Abstract
PHASTEST (PHAge Search Tool with Enhanced Sequence Translation) is the successor to the PHAST and PHASTER prophage finding web servers. PHASTEST is designed to support the rapid identification, annotation and visualization of prophage sequences within bacterial genomes and plasmids. PHASTEST also supports rapid annotation and interactive visualization of all other genes (protein coding regions, tRNA/tmRNA/rRNA sequences) in bacterial genomes. Given that bacterial genome sequencing has become so routine, the need for fast tools to comprehensively annotate bacterial genomes has become progressively more important. PHASTEST not only offers faster and more accurate prophage annotations than its predecessors, it also provides more complete whole genome annotations and much improved genome visualization capabilities. In standardized tests, we found that PHASTEST is 31% faster and 2–3% more accurate in prophage identification than PHASTER. Specifically, PHASTEST can process a typical bacterial genome in 3.2 min (raw sequence) or in 1.3 min when given a pre-annotated GenBank file. Improvements in PHASTEST’s ability to annotate bacterial genomes now make it a particularly powerful tool for whole genome annotation. In addition, PHASTEST now offers a much more modern and responsive visualization interface that allows users to generate, edit, annotate and interactively visualize (via zooming, rotating, dragging, panning, resetting), colourful, publication quality genome maps. PHASTEST continues to offer popular options such as an API for programmatic queries, a Docker image for local installations, support for multiple (metagenomic) queries and the ability to perform automated look-ups against thousands of previously PHAST-annotated bacterial genomes. PHASTEST is available online at https://phastest.ca.
Graphical Abstract
Graphical Abstract.
INTRODUCTION
Bacteriophages, also known as phages, are the most abundant biological entities on Earth (1). Phages are viruses that specifically infect and replicate in bacterial cells. They tend to fall into two categories: lytic phages and temperate phages (2). Lytic phages, such as T4, infect and replicate within bacteria leading to the eventual lysis (and death) of the infected bacterium. Temperate phages, such as phage lambda, do not always immediately lyse the infected cell. Upon infection, most phage proceed through the lytic cycle while a small fraction undergo lysogeny. Lysogeny involves the stable integration of the phage genome into the host bacterial chromosome or the stable formation of an extrachromosomal plasmid inside the bacterium. These integrated phages are called endogenous phages or prophages. Prophages may remain embedded in the genome through multiple cell divisions until activation by an external factor that leads to the production of new phage particles, causing cell lysis. In some cases, prophages can become permanently embedded into the bacterial genome and are called cryptic prophages (3). These cryptic phages are crippled and are unable to proceed through the lytic cycle. Likely, multiple cycles of replication within the bacterial genome have caused inactivation or deletion of the lytic cycle genes. However, the presence of a cryptic prophage in a bacterial genome allows the bacterium to avoid cell lysis or reinfection by the same phage as the immunity genes may yet be intact. Cryptic prophages can also give the cell a number of other selective advantages, such as antibiotic resistance, increased virulence or enhanced metabolic capacity to survive harsh environments (1, 2). In many cases, cryptic prophages function as a genetic ‘reserve’ for future evolutionary changes of the host bacterium (4). Because of their potential mutual benefits, prophages and cryptic prophages are surprisingly abundant and can account for up to 20% of the genetic material in some bacterial genomes (2). The fact that phages and prophages are so abundant and play such an important role in bacterial evolution and pathology has led to increased interest in identifying and annotating prophage sequences in bacterial genomes. As a result, prophage finding programs and web servers have become integral to many bacterial genome annotation pipelines.
Some of these prophage finding programs include so-called ‘traditional’ phage finding tools such as Phage_Finder (5), Prophage Finder (6) and Prophinder (7). These tools employ sequence comparisons (to known phage and bacterial genes), tRNA prediction and dinucleotide analysis, along with attachment site detection using a variety of pattern matching techniques. More recently, a number of ‘next-generation’ phage finding tools have appeared that employ more advanced machine learning or deep learning methods. These include Prophage Hunter (8), PPR-Meta (9) and DeepVirFinder (10), which use convolutional neural networks to identify phage features. A more recent addition is Virtifier (11), which uses an attention-based long short-term memory (LSTM) network to identify prophage. These innovations have certainly improved the accuracy of prophage identification and the detection of prophages within metagenomic data. However, even with these advances, we believe there is still room for improvement, particularly in the areas of accessibility, speed, user-friendliness and usability of phage finders. This motivated our development of two web servers for prophage annotation: PHAST (PHAge Search Tool), published in 2011 (12) and its successor PHASTER (PHAge Search Tool – Enhanced Release), released in 2016 (13). Both of these tools offered fast, visually appealing, easy-to-understand and accurate prophage annotations and both have become exceedingly popular. The PHAST paper has been cited > 1900 times and the PHASTER paper has been cited >2400 times. Together, these web servers handle over 200 000 submissions each year. Nevertheless, user feedback, ongoing algorithmic improvements and continuing advances in web technology have led us to develop a better, faster, more accurate, more comprehensive and more visually appealing tool for phage finding and general genome annotation.
Here we introduce PHASTEST (PHAge Search Tool with Enhanced Sequence Translation), the successor to previous members of the PHAST family of prophage servers. PHASTEST is a web server designed to support the rapid identification, annotation and visualization of prophage sequences within bacterial genomes and plasmids. PHASTEST not only offers faster and more accurate prophage annotations than its predecessors, it also provides more complete whole genome annotations and much improved genome visualization capabilities. These improvements in PHASTEST’s ability to annotate bacterial genomes now make it a particularly powerful tool for whole genome annotation. In addition, PHASTEST now offers a much more modern and responsive visualization interface that allows users to generate, edit, annotate and interactively visualize colourful, publication quality genome maps. These and other improvements are described in more detail below.
Back-end improvements
Algorithmic upgrades and performance optimizations
Prophage searching is a computationally intensive task that requires accurate ORF identification along with large-scale (protein or RNA) sequence comparisons and alignments. Previous versions of the PHAST family of prophage finders used GLIMMER (14) for the initial ORF identification and protein translation phase. In PHASTEST, we opted to replace GLIMMER with Prodigal (15). Comparisons between GLIMMER and Prodigal revealed that Prodigal not only had much lower false-positive and lower false-negative rates for ORF identification, it was also faster than GLIMMER. Tests conducted against 54 reference genomes showed that Prodigal had an average accuracy of 88.7% compared to 81.3% for GLIMMER. More details about the 54 reference genomes can be found by clicking the ‘About’ tab on the PHASTEST server, selecting the ‘Statistics‘ section and scrolling down to the end of the Figure 2 legend. Adopting Prodigal not only improved PHASTEST’s overall ORF identification accuracy and reduced the time taken during ORF identification, but it also had the added effect of decreasing overall runtime since fewer ORFs needed to be passed on to the sequence alignment phase.
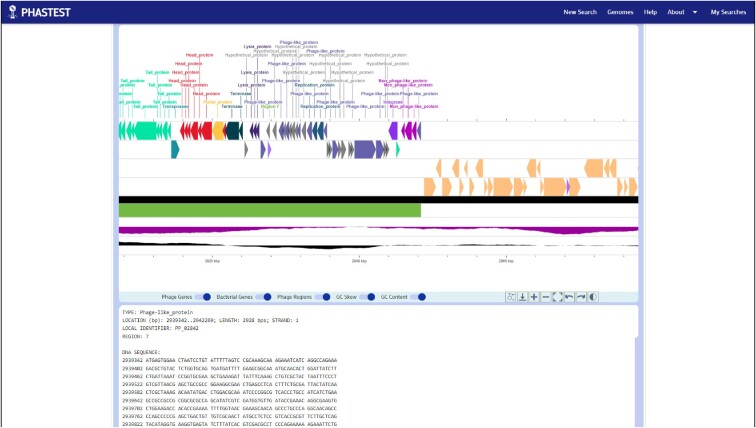
Figure 2.
PHASTEST linear genome viewer. Clicking on specific regions or specific genes on the circular genome viewer will automatically scroll the webpage down to the linear viewer.
PHASTEST has an expanded PHASTER’s protein sequence alignment pipeline to improve speed, accuracy, and user experience. PHASTEST maintains use of BLAST + with a locally curated database of 420 000 phage proteins for phage sequence alignment, but has replaced BLAST+ with Diamond BLAST (16) for faster bacterial sequence alignment. For unannotated FASTA sequence inputs, PHASTEST follows a two-step annotation process beginning with phage sequence alignment, followed by bacterial sequence alignment. For GenBank record inputs, if the query includes a set of pre-annotated CDS regions, then only the phage sequence alignment step is performed. If no pre-annotated CDS regions are present, then the two-step annotation process is followed. Additionally, for users that submitted accession numbers or FASTA sequences that had already been annotated, PHASTEST will retrieve the previously calculated output (if input was an accession number) or sequence alignment result (if input was a FASTA sequence) from its PHASTEST archive of previously annotated genomes (PHAST-ARCHIVE) directly, allowing users to bypass the time-consuming sequence alignment step altogether. This option is available to fast-track the process of annotation and to generate results for pre-annotated genomes and sequences.
Furthermore, as part of its new focus on ‘Enhanced Sequence Translation’, PHASTEST now offers two modes of bacterial sequence annotation – a ‘lite’ annotation mode that uses the Swiss-Prot database (17) with nearly 600 000 bacterial protein sequences, and a ‘deep’ annotation mode that uses a custom bacterial sequence database (PHAST-BSD) containing over 16 million bacterial protein sequences. Because of the compactness of the Swiss-Prot database, bacterial sequence alignment and annotation is 56% faster in the lite annotation mode compared to deep annotation mode with PHAST-BSD (see Table 1). Furthermore, using the Swiss-Prot database enables a more detailed predicted protein output than the PHAST-BSD database would allow. For instance, in PHASTER, a large number of proteins were generally labeled as ‘phage-like proteins’ but in PHASTEST, most of these proteins are now assigned to specific protein families such as repressors, exonucleases, kinases, endopeptidases, crossover-junction proteins, etc. The deep annotation mode, which uses the much larger PHAST-BSD database, detects and annotates 26% more proteins than the lite annotation mode.
Table 1.
Performance runtime comparison (in s) between PHAST, PHASTER and PHASTEST using identical (new) hardware, but with different databases, search algorithms and query types using E. coli O157:H7 (NC_002655.2) as the query genome
| Cumulative set of performance enhancements | BLAST vs. phage database runtime (sec) | BLAST vs. bacterial database runtime (sec) | GenBank annotated genome runtime (sec) | Unannotated genome runtime (sec) |
|---|---|---|---|---|
| PHAST (baseline) - current DBs, no other upgrades | 191 | 576 | 270 | 899 |
| PHASTER (baseline) - current DBs, no other upgrades | 116 | 83 | 162 | 277 |
| PHASTEST (upgrade 1) - BLAST+ parameter adjustment | 82 | 82 | 144 | 229 |
| PHASTEST (upgrade 2) - Whole-sequence Prodigal | 81 | 71 | 141 | 201 |
| PHASTEST (upgrade 3) - Parallel Diamond | 84 | 124 | 118 | 266 |
| PHASTEST (upgrade 4) - Swiss-Prot DB | 80 | 64 | 110 | 195 |
The speed of sequence alignment was further improved through various computing cluster optimizations. Earlier versions of the PHAST family of prophage finders employed a grid scheduler but with only minimal optimizations, resulting in frequently idle CPU cores. PHASTEST now sends its input data to a grid scheduler so that all CPU cores are used more efficiently, particularly in cases when the server is handling a single user submission and must complete it as quickly as possible. PHASTEST has also partitioned its PHAST-BSD bacterial sequence database into eight equal subsets so that during the deep annotation mode, each query sequence is now searched against the smaller sub-databases. Additionally, the query is also divided into smaller sequence fragments and then these fragments are queried against each of the smaller sub-databases. With these optimizations, smaller BLAST + jobs can be more readily distributed to available CPU cores as they become available. Table 1 compares the runtime (speed) performance of PHAST, PHASTER and PHASTEST in terms of database sizes, algorithms and query types (including raw DNA sequences and pre-annotated GenBank sequences). Accuracy assessments using a large, ‘gold standard’ set of 54 annotated genomes show that sensitivity has improved from 79.4% (PHAST) to 85.0% (PHASTER) to 85.8% (PHASTEST), while the positive predictive value (PPV) has improved from 86.5% (PHAST) to 87.3% (PHASTER) to 91.2% (PHASTEST). Additional details regarding sensitivity and specificity for PHASTEST (and other members of the PHAST suite) are available on the PHASTEST website and can be found by clicking the ‘About’ tab, selecting the ‘Statistics‘ section, and scrolling down to Table 3: PHASTEST’s evaluation (summary). As PHASTEST is a predictive tool, it is important to remember that PHASTEST predictions are not 100% accurate. For instance, the predictions of the attachment sites can be different than the actual attachment site positions in a small number of cases.
As with earlier versions of the PHAST family of prophage finders, PHASTEST continues to provide a support for contig-based queries. For the first time, PHASTEST introduces support for whole-genome shotgun (WGS) sequencing from NCBI. If a user enters a WGS master record accession number as input, PHASTEST will retrieve each sub-record associated with the master record automatically. Then, the whole record is processed and the results displayed in order of accession numbers (with their respective annotated proteins and predicted phage regions). The option to search for prophage regions in contigs assembled from metagenomic data, as with previous PHAST prophage finders, is also supported. With the metagenomic option selected, complete and partial genes are first predicted using FragGeneScan (18). Subsequently, the predicted prophages are arranged by contig in the generated results. A detailed set of contig performance data are available on the PHASTEST website as a figure (Figure 1) found under the ‘Statistics’ section under the ‘About’ tab.
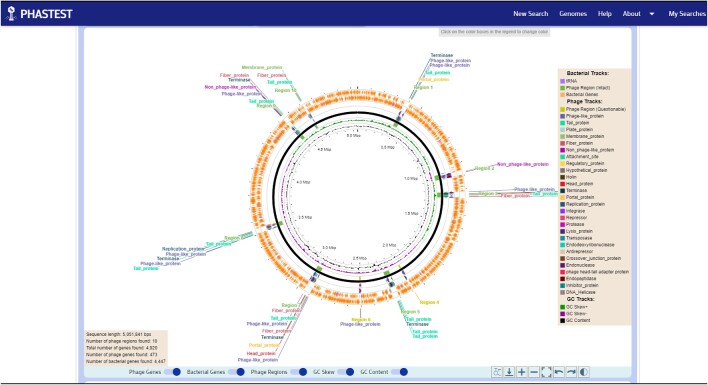
Figure 1.
PHASTEST default circular genome viewer. All the predicted phage regions, phage genes and bacterial genes can be easily viewed to see how different phage regions are positioned relative to each other across the entire genome. A genome summary table (bottom left corner of the circular genome image) can be displayed which includes information on the genome sequence length, the number of phage regions found, and the total number of genes found.
Programmatic access
Improvements in DNA sequencing technology have made it far easier to sequence multiple (complete or partial) bacterial or plasmid genomes in a short period of time. In order to support multiple whole genome submissions or multiple metagenomic submissions, PHASTEST continues to offer an Application Programming Interface (API) that supports the submission of both multiple whole genomes and multiple separate contigs without using the web interface (for more information see ‘Help’ on the PHASTEST website and scroll down to the section called ‘How to use the URLAPI’). This API allows users to upload a large number of submissions to the PHASTEST server and check the status of each job at their convenience, whether they are genomic sequences or metagenomic contigs. Results from PHASTEST API queries can be downloaded via the API or viewed on the PHASTEST web interface.
Even though PHASTEST and its predecessors were designed with speed in mind, the overwhelming popularity of these servers and the shift towards larger-scale submissions (via the API) has often meant that long submission queues develop during peak hours. To mitigate these issues, we have now created a Docker (19) image of PHASTEST that is downloadable from the PHASTEST website (under ‘About’). Docker is a containerization system that uses OS-level virtualization to create portable software in packages called containers. The containers have everything the software needs to run including libraries, databases, system tools, code and the web interface. Providing a Docker image of PHASTEST and all its accompanying databases means that users with heavy genome or prophage annotation needs can now download, install and run PHASTEST locally. The entire Docker image is nearly 5 GB in size and instructions on how to install and test the Dockerized version of PHASTEST are provided on the PHASTEST home page (under ‘About’ → Downloads).
Improved whole genome annotation
Given that the vast majority (>90%) of submissions to the PHAST family of servers are now raw DNA sequences (as opposed to annotated GenBank files), a major focus in developing PHASTEST was on improving the quality and extent of genome annotation for all genes for whole genome submissions. Historically, the PHAST family of phage finders was limited to annotating only prophage elements. As a result, other genetic elements (protein coding regions, tRNA, rRNA and tmRNA) outside these prophage regions were left mostly unannotated. In this release of PHASTEST, we have significantly improved its whole genome annotation functions. Now all protein-coding regions identified via Prodigal, BLAST+ and Diamond BLAST are given presumptive protein names, gene start/end positions, strand orientation information, GO-Lite functional categories, protein sequence length, calculated molecular weight and other data as inferred by BLAST + matches or internal protein annotation programs. A total of 14 different annotations are provided for each protein coding gene. These annotations can be downloaded as a single multi-FASTA file as well as searched or interactively viewed on the PHASTEST genome browser (as described later). In addition to providing more complete protein-coding region annotation, PHASTEST also supports non-protein coding region annotation. Now, all tRNA genes (as identified via tRNAscan-SE (20), tmRNA genes (as identified via Aragorn (21) and rRNA genes (as identified via barrnap [https://github.com/tseemann/barrnap.git]) throughout the genome are also identified, annotated and downloadable in the same multi-FASTA file. These RNA genes can also be searched or interactively viewed on the PHASTEST genome browser. As we learned from a recent user survey, most users would often turn to another tool to annotate their bacterial genome after getting the phage region predictions from PHAST/PHASTER. By upgrading PHASTEST to become a more complete genome annotation tool, we believe we have addressed this issue and it should make PHASTEST more of a ‘one-stop-shop’ for microbial researchers.
Database expansion
Like its predecessors, PHASTEST depends on the availability of high-quality sequence databases to perform most of its analyses and predictions. Three databases are used: (i) a bacterial prophage sequence database (called PHAST-PSD); (ii) a non-redundant bacterial protein sequence database (called PHAST-BSD) for deep annotations and (iii) the Swiss-Prot bacterial protein sequence database for lite annotations. Both in-house databases (PHAST-PSD and PHAST-BSD) were constructed at the time of release of PHAST in 2011. Both have been continuously improved and expanded with releases of PHASTER in 2016 and PHASTEST. The number of bacterial prophage sequences in the PHAST-PSD has steadily increased from ∼45 000 (in PHAST) to 187 000 (in PHASTER) to > 400 000 in PHASTEST. Likewise, the PHAST-BSD has grown from ∼4 million bacterial sequences (in PHAST) to 9 million (in PHASTER) to 16 million in PHASTEST. For both PHASTEST and PHASTER, we reduced the size of the PHAST-BSD by removing sequences with >70% sequence identity to any other sequence in the database, using CD-HIT (22). Certainly, as the databases have expanded, the time needed to perform sequence comparisons has also increased. These time costs have been mitigated by improving the algorithms (as described above) and upgrading the hardware (as described below).
Because we found that so many PHASTER and PHASTEST queries involve submissions of previous PHAST-annotated genomes, PHASTEST continues to perform a quick-query search to rapidly return ‘known’ results to users without performing lengthy calculations. This involves comparing the query against a local database of non-redundant, previously annotated (via PHASTER and PHASTEST) bacterial or plasmid genomes. This database (called PHAST-ARCHIVE) has grown from 14 000 sequences to more than 750 000 today. As described previously, this quick query function compares the query sequence's nucleotide frequencies and total sequence length against a database of these same statistics for all sequences in the PHAST-ARCHIVE database. Potential sequence matches are identified (often just one or two) and then aligned against the query sequence to ensure that only exact sequence matches are used. Having identified a query that is identical to an entry in the PHAST-ARCHIVE database, the annotations are transferred, and the result is returned to the user in a few seconds. As a result, while the average de novo query to PHASTEST may take 2–3 min, a significant number of user queries can be returned in 5–10 s.
Hardware upgrades
Software enhancements are not the only route to improve a web server's speed or performance. We have continued to expand the number of CPU cores in the PHAST family of servers, from 32 (in the original PHAST) to 112 (in PHASTER) to 128 (in PHASTEST). The PHASTEST cluster now has 4 Intel Xeon X5460 @ 3.16 GHz, 6 AMD Opteron 2220, and 2 AMD Opteron 6348 processing cores. We have also added more RAM to the PHASTEST server, increasing it from 400 GB to 432 GB. This additional RAM allows PHASTEST to load more of its databases into active memory, thereby decreasing the overall time spent on slower disk access operations. In addition, the front-end for the PHASTER website has been placed on a much quicker virtual server using the Google Compute Engine. This front-end server has 2 Intel Xeon CPUs @ 2.30 GHz and a local solid-state drive. The front-end performs many of PHASTEST’s other computations, and now does so approximately 50% faster. Because PHASTEST has a dedicated front-end server, it is able to accommodate multiple jobs simultaneously for the most memory-intensive portions of the data processing pipeline. This provides faster results during periods of heavier use.
Front-end improvements
Both minor and major front-end enhancements were made to PHASTEST. The minor front-end improvements were limited to the PHASTEST home page, data upload page and style sheets. These were primarily done to improve the layout and color scheme. These layout changes made the PHASTEST website look more modern, more understandable and have helped enhance the overall user experience. For instance, the sequence/file input box has been moved to the top of the web page so that it is the first item that a user sees when opening the data upload page. A more appealing color scheme, a new banner and a new logo has been designed and added to the home page to make the PHASTEST server look and behave more similar to other Wishart lab servers. Likewise, the color scheme for indicating the completeness of a predicted phage region has been changed to the more intuitive red, yellow, and green to indicate ‘incomplete’, ‘questionable’ and ‘intact’ phage regions. This colour scheme has been made consistent across both the tabulated results tabs and the genome viewer tabs. Likewise, an option has been added so that users can save their searches using a cookie-based storage mechanism by clicking on the appropriate check box. This will work for anyone returning to the PHASTEST website using the same browser on the same computer, provided that the browser has cookies enabled. Previously submitted jobs saved in this way will be available under the new ‘My Searches’ section, without any need to log in. This feature is optional, and users can still bookmark their results pages as an alternative tracking method.
The most significant front-end upgrades to PHASTEST were made to the genome viewing tools. The predecessor to PHASTEST (i.e. PHASTER) used an interactive genome viewer that was originally built using JavaScript, employing AngularPlasmid (http://angularplasmid.vixis.com) for the circular genome viewer and D3js (http://d3js.org) for the linear genome viewer. However, over the last six years a number of improvements in web technology and the quality of interactive genome viewers has occurred. Likewise, the need to improve a user's ability to see both phage and non-phage annotations (due to PHASTEST’s ‘Enhanced Sequence Translation’ tools) required substantial upgrades to the existing viewer. As a result, a complete rewrite of the old PHASTER genome viewer was undertaken using CGView.js (23). CGView.js is the JavaScript adaptation of the popular Java program known as CGView (Circular Genome View) (24). JavaScript tools are ideal for interactive visualization of images or objects on web pages. The JavaScript version of CGView supports the rendering and interactive visualization of both circular and linear genome views on the web and is capable of rendering genomes up to 10 MB with 1000’s of features. It supports smooth zooming from a simple ‘backbone’ genome image all the way down to the sequence level. CGView.js also allows users to easily generate gene-level features and plots (GC-content, GC-skew) directly from the sequence and to render and save high resolution PNG genome images of up to 8000 × 8000 pixels. To allow more user interactivity, a graphical user interface with various image control widgets (called ‘viewer control buttons’) was built around CGView using D3.js.
The default genome view for PHASTEST is the circular viewer. Through this circular view users can more easily and interactively explore their query sequence and view all the predicted phage regions, all the predicted phage genes and all the predicted bacterial genes. This allows users to easily see how different phage regions are positioned relative to each other across the entire genome (Figure 1). At the bottom left corner of the circular genome image, a genome summary table is presented. This contains information on the genome sequence length, the number of phage regions found, and the total number of genes found. Users can use their mouse or trackpad to click on specific regions or specific genes which will automatically scroll the webpage down to the ‘ultra-expanded’ linear viewer (Figure 2). The linear viewer is then automatically zoomed onto the selected region or gene. A text panel at the bottom of the linear genome viewer displays information about the selected feature in a succinct tabular format. For instance, if a user clicks on a predicted phage region, the text panel will show the predicted phage name that is most likely to be responsible for that specific cluster of phage genes, the location of the prophage region (start and stop positions), the sequence length, the GC content, the completeness level, and the DNA sequence for that region. Both the circular and linear genome viewers have a popup card that is revealed when a user hovers their cursor over any predicted region or gene (Figure 3). Users may also enter gene positions in the ‘Search box’ on the upper left corner to localize and expand the view. If multiple gene names match, they will be highlighted on the genome map in a different color and the user must manually click on a highlighted gene to expand and view it in greater detail.
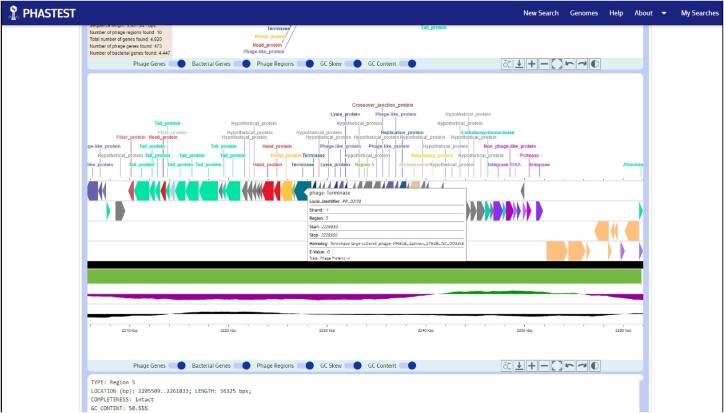
Figure 3.
Popup gene cards generated for specific regions or genes. Hovering their cursor over any predicted region in the circular or linear genome view generates a popup card with the location of the prophage region, its start and stop positions, the completeness level, the GC content and the prophage name. Hovering their cursor over any predicted gene reveals the gene name, local identifier, the strand (+ or –), the region, the start and stop positions, the highest scoring homolog and BLAST E-value.
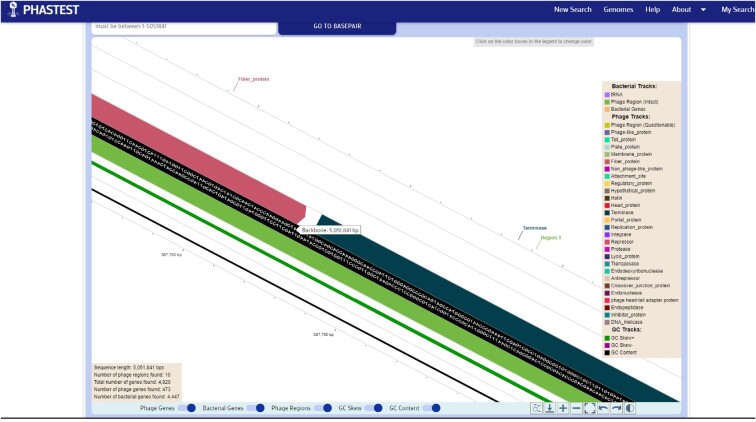
The genome maps presented on the viewers are structured identically, except one is circular and one is linear. Within the circular viewer, there are four tracks on the outside and three tracks on the inside of the genome ‘backbone’ which contains the sequence itself (Figure 4). The backbone displays the DNA sequence when a user zooms in far enough (using their mouse scroll wheel or trackpad). The two outermost tracks contain the bacterial genes, marked in orange, and separated by strand direction. The next two tracks contain the predicted phage genes, colored according to our annotations scheme, which are also separated by strand direction. All genes are shown as rectangular arcs with arrows indicating their orientation. The first track on the inside of the backbone contains the predicted phage regions which are represented by rectangular arcs and color coded according to their completeness level. The next two tracks illustrate the GC skew and the GC content of the sequence.
Figure 4.
Zoomed-in view of the circular genome viewer. This fully zoomed-in portion of the circular genome view shows how multiple tracks can be viewed. Four tracks on the outside: the two outermost tracks contain bacterial genes, marked in orange, separated by strand directions (– then +) and the next two tracks contain the predicted phage genes, also separated by strand directions (- then +). Three tracks on the inside of the circular ‘backbone’ containing the DNA sequence: the first track on the inside of the backbone contains the predicted phage regions and the next two tracks illustrate the GC skew and the GC content of the sequence.
In addition to offering the ability to click on specific features, users can drag the genome maps across the screen or zoom in and out using their mouse scroll wheel (or track pad). Users can also interact with the genome viewers using the ‘viewer control’ buttons shown below the map viewing panel. These buttons may be used to zoom in and out as well as to pan right and left on the genome map. They also can be used to reset or re-center the view. The legend box and the map annotations can also be toggled on or off using these ‘viewer control’ buttons. The legend box contains the color scheme for PHASTEST’s annotations. Users can click on the color swatches to the left of the legend names to modify the colors to their liking or modify the color scheme of protein-coding regions using GO-lite annotations, or even make certain classes of annotations invisible. Additionally, there are switches at the bottom of both the linear and circular map viewer panels that can be used to toggle the different tracks on the map. After editing the genome image (linear or circular) to their liking, users can download a high-resolution PNG file of that image, which is of publication quality.
CONCLUSIONS
Given the increasing popularity of the PHAST family of phage finding web servers, the growing demand by our user community for comprehensive genome annotation and the continuing improvements in both algorithms, hardware and data visualization tools, we decided to undertake a major update to the PHASTER web server. This work led to the creation of a new, significantly enhanced release of PHASTER called PHASTEST, which has been described in this report. In many respects, PHASTEST is faster, better, easier to use and more comprehensive than all previous members of the PHAST phage server suite (which are still being maintained for users). These performance enhancements were achieved through the addition of better genome annotation tools, through continued code optimization, through improved database preparation, and ongoing hardware upgrades. We also made PHASTEST’s web interface much more colorful, consistent, convenient and user-friendly. Despite having to handle larger databases and more complex annotation tasks, PHASTEST is still ∼31% faster than PHASTER and about 2–3% more accurate in terms of sensitivity and PPV. If users submit genome sequences that have been previously handled by PHASTEST, the server can be up to 400 times faster. These back-end changes were implemented to help handle the lengthening queues and growing demands on the PHAST suite of phage finding servers. In addition to these web server enhancements, PHASTEST is now available as a containerized (Docker) version. This will allow users to download and locally run PHASTEST on their own computers. The availability of a locally installable version of PHASTEST should further reduce the load on the server, making the PHASTEST web server more appealing to the general community. Likewise, making a Dockerized, installable version of PHASTEST available should make it more broadly appealing to ‘power-users’. While the name PHASTEST implies this is the end of the road for developments in the PHAST family of phage finders, we expect incremental improvements, such as in-house database improvements, enhancements to PHASTEST pipeline and algorithm, and improvements in the prediction accuracy of the annotated genes and attachment sites. These improvements will continue to be made and that a version numbering scheme (i.e. PHASTEST 2.0) will be used to announce and track future releases.
DATA AVAILABILITY
A Docker image of PHASTEST along with all of its accompanying databases is available for users to download, install and run locally. The entire Docker image is nearly 5 GB in size and instructions on how to install and test the Dockerized version of PHASTEST are provided on the PHASTEST home page (under ‘About’ → Downloads).
ACKNOWLEDGEMENTS
The authors wish to thank Dr Marcia LeVatte for her assistance in editing and proofing the manuscript.
Contributor Information
David S Wishart, Department of Biological Sciences, University of Alberta, Edmonton, AB, T6G 2E9, Canada; Department of Computing Science, University of Alberta, Edmonton, AB, T6G 2E8, Canada; Department of Laboratory Medicine and Pathology, University of Alberta, Edmonton, AB, T6G 2B7, Canada; Faculty of Pharmacy and Pharmaceutical Sciences, University of Alberta, Edmonton, AB, T6G 2H7, Canada.
Scott Han, Department of Biological Sciences, University of Alberta, Edmonton, AB, T6G 2E9, Canada.
Sukanta Saha, Department of Biological Sciences, University of Alberta, Edmonton, AB, T6G 2E9, Canada.
Eponine Oler, Department of Biological Sciences, University of Alberta, Edmonton, AB, T6G 2E9, Canada.
Harrison Peters, Department of Biological Sciences, University of Alberta, Edmonton, AB, T6G 2E9, Canada.
Jason R Grant, Department of Agricultural, Food and Nutritional Science, University of Alberta, Edmonton, AB, T6G 2P5, Canada.
Paul Stothard, Department of Agricultural, Food and Nutritional Science, University of Alberta, Edmonton, AB, T6G 2P5, Canada.
Vasuk Gautam, Department of Biological Sciences, University of Alberta, Edmonton, AB, T6G 2E9, Canada.
Funding
Canadian Institutes of Health Research (CIHR); Canada Foundation for Innovation (CFI); Genome Alberta, a division of Genome Canada. Funding for open access charge: Genome Canada.
Conflict of interest statement. None declared.
REFERENCES
- 1. Fortier L.C., Sekulovic O.. Importance of prophages to evolution and virulence of bacterial pathogens. Virulence. 2013; 4:354–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Casjens S. Prophages and bacterial genomics: what have we learned so far?. Mol. Microbiol. 2003; 49:277–300. [DOI] [PubMed] [Google Scholar]
- 3. Wang X., Kim Y., Ma Q., Hong S.H., Pokusaeva K., Sturino J.M., Wood T.K.. Cryptic prophages help bacteria cope with adverse environments. Nat. Commun. 2010; 1:147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bobay L.M., Touchon M., Rocha E.P.C.. Pervasive domestication of defective prophages by bacteria. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:12127–12132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Fouts D.E. Phage_Finder: automated identification and classification of prophage regions in complete bacterial genome sequences. Nucleic Acids Res. 2006; 34:5839–5851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bose M., Barber R.D.. Prophage Finder: a prophage loci prediction tool for prokaryotic genome sequences. In Silico Biol. (Gedrukt). 2006; 6:223–227. [PubMed] [Google Scholar]
- 7. Lima-Mendez G., Helden J.V., Toussaint A., Leplae R.. Prophinder: a computational tool for prophage prediction in prokaryotic genomes. Bioinformatics. 2008; 24:863–865. [DOI] [PubMed] [Google Scholar]
- 8. Song W., Sun H.X., Zhang C., Cheng L., Peng Y., Deng Z., Wang D., Wang Y., Hu M., Liu W.et al.. Prophage Hunter: an integrative hunting tool for active prophages. Nucleic Acids Res. 2019; 47:W74–W80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Fang Z., Tan J., Wu S., Li M., Xu C., Xie Z., Zhu H.. PPR-Meta: a tool for identifying phages and plasmids from metagenomic fragments using deep learning. Gigascience. 2019; 8:giz066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ren J., Song K., Deng C., Ahlgren N.A., Fuhrman J.A., Li Y., Xie X., Poplin R., Sun F.. Identifying viruses from metagenomic data using deep learning. Quant Biol. 2020; 8:64–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Miao Y., Liu F., Hou T., Liu Y.. Virtifier: a deep learning-based identifier for viral sequences from metagenomes. Bioinformatics. 2022; 38:1216–1222. [DOI] [PubMed] [Google Scholar]
- 12. Zhou Y., Liang Y., Lynch K.H., Dennis J.J., Wishart D.S.. PHAST: a Fast Phage Search Tool. Nucleic Acids Res. 2011; 39:W347–W352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Arndt D., Grant J.R., Marcu A., Sajed T., Pon A., Liang Y., Wishart D.S.. PHASTER: a better, faster version of the PHAST phage search tool. Nucleic Acids Res. 2016; 44:W16–W21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Delcher A.L., Bratke K.A., Powers E.C., Salzberg S.L.. Identifying bacterial genes and endosymbiont DNA with Glimmer. Bioinformatics. 2007; 23:673–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hyatt D., Chen G.L., LoCascio P.F., Land M.L., Larimer F.W., Hauser L.J.. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinf. 2010; 11:119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Buchfink B., Reuter K., Drost H.G.. Sensitive protein alignments at tree-of-life scale using DIAMOND. Nat. Methods. 2021; 18:366–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Boutet E., Lieberherr D., Tognolli M., Schneider M., Bairoch A.. UniProtKB/Swiss-Prot. Methods Mol. Biol. 2007; 406:89–112. [DOI] [PubMed] [Google Scholar]
- 18. Rho M., Tang H., Ye Y. FragGeneScan: predicting genes in short and error-prone reads. Nucleic Acids Res. 2010; 38:e191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Merkel D. Docker: lightweight linux containers for consistent development and deployment. Linux J. 2014; 239:2. [Google Scholar]
- 20. Chan P.P., Lowe T.M.. tRNAscan-SE: searching for tRNA genes in genomic sequences. Methods Mol. Biol. 2019; 1962:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Laslett D., Canback B.. ARAGORN, a program to detect tRNA genes and tmRNA genes in nucleotide sequences. Nucleic Acids Res. 2004; 32:11–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Fu L., Niu B., Zhu Z., Wu S., Li W.. CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics. 2012; 28:3150–3152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Stothard P., Grant J.R., Van Domselaar G.. CG View: visualizing and comparing circular genomes using the CGView family of tools. Brief Bioinform. 2019; 20:1576–1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Stothard P., Wishart D.S.. Circular genome visualization and exploration using CGView. Bioinformatics. 2005; 21:537–539. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
A Docker image of PHASTEST along with all of its accompanying databases is available for users to download, install and run locally. The entire Docker image is nearly 5 GB in size and instructions on how to install and test the Dockerized version of PHASTEST are provided on the PHASTEST home page (under ‘About’ → Downloads).