Summary
Background
Early diagnosis and appropriate treatment are essential in meningitis and encephalitis management. We aimed to implement and verify an artificial intelligence (AI) model for early aetiological determination of patients with encephalitis and meningitis, and identify important variables in the classification process.
Methods
In this retrospective observational study, patients older than 18 years old with meningitis or encephalitis at two centres in South Korea were enrolled for development (n = 283) and external validation (n = 220) of AI models, respectively. Their clinical variables within 24 h after admission were used for the multi-classification of four aetiologies including autoimmunity, bacteria, virus, and tuberculosis. The aetiology was determined based on the laboratory test results of cerebrospinal fluid conducted during hospitalization. Model performance was assessed using classification metrics, including the area under the receiver operating characteristic curve (AUROC), recall, precision, accuracy, and F1 score. Comparisons were performed between the AI model and three clinicians with varying neurology experience. Several techniques (eg, Shapley values, F score, permutation feature importance, and local interpretable model-agnostic explanations weights) were used for the explainability of the AI model.
Findings
Between January 1, 2006, and June 30, 2021, 283 patients were enrolled in the training/test dataset. An ensemble model with extreme gradient boosting and TabNet showed the best performance among the eight AI models with various settings in the external validation dataset (n = 220); accuracy, 0.8909; precision, 0.8987; recall, 0.8909; F1 score, 0.8948; AUROC, 0.9163. The AI model outperformed all clinicians who achieved a maximum F1 score of 0.7582, by demonstrating a performance of F1 score greater than 0.9264.
Interpretation
This is the first multiclass classification study for the early determination of the aetiology of meningitis and encephalitis based on the initial 24-h data using an AI model, which showed high performance metrics. Future studies can improve upon this model by securing and inputting time-series variables and setting various features about patients, and including a survival analysis for prognosis prediction.
Funding
MD-PhD/Medical Scientist Training Program through the Korea Health Industry Development Institute, funded by the Ministry of Health & Welfare, Republic of Korea.
Keywords: Meningitis, Autoimmune encephalitis, Tuberculosis, Neuroinflammation, Artificial intelligence
Research in context.
Evidence before this study
We searched PubMed and Web of Science with the terms “((meningitis) OR (encephalitis) OR (meningoencephalitis)) AND ((machine learning) OR (deep learning))” for papers published between database inception and March 31, 2023, with no language restrictions. We found that there were several previous papers in which performed binary classification with artificial intelligence (AI) model had been performed, but only one paper proposed a multi-classification model. This multi-classification model has a key limitation, mentioned in the paper, that it cannot discriminate completely between all the aetiologies. Other related studies aimed to classify only two types of aetiology. The highest performing model showed an AUC of 0.95 and was able to distinguish between aseptic meningitis and bacterial meningitis only. We aimed to advance upon these previous models with our multi-classification model.
Added value of this study
To the best of our knowledge, to date, there has been no study that simultaneously distinguishes various aetiologies of encephalitis and meningitis. Our multi-classification AI model can predict the aetiology with initial 24 h hospitalisation data. We tested it in internal test set and external validation set and compared it with clinicians including three clinicians, including two neurologists with varying experience.
Implications of all the available evidence
Our research shows that our multi-classification AI model has excellent ability to predict the aetiology of encephalitis and meningitis in various evaluation metrics, and its diagnostic performance was equivalent to, or better than, that of a senior neurologist in our comparison. The results suggest that the application of AI could help to determine the aetiology of meningitis and encephalitis, thereby potentially enabling early treatment selection. In future studies, it is expected that a better prediction model can be created by securing and inputting time-series variables and setting various features for patients.
Introduction
Meningitis and encephalitis are inflammatory conditions involving the membranes and parenchyma of the central nervous system (CNS) and are associated with high fatality rates and lifelong disability, thus demanding improved diagnosis and treatment.1 The development of antibiotics, antiviral agents, antifungal agents, and vaccines has improved the mortality rates in cases of infectious aetiology, and the elucidation of mechanisms of autoimmune encephalitis has enabled the treatment of encephalitis of unknown origin.2 Nevertheless, the aetiology of meningitis and encephalitis varies from infectious agents, including viruses, bacteria, mycobacteria, fungi, and parasites, to the autoimmune process, rendering early aetiologic diagnosis and rapid treatment a challenge, ultimately leading to a poor prognosis.2,3
Aetiologic classification is confirmed based on culture and antibody test results and requires time and specific instrumentation and facilities. Thus, researchers attempted classification by studying the differences in the initial clinical presentation, brain imaging features, electroencephalography (EEG), and cerebrospinal fluid (CSF) findings among cases with different aetiologies.4, 5, 6, 7 However, early treatment is often empirical, based on the clinician's experience, which may lead to side effects such as acute kidney injury and liver damage.8 Several studies using artificial intelligence (AI) have been conducted for the aetiologic diagnosis of meningitis; however, there are limitations, such as low performance9 or performing only binary classification.7,10, 11, 12, 13
In this study, we aimed to develop an AI classification model for early detection based on the initial 24-h data of patients with encephalitis and meningitis, verify it with an external dataset, compare the performance of the model with clinicians' judgment, and identify the important variables in the classification process.
Methods
Ethics
This study was approved by the Yonsei University Health System, Institutional Review Board (Y-2021-0960). Due to the retrospective nature and use of de-identified data, this study was approved with waiver of the requirement to obtain informed consent by the Yonsei University Health System, Institutional Review Board (Y-2021-0960). The study was performed in accordance with approved guidelines and regulations for medical research expressed in the Declaration of Helsinki.
Study design
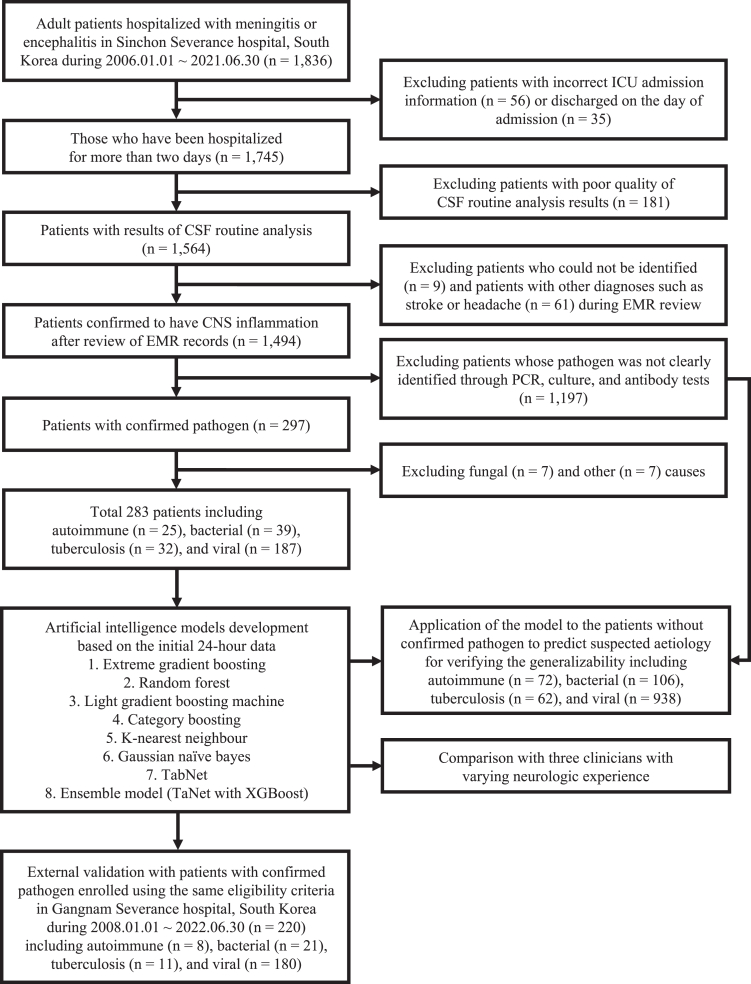
We performed a retrospective observational study at Severance Hospital in Seoul, South Korea. Patients were selected from the Severance Clinical Research Analysis Portal, which offered anonymised patient data from Severance Hospital to researchers for privacy preservation. Patients admitted to Sinchon Severance Hospital between January 2006 and June 2021 were included in this study for the train and test dataset. Patients who met the following inclusion criteria were enrolled: (1) clinically diagnosed with meningitis or encephalitis, (2) older than 18 years, and (3) CSF samples obtained through a lumbar puncture. Patients clinically diagnosed with meningitis or encephalitis were extracted based on International Classification of Diseases (ICD)-10 codes listed in Appendix 1 of Supplementary materials, and the patient selection flow chart can be found in Fig. 1. We developed AI models that predicted aetiology (autoimmunity, bacteria, tuberculosis, and virus) based on the variables available during the first 24 h of hospitalisation from patients with confirmed cause. The aetiology was determined based on the laboratory test results of cerebrospinal fluid conducted during hospitalization. To verify the generalisability of AI model, we applied the model trained on patients with a clear aetiology to patients whose pathogen was not clearly identified through laboratory tests to predict their suspected cause. Their suspected cause was determined by the consensus of three neurologists based on the electronic medical records including suspected diagnosis and treatment information documented by the attending physician at the time of hospitalisation, as well as various examination results. Patients from Gangnam Severance Hospital between January 2008 and June 2022 were enrolled using the same eligibility criteria for the external validation dataset.
Fig. 1.
Study design and workflow of patient selection. Abbreviation: ICU, Intensive care unit; CSF, Cerebrospinal fluid; EMR, Electronic medical record; CNS, Central nervous system; PCR, Polymerase chain reaction; XGBoost, Extreme gradient boosting.
The performance of the AI model was compared with the clinical judgment of clinicians. We selected three doctors from those working at Severance Hospital who had diverse levels of neurological experience and were capable of inferring causes based on patient data including one doctor who had not been trained in neurology and two neurologists (one with 5 years and the other with 15 years of clinical experience). The test set was constructed with a sample size of 100, consisting of 56 patients from the internal dataset (Sinchon Severance Hospital) and 44 from the external dataset (Gangnam Severance Hospital). Three doctors were requested to predict four aetiologies based on clinical and laboratory data collected retrospectively within 24 h of admission and to check all the variables that served as evidence for their prediction for each patient. These data were presented in the form of a tabular record.
Data pre-processing and feature selection
We collected data regarding baseline characteristics at admission, including age, sex, height, weight, and mental status. Additionally, medical history including seizures, tuberculosis, and the Charlson comorbidity index based on ICD-10 codes were collected.14 Data regarding vital signs such as blood pressure, heart rate, respiratory rate, and body temperature within 24 h and results of diagnostic modalities such as brain computed tomography (CT), chest X-ray (CXR), and EEG were also collected. Laboratory findings of CSF, blood, and urine samples were also included in the study (Table 1). The initial value was selected when there were several identical test results within the initial 24 h. All numerical variables were normalised by min–max scaling and used in the process. During the process of collecting vital signs and calculating the average and maximum values over 24 h, medically impossible values were excluded based on the clinician's judgment, as they were likely due to typographical errors. Missing values of categorical variables were replaced with −1 and those of numerical features were replaced with various imputation techniques in the AI process (Supplementary Table S1). The percentage of missing values for each variable can be found in Supplementary Tables S2 and S3.
Table 1.
Feature distribution for each aetiology at sinchon severance hospital.
| Aetiologies | Autoimmune (N = 25) | Bacterial (N = 39) | Tuberculosis (N = 32) | Viral (N = 187) | p-valuea |
|---|---|---|---|---|---|
| Age | 43.9 ± 20.7 | 56.7 ± 19.0 | 49.3 ± 18.2 | 44.5 ± 18.5 | 0.002 |
| Female sex | 14 (56.0%) | 14 (35.9%) | 16 (50.0%) | 89 (47.6%) | 0.410 |
| Body mass index | 21.8 ± 2.8 | 22.8 ± 2.6 | 21.9 ± 2.2 | 23.2 ± 2.4 | 0.004 |
| Comorbidities | |||||
| Charlson comorbidity index | 0.6 ± 1.2 | 1.1 ± 1.5 | 1.3 ± 1.9 | 0.6 ± 1.3 | 0.017 |
| Tuberculosis history | 0 (0.0%) | 1 (2.6%) | 29 (90.6%) | 2 (1.1%) | <0.001 |
| Seizure | 13 (52.0%) | 7 (17.9%) | 5 (15.6%) | 10 (5.3%) | <0.001 |
| Abnormal Brain CT | 1 (7.7%) | 5 (16.1%) | 2 (7.1%) | 8 (5.4%) | 0.229 |
| Abnormal Chest X-ray | 3 (12.5%) | 8 (20.5%) | 7 (21.9%) | 16 (9.2%) | 0.088 |
| EEG | |||||
| Abnormal | 22 (95.7%) | 11 (100.0%) | 13 (92.9%) | 30 (90.9%) | 0.714 |
| Unreactivity | 0 (0.0%) | 3 (27.3%) | 0 (0.0%) | 6 (18.2%) | 0.027 |
| Slowing | 19 (82.6%) | 11 (100.0%) | 13 (92.9%) | 30 (90.9%) | 0.423 |
| Epileptiform discharges | 5 (21.7%) | 2 (18.2%) | 0 (0.0%) | 5 (15.2%) | 0.333 |
| Abnormal mentality | 14 (56.0%) | 22 (56.4%) | 15 (46.9%) | 25 (13.4%) | <0.001 |
| Vital signs | |||||
| Mean systolic blood pressure (mmHg) | 120.6 ± 15.9 | 131.8 ± 19.1 | 132.6 ± 17.6 | 128.4 ± 15.0 | 0.022 |
| Mean diastolic blood pressure (mmHg) | 71.8 ± 9.4 | 75.2 ± 9.3 | 80.1 ± 10.6 | 76.6 ± 9.1 | 0.009 |
| Mean heart rate (rate/min) | 82.2 ± 13.4 | 93.2 ± 20.0 | 83.3 ± 15.6 | 77.2 ± 12.3 | <0.001 |
| Mean respiratory rate (rate/min) | 18.8 ± 1.0 | 19.2 ± 2.7 | 19.0 ± 2.3 | 18.5 ± 1.4 | 0.060 |
| Maximum body temperature (°C) | 37.6 ± 0.5 | 38.4 ± 0.8 | 38.0 ± 0.6 | 37.9 ± 0.6 | <0.001 |
| Mean body temperature (°C) | 37.1 ± 0.4 | 37.4 ± 0.6 | 37.2 ± 0.6 | 37.4 ± 0.5 | 0.031 |
| CSF | |||||
| WBC count (/μL) | 54.0 ± 108.4 | 1956.8 ± 2962.5 | 188.9 ± 149.3 | 254.8 ± 267.7 | <0.001 |
| Mononuclear leukocyte ratio | 35.3 ± 44.2 | 20.5 ± 23.3 | 66.5 ± 36.8 | 81.3 ± 26.7 | <0.001 |
| Polymorphonuclear leukocyte ratio | 7.2 ± 19.3 | 66.7 ± 33.3 | 17.5 ± 23.9 | 6.6 ± 17.6 | <0.001 |
| Basophil ratio | 0.0 ± 0.2 | 0.1 ± 0.2 | 0.0 ± 0.2 | 0.5 ± 1.0 | <0.001 |
| Eosinophil ratio | 0.2 ± 0.5 | 0.2 ± 0.7 | 0.0 ± 0.0 | 0.3 ± 1.2 | 0.478 |
| Protein (mg/dL) | 53.5 ± 31.1 | 462.3 ± 397.5 | 575.1 ± 1302.8 | 126.7 ± 203.7 | <0.001 |
| Glucose (mg/dL) | 67.2 ± 20.1 | 34.5 ± 44.2 | 38.1 ± 20.1 | 60.5 ± 16.8 | <0.001 |
| CSF/serum glucose ratio | 0.6 ± 0.1 | 0.2 ± 0.2 | 0.3 ± 0.2 | 0.5 ± 0.1 | <0.001 |
| RBC count >100/μL | 1 (4.0%) | 15 (38.5%) | 3 (9.4%) | 17 (9.1%) | <0.001 |
| Specific gravity | 1.0 ± 0.0 | 1.0 ± 0.0 | 1.0 ± 0.0 | 1.0 ± 0.0 | <0.001 |
| Turbidity | 0 (0.0%) | 27 (69.2%) | 8 (25.0%) | 31 (16.6%) | <0.001 |
| Adenosine deaminase (IU/L) | 3.4 ± 3.5 | 14.0 ± 24.9 | 21.6 ± 37.0 | 4.7 ± 4.1 | <0.001 |
| Abnormal colour | 0 (0.0%) | 24 (61.5%) | 13 (40.6%) | 12 (6.4%) | <0.001 |
| High pH | 14 (56.0%) | 15 (38.5%) | 16 (50.0%) | 144 (77.0%) | <0.001 |
| Cryptococcal antigen | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | 1 (0.6%) | 0.926 |
| HSV antibody IgM | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | 1 (0.6%) | 0.928 |
| VZV antibody IgM | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | 1 (0.6%) | 0.928 |
| Blood | |||||
| WBC count (109/L) | 9.7 ± 4.1 | 12.5 ± 7.5 | 8.7 ± 4.5 | 7.8 ± 3.1 | <0.001 |
| Neutrophil ratio | 75.1 ± 11.9 | 85.5 ± 9.9 | 78.6 ± 11.2 | 70.7 ± 12.0 | <0.001 |
| Monocyte ratio | 4.7 ± 2.0 | 3.4 ± 2.3 | 5.7 ± 2.5 | 5.5 ± 2.1 | <0.001 |
| Basophil ratio | 0.4 ± 0.5 | 0.2 ± 0.2 | 0.3 ± 0.2 | 0.5 ± 0.3 | <0.001 |
| Eosinophil ratio | 1.5 ± 1.2 | 0.9 ± 0.9 | 0.9 ± 1.3 | 1.6 ± 2.0 | 0.057 |
| Large unstained cells ratio | 1.3 ± 0.7 | 1.1 ± 0.7 | 1.4 ± 0.7 | 2.1 ± 1.2 | <0.001 |
| Haemoglobin (g/dL) | 13.1 ± 2.1 | 12.5 ± 2.1 | 12.7 ± 2.1 | 14.0 ± 1.4 | <0.001 |
| Platelet count (109/L) | 247.1 ± 66.0 | 181.6 ± 96.9 | 277.5 ± 99.0 | 234.1 ± 71.1 | <0.001 |
| Mean platelet volume (fL) | 7.7 ± 0.7 | 8.5 ± 1.0 | 7.5 ± 0.6 | 8.0 ± 0.9 | <0.001 |
| Red cell distribution width (%) | 13.2 ± 0.9 | 14.0 ± 2.1 | 14.4 ± 1.6 | 12.9 ± 1.2 | <0.001 |
| Delta neutrophil index (%) | 0.8 ± 1.3 | 3.4 ± 3.3 | 1.3 ± 1.3 | 0.5 ± 0.9 | <0.001 |
| Prothrombin time (INR) | 1.0 ± 0.1 | 1.1 ± 0.1 | 1.0 ± 0.1 | 1.0 ± 0.1 | <0.001 |
| aPTT (sec) | 29.1 ± 3.6 | 29.0 ± 5.0 | 29.2 ± 3.8 | 30.6 ± 3.7 | 0.028 |
| Sodium ion concentration (mmol/L) | 137.5 ± 6.1 | 137.1 ± 5.3 | 133.1 ± 3.8 | 137.5 ± 3.8 | <0.001 |
| Potassium ion concentration (mmol/L) | 4.2 ± 0.5 | 3.8 ± 0.6 | 4.0 ± 0.5 | 4.1 ± 0.4 | <0.001 |
| tCO2 (mmol/L) | 21.4 ± 3.4 | 20.7 ± 3.2 | 23.0 ± 3.8 | 23.4 ± 2.5 | <0.001 |
| Blood urea nitrogen (mg/dL) | 16.2 ± 7.3 | 21.7 ± 14.6 | 14.8 ± 9.8 | 15.2 ± 9.9 | 0.005 |
| Creatinine (mg/dL) | 0.8 ± 0.4 | 1.3 ± 1.6 | 0.8 ± 0.9 | 0.8 ± 0.3 | 0.002 |
| Glucose (mg/dL) | 123.8 ± 43.6 | 157.8 ± 70.5 | 119.2 ± 29.3 | 123.3 ± 35.5 | <0.001 |
| Albumin (g/dL) | 4.1 ± 0.5 | 3.6 ± 0.7 | 3.8 ± 0.6 | 4.3 ± 0.4 | <0.001 |
| Aspartate transaminase (IU/L) | 23.5 ± 10.9 | 35.6 ± 24.2 | 36.2 ± 37.3 | 25.3 ± 14.9 | 0.002 |
| Alanine aminotransferase (IU/L) | 23.5 ± 18.2 | 31.9 ± 28.7 | 20.6 ± 11.9 | 24.5 ± 23.7 | 0.192 |
| Total bilirubin (mg/dL) | 0.8 ± 0.6 | 1.1 ± 1.0 | 0.7 ± 0.7 | 0.7 ± 0.4 | 0.025 |
| Alkaline phosphatase (IU/L) | 68.1 ± 23.0 | 83.5 ± 62.1 | 71.2 ± 37.0 | 63.1 ± 19.8 | 0.005 |
| Uric acid (mg/dL) | 4.5 ± 1.3 | 4.5 ± 2.0 | 3.3 ± 1.8 | 4.2 ± 1.7 | 0.029 |
| Inorganic phosphorus (mg/dL) | 3.6 ± 0.6 | 2.8 ± 0.8 | 3.4 ± 0.6 | 3.3 ± 0.8 | <0.001 |
| Calcium (mg/dL) | 9.0 ± 0.6 | 8.5 ± 0.7 | 8.7 ± 0.6 | 9.0 ± 0.5 | <0.001 |
| Creatinine Kinase (IU/L) | 329.6 ± 355.7 | 299.8 ± 443.0 | 228.1 ± 323.6 | 383.2 ± 630.3 | 0.775 |
| Ammonia (μg/dL) | 49.0 ± 33.0 | 55.9 ± 27.6 | 55.7 ± 15.4 | 47.4 ± 21.3 | 0.352 |
| Total cholesterol (mg/dL) | 166.8 ± 35.5 | 146.8 ± 64.6 | 173.8 ± 38.9 | 171.1 ± 37.8 | 0.047 |
| C-reactive protein (mg/L) | 15.2 ± 43.5 | 85.5 ± 101.7 | 21.1 ± 33.9 | 11.3 ± 25.9 | <0.001 |
| Erythrocyte sedimentation rate (mm/hr) | 21.5 ± 25.2 | 50.7 ± 33.5 | 43.9 ± 31.3 | 23.0 ± 20.8 | <0.001 |
| Procalcitonin (ng/mL) | 0.1 ± 0.1 | 13.4 ± 25.0 | 0.2 ± 0.1 | 0.1 ± 0.1 | <0.001 |
| Lactate (mmol/L) | 2.3 ± 1.6 | 2.7 ± 1.3 | 0.9 ± 0.3 | 1.4 ± 0.8 | <0.001 |
| Cryptococcal antigen | 0 (0.0%) | 1 (2.8%) | 0 (0.0%) | 2 (1.2%) | 0.737 |
| Anti-HIV I/II + antigen | 0 (0.0%) | 0 (0.0%) | 1 (3.8%) | 2 (1.2%) | 0.522 |
| Urine | |||||
| Bilirubinuria | 0 (0.0%) | 3 (8.3%) | 4 (13.3%) | 12 (7.2%) | 0.343 |
| Haematuria | 3 (13.6%) | 20 (55.6%) | 13 (43.3%) | 50 (29.9%) | 0.003 |
| Glucosuria | 1 (4.5%) | 16 (44.4%) | 6 (20.0%) | 20 (12.0%) | <0.001 |
| Ketonuria | 10 (45.5%) | 20 (55.6%) | 10 (33.3%) | 86 (51.5%) | 0.254 |
| High leukocyte esterase | 4 (18.2%) | 8 (22.2%) | 14 (46.7%) | 41 (24.6%) | 0.052 |
| Albuminuria | 6 (27.3%) | 21 (58.3%) | 15 (50.0%) | 58 (34.7%) | 0.021 |
| High RBC count | 6 (27.3%) | 15 (41.7%) | 13 (43.3%) | 40 (24.0%) | 0.048 |
| High WBC count | 5 (22.7%) | 11 (30.6%) | 10 (33.3%) | 41 (24.6%) | 0.680 |
Abbreviations: CT, Computed tomography; EEG, Electroencephalography; CSF, Cerebrospinal fluid; WBC, White blood cell; RBC, Red blood cell; HSV, Herpes simplex virus; Ig, Immunoglobulin; VZV, Varicella zoster virus; aPTT, Activated partial thromboplastin time; HIV, Human immunodeficiency virus.
One-way analysis of variance (ANOVA) was used to compare continuous variables and Chi-square test was used to compare categorical variables among the four groups.
In the feature selection process, variables with a missing value percentage of more than 50% were excluded. To ensure inter-variable independence, variables were integrated into one if their Pearson correlation coefficients were higher than 0.8. Additionally, variables with a variance inflation factor >10 were eliminated to solve the problem of multicollinearity (Supplementary Table S4).15 Finally, 82 of the initial 110 variables were selected.
Classification models
We compared the performance of various classification models by utilising functions provided by the PyCaret library in Python (Supplementary Table S5). Based on the results, we adopted Extreme Gradient Boosting (XGBoost), Random Forest, and Light Gradient Boosting Machine models, which showed higher performance. We also applied the Category Boosting model with a similar structure. K-Nearest Neighbour and Gaussian Naive Bayes model were also applied because of their high interpretability.16 Additionally, we applied the TabNet for deep learning and an ensemble model with machine learning for performance improvement. When applying TabNet, we duplicated the original data 20 times and proceeded with the learning because training did not occur due to the small sample size. We checked the performance by varying the ensemble model composition ratio differently, and found that the best performance was achieved by the model composed of 80% XGBoost and 20% TabNet (Supplementary Fig. S1).
We evaluated all models with stratified K-fold cross validation (K = 2,4,5,10) and found that as the value of K increased, the training size increased, and overall performance tended to improve (Supplementary Table S6). Therefore, the final performance comparison of the models was conducted through 10-fold cross validation (Supplementary Fig. S2). Regularisation was adapted (L2 for XGBoost classification), and the model parameters were chosen to reduce model complexity with Bayesian optimisation. The optimised parameters for each model are presented in Appendix 2. We used class weights in all models to handle class imbalance and four under-sampling techniques including random sampling, Tomek links, condensed nearest neighbour, and one-sided selection; two over-sampling techniques including synthetic minority over-sampling technique (SMOTE) and adaptive synthetic sampling (ADASYN); and a combination technique called SMOTE-Tomek links (Supplementary Table S1).17 We stopped the training at the epoch where we observed the validation loss starting to increase while the training loss continued to decrease via the learning curve plot (Supplementary Fig. S3).
Model performance was assessed using classification metrics, including the area under the receiver operating characteristic curve (AUROC), recall, precision, accuracy, and F1 score. The mathematical expressions of these metrics are given in Appendix 3.
Model-agnostic methods for interpreting AI models including permutation feature importance (PIMP), local interpretable model-agnostic explanations (LIME), and Shapley additive explanations (SHAP) were adopted for verifying the explainability of AI models.18 F score, a method for measuring feature importance built into XGBoost, which is based on the number of times a variable is used to split the data (weight), the number of data points separated by that variable (cover), and the average training loss reduction achieved by using the feature (gain) was also used for verifying the explainability of model.19
Statistical analysis
We performed sample size estimation for the AI study with the Nx-subsampling scheme20 and found that each performance metric converged at around 80–100 training samples fewer than our training sample size (Supplementary Fig. S4). For the feature selection process, the Pearson correlation coefficient and variance inflation factor were used. One-way analysis of variance (ANOVA) and the Pearson χ2 test were used to compare continuous and categorical variables, respectively, among the groups. After conducting a comparison among the four aetiologies, post-hoc analysis with Bonferroni correction was performed to examine differences between each pair of groups. We also applied the AI model to predict the causes of patients whose aetiology was not clearly identified as a post-hoc analysis.
All statistical analyses were performed using R version 4.11, and the machine learning and deep learning processes were performed using Python version 3.6.8. A two-sided p-value of <0.05 was generally considered a minimum level of statistical significance.
Role of the funding source
The funders had no role in study design, data collection, data analyses, data interpretation, or writing of the report.
Results
Patients’ characteristics
Between January 1, 2006, and June 30, 2021, patients with confirmed aetiology (n = 283) were used for AI model training among patients hospitalised with meningitis and encephalitis from Sinchon Severance Hospital (n = 1836). Viral infection was the most common cause (n = 187), followed by bacterial (n = 39), tuberculous (n = 32), and autoimmune (n = 25) causes. Patients with fungal (n = 7) or other (n = 6) infections were excluded because of the small sample sizes. The details of pathogens related to each aetiology are provided in Supplementary Tables S7 and S8. The baseline characteristics according to aetiology and differences in clinical variables between the four groups are presented in Table 1.
Post-hoc analyses between the two groups were also performed and are presented in Supplementary Table S9. Variables that showed significant p values <0.001 in the four groups generally exhibited a characteristic separation of one aetiology from the other groups in the post-hoc analysis. For instance, seizures were found to occur frequently in patients with autoimmune aetiologies. Patients with bacterial infections showed distinct features compared to other groups, including faster mean heart rate, higher maximum body temperature, higher white blood cell (WBC) count and specific gravity, and more frequent turbidity or abnormal colour in CSF studies. In blood tests, patients with bacterial infection exhibited lower platelet count and inorganic phosphorus, higher mean platelet volume, delta neutrophil index, glucose, C-reactive protein, and procalcitonin. Patients with bacterial infection had the most heterogeneous variables compared to other groups. Patients with tuberculosis infection often had a history of previous tuberculosis, whereas patients with viral infection showed different CSF WBC differential counts and a higher serum large unstained cell ratio compared to other groups.
Model performance
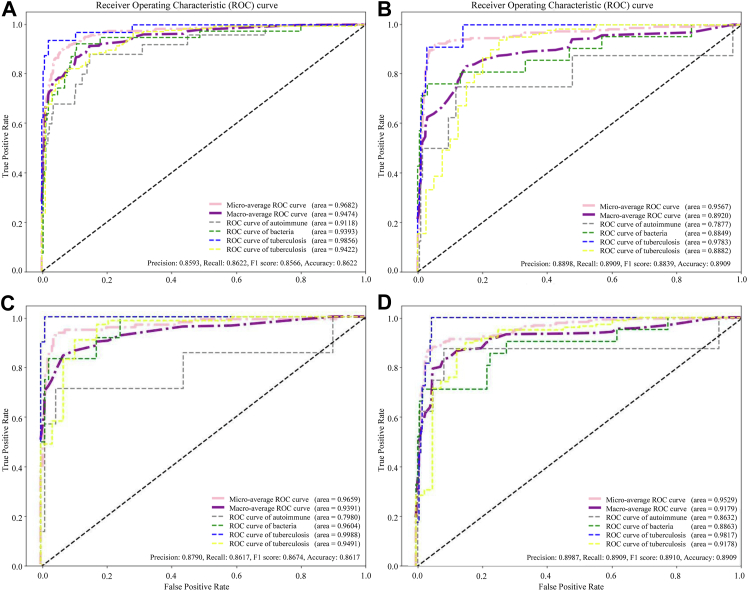
We applied different techniques in each model and conducted the analysis. The AUROC, recall, precision, accuracy, and F1 score obtained using different settings are shown in Supplementary Table S1. Patients were enrolled using the same eligibility criteria used for the external validation dataset from the Gangnam Severance Hospital. The baseline characteristics according to the validation set aetiology are presented in Supplementary Table S10. The AUROC, recall, precision, accuracy, and F1 score of the external validation set are also shown in Supplementary Table S1. The DeLong test used to compare the AUROC among the high-performance models did not show any significant differences. We placed a high emphasis on the F1 score, given the circumstance that our data had imbalanced labels when assessing model performance. When we replaced missing values of continuous variables with median values and performed oversampling using ADASYN in the ensemble model, the F1 score on the external dataset was the highest among all models. The accuracy, precision, recall, F1 score and an AUROC of 0.8909, 0.8987, 0.8909, and 0.9222, respectively, were achieved by the ensemble model (Fig. 2). Among the conventional machine learning models, the XGBoost model trained by replacing missing values with the mean value achieved the highest F1 score of 0.8839 on the external dataset (Fig. 2). The performances of the best models for each class are presented in Supplementary Table S11.
Fig. 2.
The area under the receiver operating characteristic curve (AUROC) for the models developed in the study. XGBoost (extreme gradient boosting) model (A) internal dataset (B) external validation dataset and the ensemble model composed of 80% XGBoost and 20% TabNet (C) internal dataset (D) external validation dataset.
We applied the XGBoost model to predict the causes of patients whose aetiology was not clearly identified (n = 1197). They consisted of patients suspected of autoimmune aetiology (n = 72), bacterial (n = 106), tuberculosis (n = 62), and viral infection (n = 938). The AUROC, accuracy, precision, recall, and F1 score of 0.8078, 0.7776, 0.7891, 0.7776, and 0.7793 were achieved (Supplementary Fig. S5).
Comparison between the AI model and clinicians
Test sets incorporating internal and external data sets were developed to compare our models and clinicians, and the doctors then predicted the aetiology based on the tabular data. The results of the comparisons between the AI model and clinicians are shown in Table 2. The clinician without training in neurology showed the lowest performance, whereas the performance improved with experience of the neurologists. The XGBoost model, which showed the best performance among the conventional machine learning models, and the ensemble model were used for comparison, and the performance of both AI models exceeded that of clinicians in all metrics. The AI model's predictions and those of each clinician for aetiology, as well as the true label, are presented in the form of a confusion matrix in Supplementary Fig. S6. The variables used by the clinicians to classify patients and whether they helped to correctly predict the aetiology are demonstrated in Supplementary Table S12. Overall, it can be seen that the variables including WBC, protein, differential cell count ratio, adenosine deaminase (ADA), and glucose level in the CSF, were used in this order by doctors when classifying the aetiology.
Table 2.
The performance metrics of comparing between AI model vs clinicians.
| Precision | Recall | F1 score | Accuracy | |
|---|---|---|---|---|
| XGBoost model | 0.9269 | 0.9300 | 0.9264 | 0.9300 |
| Ensemble modela | 0.9291 | 0.9300 | 0.9269 | 0.9300 |
| Doctor without training | 0.7562 | 0.3400 | 0.3651 | 0.3400 |
| Junior neurologist | 0.7528 | 0.6600 | 0.6893 | 0.6600 |
| Senior neurologist | 0.7689 | 0.7500 | 0.7582 | 0.7500 |
Abbreviations: XGBoost, extreme gradient boosting.
Ensemble model is composed of 80% of XGBoost and 20% of TabNet.
Explainability of the AI model
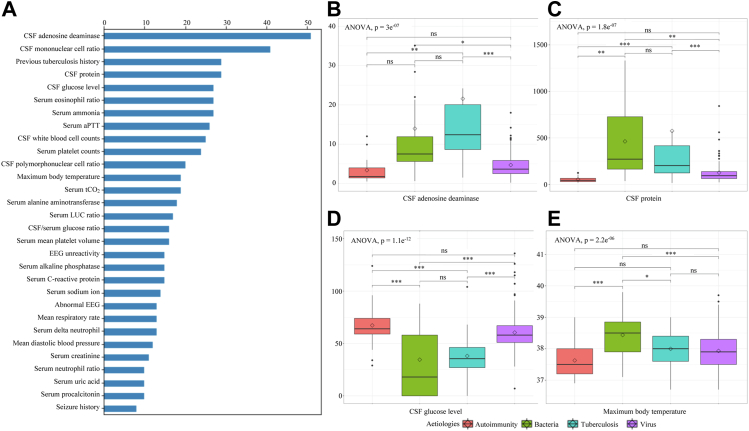
The F score of the XGBoost model was calculated based on the ‘weight’, which reflects the number of times the variable was used to split the data and is represented in Fig. 3A. Variables with high F scores were considered to play a more important role in classifying the aetiologies in this model. Box plots with statistical significance were presented in Fig. 3B–E to visualise whether the actual values of the variables with high F scores differed by aetiology and whether they were helpful in differentiating aetiology. CSF ADA, the feature with the highest F score, showed a significant difference with a p-value <0.001 in ANOVA. Post-hoc analysis showed that it had significantly higher values in patients with tuberculosis compared to those with autoimmune aetiologies (p < 0.01) and those with viral infections (p < 0.001), and in patients with bacterial infections compared to those with viral infections (p < 0.05). CSF protein, CSF glucose, and maximum body temperature also showed significant differences between the groups.
Fig. 3.
(A) Top 30 important features with F score from XGBoost classifier in internal dataset and (B–E) box plots displaying the distribution of variables for each aetiology. The diamond symbol represents the mean value and the horizontal line inside the box represents the median value. Bonferroni post-hoc analyses were done for comparing each two aetiologies. Abbreviation: CSF, Cerebrospinal fluid; aPTT, Activated partial thromboplastin time; LUC, Large unstained cells; EEG, Electroencephalography; ANOVA, Analysis of variance; ns, statistically no significant; ∗, p < 0.05; ∗∗, p < 0.01; ∗∗∗, p < 0.001.
PIMP is a method for determining feature importance by randomly shuffling a particular feature to create noise and calculating how much performance loss occurs when that feature is not used. We observed that the variables with the highest PIMP score were the previous history of tuberculosis, CSF cell differential ratio, and CSF glucose levels in this given order (Supplementary Fig. S7A). LIME weights for each aetiology are presented in Supplementary Fig. S7B–E. LIME computes the model predictions and generates a new dataset by removing a feature and keeping the rest for each feature. It estimates feature importance by calculating the prediction differences between the original and newly generated datasets. Abnormal findings in EEG were strongly associated with autoimmune causes and had a significant positive impact, the same as the case with seizures. A history of previous tuberculosis had a significantly positive impact on being classified as tuberculosis but had a negative impact on other causes.
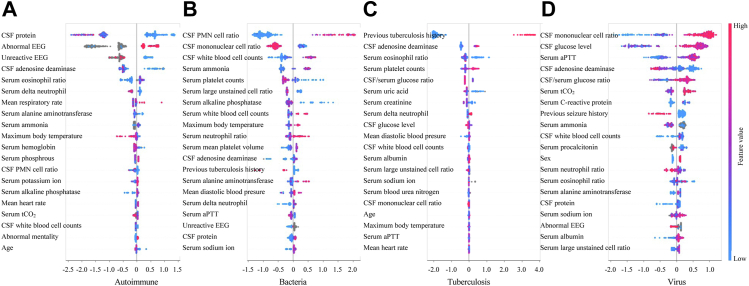
SHAP is a technique that calculates the contribution of each feature to the prediction result using the concept of Shapley values from game theory and the values for each aetiology are demonstrated in Fig. 4. The estimated SHAP value for each case is used to illustrate the influence on the X-axis. If a case's value is positive, it indicates that the associated attribute has a positive effect on the case's classification. Each point on the summary plot is a case, coloured according to the distribution of the feature's initial value along a red to blue gradient. The overall predictive power of each feature can be visually measured by its horizontal range. CSF protein, CSF polymorphonuclear cell ratio, previous tuberculosis history, and CSF mononuclear cell ratio showed the widest range, indicating they had the most considerable prediction power and can significantly impact the model's output for predicting each cause. We could observe that a decrease in the CSF protein indicated that the observed instance had a greater likelihood of being an autoimmune cause, whereas a case with abnormal findings in EEG showed the opposite trend (Fig. 4A). An increased CSF polymorphonuclear cell ratio and WBC counts, elevated serum ammonia, high WBC counts, neutrophil ratio and maximum body temperature, and a decrease in CSF mononuclear cell ratio, serum platelet counts, large unstained cells (LUC) ratio, and alkaline phosphatase were associated with a higher likelihood of bacterial classification (Fig. 4B). In addition to the history of tuberculosis, an increase in CSF ADA and serum platelet counts as well as a decrease in serum eosinophil ratio and uric acid increased the likelihood of being classified as tuberculosis (Fig. 4C). Lastly, the possibility of viral infection was increased as the CSF mononuclear cell ratio and glucose level, serum activated partial thromboplastin time (aPTT), and total carbon dioxide level increased, and the CSF ADA decreased (Fig. 4D). The SHAP values in the external validation set are presented in Supplementary Fig. S8. Previous tuberculosis history, CSF ADA level, CSF mononuclear leukocyte ratio, and CSF protein level showed the widest horizontal range for aetiology in the external validation dataset, similar to that in the internal dataset.
Fig. 4.
High-ranked Shapley values representing feature importance for each aetiology: (A) autoimmune (B) bacteria (C) tuberculosis and (D) virus. Abbreviations: CSF, Cerebrospinal fluid; EEG, Electroencephalography; PMN, polymorphonuclear; aPTT, Activated partial thromboplastin time.
Discussion
The major findings of the present study were as follows: first, we discovered that a model including the patient's initial 24-h clinical data could confirm the aetiology of meningitis and encephalitis early. Second, the performance metrics of the AI model were higher than those of clinicians. Finally, we identified the variables that were crucial to our classification model. To the best of our knowledge, this study described the first multiclass classification model for confirming the aetiology of meningitis and encephalitis.
Early diagnosis and treatment are essential for the management of meningitis and encephalitis; in actual clinical practice, it is challenging to identify the aetiology using insufficient initial data.1, 2, 3, 4 The classification models for meningitis and encephalitis developed in the present study could help combine the initial clinical features and play an auxiliary role in treatment planning. Previous studies on early diagnosis were limited to predicting CNS infection itself or classifying pathogens into two categories.7,10, 11, 12, 13 In contrast to earlier models, the model presented in this study performed multiclass classification and outperformed earlier models in general.9,21 The model with an F1 score over 0.85 performed well in both the internal test and external validation datasets. The AUROC values of aetiology classes increased in the following order: autoimmunity, bacteria, virus, and tuberculosis. In clinical practice, autoimmune encephalitis is suspected after infectious causes have been ruled out; however, its diagnosis is challenging. Therefore, the AUROC of over 0.85 for autoimmune aetiology has significant clinical value.3,5,7 In clinical practice, when bacterial meningitis is suspected, antibiotics are typically used empirically and continued until negative culture results have been reported after at least 48 h.1 With the model we proposed, if the infection is classified as being caused by other aetiologies, we might be able to make an early decision to discontinue antibiotics without waiting for culture results, thereby preventing potential liver or kidney toxicity caused by antibiotics.8 In addition, we found that the predictions of the multiclassification model were relatively consistent with the estimated diagnoses of neurologists, even in patients where the cause was not clearly identified. However, it should be noted that the true label was based on the estimated causes arrived at by consensus of neurologists using the patient's medical records and the test results, rather than actual tests, and the model only considered information that could be obtained within 24 h, which may have caused some differences. The separate dataset for external validation was developed using patient data from a different institution, and it performed well despite having somewhat different cause distributions or feature types. We found that the model was reproducible and not overfitted. We did not include the survival analysis-related content in this paper because we thought that it did not align with our research goal and the low performance of the model. In future studies, it is expected that a better prognosis prediction model can be created by securing and inputting time-series variables and setting various prognostic factors for patients.
To mitigate possible biases in research using AI, we applied the following methods at each stage.17 During the data collection and preparation stage, we maintained the quality of the training data by K-fold cross validation, and during the model development stage, we used de-biasing techniques such as oversampling to overcome the class imbalance.18 During the model evaluation, we also used relatively simple and interpretable K-Nearest Neighbour and Gaussian Naive Bayes models, and we also used LIME, SHAP, and other methods to verify explainability.19
The results of the present study revealed that the model classified meningitis and encephalitis and outperformed all clinicians. In particular, the difference in predicting ability between the model and the doctor who did not have an experience in neurology was remarkable. The model could classify the aetiology and recommend appropriate and prompt therapy in the absence of a neurologist in a primary care setting, such as an emergency room. Furthermore, the performance of the AI model was even higher than that of senior neurologists, and not just of junior neurologists. In actual clinical practice, doctors assess patients with encephalitis and meningitis through interviews and physical examinations in addition to the variables discussed in this paper. Therefore, the performance of the doctors might have been slightly worse than that in the actual clinical practice. Despite this, the high performance of AI-based predictions for each indicator suggests that AI-supported care in a clinical environment can assist neurologists in making appropriate and prompt decisions regarding treatment. Specifically, by incorporating patient characteristics and test results into the model, the AI model can help with diagnosis and treatment by considering even the items that are difficult for humans to keep track of.
We have identified important features that are AI-driven and clinically validated. For all aetiologies, the CSF test results, including those for WBC count, protein level, glucose level, and ADA level, revealed high PIMP and F scores consistent with previous research.4,5 These variables were also considered significant by the three clinicians who participated in this study when making causal judgments. Upon examining the distribution of variable values for each cause, statistically significant differences were observed in the relevant variables, which could result in their frequent use and high importance in the classification models. The CSF ADA level, which is frequently utilised in clinical settings to distinguish between infection due to tuberculosis and other pathogens, showed the highest feature importance. The presence of ADA is regarded as a hallmark of cell-mediated immunity and is primarily associated with lymphocyte proliferation and differentiation, which can result in varying levels within CSF depending on the cause of inflammation.22 Hyperammonaemia has been reported to be associated with urease-producing pathogens in infectious diseases, although the mechanism is not yet clear. In the present study, serum ammonia level also emerged as an important factor in differentiating pathogens, which could be related to the previously mentioned mechanism.23 The maximum body temperature at 24 h was an important feature for each aetiology classification. Considering the existing literature on fever patterns differing according to the pathogen, advanced research on fever patterns will be helpful in differentiation.24 Activation of platelets is a crucial underlying mechanism of inflammatory diseases. Therefore, platelet count and mean platelet volume have been reported to help differentiate meningitis causes, and they have played a significant role in the classification in the present study.25 The LUC ratio, which has been shown to help distinguish between autoimmune and infectious causes, also showed high importance.26 Furthermore, the serum eosinophil percentage, an important variable for classification, has not been reported to be directly associated with CNS pathologies but is presumed to be related as it is known to play a regulatory role in autoimmune and infectious diseases.27 The delta neutrophil index was found to help differentiate autoimmune, bacterial, and tuberculosis cases in the present study. According to previous studies, the delta neutrophil index can help predict the diagnosis and prognosis of sepsis and is considered a biomarker for autoimmune diseases such as systematic lupus erythematosus and adult-onset Still's disease.28,29 Therefore, it is thought to play a similar role in the case of CNS inflammatory conditions.
We also utilised SHAP values to explain the AI model and determine which variables are considered important in classifying each aetiology. Altered wave patterns in EEG can assist the treatment decision-making process in cases of encephalitis, particularly in the presence of altered consciousness or confusion and seizure attacks.6 Previous reports indicate that EEG abnormalities may present differently depending on the underlying cause and significantly affect autoimmune cause discrimination, and abnormal findings in EEG also showed a high SHAP value in this study.6 Patients with pleocytosis, neutrophilia, and thrombocytopenia had an increased likelihood of being classified as bacterial infection, consistent with clinical trends.1,4 A previous history of tuberculosis and the levels of glucose and ADA in the CSF are known to be important factors in determining tuberculous meningitis, and the same results were obtained with the model used in the present study.22 In cases of viral infections, a localised thrombo-inflammatory response or systemic hypercoagulability may be present. Also, in the present study, patients with increased aPTT had a greater propensity to be classified as having viral infections.30 The model we suggested showed good explanatory power when considering the SHAP values for each cause.
The present study has some limitations. First, we focused on patients with encephalitis and meningitis, and multi-label diagnosis cases were not included because of the few numbers of cases. Second, since the retrospective data were presented in a tabular format, and variables related to the diagnostic tools (brain CT, CXR, and EEG) were entered in a tabular format because the data dimension would be different if the variables were input as raw data. Third, the training and external validation data came across a rather long time; therefore, the reliability of the clinical measures over a long duration could be questionable. Fourth, a small sample size of patients with confirmed aetiology compared to the total number of registered patients may have led to a sampling bias. Moreover, classification of fungal infections could not be performed. Fifth, there may be differences when applying the results of this study to patients of other races as most of the population included in this study are of Asian ethnicity.
This is the first multiclass classification study for the early determination of the aetiology of meningitis and encephalitis based on the initial 24-h data using an AI model, which showed high performance in various evaluation metrics. The results suggest that the application of AI could help determine the aetiology of meningitis and encephalitis, thereby potentially enabling early treatment selection.
Contributors
BKC, KMK and YRP accessed and verified the underlying data. BKC, KMK and YRP conceived and designed the study. BKC, YJC, WSH, MDS and KMK analysed the data and drafted the manuscript. WJK, KH, MKC and YRP acquired the data and provided critical revision of the manuscript. All authors have critically interpreted the results and have read and approved the final manuscript. All authors read and approved the final manuscript. KMK and YRP contributed equally as the co-corresponding authors.
Data sharing statement
The data supporting the findings of this study are available from the corresponding author upon reasonable request. Some of the data may not be posted for policy purposes.
Declaration of interests
We declare no competing interests.
Acknowledgements
This research was supported by a grant of the MD-PhD/Medical Scientist Training Program through the Korea Health Industry Development Institute (KHIDI), funded by the Ministry of Health & Welfare, Republic of Korea. Funding was used for labor costs for data collection and analysis. We would like to thank the MCU (Medical informatics Collaborative Unit) members of Yonsei University College of Medicine for their assistance in data analysis.
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.eclinm.2023.102051.
Contributor Information
Kyung Min Kim, Email: penlighting11@yuhs.ac.
Yu Rang Park, Email: yurangpark@yuhs.ac.
Appendix A. Supplementary data
References
- 1.Van De Beek D., Brouwer M., Hasbun R., Koedel U., Whitney C.G., Wijdicks E. Community-acquired bacterial meningitis. Nat Rev Dis Primers. 2016;2:1171–1183. doi: 10.1038/nrdp.2016.74. [DOI] [PubMed] [Google Scholar]
- 2.Granerod J., Ambrose H.E., Davies N.W.S., et al. Causes of encephalitis and differences in their clinical presentations in England: a multicentre, population-based prospective study. Lancet Infect Dis. 2010;10:835–844. doi: 10.1016/S1473-3099(10)70222-X. [DOI] [PubMed] [Google Scholar]
- 3.Abboud H., Probasco J.C., Irani S., et al. Autoimmune encephalitis: proposed best practice recommendations for diagnosis and acute management. J Neurol Neurosurg Psychiatry. 2021;92:757–768. doi: 10.1136/jnnp-2020-325300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Negrini B., Kelleher K.J., Wald E.R. Cerebrospinal fluid findings in aseptic versus bacterial meningitis. Pediatrics. 2000;105:316–319. doi: 10.1542/peds.105.2.316. [DOI] [PubMed] [Google Scholar]
- 5.Blinder T., Lewerenz J. Cerebrospinal fluid findings in patients with autoimmune encephalitis-a systematic analysis. Front Neurol. 2019;10:804. doi: 10.3389/fneur.2019.00804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sutter R., Kaplan P.W., Cervenka M.C., et al. Electroencephalography for diagnosis and prognosis of acute encephalitis. Clin Neurophysiol. 2015;126:1524–1531. doi: 10.1016/j.clinph.2014.11.006. [DOI] [PubMed] [Google Scholar]
- 7.Xiang Y., Zeng C., Liu B., et al. Deep learning-enabled identification of autoimmune encephalitis on 3D multi-sequence MRI. J Magn Reson Imaging. 2022;55:1082–1092. doi: 10.1002/jmri.27909. [DOI] [PubMed] [Google Scholar]
- 8.Ackley E.R., Tchou M.J., Press C.A., et al. AKI in suspected meningitis/encephalitis may be “avoidable kidney injury”. Hosp Pediatr. 2021;11:e167–e169. doi: 10.1542/hpeds.2020-005770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zellweger R.M., Yacoub S., Chan Y.F.Z., et al. Disentangling etiologies of CNS infections in Singapore using multiple correspondence analysis and random forest. Sci Rep. 2020;10 doi: 10.1038/s41598-020-75088-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jeong Y.S., Jeon M., Park J.H., et al. Machine-learning-based approach to differential diagnosis in tuberculous and viral meningitis. Infect Chemother. 2021;53:53–62. doi: 10.3947/ic.2020.0104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Babenko D., Seidullayeva A., Bayesheva D., et al. Ability of procalcitonin and C-reactive protein for discriminating between bacterial and enteroviral meningitis in children using decision tree. Biomed Res Int. 2021;2021 doi: 10.1155/2021/5519436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.D'Angelo G., Pilla R., Tascini C., Rampone S. A proposal for distinguishing between bacterial and viral meningitis using genetic programming and decision trees. Soft Comput. 2019;23:11775–11791. [Google Scholar]
- 13.Guzmán E., Belmonte M.V., Lelis V.M. Ensemble methods for meningitis aetiology diagnosis. Expert Syst. 2022;39 [Google Scholar]
- 14.Glasheen W.P., Cordier T., Gumpina R., Haugh G., Davis J., Renda A. Charlson comorbidity index: ICD-9 update and ICD-10 translation. Am Health Drug Benefits. 2019;12:188–197. [PMC free article] [PubMed] [Google Scholar]
- 15.O'Brien R.M. A caution regarding rules of thumb for variance inflation factors. Qual Quant. 2007;41:673–690. [Google Scholar]
- 16.Vokinger K.N., Feuerriegel S., Kesselheim A.S. Mitigating bias in machine learning for medicine. Commun Med. 2021;1:25. doi: 10.1038/s43856-021-00028-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lemaître G., Nogueira F., Aridas C.K. Imbalanced-learn: a python toolbox to tackle the curse of imbalanced datasets in machine learning. J Mac Learn Res. 2017;18:1–5. [Google Scholar]
- 18.Giuste F., Shi W., Zhu Y., et al. Explainable artificial intelligence methods in combating pandemics: a systematic review. IEEE Rev Biomed Eng. 2023;16:5–21. doi: 10.1109/RBME.2022.3185953. [DOI] [PubMed] [Google Scholar]
- 19.Chen T., Guestrin C. Proceedings of the 22nd ACM SIGKDD international conference on knowledge discovery and data mining. 2016. XGBoost: a scalable tree boosting system; pp. 785–794. [Google Scholar]
- 20.Balki I., Amirabadi A., Levman J., et al. Sample-size determination methodologies for machine learning in medical imaging research: a systematic review. Can Assoc Radiol J. 2019;70:344–353. doi: 10.1016/j.carj.2019.06.002. [DOI] [PubMed] [Google Scholar]
- 21.van Zeggeren I.E., Bijlsma M.W., Tanck M.W., van de Beek D., Brouwer M.C. Systematic review and validation of diagnostic prediction models in patients suspected of meningitis. J Infect. 2020;80:143–151. doi: 10.1016/j.jinf.2019.11.012. [DOI] [PubMed] [Google Scholar]
- 22.Rock R.B., Olin M., Baker C.A., Molitor T.W., Peterson P.K. Central nervous system tuberculosis: pathogenesis and clinical aspects. Clin Microbiol Rev. 2008;21:243–261. doi: 10.1128/CMR.00042-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hannah W.B., Nizialek G., Dempsey K.J., Armitage K.B., McCandless S.E., Konczal L.L. A novel cause of emergent hyperammonemia: cryptococcal fungemia and meningitis. Mol Genet Metab Rep. 2021;29 doi: 10.1016/j.ymgmr.2021.100825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ogoina D. Fever, fever patterns and diseases called ‘fever’ - a review. J Infect Public Health. 2011;4:108–124. doi: 10.1016/j.jiph.2011.05.002. [DOI] [PubMed] [Google Scholar]
- 25.Cámara-Lemarroy C.R., Delgado-Garcia G., De La Cruz-Gonzalez J.G., Villarreal-Velazquez H.J., Gongora-Rivera F. Mean platelet volume in the differential diagnosis of tuberculous and bacterial meningitis. J Infect Dev Ctries. 2017;11:166–172. doi: 10.3855/jidc.7478. [DOI] [PubMed] [Google Scholar]
- 26.Vanker N., Ipp H. Large unstained cells: a potentially valuable parameter in the assessment of immune activation levels in HIV infection. Acta Haematol. 2014;131:208–212. doi: 10.1159/000355184. [DOI] [PubMed] [Google Scholar]
- 27.Gaur P., Zaffran I., George T., Rahimli Alekberli F., Ben-Zimra M., Levi-Schaffer F. The regulatory role of eosinophils in viral, bacterial, and fungal infections. Clin Exp Immunol. 2022;209:72–82. doi: 10.1093/cei/uxac038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Seok Y., Choi J.R., Kim J., et al. Delta neutrophil index: a promising diagnostic and prognostic marker for sepsis. Shock. 2012;37:242–246. doi: 10.1097/SHK.0b013e3182454acf. [DOI] [PubMed] [Google Scholar]
- 29.Li H., Yang P. Identification of biomarkers related to neutrophils and two molecular subtypes of systemic lupus erythematosus. BMC Med Genomics. 2022;15:162. doi: 10.1186/s12920-022-01306-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Iba T., Levy J.H., Levi M. Viral-induced inflammatory coagulation disorders: preparing for another epidemic. Thromb Haemost. 2021;122:8–19. doi: 10.1055/a-1562-7599. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.