Key Points
-
•
AML with RARG rearrangement is a novel subtype of AML with some unique clinical, immunophenotypic, and genetic characteristics.
-
•
AML with RARG rearrangement is insensitive to ATRA and ATO and carries a poor prognosis.
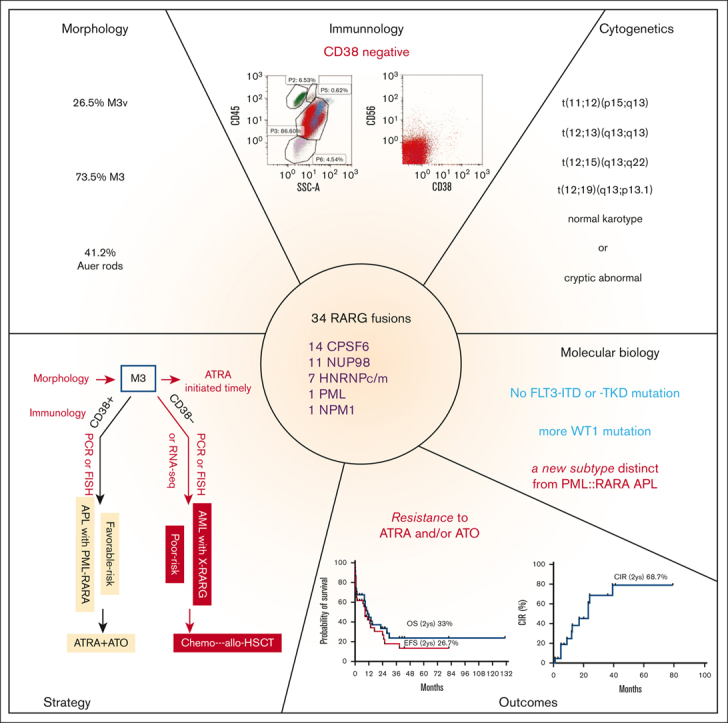
Visual Abstract
Abstract
Acute myeloid leukemia (AML) with retinoic acid receptor γ (RARG) rearrangement has clinical, morphologic, and immunophenotypic features similar to classic acute promyelocytic leukemia. However, AML with RARG rearrangement is insensitive to alltrans retinoic acid (ATRA) and arsenic trioxide (ATO) and carries a poor prognosis. We initiated a global cooperative study to define the clinicopathological features, genomic and transcriptomic landscape, and outcomes of AML with RARG rearrangements collected from 29 study groups/institutions worldwide. Thirty-four patients with AML with RARG rearrangements were identified. Bleeding or ecchymosis was present in 18 (54.5%) patients. Morphology diagnosed as M3 and M3v accounted for 73.5% and 26.5% of the cases, respectively. Immunophenotyping showed the following characteristics: positive for CD33, CD13, and MPO but negative for CD38, CD11b, CD34, and HLA-DR. Cytogenetics showed normal karyotype in 38% and t(11;12) in 26% of patients. The partner genes of RARG were diverse and included CPSF6, NUP98, HNRNPc, HNRNPm, PML, and NPM1. WT1- and NRAS/KRAS-mutations were common comutations. None of the 34 patients responded to ATRA and/or ATO. Death within 45 days from diagnosis occurred in 10 patients (∼29%). At the last follow-up, 23 patients had died, and the estimated 2-year cumulative incidence of relapse, event-free survival, and overall survival were 68.7%, 26.7%, and 33.5%, respectively. Unsupervised hierarchical clustering using RNA sequencing data from 201 patients with AML showed that 81.8% of the RARG fusion samples clustered together, suggesting a new molecular subtype. RARG rearrangement is a novel entity of AML that confers a poor prognosis. This study is registered with the Chinese Clinical Trial Registry (ChiCTR2200055810).
Introduction
Acute myeloid leukemia (AML) is a heterogeneous group of hematopoietic stem cell disorders. The revised fourth edition of the World Health Organization classification of tumors of hematopoietic and lymphoid tissues defines 19 subtypes of AML.1 With the advent of next-generation sequencing, an increasing number of novel molecular abnormalities have been found, uncovering more subtypes of AML.2
Recent use of RNA sequencing (RNA-seq) at diagnosis has led to an increased identification of cases with retinoic acid receptor γ(RARG) rearrangements. Only 1 retinoic acid receptor β(RARB) rearrangement, TBL1XR1::RARB, was identified in children with acute promyelocytic leukemia (APL).3,4 To date, 15 patients with NUP98::RARG, PML::RARG, CPSF6::RARG, NPM1::RARG, and HNRNPc::RARG fusions have been reported.3,5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17 All these patients showed strikingly similar features to those of APL, including clinical presentation and the leukemic cells’ cytomorphological and immunophenotypical features. However, these patients showed no response to standard treatment with all-trans retinoic acid (ATRA) and arsenic trioxide (ATO) and had poor outcomes. The presence of RARG but lack of RAR α(RARA) rearrangements in these patients made their disease classification and treatment options difficult.18, 19, 20, 21 Therefore, further studies of the classification and underlying molecular pathology of and therapeutic approaches to AML with RARG rearrangement are urgently needed.
The purpose of this study is to further characterize this AML subtype and provide information to facilitate its rapid diagnosis and effective treatment. Accordingly, we initiated a global cooperative study to define the clinical-biological features, transcriptomic and genomic landscape, treatment strategies, and outcomes of 34 cases of AML with RARG rearrangement diagnosed during the past decade, which were collected from 29 study groups/institutions worldwide.
Methods
Study cohort
Patients were included in the study if they were confirmed to have RARG rearrangement. Patients were identified from Chinese APL Cooperation Group, Global APL Collaborative Research Group, and publishing reports. We actively screened the database of 29 study groups/institutions from China, Spain, the United Kingdom, the Republic of Korea, and the United States for cases of AML with RARG rearrangement recorded between 2011 and 2021. Among the 34 cases included in this study, 15 have been previously reported, and their treatments and outcomes were updated.3,5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17 Because 1 patient enrolled in this study lacked updated information, the denominator of proportion calculation is 33 in some instances in this article. Data of surviving patients were updated during the study period of more than 2 years. All essential and relevant data (including laboratory features at diagnosis, type of treatment, response to therapy, and follow-up data) were collected from all participating centers. Flow cytometric analysis was performed in each center. Our previously published data, which included 221 consecutive patients with PML::RARA APL, were used as controls for flow cytometry comparisons.22 Immunophenotypic analysis was conducted using flow cytometry to assess CD3, CD13, CD14, CD19, CD33, CD34, CD38, CD56, CD64, CD117, CD11b, HLA-DR, and MPO expression. The flow cytometry thresholds for this analysis were 20%. For cytogenetic analysis, chromosomal karyotyping by G-banding or RHG-banding was performed using standard techniques, and karyotypes were described in accordance with the international system for human cytogenetic nomenclature.23 For detection of rearrangements of PML::RARA or RARA rearrangements, dual color fluorescence in situ hybridization was performed on methanol/acetic acid-fixed cells using PML/RARA and the RARA dual color break apart.
Samples from 24 patients with AML with RARG rearrangements and 142 with typical APL were used for next-generation sequencing (NGS) analysis. The NGS panel contains common mutant genes of hematological malignancies, including genes associated with activated signaling pathway (FLT3-ITD, FLT3-TKD, KRAS, NRAS, and KIT), epigenetic regulators (IDH1/2, DNMT3A, EZH2, TET2, ASXL1, and SETD2), myeloid transcription factors (WT1, NPM1, GATA2, CEBPA, ETV6, RUNX1, CBL, PTEN, and TP53).
RNA-seq preprocessing and mapping
RNA-seq preprocessing and mapping Hg38 RefSeq data were downloaded from the University of California, Santa Cruz Genome Browser. Hisat2 (version 2.0.5) and STAR (version 2.5.2b) were then used to align raw RNA-seq sequences to the reference genome. The preprocessing steps were mostly carried out per the Genome Analysis Toolkit best practices pipeline.24 Arriba version 2.1.0 was used to call fusion genes.25 To ensure the functionality and reliability of the identified fusions, we applied a strict filter in the fusion calling process. Specifically, we removed (1) fusion break points with less than 2 split reads or 3 spanning reads, (2) fusions reported in the healthy population, (3) fusions appearing in the blacklist, and (4) fusions associated with uncharacterized genes and mitochondrial genes. Out-of-frame fusions were also removed because their protein products are likely to lose function or to be rapidly degraded. A transcript-level read count matrix was generated using HTSeq-count (version 0.5.4.p3),26 based on the Gencode annotation database27 and the BAM files generated by HISAT2. Differentially expressed genes were identified using DESeq2 (version 1.18.1) based on the read count matrix. Fragments per kilobase per million values were then calculated and log-transformed to evaluate the gene expression levels.
To compare the functional characteristics of the 3 subgroups within the samples (ie, RARG rearrangement, PML::RARA, and non-APL AML), gene set enrichment analysis was performed using GSEA software (version 3.0; http://software.broadinstitute.org/gsea) with gene ontology and Kyoto encyclopedia of genes and genomes gene sets.28,29 The ward.D algorithm in R’s cluster method was used for unsupervised clustering, in which the different numbers (1% to 15%) of top-ranked highly variable genes across all samples were selected to evaluate the stability. The ComplexHeatmap R package was applied for the visualization of all samples.30
Statistical analyses and definition of outcomes
Complete remission (CR) was defined in accordance with the European Leukemia-Net AML guidelines.31 Induction death was defined as death occurring within 45 days after the start of therapy. Overall survival was calculated from the time of diagnosis to the time of death, and event-free survival was calculated from the time of diagnosis to the time of failure to enter CR, relapse, or death.
Statistical analyses were performed using SPSS software. For categorical variables, comparisons were evaluated using the Fisher exact test, and for continuous variables, the Mann-Whitney U test was used. Distributions of time-to-event variables were estimated using the Kaplan-Meier method. Univariate analysis was performed using the Cox proportional hazard model. All reported P values were two-sided.
This study was approved by the institutional review board of the First Affiliated Hospital, Zhejiang University School of Medicine (No. 037) and conducted in accordance with the principles of the Declaration of Helsinki.
Results
Clinical features
Thirty-three patients with detailed information were included for further analysis. Three patients were children 1, 10, and 13 years old. At presentation, the most common symptoms were bleeding or spontaneous ecchymosis (18 of 33 patients [54.5%]), asthenia (8 of 33 patients [24.2%]), and fever (18 of 33 patients [54.5%]); bleeding or spontaneous ecchymosis was more frequent symptoms in typical APL (P = .0114). Thirteen patients (38.2%) had white blood cells (WBC) >10 × 109/L. Coagulopathy with fibrinogen levels lower than 150 mg/dL and D-dimer levels higher than 500 μg/L occurred in 18 of 33 (54.5%) and 33 of 33 (100%) patients, respectively. Patients with typical APL had relatively higher hemoglobin level (P = .0170), longer prothrombin time (P = .0061) and partial thromboplastin time (P = .0023), and higher D-dimer level (P < .0001) than those with AML with RARG rearrangements. The initial clinical and biological features, treatments, and outcomes of the entire cohort having RARG rearrangements and control APL are shown in Table 1.
Table 1.
Baseline characteristics
| Characteristic | RARG fusions number (% or range) | Typical APL number (% or range) | P |
|---|---|---|---|
| No. of patients | 34 | 100 | |
| Age (y) median (range) | 42 (1-67) | 37 (17-74) | .7590 |
| Male sex (%) | 20 (59) | 54 s (54) | .2390 |
| Clinical presentation | n = 33 | n = 90 | |
| Bleeding | 18 (55) | 70 (78) | .0114 |
| Fever | 18 (55) | 33 (30) | .0745 |
| Blood tests (range) | n = 33 | n = 94 | |
| White blood cell count (×109/L) | 6.8 (0.2-139) | 3.63 (0.3-102.38) | .2349 |
| Hemoglobin (×g/L) | 81 (42-127) | 91 (41-139) | .0170 |
| Platelet count (×109/L) | 57.5 (7-204) | 28 (3-184) | .0004 |
| PT (s) | 14 (11.5-18.1) | 15.85(10.2-27.1) | .0061 |
| APTT (s) | 30.7 (19.5-43.1) | 35.85 (22.1-55.4) | .0023 |
| Fibrinogen (mg/dL) | 134 (40-540) | 146 (40-595) | .9417 |
| D-dimer (ug/L) | 7090 (571-94390) | 15400 (930-182310) | < .0001 |
| Bone marrow | |||
| Morphology | n = 33 | n = 79 | |
| APL-like cells (range) | 87 (27-100) | 82 (46-97) | .1204 |
| Hypergranular (%) | 25 (73.5) | 71 (90) | .0258 |
| Hypogranular (%) | 9 (26.5) | 8 (10) | .0258 |
| Auer body (%) | 14 (41) | 74 (94) | .0006 |
| Cytogenetics (%) | |||
| Normal karyotype | 14 (41) | — | |
| t(11;12)(p15;q13) | 9 (27) | — | |
| t(12;13)(q13;q13) | 1 (3) | — | |
| t(12;15)(q13;q22) | 1 (3) | — | |
| t(12;19)(q13;p13.1) | 1 (3) | — | |
| others karyotype | 7 (21) | — | |
| Not available | 1 (3) | — | |
| Partner fusion gene (%) | |||
| CPSF6 | 14 (41) | — | |
| NUP98 | 11 (32) | — | |
| HNRNPc | 6 (18) | — | |
| HNRNPm | 1 (3) | — | |
| PML | 1 (3) | — | |
| NPM1 | 1 (3) | — | |
| Sanz risk stratification (%) | |||
| Nonhigh-risk | 21 of 34 (62) | 63 of 94 (67) | .5802 |
| high-risk | 13 of 34 (38) | 31 of 94 (33) | .5802 |
APTT, partial thromboplastin time; PT, prothrombin time.
Morphology
Morphological features of leukemic blasts resembled those of blasts in typical (hypergranular) M3 in 25 (73.5%) and variant (hypogranular) M3v in and 9 (26.5%) of the 34 patients. Both typical and atypical APL features can be found in this cohort. Characteristic bundles of Auer rods, existing as bundles or single Auer rods, randomly distributed within the cytoplasm were present in 14 cases (41.2%).
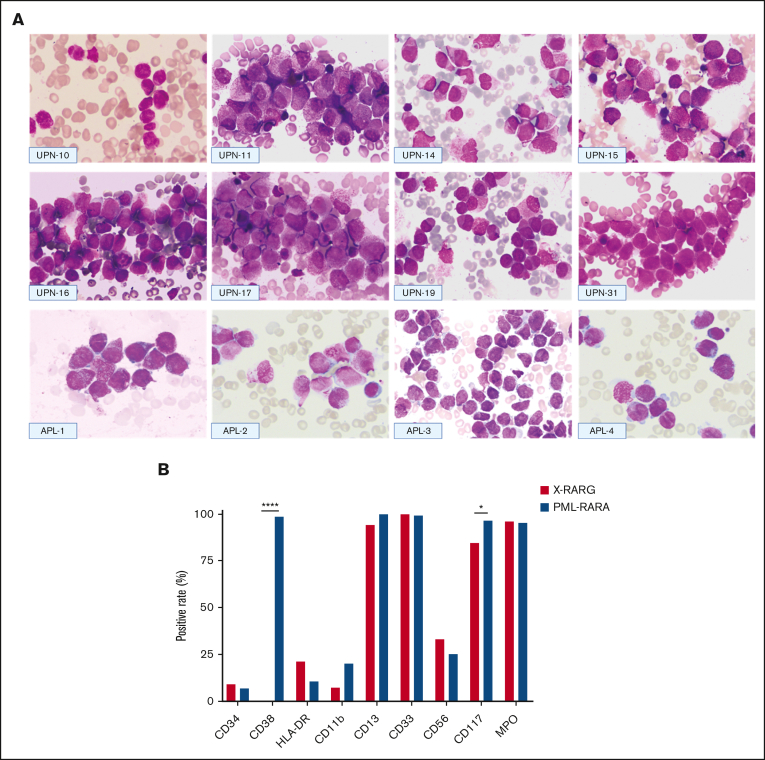
Figure 1 shows the morphological features of 8 patients included in this series (universal patient numbers [UPNs] 10, 11, 14-17,19, and 31) and 4 patients with PML::RARA serving for comparison (from the First Affiliated Hospital, Zhejiang University School of Medicine). The nuclei were often kidney-shaped, bilobed, or irregularly shaped and varied in size. Typical hypergranular promyelocytes displayed purple cytoplasm granules or stacked bundles of filaments, which completely obscured the nuclear-cytoplasmic margin (Figure 1; UPNs 10, 15-16, and 31). Microgranular promyelocytes featured cytoplasm filled with fine, dust-like granules (Figure 1; UPNs 14 and 19). Most nuclei of atypical promyelocytes were regular in shape, with slightly coarse granules (Figure 1; UPNs 11 and 17) and pseudo Chediak granules (Figure 1; UPN 17).
Figure 1.
Morphologic and immunophenotypic features of patients with AML with RARG rearrangement. (A) Morphologic features of 8 cases with RARG rearrangement (top and middle) and 4 cases with PML::RARA (bottom). Most blasts had hypergranular cytoplasm (UPNs 11, 15-17, and 31 and APL-1, -2, and -4) or hypogranular cytoplasm (UPNs 10, 14, and 19 and APL-3). Auer rods were also present in some cases (UPNs 10 and 14 and APL-1 and -2). Most blasts harbored an irregular round, oval, or bilobed nucleus that was strongly suggestive of French-American-British classification type M3. (B) Comparison of immunophenotyping features between patients with RARG rearrangement (X::RARG, n = 34) and those with PML::RARA (n = 221). The expression of surface markers was not different between the 2 groups, except for CD38 (0 vs 98.5%; P < .0001) and CD117 (84.4% vs 96.7%; P = .01).
Immunophenotype
It was understood that CD33+, CD13+, CD117+, CD34–, CD11b–, and HLA-DR– compose the immunophenotype of typical APL via flow cytometry. In the AML with RARG rearrangements, most leukemia blasts were CD33+ (33 of 33 patients [100%]), CD13+ (32 of 33 patients [97%]), and CD117+ (27 of 32 patients [84.4%]) but only a few were CD34+ (3 of 29 patients [10.3%]), CD11b+ (2 of 28 patients [7.1%]), and HLA-DR+ (7 of 32 [21.9%]); all were CD38– (0 of 26 patients). Leukemia blasts in 9 of 27 patients (33.3%) expressed CD56 (Figure 1).
The observed patterns were compared with those of the immunophenotypic profiles of 221 control cases with PML::RARA-positive APL, which we previously reported. There was no statistically significant difference in the expression of surface markers between AML with RARG rearrangement and APL cases, except for CD38 (0% vs 98.5%; P < .0001) and CD117 (84.4% vs 96.7%; P = .01; Figure 1).
Cytogenetics
Cytogenetic abnormalities were found in 19 of 33 (57.6%) patients. Translocation involving chromosome 12q13, in which the RARG gene is situated, occurred in 12 patients, including t(11;12)(p15;q13) in 9 of them, t(12;13)(q13;q13) in 1, t(12;15)(q13;q22) in 1, t(12;19)(q13;p13.1) in 1, and other karyotype abnormalities in 7 patients (Table 1; supplemental Table 1). Translocations of 12q13 were found in 0 of 14 patients with CPSF6::RARG, 9 of 11 with NUP98::RARG, 2 of 6 with HNRNPc::RARG, 1 of 1 with PML::RARG, 0 of 1 with NPM1::RARG, and 0 of 1 with HNRNPm::RARG.
Molecular biology
Fourteen patients (41%) had CPSF6::RARG, 11 (32%) had NUP98::RARG, 6 (18%) had HNRNPc::RARG, 1 had HNRNPm::RARG, 1 had PML::RARG, and 1 had NPM1::RARG. The detailed fusion sites in 22 patients with available RNA-seq data are shown in supplemental Figure 2. The fusion sites of RARG located in exon 4 and exon 1/2 accounted for 18 of 22 (81.8%) and 4 of 22 (18.2%) patients, respectively.
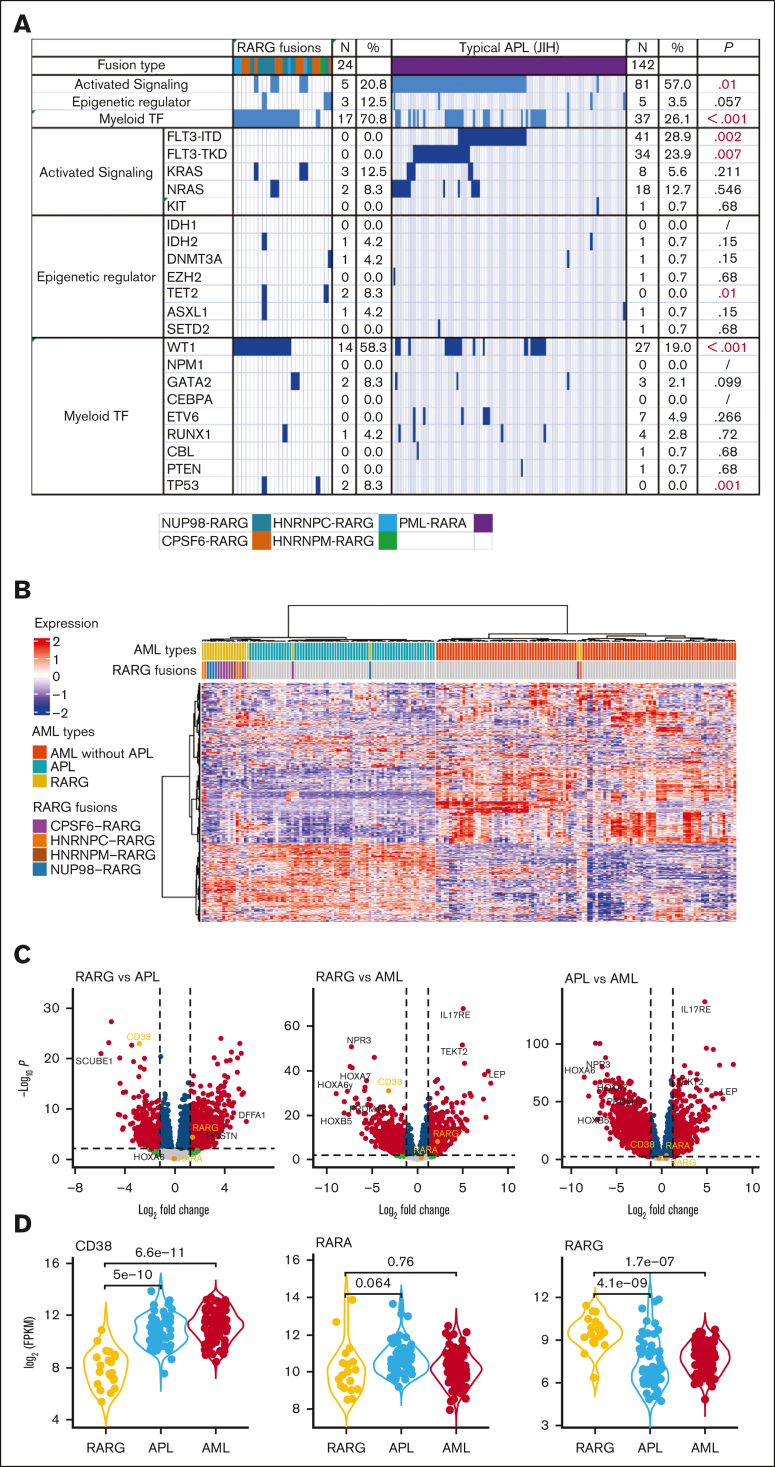
NGS results of 24 patients with RARG rearrangements showed that the concurrent mutations were WT1 (14 of 24 patients [58.3%[), KRAS (3 of 24 patients [12.5%]), NRAS (2 of 24 patients [8.3%]), TP53 (2 of 24 patients [8.3%]), TET2 (2 of 24 patients [8.3%]). No FLT3-ITD or -TKD mutations were found in patients with RARG rearrangements, which was a significantly lower incidence of these mutations compared with those in APL. Figure 2A shows the different frequencies of concurrent mutations between patients with AML with RARG rearrangements and patients with APL of Jiangsu Institute of Hematology. The frequency of WT1 and FLT3 mutations (including -ITD and -TKD) differed significantly between the 2 groups (58.3% vs 19.0% [P < .001]; 0% vs 28.9% [P = .002]; and 0% vs 23.9% [P = .007]; respectively).
Figure 2.
The genomic and transcriptomic landscape of AML with RARG rearrangement. (A) The frequency of concurrent mutation genes profile identified in 24 patients with RARG rearrangement (left) and in 142 patients APL with PML::RARA fusion (right, for comparison). (B) Unsupervised hierarchical clustering identified unique gene expression pattern of RARG rearrangement (RARG). Patients with AML with PML::RARA (APL) and others without RARG or RARA rearrangement are included as comparison. Columns indicate patients with AML and rows represent gene expression levels for each patient. Genes showing overexpression or underexpression in the heatmap are shown in red or blue, respectively. (C-D) Differentially expressed gene (DEG) comparisons between patients with AML with RARG rearrangement (RARG), PML::RARA (APL) and others without RARG or RARA rearrangement (AML). (C) Volcano plots showing DEGs among the 3 subtypes. Most significant DEGs or interested genes are labeled, and CD38, RARA, and RARG are highlighted. (D) The distribution of expression levels for representative genes shown by violin plots. The x-axis indicates the 3 subtypes, whereas the y-axis represents gene expression level (FPKM) in log scale. Each dot corresponds to 1 sample. The significance levels of difference were determined using the Wilcoxon rank-sum test. FPKM, Fragments per kilobase per million. Myeloid TF, myeloid transcription factor.
To explore whether fusion genes involving RARG constitute a unique entity of AML, unsupervised hierarchical clustering of the gene expression profile was performed to compare RARG-rearrangement samples (n = 22) with PML::RARA (n = 66) and non-APL AML (n = 113) samples from the First Affiliated Hospital Zhejiang University School of Medicine. Eighteen (81.8%) of the 22 RARG rearrangement samples clustered together, strongly suggesting a new subtype (Figure 2B). Figure 2C-D shows the differentially expressed genes among the 3 subtypes. In accordance with the immunophenotyping, CD38 expression was significantly downregulated in RARG rearrangement samples and upregulated in APL and other AML samples.
Treatment and outcomes
All 16 patients who received ATRA+ATO induction therapy (≥14 days) showed ATRA+ATO resistance, and they subsequently received AML-like induction therapy. The remaining 18 patients received ATRA/ATO (<14 days) together with AML-like induction therapy. AML-like induction therapy included cytarabine (100 mg/m2 per day for 7 days) and daunorubicin (45-60 mg/m2 for 3 days) or idarubicin (10-12 mg/m2 for 3 days) or homoharringtonine (2 mg/m2 per day for 7 days).
Of 33 patients evaluable for treatment response, 23 (69.7%) experienced CR, and approximately half of these patients achieved CR after 2 cycles of chemotherapy (supplemental Table 2). We could not evaluate CR for 1 patient who had transferred to another hospital because no detailed information was available. The early death rate within 45 days was 29.4% (10 of 34 patients), and the primary cause of death was hemorrhage (n = 6).
After achieving CR, 18 of 23 patients (78.3%) proceeded to consolidation therapy with AML-like regimens, including high-dose cytarabine (n = 18) or autologous hematopoietic stem cell transplant (HSCT; n = 1), and 11 of the 18 patients experienced relapse. Four patients underwent allogeneic HSCT (allo-HSCT) in their first CR, and no relapse or death occurred during follow-up times ranging from 3 to 12 months after HSCT. Five patients underwent allo-HSCT in their second CR: 4 experienced relapse at 44, 12, 5, and 2 months after HSCT, and 1 patient died from infection 1 month after HSCT.
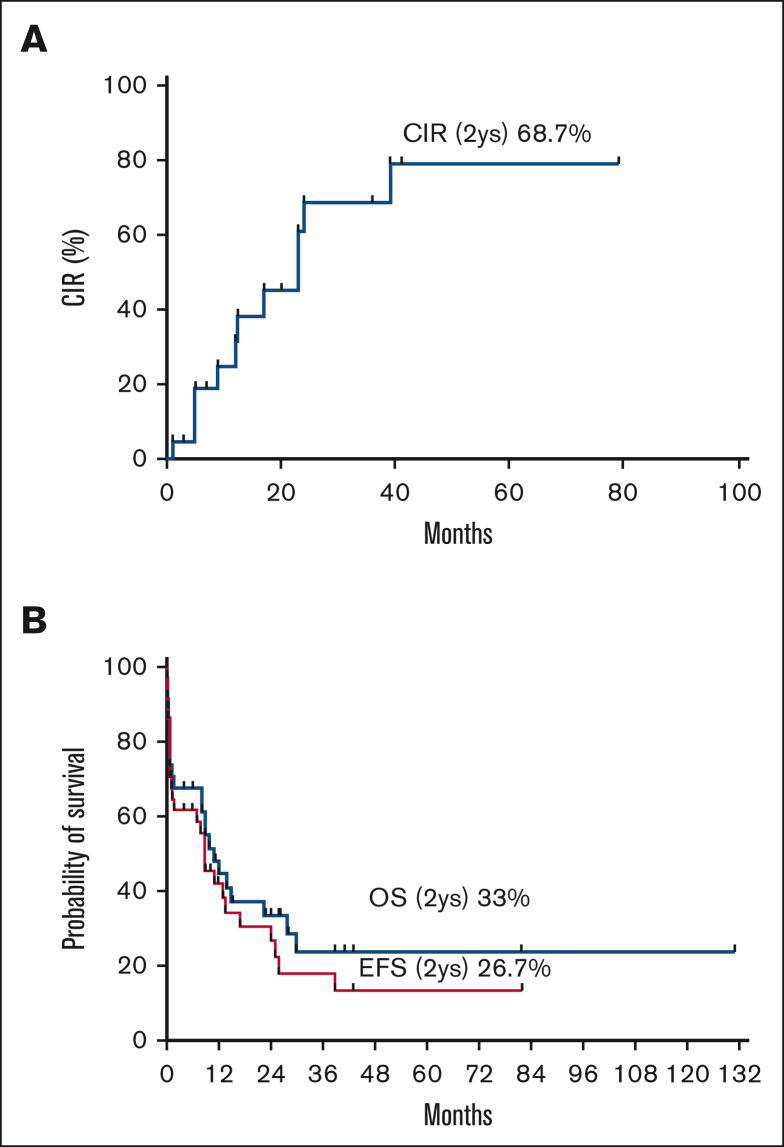
By December 2021, 10 of 33 patients were still alive, with a median follow-up time of 14 months (range 4-131 months). The estimated 2-year cumulative incidence of relapse, event-free survival, and overall survival were 68.7%, 26.7%, and 33.5%, respectively (Figure 3).
Figure 3.
The CIR, EFS, and OS of patients with RARG rearrangements (n = 34). CIR, cumulative incidence of relapse; EFS, event-free survival; OS, overall survival.
Discussion
In this study, we provided multidimensional evidence that RARG rearrangement is a nonrandom, recurrent translocation characterizing a novel subtype of AML that is ATRA/ATO–resistant and carries a dismal prognosis. The prevalence of RARG rearrangement in patients with AML or APL is unknown at present.
Patients with RARG rearrangement harbor unique characteristics, which include a leukemic blast morphology mimicking APL and distinct immunophenotypic, molecular, and gene expression profiles. Given the unfavorable prognosis, we suggest that RARG rearrangement screening should be included in the panel of molecular diagnostics in AML, especially for patients with APL morphology and those with no RARA rearrangement.
Although AML with RARG rearrangement presented with strong clinical (bleeding and coagulant), morphologic, and immunophenotypic (CD13+/CD33+/CD117+/MPO+/CD34–/HLA-DR–/CD11b–) similarities to APL, there were distinct differences. A total of 38.2% of patients with RARG rearrangements presented with WBC >10 × 109/L compared with ∼20% to 30% of those with PML-RARA–positive APL. Moreover, lack of CD38 expression status served as an excellent marker to differentiate RARG rearrangement AML (0% positive) from PML::RARA-positive APL (88% positive). CD38 expression status was also confirmed by comparison of the gene expression profiles from the RNA-seq data shown in Figure 2. The RARE motif in intron 1 of the CD38 gene functions as an enhancer to regulate CD38 expression.32 The involvement of RARG-RXR-RARE axis in CD38 downregulation of RARG fusions besidesPML::RARA needs to be explored in the future.
From the genetic perspective, NUP98::RARG fusion is easily recognized as t(11;12)(p15;q13). Because the CPSF6 and RARG loci are on chromosomes 12q15 and 12q13, respectively, this cryptic translocation is rarely identified via regular karyotyping analysis.19 Similarly, no fusions of HNRNPc, HNRNPm, or NPM1 with RARG could be detected via karyotyping analysis, which renders it difficult to provide cues for fusion genes.
The most common fusion partners of RARG in our study were CPSF6, NUP98, and HNRNPc, which accounted for 94% of RARG rearrangements. The RARG breakpoint occurs in exon 1, exon 2, or exon 4, and the key functional motifs, including DBD and LBD, are retained in the fusion protein.18,19 Our analysis also showed that concurrent mutations among RARG rearrangement AML cases showed a high frequency of WT1 mutations (58.3%) compared with those among APL cases but no FLT3 mutations. The description of these differences between the 2 subtypes of AML should be investigated in the future.
Our comparison of RNA-seq data between RARG-rearrangement AML and APL or non-APL AML revealed that RARG rearrangement clusters very similarly to APL, likely because of many shared transcripts associated with a similar promyelocyte maturation arrest. However, 18 of 22 samples (81.8%) with RARG rearrangement clustered together, strongly suggesting a new subtype distinct from PML::RARA APL. Our analysis of differentially expressed genes among the 3 subtypes showed that CD38 expression was significantly downregulated in RARG fusion samples and upregulated in APL and other AML subtypes, which is consistent with the results of immunophenotyping. This important finding will enable physicians to differentiate RARG rearrangement AML from APL because assessing CD38 expression via flow cytometry is fast and easy; the result can be available within a few hours.
Most importantly, we found that all 16 patients with RARG rearrangement who received ATRA-ATO lasting more than 2 weeks exhibited definitive ATRA and ATO resistance. ATRA resistance in primary leukemia cells from patients with NUP98::RARG has also been demonstrated in vitro,33 which is in contrast with the observation that NUP98::RARG-transformed murine primary hematopoietic stem/progenitor cell fusion was extremely sensitive to ATRA treatment.34 The underlying mechanism of ATRA resistance in AML with RARG rearrangement needs to be explored further.35
Because most cases of this novel entity of AML cannot be diagnosed in a timely manner and because of the shared morphological features with APL, extended exposure to ineffective ATRA treatment may render patients to be at high risk of early death. Moreover, overt coagulopathy could be further exacerbated by intensive chemotherapy. The early death rate of AML with RARG rearrangement was 29% in this study, which is higher than that of APL36; thus, recognizing this attribute may help avoid early deaths. AML with RARG rearrangement should be treated with intensive combinational chemotherapy and allo-HSCT per the guidelines for intermediate or adverse-risk AML. Furthermore, novel targeted treatments toned to be identified for this entity in a clinical trial.
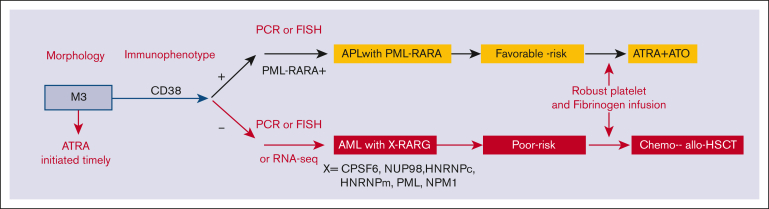
Outcomes in this AML entity can improve with an approach that include prompt diagnosis, effective therapy, and aggressive support to avoid catastrophic coagulopathy. The consistent expression of CD33, which is highly active in PML::RARA APL, prompts us to speculate that the integration of gemtuzumab ozogamicin might be of clinical utility. Given the poor outcomes observed, consideration of allo-HSCT in CR1 appears rational. To avoid delayed diagnosis, multiplex quantitative polymerase chain reaction including primers covering the 5 RARG fusions or fluorescence in situ hybridization analysis with RARG-specific fluorescence in situ hybridization probes from BAC clones is highly recommended. Although RNA-seq was mostly used in our study, it is expensive, time-consuming, and not readily used in routine clinical practice. The survival outcomes of our series indicate that the overall behavior of this entity is similar to poor-risk karyotype AML rather than to favorable-risk genetic abnormalities, such as PML::RARA. We therefore proposed a flowchart for timely evaluation and diagnosis (Figure 4).
Figure 4.
The proposed flowchart for the diagnosis and treatment of patients with AML with RARG rearrangement. FISH, fluorescence in situ hybridization; PCR, polymerase chain reaction.
Our study is limited by its small sample size, heterogeneous molecular subtypes, and different treatment regimens. Nevertheless, to our knowledge, our study represents the largest reported clinical series of AML with RARG rearrangement in the world. Indeed, despite the phenotypic and morphologic similarities with PML::RARA-positive APL, these RARG rearrangement variants show important biological diversity, which accounts for their resistance to molecularly targeted therapies known to display high antileukemic efficacy in APL. We hypothesize that this diversity might originate from differences in the defining gene rearrangement event and/or a different spectrum of cooperative mutations present in these variants. Comprehensive whole-gene sequencing and single-cell sequencing to clarify the molecular and genetic landscape are warranted. Moreover, we did not compare the features of RARG rearrangement with those of RARA variants or RARB rearrangement.4,37
In conclusion, we have presented multidimensional evidence to characterize a novel subset of AML with RARG rearrangement based on the largest case numbers and centers involved to date. This attribute is distinct from that of APL, with unique clinical features, morphology, immunotyping, cytogenetics, genomic and transcriptomic landscape, treatment response, and patient outcomes. CD38 expression detected via flow cytometry can be rapidly applicable and act as a simple diagnostic marker for excluding RARG rearrangement. Promptly initiating chemotherapy and avoiding early death and subsequent relapse is an urgent unmet need. Prospective clinical trials investigating combinatorial approaches of molecular targeted therapies with current chemotherapies are warranted for the treatment of this intractable disease.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Acknowledgments
The authors thank Xiaoxue Han (Kingmed Diagnostics), Zhuo Wang (Shanghai Children's Medical Center), Xiaoyan Han and Xiang Zhang (The First Affiliated Hospital, Zhejiang University School of Medicine), Changfeng Liao (The First Affiliated Hospital of Gannan Medical University), Shumin Xiong (Rui Jin Hospital), Dandan Liu and Mingqing Zhu (Jiangsu Institute of Hematology), Chang-Feng Liao (Department of Hematology, The First Affiliated Hospital of Gannan Medical University), Mei Jiang (Department of Clinical Laboratory, The First Affiliated Hospital of Nanchang University), Jun Peng (Qilu Hospital of Shandong University), Ran Gao (First Hospital of China Medical University), and Xin Liu (Tianjin Institute of Hematology) for analytical and technical support in this study.
This work was supported by grants from the National Key R&D Program of China (2019YFA0111000), the National Natural Science Foundation of China (81970133, 81700168, 82170158, 82160692, and 82160037), the Leading Innovative and Entrepreneur Team Introduction Program of Zhejiang (2020R01006 and 2019R01001) and Elite plan of Zhejiang (2022C03005), the National Institute for Health Research Royal Marsden/Institute of Cancer Research Biomedical Research Centre.
Authorship
Contribution: H.-H.Z., S.-N.C., and J.H. designed the research and performed clinical analysis; H.-H.Z., Y.-Z.Q., Z.-L.Z., and S.-N.C. wrote the manuscript; J.-P.H., Y.-J.L., and J.-Y.H. performed bioinformatics analysis; L.-J.W. and S.-N.C. performed molecular biological analysis; J.-H.L. performed morphology analysis; and M.J.Y., C.Z., E.S., H.L., H.-J.Y., H.-S.Z., H.-X.L., R.X., J.L., J.-H.L., J.-P.H., J.J., L.Y., J.-Y.Z., L.-P.L., L.-P.Z., R.-B.H., S.-H.S., S.-J.G., W.W., X.-J.Y., X.-Y.Z., X.D., X.-X.C., Y.-F.Y., Y.W., Y.-C.M., Y.L., Z.C., Z.S., D.C.T., S.M., E.D.B., H.-Y.W., J.S.W., C.C.Y., G.B., M.A.S., and H.M.K. provided clinical information about the patients with RARG rearrangement.
Footnotes
∗H.-H.Z, Y.-Z.Q., Z.-L.Z., and Y.-J.L. contributed equally to this study.
The raw RNA sequence data reported in this article have been deposited in the Genome Sequence Archive in BIG Data Center, Beijing Institute of Genomics, Chinese Academy of Sciences (accession number HRA003955).
Data are available on request from the corresponding authors, Su-Ning Chen (chensuning@suda.edu.cn), Jiong Hu (hj10709@rjh.com.cn), and Jin-Yan Huang (huangjinyan@zju.edu.cn).
For original data and novel insights on the topic, please contact author Hong-Hu Zhu (zhuhhdoc@163.com).
The full-text version of this article contains a data supplement. This cohort is a small but important group of patients. The authors suspect that other RARG rearrangement patients exist and welcome any reader insights, with the goal of identifying novel targeted treatments for this entity in a clinical trial, uncovering the physiological basis of this new disease.
Contributor Information
Jin-Yan Huang, Email: huangjinyan@zju.edu.cn.
Jiong Hu, Email: hj10709@rjh.com.cn.
Su-Ning Chen, Email: chensuning@suda.edu.cn.
Supplementary Material
References
- 1.Swerdlow SH, Campo E, Pileri SA, et al. The 2016 revision of the World Health Organization classification of lymphoid neoplasms. Blood. 2016;127(20):2375–2390. doi: 10.1182/blood-2016-01-643569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Duncavage EJ, Schroeder MC, O'Laughlin M, et al. Genome sequencing as an alternative to cytogenetic analysis in myeloid cancers. N Engl J Med. 2021;384(10):924–935. doi: 10.1056/NEJMoa2024534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhao J, Liang JW, Xue HL, et al. The genetics and clinical characteristics of children morphologically diagnosed as acute promyelocytic leukemia. Leukemia. 2019;33(6):1387–1399. doi: 10.1038/s41375-018-0338-z. [DOI] [PubMed] [Google Scholar]
- 4.Osumi T, Tsujimoto SI, Tamura M, et al. Recurrent RARB translocations in acute promyelocytic leukemia lacking RARA translocation. Cancer Res. 2018;78(16):4452–4458. doi: 10.1158/0008-5472.CAN-18-0840. [DOI] [PubMed] [Google Scholar]
- 5.Such E, Cervera J, Valencia A, et al. A novel NUP98/RARG gene fusion in acute myeloid leukemia resembling acute promyelocytic leukemia. Blood. 2011;117(1):242–245. doi: 10.1182/blood-2010-06-291658. [DOI] [PubMed] [Google Scholar]
- 6.Ha JS, Do YR, Ki CS, et al. Identification of a novel PML-RARG fusion in acute promyelocytic leukemia. Leukemia. 2017;31(9):1992–1995. doi: 10.1038/leu.2017.167. [DOI] [PubMed] [Google Scholar]
- 7.Liu T, Wen L, Yuan H, et al. Identification of novel recurrent CPSF6-RARG fusions in acute myeloid leukemia resembling acute promyelocytic leukemia. Blood. 2018;131(16):1870–1873. doi: 10.1182/blood-2017-11-818716. [DOI] [PubMed] [Google Scholar]
- 8.Qin YZ, Huang XJ, Zhu HH. Identification of a novel CPSF6-RARG fusion transcript in acute myeloid leukemia resembling acute promyelocytic leukemia. Leukemia. 2018;32(10):2285–2287. doi: 10.1038/s41375-018-0095-z. [DOI] [PubMed] [Google Scholar]
- 9.Miller CA, Tricarico C, Skidmore ZL, et al. A case of acute myeloid leukemia with promyelocytic features characterized by expression of a novel RARG-CPSF6 fusion. Blood Adv. 2018;2(11):1295–1299. doi: 10.1182/bloodadvances.2017014183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Luo H, Zhang S, Li K, et al. A novel entity of acute myeloid leukaemia with recurrent RARG-rearrangement resembling acute promyelocytic leukaemia. Leuk Res. 2019;77:14–16. doi: 10.1016/j.leukres.2018.12.009. [DOI] [PubMed] [Google Scholar]
- 11.Chen X, Wang F, Zhang Y, et al. A novel NPM1-RARG-NPM1 chimeric fusion in acute myeloid leukaemia resembling acute promyelocytic leukaemia but resistant to all-trans retinoic acid and arsenic trioxide. Br J Cancer. 2019;120(11):1023–1025. doi: 10.1038/s41416-019-0456-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang X, Li F, Wang J, et al. RARgamma-rearrangements resemble acute promyelocytic leukemia and benefit from 3 + 7 regimen. Leuk Lymphoma. 2019;60(7):1831–1834. doi: 10.1080/10428194.2018.1553302. [DOI] [PubMed] [Google Scholar]
- 13.Zhang Z, Jiang M, Borthakur G, et al. Acute myeloid leukemia with a novel CPSF6-RARG variant is sensitive to homoharringtonine and cytarabine chemotherapy. Am J Hematol. 2020;95(2):E48–E51. doi: 10.1002/ajh.25689. [DOI] [PubMed] [Google Scholar]
- 14.Su Z, Liu X, Xu Y, et al. Novel reciprocal fusion genes involving HNRNPC and RARG in acute promyelocytic leukemia lacking RARA rearrangement. Haematologica. 2020;105(7):e376–e378. doi: 10.3324/haematol.2019.244715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wei W, Liu Q, Song F, et al. Alkaloid-based regimen is beneficial for acute myeloid leukemia resembling acute promyelocytic leukemia with NUP98/RARG fusion and RUNX1 mutation: A case report. Medicine (Baltimore) 2020;99(40):e22488. doi: 10.1097/MD.0000000000022488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tao S, Song L, Deng Y, et al. Acute myeloid leukemia with NUP98-RARG gene fusion similar to acute promyelocytic leukemia: case report and literature review. OncoTargets Ther. 2020;13:10559–10566. doi: 10.2147/OTT.S273172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Han X, Jin C, Zheng G, et al. Acute myeloid leukemia with CPSF6-RARG fusion resembling acute promyelocytic leukemia with extramedullary infiltration. Ther Adv Hematol. 2021;12:1–8. doi: 10.1177/2040620720976984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang X, Sun J, Yu W, Jin J. Current views on the genetic landscape and management of variant acute promyelocytic leukemia. Biomark Res. 2021;9(1):33. doi: 10.1186/s40364-021-00284-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Geoffroy MC, de The H. Classic and variants apls, as viewed from a therapy response. Cancers. 2020;12(4):967. doi: 10.3390/cancers12040967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Conserva MR, Redavid I, Anelli L, Zagaria A, Specchia G, Albano F. RARG gene dysregulation in acute myeloid leukemia. Front Mol Biosci. 2019;6:114–123. doi: 10.3389/fmolb.2019.00114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Coccaro N, Zagaria A, Orsini P, et al. RARA and RARG gene downregulation associated with EZH2 mutation in acute promyelocytic-like morphology leukemia. Hum Pathol. 2018;80:82–86. doi: 10.1016/j.humpath.2018.02.023. [DOI] [PubMed] [Google Scholar]
- 22.Wang YZ, Qin YZ, Jiang B, et al. Relationship of immunophenotypic features with minimal residual disease detection and gene types in 221 cases of acute promyelocytic leukemia. Zhongguo Shi Yan Xue Ye Xue Za Zhi. 2009;17(2):271–276. [PubMed] [Google Scholar]
- 23.Brothman AR, Persons DL, Shaffer LG. Nomenclature evolution: changes in the ISCN from the 2005 to the 2009 edition. Cytogenet Genome Res. 2009;127(1):1–4. doi: 10.1159/000279442. [DOI] [PubMed] [Google Scholar]
- 24.DePristo MA, Banks E, Poplin R, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43(5):491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Uhrig S, Ellermann J, Walther T, et al. Accurate and efficient detection of gene fusions from RNA sequencing data. Genome Res. 2021;31(3):448–460. doi: 10.1101/gr.257246.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Anders S, Pyl PT, Huber W. HTSeq--a python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31(2):166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Frankish A, Diekhans M, Jungreis I, et al. 2021. Nucleic Acids Res. 2021;49(D1):D916–D923. doi: 10.1093/nar/gkaa1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gene Ontology Consortium The Gene Ontology resource: enriching a gold mine. Nucleic Acids Res. 2021;49(D1):D325–D334. doi: 10.1093/nar/gkaa1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kanehisa M, Furumichi M, Sato Y, Ishiguro-Watanabe M, Tanabe M. KEGG: integrating viruses and cellular organisms. Nucleic Acids Res. 2021;49(D1):D545–D551. doi: 10.1093/nar/gkaa970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gu Z, Eils R, Schlesner M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics. 2016;32(18):2847–2849. doi: 10.1093/bioinformatics/btw313. [DOI] [PubMed] [Google Scholar]
- 31.Dohner H, Estey E, Grimwade D, et al. Diagnosis and management of AML in adults: 2017 ELN recommendations from an international expert panel. Blood. 2017;129(4):424–447. doi: 10.1182/blood-2016-08-733196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Horenstein AL, Faini AC, Morandi F, et al. The circular life of human cd38: from basic science to clinics and back. Molecules. 2020;25(20):4844. doi: 10.3390/molecules25204844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Such E, Cordon L, Sempere A, et al. In vitro all-trans retinoic acid sensitivity of acute myeloid leukemia blasts with NUP98/RARG fusion gene. Ann Hematol. 2014;93(11):1931–1933. doi: 10.1007/s00277-014-2073-5. [DOI] [PubMed] [Google Scholar]
- 34.Qiu JJ, Zeisig BB, Li S, et al. Critical role of retinoid/rexinoid signaling in mediating transformation and therapeutic response of NUP98-RARG leukemia. Leukemia. 2015;29(5):1153–1162. doi: 10.1038/leu.2014.334. [DOI] [PubMed] [Google Scholar]
- 35.Brown G, Petrie K. The RARgamma oncogene: an Achilles heel for some cancers. Int J Mol Sci. 2021;22(7):3632. doi: 10.3390/ijms22073632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhu HH, Ma YF, Yu K, et al. APL Cooperative Group of Zhejiang Province Early death and survival of patients with acute promyelocytic leukemia in atra plus arsenic era: a population-based study. Front Oncol. 2021;11 doi: 10.3389/fonc.2021.762653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cicconi L, Testi AM, Montesinos P, et al. Characteristics and outcome of acute myeloid leukemia with uncommon retinoic acid receptor-alpha (RARA) fusion variants. Blood Cancer J. 2021;11(10):167. doi: 10.1038/s41408-021-00561-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.