Due mainly to a late stage at diagnosis and lack of effective treatment modalities, pancreatic cancer (PC) is a highly lethal malignancy with an overall 5-year survival rate of only 12%.1 Separating patients into those diagnosed with late-stage disease with distant metastases and those diagnosed with localized tumors reveals a striking difference of 3% vs. 44% 5-year survival rates, respectively. Increasing the fraction of patients detected at an early stage, when PC is most likely to be curable, could therefore be of a major benefit. While imaging-based early-detection screening methods (e.g., endoscopic ultrasound and magnetic resonance imaging) are increasingly recommended for carriers of germline pathogenic variants in cancer susceptibility genes conferring a high lifetime risk of pancreatic cancer, such screening is not practical in the general population due to the low incidence of PC and a lack of a proven benefit.
Recently, Placido et al. designed an artificial intelligence (AI) tool to predict PC up to 3 years before diagnosis based on electronic healthcare records.2 Prior to using deep learning algorithms, advanced risk assessment model building requires large and comprehensive datasets for training, development, and testing. Without enough statistical power, such prediction models are not likely to succeed. Placido et al. used disease trajectories for 9 million patients from two independent healthcare systems (the Danish National Patient Registry [DNPR] and the United States Veterans Affairs [US-VA]). The population-wide DNPR dataset is one of the largest and most comprehensive longitudinal real-world clinical datasets available containing 229 million hospital diagnoses for 8.6 million patients diagnosed in Denmark from 1977 to 2018. This resource provides unique opportunities for clinical risk assessment model building. For validation, Placido et al. used longitudinal clinical records from the US-VA dataset, which includes 16 million patients diagnosed from 1999 to 2020. After quality control, 6 and 3 million patients were selected from the DNPR and US-VA datasets for AI model training and testing, including 23,985 and 3,864 PC cases, respectively. It is worth noting that (1) the healthcare records in the DNPR dataset had longer (median 23 years in DNPR vs. 12 years in US-VA) but less dense (median 22 records patient in DNPR vs. 188 records per patient in US-VA) disease trajectories, (2) the majority of patients in the DNPR dataset were of Northern European background compared to a more diverse population in the US-VA dataset, and (3) gender distribution in the DNPR dataset was balanced (49.7% male vs. 50.3% female), while a majority of patients in the US-VA were male (85.7% male vs. 14.3% female). The heterogeneity between these two datasets highlights potential challenges for cross-applications of machine learning (ML) models trained in different healthcare systems and populations but also provided an opportunity to compare AI models in two real-life healthcare systems. Both the DNPR and US-VA datasets were randomly split into training (80%), development (10%), and testing (10%) subsets for the downstream ML and test process, respectively.
Development of deep learning methods is crucial to analyzing such large-scale but also potentially noisy clinical datasets. Placido et al. developed a sequential AI model to train longitudinal disease trajectories (including 2,000 level 3 International Classification of Diseases [ICD] codes) with time stamps to predict time-dependent (3, 6, 12, 36, or 60 months after the time of risk assessment) risk scores. Among the four ML models (bag-of-words, gated recurrent unit [GRU], multilayer perceptron [MLP], and Transformer) tested in this study, the self-attention-based Transformer model3 performed best with an area under the receiver operating characteristic (AUROC) curve of 0.88 within 3 years prior to diagnoses of PC in the DNPR dataset. Moreover, the GRU, MLP, and Transformer models outperformed a previously published non-AI-based prediction model that integrated polygenic risk scores (PRS) and clinical risk factors from 1,042 pancreatic ductal adenocarcinoma (PDAC) cases and 10,420 matched cancer-free controls drawn from the UK Biobank (AUROC of 0.83).4
An out-of-box validation is vital for any risk prediction model. Placido et al. applied the best ML model trained in the DNPR dataset to disease records from patients in the independent US-VA dataset. Compared with the DNPR dataset, prediction performance in US-VA declined from an AUROC of 0.88 to 0.71 for PC occurrence within 3 years after assessment. Due to differences between the two healthcare systems, Placido et al. also retrained and revaluated the AI model in the US-VA dataset with an improved performance (AUROC of 0.78) for PC occurring within 3 years. Validation in the US-VA dataset suggested that model generalizability can be tricky when using data from different healthcare systems, and retraining AI models from scratch for each clinical dataset is likely to improve risk prediction performance. This, however, may not always be possible due to differences in healthcare systems across regions and countries and the fact that health records may be distributed across many healthcare systems (e.g., different doctors’ offices and hospital systems). Furthermore, to assess the performance of their models in a setting more similar to a realistic PC surveillance program, Placido et al. excluded diagnoses within the 3 months prior to PC diagnoses for model training. This yielded a relative risk (RR) = 58.7 for DNPR and RR = 33.1 for US-VA for the 0.1% highest-risk individuals over the age of 50 during the 12-month prediction period.
Previous known clinical and lifestyle risk factors for PC include type 2 diabetes (T2D), pancreatitis, smoking, and obesity, as well as both common and rare germline genetic variants.5 While lifestyle and genetic risk factors were not assessed in the current study, disease codes representing 23 known PC risk factors (a subset of the 2,000 level 3 ICD codes) were chosen to test a “known risk factor” model. This reduced prediction accuracy (from AUROC of 0.88 to 0.84), indicating that as of yet unknown risk factors may improve PC risk prediction accuracy. Moreover, by a feature extraction approach, most known risk factors made a significant contribution to the ML prediction of PC occurrence.
While a deep ML algorithm typically is a black box for users, it is important to know which features contribute to the best-performing ML prediction models. The top contributing features were therefore extracted from clinical records with time to PC diagnosis within 0–6, 6–12, 12–24 and 24–36 months after assessment for all patients who developed PC. This indicated known (e.g., T2D, acute pancreatitis) but also potentially novel PC risk indicators (e.g., gallstones, gastritis, reflux disease), possibly pointing to mechanisms related to inflammation of the pancreas, in line with pancreatitis. However, more work is needed to establish if these possibly new risk indicators directly influence PC risk or not. Not surprisingly, no single diagnosis had sufficient power to predict PC.
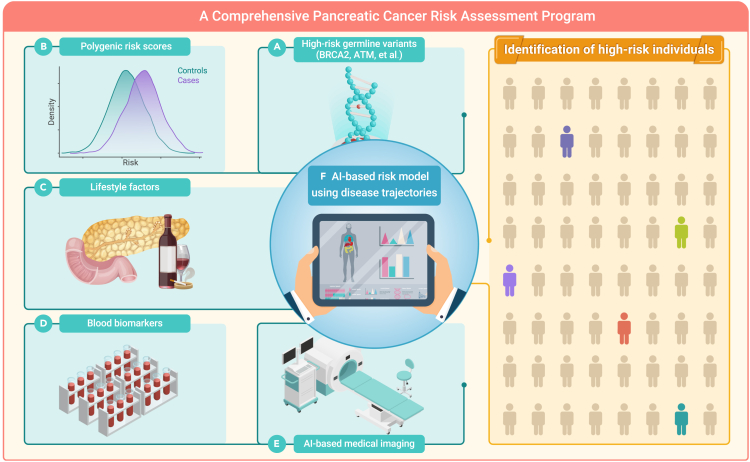
Although this study provides the best prediction model for PC risk assessment available to date in the general population, sensitivity in DNPR was only 8% and 4% for PC occurring within 1 and 3 years after assessment, respectively. Therefore, additional model improvements are important. This could be achieved by including (1) rare high-risk germline variants, (2) PRS (improvements in current PRS models are expected with larger ongoing genome-wide association studies of PDAC by the Pancreatic Cancer Cohort Consortium, the Pancreatic Cancer Case-Control Consortium, and other studies), (3) lifestyle factors, (4) blood-derived biomarkers, and (5) imaging-based indicators, as well as (6) separately analyzing patients diagnosed with the more lethal PDAC vs. the less lethal pancreatic neuroendocrine tumor subtype, when training deep ML models (Figure 1). This may significantly improve risk prediction accuracy and identify individuals at a greatly increased risk of developing PC or those already with undiagnosed early-stage PC. In addition to evaluating the performance of different AI models using AUROC and RR as done here by Placido et al., comparing the number of patients identified at a high-risk of PC using an absolute risk framework might also help.
Figure 1.
Construction of a comprehensive pancreatic cancer risk assessment program
Integrating rare high-risk germline variants, PRS, lifestyle factors, blood-derived biomarkers, AI-based medical imaging indicators, and AI-based modeling using disease trajectories may significantly improve future PC risk prediction accuracy in the general population.
In summary, the study by Placido et al. shows that cutting-edge AI methods may indeed help improve early PC risk prediction and identify individuals from the general population that would benefit from surveillance strategies now only used in carriers of germline pathogenic variants. Further AI modeling based on the above recommendations has the potential to predict PC with an even higher accuracy. An important next step before such models can be more widely applied is to assess how surveillance strategies designed for such groups of high-risk individuals facilitate early PC detection in clinical trials and improve survival while minimizing overdiagnoses and potential harm.
Acknowledgments
The authors are supported by the Intramural Research Program (IRP) of the Division of Cancer Epidemiology and Genetics (DCEG), National Cancer Institute (NCI), US National Institutes of Health (NIH). The content of this publication does not necessarily reflect the views or policies of the US Department of Health and Human Services, nor does the mention of trade names, commercial products, or organizations imply endorsement by the US Government.
Declaration of interests
The authors declare no competing interests.
Published Online: June 8, 2023
Contributor Information
Jun Zhong, Email: jun.zhong@nih.gov.
Jianxin Shi, Email: jianxin.shi@nih.gov.
Laufey T. Amundadottir, Email: amundadottirl@mail.nih.gov.
References
- 1.Siegel R.L., Miller K.D., Wagle N.S., et al. Cancer statistics, 2023. CA A Cancer J. Clin. 2023;73:17–48. doi: 10.3322/caac.21763. [DOI] [PubMed] [Google Scholar]
- 2.Placido D., Yuan B., Hjaltelin J.X., et al. A deep learning algorithm to predict risk of pancreatic cancer from disease trajectories. Nat. Med. 2023;29:1113–1122. doi: 10.1038/s41591-023-02332-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vaswani A., Shazeer N., Parmar N., et al. Attention is all you need. Adv. Neural Inf. Process. Syst. 2017:5998–6008. [Google Scholar]
- 4.Sharma S., Tapper W.J., Collins A., et al. Predicting pancreatic cancer in the UK Biobank Cohort using polygenic risk scores and diabetes mellitus. Gastroenterology. 2022;162:1665–1674.e2. doi: 10.1053/j.gastro.2022.01.016. [DOI] [PubMed] [Google Scholar]
- 5.Stolzenberg-Solomon R.Z., Amundadottir L.T. Epidemiology and inherited Predisposition for Sporadic pancreatic adenocarcinoma. Hematol Oncol Clin North Am. 2015;29:619–640. doi: 10.1016/j.hoc.2015.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]