Abstract
MiniPromoters, or compact promoters, are short DNA sequences that can drive expression in specific cells and tissues. While broadly useful, they are of high relevance to gene therapy due to their role in enabling precise control of where a therapeutic gene will be expressed. Here, we present OnTarget (http://ontarget.cmmt.ubc.ca), a webserver that streamlines the MiniPromoter design process. Users only need to specify a gene of interest or custom genomic coordinates on which to focus the identification of promoters and enhancers, and can also provide relevant cell-type-specific genomic evidence (e.g. accessible chromatin regions, histone modifications, etc.). OnTarget combines the provided data with internal data to identify candidate promoters and enhancers and design MiniPromoters. To illustrate the utility of OnTarget, we designed and characterized two MiniPromoters targeting different cell populations relevant to Parkinson Disease.
Graphical Abstract
Graphical Abstract.
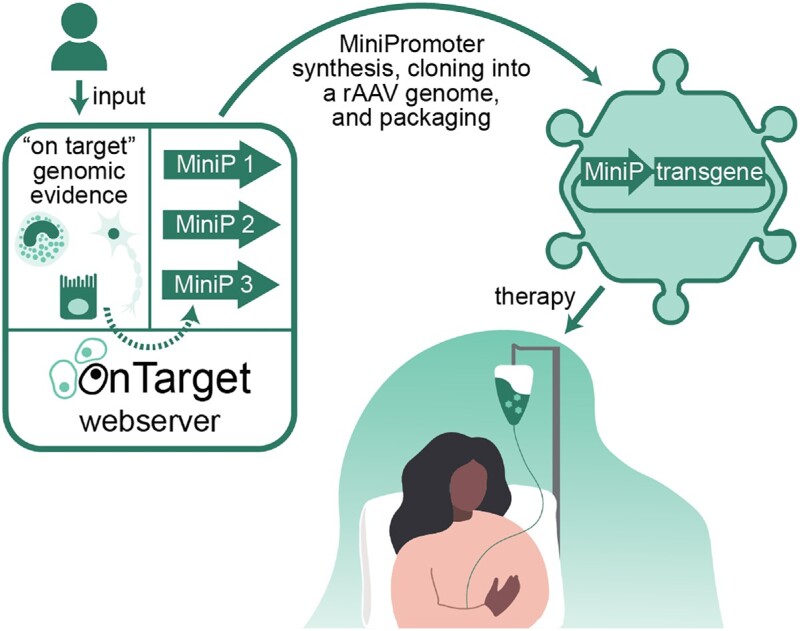
OnTarget (http://ontarget.cmmt.ubc.ca) is a webserver for the design of cell-type-specific MiniPromoters from genomic evidence provided by users. MiniP, MiniPromoter.
INTRODUCTION
MiniPromoters, or compact promoters, are short cis-regulatory sequences that control the expression of adjacent DNA in specific cells or tissues (1,2). The development of compact promoters is an active area of research in molecular biology, but recently, there has been a renewed interest due to their potential for therapeutic gene delivery (3,4). The field of gene therapy is increasingly focusing on improving expression specificity to minimize off-target expression and mitigate adverse events such as immune responses. This trend has been in part motivated by the adoption of recombinant adeno-associated virus (rAAV) vectors, which have low immunogenicity (5), allow for long-term expression (6), and can be engineered to target specific cell types, tissues, or organs (7,8). However, a limitation of rAAV vectors is their DNA payload capacity (9), highlighting the importance of promoter compactness in gene therapy research.
For decades, researchers have focused on designing compact promoters to efficiently transduce the brain, heart, liver, and retina, including specific cell types within these organs (Table S1). For instance, we have developed, and refined over the years, a manual approach to design human MiniPromoters, which were then characterized for specificity in the central nervous systems of mice and primates (1,2,10–14). The process involves selecting a gene with relevant expression in the target cells or tissue (identified from the literature or single-cell RNA-seq (15)), defining a genomic window around the gene for analysis, identifying candidate cis-regulatory regions (CRRs) within the window based on genomic evidence (from experimental assays measuring different epigenetic properties) and conservation (16), combining a subset of the resulting CRRs into a MiniPromoter, and characterizing the MiniPromoter in vivo.
To meet the increasing demand for compact promoters with user-relevant specificity, we have developed OnTarget, a webserver that streamlines the MiniPromoter design process. Users only need to specify a gene of interest or custom genomic coordinates on which to focus the identification of CRRs, and can also provide relevant cell-type-specific genomic evidence. OnTarget uses this information, as well as internal data, to identify candidate CRRs and suggest MiniPromoter designs. To illustrate the utility of OnTarget, we designed two MiniPromoters to target two cell populations relevant to Parkinson Disease. For comparison, we additionally characterized two manual MiniPromoters designed from the same genes. All MiniPromoters were experimentally validated for regional specificity, demonstrating that OnTarget designs could effectively drive on target expression.
MATERIALS AND METHODS
Implementation
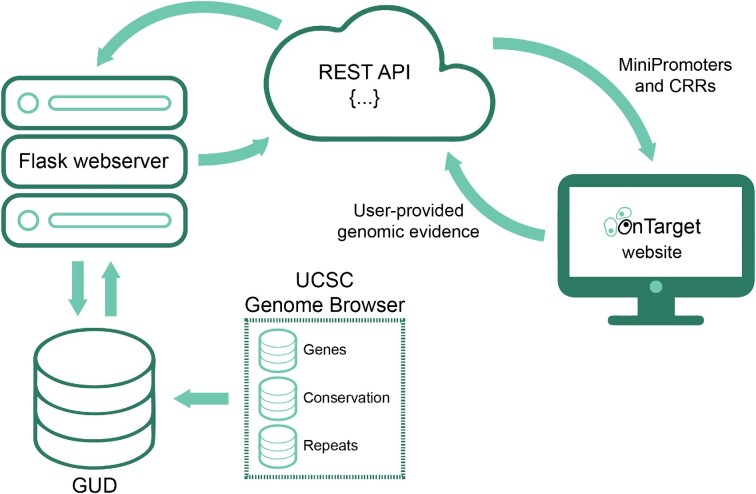
OnTarget is a webserver deployed as a Node.js application that helps users identify CRRs and design MiniPromoters. It communicates with a REST API that accesses an underlying data repository storing genomic information and a Python framework encompassing the main functionalities of OnTarget (Figure 1). Details about the identification of CRRs, MiniPromoter design process, and interaction with the webserver are provided below, and in the Results section ‘OnTarget walkthrough’.
Figure 1.
Schematic representation of the OnTarget workflow. OnTarget accepts input from the user such as genomic evidence and returns CRRs and MiniPromoters. The identification of CRRs and MiniPromoter design process is informed with internal data from GUD, a database unifying gene definitions, multi-species conservation and repetitive elements from the UCSC Genome Browser (17). CRR, cis-regulatory region; GUD, Genomic Unification Database.
The underlying data repository, named GUD (Genomic Unification Database), comprises two MySQL databases: one for storing human information related to the hg19 genome assembly and the other for mouse (mm10 genome assembly). Both databases were populated with data from the UCSC Genome Browser (17) (Table S2), which were unified for improved efficiency. These include RefSeq gene definitions (18), which, for each gene, cover its name, symbol, transcription start and end positions, coding start and end positions, exons start and end positions, and strand location, conservation values from the multiz alignment of multiple vertebrate species (16), and repetitive elements identified by RepeatMasker such as short interspersed nuclear elements (SINEs), which include ALUs, long interspersed nuclear elements (LINEs), long terminal repeat elements (LTRs), which include retroposons, DNA repeat elements, micro-satellites, low complexity repeats, satellite repeats, RNA repeats (RNA, tRNA, rRNA, snRNA, scRNA and srpRNA), and other repeats (e.g. class Rolling Circle).
The Python framework interacts with the REST API of GUD to extract genomic information and inform the identification of CRRs and MiniPromoter design process. It has three main entry points: (i) ‘gene2interval’ takes as input a gene and a genome assembly (i.e. ‘hg19’ or ‘mm10’) and returns a genomic window for analysis based on either the untranslated regions (UTRs) of the closest upstream and downstream genes or distance (in kb) around the gene body; (ii) ‘interval2regions’ takes as input a genomic window, a genome assembly, and genomic evidence (if provided), and returns a list of CRRs labelled as promoters or enhancers and (iii) ‘regions2minipromoter’ takes as input promoters and enhancers and returns MiniPromoter designs.
Identification of CRRs and conserved regions
For a given genomic window, we first initialize a NumPy array (19) of the same length using conservation values from GUD, which range from 0 to 1. If conservation is not enabled, the array is initialized with zeroes. Each array position, which corresponds to a nucleotide, is further scored positively (i.e. +1) for each type of genomic evidence that it overlaps with, and, if conservation is enabled, if it overlaps with conserved regions (details are provided below). For instance, a conserved nucleotide that is located within an accessible chromatin region, and supported by the histone modification H3K27ac (a hallmark of active enhancers (20)), additionally receives a score of 3. However, if the same nucleotide is not conserved and does not overlap any genomic evidence, it does not receive an additional score. Then, if enabled, we mask nucleotides overlapping with either coding exons or repetitive elements from GUD (i.e. their scores in the array are set to 0). Next, we normalize the array scores between 0 and 1 using the ‘MinMaxScaler’ function from scikit-learn (21). Finally, to define CRRs, we identify blocks of ≥100 consecutive nucleotides with a score in the normalized array ≥0.5 (these values can be adjusted in the webserver). To label CRRs as promoters or enhancers, we determine their overlap with gene transcription start positions from GUD, which are used as source of transcription start sites (TSSs). For genes located on the reverse strand, we use transcription end positions as source of TSSs instead. If a CRR overlaps with a TSS, it is labelled as a promoter; otherwise, it is labelled as an enhancer.
If enabled, conserved regions are identified in a similar way to CRRs. However, instead of the normalized array, we use a copy of the original NumPy array initialized with conservation values. Moreover, we set by default the number of consecutive nucleotides to identify conserved blocks to ≥10 and the score to ≥0.6 (like for CRRs, these values can be adjusted in the webserver). Finally, the system performs a second pass in which nearby conserved blocks are merged if their resulting score after merging, including the nucleotides within and between the blocks, would still be ≥ 0.6.
MiniPromoter design process
For each CRR labeled as promoter, five MiniPromoters are designed by placing it at the 3′ end and then adding CRRs labelled as enhancers, up to a maximum size of 1.9 kb (both the number of MiniPromoter designs and their size can be adjusted in the webserver). Upstream enhancers are added first, from more proximal (i.e. closer to the promoter) to more distal (i.e. farther from the promoter), followed by downstream enhancers (also from more proximal to more distal). When the gene is on the reverse strand, however, downstream enhancers are added first, followed by upstream enhancers (in both cases from more proximal to more distal). If transcription factors (TFs) are specified, CRRs are filtered out so that the final designs only include promoters and enhancers featuring binding sites for those TFs. As a source for TF binding sites (TFBSs), we relied on the JASPAR 2020 TFBS predictions (22) from the UCSC Genome Browser (17). Moreover, if any restriction enzymes are specified, MiniPromoters whose sequence include sites for those restriction enzymes are filtered out.
LiftOver
As per the recommendations from the UCSC Genome Browser, we executed LiftOver (23) to remap coordinates between the genome assemblies of two different organisms by setting the minimum nucleotide ratio for remapping (i.e. option ‘-minMatch’) to 0.1. For remapping coordinates between two genome assemblies of the same organism, we executed LiftOver with default parameters.
Manual MiniPromoter designs
We have previously described a manual bioinformatics pipeline to design MiniPromoters (2,14). The process starts with the selection of a gene with relevant expression in the target cells or tissue (e.g. a marker from single-cell RNA-seq data). Then, the promoter and enhancers of the gene are identified by integrating different types of genomic evidence associated with CRRs and visualizing them in a genome browser such as IGV (24). These include nascent transcription from CAGE (25,26) and GRO-seq (27), chromatin accessibility from DNase-seq (28) or ATAC-seq (29), ChIP-seq (30) of histone modifications and TFBSs, computational predictions from ENCODE (31), and multi-species conservation (16). Identification of promoters and enhancers can be limited by the proximity of nearby genes, regulatory confinements imposed by topologically associating domains (32), or by defining a custom genomic window. Next, MiniPromoters are designed by placing the promoter of the gene at the 3′ end and then adding enhancers as needed following the same criteria as described above for OnTarget. Finally, the MiniPromoter sequences are analyzed to detect the presence of undesired restriction sites using tools such as NEBcutter (33).
OnTarget MiniPromoter designs
As genomic evidence for designing the ADORA2A MiniPromoter, we downloaded ATAC-seq data (in bigWig format) of post-mortem human (hg19) neurons of the accumbens nucleus and putamen (34) (GEO accessions: GSM2546440, GSM2546463, GSM2546465, GSM2546489, GSM2546535). We reformatted the files from bigWig to bedGraph using bigWigToBedGraph (35) and then processed them one-by-one using MACS2 bdgpeakcall (36) (version 2.1.4) with default parameters. The resulting peaks were pooled into a single file and remapped to mm10. Next, we downloaded H3K4me1, H3K4me3, H3K36me3, and RNA polymerase II ChIP-seq data (in bigWig format) of 8 weeks-old mice (mm9) saline-treated accumbens nucleus samples (37) (GEO accessions: GSM1050343, GSM1050344, GSM1050345, GSM1050349, GSM1050350, GSM1050351, GSM1050355, GSM1050356, GSM1050357, GSM1050368, GSM1050369, GSM1050370). As with the ATAC-seq data, bigWig files were reformatted into bedGraph format and individually processed using MACS2, and the resulting peaks were pooled into individual files, one for each factor, and remapped to mm10. Finally, we downloaded H3K27ac ChIP-seq peaks of medium spiny neurons (MSNs) of the mouse striatum (mm9) (38) (GEO accession: GSM2230267), which we remapped to mm10.
For the PITX3 MiniPromoter, we downloaded DNase-seq and ChIP-seq peaks of histone marks H3K4me1, H3K4me3, H3K9ac, and H3K27ac of midbrain samples of newborn mice (mm10) (31) (ENCODE accessions: ENCFF588FLL, ENCFF741TQE, ENCFF903IBK, ENCFF770SEL, ENCFF818XLK, ENCFF991JUZ). Peaks from the two DNase-seq datasets were pooled into a single file.
In OnTarget, a key hyperparameter is the minimum score threshold that is used to identify CRRs. We set this threshold to the score of the nucleotide ranked at the top fifth percentile within the genomic window, which corresponds to ∼0.555 when designing the ADORA2A MiniPromoter and ∼0.414 for the PITX3 MiniPromoter.
Experimental validation
MiniPromoters were characterized in mice for their ability to drive expression in the D2-type MSNs of the striatum (ADORA2A) or the dopaminergic neurons of the substantia nigra (PITX3) following a well-established protocol that we have previously used to characterize hundreds of MiniPromoters (1,2,10,11,14). Each MiniPromoter was synthesized (GenScript), cloned into a rAAV genome plasmid driving the expression of emerald green fluorescent protein (EmGFP), and packaged into either AAV9 (Ple253) or rAAV-PHP.B (Ple355, Ple388, Ple389) capsids for widespread brain transduction (2,39). Mice were injected intravenously, either via the superficial temporal vein at postnatal day 4 (rAAV9), or via the tail vein in adult mice (rAAV-PHP.B). Four weeks following injection, a minimum of three mice per MiniPromoter were given a lethal dose of avertin and perfused transcardially, after which brains were harvested. Brain tissue was cryosectioned at 20 μM either sagittally (ADORA2A) or coronally (PITX3), mounted directly onto slides, and stained with anti-GFP (GFP-1020, Aves Labs; Alexa Fluor 488, Life Technologies) and Hoechst (Sigma-Aldrich). At least two mice per MiniPromoter were assessed, and unique expression patterns were determined by fluorescent microscopy and image analysis.
RESULTS
OnTarget walkthrough
The webserver guides users through the identification of candidate CRRs and MiniPromoter design process step-by-step:
Select either the human (i.e. ‘hg19’) or mouse (i.e. ‘mm10’) genome assembly. If mm10 is selected, specify whether the final CRRs should be remapped to hg19 using LiftOver.
Select a gene with an on target expression pattern with which to define a genomic window for analysis, or specify ‘Custom Coordinates’. If a gene is selected, define the genomic window based on the UTRs of the closest upstream and downstream genes (i.e. ‘Gene to Gene’) or distance (i.e. ‘±n kb from Gene’). The maximum size of genomic windows is limited to 250 kb.
Upload genomic evidence relevant to the target cells in BED format. Uploaded data will be deleted from the webserver after 1 month.
Click the button ‘Get Regulatory Regions’.
Users can additionally specify advanced parameters related to the identification of CRRs. We have set the default values for these parameters based on our experience, and expect them to be appropriate for most cases. Upon clicking the button ‘Get Regulatory Regions’, OnTarget will redirect users to a new page, where they will have to wait briefly (typically <1 min) while it identifies and returns the CRRs. The resulting CRRs are labelled as promoters or enhancers, and include useful information such as the genomic coordinates, score, and size. Next, users can design their own MiniPromoters by selecting at least one promoter from the list of CRRs and clicking the button ‘Download MiniPromoter’. Users can also click the button ‘Download Suggested MiniPromoters’ to have OnTarget automatically design and return MiniPromoters. Additionally, users can specify the number and size of the designed MiniPromoters, whether to prioritize the inclusion of CRRs with specific TFBSs, or to avoid specific restriction sites.
Case examples
To demonstrate the utility of OnTarget, we applied it to design two MiniPromoters based on the genes ADORA2A and PITX3. Both examples, including the genomic evidence used as input as well as the identified CRRs and MiniPromoter sequences, are accessible via dedicated pages on the OnTarget web server and can be downloaded by users.
ADORA2A MiniPromoter
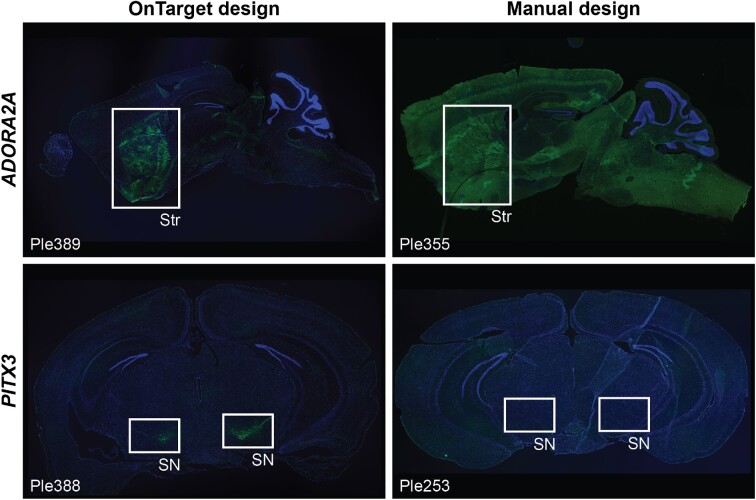
The ADORA2A gene was selected based on its well-established status as a marker of striatal D2-type MSNs (40), which are of therapeutic interest due to their contribution to the pathophysiology in Parkinson Disease (41). We downloaded publicly available ATAC-seq and ChIP-seq data of histone modifications H3K4me1, H3K4me3, H3K36me3, and H3K27ac and of RNA polymerase II from relevant samples, which were processed and remapped to mm10 (Methods). We then applied OnTarget to identify candidate CRRs of ADORA2A as follows: (i) we set the genome assembly to mm10 and specified to remap the identified CRRs to hg19; (ii) defined the genomic window within which to identify the CRRs based on a distance of ± 100 kb around the mouse gene (i.e. mm10:chr10:75216877–75434792); (iii) uploaded the genomic evidence and (iv) set the score threshold for identifying CRRs to ∼0.555 (a value that would return the top 5% of nucleotides). OnTarget returned 30 CRRs (Table S3), including RR13, one of two ADORA2A promoter-like CRRs that overlapped with a cluster of FANTOM5 TSSs characterized by robust and specific expression in striatal samples (26), and 20 enhancers overlapping with predicted TFBSs for the TFs RARB and SP9, which have been reported to regulate D2-type MSN genes (42,43). Upon examination, we applied OnTarget to combine CRRs 7, 13, 18 and 25 into the MiniPromoter ‘Ple389’, which had a length of 1344 bp and whose sequence did not contain sites for the restriction enzymes Ascl and FseI, which were used for cloning. Finally, Ple389 was synthesized, cloned into an rAAV genome plasmid, packaged into rAAV, and characterized for its ability to drive cell-type-specific EmGFP expression in the striatum (Methods). For comparison, we also characterized Ple355, a MiniPromoter that we manually designed based on ADORA2A (Methods). Intravenous delivery of the capsids rAAV9 and its derivative rAAV-PHP.B ensure widespread transduction of brain cells (2,39). Indeed, the results using the manual design Ple355 confirmed this widespread transduction, because the MiniPromoter directed ubiquitous expression throughout the brain (Figure 2, top-right panel). In contrast, the OnTarget design Ple389 showed strong and specific expression in the striatum (Figure 2, top-left panel), which is the location of the target D2-type MSNs.
Figure 2.
ADORA2A- (top panels) and PITX3-based (bottom panels) MiniPromoters designed either using OnTarget (left panels) or by manually identifying and combining CRRs potentially regulating the expression of each gene (right panels). MiniPromoters were characterized in mice for their ability to drive specific expression in the striatum (ADORA2A) or the substantia nigra (PITX3) using rAAV vectors (Methods). SN, substantia nigra; Str, striatum. Green, anti-GFP; blue, Hoechst.
PITX3 MiniPromoter
The PITX3 gene was selected because it encodes a TF that plays a crucial role in the specification of dopaminergic neurons of the substantia nigra (44), whose loss is a hallmark of Parkinson Disease (45,46), making them therapeutically relevant. As for ADORA2A, we began by downloading publicly available DNase-seq and histone ChIP-seq data (H3K4me1, H3K4me3, H3K9ac, H3K27ac) from relevant samples, which were remapped to the mm10 genome (Methods). Then, we applied OnTarget to identify candidate CRRs of PITX3. We followed the same steps as before with two minor differences: we (i) used a range of ± 50 kb around Pitx3 when defining the genomic window (i.e. mm10:chr19:46085685–46198325) and (ii) set the minimum score to identify CRRs to ∼0.414. In this example, OnTarget returned 18 CRRs (Table S3): three promoter-like CRRs, including RR8, which overlapped two TSSs associated with PITX3 (26), and 15 enhancers, 11 of which overlapped with predicted NR42A TFBSs, which is a known regulator of PITX3 in vitro and in vivo (47). Next, using OnTarget, we combined CRRs 5, 6, 8, 13, 16 and 18 into the MiniPromoter Ple388, and confirmed that its 1702 bp-long sequence did not contain any of the cloning restriction sites (i.e. Ascl and FseI). Following synthesis, cloning, and packaging, we characterized Ple388 and Ple253, the latter is a previous MiniPromoter that we had manually designed based on PITX3 (2), for their ability to drive cell-type-specific EmGFP expression in the substantia nigra (Methods). Even though the choice of delivery method and capsid ensured widespread transduction, the manual design Ple253 failed to induce any expression in the brain (Figure 2, bottom-right panel). In contrast, the OnTarget design Ple388 showed strong and specific expression in the substantia nigra (Figure 2, bottom-left panel), which is the location of the target dopaminergic neurons.
DISCUSSION
We have developed OnTarget, a webserver that enables the design of MiniPromoters for gene expression delivery in specific biological contexts. This system has been applied to two cell types with therapeutic relevance. We evaluated the specificity of the resulting promoters by comparing their expression to those of manually designed alternatives derived from the same genes, confirming the efficacy of OnTarget for designing highly specific MiniPromoters for targeted gene expression.
The utility of OnTarget extends beyond the design of MiniPromoters. It can also identify CRRs in specific genomic loci of up to 250 kb, making it a valuable tool for a broader range of users. Identification of CRRs benefits from user-provided high-quality genomic evidence relevant to their target cells. While the database underlying OnTarget supports CRR identification based on conservation alone (as in (1)), users can also provide their own genomic evidence to identify CRRs with cell-type-specificity. For instance, as multiome data becomes more accessible in single-cell studies, such as joint profiling of scATAC-seq and scRNA-seq (48), users will increasingly have access to such data relevant to their target cells.
The OnTarget parameters can affect the quality and size of the CRRs identified, most notably the minimum score threshold. During identification of CRRs, OnTarget creates an array with each position corresponding to a nucleotide in the genomic window. Each nucleotide in the array is then scored between 0 and 1, where a score of 1 indicates that the nucleotide overlaps with the greatest amount of genomic evidence, while a score of 0 indicates that the nucleotide overlaps with little (or none) of the genomic evidence. Internal benchmarks revealed that the nucleotide score distribution within the array is typically right-skewed, meaning that most nucleotides have low scores and only a few have high scores (Figure S1). We set the default threshold of 0.5 to ensure that the nucleotides included in the final CRRs would overlap, at least partially, with genomic evidence provided by the user. As properties vary greatly between genes, users may need to adjust this parameter to generate more or less candidate CRRs.
While there are supervised approaches to predict tissue-specific CRRs (e.g. (31)), OnTarget uses an unweighted scoring system: each type of data is given equal weight during the identification of CRRs. In our experience, allowing users to select their own data is crucial when designing MiniPromoters. This is because the quality of available data can vary greatly between different tissues. As such, implementing a machine learning process within OnTarget that would be suitable for all scenarios seemed ill-advised. Instead, if users wish to utilize more advanced methods, they can generate and upload their own datasets with CRR-based predictions from deep learning models.
With the advent of artificial intelligence (AI), it is becoming possible to generate compact promoters in silico. For instance, Lawler et al. used a machine learning model trained on cell-type-specific genomic evidence to prioritize enhancers based on their predicted cell-type-specificity and experimentally validated two of them in vivo (49). In a recent preprint, Taskiran et al. describe three different approaches to designing enhancers using models trained on cell-type-specific genomic evidence (50): directed evolution (i.e. inserting mutations in randomly generated sequences until their predicted cell-type-specificity is maximized), motif implantation (i.e. embedding motifs of TFs that regulate the target cells within random sequences with optimized orientation and spacing), and a generative approach (i.e. a second model is trained that creates artificial sequences predicted to be cell-type-specific by the first model). Although AI-based approaches show great potential, motif implantation still performs as well as the best methods. As generative methods continue to evolve and improve, OnTarget already provides users with the ability to identify endogenous CRRs that have the potential to deliver the desired expression characteristics.
At present, OnTarget requires a large amount of user input, from selecting a gene with a desirable expression pattern to providing cell-type-specific genomic evidence, making it one tool within a larger workflow for designing promoters. Future development will allow users to choose genes with a desirable pattern of expression, access public data repositories such as ENCODE (31) for selecting genomic evidence, and improve the visualization.
DATA AVAILABILITY
OnTarget is hosted on a virtual webserver at http://ontarget.cmmt.ubc.ca with 20 GB of RAM and 12 Intel Xeon E7540 processors (2.00 GHz, 4 cores and 8 threads) running on CentOS 7. The code for OnTarget is available on GitHub (https://github.com/wassermanlab/OnTarget and https://doi.org/10.5281/zenodo.7871189). The bash scripts that download and process the genomic evidence for the ADORA2A and PITX3 examples, the evidence itself, and the results from OnTarget are also included in the repository (https://github.com/wassermanlab/OnTarget/tree/master/data/examples; one folder per example). The code for GUD has been deposited on a separate GitHub repository (https://github.com/wassermanlab/GUD and https://doi.org/10.5281/zenodo.7871278). GUD is hosted with OnTarget for local accessibility, and runs on MariaDB Server (version 5.5.60). All MiniPromoter sequences used in this work are provided in the Supplementary Data.
Supplementary Material
ACKNOWLEDGEMENTS
We thank our collaborators Alissandra de Moura Gomes, Diane Choi, as well as Drs Terri L. Petkau, Adriana Galvan, Blair R. Leavitt, and Yoland Smith for providing useful feedback. We thank Dora Pak and Jonathan Chang for administrative and IT support, respectively.
Contributor Information
Oriol Fornes, Centre for Molecular Medicine and Therapeutics, Department of Medical Genetics, BC Children's Hospital Research Institute, University of British Columbia, Vancouver, Canada.
Tamar V Av-Shalom, Centre for Molecular Medicine and Therapeutics, Department of Medical Genetics, BC Children's Hospital Research Institute, University of British Columbia, Vancouver, Canada; Department of Cell and Systems Biology, University of Toronto, Toronto, Canada.
Andrea J Korecki, Centre for Molecular Medicine and Therapeutics, Department of Medical Genetics, BC Children's Hospital Research Institute, University of British Columbia, Vancouver, Canada.
Rachelle A Farkas, Centre for Molecular Medicine and Therapeutics, Department of Medical Genetics, BC Children's Hospital Research Institute, University of British Columbia, Vancouver, Canada.
David J Arenillas, Centre for Molecular Medicine and Therapeutics, Department of Medical Genetics, BC Children's Hospital Research Institute, University of British Columbia, Vancouver, Canada.
Anthony Mathelier, Centre for Molecular Medicine Norway (NCMM), Nordic EMBL Partnership, University of Oslo, Oslo, Norway; Department of Medical Genetics, Institute of Clinical Medicine, University of Oslo and Oslo University Hospital, Oslo, Norway.
Elizabeth M Simpson, Centre for Molecular Medicine and Therapeutics, Department of Medical Genetics, BC Children's Hospital Research Institute, University of British Columbia, Vancouver, Canada.
Wyeth W Wasserman, Centre for Molecular Medicine and Therapeutics, Department of Medical Genetics, BC Children's Hospital Research Institute, University of British Columbia, Vancouver, Canada.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Genome Canada and the Canadian Institutes of Health Research [OnTarget grants: 255ONT and BOP-149430]; National Science and Engineering Research Council of Canada [Discovery Grant: RGPIN-2017-06824]; Weston Brain Institute. Funding for open access charge: National Science and Engineering Research Council of Canada [Discovery Grant: RGPIN-2017-06824].
Conflict of interest statement. None declared.
REFERENCES
- 1. Portales-Casamar E., Swanson D.J., Liu L., de Leeuw C.N., Banks K.G., Ho Sui S.J., Fulton D.L., Ali J., Amirabbasi M., Arenillas D.J.et al.. A regulatory toolbox of MiniPromoters to drive selective expression in the brain. Proc. Natl. Acad. Sci. U.S.A. 2010; 107:16589–16594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Korecki A.J., Cueva-Vargas J.L., Fornes O., Agostinone J., Farkas R.A., Hickmott J.W., Lam S.L., Mathelier A., Zhou M., Wasserman W.W.et al.. Human MiniPromoters for ocular-rAAV expression in ON bipolar, cone, corneal, endothelial, Müller glial, and PAX6 cells. Gene Ther. 2021; 28:351–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Rodrigues G.A., Shalaev E., Karami T.K., Cunningham J., Slater N.K.H., Rivers H.M.. Pharmaceutical development of AAV-based gene therapy products for the eye. Pharm. Res. 2018; 36:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Domenger C., Grimm D. Next-generation AAV vectors-do not judge a virus (only) by its cover. Hum. Mol. Genet. 2019; 28:R3–R14. [DOI] [PubMed] [Google Scholar]
- 5. Maguire A.M., Simonelli F., Pierce E.A., Pugh E.N., Mingozzi F., Bennicelli J., Banfi S., Marshall K.A., Testa F., Surace E.M.et al.. Safety and efficacy of gene transfer for Leber's congenital amaurosis. N. Engl. J. Med. 2008; 358:2240–2248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Nathwani A.C., Reiss U.M., Tuddenham E.G.D., Rosales C., Chowdary P., McIntosh J., Della Peruta M., Lheriteau E., Patel N., Raj D.et al.. Long-term safety and efficacy of factor IX gene therapy in hemophilia B. N. Engl. J. Med. 2014; 371:1994–2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kotterman M.A., Schaffer D.V.. Engineering adeno-associated viruses for clinical gene therapy. Nat. Rev. Genet. 2014; 15:445–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Li C., Samulski R.J.. Engineering adeno-associated virus vectors for gene therapy. Nat. Rev. Genet. 2020; 21:255–272. [DOI] [PubMed] [Google Scholar]
- 9. Wu Z., Yang H., Colosi P.. Effect of genome size on AAV vector packaging. Mol. Ther. 2010; 18:80–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. de Leeuw C.N., Dyka F.M., Boye S.L., Laprise S., Zhou M., Chou A.Y., Borretta L., McInerny S.C., Banks K.G., Portales-Casamar E.et al.. Targeted CNS delivery using Human MiniPromoters and demonstrated compatibility with adeno-associated viral vectors. Mol. Ther. Methods Clin. Dev. 2014; 1:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. de Leeuw C.N., Korecki A.J., Berry G.E., Hickmott J.W., Lam S.L., Lengyell T.C., Bonaguro R.J., Borretta L.J., Chopra V., Chou A.Y.et al.. rAAV-compatible MiniPromoters for restricted expression in the brain and eye. Mol. Brain. 2016; 9:52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hickmott J.W., Chen C.-Y., Arenillas D.J., Korecki A.J., Lam S.L., Molday L.L., Bonaguro R.J., Zhou M., Chou A.Y., Mathelier A.et al.. PAX6 MiniPromoters drive restricted expression from rAAV in the adult mouse retina. Mol. Ther. Methods Clin. Dev. 2016; 3:16051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Korecki A.J., Hickmott J.W., Lam S.L., Dreolini L., Mathelier A., Baker O., Kuehne C., Bonaguro R.J., Smith J., Tan C.-V.et al.. Twenty-seven Tamoxifen-inducible iCre-driver mouse strains for eye and brain, including seventeen carrying a new inducible-first constitutive-ready Allele. Genetics. 2019; 211:1155–1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Simpson E.M., Korecki A.J., Fornes O., McGill T.J., Cueva-Vargas J.L., Agostinone J., Farkas R.A., Hickmott J.W., Lam S.L., Mathelier A.et al.. New MiniPromoter Ple345 (NEFL) drives strong and specific expression in retinal ganglion cells of mouse and primate retina. Hum. Gene Ther. 2019; 30:257–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Tang F., Barbacioru C., Wang Y., Nordman E., Lee C., Xu N., Wang X., Bodeau J., Tuch B.B., Siddiqui A.et al.. mRNA-seq whole-transcriptome analysis of a single cell. Nat. Methods. 2009; 6:377–382. [DOI] [PubMed] [Google Scholar]
- 16. Blanchette M., Kent W.J., Riemer C., Elnitski L., Smit A.F.A., Roskin K.M., Baertsch R., Rosenbloom K., Clawson H., Green E.D.et al.. Aligning multiple genomic sequences with the threaded blockset aligner. Genome Res. 2004; 14:708–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Nassar L.R., Barber G.P., Benet-Pagès A., Casper J., Clawson H., Diekhans M., Fischer C., Gonzalez J.N., Hinrichs A.S., Lee B.T.et al.. The UCSC Genome Browser database: 2023 update. Nucleic Acids Res. 2023; 51:D1188–D1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. O’Leary N.A., Wright M.W., Brister J.R., Ciufo S., Haddad D., McVeigh R., Rajput B., Robbertse B., Smith-White B., Ako-Adjei D.et al.. Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 2016; 44:D733–D745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Harris C.R., Millman K.J., van der Walt S.J., Gommers R., Virtanen P., Cournapeau D., Wieser E., Taylor J., Berg S., Smith N.J.et al.. Array programming with NumPy. Nature. 2020; 585:357–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Creyghton M.P., Cheng A.W., Welstead G.G., Kooistra T., Carey B.W., Steine E.J., Hanna J., Lodato M.A., Frampton G.M., Sharp P.A.et al.. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc. Natl. Acad. Sci. U.S.A. 2010; 107:21931–21936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Pedregosa F., Varoquaux G., Gramfort A., Michel V., Thirion B., Grisel O., Blondel M., Prettenhofer P., Weiss R., Dubourg V.et al.. Scikit-learn: machine learning in Python. J. Mach. Learn Res. 2011; 12:2825–2830. [Google Scholar]
- 22. Fornes O., Castro-Mondragon J.A., Khan A., van der Lee R., Zhang X., Richmond P.A., Modi B.P., Correard S., Gheorghe M., Baranašić D.et al.. JASPAR 2020: update of the open-access database of transcription factor binding profiles. Nucleic Acids Res. 2020; 48:D87–D92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hinrichs A.S., Karolchik D., Baertsch R., Barber G.P., Bejerano G., Clawson H., Diekhans M., Furey T.S., Harte R.A., Hsu F.et al.. The UCSC Genome Browser Database: update 2006. Nucleic Acids Res. 2006; 34:D590–D598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Robinson J.T., Thorvaldsdóttir H., Winckler W., Guttman M., Lander E.S., Getz G., Mesirov J.P.. Integrative genomics viewer. Nat. Biotechnol. 2011; 29:24–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Andersson R., Gebhard C., Miguel-Escalada I., Hoof I., Bornholdt J., Boyd M., Chen Y., Zhao X., Schmidl C., Suzuki T.et al.. An atlas of active enhancers across human cell types and tissues. Nature. 2014; 507:455–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. FANTOM Consortium and the RIKEN PMI and CLST (DGT) Forrest A.R.R., Kawaji H., Rehli M., Baillie J.K., de Hoon M.J.L., Haberle V., Lassmann T., Kulakovskiy I.V., Lizio M.et al.. A promoter-level mammalian expression atlas. Nature. 2014; 507:462–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Danko C.G., Hyland S.L., Core L.J., Martins A.L., Waters C.T., Lee H.W., Cheung V.G., Kraus W.L., Lis J.T., Siepel A.. Identification of active transcriptional regulatory elements from GRO-seq data. Nat. Methods. 2015; 12:433–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Boyle A.P., Davis S., Shulha H.P., Meltzer P., Margulies E.H., Weng Z., Furey T.S., Crawford G.E.. High-resolution mapping and characterization of open chromatin across the genome. Cell. 2008; 132:311–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Buenrostro J.D., Giresi P.G., Zaba L.C., Chang H.Y., Greenleaf W.J.. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods. 2013; 10:1213–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Johnson D.S., Mortazavi A., Myers R.M., Wold B.. Genome-wide mapping of in vivo protein-DNA interactions. Science. 2007; 316:1497–1502. [DOI] [PubMed] [Google Scholar]
- 31. ENCODE Project Consortium Moore J.E., Purcaro M.J., Pratt H.E., Epstein C.B., Shoresh N., Adrian J., Kawli T., Davis C.A., Dobin A.et al.. Expanded encyclopaedias of DNA elements in the human and mouse genomes. Nature. 2020; 583:699–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Long H.S., Greenaway S., Powell G., Mallon A.-M., Lindgren C.M., Simon M.M.. Making sense of the linear genome, gene function and tads. Epigenetics Chromatin. 2022; 15:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Vincze T., Posfai J., Roberts R.J.. NEBcutter: a program to cleave DNA with restriction enzymes. Nucleic Acids Res. 2003; 31:3688–3691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Fullard J.F., Hauberg M.E., Bendl J., Egervari G., Cirnaru M.-D., Reach S.M., Motl J., Ehrlich M.E., Hurd Y.L., Roussos P.. An atlas of chromatin accessibility in the adult human brain. Genome Res. 2018; 28:1243–1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kent W.J., Zweig A.S., Barber G., Hinrichs A.S., Karolchik D. BigWig and BigBed: enabling browsing of large distributed datasets. Bioinforma. Oxf. Engl. 2010; 26:2204–2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zhang Y., Liu T., Meyer C.A., Eeckhoute J., Johnson D.S., Bernstein B.E., Nusbaum C., Myers R.M., Brown M., Li W.et al.. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 2008; 9:R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Feng J., Wilkinson M., Liu X., Purushothaman I., Ferguson D., Vialou V., Maze I., Shao N., Kennedy P., Koo J.et al.. Chronic cocaine-regulated epigenomic changes in mouse nucleus accumbens. Genome Biol. 2014; 15:R65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. von Schimmelmann M., Feinberg P.A., Sullivan J.M., Ku S.M., Badimon A., Duff M.K., Wang Z., Lachmann A., Dewell S., Ma’ayan A.et al.. Polycomb repressive complex 2 (PRC2) silences genes responsible for neurodegeneration. Nat. Neurosci. 2016; 19:1321–1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Huang Q., Chan K.Y., Tobey I.G., Chan Y.A., Poterba T., Boutros C.L., Balazs A.B., Daneman R., Bloom J.M., Seed C.et al.. Delivering genes across the blood-brain barrier: LY6A, a novel cellular receptor for AAV-PHP.B capsids. PLoS One. 2019; 14:e0225206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Gokce O., Stanley G.M., Treutlein B., Neff N.F., Camp J.G., Malenka R.C., Rothwell P.E., Fuccillo M.V., Südhof T.C., Quake S.R.. Cellular taxonomy of the mouse striatum as revealed by single-cell RNA-seq. Cell Rep. 2016; 16:1126–1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Tozzi A., de Iure A., Di Filippo M., Tantucci M., Costa C., Borsini F., Ghiglieri V., Giampà C., Fusco F.R., Picconi B.et al.. The distinct role of medium spiny neurons and cholinergic interneurons in the D2/A2A receptor interaction in the striatum: implications for Parkinson's disease. J. Neurosci. 2011; 31:1850–1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Samad T.A., Krezel W., Chambon P., Borrelli E.. Regulation of dopaminergic pathways by retinoids: activation of the D2 receptor promoter by members of the retinoic acid receptor-retinoid X receptor family. Proc. Natl. Acad. Sci. U.S.A. 1997; 94:14349–14354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Xu Z., Liang Q., Song X., Zhang Z., Lindtner S., Li Z., Wen Y., Liu G., Guo T., Qi D.et al.. SP8 and SP9 coordinately promote D2-type medium spiny neuron production by activating Six3 expression. Dev. Camb. Engl. 2018; 145:dev165456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Nunes I., Tovmasian L.T., Silva R.M., Burke R.E., Goff S.P.. Pitx3 is required for development of substantia nigra dopaminergic neurons. Proc. Natl. Acad. Sci. U.S.A. 2003; 100:4245–4250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Fearnley J.M., Lees A.J.. Ageing and Parkinson's disease: substantia nigra regional selectivity. Biomarkers Brain Inj. Neurol. Disord. 1991; 114:2283–2301. [DOI] [PubMed] [Google Scholar]
- 46. Hornykiewicz O. Riederer P., Reichmann H., Youdim M.B.H., Gerlach M.. The discovery of dopamine deficiency in the parkinsonian brain. Parkinson’s Disease and Related Disorders. Journal of Neural Transmission. Supplementa. 2006; 70:Vienna: Springer. [DOI] [PubMed] [Google Scholar]
- 47. Volpicelli F., De Gregorio R., Pulcrano S., Perrone-Capano C., di Porzio U., Bellenchi G.C.. Direct regulation of Pitx3 expression by Nurr1 in culture and in developing mouse midbrain. PLoS One. 2012; 7:e30661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Cao J., Cusanovich D.A., Ramani V., Aghamirzaie D., Pliner H.A., Hill A.J., Daza R.M., McFaline-Figueroa J.L., Packer J.S., Christiansen L.et al.. Joint profiling of chromatin accessibility and gene expression in thousands of single cells. Science. 2018; 361:1380–1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Lawler A.J., Ramamurthy E., Brown A.R., Shin N., Kim Y., Toong N., Kaplow I.M., Wirthlin M., Zhang X., Phan B.N.et al.. Machine learning sequence prioritization for cell type-specific enhancer design. Elife. 2022; 11:e69571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Taskiran I.I., Spanier K.I., Christiaens V., Mauduit D., Aerts S.. Cell type directed design of synthetic enhancers. 2022; bioRxiv doi:27 July 2022, preprint: not peer reviewed 10.1101/2022.07.26.501466. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
OnTarget is hosted on a virtual webserver at http://ontarget.cmmt.ubc.ca with 20 GB of RAM and 12 Intel Xeon E7540 processors (2.00 GHz, 4 cores and 8 threads) running on CentOS 7. The code for OnTarget is available on GitHub (https://github.com/wassermanlab/OnTarget and https://doi.org/10.5281/zenodo.7871189). The bash scripts that download and process the genomic evidence for the ADORA2A and PITX3 examples, the evidence itself, and the results from OnTarget are also included in the repository (https://github.com/wassermanlab/OnTarget/tree/master/data/examples; one folder per example). The code for GUD has been deposited on a separate GitHub repository (https://github.com/wassermanlab/GUD and https://doi.org/10.5281/zenodo.7871278). GUD is hosted with OnTarget for local accessibility, and runs on MariaDB Server (version 5.5.60). All MiniPromoter sequences used in this work are provided in the Supplementary Data.