Abstract
Two-dimensional drawing of nucleic acid structures, particularly RNA structures, is fundamental to the communication of nucleic acids research. However, manually drawing structures is laborious and infeasible for structures thousands of nucleotides long. RNAcanvas automatically arranges residues into strictly shaped stems and loops while providing robust interactive editing features, including click-and-drag layout adjustment. Drawn elements are highly customizable in a point-and-click manner, including colours, fonts, size and shading, flexible numbering, and outlining of bases. Tertiary interactions can be drawn as draggable, curved lines. Leontis-Westhof notation for depicting non-canonical base-pairs is fully supported, as well as text labels for structural features (e.g. hairpins). RNAcanvas also has many unique features and performance optimizations for large structures that cannot be correctly predicted and require manual refinement based on the researcher's own analyses and expertise. To this end, RNAcanvas has point-and-click structure editing with real-time highlighting of complementary sequences and motif search functionality, novel features that greatly aid in the identification of putative long-range tertiary interactions, de novo analysis of local structures, and phylogenetic comparisons. For ease in producing publication quality figures, drawings can be exported in both SVG and PowerPoint formats. URL: https://rnacanvas.app.
Graphical Abstract
Graphical Abstract.
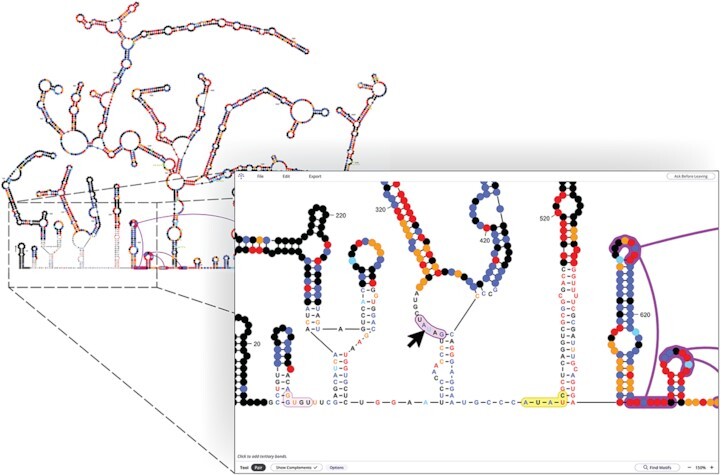
Exploring the 2,692 nt citrus yellow vein-associated virus genome structure in RNAcanvas. Currently selected sequence highlighted yellow. RNAcanvas has highlighted two putative tertiary pairings purple (hovered by cursor) and pink.
INTRODUCTION
In nucleic acids research, particularly RNA research, structures are commonly drawn in 2-dimensional (2D) format (1–3). Conventions such as the Leontis-Westhof notation (4) are also important for denoting various types of canonical and non-canonical base-pairs and interactions in RNA structures. Since manually creating 2D structure drawings is laborious for large structures (5–7), a variety of software programs have been developed to facilitate this process.
2D nucleic acid structure drawing programs can be roughly separated into two categories: (i) programs that offer flexible, force-based adjustment of drawing layouts and (ii) programs that generally apply stricter layout conventions to drawings. Programs in the first category, including forna (8) and RiboSketch (9), allow click-and-drag editing of drawings and use force-based methods (8) to help maintain the arrangement of residues into stems and loops in response to dragging. The interactivity features in this class of programs make editing drawings intuitive, and their use of force-based methods allow for great flexibility in how a drawing is arranged. These programs, however, require increased effort to (for example): (i) precisely align stems emanating from a junction or that are stacked; (ii) flatten or evenly round regions of unpaired residues and (iii) align stems with the flat base of a drawing.
Programs in the second category, including VARNA (10), XRNA (http://rna.ucsc.edu/rnacenter/xrna/xrna_faq.html), PseudoViewer (11), RnaViz (12), jViz.RNA (13), RNAView (14), RNApuzzler (15), R2R (16), R2DT (17) and RNArtist (https://github.com/fjossinet/RNArtist), generally produce drawings in a more regimented, publication-friendly manner that can more efficiently communicate secondary structure and facilitate phylogenetic comparisons (2,15–18). Importantly, programs in this category have greater functionality for precisely arranging stems and loops in a strict manner (e.g. perfectly round or flattened loops), making them preferred for the drawing of large structures, which benefit from stricter layout organization (15,17). These programs also introduce additional methods for specifying the aspects of a drawing, such as program-specific input file formats (e.g. R2R, RNArtist) and layout templates (e.g. R2DT), and also tend to offer greater customization of individual drawing elements including bases and bond notations (e.g. Leontis-Westhof). On the other hand, features for interactivity, such as click-and-drag editing of residue layout and the shapes of tertiary interaction lines, are often lacking or limited in functionality. This category of programs may also lack point-and-click editing of base-pairs and other drawing elements, thus requiring input files or application forms for edits, where bases and other elements are specified by numeric position making editing drawings cumbersome and less intuitive.
Nucleic acid structure drawings are rarely static but change as substructures are confirmed using biochemical methods or through phylogenetic comparisons, and thus drawings should also be easily editable to reflect a researcher's current knowledge. Towards this goal, RNAcanvas was designed to combine some of the best features of both types of structure drawing programs, merging the interactivity functions of the first category (e.g. click-and-drag layout adjustments) with the orderly layouts (e.g. residues maintained in strictly shaped stems and loops) and detailed drawing customizations of the second category. Furthermore, RNAcanvas provides extensive point-and-click editing features for individual components of the drawing, such as base letters, lines of bonds, and base outlines and numberings. RNAcanvas is also one of the few programs to fully support Leontis-Westhof notation for depicting canonical and non-canonical RNA base-pairs and interactions. RNAcanvas has been optimized to function smoothly with large structures, and contains unique features such as real-time highlighting of complementary sequences, motif search, and export of drawings in both scalable vector graphics (SVG) and PowerPoint (PPTX) formats, which greatly facilitates the creation of figures for scientific presentations. Exported SVG and PPTX drawings are also formatted such that individual objects (e.g. base letters, bond lines) can be altered in vector graphics editors such as Adobe Illustrator or PowerPoint. RNAcanvas was formerly named RNA2Drawer, which was initially available only in a download format when first published (19).
MATERIALS AND METHODS
Versions of software tools used for RNAcanvas are currently the most recent (2023), however continuous enhancements to the program to provide additional functionality and updating of tools will continue.
Programming languages and general tools
RNAcanvas is a single-page web application created with HTML/CSS/JavaScript. Babel v7.16.10 is used to compile JavaScript code to the ECMAScript 2009 (ES5) standard. The application is built using the webpack module bundler v4.46.0. TypeScript v3.9.10 is used to add type annotations over JavaScript code. All JavaScript application code to be received by the user is written with TypeScript type annotations. Unit testing is performed on the Node.js runtime environment v16.18.1 and using the Jest framework v24.9.0. The uuid library v8.3.2 is used to generate universally unique identifiers (UUIDs), which are assigned to a variety of components (e.g. text elements of bases) to simplify retrieval in the application code.
Interactive drawing
Within the application, the drawing itself is a scalable vector graphics (SVG) document, which is a type of element that web browsers can render. Web browsers provide built-in methods for detecting user interaction with SVG documents (e.g. mouse clicks, hovering and dehovering of elements within the SVG document). This allows user interaction with the drawing to be programmed in an event-driven manner. The SVG specification is mature and extensive, allowing additional customization of drawings. The SVG.js library v3.1.1 is also used to simplify code for manipulating the drawing and managing user interactions.
Peripheral UI elements
Peripheral user interface (UI) elements, such as the top menu, bottom information bar, and right-side forms are built using the React framework v16.14.0. The react-color library v2.19.3 is used for the colour picker UI element. The react-select library v3.2.0 is used for the UI element for picking fonts. The React Testing Library v9.5.0 and the enzyme library v3.11.0 are used for unit testing of peripheral UI elements.
Exporting drawings
The SVG.js library v3.1.1 is used to produce SVG files of drawings. (SVG files contain the textual representation of an SVG document, such as the drawing of the application.) To produce PowerPoint files of drawings, the PptxGenJS library v3.9.0 is used. Rather than convert all the SVG elements of a drawing to their nearest PowerPoint object counterpart, many drawing elements (e.g. the curved lines of tertiary bonds) are included in PPTX files as individual SVG image elements. Because of this, exported PPTX files require PowerPoint version 2016 or later. SVG image elements in exported PPTX files can still be manipulated as though they were PowerPoint objects and can often be converted to corresponding PowerPoint objects using the PowerPoint ‘Convert to Shape’ feature.
RESULTS
RNAcanvas offers extensive options to customize drawings of nucleic acid secondary structures and tertiary interactions based on graphical user interaction, with drawings edited using ‘tools’ and forms. Tools control how the user interacts with the drawing itself. For example, different tools allow the user to drag elements of the drawing, select element(s) for editing, or pair and unpair bases. Forms pop-up on the right-side of the application and allow properties of selected elements (or the drawing as a whole) to be directly edited through text and numeric inputs and various picker components and toggles (e.g. letters and fonts of bases, colours of bonds, base numbering). All structure drawing figures in this report were produced entirely within the RNAcanvas web app (unless stated otherwise).
Layout flexibility
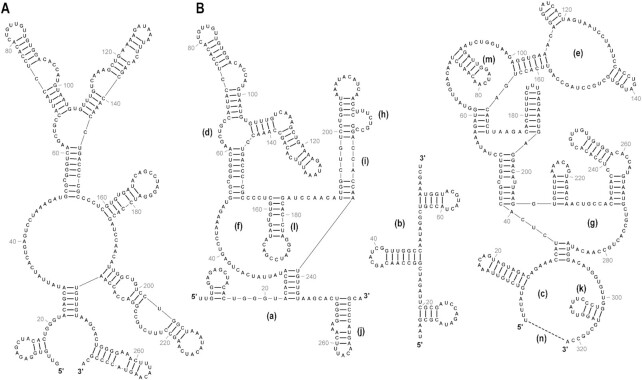
Using the Dragging Tool, the layout of a drawing can be adjusted by dragging with the mouse (Figure 1). Stems can be dragged around loops and the outermost loop can be dragged to rotate the drawing with the arrangement of bases into stems and loops strictly maintained. Using the Flattening Tool, individual loops can be flattened (or reverted to unflattened). When flattened, the outermost loop is entirely flat (Figure 1B, a-c), and inner loops with more than one child stem assume a triangular shape (Figure 1B, d–g). Flattening inner loops with multiple child stems often helps to condense a drawing and highlight conserved branching patterns in structures. Flattening an inner loop with only one child stem (Figure 1B, h and i) aligns the child stem with the parent stem, effectively ‘straightening’ the two stems. Using the Flipping Tool, individual stems can be flipped across their parent loops (Figure 1B, j–m). RNAcanvas also has a Layout form in which the rotation of the drawing can be precisely specified. The Layout form also has a field for specifying the termini gap of the drawing, which is the distance between the first and last bases when the outermost loop of a drawing is round (Figure 1B, label n and dashed line). The termini gap can be set to zero (for a fully circular outermost loop) or to larger values (e.g. for a semicircle outermost loop).
Figure 1.
Various layout options for drawings in RNAcanvas. (A) The same input structure as in (B, left) before any layout adjustments save for being rotated 90°. (B) Labels a-n and dashed line (lower right) added in Adobe Illustrator. (a) Flat outermost loop. (b) Rotated flat outermost loop. (c) Round outermost loop. (d, e) Round inner loops. (f, g) Flattened inner loops with multiple child stems. (h) Flattened inner loop with only one child stem that has been condensed. (i) Flattened inner loop with only one child stem that has been stretched. (j-m) Flipped stems. (n) Termini gap of the round outermost loop (indicated by dashed line).
Customization of drawing elements
The following customizations can be accomplished using the Editing Tool:
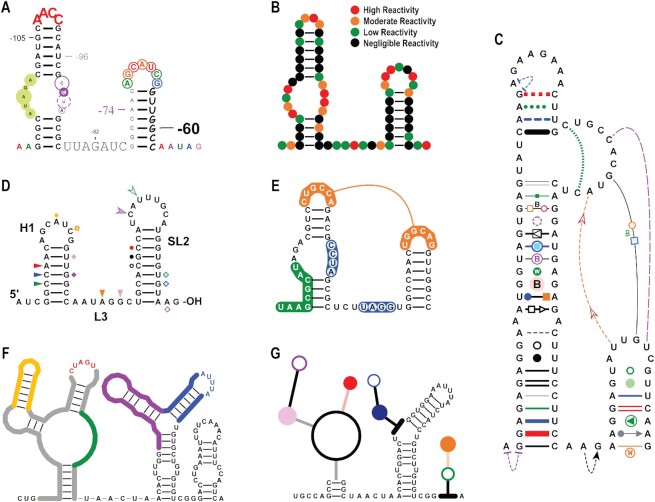
Base customizations. Fonts, font sizes and base colours can be personalised (Figure 2A). Bases can be outlined, and outlines can be adjusted for size, line thickness and line and fill colours and transparencies. Chemical probing data such as SHAPE data (20) can be easily incorporated into the structure through base and/or outline colouring. A ‘dots’ style drawing, in which only the colour-coded outlines of bases are shown, is an effective way to communicate chemical probing data (Figure 2B) and dots can be easily suppressed to reveal the underlying bases. RNAcanvas has a dedicated form to help with styling bases according to a set of data. This form allows the user to progressively select and style bases within different data ranges (e.g. low, moderate, and high SHAPE reactivities). Base letters can be directly edited, and bases can subsequently be inserted, appended, and removed. A substructure can be specified when inserting/appending a subsequence of bases. The numbering of bases can also be specified via the offset, increment and anchor properties in the Numbering form, with text and lines customizable in terms of fonts, colour, dimensions, etc. (Figure 2A). Individual bases can also be manually numbered regardless of what numbers are assigned to other bases in the drawing.
Bond customizations. In a drawing, consecutive bases are linked by a primary bond. Most primary bonds are not visible (by default) when a structure is initially drawn. Secondary bonds join base-pairs in the secondary structure of a drawing and affect the layout of the drawing. Tertiary bonds communicate base-pairs/interactions that do not affect the layout of the drawing. All bonds have a line that connects the two bases in the bond. Primary and secondary bonds have straight lines. Tertiary bonds have curved lines that can be dragged with the mouse to adjust their shape. The lines of bonds can be customized in terms of colour and thickness, as well as base padding, which is the distance between the end of a bond line and the base connected by the bond (Figure 2C). Bond lines can be dashed and their ends rounded.
Figure 2.
Various customizations of drawing elements. (A) Fonts, font sizes and colours of bases text are all customizable. Bases can be outlined with circles and numbered. Base outlines and numberings are also customizable in terms of colours, dimensions, fonts, etc. This drawing has a base numbering offset of –121 nt. (B) Drawing presented in ‘dots’ style in which only colour-coded outlines of bases are shown. Dots style is an effective way to communicate chemical probing data, such as the example SHAPE data shown in this drawing. Dots colour legend added in Adobe Illustrator. (C) Various customizations of bond lines and ‘strung’ elements. Entire Leontis-Westhof notation for depicting canonical and non-canonical base-pairs and interactions is supported. (D) Text labels and base markers. Hairpin H1, stem–loop SL2, linker region L3, the 5′end and the 3′OH were all labelled within the program. Bases also can have various markers (i.e. coloured triangles, squares, circles) associated with them. These text labels and base markers are ‘strung’ elements of nearby primary and secondary bonds that have been dragged to their current positions. Due to being strung elements, these text labels and base markers maintain both their positions and orientations relative to their parent bonds when the layout of the drawing is adjusted. (E) Shading sequences of bases. Tertiary pairing connected by a single tertiary bond is shown (orange). (F) Drawing with portions outlined. (G) Drawing with portions in the ‘stick-and-ball’ style.
Elements such as rectangles, circles, triangles, and text can be ‘strung’ onto (associated with) any bond (Figure 2C). This attachment of elements to specific bonds allows for the full Leontis-Westhof notation (4) for canonical and non-canonical base-pairs and interactions to be depicted on secondary and tertiary bonds. As with other drawing elements, strung elements can be customized in terms of colour, transparencies, dimensions, line dashing, fonts, etc. Strung elements can be dragged with the mouse and displaced from their parent bond, which adds versatility to their usage. For example, when dragged next to nearby bases, strung elements can mark the location with additional information (Figure 2D) or can be used to shade hairpins. Text strung elements can be used to name structural features as well as the 5′ and 3′ termini (Figure 2D). All strung elements maintain their position and orientation relative to their parent bond when the layout of the drawing is adjusted.
Shading a series of bases and outlining structures. Highlighting a selection of bases can be accomplished by: (i) setting primary bond base paddings to zero; (ii) making primary bonds sufficiently thick to encompass the font size of the bases; (iii) setting the line end caps of the primary bonds to round and (iv) sending the shading below the bases (Figure 2E). Tertiary pairings can also be denoted by shading the two interacting sequences and connecting one base of each sequence with a single tertiary bond (Figure 2E, in orange). It is also possible to outline structures in a drawing by shading bases using this technique and then sending base letters below the shading (Figure 2F).
‘Stick-and-ball’ drawings. ‘Stick-and-ball’ drawings (Figure 2G) can be created by making base letters invisible (e.g. by assigning them the background colour), increasing the base padding of secondary bonds to shorten them, and making secondary bonds sufficiently thick to overlap with each other (thus forming the ‘stick’ portion). The ‘ball’ portion is composed of circle strung elements that are dragged over the loops in the structure and sized to match the sizes of the loops.
Structure editing and large structure exploration
Structures thousands of nucleotides long, such as the structures of positive-strand RNA virus genomes, are increasingly studied as a whole (5,6,21). Such large structures cannot be predicted directly (5) and require manual refinement by RNA researchers based on phylogenetic comparisons, experimental results, and the researcher's own expertise. RNAcanvas has optimized performance for editing large structures, e.g. when dragging to adjust the layout.
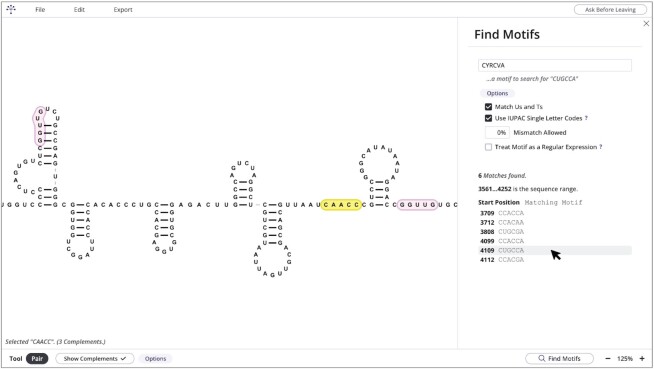
The Pairing Tool is used to pair and unpair bases (i.e. add and remove secondary and tertiary bonds) in a point-and-click manner. The Pairing Tool also has the option of highlighting sequences that are complementary to the selected sequence (Figure 3, selected sequence highlighted in yellow and complementary sequences highlighted in pink). The number of possible pairing partners within the structure can be designated, and options are available to exclude G-U/G-T pairs and permit partial mismatches between pairing partners. The Pairing Tool aids in refinement of output by structure prediction programs such as Mfold (22) and RNAfold (23) and helps to identify possible long-range base-pairings that cannot currently be predicted computationally (5). RNAcanvas also has a form for finding input motifs (Find Motifs form) (Figure 3). Both complement highlighting (using the Pairing Tool) and the Find Motifs form support partial mismatching and the use of IUPAC single letter codes (24). Complement highlighting further supports a percentage cap on the G-U/G-T base-pair composition of highlighted complements. The Find Motifs form can also treat an input motif to search for as a regular expression (as defined in Computer Science).
Figure 3.
Screenshot showing highlighting of complementary sequences with the Pairing Tool and the Find Motifs form. The 5′CAACC sequence highlighted in yellow is currently selected. Complementary sequences 5′GGUUG found by the program are highlighted in pink. (Right) Find Motifs form is open. Currently the options to match Us and Ts and to use IUPAC single letter codes are toggled. The motif 5′CYRCVA (containing the IUPAC single letter codes Y, R and V) was searched for and six matching motifs were found. The second matching motif from the bottom is being hovered with the mouse cursor (highlighted in grey). Clicking on a matching motif will centre the matching motif on the screen and flash its base letters.
Input and output formats
RNAcanvas accepts primary sequences (unstructured) as well as structures in dot-bracket notation (also called Vienna format) and in Connectivity Table (CT) files. Input structures may contain pseudoknots, which are drawn using tertiary bonds. RNAcanvas outputs drawings in SVG and PowerPoint formats. SVG files can be opened in vector graphics editors such as Adobe Illustrator and Inkscape. Drawings are exported such that all drawing elements (e.g. bases text, bond lines) are exported as individual SVG or PowerPoint objects. For instance, base letters are exported as SVG text elements or PowerPoint text boxes. This allows for further manipulation of exported drawings in vector graphics editors and PowerPoint. A scaling factor may also be specified when exporting a drawing. All elements in an exported drawing are individually scaled according to the scaling factor. Scaling elements individually on export eases the manipulation of exported drawings compared to when exported drawings are scaled as a whole after export using a separate application such as Adobe Illustrator. Additionally, RNAcanvas can save drawings in a file format unique to RNAcanvas with ‘.rnacanvas’ extension. Downloaded drawing files contain a complete representation of a drawing, allowing drawings to be reopened at later times exactly as they were when saved.
DISCUSSION
RNAcanvas has many enhancements over the free desktop-based RNA2Drawer predecessor (19). Being web-based, RNAcanvas is easier for users to access and is still available for use at no cost. The RNAcanvas web app also uses the SVG web standard to present drawings, which is expansive and permits wide customization of drawings. Other improvements in the web app include unlimited undo/redo functionality, greater layout flexibility (e.g. the ability to flatten and circularize loops individually), the availability of strung elements on bonds, support for Leontis-Westhof notation, ability to add text labels and base markers, enhanced flexibility in numbering bases, and the Find Motifs form.
Although RNAcanvas offers flexible options to customize the layout of a drawing (Figure 1), the program does not currently support certain layout features. For instance, RNAcanvas cannot currently incorporate pseudoknots into the layout of a drawing, unlike some programs such as PseudoViewer (Table 1) (11). Certain types of structures such as frameshifting elements (25) are often drawn such that pseudoknot(s) are conveyed via the layout of the drawing. The initial layouts of drawings made by RNAcanvas (especially for large structures) can also contain overlapping regions that need to be manually untangled by the user. While drawings produced by many programs often contain overlapping regions (17), some programs such as RNApuzzler/RNAturtle have made algorithmic advancements in the initial layout of an RNA drawing to avoid overlaps (15). R2DT has also demonstrated the effectiveness of template-based drawing for a wide array of small and large structures (17). Programs such as PseudoViewer, RNApuzzler and R2DT, however, do not provide the expansive features of RNAcanvas for editing drawings, such as click-and-drag layout adjustment and point-and-click editing of the structure and individual drawing elements, which allow structure drawings to be moulded beyond their initial forms. RNAcanvas is also unique in its focus on the extended exploration of structures and the continued editing of drawings that occurs as the researcher's knowledge of a structure grows.
Table 1.
Comparison of programs for 2D drawing of nucleic acid structures. Solid dots indicate that a feature is supported by a program, though the robustness of feature implementations may vary between different programs. VARNA and jViz.RNA have hollow dots for web-based use since they require a Java plugin for use in a web browser. Point-and-click structure editing refers to the ability to add and remove base-pairs and interactions in the secondary and tertiary structures of a drawing graphically using the mouse. Motif finding functionality refers to features similar to the Find Motifs form in RNAcanvas. Point-and-click element editing is the ability to select individual or groups of elements to edit by clicking on them and/or dragging a selecting box over them, rather than being required to select elements by less direct means (e.g. by inputting numeric positions). Text labels for structural features refer to the ability to place pieces of text next to structural features such as hairpins and linker regions that name them. Flexible base numbering is the ability to assign any number to any base regardless of what numbers are assigned to other bases. This is useful when a drawing contains two discontinuous portions of a larger sequence and the numbering for one portion must be offset from the other. Granular output scaling refers to the ability to scale drawings at the level of individual elements when exported, as opposed to scaling an exported drawing as a whole after export. Drawing using input files and automated drawing are grouped together since the use of input files can lend itself to automated drawing via the automated creation of input files. Automated drawing also includes the ability to generate large numbers of structure drawings in an automated way in general, such as by allowing other software written by the user to interface with the structure drawing software, such as with VARNA
| RNAcanvas | VARNA | XRNA | PseudoViewer | RnaViz | jViz.RNA | forna | RiboSketch | RNAView | R2R | R2DT | RNArtist | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Web-Based | • | ○ | • | • | ○ | • | • | • | • | |||
| Click-and-Drag Layout Adjustment | • | • | • | • | • | • | • | |||||
| Force-Based Drawing Layouts | • | • | • | |||||||||
| Draggable, Curved Tertiary Bonds | • | |||||||||||
| Incorporation of Pseudoknots into the Layout | • | • | • | • | • | • | • | • | ||||
| Point-and-Click Structure Editing | • | • | • | • | • | • | • | |||||
| Real-Time Highlighting of Complementary Sequences | • | |||||||||||
| Motif Finding Functionality | • | |||||||||||
| Point-and-Click Element Editing | • | • | • | • | • | • | ||||||
| Leontis-Westhof Notation | • | • | • | • | • | • | ||||||
| Text Labels for Structural Features | • | • | • | |||||||||
| Flexible Base Numbering | • | |||||||||||
| SVG Output | • | • | • | • | • | • | • | • | • | |||
| PPTX Output | • | |||||||||||
| Granular Output Scaling | • | |||||||||||
| Raster Image Output (e.g. PNG, TIFF, JPEG) | • | • | • | • | • | • | • | • | • | • | ||
| Template-Based Drawing | • | • | ||||||||||
| Drawing Using Input Files and Automated Drawing | • | • | • | • | • | • |
RNAcanvas is able to handle large (at least 3000 nt) structures with only modest lag times, making it significantly faster than other drawing platforms with similar click-and-drag layout adjustment. The primary performance bottleneck of RNAcanvas is the re-rendering of base letters when the layout of a drawing is changed. Note that among Chrome, Firefox and Safari, the Chrome web browser currently has the best performance when handling large RNAcanvas structures. Many biological transcripts, such as positive-strand RNA virus genomes and long noncoding RNAs, have lengths over 3 kb and thus future performance improvements will need to include new strategies for minimizing lag times when dragging portions of very large structures. Ultimately, RNAcanvas aspires to allow entire structures of any size biological transcript to be effectively drawn and edited on personal devices.
DATA AVAILABILITY
RNAcanvas web app: https://rnacanvas.app. RNAcanvas GitHub repository: https://github.com/pzhaojohnson/rnacanvas and https://doi.org/10.6084/m9.figshare.22134029.v1.
ACKNOWLEDGEMENTS
We would like to thank Dr Bruce Shapiro and Dr Wojciech Kasprzak for their invaluable help in developing RNA2Drawer, the predecessor to RNAcanvas, and all of the advice that they have provided over the years that has propelled this project forward.
Contributor Information
Philip Z Johnson, Department of Cell Biology and Molecular Genetics, University of Maryland, College Park, MD, 20742, USA.
Anne E Simon, Department of Cell Biology and Molecular Genetics, University of Maryland, College Park, MD, 20742, USA.
FUNDING
National Science Foundation [MCB-1818229]; United States Department of Agriculture [308291-00001 to A.E.S.]; P.Z.J. was partially supported by National Science Foundation Graduate Fellowship Award [DGE-1840340]. Funding for open access charge: USDA [308291-00001].
Conflict of interest statement. None declared.
REFERENCES
- 1. Ponty Y., Leclerc F.. Drawing and editing the secondary structure(s) of RNA. Methods Mol. Biol. 2015; 1269:63–100. [DOI] [PubMed] [Google Scholar]
- 2. Shabash B., Wiese K.C.. RNA visualization: relevance and the current state-of-the-art focusing on pseudoknots. IEEE/ACM Trans. Comput. Biol. Bioinform. 2017; 14:696–712. [DOI] [PubMed] [Google Scholar]
- 3. Vicens Q., Kieft J.S.. Thoughts on how to think (and talk) about RNA structure. Proc. Natl. Acad. Sci. U.S.A. 2022; 119:e2112677119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Leontis N.B., Westhof E.. Geometric nomenclature and classification of RNA base pairs. RNA. 2001; 7:499–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lu Z., Chang H.Y.. The RNA base-pairing problem and base-pairing solutions. Cold Spring Harb. Perspect. Biol. 2018; 10:a034926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Liu J., Carino E., Bera S., Gao F., May J.P., Simon A.E.. Structural analysis and whole genome mapping of a new type of plant virus subviral RNA: umbravirus-like associated RNAs. Viruses. 2021; 13:646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chkuaseli T., White K.A.. Activation of viral transcription by stepwise largescale folding of an RNA virus genome. Nucleic Acids Res. 2020; 48:9285–9300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kerpedjiev P., Hammer S., Hofacker I.L.. Forna (force-directed RNA): simple and effective online RNA secondary structure diagrams. Bioinformatics. 2015; 31:3377–3379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lu J.S., Bindewald E., Kasprzak W.K., Shapiro B.A.. RiboSketch: versatile visualization of multi-stranded RNA and DNA secondary structure. Bioinformatics. 2018; 34:4297–4299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Darty K., Denise A., Ponty Y.. VARNA: interactive drawing and editing of the RNA secondary structure. Bioinformatics. 2009; 25:1974–1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Byun Y., Han K.. PseudoViewer3: generating planar drawings of large-scale RNA structures with pseudoknots. Bioinformatics. 2009; 25:1435–1437. [DOI] [PubMed] [Google Scholar]
- 12. De Rijk P., Wuyts J., De Wachter R.. RnaViz 2: an improved representation of RNA secondary structure. Bioinformatics. 2003; 19:299–300. [DOI] [PubMed] [Google Scholar]
- 13. Shabash B., Wiese K.C.. jViz.RNA 4.0-Visualizing pseudoknots and RNA editing employing compressed tree graphs. PLoS One. 2019; 14:e0210281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yang H., Jossinet F., Leontis N., Chen L., Westbrook J., Berman H., Westhof E.. Tools for the automatic identification and classification of RNA base pairs. Nucleic Acids Res. 2003; 31:3450–3460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wiegreffe D., Alexander D., Stadler P.F., Zeckzer D. RNApuzzler: efficient outerplanar drawing of RNA-secondary structures. Bioinformatics. 2019; 35:1342–1349. [DOI] [PubMed] [Google Scholar]
- 16. Weinberg Z., Breaker R.R.. R2R–software to speed the depiction of aesthetic consensus RNA secondary structures. BMC Bioinf. 2011; 12:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sweeney B.A., Hoksza D., Nawrocki E.P., Ribas C.E., Madeira F., Cannone J.J., Gutell R., Maddala A., Meade C.D., Williams L.D.et al.. R2DT is a framework for predicting and visualising RNA secondary structure using templates. Nat. Commun. 2021; 12:3494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Muller G., Gaspin C., Etienne A., Westhof E.. Automatic display of RNA secondary structures. Comput. Appl. Biosci. 1993; 9:551–561. [DOI] [PubMed] [Google Scholar]
- 19. Johnson P.Z., Kasprzak W.K., Shapiro B.A., Simon A.E.. RNA2Drawer: geometrically strict drawing of nucleic acid structures with graphical structure editing and highlighting of complementary subsequences. RNA Biol. 2019; 16:1667–1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wilkinson K.A., Merino E.J., Weeks K.M.. Selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE): quantitative RNA structure analysis at single nucleotide resolution. Nat. Protoc. 2006; 1:1610–1616. [DOI] [PubMed] [Google Scholar]
- 21. Ziv O., Price J., Shalamova L., Kamenova T., Goodfellow I., Weber F., Miska E.A.. The short- and long-range RNA-RNA interactome of SARS-CoV-2. Mol. Cell. 2020; 80:1067–1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003; 31:3406–3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lorenz R., Bernhart S.H., Höner Zu Siederdissen C., Tafer H., Flamm C., Stadler P.F., Hofacker I.L.. ViennaRNA Package 2.0. Algorithms Mol. Biol. 2011; 6:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Cornish-Bowden A. Nomenclature for incompletely specified bases in nucleic acid sequences: recommendations 1984. Nucleic Acids Res. 1985; 13:3021–3030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Firth A.E., Brierley I.. Non-canonical translation in RNA viruses. J. Gen. Virol. 2012; 93:1385–1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
RNAcanvas web app: https://rnacanvas.app. RNAcanvas GitHub repository: https://github.com/pzhaojohnson/rnacanvas and https://doi.org/10.6084/m9.figshare.22134029.v1.