Abstract
The transcription factor Interferon regulatory factor 8 (IRF8) is involved in maintaining B cell identity. However, how IRF8 regulates T cell independent B cell responses are not fully characterized. Here, an in vivo CRISPR/Cas9 system was optimized to generate Irf8-deficient murine B cells and used to determine the role of IRF8 in B cells responding to LPS stimulation. Irf8-deficient B cells more readily formed CD138+ plasmablasts in response to LPS with the principal dysregulation occurring at the activated B cell stage. Transcriptional profiling revealed an upregulation of plasma cell associated genes prematurely in activated B cells and a failure to repress the gene expression programs of IRF1 and IRF7 in Irf8-deficient cells. These data expand on the known roles of IRF8 in regulating B cell identity by preventing premature plasma cell formation and highlight how IRF8 helps evolve TLR responses away from the initial activation towards those driving humoral immunity.
Keywords: CRISPR/Cas9, Cas9 ribonucleoprotein, Gene editing, Hematopoietic stem cells, Primary mouse B cells, IRF8
Highlights
-
•
Nucleofection is an efficient method for sgRNA screening.
-
•
Viral transduction of sgRNA into HSCs in combination with inducible Cas9 expression efficiently generates B cell-specific gene edits.
-
•
IRF8-deficient B cells generate more plasmablasts in response to LPS stimulation.
-
•
IRF8 modulates interferon response programs and prevents premature expression of the plasma cell program.
1. Introduction
The discovery of the clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated protein 9 (Cas9) system has revolutionized the ability to perform targeted gene editing. In the CRISPR/Cas9 system, the Cas9 nuclease is targeted to a genomic region by a synthetic guide RNA (sgRNA) that is complementary to a 20 bp sequence immediately downstream of the GGN protospacer-adjacent motif (PAM) sequence [1,2]. The Cas9 nuclease cleaves both DNA strands that are subsequently repaired by either the nonhomologous end joining (NHEJ) or homology-directed repair (HDR) pathway. Frequently, NHEJ repair leads to insertions and deletions (INDELs), which when occurring in the coding region of a gene, can induce a frameshift and disrupt the encoded protein resulting in a gene knockout [3,4]. Thus, the potential for rapidly editing genomes is possible.
Despite the promise of CRISPR/Cas9, challenges remain in implementing these approaches to generate genetic knockouts in vivo. For example, to establish knockout alleles in differentiated cell types, both sgRNA and Cas9 need to be active at specific cell stages to avoid disrupting developmental processes. Because hematopoietic stem cells (HSCs) can be used to reconstitute entire immune systems, this organ has unique advantages in being able to manipulate and dissect cell development and differentiation. During such a transplantation process, there is an opportunity to apply CRISPR/Cas9 techniques for gene editing and study the effects in the host in vivo. In addition, transgenic mice that permit inducible or constitutive expression of Cas9 allow for precise timing of gene editing, providing another key experimental tool [5,6]. Furthermore, sgRNAs can be delivered into the cells of interest of such mice through the use of viral expression vectors. Indeed, both retroviral and lentiviral approaches have been used to genetically engineer HSCs and differentiated immune cell types [5,[7], [8], [9], [10]]. Future work to improve the ability to screen and identify sgRNAs that lead to successful site-specific Cas9 activity prior to long-term transplantation-based experiments and validation of vectors that efficiently allow for the tracking of sgRNA targeted cells in vivo is important to continue to grow this exciting technology.
Interferon regulatory factor 8 (IRF8) is an IRF family transcription factor that is exclusively expressed in the immune system [11]. In B cells, IRF8 controls the pre-B to B cell transition through regulation of immunoglobulin light chain expression [12], although it’s activity can be partially compensated by the activity of IRF4, another IRF family member. IRF8 can bind to composite genomic cis-regulatory elements as a heterodimer with the ETS family transcription factor PU.1 at ETS-IRF composite elements (EICE) [13]. In this context, IRF8 cooperates with PU.1 to promote expression of the B cell lineage genes Spib and Ikzf1, encoding IKAROS. Intriguingly, in the absence of these factors, spontaneous leukemia is observed [14], speaking to the importance of these genes in regulating the B cell lineage. Additionally, Irf8 is highly expressed in mature and stimulated/activated B cells where it cooperates with PU.1 to promote germinal center (GC) development and to repress plasma cell differentiation [[15], [16], [17]]. As B cells differentiate into plasma cells, Irf8 is repressed and an Irf4-driven program initiates and maintains the plasma cell gene expression program [15,[18], [19], [20]]. Genomic analysis of IRF8 binding and transcriptional analysis of Irf8-deficient B cells has revealed a host of genes and pathways regulated by IRF8 [15,16,21]. However, many of these studies have focused on its interactions with PU.1 and may not have revealed the full role of IRF8 in directing B cell differentiation responses.
To begin to address the role of Irf8 in B cell differentiation, a CRISPR/Cas9 approach to validate sgRNA and rapidly generate B cell-specific gene knockouts in vivo was developed. This approach utilized lentiviral delivery of sgRNA to HSCs that allowed B cell-specific Cas9 expression under the control of the Cd19CRE allele [22], which is expressed early in the B cell lineage. Using this system, Irf8-deficient B cells were generated in vivo and the effect on B cell differentiation in response to T cell independent stimulation was measured ex vivo. These data build on the known roles of IRF8 in activated B cells, identify the transcriptional programs controlled by IRF8 in differentiating B cells in response to T cell independent stimuli, and highlight how IRF8 plays a role in repressing interferon signaling and maintaining B cell lineage transcriptional networks to prevent premature plasma cell differentiation.
2. Results
2.1. Identification of functional sgRNAs in primary murine B cells
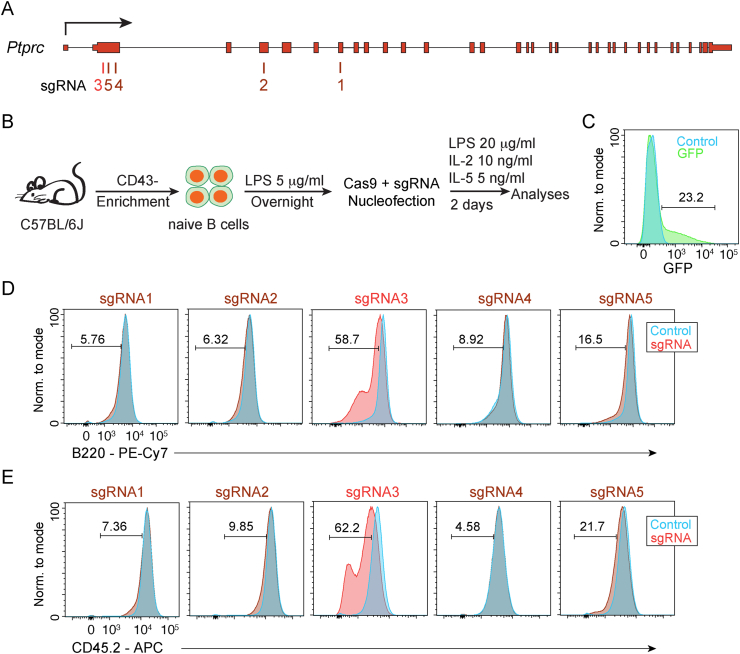
One challenge during CRISPR/Cas9 genome editing is identifying sgRNAs that successfully delete a target gene of interest. Therefore, to develop a universal approach and system for generating targeted gene knockouts in primary B cells, sgRNAs were designed to target 5′ biased exons of the B220/CD45 isoforms of the Ptprc gene, a cell surface protein that is easily detected using flow cytometry. Importantly, the CD45 isoform has previously been edited in primary murine T cells [23]. Using the CRISPOR [24] prediction track from the UCSC Genome Browser [25], five sgRNAs were chosen with a Doench/Fusi 2016 Efficiency Score greater than 55 [26] (Fig. 1A). Each sgRNA was annealed to a tracrRNA and combined with purified recombinant Cas9 to form an active complex. To increase susceptibility to genomic editing, CD43− splenic B cells were isolated by negative selection and stimulated for 18–20 h with low levels of LPS (5 μg/ml). Following stimulation, the Cas9/sgRNA ribonucleoprotein complex was electroporated by nucleoporation into prestimulated cells. B cells were then further stimulated to differentiate with 20 μg/ml LPS, 5 ng/ml IL-5, and 20 ng/ml IL-2 for 48 h [27], and the resulting cells analyzed by flow cytometry (Fig. 1B). To measure nucleoporation efficiency a GFP plasmid was also electroporated and indicated a delivery efficiency around 20–30% (Fig. 1C). Compared to the control sample, loss of both CD45.2 and B220 surface expression was observed only for sgRNA3, which targeted the second exon of Ptprc (Fig. 1D and E). These data indicate that primary B cells can be edited in ex vivo culture, with loss in expression of a surface marker detectable after 48 h.
Fig. 1.
Nucleofection of primary B cells with Cas9 and sgRNA targeting Ptprc. (A) Schematic of the Ptprc gene showing the locations of exons and sgRNAs. (B) Schematic of experimental design depicting the steps and time frames of primary B cell purification, pre-stimulation, nucleofection with Cas9/sgRNA, and analysis. (C) Histogram showing the efficiency of primary B cell nucleofection measured by GFP fluorescence. The Ptprc deletion efficiency as measured by B220 (D) and CD45 (CD45.2) (E) surface staining in cells nucleofected with each of the five sgRNAs depicted in A.
2.2. Lentiviral delivery of sgRNA for B cell specific CRISPR genome engineering
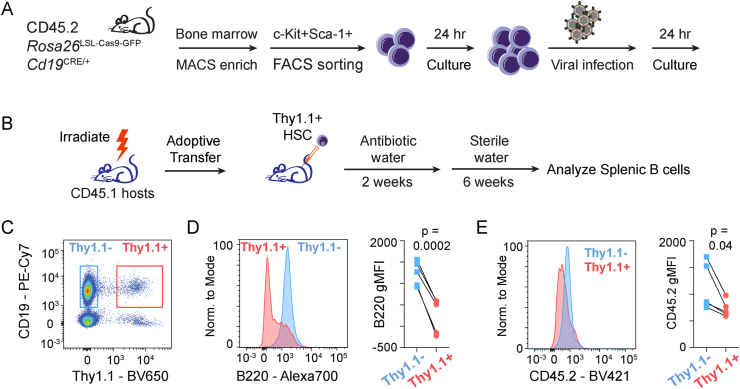
Using the sgRNA that successfully edited the Ptprc locus to reduce expression in primary B cells, we sought to optimize the system to generate B cell specific knockouts in vivo. The puromycin expression cassette of the pLentiGuide-Puro plasmid [28] was replaced with Thy1.1 to provide a unique surface marker that marked transduced cells, was easily integrated into flow cytometry panels, and could be used for antibody-directed, magnetic bead-based enrichments (Supplementary Fig. 1A). Mice with a knock-in Cas9-GFP allele placed under the control of the Rosa26 locus containing a lox-stop-lox cassette (Rosa26LSL-Cas9-GFP) allow conditional/tissue-specific Cas9 and GFP expression following CRE recombinase activity [6]. These mice were crossed to the Cd19CRE line [22] to facilitate B cell specific expression of Cas9. Indeed, GFP expression in these mice was only detected in CD19+ but not CD19− cells (Supplementary Fig. 1B). Bone marrow was isolated from CD45.2 Rosa26LSL-Cas9-GFPCd19CRE/+ mice and the hematopoietic stem cells (HSCs) enriched by magnetic column depletion of lineage committed cells followed by purification of c-Kit+Sca-1+ cells by FACS (Supplementary Fig. 1C). HSCs were initially cultured for 24 h, transduced with lentiviruses containing Ptprc sgRNA and Thy1.1, and cultured for an additional 24 h (Fig. 2A). At 24 h following transduction, greater than 99% of HSCs expressed the Thy1.1 transgene (Supplementary Fig. 1D). After confirmation of transduction efficiency, transduced HSCs were adoptively transferred into lethally irradiated CD45.1 mice and the immune system was allowed to reconstitute over 8 weeks (Fig. 2B). After immune reconstitution, animals were euthanized and the ability to delete Ptprc in splenic B cells was assessed by flow cytometry. After gating on CD19+ B cells, a distinct population of transduced B cells expressing the Thy1.1 transgene was detected (Fig. 2C). The presence of both Thy1.1 negative and positive cells provided an internal control for comparison of transduced versus non-transduced cells. As expected, both Thy1.1+ and Thy1.1− cells expressed equal amounts of GFP (Supplementary Fig. 1E), indicating the genetic induction of Cas9 expression was independent of lentiviral delivery and expression of Thy1.1 and sgRNAs. A significant reduction in the surface expression of the cells expressing the Ptprc gene was observed as measured by using antibodies to the common (CD45) and B cell specific isoforms (B220) (Fig. 2D and E). These data establish an in vivo “CRISPRgenic” system to rapidly generate B cell specific genetic knockouts to assess gene function.
Fig. 2.
In vivo editing of the Ptprc locus in B cells. (A) Schematic of the experimental design showing the isolation and viral infection of HSCs with lentiviral vectors expressing sgRNA. (B) Schematic of the experimental design depicting the construction of CRISPRgenic mice using transduced HSCs from A and time frames for phenotypic analysis. (C) Flow cytometry analysis showing the detection of transduced Thy1.1+ splenic B cells from CRISPRgenic mice 8 weeks post adoptive transfer. Histogram showing the expression of the Ptprc isoform B220 (D) or CD45.2 (E) in Thy1.1− and Thy1.1+ cells from C and quantitation of geometric mean fluorescence intensity (gMFI) in each population. Significance determined by paired Student’s t-test.
2.3. Selection of sgRNAs for IRF8
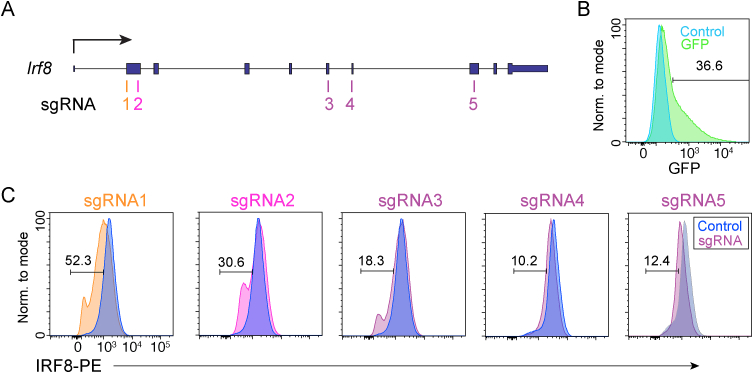
To apply this approach to studying the role of key transcription factors that regulate B cell differentiation into plasma cells, sgRNAs were designed to target the transcription factor Irf8, which is important in B cell development and restraining plasma cell formation [[15], [16], [17]]. As above, five sgRNAs were designed targeting a range of Irf8 exons (Fig. 3A), complexed with tracrRNA and Cas9 protein, and electroporated into pre-stimulated CD43− splenic B cells. After 48 h, B cells were harvested, permeabilized, and intracellularly stained to quantitate IRF8 protein levels. Similar to the Ptprc experiments above, 37% electroporation efficiency was observed using a GFP vector (Fig. 3B). Assessing IRF8 expression, sgRNAs 1 and 2 demonstrated a reduction in protein levels indicating gene editing had occurred (Fig. 3C). These experiments establish a system to identify functional sgRNAs for genome engineering of primary murine B cells to target both surface and intracellular proteins.
Fig. 3.
Assessment of sgRNA targeting Irf8 in primary B cells. (A) Schematic of the Irf8 gene showing the locations of exons and sgRNAs. (B) Histogram showing the efficiency of primary B cell nucleofection measured by GFP fluorescence. (C) The sgRNA editing efficiency as measured by IRF8 intracellular staining in cells nucleofected with each of the five sgRNAs depicted in A.
2.4. Deletion of Irf8 leads to an increase in plasmablasts following LPS stimulation
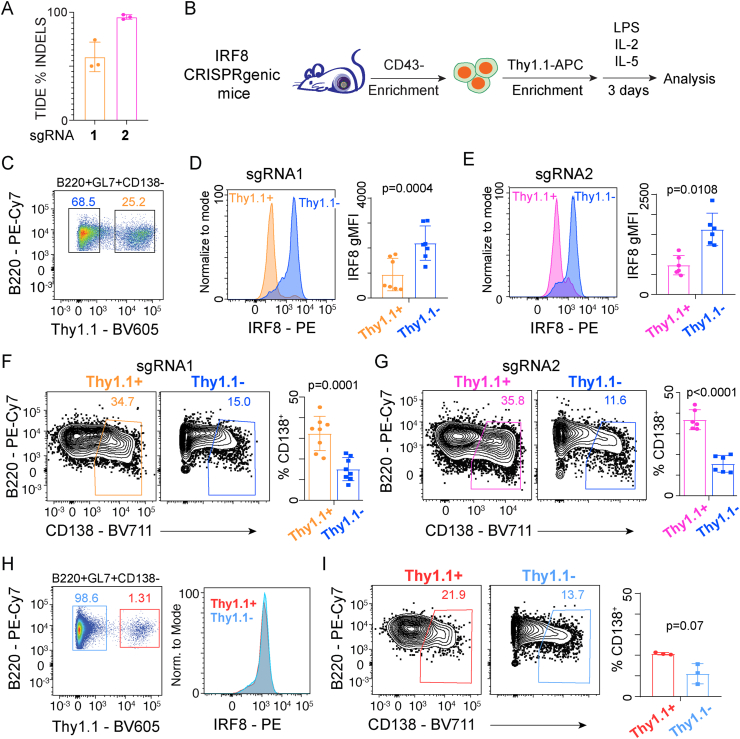
Despite the extensive literature about the role of IRF8 in B cell development and in GC B cells [[16], [17], [18]], the gene expression programs regulated by IRF8 in response to TLR stimulation of B cells has not been completely characterized. Therefore, sgRNA1 and sgRNA2 that successfully targeted Irf8 were cloned into the lentiviral CRISPR expression vector and CRISPRgenic mice generated as described above. To confirm the specificity of editing, Thy1.1+ and Thy1.1− naïve B cells, representing edited and WT cells, respectively, were FACS isolated from a cohort of CRISPRgenic mice, DNA purified, and subjected to the Tracking of INDELS by Decomposition (TIDE) assay [29] (Supplementary Figs. 2A–C). The TIDE assay estimates INDEL percentages using Sanger sequencing spectral traces surrounding the PAM sequence by comparing control versus Cas9/sgRNA targeted samples. This analysis predicted that the Irf8 locus that was targeted by sgRNA1 was edited with 55% efficiency and sgRNA2 achieved 92% efficiency (Fig. 4A), indicating that the Irf8 locus can be successfully targeted for disruption in vivo.
Fig. 4.
IRF8 restrains plasmablast formation in response to LPS. (A) Bar plot showing the deletion efficiency as determined by TIDE assay in Thy1.1+ and Thy1.1− B cells from Irf8 CRISPRgenic mice generated with the indicated sgRNA. Data represent cells isolated from three independent CRISPRgenic mice. (B) Schematic of the workflow to enrich transduced B cells, ex vivo stimulation with LPS, IL-2, and IL-5 for 3 days to induce B cell differentiation, followed by analysis. (C) Flow cytometry showing the percentage of Thy1.1− and Thy1.1+ activated B cells (B220+GL7+CD138−) from a representative IRF8 CRISPRgenic mouse stimulated as in B. Histograms showing the expression of IRF8 by intracellular staining in activated Thy1.1− and Thy1.1+ B cell populations from stimulations defined in B in CRISPRgenic mice from sgRNA1 (D) or sgRNA2 (E) and quantitation of geometric mean fluorescence intensity (gMFI) in each population, respectively. B cells from sgRNA1 (F) or sgRNA2 (G) were stimulated with LPS, IL-2, and IL-5 and analyzed after 3 days. Representative flow cytometry plots (left) and quantification (right) of the percentage of CD138+ plasmablasts from Thy1.1− and Thy1.1+ cells for the indicated sgRNA. (H) Flow cytometry (left) showing the percentage of Thy1.1− and Thy1.1+ activated B cells (B220+GL7+CD138−) and histogram (right) of IRF8 expression as measured by intracellular staining for both Thy1.1− and Thy1.1+ activated B cells from a representative control CRISPRgenic mouse. (I) B cells from control CRISPRgenic mice were stimulated to differentiate as in F and G and analyzed after 3 days. Flow cytometry (left) showing the gating and bar plots (right) quantitating the percentage of CD138+ plasmablasts from Thy1.1− and Thy1.1+ cells. Significance was determined by paired Student’s t-tests in D-G and I. Data from D and F represent 7–8 independent animals from 2 experiments. Data from E and G represent 6 independent animals from 2 experiments. Data from H and I represent 3 independent animals from 1 experiment. The strategy for FACS isolation of each population in A and the gating strategy for C–I is depicted in Supplementary Fig. 2.
To further explore the phenotypic consequences of disrupting Irf8, CD43− splenic B cells were purified from CRISPRgenic mice and Thy1.1+ transduced B cells were enriched using anti-Thy1.1-APC antibodies and anti-APC magnetic beads. The enriched cells were then stimulated to differentiate ex vivo using LPS, IL-2 and IL-5 as above [27] and analyzed by flow cytometry after 72 h (Fig. 4B, Supplementary Fig. 2D). Irf8 expression is known to increase in activated B cells following stimulation [15]; therefore, the intracellular levels of IRF8 protein were analyzed in Thy1.1+ and Thy1.1− activated B cells (B220+GL7+CD138−) (Fig. 4C). A significant decrease was observed in IRF8 protein levels in Thy1.1+ activated B cells that received either of the two sgRNA targeting Irf8 compared to untransduced Thy1.1− cells (Fig. 4D and E, Supplementary Figs. 2E–G). The contrast between the reduction in IRF8 protein levels and TIDE assay predicted efficiency of sgRNA1 highlights the importance of confirming the activity of sgRNAs using multiple approaches. Hereafter, we will refer to the Thy1.1+ as the knockout (KO) cells and the Thy1.1− cells as WT. Consistent with previously published data from CD40L stimulations [15], deletion of Irf8 in KO cells led to a significant increase in CD138+ plasmablasts in both sets of sgRNAs compared to WT cells (Fig. 4F and G). Importantly, the comparison of WT and KO B cells from the same animal also controlled for any effects of hemizygosity of Cd19 introduced by the CRE allele between the two populations. Moreover, in a set of CRISPRgenic animals that received a control sgRNA vector, there was no difference in IRF8 levels between Thy1.1+ and Thy1.1− activated B cells (Fig. 4H), and although there appear to be more CD138+ plasmablasts in the KO, this was not statistically significant (Fig. 4I). Importantly, there was a significant increase in CD138+ plasmablasts from Thy1.1+ KO cells derived from both sets of sgRNAs compared to the control sgRNA Thy1.1+ cells (sgRNA1, p = 0.005; sgRNA2, p < 0.0001; two-tailed Student’s t-test).
2.5. Transcriptional analysis of differentiating Irf8-deficient B cells
To investigate the transcriptional programs regulated by Irf8, RNA-seq was performed on naïve CD43− splenic B cells (nB) from CRISPRgenic mice for both Irf8 sgRNA1 and sgRNA2, representing the KO and WT B cells. Comparison between the naïve B cells (KO versus WT) revealed only 7 and 4 differentially expressed genes (DEG, absolute log2FC > 1 and FDR < 0.05) for sgRNA1 and sgRNA2, respectively (Fig. 5A, Supplemental Table 1). These results are consistent with previous microarray profiling of follicular and marginal zone B cells from Irf8-KO mice that exhibited a small number of genes dysregulated in unstimulated nB [21]. These data suggest that IRF8’s regulation of the transcriptional network in developing splenic B cells is minimal or alternatively can be complemented by IRF4, as is the case for some dendritic cell populations [30].
Fig. 5.
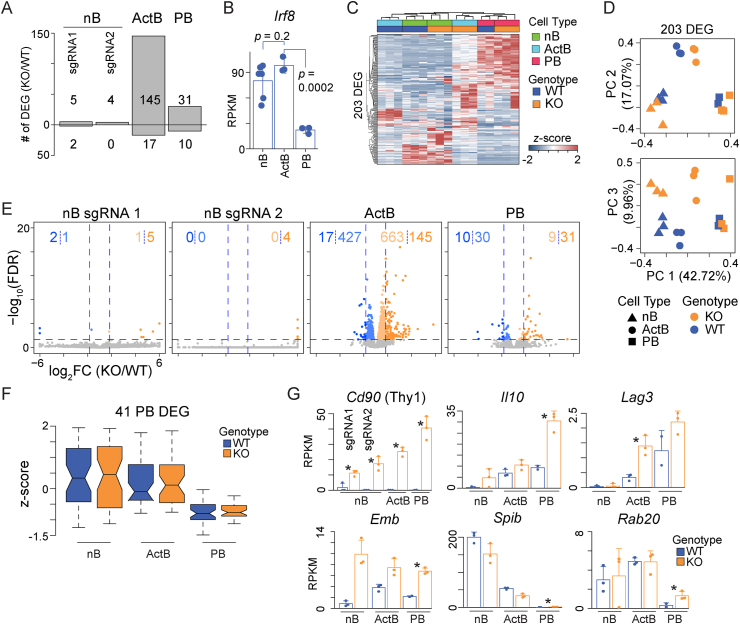
IRF8 controls gene expression programs in activated B cells. RNA-seq was performed on Thy1.1− (WT) and Thy1.1+ (KO) naïve B cells (nB) from sgRNA1 and sgRNA2 Irf8 CRISPRgenic mice and activated B cells (ActB; B220+GL7+CD138−) and plasmablasts (PB; CD138+) isolated after 72 h of ex vivo culture with LPS, IL-2, and IL-5. (A) Bar plot showing the number of differentially expressed genes (DEG: absolute log2FC > 1 and FDR < 0.05) for each of the indicated populations. ActB and PB for KO and WT samples are from sgRNA1 Irf8 CRISPRgenic mice. (B) Bar plot of Irf8 in reads per kilobase per million (RPKM) normalized expression for the indicated Thy1.1− (WT) cell types. sgRNA1 and sgRNA2 data are combined for nB. Significance determined by two-tailed Student’s t-test. (C) Heatmap and hierarchical clustering of 203 unique DEG from A in the nB, ActB, and PB for KO and WT samples from sgRNA1 Irf8 CRISPRgenic mice. The grouping of samples by cell type and IRF8 status is indicated by a color bar at the top of the heatmap. (D) Principal component analysis of the same DEG and samples from C. The percentage of variation captured by each principal component is indicated on the axis. Shapes designate the cell type and colors designate the KO or WT status of each sample. (E) Volcano plots for each of the comparisons in A. The number of DEG (darker colors) and genes that meet an FDR < 0.05 threshold but displayed an absolute log2FC ≤ 1 (lighter colors) for each comparison is annotated. (F) Box plots showing the z-score normalized mean expression for 41 DEG in PB from A in nB, ActB, and PB from WT and KO cells. For each group, the mean expression value for each gene was used. Boxes represent the interquartile range, and notches indicate the data median. (G) Bar plots depicting the reads per kilobase per million (RPKM) normalized expression for the indicated genes. * Signifies the gene is a DEG for the indicated comparison. Error bars represent SD and dots represent individual data points. Data represent three independent samples for each group except WT PB, which represents two independent samples. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
In T cell dependent immune responses, Irf8 is expressed at the highest levels in GC B cells [31]. To determine the role for IRF8 in controlling the gene expression program of activated B cells and plasma cells, CD43−Thy1.1+ KO transduced splenic B cells were enriched as above and stimulated to differentiate with LPS, IL-2, and IL-5. After 72 h, KO and WT cells representing B220+GL7+CD138− activated B cells (ActB) and CD138+ plasmablasts (PB) were FACS isolated and RNA-seq was performed. Irf8 transcripts were highest in nB and ActB and were significantly repressed during differentiation in PB (Fig. 5B). In contrast to the naïve B cell data, 162 DEG were observed in ActB cells and 41 in PB (Fig. 5A, Supplemental Table 1). Hierarchical clustering of WT and KO cell types spanning nB, ActB, and PB using the unique 203 DEG identified in any of the three populations showed that PB and nB samples largely clustered together with ActB being the most distinct. Additionally, while Irf8-sufficient ActB clustered together with nB, Irf8-deficient ActB clustered more closely with PB (Fig. 5C). Principal component analysis (PCA) using the same set of DEG revealed that principal components (PC) 1 and 2 mainly separated cells based on cell type (nB, ActB, PB) (Fig. 5D). PC 3 revealed the largest separation between KO and WT nB and ActB cell types, with PB mostly clustering together. The differences between WT and KO cells were explored using volcano plots to determine if there was a larger skewing in gene expression below the DEG thresholds. Consistent with the DEG results, ActB cells showed 1090 additional genes that were significantly differentially expressed (FDR < 0.05) with small fold changes in expression, with 61% of the genes being upregulated in the KO ActB cells (Fig. 5E). These data suggest IRF8 mainly functions to repress gene expression networks in ActB cells in response to LPS.
During the final stages of differentiation, when B cells transition to PB, Irf8 is repressed (Fig. 5B) [15]. The RNA-seq data indicated that PB harbored 41 DEG that displayed differential expression in KO cells. Given that Irf8 is not expressed in PB, we hypothesized that the observed differences resulted from dysregulation in the previous ActB differentiation stage. For all 41 DEG, the expression levels in WT and KO cells were plotted in nB, ActB, and PB to assess the changes across differentiation. Overall, the group of 41 DEG were expressed at lower levels in PB compared to nB and ActB (Fig. 5F). For example, the B cell transcription factor Spib was most highly expressed in nB and repressed in both KO and WT PB (Fig. 5G). A similar trend was observed for other B cell specific genes, such as the GTPase Rab20, Ciita, and Cd22 (Fig. 5G, Supplemental Table 1). This suggested a failure to fully repress the B cell program. A second set of genes were more highly expressed in KO PB and included Il10, Emb, and Lag3 (Fig. 5G). Lag3 expression has been shown to mark immunosuppressive plasma cells that secrete IL-10 [32]. Emb encodes Embigin, an adhesion molecule that is also repressed by PAX5 during B cell lineage commitment [33]. Emb was upregulated in all B cell subsets suggesting IRF8 also participates in the repression of Emb in developing B cells.
2.6. IRF8 represses interferon and the plasma cell gene expression signatures in ActB cells
Given that the effect of Irf8-deficiency was primarily observed in the ActB cell stage, this set of DEG was explored further. The change in expression for the 162 DEG was assessed as B cells differentiated from nB to PB. The largest difference in expression was observed in ActB cells with minimal expression changes in nB and PB (Fig. 6A), suggesting these genes are uniquely regulated by IRF8 in ActB. Previous expression profiling identified a set of genes regulated by IRF8 and IRF4 in LPS activated B cells [16] as well as genes jointly regulated by PU.1 and IRF8 [15]. The overlap with previous datasets was explored using gene set enrichment analysis (GSEA) [34,35] and a significant down regulation of IRF8 regulated genes in KO B cells was observed (Fig. 6B). In contrast, IRF4 regulated genes were significantly upregulated in KO B cells. Similarly, genes regulated by PU.1 and IRF8 in naïve and activated B cells were downregulated (Fig. 6B). Given the observed phenotype of increased plasmablast differentiation resulting from KO B cells, this suggested that these cells may upregulate a plasmablast gene signature early in ActB when IRF8 is absent. These data are consistent with previous results from IRF4hiIRF8low GC B cells [16]. To address this informatically, GSEA was performed using an antibody secreting cell (ASC) specific gene set [36] and the enrichment tested between KO and WT ActB. This analysis showed that ActB KO cells strongly upregulated ASC signature genes (Fig. 6C), further suggesting that in the absence of IRF8, the plasmablast program is engaged prematurely.
Fig. 6.
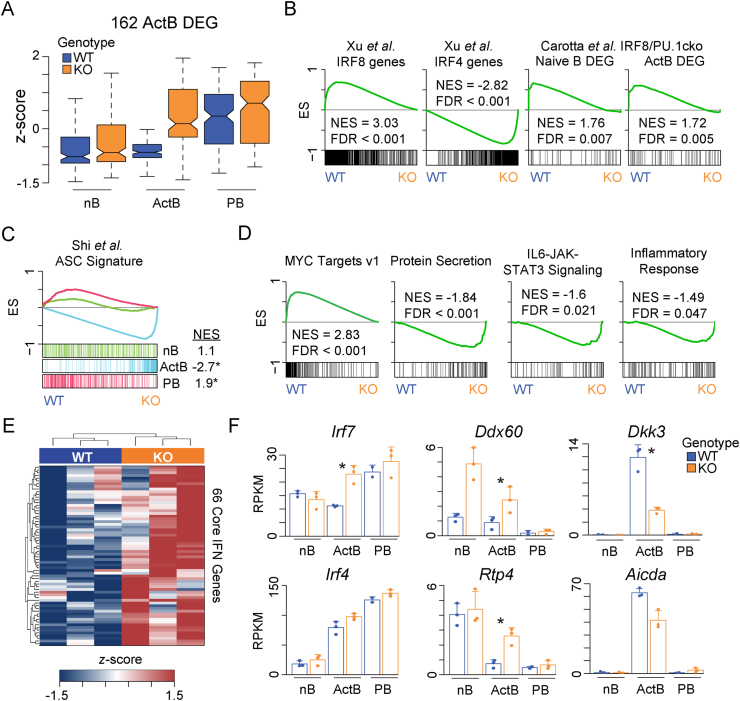
Irf8-deficient activated B cells upregulate plasmablast and IFN gene signatures. (A) Box plots showing the z-score normalized mean expression for 162 DEG in ActB from Fig. 5A in nB, ActB, and PB from WT and KO cells. For each group, the mean expression value for each gene was used. Boxes represent the interquartile range and notches the data median. (B) GSEA showing the enrichment of Xu et al. IRF8 and IRF4 target gene data sets [16] and PU.1-IRF8 regulated genes in naive B cells or activated B cells from Carotta et al. [15] in the KO versus WT ActB cells. ES, enrichment score; NES, normalized enrichment score. (C) GSEA showing the enrichment of Shi et al. Antibody-Secreting Cell (ASC) signature gene set [36] in the KO versus WT ActB cells. (D) GSEA showing the enrichment of the indicated HALLMARK MSigDB gene sets in the KO versus WT ActB cells. (E) Gene expression heatmap of 66 genes in WT and KO ActB cells that were derived from a GSEA Leading Edge analysis of the HALLMARK Inflammatory Response, Interferon Alpha Response, and Interferon Gamma Response gene sets. (F) Bar plots depicting the reads per kilobase per million (RPKM) normalized expression for the indicated genes. * Signifies the gene is a DEG for the indicated comparison. Error bars represent SD and dots represent individual data points.
The dysregulation in KO ActB was further explored by GSEA using the HALLMARK MSigDB database of gene sets. The top pathway enriched in WT ActB was the MYC Targets v1 gene set (Fig. 6D). Consistent with KO ActB cells engaging the plasmablast gene expression program early, these cells displayed significant enrichment of Protein Secretion and IL6-JAK-STAT3 signaling gene sets. STAT3 signaling has been shown to induce expression of Prdm1 [37], which encodes BLIMP1, the master transcription factor for plasmablast differentiation. In addition, multiple HALLMARK gene sets including the Interferon Alpha Response, Interferon Gamma Response, and Inflammatory Response were also upregulated in KO ActB. To further highlight the upregulation of these pathways in KO ActB, a core set of 66 interferon genes from all three gene sets were derived using a GSEA Leading Edge analysis, plotted as a heatmap, and included Irf7, Ddx60, and Rtp4 (Fig. 6E and F, Supplemental Table 2). Upregulation of antiviral genes in the absence of Irf8 was also observed in studies that mapped IRF8 target genes by ChIP-chip [38]. In addition, to the above genes and pathways, a reduction in Aicda and Dkk3 expression was observed. The Aicda locus, which encodes AID, has been shown to be directly bound by IRF8 in LPS activated B cells [16]. Dkk3 is a dickkopf family gene that antagonizes WNT signaling and has been shown to negatively regulate antibody secretion and B cell differentiation [39], suggesting IRF8 promotes Dkk3 expression to maintain B cell homeostasis and identity. Other IRF family members, such as Irf4, were increased in expression but this was not significant between WT and KO cells.
2.7. Irf8-deficient B cells exhibit alterations in IRF and IFN gene expression networks
IRF8 has low inherent affinity for DNA and largely relies on cooperative interactions with ETS and AP-1 family transcription factors to bind DNA and regulate gene expression at composite EICE and AICE elements, respectively [30]. At high enough concentrations, IRF8 can form homodimers to bind canonical ISRE motifs. To determine which mode of IRF8 activity dominated in LPS activated B cells, the assay for transposase accessible chromatin sequencing (ATAC-seq) was performed on WT ActB. Accessible regions that mapped to any gene with a change in expression that was below the FDR <0.05 threshold in ActB were determined and the presence of an IRF half-site annotated using HOMER [40]. The IRF half-site was chosen to increase sensitivity to capture all IRF binding modes. Of these regions, 56% were associated with at least one IRF binding mode (Fig. 7A). Furthermore, the presence of EICE, AICE, and ISRE was determined and revealed that 45.1% of IRF sites in ActB DEG were the EICE composite element. This data is consistent with other IRF8 ChIP studies in B cells that have shown IRF8 predominantly interacting with PU.1 to regulate gene expression [14,15].
Fig. 7.
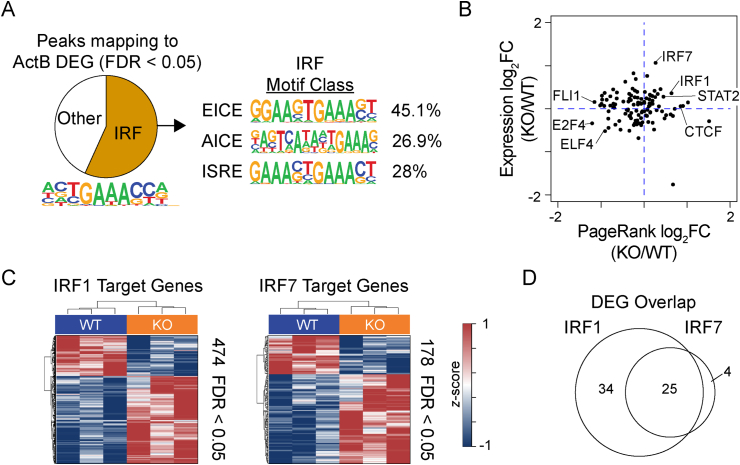
IRF8 modulates the transcriptional networks of IRF1 and IRF7. (A) ATAC-seq was performed on ActB formed after 72 h stimulation with LPS, IL-2, and IL-5; and accessible peaks mapping to 1252 genes with an FDR < 0.05 (Fig. 5E, ActB panel) were annotated for the presence of an IRF monomer binding sequence. Pie chart (left) showing the percentage of genes with an IRF motif and the percentage (right) that match the three composite binding modes for IRF8 is indicated. (B) Scatter plot comparing the log2FC in Taiji PageRank transcription factor activity scores versus log2FC change in gene expression between KO versus WT ActB cells. Selected transcription factors are indicated. (C) Heatmap and hierarchical clustering of IRF1 (left) and IRF7 (right) target genes determined by Taiji exhibiting a change in expression between WT and KO ActB cells with an FDR < 0.05. (D) Venn diagram showing the overlap in DEG that are IRF1 and IRF7 target genes from C.
To determine if dysregulation of other transcription factor networks occurred in the absence of IRF8, the Taiji analysis was used [41]. Taiji integrates gene expression data from WT and KO ActB with chromatin accessibility data from WT ActB to compute how important an individual transcription factor is for cell type gene expression using the PageRank algorithm. PageRank scores can be compared between WT and KO ActB to determine differences in activity for individual transcription factors. Correlation of the change in expression for each transcription factor with the change in PageRank activity score between KO versus WT ActB revealed an increased activity of IRF7, IRF1, STAT2, and CTCF transcription factors in KO ActB and a decrease in activity for numerous ETS factors, including FLI1 and ELF4, and the E2F family factor E2F4 (Fig. 7B). Further examination of IRF1 and IRF7 target genes revealed 474 and 171 target genes, respectively, that were significantly altered in expression (FDR <0.05) in the absence of Irf8, with the majority of genes upregulated in KO ActB (Fig. 7C). When only DEG were considered, the IRF7 target genes largely overlapped with IRF1, yet IRF1 appeared to drive a small but unique set of DEG independently or in parallel with IRF7 (Fig. 7D). These data suggest that one role of IRF8 is to modulate the transcriptional networks of other IRF family members.
3. Discussion
The CRISPRgenic approach developed in this study lays the foundation to rapidly generate B cell-specific knockouts. As CRISPR-based approaches are increasingly applied in vivo to study the immune system and have become more robust [5,7,8,23], it is now possible to study the function of newly identified genes and pathways from high-throughput genomic approaches such as ATAC-seq and RNA-seq. This study used lentiviral delivery of sgRNA expressing constructs into HSCs carrying the Cas9 transgene that was specifically expressed in CD19+ B cells. Using this approach, a role for the transcription factor IRF8 in regulating B cell responses to LPS, a T cell-independent type I stimulation, was determined. It is important to note that these observations were based on co-culture experiments of KO and WT B cells and cannot rule out the possibility that upregulation of Il10 and/or Il15 by KO B cells influenced the phenotype of WT B cells by paracrine signaling. Nevertheless, and consistent with previous literature [15], Irf8-deficient B cells more rapidly formed PB upon stimulation and upregulated a set of ASC signature genes prematurely in the ActB cell stage. Additionally, an upregulation in the transcriptional targets of IRF1 and IRF7 was observed, indicating IRF8 partly functions to modulate the gene expression programs of other IRF family transcription factors.
IRF8 and IRF4 are closely related and unique among the IRF family of transcription factors because their expression is restricted to the immune system, and they utilize distinct DNA binding partners to regulate transcription [42]. Using ATAC-seq data from ActB cells to identify transcription factor motifs enriched in the accessible chromatin surrounding KO genes with altered expression, an enrichment of the EICE motif, which is a composite site that binds IRF:ETS heterodimers, was identified. This is consistent with other data indicating that IRF8 primarily cooperates with the ETS factor PU.1 to regulate target gene expression [14,17,38]. Although a smaller percentage compared to EICE, AICE and ISRE sites were also identified surrounding KO DEG. Irf1 and Irf7, which were upregulated in the absence of Irf8, are two factors that bind ISRE sequences and were predicted by PageRank analysis to have increased influence over the KO transcriptional network. The Irf7 promoter is directly bound by IRF8 in mouse GC B cell lines and Irf7 is upregulated following transfection of Irf8 targeting siRNAs [38]. These data suggest Irf7, but not necessarily Irf1, is directly regulated by IRF8. Both IRF1 and IRF7 can be activated downstream of TLR receptors that signal through Myd88 [43]. In the absence of Irf8, inflammatory gene sets were clearly upregulated following TLR stimulation with LPS. These data suggest that one role for IRF8 may be to fine-tune the IRF activity away from an IFN-driven network towards one more conducive for ActB, and in response to T cell dependent antigens, the GC response.
The majority of focus has been on the role of IRF8 in promoting activated, GC B cells in response to T cell-dependent antigens [[15], [16], [17]]. In response to T cell-independent antigens, ActB cells represent a heterogeneous state of proliferating B cells that undergo progressive epigenetic and transcriptional remodeling [44]. In vivo, B cells progressively lose their identity as they undergo at least eight cellular divisions before ultimately differentiating into antibody secreting plasmablasts [45]. The lack of IRF8 in the CRISPRgenic ActB cells may allow the low levels of IRF4 to accumulate at EICE and AICE targets thereby promoting the ASC program, resulting in more plasmablasts formed. Multiple targets of IRF8 identified here were consistent with the role of IRF8 in the repression of plasma cell differentiation in ActB. Consistent with ChIP-seq data from cell lines representing GC derived B cells [38], IRF8 was found to regulate the expression of the same genes in ActB following LPS stimulation, including activation of the ETS family factor ELF4 and repression of IRF7. FLI1, another ETS family TF, was predicted by PageRank analysis to be dysregulated and less important/active in the KO transcriptional network. B cells expressing a Fli1 allele that is deficient in the transcriptional activation domain have heightened IgM responses to T cell independent antigens [46], suggesting that IRF8 may cooperate with other ETS factors such as FLI1 to repress plasma cell differentiation. Moreover, KO ActB expressed lower levels of Aicda, which encodes AID. The Aicda locus is directly bound by IRF8; however, Aicda was not dysregulated in PU.1/IRF8 double knockout mice [15,17], indicating a redundant role for another factor when both PU.1 and IRF8 are absent. In the absence of IRF4, Aicda was also reduced in expression in response to LPS [47], suggesting that both IRF8 and IRF4 are necessary for full expression and in response to T cell independent antigens.
In summary, we present a path to explore specific stages of regulation during the differentiation of B cells using in vivo approaches and cell-specific CRISPR/Cas9 expression technologies. Irf8 editing confirmed the expected phenotypic outcomes, such as the increased frequency of plasmablasts, as well as revealed novel observations regarding gene expression pathways and potential exchangeable roles of the IRF transcription factors.
4. STAR methods
4.1. Mice
C57BL/6J (JAX; 000664), CD45.1 (JAX; 002014), Cd19CRE (JAX; 006785) and Rosa26LSL-Cas9-GFP mice (JAX; 026175) were purchased from The Jackson Laboratory and bred on site. Mice used in experiments were between 7 and 12 weeks of age and all genders were equally represented throughout the experiments. Animals were randomly assigned to experimental groups, and independent biological replicates of three or more were used for all phenotyping experiments. Experimental mice were euthanized via carbon dioxide asphyxiation in accordance with AVMA Guidelines, 2020 edition. Animals were housed in specific pathogen-free caging with 12 h light/dark cycles by the Emory Division of Animal Resources.
4.2. Cell purification and culture
For adoptive transfer assays, bone marrow hematopoietic stem cells were first isolated with the Direct lineage depletion Kit (Miltenyi; 130-110-470), and stained with c-Kit-PE, Sca-1-PE-Cy7, and non-HSC lineage makers (B220-APC-Cy7, CD11b-APC-Cy7, and Thy1.2-APC-Cy7) (BioLegend) for cell sorting. FACS isolated Lin−Sca-1+c-Kit+/low cells (LSK) were cultured for 24 h in Stem Cell Media: StemPro-34 SFM media (ThermoFisher; 10639011) with 100 ng/ml each of the following supplements: Thrombopoietin (BioLegend; 593302), SCF (BioLegend; 579702), FMS-related tyrosine kinase 3 ligand (BioLegend; 550702), and IL-7 (BioLegend; 577802). For nucleofection experiments, splenic CD43− B cells were isolated using the B-cell Isolation Kit (Miltenyi; 130-090-862). Isolated B cells were cultured in B cell media: RPMI 1640 (Corning Cellgro; 50-020-PC), 10% heat-inactivated FBS (Sigma-Aldrich), 10 mM HEPES (HyClone; SH30237), 1% MEM nonessential amino acids (Sigma-Aldrich; ENBF3930-01), 10 μM Sodium Pyruvate (Sigma-Aldrich; RNBF6686), 1× penicillin– streptomycin–glutamine (10378016; Life Technologies), and 0.0035% 2-mercaptoethanol (Sigma-Aldrich). For enrichment of adoptively transferred Thy1.1 congenically marked cells, CD43− B cells were stained with Thy1.1-APC, followed by immunomagnetic enrichment using anti-APC beads (Miltenyi; 130-097-143) according to the manufacturer’s protocol.
4.3. Flow and FACS sorting
For antibody staining, cells were resuspended at a concentration of 1 × 106/100 μl in FACS buffer (1 × PBS, 2 mM EDTA, and 1% BSA), stained with Fc Block (BD; 553141) for 10–15 min, stained with fluorophore-conjugated antibodies for 30 min on ice, and then washed with 10 vol of FACS buffer. For flow cytometry analyses all cells were fixed using 1% paraformaldehyde. The following specific fluorescent antibodies, with specific clones indicated in parentheses, were used: BioLegend: B220-PE-Cy7 (RA3-6B2), CD45.1-APC (A20), CD45.2-BV421, GL-7-APC, B220-A700 (RA3-6B2), Thy1.1-APC, Thy1.1-BV650, CD45.2-PE, CD11b-APC-Cy7 (M1/70), F4/80-APC-Cy7 (BM8), CD90.2-APC-Cy7 (30H12), B220-APC-Cy7 (RA3-6B2). Zombie Yellow Fixable Viability Dye (423104); BD Biosciences, Inc.: CD138-BV711 (281-2); IRF8-PE (V3GYWCH), c-Kit-PE (2B8), Sca-1-PE-Cy7 (E13-161.7). For intracellular staining of IRF8, the FIX & PERM Cell Permeabilization Kit (ThermoFisher; GAS003) was used per the manufacturer’s protocol. For all flow cytometry analysis, the gating strategy is depicted in Supplemental Fig. 1 and Supplemental Fig. 2. Generally, the gating scheme progressed through the following gates: lymphocytes based on SSC-A and FSC-A, singlets based on FSC-H and FSC-W, live cells based on the exclusion of Zombie Yellow Fixable Viability dye, and dump-negative cells using the markers CD11b−F4/80−CD90.2− to remove non-B cell lineage cells. All flow cytometry data were collected on a LSR II or LSRFortessa (BD Biosciences) and analyzed using FlowJo v10.6.2 software. Cell sorting was performed at the Emory Flow Cytometry Core using a FACSAria II (BD Biosciences) and BD FACSDiva software v8.0 (BD Biosciences).
4.4. sgRNA design and nucleofection
Synthetic guide RNAs were designed using the CRISPOR [24] track from the University of California, Santa Cruz Genome Browser and synthesized by Integrated DNA Technologies (See Table 1). A 100 μM working concentration was generated by combining an equal volume of 200 μM crRNA with 200 μM tracrRNA (1:1 ratio), incubated at 95 °C for 5 min, and slowly cooled to room temperature. Annealed sgRNA were stored at −80 °C. CD43− B cells were pre-stimulated with 5 μg/ml lipopolysaccharide for 18–20 h. 2 μl Cas9 protein (Invitrogen; A36499) was incubated with 1.2 μl 100 μM sgRNA at room temperature for 10 min. The primary B cell nucleofection kit (Lonza; V4XP-4024) and 4D-Nucleofector core X Unit (Lonza; AAF-1002B, AAF-1002X) were used to electroporate Cas9/sgRNA complexes into pre-stimulated B cells according to the manufacture’s protocol. B cells then were cultured in B cell media supplemented with 20 μg/ml LPS, 20 ng/ml IL-2, 5 ng/ml IL-5 for 2 days as previously described [27].
Table 1.
Summary of sgRNA sequences and cloning oligos.
| Ptprc | Sequence (5′-3′) | Irf8 | Sequence (5′-3′) |
|---|---|---|---|
| gRNA1 | GGCTAATACTTCAATTTGTT | gRNA1 | TCGACAGCAGCATGTACCCG |
| gRNA2 | AACTCTTACACCATCCACTC | gRNA2 | CAAGCAGGATTACAATCAGG |
| gRNA3 | GTCCAGAAGGGCAAATCCAA | gRNA3 | AGTACATGGGTATGACCAAG |
| gRNA4 | GACACAGAAGTCTTTGTCAC | gRNA4 | GCGGCATATCCGGTCACCAG |
| gRNA5 | AAAGACTTCTGTGTCCAGAA | gRNA5 | TCGGCCGTCGGAAAGCACAC |
| gRNA3 oligo F | CACCGGTCCAGAAGGGCAAATCCAA | gRNA2 oligo F | CACCGCAAGCAGGATTACAATCAGG |
| gRNA3 oligo R | AAACTTGGATTTGCCCTTCTGGACC | gRNA2 oligo R | AAACCCTGATTGTAATCCTGCTTGC |
| gRNA3 oligo F | CACCGAGTACATGGGTATGACCAAG | ||
| gRNA3 oligo R | AAACCTTGGTCATACCCATGTACTC |
4.5. Lentiviral production
The pLentiGuide-Puro plasmid was a gift from Feng Zhang (Addgene plasmid # 52963) [28]. The Puromycin resistance cassette was replaced with Thy1.1, including the insertion of a silent mutation to eliminate a BsmBI site in Thy1.1, by the Custom Cloning Core of the Emory Integrated Genomics Core. sgRNAs were cloned into the BsmBI site according to previously described protocols [48]. LentiX-293T cells (Takara; 632180) were cultured with high glucose (4.5 g/L) DMEM supplemented with 10% heat-inactivated FBS, 2 mM l-glutamine, 1 mM sodium pyruvate, and 100 U/ml penicillin/streptomycin. For lentiviral production, LentiX-293T cells were seeded at 5 million/10 ml in a 10 cm dish for 20–22 h. The pCMV delta R8.2 (Addgene; 12263), pCMV-VSV-G (Addgene; 8454), pLentiGuide-Thy1.1 plasmid were mixed at a 4:1:5.4 M ratio in Opti-MEM I medium (Gibco; 31985070) to make a 500 μl suspension. 50 μl TransIT-293 reagent (Mirus; MIR 2700) was then added, mixed, and allowed to sit at room temperature for 30 min. The suspension was added dropwise into each plate and mixed by gentle shaking of the plate. Following a 16–18 h incubation, the media was replaced with 15 ml DMEM growth medium supplemented with 1% BSA. Lentiviruses were harvested 24 and 48 h later and purified by ultracentrifugation using a SW28 and SW41 rotor (Beckman; 342207 and 331362, respectively).
4.6. LSK transduction and adoptive transfer
Flat bottom 96-well-plate was coated with 50 μl 100 ng/μl RetroNectin (Takara; T100A) overnight, then incubated with 100 μl 2% BSA for 30 min and washed with 150 μl PBS. After isolation and 24 h culture in StemPro-34 SFM media, LSKs were resuspended in SFM34 stem-pro media containing polybrene and seeded in a RetroNectin coated flat bottom 96-well-plate at 50,000 cells/well. Lentivirus was added into each well, cells were centrifuged at 650×g for 1.5 h at 32 °C, followed by recovery at 37 °C for 1 h. The supernatant was removed, cells were resuspended in Stem Cell Media, transferred to a round bottom plate, and cultured for 24 h at 37 °C. LSKs were harvested with 2 mM EDTA and PBS and transferred intravenously into CD45.1 mice that were lethally irradiated with two doses of 450 rad, 6 h apart. Host mice were given water containing 2% sucrose, 10 μg/ml Neomycin (N5285; Sigma-Aldrich), and 125 ng/ml polymyxin B (P0972; Sigma-Aldrich) for two weeks and were euthanized eight weeks post-irradiation.
4.7. RNA-seq
For each sample, 1000 cells were sorted directly into 300 μl of RLT buffer (Qiagen; 79216) containing 1% BME by FACS. RNA was extracted using the Quick-RNA Microprep kit (Zymo Research; R1050) and all purified RNA was used as input for the SMART-seq v4 cDNA synthesis kit (Takara; 634894) with 12 cycles of PCR amplification. 200 pg of cDNA was used as input for the NexteraXT kit (Illumina, FC-131-1096) using 12 cycles of PCR amplification. Final libraries were quantitated by qPCR, size distributions determined by bioanalyzer, pooled at equimolar ratios, and sequenced at the Emory Non-human Primate Genomics Core on a NovaSeq6000 using a PE100 run or a NextSeq550 using a PE75 run. Raw fastq reads were mapped to the mm10 genome using STAR v2.7.6a [49] with the Gencode v27 reference transcriptome. Duplicate reads were removed from downstream analysis using the PICARD markduplicates v2.23.8 function (http://broadinstitute.github.io/picard/). Reads mapping to exons for all unique ENTREZ genes were compiled and normalized using GenomicRanges v1.38.0 [50] and all genes expressed at 3 or more reads per million in all samples of one group were considered expressed. DESeq2 v1.26.0 [51] was used to assess differential expression between group and all genes with a false-discovery rate corrected p-value < 0.05 and absolute log2 fold-change >1 were considered significant. For the GSEA analysis genes were ranked by multiplying the sign of the fold change (+ or −) by the -log10 of the DESeq2 derived p-value for each comparison. GSEA v4.2.3 was used for PreRanked and Leading Edge Analysis as indicated. Downstream analysis and plotting was performed in R v3.6.3 and v4.1.0.
4.8. ATAC-seq
For each sample, 10,000 cells were sorted into FACS buffer and transposition was performed as previously described [52]. Briefly, cells were resuspended in 12.5 μl 2x TD Buffer, 2.5 μl Tn5, 2.5 μl 1% Tween-20, 2.5 μl 0.2% Digitonin, and 5 μl H2O and incubated at 37°C for 1 h. Cells were lysed by addition of 2 μl 10 mg/ml Proteinase-K, 23 μl Tagmentation Clean-up buffer (326 mM NaCl, 109 mM EDTA, 0.63% SDS), and incubated at 40 °C for 30 min. DNA was extracted and size-selected for small fragments using AMPureXP beads (Beckman Coulter, A63881) and PCR amplified into a sequencing library using NexteraXT indexing primers (Illumina, FC-131-2004) and KAPA HiFi HotStart Ready Mix (Roche, KK2602). Final libraries were again purified and size selected using AMPureXP beads, quantitated by qPCR, size distributions determined by bioanalyzer, pooled at equimolar ratios, and sequenced at the Emory Non-human Primate Genomics Core on a NovaSeq6000 using a PE100 run. Raw fastq reads were mapped to the mm10 genome using Bowtie2 v2.4.2 [53] and accessible chromatin regions identified using MACS2 2.2.7.1 [54]. Target motif enrichment was assessed using HOMER v4.11.1 [40] and Taiji PageRank v1.2.1.3 [41] analysis performed using ActB ATAC-seq and WT and KO RNA-seq datasets.
Ethics statement
All animal procedures were approved by the Emory Institutional Animal Care and Use Committee under protocol number PROTO201900113.
Author contribution statement
Zhihong Zuo: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Wrote the paper.
Anna Kania: Performed the experiments; Analyzed and interpreted the data.
Dillon Patterson; Sakeenah Hicks: Performed the experiments.
Jeffrey Maurer; Mansi Gupta: Contributed reagents, materials, analysis tools or data.
Jeremy Boss: Conceived and designed the experiments; Wrote the paper.
Christopher Scharer: Conceived and designed the experiments; Analyzed and interpreted the data; Wrote the paper.
Data availability statement
Data associated with this study has been deposited at NCBI GEO under the accession number GSE208254 and GSE208255.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper
Acknowledgements
We thank members of the Scharer and Boss labs for critical discussions and reading of the manuscript. This work was supported by the Emory Integrated Flow Cytometry Core and the Emory Integrated Genomics Core. This work was supported by NIH R01 AI123733 to J.M.B, P01 AI125180 to J.M.B. and C.D.S., U19 AI110483 to J.M.B., T32 GM008490 to J.M.B. and R01 AI148471 to C.D.S.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2023.e17527.
Appendix A. Supplementary data
The following are the Supplementary data to this article.
References
- 1.Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J.A., Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gasiunas G., Barrangou R., Horvath P., Siksnys V. Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc. Natl. Acad. Sci. U. S. A. 2012;109:E2579–E2586. doi: 10.1073/pnas.1208507109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mali P., Yang L., Esvelt K.M., Aach J., Guell M., DiCarlo J.E., Norville J.E., Church G.M. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shalem O., Sanjana N.E., Hartenian E., Shi X., Scott D.A., Mikkelson T., Heckl D., Ebert B.L., Root D.E., Doench J.G., Zhang F. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science. 2014;343:84–87. doi: 10.1126/science.1247005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chu V.T., Graf R., Wirtz T., Weber T., Favret J., Li X., Petsch K., Tran N.T., Sieweke M.H., Berek C., et al. Efficient CRISPR-mediated mutagenesis in primary immune cells using CrispRGold and a C57BL/6 Cas9 transgenic mouse line. Proc. Natl. Acad. Sci. U. S. A. 2016;113:12514–12519. doi: 10.1073/pnas.1613884113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Platt R.J., Chen S., Zhou Y., Yim M.J., Swiech L., Kempton H.R., Dahlman J.E., Parnas O., Eisenhaure T.M., Jovanovic M., et al. CRISPR-Cas9 knockin mice for genome editing and cancer modeling. Cell. 2014;159:440–455. doi: 10.1016/j.cell.2014.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Laidlaw B.J., Duan L., Xu Y., Vazquez S.E., Cyster J.G. The transcription factor Hhex cooperates with the corepressor Tle3 to promote memory B cell development. Nat. Immunol. 2020;21:1082–1093. doi: 10.1038/s41590-020-0713-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Greiner V., Bou Puerto R., Liu S., Herbel C., Carmona E.M., Goldberg M.S. CRISPR-mediated editing of the B cell receptor in primary human B cells. iScience. 2019;12:369–378. doi: 10.1016/j.isci.2019.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.LaFleur M.W., Nguyen T.H., Coxe M.A., Miller B.C., Yates K.B., Gillis J.E., Sen D.R., Gaudiano E.F., Al Abosy R., Freeman G.J., et al. PTPN2 regulates the generation of exhausted CD8(+) T cell subpopulations and restrains tumor immunity. Nat. Immunol. 2019;20:1335–1347. doi: 10.1038/s41590-019-0480-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.LaFleur M.W., Nguyen T.H., Coxe M.A., Yates K.B., Trombley J.D., Weiss S.A., Brown F.D., Gillis J.E., Coxe D.J., Doench J.G., et al. A CRISPR-Cas9 delivery system for in vivo screening of genes in the immune system. Nat. Commun. 2019;10:1668. doi: 10.1038/s41467-019-09656-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tamura T., Yanai H., Savitsky D., Taniguchi T. The IRF family transcription factors in immunity and oncogenesis. Annu. Rev. Immunol. 2008;26:535–584. doi: 10.1146/annurev.immunol.26.021607.090400. [DOI] [PubMed] [Google Scholar]
- 12.Lu R., Medina K.L., Lancki D.W., Singh H. IRF-4,8 orchestrate the pre-B-to-B transition in lymphocyte development. Genes Dev. 2003;17:1703–1708. doi: 10.1101/gad.1104803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tamura T., Thotakura P., Tanaka T.S., Ko M.S., Ozato K. Identification of target genes and a unique cis element regulated by IRF-8 in developing macrophages. Blood. 2005;106:1938–1947. doi: 10.1182/blood-2005-01-0080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pang S.H., Minnich M., Gangatirkar P., Zheng Z., Ebert A., Song G., Dickins R.A., Corcoran L.M., Mullighan C.G., Busslinger M., et al. PU.1 cooperates with IRF4 and IRF8 to suppress pre-B-cell leukemia. Leukemia. 2016;30:1375–1387. doi: 10.1038/leu.2016.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Carotta S., Willis S.N., Hasbold J., Inouye M., Pang S.H., Emslie D., Light A., Chopin M., Shi W., Wang H., et al. The transcription factors IRF8 and PU.1 negatively regulate plasma cell differentiation. J. Exp. Med. 2014;211:2169–2181. doi: 10.1084/jem.20140425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xu H., Chaudhri V.K., Wu Z., Biliouris K., Dienger-Stambaugh K., Rochman Y., Singh H. Regulation of bifurcating B cell trajectories by mutual antagonism between transcription factors IRF4 and IRF8. Nat. Immunol. 2015;16:1274–1281. doi: 10.1038/ni.3287. [DOI] [PubMed] [Google Scholar]
- 17.Wang H., Jain S., Li P., Lin J.X., Oh J., Qi C., Gao Y., Sun J., Sakai T., Naghashfar Z., et al. Transcription factors IRF8 and PU.1 are required for follicular B cell development and BCL6-driven germinal center responses. Proc. Natl. Acad. Sci. U. S. A. 2019;116:9511–9520. doi: 10.1073/pnas.1901258116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ochiai K., Maienschein-Cline M., Simonetti G., Chen J., Rosenthal R., Brink R., Chong A.S., Klein U., Dinner A.R., Singh H., Sciammas R. Transcriptional regulation of germinal center B and plasma cell fates by dynamical control of IRF4. Immunity. 2013;38:918–929. doi: 10.1016/j.immuni.2013.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sciammas R., Shaffer A.L., Schatz J.H., Zhao H., Staudt L.M., Singh H. Graded expression of interferon regulatory factor-4 coordinates isotype switching with plasma cell differentiation. Immunity. 2006;25:225–236. doi: 10.1016/j.immuni.2006.07.009. [DOI] [PubMed] [Google Scholar]
- 20.Low M.S.Y., Brodie E.J., Fedele P.L., Liao Y., Grigoriadis G., Strasser A., Kallies A., Willis S.N., Tellier J., Shi W., et al. IRF4 activity is required in established plasma cells to regulate gene transcription and mitochondrial homeostasis. Cell Rep. 2019;29 doi: 10.1016/j.celrep.2019.10.097. 2634-2645 e2635. [DOI] [PubMed] [Google Scholar]
- 21.Feng J., Wang H., Shin D.M., Masiuk M., Qi C.F., Morse H.C., 3rd IFN regulatory factor 8 restricts the size of the marginal zone and follicular B cell pools. J. Immunol. 2011;186:1458–1466. doi: 10.4049/jimmunol.1001950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rickert R.C., Roes J., Rajewsky K. B lymphocyte-specific, cre-mediated mutagenesis in mice. Nucleic Acids Res. 1997;25:1317–1318. doi: 10.1093/nar/25.6.1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kornete M., Marone R., Jeker L.T. Highly efficient and versatile plasmid-based gene editing in primary T cells. J. Immunol. 2018;200:2489–2501. doi: 10.4049/jimmunol.1701121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Haeussler M., Schonig K., Eckert H., Eschstruth A., Mianne J., Renaud J.B., Schneider-Maunoury S., Shkumatava A., Teboul L., Kent J., et al. Evaluation of off-target and on-target scoring algorithms and integration into the guide RNA selection tool CRISPOR. Genome Biol. 2016;17:148. doi: 10.1186/s13059-016-1012-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kent W.J., Sugnet C.W., Furey T.S., Roskin K.M., Pringle T.H., Zahler A.M., Haussler D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Doench J.G., Fusi N., Sullender M., Hegde M., Vaimberg E.W., Donovan K.F., Smith I., Tothova Z., Wilen C., Orchard R., et al. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat. Biotechnol. 2016;34:184–191. doi: 10.1038/nbt.3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yoon H.S., Scharer C.D., Majumder P., Davis C.W., Butler R., Zinzow-Kramer W., Skountzou I., Koutsonanos D.G., Ahmed R., Boss J.M. ZBTB32 is an early repressor of the CIITA and MHC class II gene expression during B cell differentiation to plasma cells. J. Immunol. 2012;189:2393–2403. doi: 10.4049/jimmunol.1103371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sanjana N.E., Shalem O., Zhang F. Improved vectors and genome-wide libraries for CRISPR screening. Nat. Methods. 2014;11:783–784. doi: 10.1038/nmeth.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brinkman E.K., Chen T., Amendola M., van Steensel B. Easy quantitative assessment of genome editing by sequence trace decomposition. Nucleic Acids Res. 2014;42:e168. doi: 10.1093/nar/gku936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim S., Bagadia P., Anderson D.A., 3rd, Liu T.T., Huang X., Theisen D.J., O’Connor K.W., Ohara R.A., Iwata A., Murphy T.L., Murphy K.M. High amount of transcription factor IRF8 engages AP1-IRF composite elements in enhancers to Direct type 1 conventional dendritic cell identity. Immunity. 2020;53:759–774 e759. doi: 10.1016/j.immuni.2020.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee C.H., Melchers M., Wang H., Torrey T.A., Slota R., Qi C.F., Kim J.Y., Lugar P., Kong H.J., Farrington L., et al. Regulation of the germinal center gene program by interferon (IFN) regulatory factor 8/IFN consensus sequence-binding protein. J. Exp. Med. 2006;203:63–72. doi: 10.1084/jem.20051450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lino A.C., Dang V.D., Lampropoulou V., Welle A., Joedicke J., Pohar J., Simon Q., Thalmensi J., Baures A., Fluhler V., et al. LAG-3 inhibitory receptor expression identifies immunosuppressive natural regulatory plasma cells. Immunity. 2018;49 doi: 10.1016/j.immuni.2018.06.007. 120-133 e129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pridans C., Holmes M.L., Polli M., Wettenhall J.M., Dakic A., Corcoran L.M., Smyth G.K., Nutt S.L. Identification of Pax5 target genes in early B cell differentiation. J. Immunol. 2008;180:1719–1728. doi: 10.4049/jimmunol.180.3.1719. [DOI] [PubMed] [Google Scholar]
- 34.Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., Mesirov J.P. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U. S. A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Subramanian K., Jia D., Kapoor-Vazirani P., Powell D.R., Collins R.E., Sharma D., Peng J., Cheng X., Vertino P.M. Regulation of estrogen receptor alpha by the SET7 lysine methyltransferase. Mol. Cell. 2008;30:336–347. doi: 10.1016/j.molcel.2008.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shi W., Liao Y., Willis S.N., Taubenheim N., Inouye M., Tarlinton D.M., Smyth G.K., Hodgkin P.D., Nutt S.L., Corcoran L.M. Transcriptional profiling of mouse B cell terminal differentiation defines a signature for antibody-secreting plasma cells. Nat. Immunol. 2015;16:663–673. doi: 10.1038/ni.3154. [DOI] [PubMed] [Google Scholar]
- 37.Diehl S.A., Schmidlin H., Nagasawa M., van Haren S.D., Kwakkenbos M.J., Yasuda E., Beaumont T., Scheeren F.A., Spits H. STAT3-mediated up-regulation of BLIMP1 Is coordinated with BCL6 down-regulation to control human plasma cell differentiation. J. Immunol. 2008;180:4805–4815. doi: 10.4049/jimmunol.180.7.4805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shin D.M., Lee C.H., Morse H.C., 3rd IRF8 governs expression of genes involved in innate and adaptive immunity in human and mouse germinal center B cells. PLoS One. 2011;6 doi: 10.1371/journal.pone.0027384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ludwig J., Federico G., Prokosch S., Kublbeck G., Schmitt S., Klevenz A., Grone H.J., Nitschke L., Arnold B. Dickkopf-3 acts as a modulator of B cell fate and function. J. Immunol. 2015;194:2624–2634. doi: 10.4049/jimmunol.1402160. [DOI] [PubMed] [Google Scholar]
- 40.Heinz S., Benner C., Spann N., Bertolino E., Lin Y.C., Laslo P., Cheng J.X., Murre C., Singh H., Glass C.K. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang K., Wang M., Zhao Y., Wang W. Taiji: system-level identification of key transcription factors reveals transcriptional waves in mouse embryonic development. Sci. Adv. 2019;5:eaav3262. doi: 10.1126/sciadv.aav3262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Taniguchi T., Ogasawara K., Takaoka A., Tanaka N. IRF family of transcription factors as regulators of host defense. Annu. Rev. Immunol. 2001;19:623–655. doi: 10.1146/annurev.immunol.19.1.623. [DOI] [PubMed] [Google Scholar]
- 43.Negishi H., Fujita Y., Yanai H., Sakaguchi S., Ouyang X., Shinohara M., Takayanagi H., Ohba Y., Taniguchi T., Honda K. Evidence for licensing of IFN-gamma-induced IFN regulatory factor 1 transcription factor by MyD88 in Toll-like receptor-dependent gene induction program. Proc. Natl. Acad. Sci. U. S. A. 2006;103:15136–15141. doi: 10.1073/pnas.0607181103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wiggins K.J., Scharer C.D. Roadmap to a plasma cell: epigenetic and transcriptional cues that guide B cell differentiation. Immunol. Rev. 2021;300:54–64. doi: 10.1111/imr.12934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Scharer C.D., Patterson D.G., Mi T., Price M.J., Hicks S.L., Boss J.M. Antibody-secreting cell destiny emerges during the initial stages of B-cell activation. Nat. Commun. 2020;11:3989. doi: 10.1038/s41467-020-17798-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang X.K., Moussa O., LaRue A., Bradshaw S., Molano I., Spyropoulos D.D., Gilkeson G.S., Watson D.K. The transcription factor Fli-1 modulates marginal zone and follicular B cell development in mice. J. Immunol. 2008;181:1644–1654. doi: 10.4049/jimmunol.181.3.1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Patterson D.G., Kania A.K., Price M.J., Rose J.R., Scharer C.D., Boss J.M. An IRF4-MYC-mTORC1 integrated pathway controls cell growth and the proliferative capacity of activated B cells during B cell differentiation in vivo. J. Immunol. 2021;207:1798–1811. doi: 10.4049/jimmunol.2100440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ran F.A., Hsu P.D., Wright J., Agarwala V., Scott D.A., Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 2013;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dobin A., Gingeras T.R. Mapping RNA-seq reads with STAR. Curr Protoc Bioinformatics. 2015;51 doi: 10.1002/0471250953.bi1114s51. 11 14 11-11 14 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lawrence M., Huber W., Pages H., Aboyoun P., Carlson M., Gentleman R., Morgan M.T., Carey V.J. Software for computing and annotating genomic ranges. PLoS Comput. Biol. 2013;9 doi: 10.1371/journal.pcbi.1003118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Guo M., Price M.J., Patterson D.G., Barwick B.G., Haines R.R., Kania A.K., Bradley J.E., Randall T.D., Boss J.M., Scharer C.D. EZH2 represses the B cell transcriptional program and regulates antibody-secreting cell metabolism and antibody production. J. Immunol. 2018;200:1039–1052. doi: 10.4049/jimmunol.1701470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang Y., Liu T., Meyer C.A., Eeckhoute J., Johnson D.S., Bernstein B.E., Nusbaum C., Myers R.M., Brown M., Li W., Liu X.S. Model-based analysis of ChIP-seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data associated with this study has been deposited at NCBI GEO under the accession number GSE208254 and GSE208255.