Abstract
Intrinsic disorder (ID) in proteins is well-established in structural biology, with increasing evidence for its involvement in essential biological processes. As measuring dynamic ID behavior experimentally on a large scale remains difficult, scores of published ID predictors have tried to fill this gap. Unfortunately, their heterogeneity makes it difficult to compare performance, confounding biologists wanting to make an informed choice. To address this issue, the Critical Assessment of protein Intrinsic Disorder (CAID) benchmarks predictors for ID and binding regions as a community blind-test in a standardized computing environment. Here we present the CAID Prediction Portal, a web server executing all CAID methods on user-defined sequences. The server generates standardized output and facilitates comparison between methods, producing a consensus prediction highlighting high-confidence ID regions. The website contains extensive documentation explaining the meaning of different CAID statistics and providing a brief description of all methods. Predictor output is visualized in an interactive feature viewer and made available for download in a single table, with the option to recover previous sessions via a private dashboard. The CAID Prediction Portal is a valuable resource for researchers interested in studying ID in proteins. The server is available at the URL: https://caid.idpcentral.org.
Graphical Abstract
Graphical Abstract.
INTRODUCTION
The study of intrinsically disordered proteins and regions (IDPs/IDRs), which do not adopt a fixed three-dimensional fold in isolation under physiological conditions, is now a well-established field in structural biology. Over the past two decades, there has been increasing evidence for the involvement of IDPs and IDRs in a variety of essential biological processes, making them promising novel targets for drug discovery (1). While experimental methods can detect intrinsic structural disorder, such as X-ray crystallography, nuclear magnetic resonance spectroscopy, small-angle X-ray scattering, circular dichroism, and Förster resonance energy transfer, directly measuring their dynamic behavior and their context-dependent structural disorder remains difficult (2). Furthermore, various types of experiments emphasize distinct functional mechanisms of IDPs, commonly identified as disorder ‘flavors’, including flexibility, folding-upon-binding and conformational heterogeneity (3).
Dozens of ID prediction methods have been published, and both predicted and experimentally derived properties of IDRs, as well as annotations related to their function, are stored in dedicated databases (4). However, the large variety of available predictors makes it difficult to compare their performance, which can confound biologists wanting to make an informed choice.
To address this issue, the Critical Assessment of Protein Intrinsic Disorder (CAID) (2) was introduced to benchmark ID and binding predictors on a community-curated dataset of novel proteins obtained from the DisProt database (5). In CAID, participants submit their implemented prediction software to the organizers, who generate predictions by executing the software on selected protein targets whose disorder annotations were not previously available. Given a new protein sequence, the task of an IDR predictor is to assign a score to each residue for the tendency to be intrinsically disordered at any stage of the protein life. In CAID, both the accuracy of prediction methods and technical aspects related to software implementation are evaluated. However, accessing the prediction power of the tools is not always possible. Often, the software is not publicly available, exists solely as a stand-alone executable, or is available as a web server with limitations. Moreover, publicly available methods are not standardized and require informed use, often entailing careful reading of the corresponding publication and interpreting predictors' output.
To address these issues, we present the CAID Prediction Portal, a web server that executes all CAID methods with a single click on a user-defined input sequence. The server generates a standardized output and facilitates comparing methods, and it produces a consensus prediction that highlights high-confidence disordered regions. Disordered (or binding) residues are identified by selecting a threshold on the prediction score. Depending on the type of benchmark, different thresholds can be selected, leading to different results. To guide the user in selecting the best parameters, the website is accompanied by extended documentation that explains the meaning of the different statistics presented in CAID and provides a brief description of all the methods. The predictors’ output is rendered in a feature viewer and made available for download in a single table. While anonymous usage of the CAID Prediction Portal is always permitted, interested users can choose to use an optional log in to recover previous sessions via a private dashboard.
IMPLEMENTATION
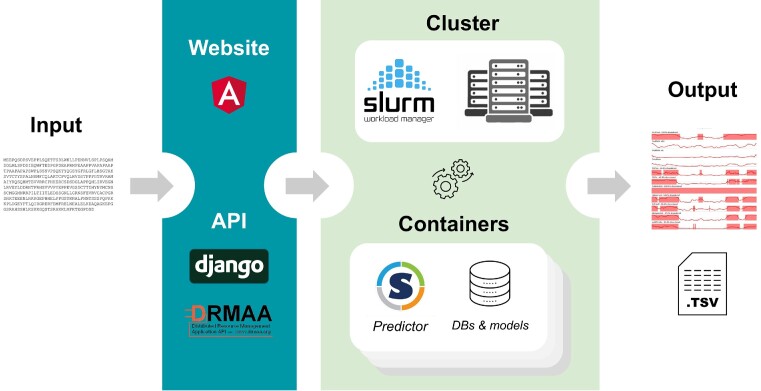
An overview of the CAID Prediction Portal is provided in Figure 1. The CAID Prediction Portal needs to execute many different predictors on the same input sequence, provided by the user. To do so, we implemented a back-end interface using the Django REST framework (DRF, https://www.django-rest-framework.org) that interacts with the scheduler controller of a computing cluster through the Distributed Resource Management Application API (DRMAA) (6), a high-level API that provides a standardized interface for submitting and managing jobs on a wide range of cluster systems. In our specific implementation, we used the Slurm Workload Manager (https://slurm.schedmd.com) as a job scheduler for the cluster. The purpose of this implementation is to allow users to submit, monitor and manage jobs on the computing cluster through a friendly web interface which exploits the RESTful API provided by the DRF. We also implemented various management features, such as the ability to stop or delete jobs, and to retrieve the job state, history and outputs for a particular user.
Figure 1.
Overview of the CAID Prediction Portal implementation.
The server provides OAuth 2.0 authentication for ORCID users. When authenticated the user is able to recover previous sessions via a private dashboard. Non-authenticated users are allowed to create new jobs and access the results. However, the amount of resources available to a single non-authenticated user is more limited, meaning that the number of daily and burst requests allowed is reduced.
The DRF back-end is also responsible for managing all the possible jobs that can be submitted to the cluster, the resources to allocate for each specific job (e.g. CPUs, random access memory), and the dependencies that can be created between different jobs.
For the CAID Prediction Portal, we created separate jobs for each of the available predictors, and a few additional jobs for creating input data for some predictors such as PSI-BLAST (7), HHBlits (8), SPIDER2 (9). This separation of predictors into different jobs is crucial as it provides flexibility to execute only the predictors of interest and display the results of fast predictors without waiting for others to finish.
The CAID Prediction Portal includes a server (dark background), which accepts a protein sequence as input, and a computing cluster (pale background), which generates the output, which is available as a table (TSV format) and rendered in a dynamic feature viewer on the web interface.
Standardization
We used Singularity (https://sylabs.io) containers to containerize all the predictor software in order to standardize the input and output data, and ensure reproducible results. By containerizing the software, we can ensure that the software runs consistently across different machines, and most importantly it is not needed to install it manually in each machine. Furthermore, containerizing the predictors enables us to package all the necessary software and dependencies together, making it easier to deploy and update the predictors. With the creation of the container we also included scripts that are executed before and after the predictor, in order to standardize the input and output of the container, creating an interface with the predictor software. The input of the predictor is a FASTA file containing multiple sequences, and the predictor is executed on each sequence, producing one output per sequence (please note that this should not be confused with the input of the CAID server, which is restricted to a single sequence). The execution time of the predictor for each sequence is also recorded. If the predictor generates multiple outputs, each output will be stored in a distinct directory corresponding to the different variations, or ‘flavors,’ of the predictor.
Some software present in the CAID Prediction Portal requires additional inputs, such as the results of PSI-BLAST, HHblits, or SPIDER2, to make their predictions. These additional inputs can be created inside the software's container itself, but they can also be provided in most of the cases as an additional parameter. This ensures that the computation of common inputs is not duplicated, leading to faster and more efficient predictions.
We used Singularity containers over Docker (https://www.docker.com) containers because Singularity is designed specifically for high-performance computing environments and has several advantages in the context of computing clusters. Firstly, Singularity does not require root access, making it easier to deploy and manage in a shared computing environment. Secondly, Singularity is optimized for running scientific workloads, with features such as support for MPI (Message Passing Interface) and GPUs (Graphical Processing Units). Thirdly, Singularity images can be easily hosted on a variety of storage systems, such as local filesystems, networked file systems, and cloud storage.
To make the container size smaller, some large datasets such as UniRef90 (10), Uniclust30 (11) or large machine learning models are mounted inside the container at runtime. This approach allows the container to access these datasets only when needed, rather than including them in the container itself. However, it is important to note that if these mounts are not created, the script that runs the predictor inside the container will fail with an error, since it will not be able to access the required data.
In order to provide a comparison baseline, we also integrate the AlphaFold-disorder (12) method that infers disorder and binding predictions by exploiting AlphaFold predicted structures available in public databases (13).
As the last step of our standardization process, we opted to create individualized tasks for each predictor that can be conveniently executed through the CAID Prediction Portal. This implementation grants users a heightened level of flexibility in their selection of methods, allowing them to make informed decisions that best suit their specific needs. Each predictor execution is linked to an API call through the portal's front-end interface, while also remaining compatible with stand-alone usage for batch executions. The API is publicly available and lets third party services request specific predictions on demand. Full documentation is available on the website.
Benchmarking
The CAID Prediction Portal includes a CAID page (https://caid.idpcentral.org/challenge) which contains information about how the challenge is organized, a detailed description of the methods, and the main benchmarking results. In Table 1, we reported all methods available in the CAID server along with the corresponding publication when available. These methods are a subset of those evaluated in the second round of the CAID challenge, i.e. those for which the authors gave permission or those that were already publicly available and licensed for free use. Some of the methods can include more than one predictor (disorder and binding) and the same predictor can generate more than one output (different flavors) representing different implementations (fast, slow), training strategies (dataset), or prediction features (DNA/RNA/protein binding, linker, short/long region, etc.). Given the repertoire of different flavors predicted by the various methods, in the CAID Prediction Portal, we divided them into two broad disorder and binding categories. Users interested in specific subcategories or flavors are invited to read the description of the methods as reported on the website.
Table 1.
Predictors included in the CAID prediction portal
| Name | Type (flavour) * | Authors | Reference |
|---|---|---|---|
| AIUPred-0.5 | Disorder | Gábor Erdős, Zsuzsanna Dosztányi | |
| AlphaFold-disorder | Disorder (Disorder, RSA), Binding | Damiano Piovesan, Alexander Miguel Monzon, Silvio C E Tosatto | (12) |
| ANCHOR2 | Binding | Bálint Mészáros, Gábor Erdős, Zsuzsanna Dosztányi | (14) |
| APOD | Disorder | Zhenling Peng, Qian Xing, Lukasz Kurgan | (15) |
| AUCpred | Disorder | Sheng Wang, Jianzhu Ma, Jinbo Xu | (16) |
| bindEmbed21IDR | Binding (idrGeneral, idrNuc, rawGeneral, rawNuc) | Burkhard Rost | (17) |
| DeepDISObind | Binding | Fuhao Zhang, Bi Zhao, Wenbo Shi, Min Li, Lukasz Kurgan | (18) |
| DeepIDP-2L | Disorder | Yi Jun Tang, Yi-He Pang, Bin Liu | (19) |
| DisEMBL | Disorder (dis465, disHL) | Rune Linding, Lars Juhl Jensen, Francesca Diella, Peer Bork, Toby J Gibson, Robert B Russell | (20) |
| DisoMine | Disorder | Gabriele Orlando, Daniele Raimondi, Francesco Codicè, Francesco Tabaro, Adrián Díaz, Wim Vranken | (21) |
| DisoPred | Disorder | Min Li, Yida Wang, Fuhao Zhang | |
| DISOPRED3 | Disorder, Binding | David T Jones, Domenico Cozzetto | (22) |
| DisPredict2 | Disorder | Sumaiya Iqbal, Md Tamjidul Hoque | (23) |
| DisPredict3 | Disorder | Md Wasi Ul Kabir, Md Tamjidul Hoque | |
| DRPBind | Binding (DNA, RNA, Protein, DeepDNA, DeepRNA, DeepProtein) | Alok Sharma, Ronesh Sharma, Tatsuhiko Tsunoda | (24) |
| ENSHROUD | Binding (all, nucleic, protein) | Min Li, Fuhao Zhang, Pengzhen Jia | |
| ESpritz | Disorder (D, N, X) | Ian Walsh, Alberto J M Martin, Tomàs Di Domenico, Silvio Tosatto | (25) |
| flDPlr | Disorder | Gang Hu, Akila Katuwawala, Kui Wang, Zhonghua Wu, Sina Ghadermarzi, Jianzhao Gao, Lukasz Kurgan | (26) |
| flDPnn | Disorder | Gang Hu, Akila Katuwawala, Kui Wang, Zhonghua Wu, Sina Ghadermarzi, Jianzhao Gao, Lukasz Kurgan | (26) |
| FoldUnfold | Disorder | Oxana V Galzitskaya, Sergiy O Garbuzynskiy, Michail Yu Lobanov | (27) |
| IDP-Fusion | Disorder | Yi Jun Tang, Bin Liu | |
| IsUnstruct | Disorder | Oxana V Galzitskaya, Michail Yu Lobanov | (28) |
| IUPred3 | Disorder | Gábor Erdős, Mátyás Pajkos, Zsuzsanna Dosztányi | (29) |
| Metapredict (V2) | Disorder | Ryan J Emenecker, Daniel Griffith, Alex S Holehouse | (30) |
| MobiDB-lite | Disorder | Marco Necci, Damiano Piovesan, Zsuzsanna Dosztányi, Silvio C E Tosatto | (31) |
| MoRFchibi | Binding (web, light) | Nawar Malhis, Matthew Jacobson, Jörg Gsponer | (32) |
| OPAL | Binding | Ronesh Sharma, Gaurav Raicar, Tatsuhiko Tsunoda, Ashwini Patil, Alok Sharma | (33) |
| PredIDR | Disorder (long, short) | Kun-Sop Han, Chol-Song Kim, Myong-Chol Ma | |
| PreDisorder | Disorder | Xin Deng, Jesse Eickholt, Jianlin Cheng | (34) |
| ProBiPred | Binding (nucleic, protein) | Lea I M Krautheimer, Michael Bernhofer, Burkhard Rost | |
| pyHCA | Disorder | Isabelle Callebaut, Tristan Bitard Feildel | |
| rawMSA | Disorder | Claudio Mirabello, Björn Wallner | (35) |
| RONN | Disorder | Zheng Rong Yang, Rebecca Thomson, Philip McNeil, Robert M Esnouf | (36) |
| s2D-2 | Disorder | Pietro Sormanni, Carlo Camilloni, Piero Fariselli, Michele Vendruscolo | (37) |
| SETH_0 | Disorder | Dagmar Ilzhöfer, Michael Heinzinger, Burkhard Rost | (38) |
| SETH_1 | Disorder | Dagmar Ilzhöfer, Michael Heinzinger, Burkhard Rost | (38) |
| SPOT-Disorder | Disorder | Jack Hanson, Yuedong Yang, Kuldip Paliwal, Yaoqi Zhou | (39) |
| SPOT-Disorder-Single | Disorder | Jack Hanson, Kuldip Paliwal, Yaoqi Zhou | (40) |
| SPOT-Disorder2 | Disorder | Jack Hanson, Kuldip Paliwal, Thomas Litfin, Yaoqi Zhou | (41) |
| VSL2 | Disorder | Kang Peng, Predrag Radivojac, Slobodan Vucetic, A Keith Dunker, Zoran Obradovic | (42) |
All methods generate predictions from the protein sequence. Some methods require additional input which is generated by helper methods, e.g. BLAST or HHblits for sequence profiles. In those cases, the additional input is generated once and shared with all dependent methods.
The AlphaFold-disorder (12) method, instead of using the sequence, takes as input the protein structure predicted by AlphaFold. In the CAID Prediction Portal the structure is retrieved directly from the AlphaFoldDB (13) database by searching the UniProtKB accession number. The server tries to retrieve the accession number by querying the UniProtKB mapping service with the provided sequence encoded with the CRC64 algorithm, and selecting the first result. If the protein sequence is not present in the UniprotKB, no structure can be downloaded and the predictor will fail to execute.
Methods are listed in alphabetical order. (*) The same package can include multiple predictors, each generating multiple outputs. The Type column indicates the type of output and the values in parentheses indicate the predictor name suffixes which correspond to different flavors or different implementations. When available, the corresponding publication is provided along with the corresponding authors. For new methods, authors are those that submitted the method to CAID.
Website
The CAID Prediction Portal website allows users to execute the available predictors on a provided protein sequence. The server can process only one sequence at a time. The predictors that are going to be executed can be configured, with some pre-made settings (e.g. running only disorder, binding or quick predictors), or manually, selecting the predictors of interest. When submitting a new job, the user can also decide to associate a description to the job and an email address that will be used to send a notification when all the predictors will finish executing. The job name is helpful to attach a text description or just a meaningful identifier to the input sequence, while the user email can be used to receive a notification when the calculation is done.
After the submission, the user will be redirected to the results page. At the top of the page, a header card will be displayed, this contains various information about the execution status of the predictors, along with a control for stopping the jobs still executing, and a button to download all the currently available results in tab-separated values (TSV) format.
The result page will poll the back-end server to update the status of the jobs that did not finish yet, to retrieve their current status and download the results from the server when available. These results will be used to create and update a feature viewer, to display the outputs of the predictors. These outputs are all aligned to the protein sequence that was submitted, and they can be of two different types, a binary score and a probability score.
The feature viewer offers various controls to manipulate the display of the results. The predictions can be filtered based on their type (disorder or binding), the threshold for the binary score can be changed from the predictor's default to optimized thresholds as provided by CAID. Optimized thresholds correspond to a selection of metrics reported by the CAID challenge. The optimization strategy depends on the type of metric and validation dataset, those available in the CAID Prediction Portal are described in the website documentation, while we refer to the CAID paper (2) for a full description of all possible benchmarks. The methods can be sorted based on their performance in CAID, disorder (or binding) content, or alphabetically based on their names.
In the feature viewer, a consensus is also computed with the prediction of the available predictors, divided in the two categories, disordered and binding. This consensus is calculated as a majority vote of the binary predictions available. The consensus will also be influenced by the chosen threshold. In order to compare predictions with structural and functional domains, Pfam (43) and Gene3D (44) assignments from the InterProScan (45) output are reported. These annotations are calculated in parallel on a separate job, and shown as separate tracks on the feature viewer when available.
While anonymous usage of the CAID Prediction Portal is always permitted, interested users can choose to recover previous sessions via a private dashboard after a login using their ORCID credentials, where all the previously submitted jobs can be accessed. An anonymous user can recover a previous job by saving its UUID and later use it to access the results again.
CONCLUSIONS
The CAID Prediction Portal is a valuable resource for researchers and scientists working in the field of protein structure and intrinsic disorder prediction. By combining state-of-the-art ID and binding prediction methods with the CAID optimization strategy, the portal allows users to calculate and compare different predictions in a single view. Predictions can be dynamically adapted on the fly by choosing different CAID optimization strategies. For example, the user can focus on precision over recall, or on the contrary, can relax the optimization cutoffs to expand disorder detection.
One of the key advantages of the portal is its speed and dynamic nature, as the server displays the results of a method as soon as the calculation is completed. Additionally, the portal's modular and extensible design makes it easy to add or remove prediction methods at any time, providing maintainers with the flexibility to adapt to new developments in the field. Finally, all methods are standardized and their output is made available in the same format.
The CAID section of the portal provides benchmarking results and statistics that can guide users in the evaluation of the performance of the predictors. This information is particularly useful for researchers who are looking to improve their methods and algorithms.
Moreover, the CAID Prediction Server is integrated into the OpenEBench (46) infrastructure for community benchmarking experiments of computational methods in the life sciences, which displays the results of various CAID editions in a dedicated section. This integration allows for the prediction output generated by the portal to be used in generating assessment results, thereby facilitating a transition from a timeframe-based challenge (as was the case for CAID rounds 1 and 2) into a continuous assessment.
Last but not least, the CAID portal will help inform and improve the selection ID predictors available in the MobiDB database (47) for large-scale annotation of ID in proteins. The latter is the main source of ID data for core data resources such as InterPro (48) and UniProtKB (49). Any small improvement in ID prediction performance documented in the CAID Portal therefore has a large potential knock-on effect in improving ID annotations across the known protein universe.
In summary, the CAID Prediction Portal is a valuable resource that can help researchers develop more accurate and effective methods for predicting intrinsic protein disorder and their binding regions. By enabling continuous assessment and benchmarking of different prediction methods, the portal can help accelerate progress in this important field and benefit the scientific community at large.
DATA AVAILABILITY
The CAID Prediction Portal is freely available at https://caid.idpcentral.org.
ACKNOWLEDGEMENTS
This publication is part of a project that has received funding from the European Union's Horizon 2020 research and innovation programme under grant agreements No 778247 and No 823886. This work was supported by ELIXIR, the research infrastructure for life-science data. The authors are grateful to members of the BioComputing UP group for insightful discussions.
APPENDIX
CAID predictors
Alex S Holehouse3,4, Daniel Griffith3,4, Ryan J Emenecker3,4, Ashwini Patil5, Ronesh Sharma6, Tatsuhiko Tsunoda7,8,9, Alok Sharma9,10, Yi Jun Tang11, Bin Liu11, Claudio Mirabello12, Björn Wallner12, Burkhard Rost13, Dagmar Ilzhöfer13, Maria Littmann13, Michael Heinzinger13, Lea I M Krautheimer13, Michael Bernhofer13, Liam J McGuffin14, Isabelle Callebaut15, Tristan Bitard Feildel16, Jian Liu17, Jianlin Cheng17, Zhiye Guo17, Jinbo Xu18, Sheng Wang18,19, Nawar Malhis20, Jörg Gsponer21, Chol-Song Kim22, Kun-Sop Han22, Myong-Chol Ma22, Lukasz Kurgan23, Sina Ghadermarzi23, Akila Katuwawala23,24, Bi Zhao25, Zhenling Peng26, Zhonghua Wu27, Gang Hu28, Kui Wang28, Md Tamjidul Hoque29, Md Wasi Ul Kabir29, Michele Vendruscolo30, Pietro Sormanni30, Min Li31, Fuhao Zhang31, Pengzhen Jia31, Yida Wang32, Michail Yu Lobanov33, Oxana V Galzitskaya33,34, Wim Vranken35,36, Adrián Díaz35,36, Thomas Litfin37, Yaoqi Zhou37,38, Jack Hanson39, Kuldip Paliwal39, Zsuzsanna Dosztányi40, Gábor Erdős40.
3Department of Biochemistry and Molecular Biophysics, Washington University School of Medicine, St. Louis, Missouri
4Center for Biomolecular Condensates, Washington University in St. Louis, St. Louis, MO, USA
5Combinatics Inc. Ichikawa-shi, Chiba 272-0824, Japan
6Fiji National University, Suva, Fiji
7Laboratory for Medical Science Mathematics, Department of Biological Sciences, School of Science, The University of Tokyo, Tokyo,113-0033, Japan
8Laboratory for Medical Science Mathematics, Department of Computational Biology and Medical Sciences, Graduate School of Frontier Sciences, The University of Tokyo, Tokyo 113–0033, Japan
9Laboratory for Medical Science Mathematics, RIKEN Center for Integrative Medical Sciences, Yokohama 230-0045, Japan
10Institute for Integrated and Intelligent Systems, Griffith University, Nathan, Brisbane, QLD 4111, Australia
11School of Computer Science and Technology, Beijing Institute of Technology, Beijing 100081, China
12Division of Bioinformatics, Department of Physics, Chemistry, and Biology, Linköping University
13TUM School of Computation, Information and Technology, Department of Computer Science, TUM (Technical University of Munich), Garching/Munich 85748, Germany
14School of Biological Sciences, University of Reading, Whiteknights, Reading RG6 6AS, UK
15Sorbonne Université, Muséum National d’Histoire Naturelle, UMR CNRS 7590, IMPMC, 75005 Paris, France
16DGA Maîtrise de l’information, 35170 Bruz, France
17Department of Electrical Engineering and Computer Science, University of Missouri – Columbia, Columbia, MO 65211, USA
18Toyota Technological Institute at Chicago, Chicago, IL, USA
19Department of Human Genetics, University of Chicago, Chicago, IL, USA
20Michael Smith Laboratories, The University of British Columbia, Vancouver, BC V6T 1Z4, Canada
21Michael Smith Laboratories, Department of Biochemistry and Molecular Biology, The University of British Columbia, Vancouver, BC V6T 1Z4, Canada
22University of Sciences, Pyongyang, D.P.R. of Korea
23Department of Computer Science, Virginia Commonwealth University, Richmond, VA, USA
24Adimab LLC,Computational Biology, Palo Alto, CA, USA
25Genomics program, College of Public Health, University of South Florida, Tampa, FL, USA
26Research Center for Mathematics and Interdisciplinary Sciences, Shandong University, Qingdao, 266237, China
27School of Mathematical Sciences and LPMC, Nankai University, Tianjin 300071, China
28School of Statistics and Data Science, LPMC and KLMDASR, Nankai University, Tianjin, China
29Department of Computer Science, University of New Orleans, New Orleans, LA, USA
30Centre for Misfolding Diseases, Yusuf Hamied Department of Chemistry, University of Cambridge, Cambridge CB2 1EW, UK
31Hunan Provincial Key Lab on Bioinformatics, School of Computer Science and Engineering, Central South University, Changsha, 410083, China
32Department of Computer Science and Engineering, School of Electronic Information and Electrical Engineering, Shanghai Jiao Tong University, Shanghai 200240, China
33Institute of Protein Research of the Russian Academy of Sciences, 4 Institutskaya str., Pushchino, Moscow Region 142290, Russia
34Institute of Theoretical and Experimental Biophysics, Russian Academy of Sciences, 142290 Pushchino, Russia
35Interuniversity Institute of Bioinformatics in Brussels, ULB-VUB, Brussels 1050, Belgium
36Structural Biology Brussels, Vrije Universiteit Brussel, Brussels 1050, Belgium
37Institute for Glycomics, Griffith University, Parklands Dr. Southport, QLD 4222, Australia
38Institute of Systems and Physical Biology, Shenzhen Bay Laboratory, Shenzhen 518107, China
39Signal Processing Laboratory, School of Engineering and Built Environment, Griffith University, Brisbane, QLD 4111, Australia
40Department of Biochemistry, Eötvös Loránd University, Pázmány Péter stny 1/c, Budapest H-1117, Hungary
Contributor Information
Alessio Del Conte, Department of Biomedical Sciences, University of Padova, via Ugo Bassi 58b, 35121, Padova, Italy.
Adel Bouhraoua, Department of Biomedical Sciences, University of Padova, via Ugo Bassi 58b, 35121, Padova, Italy.
Mahta Mehdiabadi, Department of Biomedical Sciences, University of Padova, via Ugo Bassi 58b, 35121, Padova, Italy.
Damiano Clementel, Department of Biomedical Sciences, University of Padova, via Ugo Bassi 58b, 35121, Padova, Italy.
Alexander Miguel Monzon, Department of Information Engineering, University of Padova, via Giovanni Gradenigo 6/B, 35131, Padova, Italy.
Silvio C E Tosatto, Department of Biomedical Sciences, University of Padova, via Ugo Bassi 58b, 35121, Padova, Italy.
Damiano Piovesan, Department of Biomedical Sciences, University of Padova, via Ugo Bassi 58b, 35121, Padova, Italy.
CAID predictors:
Alex S Holehouse, Daniel Griffith, Ryan J Emenecker, Ashwini Patil, Ronesh Sharma, Tatsuhiko Tsunoda, Alok Sharma, Yi Jun Tang, Bin Liu, Claudio Mirabello, Björn Wallner, Burkhard Rost, Dagmar Ilzhöfer, Maria Littmann, Michael Heinzinger, Lea I M Krautheimer, Michael Bernhofer, Liam J McGuffin, Isabelle Callebaut, Tristan Bitard Feildel, Jian Liu, Jianlin Cheng, Zhiye Guo, Jinbo Xu, Sheng Wang, Nawar Malhis, Jörg Gsponer, Chol-Song Kim, Kun-Sop Han, Myong-Chol Ma, Lukasz Kurgan, Sina Ghadermarzi, Akila Katuwawala, Bi Zhao, Zhenling Peng, Zhonghua Wu, Gang Hu, Kui Wang, Md Tamjidul Hoque, Md Wasi Ul Kabir, Michele Vendruscolo, Pietro Sormanni, Min Li, Fuhao Zhang, Pengzhen Jia, Yida Wang, Michail Yu Lobanov, Oxana V Galzitskaya, Wim Vranken, Adrián Díaz, Thomas Litfin, Yaoqi Zhou, Jack Hanson, Kuldip Paliwal, Zsuzsanna Dosztányi, and Gábor Erdős
FUNDING
The European Union's Horizon 2020 research and innovation programme MSCA-RISE [778 247, 823 886, 952 334]; ELIXIR, the research infrastructure for life-science data; COST Action ML4NGP [CA21160] is supported by COST (European Cooperation in Science and Technology) under the EU Framework Programme Horizon Europe; Italian Ministry of University and Research (MIUR) – PRIN [2017483NH8]; NextGenerationEU, PNRR – ‘ELIXIR × NextGenerationIT: Consolidamento dell’Infrastruttura Italiana per i Dati Omici e la Bioinformatica – ElixirxNextGenIT’ [IR0000010]. Funding for open access charge: University of Padova.
Conflict of interest statement. None declared.
REFERENCES
- 1. Piovesan D., Arbesú M., Fuxreiter M., Pons M.. Editorial: fuzzy interactions: many facets of protein binding. Front. Mol. Biosci. 2022; 9:947215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. CAID Predictors, DisProt Curators Necci M., Piovesan D., Tosatto S.C.E.. Critical assessment of protein intrinsic disorder prediction. Nat. Methods. 2021; 18:472–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Necci M., Piovesan D., Tosatto S.C.E.. Large-scale analysis of intrinsic disorder flavors and associated functions in the protein sequence universe. Protein Sci. Publ. Protein Soc. 2016; 25:2164–2174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Piovesan D., Monzon A.M., Quaglia F., Tosatto S.C.E.. Databases for intrinsically disordered proteins. Acta Crystallogr. Sect. Struct. Biol. 2022; 78:144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Quaglia F., Mészáros B., Salladini E., Hatos A., Pancsa R., Chemes L.B., Pajkos M., Lazar T., Peña-Díaz S., Santos J.et al.. DisProt in 2022: improved quality and accessibility of protein intrinsic disorder annotation. Nucleic Acids Res. 2021; 50:D480–D487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Troger P., Rajic H., Haas A., Domagalski P.. Standardization of an API for distributed resource management systems. Seventh IEEE International Symposium on Cluster Computing and the Grid (CCGrid ’07). 2007; 619–626. [Google Scholar]
- 7. Schäffer A.A., Aravind L., Madden T.L., Shavirin S., Spouge J.L., Wolf Y.I., Koonin E.V., Altschul S.F.. Improving the accuracy of PSI-BLAST protein database searches with composition-based statistics and other refinements. Nucleic Acids Res. 2001; 29:2994–3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Steinegger M., Meier M., Mirdita M., Vöhringer H., Haunsberger S.J., Söding J.. HH-suite3 for fast remote homology detection and deep protein annotation. BMC Bioinf. 2019; 20:473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Yang Y., Heffernan R., Paliwal K., Lyons J., Dehzangi A., Sharma A., Wang J., Sattar A., Zhou Y.. Zhou Y., Kloczkowski A., Faraggi E., Yang Y.. SPIDER2: a package to predict secondary structure, accessible surface area, and main-chain torsional angles by deep neural networks. Prediction of Protein Secondary Structure, Methods in Molecular Biology. 2017; New York, NY: Springer; 55–63. [DOI] [PubMed] [Google Scholar]
- 10. UniProt Consortium Suzek B.E., Wang Y., Huang H., McGarvey P.B., Wu C.H.. UniRef clusters: a comprehensive and scalable alternative for improving sequence similarity searches. Bioinforma. Oxf. Engl. 2015; 31:926–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Mirdita M., von den Driesch L., Galiez C., Martin M.J., Söding J., Steinegger M.. Uniclust databases of clustered and deeply annotated protein sequences and alignments. Nucleic Acids Res. 2017; 45:D170–D176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Piovesan D., Monzon A.M., Tosatto S.C.E.. Intrinsic protein disorder and conditional folding in AlphaFoldDB. Protein Sci. 2022; 31:e4466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Varadi M., Anyango S., Deshpande M., Nair S., Natassia C., Yordanova G., Yuan D., Stroe O., Wood G., Laydon A.et al.. AlphaFold Protein Structure Database: massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res. 2022; 50:D439–D444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mészáros B., Erdős G., Dosztányi Z.. IUPred2A: context-dependent prediction of protein disorder as a function of redox state and protein binding. Nucleic Acids Res. 2018; 46:W329–W337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Peng Z., Xing Q., Kurgan L.. APOD: accurate sequence-based predictor of disordered flexible linkers. Bioinformatics. 2020; 36:i754–i761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wang S., Ma J., Xu J.. AUCpreD: proteome-level protein disorder prediction by AUC-maximized deep convolutional neural fields. Bioinformatics. 2016; 32:i672–i679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Littmann M., Heinzinger M., Dallago C., Weissenow K., Rost B.. Protein embeddings and deep learning predict binding residues for various ligand classes. Sci. Rep. 2021; 11:23916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhang F., Zhao B., Shi W., Li M., Kurgan L.. DeepDISOBind: accurate prediction of RNA-, DNA- and protein-binding intrinsically disordered residues with deep multi-task learning. Brief. Bioinform. 2022; 23:bbab521. [DOI] [PubMed] [Google Scholar]
- 19. Tang Y.-J., Pang Y.-H., Liu B.. DeepIDP-2L: protein intrinsically disordered region prediction by combining convolutional attention network and hierarchical attention network. Bioinformatics. 2022; 38:1252–1260. [DOI] [PubMed] [Google Scholar]
- 20. Linding R., Jensen L.J., Diella F., Bork P., Gibson T.J., Russell R.B.. Protein disorder prediction: implications for structural proteomics. Structure. 2003; 11:1453–1459. [DOI] [PubMed] [Google Scholar]
- 21. Orlando G., Raimondi D., Codicè F., Tabaro F., Vranken W.. Prediction of disordered regions in proteins with recurrent neural networks and protein dynamics. J. Mol. Biol. 2022; 434:167579. [DOI] [PubMed] [Google Scholar]
- 22. Jones D.T., Cozzetto D.. DISOPRED3: precise disordered region predictions with annotated protein-binding activity. Bioinformatics. 2015; 31:857–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Iqbal S., Hoque M.T.. Estimation of position specific energy as a feature of protein residues from sequence alone for structural classification. PLoS One. 2016; 11:e0161452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sharma R., Tsunoda T., Sharma A.. DRPBind: prediction of DNA, RNA and protein binding residues in intrinsically disordered protein sequences. 2023; bioRxiv doi:23 March 2023, preprint: not peer reviewed 10.1101/2023.03.20.533427. [DOI]
- 25. Walsh I., Martin A.J.M., Di Domenico T., Tosatto S.C.E.. ESpritz: accurate and fast prediction of protein disorder. Bioinformatics. 2012; 28:503–509. [DOI] [PubMed] [Google Scholar]
- 26. Hu G., Katuwawala A., Wang K., Wu Z., Ghadermarzi S., Gao J., Kurgan L.. flDPnn: accurate intrinsic disorder prediction with putative propensities of disorder functions. Nat. Commun. 2021; 12:4438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Galzitskaya O.V., Garbuzynskiy S.O., Lobanov M.Y.. FoldUnfold: web server for the prediction of disordered regions in protein chain. Bioinformatics. 2006; 22:2948–2949. [DOI] [PubMed] [Google Scholar]
- 28. Lobanov M.Y., Sokolovskiy I.V., Galzitskaya O.V.. IsUnstruct: prediction of the residue status to be ordered or disordered in the protein chain by a method based on the Ising model. J. Biomol. Struct. Dyn. 2013; 31:1034–1043. [DOI] [PubMed] [Google Scholar]
- 29. Erdős G., Pajkos M., Dosztányi Z.. IUPred3: prediction of protein disorder enhanced with unambiguous experimental annotation and visualization of evolutionary conservation. Nucleic Acids Res. 2021; 49:W297–W303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Emenecker R.J., Griffith D., Holehouse A.S.. Metapredict: a fast, accurate, and easy-to-use predictor of consensus disorder and structure. Biophys. J. 2021; 120:4312–4319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Necci M., Piovesan D., Clementel D., Dosztányi Z., Tosatto S.C.E.. MobiDB-lite 3.0: fast consensus annotation of intrinsic disorder flavours in proteins. Bioinformatics. 2020; 36:5533–5534. [DOI] [PubMed] [Google Scholar]
- 32. Malhis N., Jacobson M., Gsponer J.. MoRFchibi SYSTEM: software tools for the identification of MoRFs in protein sequences. Nucleic Acids Res. 2016; 44:W488–W493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sharma R., Raicar G., Tsunoda T., Patil A., Sharma A.. OPAL: prediction of MoRF regions in intrinsically disordered protein sequences. Bioinformatics. 2018; 34:1850–1858. [DOI] [PubMed] [Google Scholar]
- 34. Deng X., Eickholt J., Cheng J.. PreDisorder: ab initio sequence-based prediction of protein disordered regions. BMC Bioinf. 2009; 10:436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Mirabello C., Wallner B.. rawMSA: end-to-end deep learning using raw multiple sequence alignments. PLoS One. 2019; 14:e0220182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Yang Z.R., Thomson R., McNeil P., Esnouf R.M.. RONN: the bio-basis function neural network technique applied to the detection of natively disordered regions in proteins. Bioinformatics. 2005; 21:3369–3376. [DOI] [PubMed] [Google Scholar]
- 37. Sormanni P., Camilloni C., Fariselli P., Vendruscolo M.. The s2D method: simultaneous sequence-based prediction of the statistical populations of ordered and disordered regions in proteins. J. Mol. Biol. 2015; 427:982–996. [DOI] [PubMed] [Google Scholar]
- 38. Ilzhöfer D., Heinzinger M., Rost B.. SETH predicts nuances of residue disorder from protein embeddings. Front. Bioinforma. 2022; 2:1019597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hanson J., Yang Y., Paliwal K., Zhou Y.. Improving protein disorder prediction by deep bidirectional long short-term memory recurrent neural networks. Bioinforma. Oxf. Engl. 2017; 33:685–692. [DOI] [PubMed] [Google Scholar]
- 40. Hanson J., Paliwal K., Zhou Y.. Accurate single-sequence prediction of protein intrinsic disorder by an ensemble of deep recurrent and convolutional architectures. J. Chem. Inf. Model. 2018; 58:2369–2376. [DOI] [PubMed] [Google Scholar]
- 41. Hanson J., Paliwal K.K., Litfin T., Zhou Y.. SPOT-Disorder2: improved protein intrinsic disorder prediction by ensembled deep learning. Genomics Proteomics Bioinformatics. 2019; 17:645–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Peng K., Radivojac P., Vucetic S., Dunker A.K., Obradovic Z.. Length-dependent prediction of protein intrinsic disorder. BMC Bioinf. 2006; 7:208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Mistry J., Chuguransky S., Williams L., Qureshi M., Salazar G.A., Sonnhammer E.L.L., Tosatto S.C.E., Paladin L., Raj S., Richardson L.J.et al.. Pfam: the protein families database in 2021. Nucleic Acids Res. 2021; 49:D412–D419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Lewis T.E., Sillitoe I., Dawson N., Lam S.D., Clarke T., Lee D., Orengo C., Lees J.. Gene3D: extensive prediction of globular domains in proteins. Nucleic Acids Res. 2018; 46:D1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Paysan-Lafosse T., Blum M., Chuguransky S., Grego T., Pinto B.L., Salazar G.A., Bileschi M.L., Bork P., Bridge A., Colwell L.et al.. InterPro in 2022. Nucleic Acids Res. 2023; 51:D418–D427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Capella-Gutierrez S., Iglesia D.d., Haas J., Lourenco A., Fernández J.M., Repchevsky D., Dessimoz C., Schwede T., Notredame C., Gelpi J.L.et al.. Lessons learned: recommendations for establishing critical periodic scientific benchmarking. 2017; bioRxiv doi:31 August 2017, preprint: not peer reviewed 10.1101/181677. [DOI]
- 47. Piovesan D., Del Conte A., Clementel D., Monzon A.M., Bevilacqua M., Aspromonte M.C., Iserte J.A., Orti F.E., Marino-Buslje C., Tosatto S.C.E.. MobiDB: 10 years of intrinsically disordered proteins. Nucleic Acids Res. 2023; 51:D438–D444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Blum M., Chang H.-Y., Chuguransky S., Grego T., Kandasaamy S., Mitchell A., Nuka G., Paysan-Lafosse T., Qureshi M., Raj S.et al.. The InterPro protein families and domains database: 20 years on. Nucleic Acids Res. 2021; 49:D344–D354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. The UniProt Consortium UniProt: the universal protein knowledgebase in 2021. Nucleic Acids Res. 2021; 49:D480–D489. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The CAID Prediction Portal is freely available at https://caid.idpcentral.org.