed by an α-crystallin fold. There are 3 β-strands in each of the 9 rungs in the β-helix, stacked into 3 parallel β-sheets (Fig. 3d). The α-crystallin domain consists of 7 β-strands forming 2 antiparallel β-sheets, adopting an immunoglobulin-like β-sandwich fold (Fig. 3e) (Koteiche and Mchaourab 1999; Stamler et al. 2005).
Fig. 3.
Predicted structures of the Hyphal_reg_CWP domain in 2 C. auris Hil proteins are similar to yeast and bacterial adhesins. a) and (b) AlphaFold2 (AF2)–predicted structures of the Hyphal_reg_CWP domains from C. auris Hil1 and Hil7, which consist of a β-helix followed by a α-crystallin domain, with the C-terminal loop linked to the β-helix via 2 disulfide bonds. c) Crystal structure of the C. glabrata Awp1 effector domain, which is highly similar to C. auris Hil1 and Hil7, but with the disulfide bond in a different location. d) Cross-section of the first 2 rungs of the β-helix in (a), showing the 3 β-strands per rung. e) α-Crystallin domain in (a), showing the 7 β-strands forming 2 antiparallel β-sheets. f) Crystal structure of the serine-rich repeat protein binding region (SRRP-BR) from the gram-positive bacterium L. reuteri, which adopts a β-helix fold.
The β-strand–rich structure is typical of effector domains in known yeast adhesins, but the β-helix fold at the N-terminus is uncommon (Willaert 2018). Proteins with a β-helix domain often have carbohydrate-binding capabilities and act as enzymes, e.g. hydrolase and pectate lyase (SCOP ID: 3001746). To gain further insight into Hyphal_reg_CWP domain's function, we searched the PDB50 database for structures similar to what was predicted for C. auris Hil1 using DALI (Holm 2022). We identified a number of bacterial adhesins with a highly similar β-helix fold but no α-crystallin domain (Supplementary Table 3), e.g. Hmw1 from Haemophilus influenzae (PDB: 2ODL), tāpirins from Caldicellulosiruptor hydrothermalis (PDB: 6N2C), TibA from enterotoxigenic E. coli (PDB: 4Q1Q), and SRRP from Limosilactobacillus reuteri (PDB: 5NY0). For comparison, the binding region of the serine-rich repeat protein 100-23 (SRRP100-23) from L. reuteri was shown in Fig. 3f (Sequeira et al. 2018). Together, these results strongly suggest that the Hyphal_reg_CWP domain in the C. auris Hil family genes mediates adhesion. Additionally, the low sequence identity (12–15%) between the yeast Hyphal_reg_CWP domain and the bacterial adhesins’ binding regions further suggests the 2 groups have convergently evolved a similar structure to achieve adhesion functions.
Rapid divergence of the repeat-rich central domain in Hil family proteins in C. auris
While the overall domain architecture is well conserved, the 8 Hil family proteins in C. auris differ significantly in length and sequence of their central domains (Fig. 2b). While not involved in ligand binding, central domains in yeast adhesins are known to play a critical role in mediating adhesion: the length and stiffness of the central domain are essential for elevating and exposing the effector domain (Frieman et al. 2002; Boisramé et al. 2011), and the tandem repeats and β-aggregation sequences within them directly contribute to adhesion by mediating homophilic binding and amyloid formation (Rauceo et al. 2006; Otoo et al. 2008; Frank et al. 2010; Wilkins et al. 2018). Thus, divergence in the central domain of the Hil family has the potential to lead to phenotypic diversity, as shown in S. cerevisiae (Verstrepen et al. 2004, 2005).
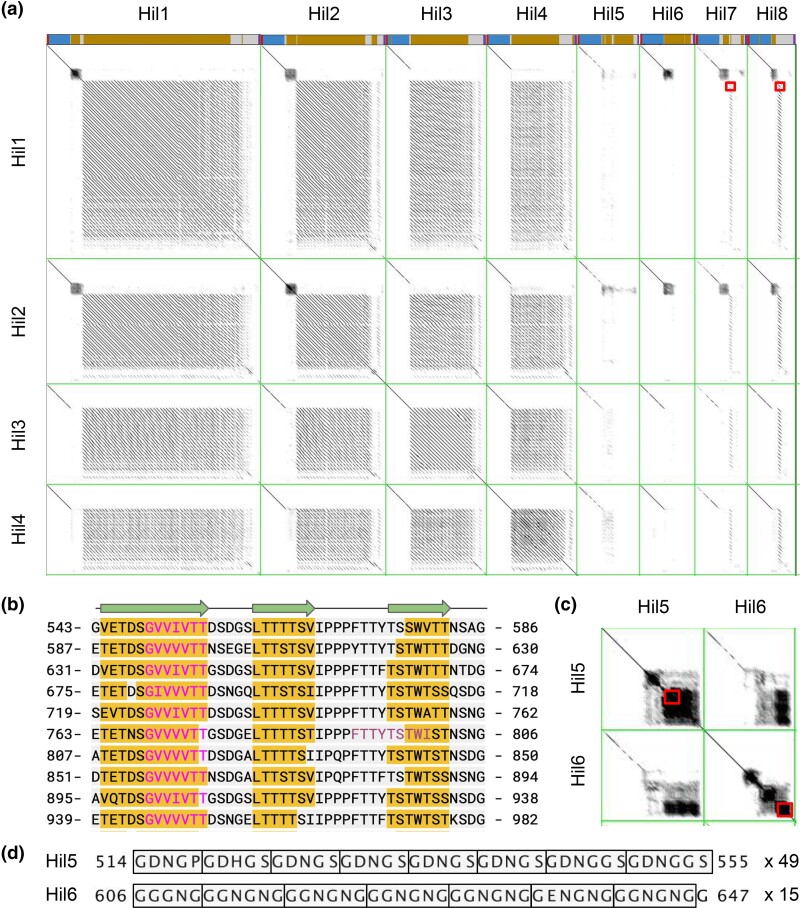
To determine how the central domain sequences evolved in the C. auris Hil family, we used dot plots both to reveal the tandem repeat structure within each protein and to examine the similarity among the paralogs. A “dot” on the x–y plot indicates that the corresponding segments (window size = 50 a.a.) from the 2 proteins on the x- and y-axes share similarity, with the gray scale being proportional to the degree of similarity (Brodie et al. 2004). We found that C. auris Hil1, Hil2, Hil3, and Hil4 share a ∼44 aa repeat unit, whose copy number varies between 15 and 46, driving differences in their protein lengths (Fig. 4a). These repeats have conserved periodicity as well as sequence (Fig. 4b, Supplementary Fig. 4). There are 2 interesting features of this 44 aa repeat unit: (1) it contains a heptapeptide “GVVIVTT” that is predicted to be strongly β-aggregation-prone, which explains the large number of regularly spaced β-aggregation motifs in Hil1–Hil4 (Fig. 2b); (2) it is predicted to form 3 β-strands in the same orientation (Fig. 4b), raising an interesting question of whether the tandem repeats may adopt a β-structure similar to that of the effector domain. Hil7 and Hil8 encode the same repeat unit but have only one copy (Fig. 4a, red boxes). By contrast, Hil5 and Hil6 encode very different low-complexity repeats with a unit length of ∼5 aa. Their copy numbers range between 15 and 49 (Fig. 4, c and d), and their sequences have relatively low Ser/Thr frequencies (Fig. 2c). Another consequence of encoding only 1 or 0 copies of the 44 aa repeat unit found in Hil1–Hil4 is that Hil5–Hil8 are predicted to have 2–4 β-aggregation–prone sequences in contrast to 21–50 in Hil1–Hil4. For comparison, characterized yeast adhesins contain 1–3 such sequences at a cut-off of >30% β-aggregation potential predicted by TANGO (Fernandez-Escamilla et al. 2004; Ramsook et al. 2010; Lipke 2018). The variable lengths, Ser/Thr frequencies, and distribution of β-aggregation sequences, all resulting from the evolution of the tandem repeats, suggest the intriguing possibility that the 8 different Hil proteins in C. auris are nonredundant, playing distinct roles in cell adhesion and other cell wall–related phenotypes.
Fig. 4.
Dotplot shows the tandem repeat structure within and similarity between C. auris Hil proteins. a) Hil1–Hil4 are compared to all 8 Hil proteins in C. auris including themselves in dotplots with a sliding window of 50 aa and Grey Map set to 60–245 (min–max). A schematic for each protein is shown above each column (colors same as in Fig. 2). The regions highlighted by the boxes in row 1 reveal the presence of a single copy of the 44 aa repeat unit in Hil7 and Hil8. b) Wrapped sequence of aa 543–982 from Hil1 showing the conserved period and sequence of the 44 aa tandem repeat. Variants of the GVVIVTT motif have strong (probability > 90%) predicted β-aggregation potential while the FTTYTSTWI motif is predicted to have moderate (30–90%) β-aggregation potential. The highlighted regions are predicted by PSIPred to form β-strands, with cartoons shown above. c) Dotplots between Hil5 and Hil6 with the same settings as in (a), showing the low-complexity repeats unique to these two proteins. Regions within the 2 boxes are shown in (d), with residue numbers shown on both ends. The rectangles delineate individual repeats, with the number of copies for each repeat shown to the right.
Because tandem repeats are prone to recombination-mediated expansions and contractions, we asked if there are variable numbers of tandem repeats (VNTR) among strains in C. auris, which could generate diversity in cell adhesive properties as shown in S. cerevisiae (Verstrepen et al. 2005). To answer this question, we identified homologs of Hil1–Hil4 in 9 C. auris strains from 3 geographically stratified clades (Muñoz et al. 2018, 2021). The genomes of these strains were de novo–assembled using long-read technologies (Supplementary Table 4), which allowed us to confidently assess copy number variations within tandem repeats. We identified a total of 8 indel polymorphisms in Hil1–Hil4 (Supplementary Table 5, example alignments in Supplementary Fig. 5). Except for 1 16 aa deletion that is in a single Clade III strain, all 7 other indels span 1 or multiples of the repeat unit and affect all strains within a clade. This is consistent with them being driven by recombination between repeats. The agreement within clades additionally shows the indels are not due to sequencing/assembly artifacts, which are not expected to follow the clade labels. As previously reported, Clade II strains lack 5 of the 8 Hil family proteins, including Hil1-4 (Muñoz et al. 2021). Our phylogenetic analysis further showed that this was due to gene losses within Clade II (Supplementary Fig. 6). The potential relationship between the Hil family size and the virulence profiles of Clade II strains is discussed later.
Natural selection on the effector domain during the Hil family expansion in C. auris
Gene duplication provides raw materials for natural selection and is often followed by a period of relaxed functional constraints on one or both copies, allowing for sub- or neo-functionalization (Zhang 2003; Innan and Kondrashov 2010). Positive selection can be involved in this process, which can lead to a ratio of nonsynonymous to synonymous substitution rates dN/dS > 1 (Yang 1998). Here, we ask if the Hyphal_reg_CWP domain in C. auris Hil1–Hil8 experienced relaxed selective constraints and/or positive selection following gene duplications, the latter of which would suggest functional diversification. We chose to focus on the Hyphal_reg_CWP domain because of its functional importance and because the high-quality alignment in this domain allowed us to make confident evolutionary inferences (Supplementary Fig. 7).
Because gene conversion between paralogs can cause distinct genealogical histories for different parts of the alignment and mislead evolutionary inferences (Casola and Hahn 2009), we first identified putatively nonrecombining partitions using GARD (Kosakovsky Pond et al. 2006) (Supplementary Fig. 8) and chose 2 partitions, P1-414 and P697-981, for maximum likelihood–based analyses using PAML (Yang 2007) (Fig. 5a).
Fig. 5.
Maximum likelihood–based analyses for selective pressure variation and role of positive selection on the Hyphal_reg_CWP domain in C. auris. a) Schematic showing the putative nonrecombining partitions within the Hyphal_reg_CWP domain determined by GARD (see Supplementary Fig. 8). The 2 partitions labeled in gray were studied separately. The numbers refer to the columns in the coding sequence alignment. b, c) Phylogenetic trees were reconstructed for the 2 partitions and are shown as a cladogram. The vertical bar next to the Hil1/Hil2 pair indicates the difference in topology between the 2 trees. Branch colors are based on the dN/dS values estimated from a free-ratio model in PAML. “FG” designate foreground branches, whose dN/dS were greater than 10, except for branch 14..16 in (c), which was selected instead of 16..Hil4 because this would require fewer evolutionary changes in selective forces. We also analyzed the scenario with 16..Hil4 as the foreground, and the conclusions remained the same with slightly different P-values. d) Summary of the maximum likelihood–based tests for selective force heterogeneity and for positive selection. “Insig.” means P-value > 0.05. In the branch-site test, P(ω>1) is the total proportion of sites with dN/dS > 1 on the FG branches and ω2 their estimated dN/dS. The listed sites were identified as being under positive selection with a posterior probability greater than 0.99 by the Bayes Empirical Bayes (BEB). The one-letter code and number refer to the amino acid in the OG sequence and the alignment column (Supplementary Fig. 7).
We first tested if a subset of the sites evolved under positive selection consistently on all branches. We found moderate evidence supporting the hypothesis for the P697-981 partition, where the M8 vs M7 and M8 vs M8a tests were significant at a 0.01 level, but the more conservative test M2a vs M1a was not (Supplementary Table 6). All 3 tests were insignificant for the P1-414 partition. Next, we tested for elevated dN/dS on selected branches of the tree, sign of relaxed selective constraints, or positive selection. We first estimated the dN/dS for each branch using a free-ratio model and designated those with dN/dS greater than 10 as the “foreground” (Fig. 5, b and c, “FG”). We found strong evidence for the FG branches to have a higher dN/dS than the remainder of the tree (log–likelihood ratio test P < 0.01, Fig. 5d). There is no evidence, however, for the dN/dS across the entire domain on the FG branches to be greater than one (Fig. 5d, a, row 2). We then tested the more realistic scenario, where a subset of the sites on the FG branches was subject to positive selection. Using the branch-site test 2 as defined in Zhang et al. (2005), we found evidence for positive selection on a subset of the sites on the FG branches for both partitions (log–likelihood ratio test P < 0.01) and identified residues in both as candidate targets of positive selection with a posterior probability greater than 0.99 (Fig. 5d). We conclude that there is strong evidence for relaxed selective constraint on the Hyphal_reg_CWP domain on some branches following gene duplications; there is also evidence for positive selection acting on a subset of the sites on those branches. However, as the free-ratio model estimates were noisy and the Empirical Bayes method used to identify the residues under selection lacks power (Zhang et al. 2005) and can produce false positives (Nozawa et al. 2009), the specific branches and residues implicated must be interpreted with caution.
The yeast Hil family has adhesin-like domain architecture with rapidly diverging central domain sequences
We next examined the entire yeast Hil family to reveal the broader patterns of its evolution. We found that the Hil family in general has elevated Ser/Thr content compared with the rest of the proteome (Supplementary Fig. 9). Moreover, the majority of family members encode tandem repeats in the central domain (Fig. 6a) and contain predicted β-aggregation–prone sequences (Fig. 6b). Together, these features further suggest that most yeast Hil family members encode fungal adhesins. While these key features typical of yeast adhesins are conserved, the yeast Hil family exhibits extreme variation in protein length, in tandem repeat content, as well as in β-aggregation potential (Fig. 6, a and b, Supplementary Fig. 10), extending the pattern seen in C. auris (Fig. 2). The length of the protein outside of the Hyphal_reg_CWP domain has a mean ± standard deviation of 822.4 ± 785.8 aa and a median of 608.5 aa. This large variation in protein length is almost entirely driven by the tandem repeats (Fig. 6c, linear regression slope = 1.0, r 2 = 0.83). A subset of the Hil proteins (vertical bar in Fig. 6, a and b) stand out in that they are both longer than the rest of the family (1,745 vs 770 aa, median protein length) and have an unusually large number of β-aggregation–prone motifs (25 vs 6, median number of TANGO hits per protein). The motifs in this group of proteins are regularly spaced as a result of being part of the tandem repeat unit (median absolute deviation (MAD) of distances between adjacent TANGO hits less than 5 aa, Fig. 6d). The motif “GVVIVTT” and its variants account for 61% of all hits in this subset and is not found in significant number in the rest of the family. Together, these observations combined with previous experimental studies showing a direct impact of adhesin length and β-aggregation potential on function (Verstrepen et al. 2005; Lipke et al. 2012) lead us to propose that the rapid divergence of the Hil family members following the parallel expansion of the family led to functional diversification in adhesion in pathogenic yeasts and may have contributed to their enhanced virulence.
Fig. 6.
Evolution of protein length and β-aggregation potential in the yeast Hil family. a) Domain schematic shows that most homologs have a signal peptide at the N-terminus, then the Hyphal_reg_CWP domain, and a highly repetitive region central domain, followed by the C-terminal GPI anchor peptide. Homologs from M. bicuspidata were not included because many were annotated as incomplete. They were also excluded from other results in this figure. b) Distribution of TANGO-predicted β-aggregation sequences. The score for each sequence is shown as a color gradient and represents the median of the per-residue probability of aggregation. A vertical bar marks a group of MDR clade sequences that have a large number of β-aggregation–prone sequences arranged in regular intervals. c) X–Y plot showing the relationship between total protein length and tandem repeat sequence length for Hil family homologs. The linear regression line is shown in blue, with coefficients and r 2 values below. d) The species tree on the left is the same as in Fig. 1. The middle panel shows the number of Hil homologs per species. M. bicuspidata homologs were excluded; S. cerevisiae was included in the species tree, but no Hil homolog was identified in it (see text). The right panel shows the number of predicted β-aggregation–prone motifs per Hil homolog. Only motifs with a median probability ≥30% were counted. Proteins are colored in gold if they have 5 or more such motifs and if the MAD of the inter-motif distances is < 5 aa.
The yeast Hil family genes are preferentially located near chromosome ends
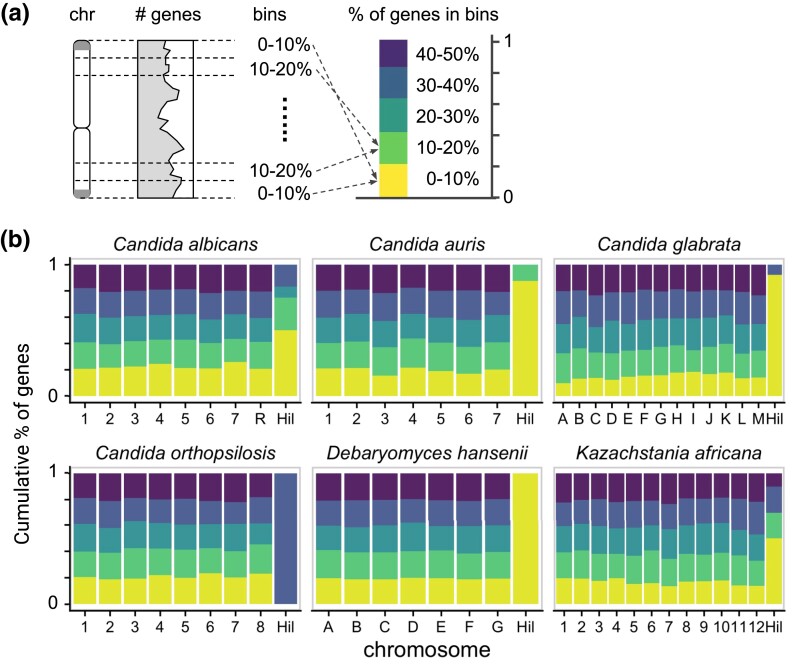
Several well-characterized yeast adhesin families, including the Flo family in S. cerevisiae and the Epa family in C. glabrata, are enriched in the subtelomeres (Teunissen and Steensma 1995; De Las Peñas et al. 2003; Xu et al. 2020, 2021). This region is associated with high rates of single-nucleotide polymorphisms (SNPs), indels, and copy number variations and can undergo ectopic recombination that enables the spread of genes between chromosome ends or their losses (Mefford and Trask 2002; Anderson et al. 2015). To determine if the Hil family is also enriched in the subtelomeric region, we compared their chromosomal locations with the background gene density distribution (Fig. 7a) in species with a chromosomal level assembly (Supplementary Table 7). To account for the shared evolutionary history, we selected one species per closely related group such that the Hil family homologs in these species were mostly derived through independent duplications based on our gene tree (Supplementary Fig. 2). The result showed that the Hil family genes are indeed enriched at chromosomal ends (Fig. 7b). A goodness-of-fit test confirmed that the difference between the chromosomal locations of the Hil family and the genome background is highly significant (P = 1.3 × 10−12). As ectopic recombination between subtelomeres has been suggested to underlie the spread of gene families (Anderson et al. 2015), we hypothesize that the enrichment of the Hil family toward the chromosome ends is both a cause and consequence of its parallel expansion in different Candida lineages.
Fig. 7.
Hil family genes are preferentially located near the chromosome ends. a) Schematic of the analysis: each chromosome (chr) is folded in half and divided into 5 equal-length “bins,” ordered by their distance to the nearest telomere. The cumulative bar graph on the right summarizes the gene density distribution in the 5 bins. b) Folded gene density distribution for 6 species with a chromosomal level assembly and more than 2 Hil family genes. The bin colors are as shown in (a). The Hil homologs in each species are plotted as a separate group. A goodness-of-fit test comparing the distribution of the Hil family genes to the genome background yielded a P-value of 1.3 × 10−12.
Discussion
The repeated emergence of human pathogens in the Saccharomycetes class poses serious health threats, as many emerging pathogenic species are resistant or quickly gain resistance to available antifungal drugs (Lamoth et al. 2018; Srivastava et al. 2018). This raises an evolutionary question: are there shared genomic changes in independently derived Candida pathogens, which could be key factors in host adaptation? Yeast adhesin families were among the most enriched gene families in pathogenic lineages relative to the low pathogenic potential relatives (Butler et al. 2009). It has been proposed that expansion of adhesin families could be a key step in the emergence of novel yeast pathogens (Gabaldón et al. 2016). However, detailed phylogenetic studies supporting this hypothesis are rare (Gabaldón et al. 2013), and far less is known about how their sequences diverge and what selective forces are involved during the expansions (Xie et al. 2011; Muñoz et al. 2021). In this study, we found that the Hyr/Iff-like (Hil) family, defined by the conserved Hyphal_reg_CWP domain, is significantly enriched among distantly related pathogenic clades (Fig. 1, a and b). This resulted from independent expansion of the family in these clades, including among closely related species (Fig. 1, c and d). We also showed that the protein sequences diverged extremely rapidly after duplications, driven mostly by the evolution of the tandem repeats and resulting in large variations in protein length, Ser/Thr content, and β-aggregation potential (Figs. 2b, c, and 6). Our evolutionary analyses revealed evidence of relaxed selective constraint and a potential role of positive selection acting on the Hyphal_reg_CWP domain during the family's expansion in C. auris (Fig. 5). We also found the Hil family to be strongly enriched near chromosomal ends (Fig. 7). Overall, our results support the hypothesis that expansion and diversification of adhesin families is a key step toward the emergence of yeast pathogens.
Genome assembly quality limits gene family evolution studies
Like any study of multigene family evolution, our work relies on and is limited by the quality of the genome assemblies. Two additional challenges in our study are due to the fact that Hil family genes are rich in tandem repeats (Figs. 2b and 6a), and many are located near chromosome ends (Fig. 7b), both of which pose problems for genome assemblies. For example, we found significant disagreement in length for 8 of the 16 Hil proteins in C. tropicalis between a long-read assembly and the RefSeq assembly, consistent with a recent study (Oh et al. 2020) (Supplementary Table 8) and in C. glabrata, we identified 13 Hil family genes in a long-read assembly (GCA_010111755.1) vs 3 in the RefSeq assembly (GCF_000002545.3); 12 of the 13 genes were in the subtelomeres (Xu et al. 2020). However, similar analyses in additional species did not reveal these problems, suggesting that the issues were at least in part due to difficulties in some genomes (Supplementary Text 1). Nonetheless, we acknowledge the possibility of missing homologs and inaccurate sequences, especially in the tandem repeat region. We thus believe the expected improvements in genome assemblies due to advances in long-read sequencing technologies will be crucial for future studies of the adhesin gene family in yeasts. It is worth noting that our main conclusions about the parallel expansion of the Hil family and its rapid divergence patterns are robust with respect to isolated problems as described above. Also, the long-read technology–based and de novo–assembled genomes for C. auris strains allowed us to confidently assess variation in the Hil family size and tandem repeat copy number between paralogs and among individual strains (Supplementary Table 4). The accuracy of the tandem repeat sequences in multiple strains in this species is supported by the conservation of repeat copy numbers within clades (Supplementary Table 5).
Evidence for adhesin functions in the Hil family
A few members of the Hil family, e.g. Iff4 in C. albicans and Awp2 in C. glabrata, were shown to mediate adhesiveness to polystyrene (Fu et al. 2008; Kempf et al. 2009; Reithofer et al. 2021). While further experimental studies are needed to establish the adhesin functions of other Hil family members, our work provides bioinformatic support for this hypothesis (Figs. 2 and 6). The predicted β-helix fold of the Hyphal_reg_CWP domain (Fig. 3), while unusual among characterized yeast adhesins (Willaert 2018), is found in many virulence factors residing on the surface of bacteria or viruses as well as enzymes that degrade or modify polysaccharides (Supplementary Table 3) (Kajava and Steven 2006). The elongated shape and rigid structure of the β-helix are consistent with the functional requirements of adhesins, including the need to protrude from the cell surface and the capacity for multiple binding sites along its length that facilitate adhesion. In a bacterial adhesin—the serine-rich repeat protein (SRRP) from the gram-positive bacterium, L. reuteri—a protruding, flexible loop in the β-helix was proposed to serve as a binding pocket for its ligand (Sequeira et al. 2018). Such a feature is not apparent in the predicted structure of the Hyphal_reg_CWP domain. Further studies are needed to elucidate the mechanism of action of this domain and its potential substrates.
The cross-kingdom similarity in adhesin effector domain structure is intriguing in several ways. First, it suggests convergent evolution in bacteria and yeasts. Second, it suggests that what is known about the structure–function relationship in bacteria can provide insight into the Hyphal_reg_CWP domain in yeast. Notably, the LrSRRP shows a pH-dependent substrate specificity that is potentially adapted to distinct host niches (Sequeira et al. 2018). Finally, the similar structure and function of the bacterial and yeast adhesins could mediate cross-kingdom interactions in natural and host environments (Uppuluri et al. 2018).
However, not all Hil family homologs are likely to encode adhesins. Sequence features suggest some Hil family proteins may have non-adhesin functions. For example, 39 of 193 Hil proteins (homologs labeled as incomplete were excluded) have the requisite signal sequence (SP+) but lack a GPI anchor attachment site (gpi-, Supplementary Fig. 1b). One, Iff11 in C. albicans, was shown to be secreted, and a null mutant was found to be hypersensitive to cell wall–damaging agents and less virulent in a murine systemic infection model (Bates et al. 2007). Moreover, 75% of these “SP+, gpi-” proteins are shorter than 600 amino acids, in contrast to only 4% of the 117 proteins having both a signal peptide and a GPI anchor attachment site. Such short, secreted proteins with tandem repeat sequences identical or similar to those present in the cell wall–associated Hil protein counterparts may serve an important regulatory function by bundling with wall-associated adhesins as previously suggested for similar subclass of proteins within the Als family (Oh et al. 2019). It is possible that the Hil family has evolved diverse functions broadly related to cell adhesion.
Ongoing diversification of the Hil family within species
In addition to the parallel expansion and the subsequent rapid sequence divergence in the Hil family between species, we and others also revealed population-level variation in both the family size and sequences within C. auris (Supplementary Figs. 5 and 6, and Supplementary Table 5) (Muñoz et al. 2021). Notably, among the 4 geographically stratified clades, Clade II strains lost 5 of the 8 Hil family members (Supplementary Fig. 6). Besides missing members of the Hil family, Clade II strains also lack 7 of the 8 members of another GPI anchor family that is specific to C. auris (Muñoz et al. 2021). These coincide with the finding that Clade II strains were mostly associated with ear infections (57/61 isolates (Kwon et al. 2019)) rather than hospital outbreaks, as reported for strains from the other clades, and that they were generally less resistant to antifungal drugs (Kwon et al. 2019; Welsh et al. 2019). This raises the question of whether the smaller adhesin repertoire in Clade II strains limits their adhesive capability and results in a different pathology. Similar expansion and contraction of adhesin families have been shown for the C. glabrata Hil family (AWP Cluster V) and Epa family (Marcet-Houben et al. 2022), suggesting that dynamic evolution of adhesin families in pathogenic yeasts could be a common pattern. Variation in the tandem repeat copy number in Hil1–Hil4 among C. auris strains is also intriguing (Supplementary Fig. 5). Prior studies of the S. cerevisiae Flo proteins have shown that protein length directly impacts cellular adhesion phenotypes (Verstrepen et al. 2005) and thus population-level variation in adhesin length could further contribute to phenotypic diversity. Lastly, scans for selective sweeps in C. auris identified Hil and Als family members as being among the top 5% of all genes, suggesting that adhesins are targets of natural selection in the recent evolutionary history of this newly emerged pathogen (Muñoz et al. 2021).
Diversification of the adhesin repertoire within a strain can arise from a variety of molecular mechanisms. For example, chimeric proteins generated through recombination between Als family members or between an Als protein's N-terminal effector domain and an Hyr/Iff protein's repeat region have been shown (Butler et al. 2009; Zhao et al. 2011; Oh et al. 2019). Some of the adhesins with highly diverged central domains may have arisen in this manner (Supplementary Fig. 10). Gene conversion between members of the same family can also drive the evolution of adhesin families within a species, as shown in S. cerevisiae and C. glabrata (Verstrepen et al. 2004; Marcet-Houben et al. 2022). Evidence of this in the Hil family was revealed in our analysis of recombination within the effector domain in C. auris (Supplementary Fig. 8).
Special properties of the central domain in C. auris Hil1–Hil4 and related Hil proteins
A subset of Hil proteins represented by C. auris Hil1–Hil4 (Fig. 6, a and b, vertical bar) stands out in that they are much longer on average and encode a large number of β-aggregation–prone sequences compared with the rest of the family (Fig. 6, b and d). Behind these properties is a conserved ∼44 aa repeat unit containing a highly β-aggregation–prone sequence (“GVVIVTT” and its variants) (Fig. 4b). β-aggregation–prone sequences and the amyloid-like interaction they mediate have been extensively studied, especially in the Als protein family in C. albicans: they were experimentally shown to mediate aggregation (Otoo et al. 2008; Ramsook et al. 2010) and were crucial for forming protein clusters on cell surfaces known as nanodomains in response to physical tension or sheer forces (Alsteens et al. 2010; Lipke et al. 2012). Recently, they were also shown to mediate cell–cell trans interactions via homotypic protein binding (Dehullu et al. 2019; Ho et al. 2019). This may underlie biofilm formation and kin discrimination (Smukalla et al. 2008; Brückner et al. 2020; Lipke et al. 2021). Most known yeast adhesins, including the Als family proteins, encode between 1 and 3 β-aggregation–prone sequences (Ramsook et al. 2010). C. auris Hil1–Hil4 and their close relatives are unusual in that they have as many as 50 such sequences, with each predicted by TANGO to have ∼90% probability of aggregation, whereas the positive threshold for the algorithm is only >5% over 5–6 residues (Fernandez-Escamilla et al. 2004). The structural implications of the vast number of β-aggregation–prone motifs may be that such tandem repeat domains are constitutively amyloid in nature, rather than requiring force or other stimuli as required by the Als proteins. The functional implications are unclear without the requisite experimental tests. However, we speculate that variations in protein length and β-aggregation potential resulting from the central domain divergence could directly impact the adhesion functions as previously suggested (Verstrepen et al. 2005; Boisramé et al. 2011; Lipke et al. 2012).
Structural predictions of the tandem repeat region in C. auris Hil1 and Hil2
Given the large number of ∼44 aa repeats in the central domain of C. auris Hil1–Hil4 and the prediction that each repeat encodes 3–4 short consecutive β-strands (Fig. 4b), we wondered what structural properties this region may have and how these features might contribute to the adhesion function. We explored this question using threading-based structural prediction tools such as I-TASSER (Yang et al. 2015) and pDOMThreader (Lobley et al. 2009). For the tandem repeat region in the central domain of Hil1, I-TASSER identified (S)-layer protein (SLP) structures (e.g. RsaA from Caulobacter crescentus and SbsA and SbsC from Geobacillus stearothermophilus) as among the top structural analogs. These β-strand–rich structures are known to self-assemble to form a 2-dimensional array on the surface of bacteria, mediating a range of functions including adhesion to host cells in pathogens (Fagan and Fairweather 2014). pDOMThreader analyses of the central domains in Hil1 and Hil2 identified a different set of templates, namely, bacterial self-associating proteins including Ag43a from uropathogenic E. coli, pertactin from B. pertussis, and the H. influenzae Hap adhesin. Interestingly, these proteins have β-helical structures like the Hyphal_reg_CWP domain, with the β-helices being involved in cell–cell interaction via an interface along the long solenoidal axis for homotypic interactions, mediate bacterial clumping (Heras et al. 2014), and lead to biofilm formation in H. influenzae (Meng et al. 2011). We speculate that the long repeat regions in Hil1 and Hil2 may similarly mediate cell–cell interactions in C. auris.
The possibility that the central domains in Hil1 and Hil2 form a β-helix is interesting in that β-helix is one of the commonly described structural motifs in functional amyloids, e.g. HET-s from the fungus Podospora anserina (Wasmer et al. 2008). Such a solenoid-type amyloid is distinguished from other amyloid types in that the β-helices formed by repeats within the same protein, rather than among distinct monomeric proteins, are suggested to be stabilized not only by polar zippers and hydrophobic contacts but also by electrostatic interactions between the alternating β-strands (Willbold et al. 2021). Other examples of amyloid-forming proteins with a predicted β-helix structure include the imperfect repeat domain in the human premelanosome protein Pmel17 (Louros et al. 2016) and the extracellular curli proteins of Enterobacteriaceae that are involved in biofilm formation and adhesion to host cells (Shewmaker et al. 2009). The proposed solenoidal structure of the central domain of Hil1–Hil4 like proteins, if true, would have 2 significant implications. First, it confers the necessary rigidity and extended conformation required for cell wall–anchored adhesins to extend into the surrounding extracellular milieu. Second, the numerous β-strand–rich repeats each containing a highly amyloid-prone heptameric sequence and capable of wrapping into a solenoidal shaped stack are likely to substantially reduce the rate-limiting nucleation step, which limits the formation of, e.g., an Aβ amyloid fiber. This would allow the formation of extracellular extensions at low protein concentrations without the need for an extensive fiber-lengthening process via the incorporation of additional monomeric units. Finally, the observation of solenoid-mediated intercellular interactions in the Hap adhesins suggests that Hil proteins may likewise have a biofilm-related function.
Genomic context
As reported by Muñoz et al. (2021), we found that the Hil family genes are preferentially located near chromosomal ends in C. auris and also in other species examined (Fig. 7). This is similar to previous findings for the Flo and Epa families (Teunissen and Steensma 1995; De Las Peñas et al. 2003; Xu et al. 2020, 2021), as well as the Als genes in some species (Oh et al. 2021). This location bias of the Hil and other adhesin families is likely a key mechanism for their dynamic expansion and sequence evolution via ectopic recombination (Anderson et al. 2015) and by break-induced replication (Bosco and Haber 1998; Sakofsky and Malkova 2017; Xu et al. 2021). Another potential consequence of the Hil family genes being located in subtelomeres is that they may be subject to epigenetic silencing as an additional regulatory mechanism, which can be derepressed in response to stress (Ai et al. 2002). Such epigenetic regulation of the adhesin genes was found to generate cell surface heterogeneity in S. cerevisiae (Halme et al. 2004) and leads to hyperadherent phenotypes in C. glabrata (Castaño et al. 2005).
Conclusion
To address the lack of candidate adhesins in C. auris, we identified and characterized the Hil family in this species and all yeasts. Based on our results, we hypothesize that expansion and diversification of adhesin gene families is a key step toward the evolution of fungal pathogenesis and that variation in the adhesin repertoire contributes to within- and between-species differences in the adhesive and virulence properties. Future experimental tests of these hypotheses will be important biologically for improving our understanding of the fungal adhesin repertoire, biotechnologically for inspiring additional nanomaterials, and biomedically for advancing the development of C. auris–directed therapeutics.
Supplementary Material
Acknowledgments
This study was inspired by work conducted during a bioinformatics course (BIOL:4213) in Fall of 2019, whose semester theme was the recently published C. auris genomes. Graduate students, LS and RS, were lead investigators on a group project centered on the genes containing the Hyphal_reg_CWP (PF11765) domain during that course and were happy to be invited to continue working on these genes long after the semester ended. We would also like to thank members of the Gene Regulatory Evolution lab for discussion. We thank Drs. Yong Zhang (CAS Zoology Institute), Peter Lipke (Brooklyn College), John McCutcheon (ASU), Matthew Anderson (OSU), Matthew Hahn (Indiana University), Rich Lenski (MSU), Josep Comeron (University of Iowa), Daniel Weeks (University of Iowa), Lois Hoyer (UIUC) and 2 other anonymous reviewers for providing useful suggestions and critical comments.
Contributor Information
Rachel A Smoak, Civil and Environmental Engineering, The University of Iowa, Iowa City, IA 52242, USA.
Lindsey F Snyder, Interdisciplinary Graduate Program in Genetics, The University of Iowa, Iowa City, IA 52242, USA.
Jan S Fassler, Interdisciplinary Graduate Program in Genetics, The University of Iowa, Iowa City, IA 52242, USA; Department of Biology, The University of Iowa, Iowa City, IA 52242, USA.
Bin Z He, Interdisciplinary Graduate Program in Genetics, The University of Iowa, Iowa City, IA 52242, USA; Department of Biology, The University of Iowa, Iowa City, IA 52242, USA.
Data availability
Raw data and analysis scripts are available at https://doi.org/10.5281/zenodo.7631150. The version and reference of the main analysis software are listed in Table 1.
Supplemental material available at GENETICS online.
Funding
BZH is supported by NIH R35GM137831. JSF is supported by NSF-IOS-1917169. LFS was supported by the NIH Predoctoral Training grant T32GM008629. RAS is supported by an NSF Graduate Research Fellowship Program under Grant No. 1546595, with additional support through the NSF Division of Graduate Education under Grant No. 1633098.
Literature cited
- Ai W, Bertram PG, Tsang CK, Chan TF, Zheng XFS. Regulation of subtelomeric silencing during stress response. Mol Cell. 2002;10(6):1295–1305. doi: 10.1016/S1097-2765(02)00695-0 [DOI] [PubMed] [Google Scholar]
- Alsteens D, Garcia MC, Lipke PN, Dufrêne YF. Force-induced formation and propagation of adhesion nanodomains in living fungal cells. Proc Natl Acad Sci USA. 2010;107(48):20744–20749. doi: 10.1073/pnas.1013893107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson MZ, Wigen LJ, Burrack LS, Berman J. Real-time evolution of a subtelomeric gene family in Candida albicans. Genetics. 2015;200(3):907–919. doi: 10.1534/genetics.115.177451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey DA, Feldmann PJ, Bovey M, Gow NA, Brown AJ. The Candida albicans HYR1 gene, which is activated in response to hyphal development, belongs to a gene family encoding yeast cell wall proteins. J Bacteriol. 1996;178(18):5353–5360. doi: 10.1128/jb.178.18.5353-5360.1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates S, de la Rosa JM, MacCallum DM, Brown AJP, Gow NAR, Odds FC. Candida albicans Iff11, a secreted protein required for cell wall structure and virulence. Infect Immun. 2007;75(6):2922–2928. doi: 10.1128/IAI.00102-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger SA, Krompass D, Stamatakis A. Performance, accuracy, and web server for evolutionary placement of short sequence reads under maximum likelihood. Syst Biol. 2011;60(3):291–302. doi: 10.1093/sysbio/syr010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boisramé A, Cornu A, Da Costa G, Richard ML. Unexpected role for a serine/threonine-rich domain in the Candida albicans iff protein family. Eukaryot Cell. 2011;10(10):1317–1330. doi: 10.1128/EC.05044-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosco G, Haber JE. Chromosome break-induced DNA replication leads to nonreciprocal translocations and telomere capture. Genetics. 1998;150(3):1037–1047. doi: 10.1093/genetics/150.3.1037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley PH, Nayfach S, Pollard KS. Phylogeny-corrected identification of microbial gene families relevant to human gut colonization. PLoS Comput Biol. 2018;14(8):e1006242. doi: 10.1371/journal.pcbi.1006242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodie R, Roper RL, Upton C. JDotter: a Java interface to multiple dotplots generated by dotter. Bioinformatics. 2004;20(2):279–281. doi: 10.1093/bioinformatics/btg406 [DOI] [PubMed] [Google Scholar]
- Brückner S, Schubert R, Kraushaar T, Hartmann R, Hoffmann D, Jelli E, Drescher K, Müller DJ, Oliver Essen L, Mösch H-U. Kin discrimination in social yeast is mediated by cell surface receptors of the Flo11 adhesin family. eLife. 2020;9:e55587. doi: 10.7554/eLife.55587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchan DWA, Jones DT. The PSIPRED protein analysis workbench: 20 years on. Nucleic Acids Res. 2019;47(W1):W402–W407. doi: 10.1093/nar/gkz297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler G, Rasmussen MD, Lin MF, Santos MAS, Sakthikumar S, Munro CA, Rheinbay E, Grabherr M, Forche A, Reedy JL, et al. Evolution of pathogenicity and sexual reproduction in eight Candida genomes. Nature. 2009;459(7247):657–662. doi: 10.1038/nature08064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL. BLAST+: architecture and applications. BMC Bioinformatics. 2009;10(1):421. doi: 10.1186/1471-2105-10-421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casola C, Hahn MW. Gene conversion among paralogs results in moderate false detection of positive selection using likelihood methods. J Mol Evol. 2009;68(6):679–687. doi: 10.1007/s00239-009-9241-6 [DOI] [PubMed] [Google Scholar]
- Castaño I, Pan S-J, Zupancic M, Hennequin C, Dujon B, Cormack BP. Telomere length control and transcriptional regulation of subtelomeric adhesins in Candida glabrata. Mol Microbiol. 2005;55(4):1246–1258. doi: 10.1111/j.1365-2958.2004.04465.x [DOI] [PubMed] [Google Scholar]
- CDC . 2019. Antibiotic resistance threats in the United States, 2019. US Dep. Health Hum. Serv. CDC. 10.15620/cdc:82532. [DOI]
- Chaudhuri R, Ansari FA, Raghunandanan MV, Ramachandran S. FungalRV: adhesin prediction and immunoinformatics portal for human fungal pathogens. BMC Genomics. 2011;12(1):192. doi: 10.1186/1471-2164-12-192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Groot PWJ, Bader O, de Boer AD, Weig M, Chauhan N. Adhesins in human fungal pathogens: glue with plenty of stick. Eukaryot Cell. 2013;12(4):470–481. doi: 10.1128/EC.00364-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehullu J, Valotteau C, Herman-Bausier P, Garcia-Sherman M, Mittelviefhaus M, Vorholt JA, Lipke PN, Dufrêne YF. Fluidic force microscopy demonstrates that homophilic adhesion by Candida albicans als proteins is mediated by amyloid bonds between cells. Nano Lett. 2019;19(6):3846–3853. doi: 10.1021/acs.nanolett.9b01010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Las Peñas A, Pan S-J, Castaño I, Alder J, Cregg R, Cormack BP. Virulence-related surface glycoproteins in the yeast pathogen Candida glabrata are encoded in subtelomeric clusters and subject to RAP1- and SIR-dependent transcriptional silencing. Genes Dev. 2003;17(18):2245–2258. doi: 10.1101/gad.1121003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dujon B, Sherman D, Fischer G, Durrens P, Casaregola S, Lafontaine I, de Montigny J, Marck C, Neuvéglise C, Talla E, et al. Genome evolution in yeasts. Nature. 2004;430(6995):35–44. doi: 10.1038/nature02579 [DOI] [PubMed] [Google Scholar]
- Eberlein C, Nielly-Thibault L, Maaroufi H, Dubé AK, Leducq J-B, Charron G, Landry CR. The rapid evolution of an ohnolog contributes to the ecological specialization of incipient yeast species. Mol Biol Evol. 2017;34(9):2173–2186. doi: 10.1093/molbev/msx153 [DOI] [PubMed] [Google Scholar]
- Engels B. XNomial: exact goodness-of-fit test for multinomial data with fixed probabilities. Vienna (Austria): CRAN; ; 2015. [Google Scholar]
- Fagan RP, Fairweather NF. Biogenesis and functions of bacterial S-layers. Nat Rev Microbiol. 2014;12(3):211–222. doi: 10.1038/nrmicro3213 [DOI] [PubMed] [Google Scholar]
- Fernandez-Escamilla A-M, Rousseau F, Schymkowitz J, Serrano L. Prediction of sequence-dependent and mutational effects on the aggregation of peptides and proteins. Nat Biotechnol. 2004;22(10):1302–1306. doi: 10.1038/nbt1012 [DOI] [PubMed] [Google Scholar]
- Frank AT, Ramsook CB, Otoo HN, Tan C, Soybelman G, Rauceo JM, Gaur NK, Klotz SA, Lipke PN. Structure and function of glycosylated tandem repeats from Candida albicans als adhesins. Eukaryot Cell. 2010;9(3):405–414. doi: 10.1128/EC.00235-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frieman MB, McCaffery JM, Cormack BP. Modular domain structure in the Candida glabrata adhesin Epa1p, a beta1,6 glucan-cross-linked cell wall protein. Mol Microbiol. 2002;46(2):479–492. doi: 10.1046/j.1365-2958.2002.03166.x [DOI] [PubMed] [Google Scholar]
- Fu Y, Luo G, Spellberg BJ, Edwards JE, Ibrahim AS. Gene overexpression/suppression analysis of candidate virulence factors of Candida albicans. Eukaryot Cell. 2008;7(3):483–492. doi: 10.1128/EC.00445-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabaldón T, Martin T, Marcet-Houben M, Durrens P, Bolotin-Fukuhara M, Lespinet O, Arnaise S, Boisnard S, Aguileta G, Atanasova R, et al. Comparative genomics of emerging pathogens in the Candida glabrata clade. BMC Genomics. 2013;14(1):623. doi: 10.1186/1471-2164-14-623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabaldón T, Naranjo-Ortíz MA, Marcet-Houben M. Evolutionary genomics of yeast pathogens in the saccharomycotina. FEMS Yeast Res. 2016;16(6):fow064. doi: 10.1093/femsyr/fow064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon JL, Armisén D, Proux-Wéra E, ÓhÉigeartaigh SS, Byrne KP, Wolfe KH. Evolutionary erosion of yeast sex chromosomes by mating-type switching accidents. Proc Natl Acad Sci USA. 2011;108(50):20024–20029. doi: 10.1073/pnas.1112808108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta R, Brunak S. Prediction of glycosylation across the human proteome and the correlation to protein function. Pac Symp Biocomput. 2002:310–322. [PubMed] [Google Scholar]
- Hall SR, Becker CR, Duffy MA, Cáceres CE. Variation in resource acquisition and use among host clones creates key epidemiological trade-offs. Am Nat. 2010;176(5):557–565. doi: 10.1086/656523 [DOI] [PubMed] [Google Scholar]
- Halme A, Bumgarner S, Styles C, Fink GR. Genetic and epigenetic regulation of the FLO gene family generates cell-surface variation in yeast. Cell. 2004;116(3):405–415. doi: 10.1016/S0092-8674(04)00118-7 [DOI] [PubMed] [Google Scholar]
- Heras B, Totsika M, Peters KM, Paxman JJ, Gee CL, Jarrott RJ, Perugini MA, Whitten AE, Schembri MA. The antigen 43 structure reveals a molecular Velcro-like mechanism of autotransporter-mediated bacterial clumping. Proc Natl Acad Sci USA. 2014;111(1):457–462. doi: 10.1073/pnas.1311592111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho V, Herman-Bausier P, Shaw C, Conrad KA, Garcia-Sherman MC, Draghi J, Dufrene YF, Lipke PN, Rauceo JM. An amyloid core sequence in the major Candida albicans adhesin Als1p mediates cell-cell adhesion. mBio. 2019;10(5):e01766–19. doi: 10.1128/mBio.01766-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho L, Si T, Ané C. A linear-time algorithm for Gaussian and non-Gaussian trait evolution models. Syst Biol. 2014;63(3):397–408. doi: 10.1093/sysbio/syu005 [DOI] [PubMed] [Google Scholar]
- Holm L. Dali server: structural unification of protein families. Nucleic Acids Res. 2022;50(W1):W210. doi: 10.1093/nar/gkac387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyer LL. The ALS gene family of Candida albicans. Trends Microbiol. 2001;9(4):176–180. doi: 10.1016/S0966-842X(01)01984-9 [DOI] [PubMed] [Google Scholar]
- Hoyer LL, Green CB, Oh S-H, Zhao X. Discovering the secrets of the Candida albicans agglutinin-like sequence (ALS) gene family–a sticky pursuit. Med Mycol. 2008;46(1):1–15. doi: 10.1080/13693780701435317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Innan H, Kondrashov F. The evolution of gene duplications: classifying and distinguishing between models. Nat Rev Genet. 2010;11(2):97–108. doi: 10.1038/nrg2689 [DOI] [PubMed] [Google Scholar]
- Ives AR, Garland T. Phylogenetic logistic regression for binary dependent variables. Syst Biol. 2010;59(1):9–26. doi: 10.1093/sysbio/syp074 [DOI] [PubMed] [Google Scholar]
- Jenull S, Tscherner M, Kashko N, Shivarathri R, Stoiber A, Chauhan M, Petryshyn A, Chauhan N, Kuchler K. Transcriptome signatures predict phenotypic variations of Candida auris. Front Cell Infect Microbiol. 2021;11. doi: 10.3389/fcimb.2021.662563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H, Bao J, Xing Y, Li X, Chen Q. Comparative genomic analyses provide insight into the pathogenicity of metschnikowia bicuspidata LNES0119. Front Microbiol. 2022;13:939141. doi: 10.3389/fmicb.2022.939141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jumper J, Evans R, Pritzel A, Green T, Figurnov M, Ronneberger O, Tunyasuvunakool K, Bates R, Žídek A, Potapenko A, et al. Highly accurate protein structure prediction with AlphaFold. Nature. 2021;596(7873):583–589. doi: 10.1038/s41586-021-03819-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajava AV, Steven AC. The turn of the screw: variations of the abundant beta-solenoid motif in passenger domains of type V secretory proteins. J Struct Biol. 2006;155(2):306–315. doi: 10.1016/j.jsb.2006.01.015 [DOI] [PubMed] [Google Scholar]
- Kean R, Delaney C, Sherry L, Borman A, Johnson EM, Richardson MD, Rautemaa-Richardson R, Williams C, Ramage G. Transcriptome assembly and profiling of Candida auris reveals novel insights into biofilm-mediated resistance. mSphere. 2018;3(4). doi: 10.1128/mSphere.00334-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kempf M, Cottin J, Licznar P, Lefrançois C, Robert R, Apaire-Marchais V. Disruption of the GPI protein-encoding gene IFF4 of Candida albicans results in decreased adherence and virulence. Mycopathologia. 2009;168(2):73–77. doi: 10.1007/s11046-009-9201-0 [DOI] [PubMed] [Google Scholar]
- Kosakovsky Pond SL, Posada D, Gravenor MB, Woelk CH, Frost SDW. Automated phylogenetic detection of recombination using a genetic algorithm. Mol Biol Evol. 2006;23(10):1891–1901. doi: 10.1093/molbev/msl051 [DOI] [PubMed] [Google Scholar]
- Koteiche HA, Mchaourab HS. Folding pattern of the alpha-crystallin domain in alphaA-crystallin determined by site-directed spin labeling. J Mol Biol. 1999;294(2):561–577. doi: 10.1006/jmbi.1999.3242 [DOI] [PubMed] [Google Scholar]
- Kozlov AM, Darriba D, Flouri T, Morel B, Stamatakis A. RAxML-NG: a fast, scalable and user-friendly tool for maximum likelihood phylogenetic inference. Bioinformatics. 2019;35(21):4453–4455. doi: 10.1093/bioinformatics/btz305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuang MC, Hutchins PD, Russell JD, Coon JJ, Hittinger CT. Ongoing resolution of duplicate gene functions shapes the diversification of a metabolic network. eLife. 2016;5:e19027. doi: 10.7554/eLife.19027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon YJ, Shin JH, Byun SA, Choi MJ, Won EJ, Lee D, Lee SY, Chun S, Lee JH, Choi HJ, et al. Candida auris clinical isolates from South Korea: identification, antifungal susceptibility, and genotyping. J Clin Microbiol. 2019;57(4):e01624–18. doi: 10.1128/JCM.01624-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamoth F, Lockhart SR, Berkow EL, Calandra T. Changes in the epidemiological landscape of invasive candidiasis. J Antimicrob Chemother. 2018;73(suppl_1):i4–i13. doi: 10.1093/jac/dkx444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy A, Salas Gonzalez I, Mittelviefhaus M, Clingenpeel S, Herrera Paredes S, Miao J, Wang K, Devescovi G, Stillman K, Monteiro F, et al. Genomic features of bacterial adaptation to plants. Nat Genet. 2018;50(1):138–150. doi: 10.1038/s41588-017-0012-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linder T, Gustafsson CM. Molecular phylogenetics of ascomycotal adhesins—a novel family of putative cell-surface adhesive proteins in fission yeasts. Fungal Genet Biol. 2008;45(4):485–497. doi: 10.1016/j.fgb.2007.08.002 [DOI] [PubMed] [Google Scholar]
- Lipke PN. What we do not know about fungal cell adhesion molecules. J Fungi. 2018;4(2):59. doi: 10.3390/jof4020059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipke PN, Garcia MC, Alsteens D, Ramsook CB, Klotz SA, Dufrêne YF. Strengthening relationships: amyloids create adhesion nanodomains in yeasts. Trends Microbiol. 2012;20(2):59–65. doi: 10.1016/j.tim.2011.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipke PN, Mathelié-Guinlet M, Viljoen A, Dufrêne YF. A new function for amyloid-like interactions: cross-beta aggregates of adhesins form cell-to-cell bonds. Pathogens. 2021;10(8):1013. doi: 10.3390/pathogens10081013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobley A, Sadowski MI, Jones DT. pGenTHREADER and pDomTHREADER: new methods for improved protein fold recognition and superfamily discrimination. Bioinformatics. 2009;25(14):1761–1767. doi: 10.1093/bioinformatics/btp302 [DOI] [PubMed] [Google Scholar]
- Lockhart SR, Etienne KA, Vallabhaneni S, Farooqi J, Chowdhary A, Govender NP, Colombo AL, Calvo B, Cuomo CA, Desjardins CA, et al. Simultaneous emergence of multidrug-resistant Candida auris on 3 continents confirmed by whole-genome sequencing and epidemiological analyses. Clin Infect Dis. 2017;64(2):134–140. doi: 10.1093/cid/ciw691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louros NN, Baltoumas FA, Hamodrakas SJ, Iconomidou VA. A β-solenoid model of the Pmel17 repeat domain: insights to the formation of functional amyloid fibrils. J Comput Aided Mol Des. 2016;30(2):153–164. doi: 10.1007/s10822-015-9892-x [DOI] [PubMed] [Google Scholar]
- Luo G, Ibrahim A, Spellberg B, Nobile C, Mitchell A, Fu Y. Candida albicans Hyr1p confers resistance to neutrophil killing and is a potential vaccine target. J Infect Dis. 2010;201(11):1718–1728. doi: 10.1086/652407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcet-Houben M, Alvarado M, Ksiezopolska E, Saus E, de Groot PWJ, Gabaldón T. Chromosome-level assemblies from diverse clades reveal limited structural and gene content variation in the genome of Candida glabrata. BMC Biol. 2022;20(1):226. doi: 10.1186/s12915-022-01412-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mefford HC, Trask BJ. The complex structure and dynamic evolution of human subtelomeres. Nat Rev Genet. 2002;3(2):91–102. doi: 10.1038/nrg727 [DOI] [PubMed] [Google Scholar]
- Meng G, Spahich N, Kenjale R, Waksman G, St Geme JW. Crystal structure of the Haemophilus influenzae hap adhesin reveals an intercellular oligomerization mechanism for bacterial aggregation. EMBO J. 2011;30(18):3864–3874. doi: 10.1038/emboj.2011.279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morel B, Kozlov AM, Stamatakis A, Szöllősi GJ. GeneRax: a tool for species-tree-aware maximum likelihood-based gene family tree inference under gene duplication, transfer, and loss. Mol Biol Evol. 2020;37(9):2763–2774. doi: 10.1093/molbev/msaa141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muñoz JF, Gade L, Chow NA, Loparev VN, Juieng P, Berkow EL, Farrer RA, Litvintseva AP, Cuomo CA. Genomic insights into multidrug-resistance, mating and virulence in Candida auris and related emerging species. Nat Commun. 2018;9(1):5346. doi: 10.1038/s41467-018-07779-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muñoz JF, Welsh RM, Shea T, Batra D, Gade L, Howard D, Rowe LA, Meis JF, Litvintseva AP, Cuomo CA. Clade-specific chromosomal rearrangements and loss of subtelomeric adhesins in Candida auris. Genetics. 2021;218(1). doi: 10.1093/genetics/iyab029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman AM, Cooper JB. XSTREAM: a practical algorithm for identification and architecture modeling of tandem repeats in protein sequences. BMC Bioinformatics. 2007;8(1):382. doi: 10.1186/1471-2105-8-382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nozawa M, Suzuki Y, Nei M. Reliabilities of identifying positive selection by the branch-site and the site-prediction methods. Proc Natl Acad Sci. 2009;106(16):6700–6705. doi: 10.1073/pnas.0901855106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh S-H, Isenhower A, Rodriguez-Bobadilla R, Smith B, Jones J, Hubka V, Fields C, Hernandez A, Hoyer LL. Pursuing advances in DNA sequencing technology to solve a complex genomic jigsaw puzzle: the agglutinin-like sequence (ALS) genes of Candida tropicalis. Front Microbiol. 2020;11:594531. doi: 10.3389/fmicb.2020.594531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh S-H, Schliep K, Isenhower A, Rodriguez-Bobadilla R, Vuong VM, Fields CJ, Hernandez AG, Hoyer LL. Using genomics to shape the definition of the agglutinin-like sequence (ALS) family in the saccharomycetales. Front Cell Infect Microbiol. 2021;11:794529. doi: 10.3389/fcimb.2021.794529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh S-H, Smith B, Miller AN, Staker B, Fields C, Hernandez A, Hoyer LL. Agglutinin-like sequence (ALS) genes in the Candida parapsilosis species complex: blurring the boundaries between gene families that encode cell-wall proteins. Front Microbiol. 2019;10:781. doi: 10.3389/fmicb.2019.00781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otoo HN, Lee KG, Qiu W, Lipke PN. Candida albicans als adhesins have conserved amyloid-forming sequences. Eukaryot Cell. 2008;7(5):776–782. doi: 10.1128/EC.00309-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierleoni A, Martelli PL, Casadio R. PredGPI: a GPI-anchor predictor. BMC Bioinformatics. 2008;9(1):392. doi: 10.1186/1471-2105-9-392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potter SC, Luciani A, Eddy SR, Park Y, Lopez R, Finn RD. HMMER web server: 2018 update. Nucleic Acids Res. 2018;46(W1):W200–W204. doi: 10.1093/nar/gky448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian W, Zhang JG. Genomic evidence for adaptation by gene duplication. Genome Res. 2014;24(8):1356–1362. 10.1101/gr.172098.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsook CB, Tan C, Garcia MC, Fung R, Soybelman G, Henry R, Litewka A, O'Meally S, Otoo HN, Khalaf RA, et al. Yeast cell adhesion molecules have functional amyloid-forming sequences. Eukaryot Cell. 2010;9(3):393–404. doi: 10.1128/EC.00068-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauceo JM, De Armond R, Otoo H, Kahn PC, Klotz SA, Gaur NK, Lipke PN. Threonine-rich repeats increase fibronectin binding in the Candida albicans adhesin Als5p. Eukaryot Cell. 2006;5(10):1664–1673. doi: 10.1128/EC.00120-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team . R: a language and environment for statistical computing. Vienna (Austria): R Foundation for Statistical Computing; 2021. [Google Scholar]
- Reid SD, Herbelin CJ, Bumbaugh AC, Selander RK, Whittam TS. Parallel evolution of virulence in pathogenic Escherichia coli. Nature. 2000;406(6791):64–67. doi: 10.1038/35017546 [DOI] [PubMed] [Google Scholar]
- Reithofer V, Fernández-Pereira J, Alvarado M, de Groot P, Essen L-O. A novel class of Candida glabrata cell wall proteins with β-helix fold mediates adhesion in clinical isolates. PLoS Pathog. 2021;17(12):e1009980. doi: 10.1371/journal.ppat.1009980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice P, Longden I, Bleasby A. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet. 2000;16(6):276–277. doi: 10.1016/S0168-9525(00)02024-2 [DOI] [PubMed] [Google Scholar]
- Richard ML, Plaine A. Comprehensive analysis of glycosylphosphatidylinositol-anchored proteins in Candida albicans. Eukaryot Cell. 2007;6(2):119–133. doi: 10.1128/EC.00297-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosiana S, Zhang L, Kim GH, Revtovich AV, Uthayakumar D, Sukumaran A, Geddes-McAlister J, Kirienko NV, Shapiro RS. Comprehensive genetic analysis of adhesin proteins and their role in virulence of Candida albicans. Genetics. 2021;217(2). doi: 10.1093/genetics/iyab003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- RStudio Team . RStudio: integrated development environment for R. Boston: (MA: ): RStudio, PBC; 2021. [Google Scholar]
- Sakofsky CJ, Malkova A. Break induced replication in eukaryotes: mechanisms, functions, and consequences. Crit Rev Biochem Mol Biol. 2017;52(4):395–413. doi: 10.1080/10409238.2017.1314444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrödinger LLC. The PyMOL Molecular Graphics System, Version 2.5.2. New York: Schrödinger LLC; 2021. [Google Scholar]
- Sequeira S, Kavanaugh D, MacKenzie DA, Šuligoj T, Walpole S, Leclaire C, Gunning AP, Latousakis D, Willats WGT, Angulo J, et al. Structural basis for the role of serine-rich repeat proteins from Lactobacillus reuteri in gut microbe–host interactions. Proc Natl Acad Sci. 2018;115(12):E2706–E2715. doi: 10.1073/pnas.1715016115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen X-X, Opulente DA, Kominek J, Zhou X, Steenwyk JL, Buh KV, Haase MAB, Wisecaver JH, Wang M, Doering DT, et al. Tempo and mode of genome evolution in the budding yeast subphylum. Cell. 2018;175(6):1533–1545.e20. doi: 10.1016/j.cell.2018.10.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shewmaker F, Mcglinchey RP, Thurber KR, Mcphie P, Dyda F, Tycko R, Wickner RB. The functional curli amyloid is not based on in-register parallel beta-sheet structure. J Biol Chem. 2009;284:25065–25076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Söding J, et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 2011;7(1):539. doi: 10.1038/msb.2011.75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh S, Uppuluri P, Mamouei Z, Alqarihi A, Elhassan H, French S, Lockhart SR, Chiller T, Edwards JE, Ibrahim AS. The NDV-3A vaccine protects mice from multidrug resistant Candida auris infection. PLOS Pathog. 2019;15(8):e1007460. doi: 10.1371/journal.ppat.1007460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smukalla S, Caldara M, Pochet N, Beauvais A, Guadagnini S, Yan C, Vinces MD, Jansen A, Prevost MC, Latgé J-P, et al. FLO1 is a variable green beard gene that drives biofilm-like cooperation in budding yeast. Cell. 2008;135(4):726–737. doi: 10.1016/j.cell.2008.09.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava V, Singla RK, Dubey AK. Emerging virulence, drug resistance and future anti-fungal drugs for Candida pathogens. Curr Top Med Chem. 2018;18(9):759–778. doi: 10.2174/1568026618666180528121707 [DOI] [PubMed] [Google Scholar]
- Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30(9):1312–1313. doi: 10.1093/bioinformatics/btu033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamler R, Kappé G, Boelens W, Slingsby C. Wrapping the α-crystallin domain fold in a chaperone assembly. J Mol Biol. 2005;353(1):68–79. doi: 10.1016/j.jmb.2005.08.025 [DOI] [PubMed] [Google Scholar]
- Steentoft C, Vakhrushev SY, Joshi HJ, Kong Y, Vester-Christensen MB, Schjoldager KT-BG, Lavrsen K, Dabelsteen S, Pedersen NB, Marcos-Silva L, et al. Precision mapping of the human O-GalNAc glycoproteome through SimpleCell technology. EMBO J. 2013;32(10):1478–1488. doi: 10.1038/emboj.2013.79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steenwyk JL, Buida TJ, Li Y, Shen X-X, Rokas A. ClipKIT: a multiple sequence alignment trimming software for accurate phylogenomic inference. PLoS Biol. 2020;18(12):e3001007. doi: 10.1371/journal.pbio.3001007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suyama M, Torrents D, Bork P. PAL2NAL: robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res. 2006;34(Web Server):W609–W612. doi: 10.1093/nar/gkl315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teufel F, Almagro Armenteros JJ, Johansen AR, Gíslason MH, Pihl SI, Tsirigos KD, Winther O, Brunak S, von Heijne G, Nielsen H. Signalp 6.0 predicts all five types of signal peptides using protein language models. Nat Biotechnol. 2022;40(7):1023–1025. doi: 10.1038/s41587-021-01156-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teunissen AW, Steensma HY. Review: the dominant flocculation genes of saccharomyces cerevisiae constitute a new subtelomeric gene family. Yeast. 1995;11(11):1001–1013. doi: 10.1002/yea.320111102 [DOI] [PubMed] [Google Scholar]
- Uppuluri P, Lin L, Alqarihi A, Luo G, Youssef EG, Alkhazraji S, Yount NY, Ibrahim BA, Bolaris MA, Edwards JE, et al. The Hyr1 protein from the fungus Candida albicans is a cross kingdom immunotherapeutic target for Acinetobacter bacterial infection. PLoS Pathog. 2018;14(5):e1007056. doi: 10.1371/journal.ppat.1007056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verstrepen KJ, Jansen A, Lewitter F, Fink GR. Intragenic tandem repeats generate functional variability. Nat Genet. 2005;37(9):986–990. doi: 10.1038/ng1618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verstrepen KJ, Reynolds TB, Fink GR. Origins of variation in the fungal cell surface. Nat Rev Microbiol. 2004;2(7):533–540. doi: 10.1038/nrmicro927 [DOI] [PubMed] [Google Scholar]
- Wang L-G, Lam TT-Y, Xu S, Dai Z, Zhou L, Feng T, Guo P, Dunn CW, Jones BR, Bradley T, et al. Treeio: an R package for phylogenetic tree input and output with richly annotated and associated data. Mol Biol Evol. 2020;37(2):599–603. doi: 10.1093/molbev/msz240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasmer C, Lange A, Van Melckebeke H, Siemer AB, Riek R, Meier BH. Amyloid fibrils of the HET-s(218–289) prion form a beta solenoid with a triangular hydrophobic core. Science. 2008;319(5869):1523–1526. doi: 10.1126/science.1151839 [DOI] [PubMed] [Google Scholar]
- Waterhouse AM, Procter JB, Martin DMA, Clamp M, Barton GJ. Jalview version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25(9):1189–1191. doi: 10.1093/bioinformatics/btp033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsh RM, Sexton DJ, Forsberg K, Vallabhaneni S, Litvintseva A. Insights into the unique nature of the east Asian clade of the emerging pathogenic yeast Candida auris. J Clin Microbiol. 2019;57(4):e00007–19. doi: 10.1128/JCM.00007-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkins M, Zhang N, Schmid J. Biological roles of protein-coding tandem repeats in the yeast Candida albicans. J Fungi. 2018;4(3):78. doi: 10.3390/jof4030078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willaert R. Adhesins of yeasts: protein structure and interactions. J Fungi. 2018;4(4):119. doi: 10.3390/jof4040119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willbold D, Strodel B, Schröder GF, Hoyer W, Heise H. Amyloid-type protein aggregation and prion-like properties of amyloids. Chem Rev. 2021;121(13):8285–8307. doi: 10.1021/acs.chemrev.1c00196 [DOI] [PubMed] [Google Scholar]
- Winter DJ. Rentrez: an R package for the NCBI eUtils API. R J. 2017;9(2):520–526. doi: 10.32614/RJ-2017-058 [DOI] [Google Scholar]
- Xie X, Qiu W-G, Lipke PN. Accelerated and adaptive evolution of yeast sexual adhesins. Mol Biol Evol. 2011;28(11):3127–3137. doi: 10.1093/molbev/msr145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z, Green B, Benoit N, Schatz M, Wheelan S, Cormack B. De novo genome assembly of Candida glabrata reveals cell wall protein complement and structure of dispersed tandem repeat arrays. Mol Microbiol. 2020;113(6):1209–1224. doi: 10.1111/mmi.14488 [DOI] [PubMed] [Google Scholar]
- Xu Z, Green B, Benoit N, Sobel JD, Schatz MC, Wheelan S, Cormack BP. Cell wall protein variation, break-induced replication, and subtelomere dynamics in Candida glabrata. Mol Microbiol. 2021;116(1):260–276. doi: 10.1111/mmi.14707 [DOI] [PubMed] [Google Scholar]
- Yang Z. Likelihood ratio tests for detecting positive selection and application to primate lysozyme evolution. Mol Biol Evol. 1998;15(5):568–573. doi: 10.1093/oxfordjournals.molbev.a025957 [DOI] [PubMed] [Google Scholar]
- Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24(8):1586–1591. doi: 10.1093/molbev/msm088 [DOI] [PubMed] [Google Scholar]
- Yang J, Yan R, Roy A, Xu D, Poisson J, Zhang Y. The I-TASSER suite: protein structure and function prediction. Nat Methods. 2015;12(1):7–8. doi: 10.1038/nmeth.3213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G. Using ggtree to visualize data on tree-like structures. Curr Protoc Bioinforma. 2020;69(1):e96. doi: 10.1002/cpbi.96 [DOI] [PubMed] [Google Scholar]
- Zhang J. Evolution by gene duplication: an update. Trends Ecol Evol. 2003;18(6):292–298. doi: 10.1016/S0169-5347(03)00033-8 [DOI] [Google Scholar]
- Zhang J, Nielsen R, Yang Z. Evaluation of an improved branch-site likelihood method for detecting positive selection at the molecular level. Mol Biol Evol. 2005;22(12):2472–2479. doi: 10.1093/molbev/msi237 [DOI] [PubMed] [Google Scholar]
- Zhao X, Oh S-H, Coleman DA, Hoyer LL. ALS51, a newly discovered gene in the Candida albicans ALS family, created by intergenic recombination: analysis of the gene and protein, and implications for evolution of microbial gene families. FEMS Immunol Med Microbiol. 2011;61(3):245–257. doi: 10.1111/j.1574-695X.2010.00769.x [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw data and analysis scripts are available at https://doi.org/10.5281/zenodo.7631150. The version and reference of the main analysis software are listed in Table 1.
Supplemental material available at GENETICS online.