Abstract
Non-self epitopes, whether originated from foreign substances or somatic mutations, trigger immune responses when presented by major histocompatibility complex (MHC) molecules and recognized by T cells. Identification of immunogenically active neoepitopes has significant implications in cancer and virus medicine. However, current methods are mostly limited to predicting physical binding of mutant peptides and MHCs. We previously developed a deep-learning based model, DeepNeo, to identify immunogenic neoepitopes by capturing the structural properties of peptide-MHC pairs with T cell reactivity. Here, we upgraded our DeepNeo model with up-to-date training data. The upgraded model (DeepNeo-v2) was improved in evaluation metrics and showed prediction score distribution that better fits known neoantigen behavior. The immunogenic neoantigen prediction can be conducted at https://deepneo.net.
Graphical Abstract
Graphical Abstract.
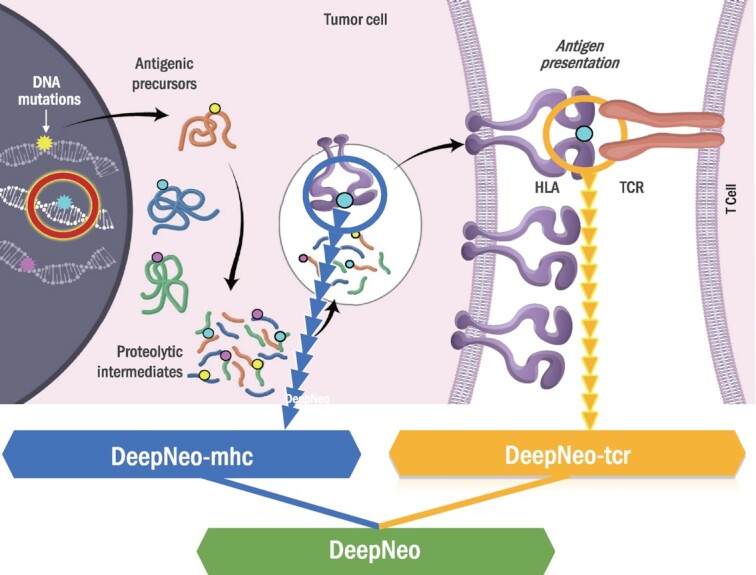
DeepNeo integrates two deep learning algorithms to predict peptide-MHC binding and immunogenicity.
INTRODUCTION
Major histocompatibility complex (MHC) is a cell surface protein that can present processed peptides to immune cells. Neoantigens derived from somatic mutations in cancer cells stimulate anticancer immune responses when they form a complex with MHCs that T cells recognize as foreign objects. Therefore, neoantigens are regarded as potential targets for personalized cancer vaccines, and the amount of neoantigens is used as a biomarker for cancer immunotherapy such as immune checkpoint blockade (ICB) (1–3). However, current methods of neoantigen identification are mostly restricted to predicting physical binding of peptides and MHCs, which is necessary but not sufficient for inducing immune responses (4–6). This prompted us to develop DeepNeo, a pipeline integrating DeepNeo-mhc (7), a tool for predicting peptide-MHC binding, and DeepNeo-tcr (8), a tool for predicting T cell reactivity of a pMHC complex. DeepNeo predicts immunogenic neoantigens for MHC I, which are presented to CD8 + T cells, and MHC II, which are presented to CD4 + T cells, in human and mouse. The model accuracy was extensively validated using different datasets and have shown signals of neoantigen depletion in both ICB therapy cohort and treatment naïve TCGA samples. Here, we upgraded DeepNeo to enhance reliability and developed a web server for user convenience.
MATERIAL AND METHODS
Data collection
Additional data used in constructing the models was obtained from IEDB as of August 30th, 2022 (9). All data were filtered using the following criteria. First, the length of peptides must be 9 for MHC I and 15 for MHC II. Second, four-digit HLA type must be present. Third, for DeepNeo-tcr, only experimental data measuring Interferon-γ (IFN-γ) release upon pMHC challenge was used. Fourth, if contradictory results were collected from different sources, the pMHC was labelled as positive. If a pMHC can bind or trigger immune response under one condition but not under another, we concluded that the pair are reactive.
For DeepNeo-tcr (MHC I) training set, we kept all pairs with positive labels (N = 8148) and randomly selected 11001 pairs with negative labels so that the model could be trained without severe data imbalance. We also expanded the DeepNeo-tcr (MHC II) training set to have a significant proportion of the DeepNeo-tcr (MHC II) positive binding set by adding 1705 randomly selected pairs with negative MHC binding data. As a result, 340987 data for DeepNeo-mhc (MHC I), 106908 for DeepNeo-mhc (MHC II), 19149 for DeepNeo-tcr (MHC I), and 12913 for DeepNeo-tcr (MHC II) model were used for learning each model. For each model, the data was randomly divided into test set and training set, which was further divided into training and validation sets using three-fold cross validation.
Upgrades in MHC binding model
DeepNeo-mhc has been upgraded to predict neoantigens for MHC I and II for human and mouse alleles. As the role of CD4 in cancer immunity is being actively researched, there has been increasing demands for prediction of MHC II epitopes (10). Moreover, many tools only cover human alleles, but prediction on mouse alleles is also important in experimental settings. To bridge the gap, we expanded the model to predict MHC I and II epitopes for human and mouse alleles.
The architecture of the model is identical to DeepNeo-mhc-v1 model but trained with a larger dataset, leading to different set of hyperparameters yielding best performance. To briefly summarize, the model is generated with convolutional neural network, with two convolution layers to learn features from MHC and peptide using rectified linear (ReLu) function, a fully connected layer, and a sigmoid layer that transforms prediction values within range of 0 and 1.
Upgrades in T cell reactivity model
In the upgraded DeepNeo-tcr model, the activation functions of the two convolution layers were changed. DeepNeo-tcr-v1 uses ReLU activation function. Because the function converts all negative weights to zero, it is prone to dying ReLU problem (11). Although this property can prevent overfitting as sparsity inducing regularization method, DeepNeo-tcr has relatively small dimensions with large filter size, requiring all the nodes to have weights. Thus, we replaced ReLU function with linear function to include all feature importance of the convolution layers. Similar to DeepNeo-mhc, hyperparameter tuning was conducted to determine the set of weights that maximizes model performance.
Prediction scores above the threshold (0.5) are defined as positive calls for both MHC binding and T cell reactivity model. In evaluation of the external validation set, which requires consideration of both models, the scores were multiplied to assess model performance.
Conversion of deep learning framework
We developed DeepNeo-v2 based on Keras (https://github.com/fchollet/keras) framework v2.4.0 in Python 3.7 environment, whereas DeepNeo-v1 was implemented with Theano library (https://github.com/Theano/Theano) v1.0.4 on Python 2.7, which are no longer actively maintained. The architecture of the model was transferred with a minor change in the loss function; negative log-likelihood (NLL) was used in DeepNeo-v1 and binary cross entropy (BCE) was used in DeepNeo-v2. The two loss functions are often used interchangeably (12) because they have the same concept, but the BCE function makes computation more straightforward and have lower probability of numerical instabilities. Finally, the codes were optimized to reduce prediction runtime to be efficiently implemented in the webserver (Supplementary Table 1). The runtime for DeepNeo-v1 and -v2 was calculated as the mean of ten trials per input data size.
Implementation of web server
DeepNeo-v2 utilizes the Web Server Gateway Interface (WSGI) as the gateway interface between Apache2 HTTP server (https://httpd.apache.org) and Flask web application framework (https://flask.palletsprojects.com/). The front-end of the web-application is written with HTML5 (https://www.w3.org/TR/html5/), CSS (https://www.w3.org/Style/CSS/), JavaScript (https://javascript.com) and jQuery (https://jquery.com).
RESULTS
Model evaluation
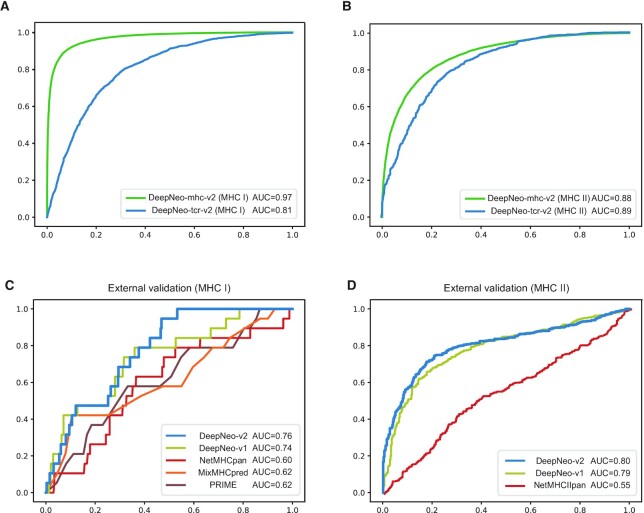
DeepNeo integrates DeepNeo-mhc and DeepNeo-tcr to predict peptide-MHC binding and T cell reactivity, respectively. Both models have been upgraded since the publication as additional data were obtained. Compared to DeepNeo-v1 models, data size for generating MHC binding model increased by 7.8 fold and T cell reactivity model by 1.2 fold. With the updated dataset, the models were reconstructed with different hyperparameters and architectures to obtain best performance. Then, the predictive performance was compared to that of DeepNeo-v1. Both DeepNeo-mhc-v2 and DeepNeo-tcr-v2 achieved higher ROC AUC than previous versions for both classes (Table 1, Figure 1a,b). Although PR AUC and F1 scores could not be directly compared due to different data label composition, the prediction score distribution indicated that the upgraded models can predict with higher discriminatory power.
Table 1.
Measures of model performance. The accuracy metrics including ROC AUC, PR AUC, and F1 scores are compared between DeepNeo-v1 and DeepNeo-v2
| DeepNeo-v1 | DeepNeo-v2 | |||||||
|---|---|---|---|---|---|---|---|---|
| Data(Positive %) | ROC AUC | PR AUC | F1 score | Data (Positive %) | ROC AUC | PR AUC | F1 score | |
| MHC I | 57173 (24.9) | 0.875 | 0.456 | 0.766 | 340987 (75.0) | 0.969 | 0.969 | 0.948 |
| MHC II | – | – | – | – | 106908 (57.2) | 0.882 | 0.874 | 0.827 |
| TCR I | 12128 (50) | 0.778 | 0.781 | 0.727 | 19149 (42.6) | 0.805 | 0.713 | 0.687 |
| TCR II | 6260 (50) | 0.805 | 0.798 | 0.751 | 12913 (42.4) | 0.826 | 0.756 | 0.717 |
Figure 1.
Model performance metrics. ROC AUC of MHC binding model (green) and T cell reactivity model (blue) for MHC I (A) and II (B) indicate reliable prediction performance. ROC AUC of external validation set for MHC I (C) and MHC II (D) show that DeepNeo-v2 outperforms other tools.
Next, we evaluated DeepNeo in predicting external validation sets for MHC I and II, which were not included in model construction, in comparison with other prediction tools. More specifically, PRIME (13), a tool for predicting human MHC I immunogenic neoantigens, and NetMHCpan (4), MixMHCpred (5), tools for predicting MHC I and II neoantigens without regard to immunogenicity, were included. For MHC I, in vitro assay data measuring immunogenicity of pMHC-I complex was retrieved from the PRIME publication (13). DeepNeo-v2 outperformed other tools as well as DeepNeo-v1 in ROC AUC as well as other statistical metrics (Figure 1c, Supplementary Figure S1a). Moreover, a total of 812 independent immunogenicity data of pMHC-II was retrieved from multiple publications (14–18). Here, DeepNeo-v2 also exhibited superior power in prediction compared to NetMHCpan and DeepNeo-v1 (Figure 1d, Supplementary Figure S1b).
Resolving prediction score skewness
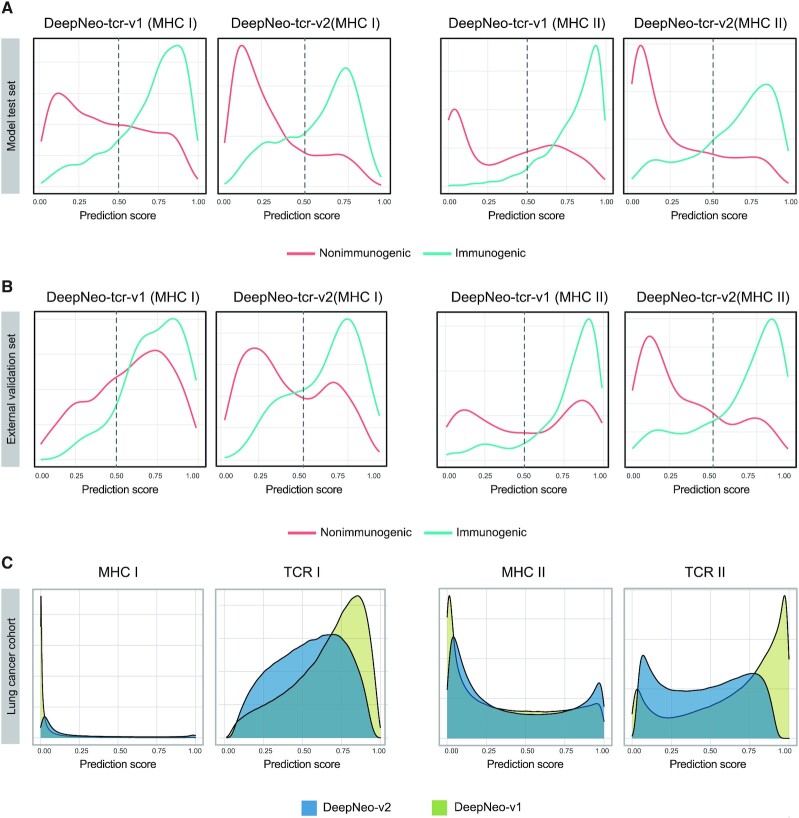
Imbalance in training data labels can lead to prediction score skewness, especially if data size is limited. Because DeepNeo-tcr models had relatively small data size (<20000) and data imbalance (25:75), the models could not efficiently learn features of immunogenic pMHCs (data with positive labels). DeepNeo-tcr-v1 sought to deal with this problem by adjusting label balance (50:50) by random selection and training to maximize sensitivity. However, this approach led to undermining prediction for negative label data and overall skewness to positive (>0.5) prediction scores. Here, we employed label balancing with increased proportion of negative label (43:57) and changing convolution layer activation functions to enhance specificity and modify prediction score distribution. The optimal label ratio was determined as that yielding best prediction performance in both test set and external validation set (Supplementary Table 2). Then, the score distributions of the two versions were evaluated to check if the upgraded model better represents labels and known neoantigen behavior.
First, the score distribution of each label was analyzed in DeepNeo-tcr test sets. As expected, the density peak of nonimmunogenic neoantigens (red) were centered below threshold while that of immunogenic neoantigens (blue) were above the threshold of 0.5 (Figure 2a). The discriminatory score distribution indicate that the model has learned distinctive features of immunogenicity. Notably, the power of predicting negative values was dramatically improved in DeepNeo-tcr-v2 (MHC I) and DeepNeo-tcr-v2 (MHC II).
Figure 2.
Score distribution of DeepNeo-tcr-v1 and DeepNeo-tcr-v2 models. In model test set (A), external validation set (B), cancer patient cohort (C), score distributions indicate that DeepNeo-tcr-v2 of both classes can better discriminate spatial interaction of pMHC that elicit immune response and represent known neoantigen behavior.
Moreover, the score distribution in the external validation set was examined. In consensus with the test set analysis, DeepNeo-tcr-v2 show that they can yield prediction scores that match labels compared to DeepNeo-tcr-v1 (Figure 2b). Importantly, the score distributions of nonimmunogenic pMHCs of both classes show distinctive peak below the threshold in DeepNeo-tcr-v2, as more negative data were included and the models were trained to maximize specificity.
Finally, the prediction score distribution was examined in patient data. From 335 lung cancer patients, >900000 putative pMHCs were obtained and subjected to DeepNeo prediction (8). Although somatic mutations can give rise to many neoantigen sequence combinations, only a fraction can bind to MHC complex, and only a fraction of the pMHC can exert immune response. Due to the structure of MHC complex and length of recognizable peptides, MHC I, which have closed binding groove and bind shorter peptides, tend to be more specific in antigen recognition compared to MHC II, which have open binding groove and bind longer peptides. As expected, the distribution of peptide-MHC I binding was mostly negative, while the distribution of MHC II yielded more positive values. Compared to DeepNeo-mhc-v1, DeepNeo-mhc-v2 show peak in both negative and positive area, signifying that the model has confidently learned features of each label. In DeepNeo-tcr-v2, the negative shift in score distribution and smaller number of pMHC above threshold supports that only a fraction can induce T cell reaction.
Webserver
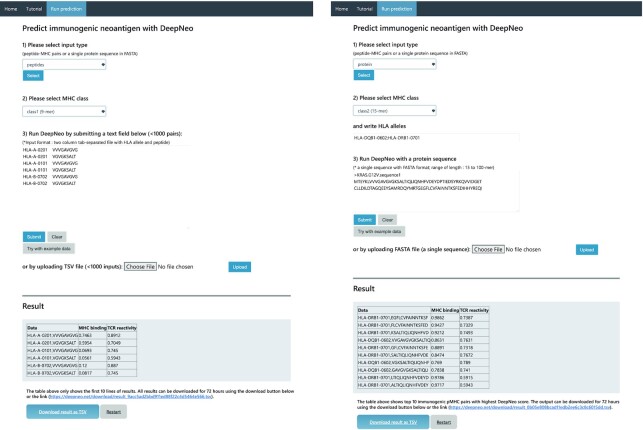
The webserver is free and open to all and there is no login requirement. Upon running the prediction, DeepNeo will provide prediction score of MHC binding and T cell reactivity (Figure 3). The output is immediately shown on the same page or can be revisited within 72 hours using the URL provided. Estimated prediction times for various input length is provided in Supplementary Table 1.
Figure 3.
Example webserver result of peptides mode (left) and protein mode (right).
Input format description
For user convenience, DeepNeo can be performed in two modes: peptides and protein mode. The peptides mode deals with a list of 9- or 15-mers usually containing substitution mutations whereas the protein mode identifies potential neoepitopes from a novel sequence usually derived by frameshift mutations or aberrant splicing. The pMHC pairs should be parsed as tab delimited format as detailed in the tutorial section. In essence, neoantigens derived from somatic mutation should be annotated, translated, and clipped to generate putative neoantigen candidates. Furthermore, DeepNeo prediction can be performed against full protein sequence provided as fasta format.
Another component of the input is MHC genotype. For human samples, the MHC genotype can be typed by multiple human leukocyte antigen (HLA) typing tools. The mouse genotypes are documented for each strain. The supported alleles of the program include HLA-A, -B, -DQB1, -DRB1 for human alleles and most mouse alleles. In peptides mode, the HLA allele and peptides should be combined as tab-separated file. In protein mode, the HLA alleles should be listed in the HLA allele textbox and separated with semicolon.
Output format description
DeepNeo output is composed of three columns: data, MHC binding prediction score, and T cell reactivity prediction score. By definition, score above 0.5 for both MHC binding prediction score and T cell reactivity score is interpreted as immunogenic neoantigen. If the pMHC has positive prediction score for MHC binding, but not T cell reactive, the pMHC can be interpreted as nonimmunogenic neoantigen. If the pMHC is predicted to be T cell reactive without MHC binding, the complex is predicted to have no biological significance, as MHC binding is a mandatory precursor of peptide derived immune reaction. In peptides mode, ten pMHC scores are shown immediately and the full result can be retrieved by downloading, while 'protein’ mode returns top ten candidates (>0.5 for both MHC binding and T cell reactivity) with highest combined score.
DISCUSSION
With the outbreak of SARS-CoV-2, the concept of mRNA vaccine has become widely accepted. The SARS-CoV-2 vaccines contain fragment of virus mRNA that can stimulate immune response in human (19,20). The neoantigens, or the translated mRNA, can also have similar vaccination effect. Indeed, neoantigen vaccine is considered as a promising next generation cancer treatment method that is tailored to each patient (21). Although its clinical effectiveness in tumor regression and prevention of relapse have been reported by multiple trials, prioritizing vaccine target neoantigens still remains as a major challenge (22,23). In fact, most pMHC pairs were incapable of stimulating T cell response, emphasizing need for immunogenicity prediction (24). Furthermore, the role of CD4 + mediated immunogenicity has been reported on various neoantigen vaccine clinical trials (25–27), even when pMHC-I was targeted (28). DeepNeo-v2 overcomes these limitations by providing comprehensive prediction of MHC binding and T cell reactivity of both MHC I and II. Furthermore, simple, fast, and user-friendly implementation of web service will be able to reach wider audience.
Although this work has made significant upgrades to DeepNeo, there is room for improvement. Compared to DeepNeo-mhc, DeepNeo-tcr has limited data size, which is expected to increase as more public data on IFN-γ assay will be available in the future. As more data become available, we anticipate that prediction score skewness as well as model evaluation metrics will improve.
DATA AVAILABILITY
DeepNeo-v2 can be accessed with https://deepneo.net.
Supplementary Material
Contributor Information
Jeong Yeon Kim, Department of Bio and Brain Engineering, KAIST, Daejeon, Republic of Korea.
Hyoeun Bang, PentaMedix Co., Ltd., Seongnam-si, Gyeonggi-do, Republic of Korea.
Seung-Jae Noh, PentaMedix Co., Ltd., Seongnam-si, Gyeonggi-do, Republic of Korea.
Jung Kyoon Choi, Department of Bio and Brain Engineering, KAIST, Daejeon, Republic of Korea.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
This work was supported by the Basic Research Laboratory Program (NRF-2022R1A4A5028131), by the Korea Drug Development Fund (RS-2022-00166187), and by the Technology Development Program (S3046270). Funding for open access charge: Basic Research Laboratory Program.
Conflict of interest statement. None declared.
REFERENCES
- 1. Peng M., Mo Y., Wang Y., Wu P., Zhang Y., Xiong F., Guo C., Wu X., Li Y., Li X.et al.. Neoantigen vaccine: an emerging tumor immunotherapy. Mol. Cancer. 2019; 18:128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Van Allen E.M., Miao D., Schilling B., Shukla S.A., Blank C., Zimmer L., Sucker A., Hillen U., Geukes Foppen M.H., Goldinger S.M.et al.. Genomic correlates of response to CTLA-4 blockade in metastatic melanoma. Science. 2015; 350:207–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Anagnostou V., Smith K.N., Forde P.M., Niknafs N., Bhattacharya R., White J., Zhang T., Adleff V., Phallen J., Wali N.et al.. Evolution of neoantigen landscape during immune checkpoint blockade in non-small cell lung cancer. Cancer Discov. 2017; 7:264–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Reynisson B., Alvarez B., Paul S., Peters B., Nielsen M.. NetMHCpan-4.1 and NetMHCIIpan-4.0: improved predictions of MHC antigen presentation by concurrent motif deconvolution and integration of MS MHC eluted ligand data. Nucleic Acids Res. 2020; 48:W449–W454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bassani-Sternberg M., Chong C., Guillaume P., Solleder M., Pak H., Gannon P.O., Kandalaft L.E., Coukos G., Gfeller D.. Deciphering HLA-I motifs across HLA peptidomes improves neo-antigen predictions and identifies allostery regulating HLA specificity. PLOS Comput. Biol. 2017; 13:e1005725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Racle J., Michaux J., Rockinger G.A., Arnaud M., Bobisse S., Chong C., Guillaume P., Coukos G., Harari A., Jandus C.et al.. Robust prediction of HLA class II epitopes by deep motif deconvolution of immunopeptidomes. Nat. Biotechnol. 2019; 37:1283–1286. [DOI] [PubMed] [Google Scholar]
- 7. Kim K., Kim H.S., Kim J.Y., Jung H., Sun J.-M., Ahn J.S., Ahn M.-J., Park K., Lee S.-H., Choi J.K.. Predicting clinical benefit of immunotherapy by antigenic or functional mutations affecting tumour immunogenicity. Nat. Commun. 2020; 11:951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kim J.Y., Cha H., Kim K., Sung C., An J., Bang H., Kim H., Yang J.O., Chang S., Shin I.et al.. MHC II immunogenicity shapes the neoepitope landscape in human tumors. Nat. Genet. 2023; 55:221–231. [DOI] [PubMed] [Google Scholar]
- 9. Vita R., Overton J.a., Greenbaum J.a., Ponomarenko J., Clark J.D., Cantrell J.R., Wheeler D.K., Gabbard J.L., Hix D., Sette A.et al.. The immune epitope database (IEDB) 3.0. Nucleic Acids Res. 2015; 43:D405–D412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Weaver J.M., Lazarski C.A., Richards K.A., Chaves F.A., Jenks S.A., Menges P.R., Sant A.J.. Immunodominance of CD4 T cells to foreign antigens is peptide intrinsic and independent of molecular context: implications for vaccine design. J. Immunol. 2008; 181:3039–3048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lu L., Shin Y., Su Y., Karniadakis G.. Dying ReLU and initialization: theory and numerical examples. Commun. Comput. Phys. 2020; 28:1671–1706. [Google Scholar]
- 12. Zhao G., Yang W., Ren X., Li L., Wu Y., Sun X.. Well-classified examples are underestimated in classification with deep Neural networks. Proc. AAAI Conf. Artif. Intell. 2022; 36:9180–9189. [Google Scholar]
- 13. Schmidt J., Smith A.R., Magnin M., Racle J., Devlin J.R., Bobisse S., Cesbron J., Bonnet V., Carmona S.J., Huber F.et al.. Prediction of neo-epitope immunogenicity reveals TCR recognition determinants and provides insight into immunoediting. Cell Reports Med. 2021; 2:100194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lee M.N., Meyerson M.. Antigen identification for HLA class I- and HLA class II-restricted T cell receptors using cytokine-capturing antigen-presenting cells. Sci. Immunol. 2021; 6:eabf4001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Stryhn A., Kongsgaard M., Rasmussen M., Harndahl M.N., Østerbye T., Bassi M.R., Thybo S., Gabriel M., Hansen M.B., Nielsen M.et al.. A systematic, unbiased mapping of CD8 + and CD4 + T cell epitopes in Yellow fever vaccinees. Front. Immunol. 2020; 11:1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Grifoni A., Moore E., Voic H., Sidney J., Phillips E., Jadi R., Mallal S., De Silva A.D., De Silva A.M., Peters B.et al.. Characterization of magnitude and antigen specificity of HLA-DP, DQ, and DRB3/4/5 restricted DENV-specific CD4+ T cell responses. Front. Immunol. 2019; 10:1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ferrari V., Tarke A., Fields H., Ferrari L., Conley T., Ferrari F., Koşaloğlu-Yalçın Z., Sette A., Peters B., McCarthy C.L.et al.. In vitro induction of neoantigen-specific T cells in myelodysplastic syndrome, a disease with low mutational burden. Cytotherapy. 2021; 23:320–328. [DOI] [PubMed] [Google Scholar]
- 18. Parn S., Jabbour G., Nguyenkhoa V., Dakshanamurthy S.. Design of peptide Vaccine for COVID19: CD8+ and CD4+ T cell epitopes from SARS-CoV-2 open-reading-frame protein variants. 2021; bioRxiv doi:22 September 2021, preprint: not peer reviewed 10.1101/2021.09.21.461301. [DOI]
- 19. Krammer F. SARS-CoV-2 vaccines in development. Nature. 2020; 586:516–527. [DOI] [PubMed] [Google Scholar]
- 20. Kyriakidis N.C., López-Cortés A., González E.V., Grimaldos A.B., Prado E.O.. SARS-CoV-2 vaccines strategies: a comprehensive review of phase 3 candidates. NPJ Vaccines. 2021; 6:28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Blass E., Ott P.A.. Advances in the development of personalized neoantigen-based therapeutic cancer vaccines. Nat. Rev. Clin. Oncol. 2021; 18:215–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Redwood A.J., Dick I.M., Creaney J., Robinson B.W.S.. What's next in cancer immunotherapy? - the promise and challenges of neoantigen vaccination. Oncoimmunology. 2022; 11:2038403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chen I., Chen M.Y., Goedegebuure S.P., Gillanders W.E.. Challenges targeting cancer neoantigens in 2021: a systematic literature review. Expert Rev. Vaccines. 2021; 20:827–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wells D.K., van Buuren M.M., Dang K.K., Hubbard-Lucey V.M., Sheehan K.C.F., Campbell K.M., Lamb A., Ward J.P., Sidney J., Blazquez A.B.et al.. Key parameters of tumor epitope immunogenicity revealed through a consortium approach improve neoantigen prediction. Cell. 2020; 183:818–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sahin U., Derhovanessian E., Miller M., Kloke B.P., Simon P., Löwer M., Bukur V., Tadmor A.D., Luxemburger U., Schrörs B.et al.. Personalized RNA mutanome vaccines mobilize poly-specific therapeutic immunity against cancer. Nature. 2017; 547:222–226. [DOI] [PubMed] [Google Scholar]
- 26. Keskin D.B., Anandappa A.J., Sun J., Tirosh I., Mathewson N.D., Li S., Oliveira G., Giobbie-Hurder A., Felt K., Gjini E.et al.. Neoantigen vaccine generates intratumoral T cell responses in phase ib glioblastoma trial. Nature. 2019; 565:234–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hilf N., Kuttruff-Coqui S., Frenzel K., Bukur V., Stevanović S., Gouttefangeas C., Platten M., Tabatabai G., Dutoit V., van der Burg S.H.et al.. Actively personalized vaccination trial for newly diagnosed glioblastoma. Nature. 2019; 565:240–245. [DOI] [PubMed] [Google Scholar]
- 28. Ott P.A., Hu Z., Keskin D.B., Shukla S.A., Sun J., Bozym D.J., Zhang W., Luoma A., Giobbie-Hurder A., Peter L.et al.. An immunogenic personal neoantigen vaccine for patients with melanoma. Nature. 2017; 547:217–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
DeepNeo-v2 can be accessed with https://deepneo.net.