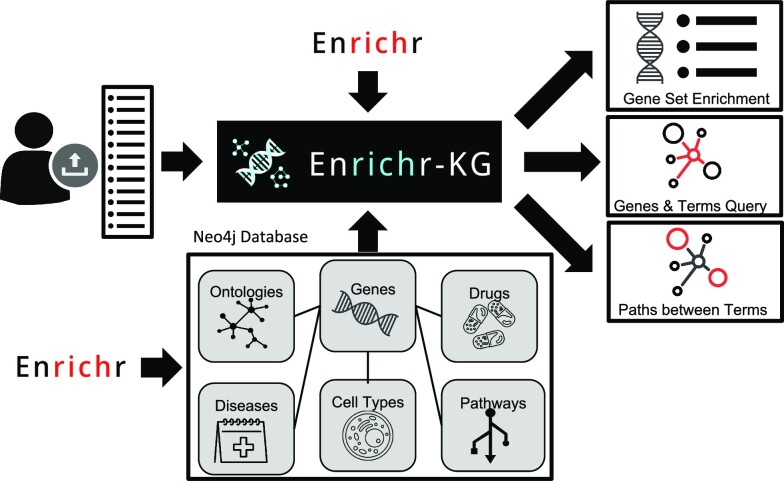
Abstract
Gene and protein set enrichment analysis is a critical step in the analysis of data collected from omics experiments. Enrichr is a popular gene set enrichment analysis web-server search engine that contains hundreds of thousands of annotated gene sets. While Enrichr has been useful in providing enrichment analysis with many gene set libraries from different categories, integrating enrichment results across libraries and domains of knowledge can further hypothesis generation. To this end, Enrichr-KG is a knowledge graph database and a web-server application that combines selected gene set libraries from Enrichr for integrative enrichment analysis and visualization. The enrichment results are presented as subgraphs made of nodes and links that connect genes to their enriched terms. In addition, users of Enrichr-KG can add gene-gene links, as well as predicted genes to the subgraphs. This graphical representation of cross-library results with enriched and predicted genes can illuminate hidden associations between genes and annotated enriched terms from across datasets and resources. Enrichr-KG currently serves 26 gene set libraries from different categories that include transcription, pathways, ontologies, diseases/drugs, and cell types. To demonstrate the utility of Enrichr-KG we provide several case studies. Enrichr-KG is freely available at: https://maayanlab.cloud/enrichr-kg.
Graphical Abstract
Graphical Abstract.
INTRODUCTION
Gene and protein set enrichment analysis provides context for genes and proteins identified in omics experiments using prior knowledge (1). Enrichment analysis involves querying a gene set against a catalog of annotated gene sets to find significant overlap between the input set and the annotated prior-knowledge gene sets. The results are ranked associated terms such as pathways, transcription factors, small molecules, diseases and other phenotypes, cell lines, cell types and tissues, and other biological and biomedical terms.
Enrichr (2–4) is a widely popular search engine for gene sets, performing enrichment analysis instantly against many annotated gene sets. In the past 10 years, over 59 million gene sets have been submitted as queries to Enrichr; and as of mid-2023, Enrichr has grown to host over ∼400 000 annotated gene sets from ∼200 gene set libraries. Such a resource provides a comprehensive collection of knowledge about genes, including their transcriptional and translational regulation, membership in pathways and biological processes, regulation and binding to drugs, association with diseases and other phenotypes, and expression across cell types, tissues, and cell lines. While Enrichr has been a valuable resource for hypothesis generation for many studies, there is still an opportunity to improve its functionality by, for example, integrating enrichment results across libraries and domains of knowledge. This can be achieved by viewing results of the enrichment analysis across libraries as an integrated network of genes and their annotations.
Network representation of biological molecular systems have been widely applied in biomedical research for abstracting connections between molecular entities (5–10). At the same time, many widely used web-based tools have been developed for network visualization and analysis. For example, STRING provides network visualizations of known and predicted associations between proteins, including physical protein-protein interactions (11). Genes2Networks (G2N) returns a protein interaction subnetwork that connects a set of input genes/proteins based on known protein-protein interactions (12). Another example is GeneMania (13) which visualizes associations between genes using evidence from across domains of knowledge such as co-expression, physical interaction, pathway membership, and shared structural domains. Other notable examples are HumanNet (14) and the DisGeNet Cytoscape app (15) which provide integrated network visualizations centered on disease genes and include predictions and prioritization of gene-disease associations.
Recently, knowledge graphs have gained popularity for integrating and generating hypotheses from connected data (16,17). Knowledge graphs have been used for studying disease mechanisms (18,19), mining small molecules for drug discovery (20,21), and analyzing connections between authors and biomedical entities using PubMed (22). Recently, there was an attempt to create a massive knowledge graph that integrates biomedical data for precision medicine (23). Within knowledge graphs data is stored as triples that describe how a subject entity is related to an object entity. For example, in the statement ‘Drug A’ targets ‘Protein B’, ‘Drug A’ is the subject, and ‘Protein B’ is the object, and the connection between them is described by the verb ‘targets’. Generating a collection of these triples made of different types of entities forms a network of knowledge that can be navigated, becoming the subject for application of graph traversal algorithms, and graph completion prediction algorithms. However, one of the challenges with knowledge graphs is that their size grows rapidly and querying the graph for useful applications becomes challenging. At the same time, biomedical and biological knowledge about genes and proteins, as well as other molecular entities, can be stored as annotated gene sets. Such gene sets are useful for performing gene set enrichment analysis (1). Many tools and databases have been developed for performing gene set enrichment analysis, for example, DAVID (24), g:Profiler (25), WebGestalt (26), MSigDB-GSEA (27) and Enrichr (3). Currently, most enrichment analysis tools and databases store knowledge as gene set libraries. While such a storage schema has benefits, for example, performing fast overlap analysis across thousands of gene sets instantly, the comparison of enrichment results across multiple gene set libraries is not trivial. To solve this, tools such as EnrichmentMaps (28) visualize gene set enrichment analysis results as ball-and-stick subgraphs that connect genes to their enriched terms. Hence, several gene set enrichment analysis tools with network visualization already exist, each providing different features and advantages. A collection of such tools with a comparison of their features is provided (Table 1).
Table 1.
A comparison of features from resources providing enrichment analysis with network representations. If a resource had a broken URL, its features were taken from the relevant literature. Column values are: A: Interactive web server, B: Number of libraries, C: Statistical method, D: Cytoscape enabled, E: Gene set augmentation/predictions, F: URL to site works, G: PPIs, H: Co-expression correlations, I: Multiple edge types, J: Different node types in the same graph, K: Provides enrichment analysis
| Resource | URL | PMID | A | B | C | D | E | F | G | H | I | J | K |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Enrichr-KG | maayanlab.cloud/enrichr-kg | ✓ | 24 | Fisher exact test | ✓ | ✓ | ✓ | ✓ | ✓ | × | ✓ | ✓ | |
| EnrichmentMap | baderlab.org/Software/ EnrichmentMap | 21085593 | × | NA | NA | ✓ | × | ✓ | × | × | × | ✓ | × |
| BioGraph | biograph.pa.icar.cnr.it | 30458802 | ✓ | 9 | Fisher exact test | ✓ | × | × | × | × | × | ✓ | ✓ |
| MELODI | melodi.biocompute.org.uk | 29342271 | ✓ | 5 | Fisher exact test | × | × | ✓ | × | × | ✓ | ✓ | ✓ |
| Reactome graph database | reactome.org/dev/graph-database | 29377902 | × | 1 | NA | × | × | ✓ | × | × | × | × | × |
| GREG | www.moralab.science/GREG | 32055858 | ✓ | 6 | NA | × | × | × | ✓ | × | ✓ | ✓ | × |
| Bio4j | bio4j.github.io | NA | × | 5 | NA | ✓ | × | ✓ | ✓ | × | ✓ | ✓ | × |
| cyNeo4j | apps.cytoscape.org/apps/cyneo4j | 26272981 | × | NA | NA | ✓ | × | ✓ | × | × | ✓ | ✓ | × |
| DGLinker | dglinker.rosalind.kcl.ac.uk | 34125897 | ✓ | 12 | Fisher exact test | × | ✓ | ✓ | ✓ | ✓ | × | × | ✓ |
| AmiGO | amigo.geneontology.org/amigo | 19033274 | ✓ | 2 | Hypergeometric | ✓ | × | ✓ | × | × | × | × | ✓ |
| Genes2FANs | actin.pharm.mssm.edu/genes2FANs | 22748121 | ✓ | 15 | NA | ✓ | × | × | ✓ | ✓ | × | ✓ | × |
| STRING | string-db.org | 36370105 | ✓ | 12 | Kolmogorov–Smirnov | ✓ | × | ✓ | ✓ | ✓ | ✓ | × | ✓ |
| GeneMANIA | genemania.org | 29912392 | ✓ | 20 | Fisher | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | × | ✓ |
| DAVID | david.ncifcrf.gov | 35325185 | ✓ | 16 | EASE score | ✓ | × | ✓ | × | × | × | × | ✓ |
| ClueGO | apps.cytoscape.org/apps/cluego | 19237447 | × | 3 | Hypergeometric | ✓ | × | ✓ | × | × | × | ✓ | ✓ |
| Metascape | metascape.org | 30944313 | ✓ | 24 | Custom* | ✓ | × | ✓ | × | × | × | ✓ | ✓ |
| NetworkAnalyst | www.networkanalyst.ca | 30931480 | ✓ | 11 | GSEA | × | × | ✓ | ✓ | × | × | ✓ | ✓ |
| MSigDB-GSEA | www.gsea-msigdb.org | 26771021 | ✓ | 15 | GSEA | × | × | ✓ | ✓ | × | × | × | ✓ |
*Explained in this blog post: https://metascape.org/blog/?p=122
Here we describe a web-server application called Enrichr Knowledge Graph (Enrichr-KG) which combines enrichment analysis with a knowledge graph data representation to query a large collection of processed datasets made of associations between genes and many biological and biomedical terms. To create Enrichr-KG we converted selected gene set libraries from Enrichr into triples for ingestion into a knowledge graph database. Each triple represents the membership of a gene in an annotated gene set. Thus, performing enrichment analysis with Enrichr-KG returns an integrated network containing the top enriched terms from across multiple libraries connected to their overlapping genes. To preserve the breadth of knowledge offered by the Enrichr libraries, libraries were selected by their diversity, level of prior use, and uniqueness (Table 2).
Table 2.
Gene set libraries stored in the Enrichr-KG database, their category, and term and gene coverage counts
| Resource | Category | Terms | Gene coverage |
|---|---|---|---|
| Achilles (44) | Diseases/Drugs | 216 | 4779 |
| ARCHS4 (29) | Transcription | 1724 | 22 226 |
| ASCT + B (103) | Cell Types | 777 | 12 531 |
| CCLE (47) | Cell Types | 378 | 11 710 |
| ChEA3 (30) | Transcription | 757 | 18 364 |
| Descartes (48) | Cell Types | 172 | 9515 |
| DisGeNET (104) | Diseases/Drugs | 9828 | 17 266 |
| FANTOM6 (31) | Transcription | 206 | 13 682 |
| Gene Ontology (39,40) | Ontologies | 6036 | 14 929 |
| GWAS Catalog (105) | Diseases/Drugs | 1737 | 15 296 |
| Human Gene Atlas (49) | Cell Types | 84 | 12 087 |
| Human Phenotype Ontology (41) | Ontologies | 1779 | 3077 |
| DISEASES (42) | Ontologies | 1811 | 15 141 |
| KEGG (33,34) | Pathways | 320 | 8073 |
| LINCS (CRISPR KO) (45) | Diseases/Drugs | 5212 | 9440 |
| LINCS (Small Molecule) (45) | Diseases/Drugs | 5425 | 9525 |
| MGI Mammalian Phenotype (43) | Ontologies | 4601 | 9756 |
| Pfam (52) | Misc | 608 | 8975 |
| PFOCR (35) | Pathways | 17 326 | 12 765 |
| Reactome (36) | Pathways | 1818 | 10 489 |
| Tabula Muris (50) | Cell Types | 106 | 3857 |
| Tabula Sapiens (51) | Cell Types | 469 | 1509 |
| TRRUST (32) | Transcription | 571 | 3126 |
| WikiPathways (38) | Pathways | 622 | 7151 |
MATERIALS AND METHODS
Collecting and processing the Enrichr libraries for knowledge graph database ingestion
We first selected 26 gene set libraries from Enrichr (2–4) for ingestion into the knowledge graph (Table 2). To preserve the variety of library types, we selected representative libraries from each Enrichr category as follows: transcription (ARCHS4 TFs (29), ChEA3 (30), FANTOM6 (31) and TRRUST (32)); pathways (KEGG (33,34), PFOCR (35), Reactome (36), the Kinase Library (37), and WikiPathways (38)); ontologies (Gene Ontology (39,40), Human Phenotype Ontology (41), Jensen DISEASES (42), and MGI Mammalian Phenotypes (43)); diseases/drugs (Project Achilles (44), LINCS L1000 perturbation signatures (45), and Drug Perturbation Proteome Atlas (46)), cell types (CCLE (47), Descartes (48), Human Gene Atlas (49), Tabula Muris (50), and Tabula Sapiens (51)); and other (Pfam (52)). Persistent IDs were then assigned to each term and gene. For genes, gene names were mapped to Entrez gene symbols (53). Genes that do not have a matching Entrez gene symbol, or a synonym, were discarded. The following method was employed for obtaining persistent identifiers for a term: (i) parsing the gene set term with a regular expression to extract the ID from the term; (ii) parsing data tables from the resource that was used to create the gene set library utilizing available APIs or downloadable mapping files; (iii) using ontologies and controlled vocabularies such as UBERON (54), Cell Ontology (55), Cellosaurus (55,56), PubChem (57) and Entrez Gene (53) and (iv) using the term as the persistent ID for those terms that failed the mapping methods described above. When possible, additional metadata elements were resolved from the gene set terms using regular expressions.
Building the graph database
Enrichr-KG visualizes the connections between enriched terms and genes across multiple selected gene set libraries. Such visualization is achieved by storing the serialized processed gene set libraries in a Neo4j database (58). Specifically, the gene set libraries from Enrichr are converted into nodes and links where the nodes are either gene set terms or genes. If a gene is part of a gene set, then an edge is added to the network. Such information is stored in separate CSV files. The conversion of a gene set library into nodes and edges CSV files requires the construction of three files: (i) a term node CSV file containing the term identifiers, the term string, and any additional metadata; (ii) a unified gene node CSV file containing all the genes that appear in the gene set libraries and their respective identifiers and (iii) an edge CSV file that contains information on the connection between the genes and the terms, not including the weights which are associated with the enrichment results. These files are ingested into the Neo4j database using the py2neo bulk import function. Querying the database is achieved via the Cypher query language (59).
Creating the web-server application
While Neo4j comes with a console to query the database, it is not very customizable, and it is difficult to export as an open web-server application that can be shared publicly without a login requirement. To provide a public facing open customizable interface that enables users to interact with the data, we constructed a web application that uses Cytoscape.js (60) to visualize the results from the Cypher queries. Next.js and React were used to build the Enrichr-KG website. To provide enrichment analysis, Enrichr-KG communicates with the Enrichr API (2–4) to perform enrichment analysis. The results are then queried against the Neo4j database and visualized as a network.
Adding protein-protein interactions and gene-gene co-expression correlations
To include protein-protein interactions as an option for inclusion in subnetworks, human protein-protein interactions were downloaded from the STRING database (11). The top-scored 150 000 protein-protein interactions (PPIs) that share a physical complex ranked by the combined score are included in the Enrichr-KG database. To include gene-gene co-expression correlations, the co-expression correlation matrix from ARCHS4 (29) was used. The top 10 co-expressed genes for each gene are extracted and included in the database. Correlations are computed and ranked by the Pearson correlation coefficient.
Augmenting subnetworks with predicted genes
To augment the genes in the subnetworks with additional genes based on co-expression, the genes in the subnetwork are used as the input. These genes are submitted to the Geneshot API (61) to obtain genes that on average are mostly co-expressed with the genes in the subnetwork using the co-expression matrix from ARCHS4 (29). The Cypher query is then updated to identify connections between the augmented co-expressed genes and the enriched terms.
Free text descriptions of subgraphs
To produce free text descriptions of the visualized subgraph, we developed templates that describe each type of association. The templates are then filled with the text describing genes and gene sets. For example, for gene-structural domain associations from Pfam (52), the template is: ‘the gene products ${genes} have the structural domain ${term}’.
RESULTS
Interacting with the Enrichr-KG web-server application
Enrichr-KG is a gene set enrichment analysis tool that visualizes enrichment results as an interactive web-based network that connects genes to enriched terms, for example, pathways, biological processes, or phenotypes. To create Enrichr-KG, we serialized gene set libraries into CSV files that are ingested into a Neo4j database. The Enrichr-KG web interface is a customizable general-purpose UI that is built on top of the Neo4j database. As such, the UI component of Enrichr-KG can be reused for other related bioinformatics projects. The Enrichr-KG UI enables users to interact with the underlying data stored in the Neo4j database by performing gene set enrichment analyses on their input gene sets with various customization and interactive features (Figure 1). First, users can submit gene sets to perform enrichment analysis against a maximum of five selected gene set libraries. Input genes are validated against a dictionary of Entrez gene symbols and users are informed in real-time whether the gene is in the database (Supplementary Figure S1). Upon pressing the submit button, Enrichr-KG returns a subgraph that displays the top enriched terms per library as well as the genes that overlap across these terms (Supplementary Figure S2). Users can tweak the settings to control the subgraph content by adjusting various parameters such as the maximum node degree, maximum subgraph size, the gene set libraries to use, and the number of top terms to include from each library. The subnetwork layout can be changed to force-directed, circular, or hierarchical layouts. Additionally, the subnetwork can be downloaded as an image, as a story described in free-text, or as a serialized CSV file. Users can also view the enrichment results in a table or a bar chart that summarizes the results across libraries (Supplementary Figure S3). In addition, users can augment the subnetworks with additional predicted genes based on co-expression correlations; and add to the subnetworks known gene-gene links based on protein-protein interactions and/or co-expression correlations.
Figure 1.
The Enrichr-KG workflow. Enrichr gene set libraries were serialized to nodes and edges where a gene is connected to a term if it is a member of the annotated gene set. The resulting network is made of links that connect genes to drugs, diseases, cell types, pathways, transcription factor regulators, and biological processes. A public facing web interface is provided to interact with the data. The interface enables users to input gene sets to perform integrated gene set enrichment analysis. The returned result is a subnetwork containing the top enriched terms from the libraries selected connected to the overlapping genes across libraries. Bar graphs and tables are also provided to visualize the top enriched terms. Gene and term search functionality can be used to query the immediate neighbors of a node. This is useful for finding shortest paths between two genes, two terms, or a gene and a term. All subnetworks can be downloaded as CSV files containing the nodes and edges of the subnetwork, as well as a free text description of the contents of the subnetwork.
The term and gene search tab in Enrichr-KG enables users to query the database to identify specific genes or terms. The single term or single gene search queries display the immediate neighbors of that node. For example, known annotations for the gene APOE are displayed as a star subnetwork (Supplementary Figure S4). APOE is known to be associated with Alzheimer's disease and cholesterol metabolism (62–66). The two-term search feature of Enrichr-KG returns a subgraph that contains the shortest paths between two nodes. Shortest paths can be used to find connections between pairs of gene-gene, term-term, or gene-term nodes. This type of query returns the shared genes between two gene sets, or shared annotations between two genes. Such queries can illuminate connections between a gene and a gene set even if the gene is not a member of the set. For example, the subgraph that connects the two genes HNF1B and KCNJ11 shows their shared annotations such as decreased ß-cell function and decreased insulin sensitivity (67) (Supplementary Figure S5). Like the enrichment analysis subgraphs, the layout of these subgraphs can be changed, the subgraph can be augmented with additional genes, enriched with known protein-protein interactions, and made available for download as a CSV file, or as a story written in free text.
Case study 1: exploring knowledge about the APOE4 variant
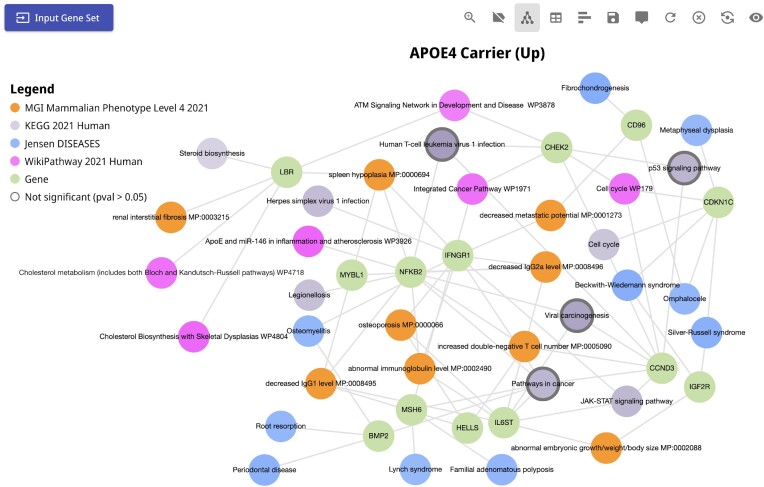
APOE is a polymorphic gene associated with the risk of late-onset Alzheimer's disease (AD) (62–65). To understand the mechanisms of APOE4, the highest risk polymorphic form of APOE, Blanchard et al. (66) performed single cell RNA-seq profiling of post-mortem human brains of APOE4 carriers vs. non-carriers. Their findings show altered cell signaling of pathways involved in cholesterol homeostasis and transport. From this study, we submitted the top 100 up-regulated genes in the APOE4 carriers compared to the non-carriers for analysis with Enrichr-KG (Figure 2). Consistent with the reported findings, Enrichr-KG also identifies cholesterol-related enriched pathways from KEGG and WikiPathways. We also found enrichment for terms related to regulation of the cell cycle and immune activation. It is well accepted that inflammation and immune response activation is the key molecular mechanism of AD (68). The Enrichr-KG subnetwork can be used to narrow down mechanisms to the specific genes, pathways, transcription factors, and cell types that may be involved.
Figure 2.
Subnetwork from the APOE4 case study. Enrichment analysis was performed on the top 100 up-regulated genes of postmortem brains from APOE4 carriers compared with the non-carriers extracted from the Blanchard et al. study. The KEGG Pathways, WikiPathways, DISEASES, DisGeNET and MGI Mammalian Phenotype gene set libraries were selected for the analysis. A subnetwork that connects the top enriched terms and the up-regulated genes that appear in at least two libraries is shown. The query can be accessed via the following URL: https://maayanlab.cloud/turl/199918c6.
Case study 2: exploring molecular mechanisms of diabetic nephropathy
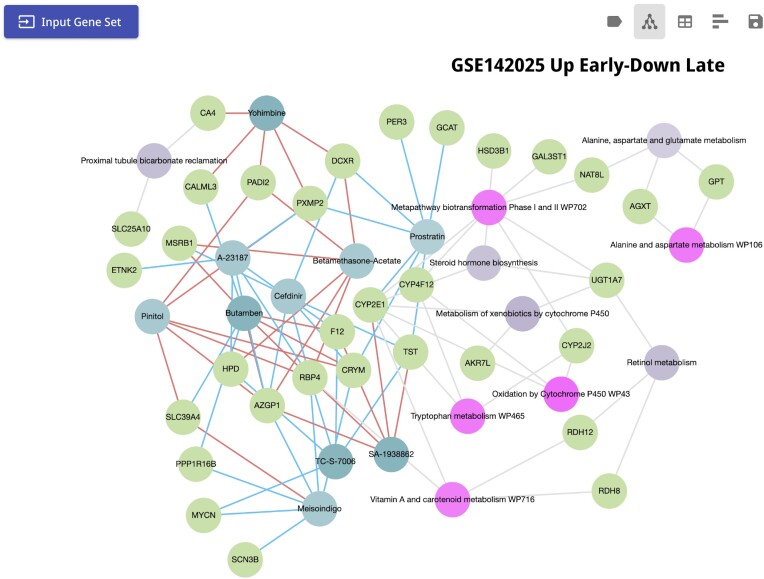
In the US, diabetic nephropathy (DN) is a common complication of diabetes that often leads to end-stage renal disease (ESRD) (69). Several studies examined the progression of DN in hopes of finding targets and therapeutics to intervene with the progression of the disease to prevent or postpone ESRD (70–74). In a recent study, Fan et. al. (70) compared the transcriptomics profiles of early stage DN to advanced DN. 270 genes were identified to have lower expression level during early stage DN compared to the advanced stage, while 148 genes are up-regulated in early DN but are lowly expressed in advanced DN. The authors concluded that up-regulated genes in late DN are mainly related to inflammation and increased immune response. On the other hand, several genes that are downregulated in the late stage are reno-protective with RDH8, RDH12 and RBP4 being part of the retinoic acid pathway. The results from this study suggest that increasing the expression of these genes may help in preventing the progression of diabetic nephropathy. To further demonstrate the functionality of Enrichr-KG, we submitted the 148 genes that are down-regulated in advanced DN compared with early DN for analysis with Enrichr-KG. To perform the pathway enrichment analysis of these genes, we selected the KEGG and WikiPathways gene set libraries. Consistent with the published study, we find that the genes RDH8, RDH12 and RBP4, as well as CYP2E1 are enriched for the Vitamin A and carotenoid metabolism pathways; meanwhile RDH8, RDH12 and UGT1A7 overlapped with the retinol metabolism pathway in KEGG (Figure 3). Interestingly, we found that the 148 genes are also enriched for alanine, aspartate, and glutamate metabolism (GPT, NAT8L and AGXT), alanine and aspartate metabolism (GPT, and AGXT), tryptophan metabolism (CYP2E1, CYP4F12, and CYP2J2), and steroid hormone biosynthesis (CYP2E1, UGT1A7, and HSD3B1). It has been shown that a high ratio between aspartate aminotransferase to alanine aminotransferase can be a risk factor for DN (75). Furthermore, low levels of tryptophan were also identified as a prognostic marker for DN (76). Another study reported that impairment of renal steroidogenesis has a probable role in diabetes related kidney damage in rats (77). Lastly, selecting the SigCom LINCS (45) L1000 chemical perturbation consensus signatures resource, Enrichr-KG reports small molecules that may up-regulate the reno-protective genes. These genes include RDH8, RDH12, RBP4 and GLP1R from the original paper, as well as CYP2E1 and UGT1A7 that also overlap with the vitamin A and retinol related pathways. We found three small molecules that up-regulate some of these genes (Figure 3). One of them is Pinitol, a known anti-diabetic agent extracted from the plant Bougainvillea spectabilis. It has been shown to improve glycaemic control in mice (78). Recently, it has been shown to also have a reno-protective effect on diabetic rats (79). Another compound that up-regulates the reno-protective genes is the ChEBI classified SA-1938862 (CHEBI:126863, BRD-K21368140-001-01-0) which is a harmala alkaloid (CHEBI:61379) (80). Harmala alkaloids are alkaloids extracted from the Peganum harmala plant (81). It has been shown that seed extracts from Peganum harmala mitigate kidney damage in diabetic rats (82). In addition, the anti-inflammatory drug bethametasone is also enriched for up regulating the reno-protective genes. It is known that bethametasone causes spikes in blood sugar (83). However, in general, glucocorticoids have been used alone or in combination with other drugs to treat glomerular diseases (84).
Figure 3.
Enriched terms for the diabetic nephropathy case study. 148 genes that are down-regulated in advanced stage DN compared with early stage were submitted to Enrichr-KG with the selected gene set libraries: KEGG pathways, WikiPathways and LINCS L1000 consensus chemical perturbations. Genes appear as green nodes, WikiPathways pathways are pink nodes, KEGG pathways are in light purple, and LINCS L1000 compounds are in turquoise and light blue. Small molecules that up regulate a gene are linked to the gene via a red link, and those that down-regulate the genes are in blue. The query can be accessed via the following URL: https://maayanlab.cloud/turl/d9e0a6f3.
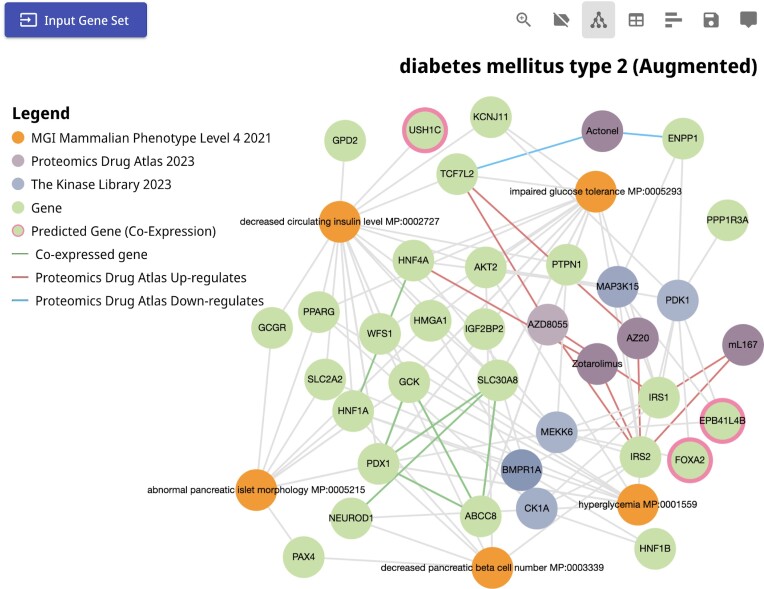
Case study 3: exploring phenotypes, kinases, and drugs related to type 2 diabetes mellitus utilizing the gene set augmentation feature of Enrichr-KG
Type 2 diabetes mellitus is a common metabolic disease characterized by an inability to secrete insulin by pancreatic beta cells, and/or a loss in the ability of cells and tissues to respond to insulin (85). We can examine genes associated with type 2 diabetes mellitus with Enrichr-KG by using the gene set search feature in the input form of the application. Searching for ‘type 2 diabetes’ in the term search feature of Enrichr-KG, we identified a gene set sourced from ClinVar (86) which provides a list of genes with mutations and other various are known to be associated with the disease. Next, we selected the MGI Mammalian Phenotype Library (43) to view mouse phenotypes associated with this gene set, the Kinase Library (37) to view related kinases that phosphorylate gene products from the gene set, and the Proteomics Drug Atlas (46) to prioritize drugs that may induce or suppress the expression of the genes in the set (Figure 4). Examining the resultant subgraph, we observe phenotypes such as decreased pancreatic beta cell number, hyperglycemia, and impaired glucose tolerance. These terms are connected to the IRS2 gene, which encodes the insulin receptor substrate. IRS2 is also the substrate of multiple kinases including MEKK6, MAP3K15, PDK1 and CK1A. Impaired IRS2 function is crucial in the development of type 2 diabetes (87,88). Additionally, the drug AZD8055 is identified as an up-regulator of IRS2, IRS1 and TCF7L2 at the protein level. AZD8055 is an mTOR inhibitor that was shown to induce insulin resistance in vivo (89). It is unclear how this seemingly conflicting evidence is resolved. Selecting the ‘augment gene set’ feature of Enrichr-KG, we can add co-expressed genes into the displayed subnetwork. One of the genes that are added to the network is USH1C. This gene was identified to play a role in hearing and vision (90), but the observation that it is highly co-expressed with the genes in the subnetwork, and the observation that mice with this gene knocked out display a decrease in circulating insulin levels, suggest that it is likely also playing an important role in diabetes. Vision impairments are a known phenotype of type 2 diabetes, and USH1C function is likely involved.
Figure 4.
Subnetwork for the type 2 diabetes mellitus case study. A gene set made of 28 genes was fetched from the ClinVar 2019 gene set library with the annotation term ‘diabetes mellitus type 2’ and was submitted to Enrichr-KG. The MGI Mammalian Phenotype, the Kinase Library, and the Proteomics Drug Atlas gene set libraries were selected for the analysis. The resultant subnetwork that connects the top enriched terms and the up-regulated genes that appear in at least two libraries is visualized. Gene set augmentation was selected to add highly co-expressed genes. The query can be accessed via the following URL: https://maayanlab.cloud/turl/8b005a41.
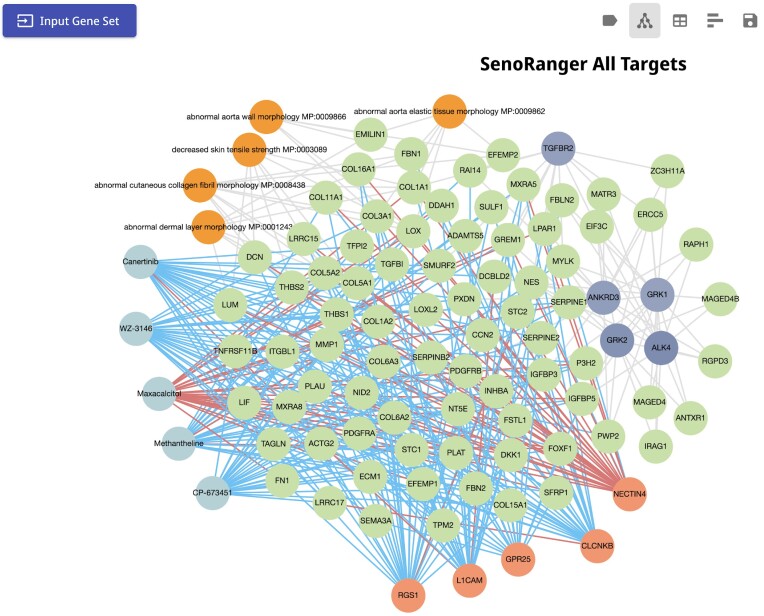
Case study 4: exploring phenotypes, drugs, genes and kinases related to cellular senescence
Cellular senescence is a state of permanent cell cycle arrest of somatic cells (91) and is implicated in aging-related pathologies and cancer (92,93). A gene set containing 301 genes called SenoRanger was established by identifying genes upregulated in RNA-seq profiles of senescent cells from a variety of studies compared to expression from multiple atlases containing normal expression, retaining genes identified in multiple of these comparisons (94). To identify mouse phenotypes enriched in the SenoRanger gene set, we submitted it to Enrichr-KG (Figure 5) and selected the MGI mouse phenotypes (43) gene set library. Several phenotypes related to skin appear in the subnetwork, namely, ‘abnormal cutaneous collagen fibril morphology MP:0008438’, ‘decreased skin tensile strength MP:0003089’, and ‘abnormal dermal layer morphology MP:0001243’. At the same time, using the SigCom LINCS resource (45), maxacalcitol, a derivative of vitamin D used to treat skin disorders (95) is identified as the only drug that up-regulates many of the genes in the subnetwork. This observation is in concordance with prior literature where vitamin D analogs have been shown to cause DNA damage and cellular senescence in epithelial type II cells (96).
Figure 5.
Subnetwork for the cellular senescence case study. A gene set containing 301 genes called SenoRanger [Deng et al. in press] was established by identifying genes upregulated in RNA-seq profiles of senescent cells from several in-vitro studies. The set of 301 genes was submitted to Enrichr-KG for analysis. The MGI Mammalian Phenotype, the Kinase Library, and SigCom LINCS gene set libraries were selected for the analysis. A subnetwork that connects the top enriched terms and the up-regulated genes that appear in at least two libraries is automatically generated. The query can be accessed via the following URL: https://maayanlab.cloud/turl/43025ee5.
The SenoRanger genes were also enriched for genes downregulated in several CRISPR KO signatures including those of GPR25, RGS1, CLCNKB, and L1CAM. RGS1 is a regulator of T-cell migration and exhaustion and has been investigated as a target for treating multiple cancers (97). Its knockdown in cervical cancer cell lines led to increased apoptosis and inhibition of cell proliferation and migration (98). L1CAM, a cell adhesion molecule, has been previously identified as an overrepresented cell surface maker in senescence cells. Additionally, its expression is associated with metabolic changes and enhanced migration and adhesion (99). Finally, selecting the Kinase Library (37) we observed multiple significantly enriched kinases including TGFBR2, ANKRD3, GRK1, GRK2 and ALK4. GRK2 is involved in cell cycle regulation and progression and its increased expression may induce cellular senescence through cell cycle arrest mediated by increased p53 phosphorylation (100,101). Additionally, TGFRBR2, the TGF-β receptor, was identified and included in the subnetwork. TGF-β signaling plays an important role in cellular senescence as well as age-related pathologies such as obesity and Alzheimer's disease (102). Overall, this and the other use cases demonstrate how Enrichr-KG can be used to confirm existing knowledge and to form new hypotheses via integrative analysis and visualization.
DISCUSSION
Here, we present Enrichr-KG, a web-server application that extends Enrichr's gene set enrichment analysis by bridging results from across multiple gene set libraries. To achieve this, we converted gene set libraries into a bipartite graph where genes are connected to their annotation terms. Such a representation can be ingested into a knowledge graph database for fast querying. Importantly, to distill the most useful information from this knowledge graph, the queries are coupled with gene set enrichment analysis results. The networked approach facilitates querying for paths between genes and annotation terms that might otherwise be difficult to extract. Some of the features available from Enrichr-KG that are not part of Enrichr are infusion of known gene-gene associations from protein interactions and co-expression resources, augmentation with additional relevant genes based on co-expression, and textual summaries of the contents of the subnetworks produced by Enrichr-KG.
Because Enrichr-KG relies on Enrichr for the gene set enrichment analysis component, it also shares some of the limitations of Enrichr. First, human and mouse genes are merged to simplify the gene search space, which could be a disadvantage for some analysis contexts. While the Enrichr API supports the upload of a gene set background, this feature is not currently implemented for Enrichr-KG. In addition, Enrichr-KG currently contains a small subset of all the gene set libraries available from Enrichr. Apart from these limitations, which will be mitigated in future releases, we also plan on extending the knowledge graph's functionality by better predicting additional links using more sophisticated graph completion machine learning algorithms. Such functionality will further assist with hypothesis generation. In addition, we also plan on utilizing existing large language models (LLM) to produce improved textual descriptions with references to better describe the resultant subnetworks extracted from the enrichment analysis results.
DATA AVAILABILITY
The Enrichr-KG web-server application is available at: https://maayanlab.cloud/enrichr-kg. The processed datasets for ingestions into a knowledge graph database are available as CSV files for download from: https://maayanlab.cloud/enrichr-kg/downloads.
Supplementary Material
Contributor Information
John Erol Evangelista, Department of Pharmacological Sciences, Mount Sinai Center for Bioinformatics, Icahn School of Medicine at Mount Sinai, NY, NY, USA.
Zhuorui Xie, Department of Pharmacological Sciences, Mount Sinai Center for Bioinformatics, Icahn School of Medicine at Mount Sinai, NY, NY, USA.
Giacomo B Marino, Department of Pharmacological Sciences, Mount Sinai Center for Bioinformatics, Icahn School of Medicine at Mount Sinai, NY, NY, USA.
Nhi Nguyen, Department of Pharmacological Sciences, Mount Sinai Center for Bioinformatics, Icahn School of Medicine at Mount Sinai, NY, NY, USA.
Daniel J B Clarke, Department of Pharmacological Sciences, Mount Sinai Center for Bioinformatics, Icahn School of Medicine at Mount Sinai, NY, NY, USA.
Avi Ma’ayan, Department of Pharmacological Sciences, Mount Sinai Center for Bioinformatics, Icahn School of Medicine at Mount Sinai, NY, NY, USA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
NIH [U24CA264250, U24CA224260, R01DK131525, OT2OD030160, RC2DK131995, U24CA271114]. Funding for open access charge: NIH [U24CA224260].
Conflict of interest statement. None declared.
REFERENCES
- 1. Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S.et al.. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U.S.A. 2005; 102:15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Chen E.Y., Tan C.M., Kou Y., Duan Q., Wang Z., Meirelles G.V., Clark N.R., Ma’ayan A.. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinf. 2013; 14:128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kuleshov M.V., Jones M.R., Rouillard A.D., Fernandez N.F., Duan Q., Wang Z., Koplev S., Jenkins S.L., Jagodnik K.M., Lachmann A.et al.. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016; 44:W90–W97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Xie Z., Bailey A., Kuleshov M.V., Clarke D.J.B., Evangelista J.E., Jenkins S.L., Lachmann A., Wojciechowicz M.L., Kropiwnicki E., Jagodnik K.M.et al.. Gene set knowledge discovery with Enrichr. Curr. Protoc. 2021; 1:e90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Barabási A.L., Oltvai Z.N.. Network biology: understanding the cell's functional organization. Nat. Rev. Genet. 2004; 5:101–113. [DOI] [PubMed] [Google Scholar]
- 6. Pavlopoulos G.A., Secrier M., Moschopoulos C.N., Soldatos T.G., Kossida S., Aerts J., Schneider R., Bagos P.G.. Using graph theory to analyze biological networks. BioData Mining. 2011; 4:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hu J.X., Thomas C.E., Brunak S.. Network biology concepts in complex disease comorbidities. Nat. Rev. Genet. 2016; 17:615–629. [DOI] [PubMed] [Google Scholar]
- 8. Gosak M., Markovič R., Dolenšek J., Slak Rupnik M., Marhl M., Stožer A., Perc M.. Network science of biological systems at different scales: a review. Phys Life Rev. 2018; 24:118–135. [DOI] [PubMed] [Google Scholar]
- 9. Ma’ayan A., Jenkins S.L., Neves S., Hasseldine A., Grace E., Dubin-Thaler B., Eungdamrong N.J., Weng G., Ram P.T., Rice J.J.et al.. Formation of regulatory patterns during signal propagation in a Mammalian cellular network. Science. 2005; 309:1078–1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ma’ayan A. Insights into the organization of biochemical regulatory networks using graph theory analyses. J. Biol. Chem. 2009; 284:5451–5455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Szklarczyk D., Gable A.L., Nastou K.C., Lyon D., Kirsch R., Pyysalo S., Doncheva N.T., Legeay M., Fang T., Bork P.et al.. The STRING database in 2021: customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2021; 49:D605–D612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Berger S.I., Posner J.M., Ma’ayan A.. Genes2Networks: connecting lists of gene symbols using mammalian protein interactions databases. BMC Bioinf. 2007; 8:372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Franz M., Rodriguez H., Lopes C., Zuberi K., Montojo J., Bader G.D., Morris Q.. GeneMANIA update 2018. Nucleic Acids Res. 2018; 46:W60–W64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kim C.Y., Baek S., Cha J., Yang S., Kim E., Marcotte E.M., Hart T., Lee I.. HumanNet v3: an improved database of human gene networks for disease research. Nucleic Acids Res. 2022; 50:D632–D639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Piñero J., Saüch J., Sanz F., Furlong L.I.. The DisGeNET cytoscape app: exploring and visualizing disease genomics data. Comput. Struct. Biotechnol. J. 2021; 19:2960–2967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chaudhri V., Baru C., Chittar N., Dong X., Genesereth M., Hendler J., Kalyanpur A., Lenat D., Sequeda J., Vrandečić D.. Knowledge graphs: introduction, history and, perspectives. AI Mag. 2022; 43:17–29. [Google Scholar]
- 17. Mohamed S.K., Nounu A., Nováček V.. Biological applications of knowledge graph embedding models. Briefings Bioinf. 2021; 22:1679–1693. [DOI] [PubMed] [Google Scholar]
- 18. Evangelista J.E., Clarke D.J.B., Xie Z., Marino G.B., Utti V., Ahooyi T.M., Jenkins S.L., Taylor D., Bologa C.G., Yang J.J.. ReproTox-KG: toxicology knowledge graph for structural birth defects. 2022; bioRxiv doi:17 September 2022, preprint: not peer reviewed 10.1101/2022.09.15.508198. [DOI] [PMC free article] [PubMed]
- 19. Hu J., Lepore R., Dobson R.J.B., Al-Chalabi A., D M.B., Iacoangeli A.. DGLinker: flexible knowledge-graph prediction of disease-gene associations. Nucleic Acids Res. 2021; 49:W153–W161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zeng X., Tu X., Liu Y., Fu X., Su Y.. Toward better drug discovery with knowledge graph. Curr. Opin. Struct. Biol. 2022; 72:114–126. [DOI] [PubMed] [Google Scholar]
- 21. MacLean F. Knowledge graphs and their applications in drug discovery. Expert Opin. Drug Discov. 2021; 16:1057–1069. [DOI] [PubMed] [Google Scholar]
- 22. Xu J., Kim S., Song M., Jeong M., Kim D., Kang J., Rousseau J.F., Li X., Xu W., Torvik V.I.et al.. Building a PubMed knowledge graph. Sci Data. 2020; 7:205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Morris J.H., Soman K., Akbas R.E., Zhou X., Smith B., Meng E.C., Huang C.C., Cerono G., Schenk G., Rizk-Jackson A.et al.. The scalable precision medicine open knowledge engine (SPOKE): a massive knowledge graph of biomedical information. Bioinformatics. 2023; 39:btad080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sherman B.T., Hao M., Qiu J., Jiao X., Baseler M.W., Lane H.C., Imamichi T., Chang W.. DAVID: a web server for functional enrichment analysis and functional annotation of gene lists (2021 update). Nucleic Acids Res. 2022; 50:W216–W221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Raudvere U., Kolberg L., Kuzmin I., Arak T., Adler P., Peterson H., Vilo J.. g:profiler: a web server for functional enrichment analysis and conversions of gene lists (2019 update). Nucleic Acids Res. 2019; 47:W191–W198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Liao Y., Wang J., Jaehnig E.J., Shi Z., Zhang B.. WebGestalt 2019: gene set analysis toolkit with revamped uis and apis. Nucleic Acids Res. 2019; 47:W199–W205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Liberzon A., Birger C., Thorvaldsdóttir H., Ghandi M., Mesirov J.P., Tamayo P.. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 2015; 1:417–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Reimand J., Isserlin R., Voisin V., Kucera M., Tannus-Lopes C., Rostamianfar A., Wadi L., Meyer M., Wong J., Xu C.et al.. Pathway enrichment analysis and visualization of omics data using g:profiler, GSEA, Cytoscape and EnrichmentMap. Nat. Protoc. 2019; 14:482–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lachmann A., Torre D., Keenan A.B., Jagodnik K.M., Lee H.J., Wang L., Silverstein M.C., Ma’ayan A.. Massive mining of publicly available RNA-seq data from human and mouse. Nat. Commun. 2018; 9:1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Keenan A.B., Torre D., Lachmann A., Leong A.K., Wojciechowicz M.L., Utti V., Jagodnik K.M., Kropiwnicki E., Wang Z., Ma’ayan A.. ChEA3: transcription factor enrichment analysis by orthogonal omics integration. Nucleic Acids Res. 2019; 47:W212–W224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Abugessaisa I., Ramilowski J.A., Lizio M., Severin J., Hasegawa A., Harshbarger J., Kondo A., Noguchi S., Yip C.W., Ooi J.L.C.et al.. FANTOM enters 20th year: expansion of transcriptomic atlases and functional annotation of non-coding rnas. Nucleic Acids Res. 2021; 49:D892–D898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Han H., Cho J.W., Lee S., Yun A., Kim H., Bae D., Yang S., Kim C.Y., Lee M., Kim E.et al.. TRRUST v2: an expanded reference database of human and mouse transcriptional regulatory interactions. Nucleic Acids Res. 2018; 46:D380–D386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kanehisa M., Goto S.. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000; 28:27–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kanehisa M. Toward understanding the origin and evolution of cellular organisms. Protein Sci. 2019; 28:1947–1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hanspers K., Riutta A., Summer-Kutmon M., Pico A.R.. Pathway information extracted from 25 years of pathway figures. Genome Biol. 2020; 21:273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Gillespie M., Jassal B., Stephan R., Milacic M., Rothfels K., Senff-Ribeiro A., Griss J., Sevilla C., Matthews L., Gong C.et al.. The reactome pathway knowledgebase 2022. Nucleic Acids Res. 2022; 50:D687–D692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Johnson J.L., Yaron T.M., Huntsman E.M., Kerelsky A., Song J., Regev A., Lin T.Y., Liberatore K., Cizin D.M., Cohen B.M.et al.. An atlas of substrate specificities for the human serine/threonine kinome. Nature. 2023; 613:759–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Martens M., Ammar A., Riutta A., Waagmeester A., Slenter D.N., Hanspers K.R.A.M., Digles D., Lopes E.N., Ehrhart F.et al.. WikiPathways: connecting communities. Nucleic Acids Res. 2021; 49:D613–D621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T.et al.. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000; 25:25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. The Gene ontology resource: enriching a GOld mine. Nucleic Acids Res. 2021; 49:D325–D334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Köhler S., Gargano M., Matentzoglu N., Carmody L.C., Lewis-Smith D., Vasilevsky N.A., Danis D., Balagura G., Baynam G., Brower A.M.et al.. The Human phenotype ontology in 2021. Nucleic Acids Res. 2021; 49:D1207–D1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Pletscher-Frankild S., Pallejà A., Tsafou K., Binder J.X., Jensen L.J.. DISEASES: text mining and data integration of disease-gene associations. Methods. 2015; 74:83–89. [DOI] [PubMed] [Google Scholar]
- 43. Smith C.L., Eppig J.T.. The mammalian phenotype ontology: enabling robust annotation and comparative analysis. Wiley Interdiscip. Rev. Syst. Biol. Med. 2009; 1:390–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Tsherniak A., Vazquez F., Montgomery P.G., Weir B.A., Kryukov G., Cowley G.S., Gill S., Harrington W.F., Pantel S., Krill-Burger J.M.et al.. Defining a cancer dependency map. Cell. 2017; 170:564–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Evangelista J.E., Clarke D.J.B., Xie Z., Lachmann A., Jeon M., Chen K., Jagodnik K.M., Jenkins S.L., Kuleshov M.V., Wojciechowicz M.L.et al.. SigCom LINCS: data and metadata search engine for a million gene expression signatures. Nucleic Acids Res. 2022; 50:W697–W709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Mitchell D.C., Kuljanin M., Li J., Van Vranken J.G., Bulloch N., Schweppe D.K., Huttlin E.L., Gygi S.P.. A proteome-wide atlas of drug mechanism of action. Nat. Biotechnol. 2023; 10.1038/s41587-022-01539-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Nusinow D.P., Szpyt J., Ghandi M., Rose C.M., McDonald E.R. 3rd, Kalocsay M., Jané-Valbuena J., Gelfand E., Schweppe D.K., Jedrychowski M.et al.. Quantitative proteomics of the cancer Cell Line encyclopedia. Cell. 2020; 180:387–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Cao J., O’Day D.R., Pliner H.A., Kingsley P.D., Deng M., Daza R.M., Zager M.A., Aldinger K.A., Blecher-Gonen R., Zhang F.et al.. A human cell atlas of fetal gene expression. Science (New York, N.Y.). 2020; 370:eaba7721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Su A.I., Wiltshire T., Batalov S., Lapp H., Ching K.A., Block D., Zhang J., Soden R., Hayakawa M., Kreiman G.et al.. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc. Natl. Acad. Sci. U.S.A. 2004; 101:6062–6067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. A single-cell transcriptomic atlas characterizes ageing tissues in the mouse. Nature. 2020; 583:590–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Jones R.C., Karkanias J., Krasnow M.A., Pisco A.O., Quake S.R., Salzman J., Yosef N., Bulthaup B., Brown P., Harper W.et al.. The Tabula Sapiens: a multiple-organ, single-cell transcriptomic atlas of humans. Science. 2022; 376:eabl4896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Finn R.D., Bateman A., Clements J., Coggill P., Eberhardt R.Y., Eddy S.R., Heger A., Hetherington K., Holm L., Mistry J.et al.. Pfam: the protein families database. Nucleic Acids Res. 2014; 42:D222–D230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Maglott D., Ostell J., Pruitt K.D., Tatusova T.. Entrez Gene: gene-centered information at NCBI. Nucleic Acids Res. 2011; 39:D52–D57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Mungall C.J., Torniai C., Gkoutos G.V., Lewis S.E., Haendel M.A.. Uberon, an integrative multi-species anatomy ontology. Genome Biol. 2012; 13:R5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Diehl A.D., Meehan T.F., Bradford Y.M., Brush M.H., Dahdul W.M., Dougall D.S., He Y., Osumi-Sutherland D., Ruttenberg A., Sarntivijai S.et al.. The Cell Ontology 2016: enhanced content, modularization, and ontology interoperability. J. Biomed. Semantics. 2016; 7:44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Bairoch A. The Cellosaurus, a cell-line knowledge resource. J. Biomol. Tech. 2018; 29:25–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Kim S., Chen J., Cheng T., Gindulyte A., He J., He S., Li Q., Shoemaker B.A., Thiessen P.A., Yu B.et al.. PubChem in 2021: new data content and improved web interfaces. Nucleic Acids Res. 2021; 49:D1388–D1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Lyon W. Full Stack GraphQL Applications: With React, Node.Js, and Neo4j. 2022; Shelter Island, NY: Manning Publications. [Google Scholar]
- 59. Francis N., Green A., Guagliardo P., Libkin L., Lindaaker T., Marsault V., Plantikow S., Rydberg M., Selmer P., Taylor A.. Cypher: an evolving query language for property graphs. Proceedings of the 2018 international conference on management of data. 2018; 1433–1445. [Google Scholar]
- 60. Franz M., Lopes C.T., Huck G., Dong Y., Sumer O., Bader G.D.. Cytoscape.Js: a graph theory library for visualisation and analysis. Bioinformatics. 2016; 32:309–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Lachmann A., Schilder B.M., Wojciechowicz M.L., Torre D., Kuleshov M.V., Keenan A.B., Ma’ayan A.. Geneshot: search engine for ranking genes from arbitrary text queries. Nucleic Acids Res. 2019; 47:W571–W577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Yamazaki Y., Zhao N., Caulfield T.R., Liu C.C., Bu G.. Apolipoprotein E and Alzheimer disease: pathobiology and targeting strategies. Nat. Rev. Neurol. 2019; 15:501–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Lambert J.C., Ibrahim-Verbaas C.A., Harold D., Naj A.C., Sims R., Bellenguez C., DeStafano A.L., Bis J.C., Beecham G.W., Grenier-Boley B.et al.. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer's disease. Nat. Genet. 2013; 45:1452–1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Corder E.H., Saunders A.M., Strittmatter W.J., Schmechel D.E., Gaskell P.C., Small G.W., Roses A.D., Haines J.L., Pericak-Vance M.A.. Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer's disease in late onset families. Science. 1993; 261:921–923. [DOI] [PubMed] [Google Scholar]
- 65. Strittmatter W.J., Saunders A.M., Schmechel D., Pericak-Vance M., Enghild J., Salvesen G.S., Roses A.D.. Apolipoprotein E: high-avidity binding to beta-amyloid and increased frequency of type 4 allele in late-onset familial Alzheimer disease. Proc. Natl. Acad. Sci. U.S.A. 1993; 90:1977–1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Blanchard J.W., Akay L.A., Davila-Velderrain J., von Maydell D., Mathys H., Davidson S.M., Effenberger A., Chen C.Y., Maner-Smith K., Hajjar I.et al.. APOE4 impairs myelination via cholesterol dysregulation in oligodendrocytes. Nature. 2022; 611:769–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Bonnefond A., Froguel P., Vaxillaire M.. The emerging genetics of type 2 diabetes. Trends Mol. Med. 2010; 16:407–416. [DOI] [PubMed] [Google Scholar]
- 68. Kinney J.W., Bemiller S.M., Murtishaw A.S., Leisgang A.M., Salazar A.M., Lamb B.T.. Inflammation as a central mechanism in Alzheimer's disease. Alzheimers Dementia. 2018; 4:575–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Collins A.J., Foley R.N., Chavers B., Gilbertson D., Herzog C., Ishani A., Johansen K., Kasiske B.L., Kutner N., Liu J.et al.. US Renal Data System 2013 annual data report. Am. J. Kidney Dis. 2014; 63:A7. [DOI] [PubMed] [Google Scholar]
- 70. Fan Y., Yi Z., D’Agati V.D., Sun Z., Zhong F., Zhang W., Wen J., Zhou T., Li Z., He L.et al.. Comparison of kidney transcriptomic profiles of early and advanced diabetic nephropathy reveals potential new mechanisms for disease progression. Diabetes. 2019; 68:2301–2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Schmid H., Boucherot A., Yasuda Y., Henger A., Brunner B., Eichinger F., Nitsche A., Kiss E., Bleich M., Gröne H.J.et al.. Modular activation of nuclear factor-kappaB transcriptional programs in human diabetic nephropathy. Diabetes. 2006; 55:2993–3003. [DOI] [PubMed] [Google Scholar]
- 72. Lindenmeyer M.T., Kretzler M., Boucherot A., Berra S., Yasuda Y., Henger A., Eichinger F., Gaiser S., Schmid H., Rastaldi M.P.et al.. Interstitial vascular rarefaction and reduced VEGF-A expression in human diabetic nephropathy. J. Am. Soc. Nephrol. 2007; 18:1765–1776. [DOI] [PubMed] [Google Scholar]
- 73. Berthier C.C., Zhang H., Schin M., Henger A., Nelson R.G., Yee B., Boucherot A., Neusser M.A., Cohen C.D., Carter-Su C.et al.. Enhanced expression of Janus kinase-signal transducer and activator of transcription pathway members in human diabetic nephropathy. Diabetes. 2009; 58:469–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Woroniecka K.I., Park A.S., Mohtat D., Thomas D.B., Pullman J.M., Susztak K.. Transcriptome analysis of human diabetic kidney disease. Diabetes. 2011; 60:2354–2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Xu J., Shi X., Pan Y.. The Association of aspartate aminotransferase/alanine aminotransferase ratio with diabetic nephropathy in patients with type 2 diabetes. Diabetes Metab. Syndr. Obes. 2021; 14:3831–3837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Chou C.A., Lin C.N., Chiu D.T., Chen I.W., Chen S.T.. Tryptophan as a surrogate prognostic marker for diabetic nephropathy. J. Diabetes Investig. 2018; 9:366–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Pagotto M.A., Roldán M.L., Molinas S.M., Raices T., Pisani G.B., Pignataro O.P., Monasterolo L.A.. Impairment of renal steroidogenesis at the onset of diabetes. Mol. Cell. Endocrinol. 2021; 524:111170. [DOI] [PubMed] [Google Scholar]
- 78. Bates S.H., Jones R.B., Bailey C.J.. Insulin-like effect of pinitol. Br. J. Pharmacol. 2000; 130:1944–1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Sousa L.G.F., Cortez L., Evangelista J., Xavier-Júnior F.A.F., Heimark D.B., Fonteles M.C., Santos C.F., Nascimento N.R.F.. Renal protective effect of pinitol in experimental diabetes. Eur. J. Pharmacol. 2020; 880:173130. [DOI] [PubMed] [Google Scholar]
- 80. Hastings J., Owen G., Dekker A., Ennis M., Kale N., Muthukrishnan V., Turner S., Swainston N., Mendes P., Steinbeck C.. ChEBI in 2016: improved services and an expanding collection of metabolites. Nucleic Acids Res. 2016; 44:D1214–D1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Lamchouri F., Settaf A., Cherrah Y., Hassar M., Zemzami M., Atif N., Nadori E.B., Zaid A., Lyoussi B.. In vitro cell-toxicity of Peganum harmala alkaloids on cancerous cell-lines. Fitoterapia. 2000; 71:50–54. [DOI] [PubMed] [Google Scholar]
- 82. Kajbaf F., Oryan S., Ahmadi R., Eidi A.. Harmine, a natural β-carboline alkaloid, ameliorates apoptosis by decreasing the expression of caspase-3 in the kidney of diabetic male Wistar rats. Gene Rep. 2020; 21:100863. [Google Scholar]
- 83. Kakoulidis I., Ilias I., Linardi A., Michou A., Milionis C., Petychaki F., Venaki E., Koukkou E.. Glycemia after betamethasone in pregnant women without diabetes-impact of marginal values in the 75-g OGTT. Healthcare. 2020; 8:40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Ponticelli C., Locatelli F.. Glucocorticoids in the treatment of glomerular diseases: pitfalls and pearls. Clin. J. Am. Soc. Nephrol. 2018; 13:815–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Galicia-Garcia U., Benito-Vicente A., Jebari S., Larrea-Sebal A., Siddiqi H., Uribe K.B., Ostolaza H., Martín C.. Pathophysiology of type 2 diabetes mellitus. Int. J. Mol. Sci. 2020; 21:6275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Landrum M.J., Lee J.M., Benson M., Brown G.R., Chao C., Chitipiralla S., Gu B., Hart J., Hoffman D., Jang W.et al.. ClinVar: improving access to variant interpretations and supporting evidence. Nucleic Acids Res. 2018; 46:D1062–D1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Withers D.J., Gutierrez J.S., Towery H., Burks D.J., Ren J.M., Previs S., Zhang Y., Bernal D., Pons S., Shulman G.I.et al.. Disruption of IRS-2 causes type 2 diabetes in mice. Nature. 1998; 391:900–904. [DOI] [PubMed] [Google Scholar]
- 88. Brady M.J. IRS2 takes center stage in the development of type 2 diabetes. J. Clin. Invest. 2004; 114:886–888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Kleinert M., Sylow L., Fazakerley D.J., Krycer J.R., Thomas K.C., Oxbøll A.J., Jordy A.B., Jensen T.E., Yang G., Schjerling P.et al.. Acute mTOR inhibition induces insulin resistance and alters substrate utilization in vivo. Mol. Metab. 2014; 3:630–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Schäfer J., Wenck N., Janik K., Linnert J., Stingl K., Kohl S., Nagel-Wolfrum K., Wolfrum U.. The Usher syndrome 1C protein harmonin regulates canonical wnt signaling. Front. Cell Dev. Biol. 2023; 11:1130058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Campisi J., d’Adda di Fagagna F.. Cellular senescence: when bad things happen to good cells. Nat. Rev. Mol. Cell Biol. 2007; 8:729–740. [DOI] [PubMed] [Google Scholar]
- 92. Childs B.G., Durik M., Baker D.J., van Deursen J.M.. Cellular senescence in aging and age-related disease: from mechanisms to therapy. Nat. Med. 2015; 21:1424–1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. van Deursen J.M. The role of senescent cells in ageing. Nature. 2014; 509:439–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Deng E.Z., Fleishman R.H., Xie Z., Marino G.B., Clarke D.J.B., Ma'ayan A.. Computational screen to identify potential targets for immunotherapeutic identification and removal of senescence cells. Aging Cell. 2023; e13809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Barker J.N., Ashton R.E., Marks R., Harris R.I., Berth-Jones J.. Topical maxacalcitol for the treatment of psoriasis vulgaris: a placebo-controlled, double-blind, dose-finding study with active comparator. Br. J. Dermatol. 1999; 141:274–278. [DOI] [PubMed] [Google Scholar]
- 96. Magro-Lopez E., Chamorro-Herrero I., Zambrano A.. Effects of hypocalcemic vitamin D analogs in the expression of DNA damage induced in minilungs from hESCs: implications for lung fibrosis. Int. J. Mol. Sci. 2022; 23:4921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Bai Y., Hu M., Chen Z., Wei J., Du H.. Single-cell transcriptome analysis reveals RGS1 as a new marker and promoting factor for T-cell exhaustion in multiple cancers. Front. Immunol. 2021; 12:767070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Zhang S., Wang H., Liu J., Tao T., Zeng Z., Wang M.. RGS1 and related genes as potential targets for immunotherapy in cervical cancer: computational biology and experimental validation. J. Transl. Med. 2022; 20:334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Mrazkova B., Dzijak R., Imrichova T., Kyjacova L., Barath P., Dzubak P., Holub D., Hajduch M., Nahacka Z., Andera L.et al.. Induction, regulation and roles of neural adhesion molecule L1CAM in cellular senescence. Aging. 2018; 10:434–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Penela P., Rivas V., Salcedo A., Mayor F. Jr. G protein-coupled receptor kinase 2 (GRK2) modulation and cell cycle progression. Proc. Natl. Acad. Sci. U.S.A. 2010; 107:1118–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Wei Z., Hurtt R., Ciccarelli M., Koch W.J., Doria C.. Growth inhibition of human hepatocellular carcinoma cells by overexpression of G-protein-coupled receptor kinase 2. J. Cell. Physiol. 2012; 227:2371–2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Tominaga K., Suzuki H.I.. TGF-β signaling in cellular senescence and aging-related pathology. Int. J. Mol. Sci. 2019; 20:5002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. The human body at cellular resolution: the NIH Human Biomolecular Atlas Program. Nature. 2019; 574:187–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Piñero J., Ramírez-Anguita J.M., Saüch-Pitarch J., Ronzano F., Centeno E., Sanz F., Furlong L.I.. The DisGeNET knowledge platform for disease genomics: 2019 update. Nucleic Acids Res. 2020; 48:D845–D855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Sollis E., Mosaku A., Abid A., Buniello A., Cerezo M., Gil L., Groza T., Güneş O., Hall P., Hayhurst J.et al.. The NHGRI-EBI GWAS Catalog: knowledgebase and deposition resource. Nucleic Acids Res. 2023; 51:D977–D985. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The Enrichr-KG web-server application is available at: https://maayanlab.cloud/enrichr-kg. The processed datasets for ingestions into a knowledge graph database are available as CSV files for download from: https://maayanlab.cloud/enrichr-kg/downloads.