Summary
Chromatin accessibility is critical for cell identity. Conventional ATAC-seq can examine chromatin accessibility on freshly prepared muscle stem cells or satellite cells (SCs); however, isolating SCs in mice remains challenging. Here, we present a protocol to preserve the in vivo chromatin profile of SCs by applying paraformaldehyde (PFA) perfusion throughout the mouse before SC isolation. We describe steps for PFA perfusion and FACS sorting of SCs. We then detail library preparation for ATAC-seq.
For complete details on the use and execution of this protocol, please refer to Dong et al.1
Subject areas: Cell Biology, Cell isolation, Molecular Biology, Stem Cells
Graphical abstract

Highlights
-
•
Optimized PFA perfusion protocol for improved performance and consistency
-
•
Detailed procedure for ATAC-seq library generation from PFA-perfused muscle stem cells
-
•
Evaluation of the quality of the PFA-perfused cells using FACS plots
Publisher’s note: Undertaking any experimental protocol requires adherence to local institutional guidelines for laboratory safety and ethics.
Chromatin accessibility is critical for cell identity. Conventional ATAC-seq can examine chromatin accessibility on freshly prepared muscle stem cells or satellite cells (SCs); however, isolating SCs in mice remains challenging. Here, we present a protocol to preserve the in vivo chromatin profile of SCs by applying paraformaldehyde (PFA) perfusion throughout the mouse before SC isolation. We describe steps for PFA perfusion and FACS sorting of SCs. We then detail library preparation for ATAC-seq.
Before you begin
The Assay for Transposase-Accessible Chromatin with high-throughput sequencing (ATAC-seq) enables the global examination of chromatin accessibility. Most current protocols for ATAC-seq describe the library generation on freshly prepared/isolated cells2,3 However, the SC isolation process damages the local environment where the SCs reside, inducing activation from the quiescence state and changes in the global chromatin accessibility, transcriptomes, and proteomes.1,4,5 To obtain the in vivo profile of SCs, we applied Paraformaldehyde (PFA)-perfusion to preserve the signature of the quiescent SCs and further optimized the ATAC-seq protocol for better library generation.
We have previously described the protocol for isolating the In situ fixed quiescent muscle stem cells.6 We have further optimized the protocol using a perfusion pump for easier adaptation and have improved the consistency of the fixation outcome.
The protocol below describes the procedure of PFA perfusion and the downstream ATAC-seq library preparation of SCs from the transgenic mouse line Tg: Pax7nGFP. Other reporter mouse lines which enable specific labeling of SCs (e.g., Pax7CreER::Rosa26YFP) are also applicable.
Institutional permissions
The experiments were approved by the Hong Kong University of Science and Technology Animal Ethics Committee.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Chemicals, peptides, and recombinant proteins | ||
| 2,2,2-Tribromoethanol, 99% (Avertin) | Acros Organics | Catalog #: 421430100 |
| Phosphate buffered saline | Sigma-Aldrich | Catalog #: P3813 |
| 32% Paraformaldehyde aqueous solution, EM Grade | Electron Microscopy Sciences | Catalog #: 15714 |
| Glycine, for electrophoresis, 99% | Sigma-Aldrich | Catalog #: G8898 |
| Nutrient mixture F-10 Ham | Sigma-Aldrich | Catalog #: N6635 |
| Horse serum | Invitrogen | Catalog #: 16050114 |
| Penicillin Streptomycin (10,000 U/mL) | Thermo Fisher Scientific | Catalog #: 15140122 |
| Collagenase, type 2 | Worthington Biochemical | Catalog #: LS004177 |
| Dispase II, powder | Thermo Fisher Scientific | Catalog #: 17105041 |
| Sodium chloride, puriss. P.a., ACS reagent, reag. ISO, reag. Ph. Eur., 99.8% | Sigma-Aldrich | Catalog #: 31434 |
| IGEPAL® CA-630, viscous liquid | Sigma-Aldrich | Catalog #: I3021-100ML |
| Tris(2-carboxyethyl)phosphine hydrochloride | Sigma-Aldrich | Catalog #: C4706-10G |
| Hydrochloric acid 37%, AnalaR NORMAPUR Reag. Ph. Eur. analytical reagent | VWR Chemicals | Catalog #: 20252.290 |
| UltraPure DNase/RNase-Free Distilled Water | Thermo Fisher Scientific | Catalog #: 10977015 |
| EDTA (0.5 M), pH 8.0, RNase-free | Thermo Fisher Scientific | Catalog #: AM9261 |
| Sodium dodecyl sulfate, BioReagent, suitable for electrophoresis, for molecular biology, 98.5% (GC) | Sigma-Aldrich | Catalog #: L3771-500G |
| Proteinase K from Tritirachium album, lyophilized powder, Bioultra, 30 units/mg protein, for molecular biology | Sigma-Aldrich | Catalog #: P2308-500MG |
| Membrane binding solution | Promega | Catalog #: A9301 |
| LightCycler® 480 SYBR® Green I Master | Roche | Catalog #: 04887352001 |
| Agencourt AMPure XP | Beckman Coulter | Catalog #: A63881 |
| Ethanol absolute | VWR Chemicals | Catalog #: 20821.330 |
| Critical commercial assays | ||
| TruePrep DNA Library Prep Kit V2 for Illumina | Vazyme | Catalog #: TD501-02 |
| Kapa HiFi PCR Kit, with dNTPs, 250 units | Kapa Biosystems | Catalog #: KK2102 |
| Experimental models: Organisms/strains | ||
| Mouse: Tg: Pax7nGFP (male, 8–10 weeks old) | A kind gift from Shahragim Tajbakhsh (Institut Pasteur) | N/A |
| Other | ||
| Precise peristaltic pump | Longer Precision Pump | BT100-2J |
| Surflo Winged Infusion Set | Terumo | Catalog #: SV∗23NL30 |
| Filter upper cup, PES, 0.22 μm, 500 mL, 75 mm | JET BIOFIL | Catalog #: FPE214150 |
| Syringe filter, minisart high flow, 0.22 μm, PES membrane, sterile, 28 mm | Sartorius | Catalog #: 1209Z85 |
| 30 mL syringe | Terumo | Catalog #: SS∗30LE1 |
| 50 mL syringe | Terumo | Catalog #: SS+50L1 |
| Shaking water bath | Memmert | Catalog #: WNB 22 |
| Eppendorf Centrifuge 5804R | Eppendorf | Catalog #: 5804R |
| Eppendorf Centrifuge 5427R | Eppendorf | Catalog #: 5427R |
| Eppendorf ThermoMixer C | Eppendorf | Catalog #: 5382000074 |
| Disposable needle 20G × 11/2 inch | Terumo | Catalog #: NN+2038R |
| 40 μm cell strainer | SPL Life Science | Catalog #: 93040 |
| Round-bottom polypropylene tube, 5 mL | Falcon | Catalog #: 352063 |
| Round-bottom polystyrene test tube, with cell strainer snap cap, 5 mL | Falcon | Catalog #: 352235 |
| BD Influx Cell Sorter | Biosciences | N/A |
| EconoSpin Micro DNA/RNA Columns, 5 μL Elution Volume w/ lid | Epoch Life Science | Catalog #: NC0857585 |
| PCR Strip Magnetic Rack | N/A | N/A |
Materials and equipment
0.5% Paraformaldehyde (PFA)
| Solution | Volume |
|---|---|
| 32% PFA | 468.75 μL |
| Cold 1× PBS | 29.5 mL |
| Total | 30 mL |
To be used immediately after making at 4°C.
CRITICAL: PFA is water-soluble and should always be used with adequate ventilation, preferably in a fume hood. Eyes and skin exposure should be avoided. Follow the safety data sheet when handling PFA.
2 M Glycine
| Solution | Amount |
|---|---|
| Glycine | 75 g |
| 1× PBS | 500 mL |
| Total | 500 mL |
Store at 4°C for up to one month.
Note: Filter the Glycine solution with a 0.22 μm filter cup before use.
Wash Medium
| Solution | Volume |
|---|---|
| Ham’s F10 | 445 mL |
| Horse Serum (HS) | 50 mL |
| Penicillin and Streptomycin (P/S) | 5 mL |
| Total | 500 mL |
Store at 4°C for up to one month.
Note: Filter the Horse Serum with a syringe filter before mixing with Ham’s F10.
Muscle Dissociation Buffer (per mouse)
| Solution | Volume |
|---|---|
| Wash Medium | 10 mL |
| Collagenase, Type II | 2,000 U/mL |
| Total | 10 mL |
To be used immediately after making at 4°C.
Stock Collagenase II Solution (per mouse)
| Solution | Volume |
|---|---|
| PBS | 1 mL |
| Collagenase II | 3000 U/mL |
| Total | 1 mL |
Store at −20°C for up to one year.
Stock Dispase Solution (per mouse)
| Solution | Volume |
|---|---|
| PBS | 1 mL |
| Dispase | 33 U/mL |
| Total | 1 mL |
Store at −20°C for up to one year.
Note: Filter the Dispase solution through a 0.45 μm filter before storage at −20°C.
Lysis Buffer
| Solution | Final Concentration |
|---|---|
| Sodium Chloride (NaCl) | 10 mM |
| Magnesium Chloride (MgCl2) | 3 mM |
| Tris-Cl, pH 7.4 | 10 mM |
| Igepal CA-630 (0.05% v/v) | 0.05% (v/v) |
| Total Volume | 50 μL |
To be used immediately after making at 4°C.
Tagmentation Reaction Mix
| Solution | Volume |
|---|---|
| 5× TTBL buffer | 10 μL |
| TTE Mix | 5 μL |
| ddH2O | 35 μL |
| Total | 50 μL |
To be used immediately after making at 4°C.
Step-by-step method details
Mouse perfusion
Timing: 30 min per mouse
PFA perfusion is critical for isolating cells while maintaining their in vivo cell states.
-
1.
Prepare 30 mL each of ice-cold PBS, 0.5% PFA, and 2 M Glycine solution in 50 mL falcon tubes for one mouse and label them with “PBS”, “PFA”, and “Glycine”, respectively. Store the tubes on ice.
Note: If processing multiple mice, a larger volume of solutions can be prepared in a beaker and stored on ice.
-
2.
Set up the perfusion pump with a speed of 70–80 rpm.
Note: The pump is specifically designed to mimic the contraction of the heart so that the solution is not continuously pumped into the mouse.
-
3.
Anesthetize the mouse with 2,2,2-Tribromoethanol (Avertin) (250 mg/kg) through intraperitoneal injection.
Note: Avertin is 1.25% in PBS. 125–300 mg is injected per kg of a mouse through intraperitoneal injection. We administer 600–800 μL of Avertin solution for a 2-month-old mouse. The mouse that weighs heavier or older may require more anesthetization reagent.
Alternatives: Other anesthetization reagents can also be used. We used Pentobarbitone (20 mg/mL) as an alternative. 50–90 mg is injected per kg of a mouse through intraperitoneal injection.
CRITICAL: Only proceed to the next step when the mouse stops moving. Pinch the mouse on the sole to check whether it is fully anesthetized. Mouse that is not fully anesthetized may struggle during the perfusion process, influencing your operation. Wait longer or administer an additional dose of anesthetization reagent if needed.
-
4.
Immobilize the mouse with pins/needles on plastic foam. Open the chest and expose the heart for perfusion.
-
5.
Make an incision on the right atrium for fluid removal. Hold the heart with blunt-end forceps and pierce the heart near the bottom of the left ventricle with the butterfly needle. (Figure 1).
-
6.
Perfuse the mouse with PBS for 3 min or until the pumped-out solution becomes clear.
-
7.
Perfuse the mouse with 0.5% PFA for 5 min.
Note: The heart will stop beating once perfused with PFA. The tail of the mouse will become rigid during the PFA perfusion.
-
8.
Perfuse the mouse with 2 M Glycine for 3 min to quench the PFA fixation.
Figure 1.
Example of stabilization of the perfusion setup
FACS sorting of SCs from PFA-perfused mouse
Timing: 4 h
-
9.
Prepare a 10 cm glass petri-dish with 10 mL of wash medium for each mouse.
-
10.
Dissect the hindlimb muscles and put them into the petri-dish.
-
11.
Transfer all muscles of a mouse to a fresh Petri dish (or use the lid of the initial one) and mince the muscles in a the dissociation buffer until no large pieces can be seen.
-
12.
Transfer the minced muscle pieces to a 50 mL falcon tube with 10 mL of muscle dissociation buffer.
-
13.
Seal the tube with Parafilm and incubate them in a 37°C shaking water bath with agitation at 65–70 rpm for 90 min.
-
14.
After digestion, top up the tube to 50 mL with the cold wash medium.
-
15.
Centrifuge the cells in a swing-bucket rotor at 500 × g for 10 min at 4°C. Aspirate the supernatant down to 10 mL.
-
16.
Add 1 mL of stock Collagenase II solution and 1 mL of stock Dispase solution. Resuspend the pellet with a 10 mL serological pipette. Triturate until the pellet is fully suspended.
-
17.
Fill the tube to 30 mL with the cold wash medium. Seal the tube with Parafilm and incubate in a 37°C water bath with agitation at 65–70 rpm for 30 min.
-
18.
After digestion, use a 30 mL syringe and a 20-gauge needle to aspirate and eject the muscle suspension 10 times.
-
19.
Place a 40 μm nylon cell strainer on a new 50 mL falcon tube. Transfer the cell suspension to the new 50 mL falcon tube through the cell strainer.
-
20.
Add 10 mL of wash medium to the original tube. Swirl to rinse and transfer it through the same cell strainer. Collect any remaining liquid on the underside of the strainer using a 200 μL pipette.
-
21.
Top up the tube to 50 mL with the cold wash medium.
-
22.
Centrifuge the tubes in a swing-bucket rotor at 500 × g for 10 min at 4°C. Remove all the supernatant by aspiration immediately after centrifugation.
-
23.
Resuspend the cell pellet in 1 mL of wash medium. Transfer to the strainer cap attached to a 5 mL FACS tube. Gently tap the tube to facilitate flow-through.
-
24.
Set up the cell sorter following the manufacturer’s specifications with the 70-μm nozzle. The sorting does not need to be done at 4°C.
-
25.
Create gates to collect the singlet GFP+ population into 500 μL of PBS in a 5 mL round-bottom FACS tube (Figure 2A). The cells are ready for library generation.
CRITICAL: Do not snap-freeze the cell pellet for ATAC-seq. Proceed the cells directly to the tagmentation steps. Cell pellets may be stored at 4°C for up to 3 days but it is not recommended. A comparison between ATAC-seq libraries generated from PFA-perfused SCs stored under different conditions is shown in Figures 2B–2D.
Figure 2.
ATAC-seq libraries generated from cells stored under different conditions
(A) FACS sorting plot showing the GFP signal of the PFA-perfused cells.
(B) Pearson correlation between ATAC-seq libraries generated from perfused SCs that are directly proceed to Tn5 tagmentation (freshly prepared) and stored at 4°C for 3 days (4°C storage).
(C) Pearson correlation between ATAC-seq libraries generated from perfused SCs that are directly proceed to Tn5 tagmentation (freshly prepared) and snap-frozen and stored at −80°C for 3 days (snapfreeze).
(D) Genome tracks showing ATAC-seq signal coverage across Pax7 locus with different storage conditions.
ATAC-seq library generation from PFA-perfused SCs
Timing: ∼20 h
100,000 cells were collected for the tagmentation reaction.
Note: Pre-cool PBS and Lysis Buffer on ice before starting.
Note: The number of cells can be reduced to 50,000.
-
26.
Centrifuge the sorted cells at 2000 × g for 10 min at 4°C.
-
27.
(Optional) Gently remove the supernatant using a 200 μL pipette. Resuspend the cells with 50 μL cold PBS. Centrifuge at 2000 × g for 10 min at 4°C.
-
28.
Gently remove the supernatant using a 200 μL pipette. Resuspend the cells with 50 μL Lysis buffer and centrifuge at 2000 × g for 10 min at 4°C.
CRITICAL: The cell pellet is too little to be seen. Remember the orientation of the tube when you put it in the centrifuge. Do not touch the side where the pellet should be.
Note: Using a white tip on top of the yellow tip on the 200 μL -pipette helps minimize cell loss.
-
29.
Gently remove the supernatant using a 200 μL pipette. Resuspend the cells with a 50 μL Tagmentation reaction mix.
-
30.
Incubate the reaction mix at 37°C for 30 min in a thermomixer.
-
31.
Add reverse crosslinking solution to the tagmented mixture. Incubate at 65°C with agitation at 1000 rpm in a thermomixer for 12–16 h.
Reverse-crosslinking solution
| Reagent | Final Concentration |
|---|---|
| Tris-Cl, pH 7.4 | 50 mM |
| EDTA | 1 mM |
| Sodium Dodecyl Sulfate (SDS) | 1% |
| Sodium Chloride (NaCl) | 0.2 M |
| Proteinase K | 5 ng/mL |
| Tagmented Sample | 50 μL |
| ddH2O | Top up to 200 μL |
| Total Volume | 200 μL |
Note: Incubate with shaking allows better reverse crosslinking.
-
32.
Purify the tagmented DNA with a PCR Cleanup kit and elute in 10 μL elution buffer.
Pause point: Purified DNA can be stored at −20°C for days.
-
33.
Amplify the purified DNA for 5 cycles.
PCR amplification reaction mix (from Kapa HiFi PCR Kit, with dNTPs)
| Reagent | Amount |
|---|---|
| KAPA HiFi Fidelity Buffer (5×) | 10 μL |
| KAPA dNTP Mix (10 mM) | 1.5 μL |
| KAPA HiFi DNA Polymerase (1 U/ μL) | 3 μL |
| Tagmented DNA | 10 μL |
| TM-Tn5-F Primer (10 μM) | 2 μL |
| TM-Tn5-R Primer (10 μM) | 2 μL |
| Nuclease-free Water | 23.5 μL |
| Total Volume | 50 μL |
PCR cycling condition
| Steps | Temperature | Time | Cycles |
|---|---|---|---|
| Pre-incubation | 72°C | 5 min | 1 |
| Initial Denaturation | 98°C | 30 s | 1 |
| Denaturation | 98°C | 10 s | 5 cycles |
| Annealing | 63°C | 30 s | |
| Extension | 72°C | 1 min | |
| Hold | 4°C | Infinity | |
Alternatives: Other PCR Mix reagents such as NEBNext High-Fidelity 2× PCR Master Mix can also be used.
-
34.
Determine the cycle number with qPCR for library amplification (Figure 3).
| Reagent | Amount |
|---|---|
| Previously PCR-amplified DNA | 5 μL |
| TM-Tn5-F Primer (10 μM) | 0.2 μL |
| TM-Tn5-R Primer (10 μM) | 0.2 μL |
| 2× SYBR Green Mix | 5 μL |
| Nuclease-free Water | 4.6 μL |
| Total Volume | 15 μL |
-
35.
Amplify the rest of the previously PCR-amplified DNA with the additional number of cycles you chose based on the fluorescence intensity.
-
36.Purify the DNA using AMPure XP beads with a size selection of 200 bp to 1 kb.
-
a.Equilibrate AMPure XP beads to 20°C–22°C for at least 30 min beforehand and mix the beads thoroughly.
-
b.Top up the PCR product to 50 μL with Nuclease-free water and add 27.5 μL of beads (0.55×). Mix well and incubate at 20°C–22°C for 10 min.
-
c.Centrifuge briefly, then place the tube onto a magnetic rack for 2 min until the liquid becomes clear. Carefully transfer the supernatant to a new tube and discard the beads.
-
d.Add 32.5 μL of beads (1.2×) to the supernatant from the previous step. Mix well and incubate at 20°C–22°C for 10 min.
-
e.Centrifuge briefly, then place the tube onto a magnetic rack for 2 min until the liquid becomes clear. Carefully remove and discard the supernatant.
-
f.Add 200 μL freshly prepared 80% ethanol to the tube while on the magnetic rack without disturbing it. Incubate at 20°C–22°C for 30 s and discard the supernatant. Repeat this step once.
-
g.Completely remove the residual ethanol, and air-dry beads for 2 min while the tube is on the magnetic rack with the lid open.
-
h.Remove the tube from the magnetic rack and add 21 μL of nuclease-free water or elution buffer to elute DNA. Mix thoroughly and incubate at 20°C–22°C for 5 min.
-
i.Centrifuge briefly and then place the tube back onto the magnetic rack until the liquid becomes clear. Transfer 20 μL of supernatant to a new tube with no beads carry-over.
-
a.
-
37.
The library is sequenced in 2 × 100 bp paired-end mode with at least 50 M read-depth.
Alternatives: The library can also be sequenced in 2 × 75 bp paired-end mode.
Figure 3.
Example of PCR cycle determination
The cycle number is chosen when the fluorescence intensity reaches 1/3 of the maximum intensity. Alternatively, the Ct value can also be used as the final cycle number. Usually 17–19 cycles are used for final library amplification.
Expected outcomes
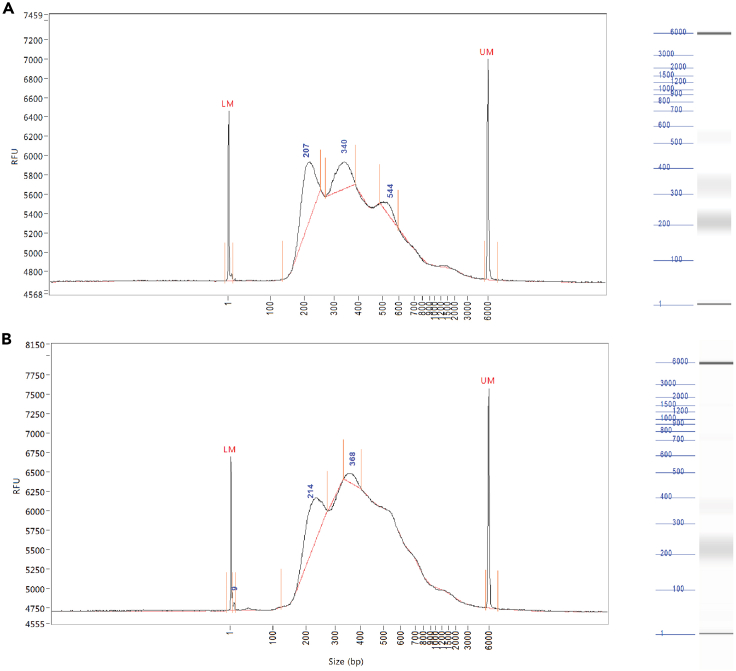
The protocol described how to prepare the ATAC-seq library from fixed muscle stem cells. Figure 4 represents the library size distribution for the ATAC-seq library. The first peak corresponds to the nucleosome-free fragments, the second peak represents the mono-nucleosome fragments, and the third peak represents the di-nucleosome fragments. The average size of the ATAC-seq library fragments is around 400 bp (Figure 4). This protocol not only applies to generating the ATAC-seq library from the quiescent muscle stem cells, but it can also apply to activated muscle stem cells induced by muscle injury. The relative height of peaks may occasionally vary between different ATAC-seq libraries but the size distribution remains similar (Figure 4).
Figure 4.
Size distribution of ATAC-seq libraries
(A and B) ATAC-seq libraries display a distinct nucleosomal pattern. The relative height of each nucleosomal peaks differ between different libraries (A and B).
Limitations
In the current protocol, we utilized a perfusion pump to increase the stability of the perfusion step. However, the insertion site of the needle in the heart, as well as the depth of the insertion differs every time, leading to occasionally inconsistent perfusion results, such as over-fixation or incomplete fixation (Figure 5). Over-fixation will decrease the total sorted cell numbers, while incomplete fixation will not preserve the chromatin signature in vivo. Fortunately, the degree of fixation can be inferred from the FACS plots (Figure 5), which can help to decide whether to process the samples further.
Figure 5.
Examples of over-fixation and incomplete fixation
(A–C) Examples of FACS plots and cell shape differences among successful fixation (A), over-fixation (B) and incomplete fixation (C). Cells that are over-fixed have smaller FSC value while cells that are not fixed completely are usually with larger FSC value.
This protocol is only applicable for isolating cells labeled with reporters. For cell isolation with specific cell surface marker staining, a test trial should be performed to validate the applicability of the perfusion-based cell isolation.
Troubleshooting
Problem 1
Over fixation or Incomplete fixation (step 5).
Potential solution
-
•
Make sure the 0.5% PFA is freshly made and mixed well.
-
•
Make sure the needle does not go through the left ventricle to the right ventricle and no liquid is coming out from the lung or the nose/mouse of the mouse during perfusion.
-
•
Make sure the mouse is completely anesthetized and remain stable during the PBS perfusion process.
-
•
Make sure the needle is stabilized during the perfusion. For example, use another tweezer/scissor to stabilize the needle (Figure 1).
-
•
Optimize the time and flow rate of the perfusion if you are using other models of the perfusion pump.
Problem 2
Over-tagmentation or incomplete tagmentation (step 29).
Potential solution
Optimize the input cell number for the transposition reaction. Typically, 1 unit of Tn5 works for 50 ng of DNA. We recommend calibrating the number of cell input for your experiment.
Problem 3
ATAC-seq library does not have the right size distribution (open chromatin, single nucleosome, di-nucleosome, and tri-nucleosome patterns) (step 30).
Potential solution
Optimize the reverse crosslinking step. Incubation with shaking is highly recommended if encountered with this problem. Increasing the incubation time with Proteinase K is also recommended.
Problem 4
Blockage in the column during DNA purification (step 32).
Potential solution
Warm the reverse-crosslinked sample before purifying the DNA to prevent SDS precipitation.
Problem 5
Poor library quality or high mitochondrial reads (step 37).
Potential solution
Make sure to aspirate all the supernatants during the sorting preparation step to avoid mitochondria contamination.
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Prof. Tom Cheung, tcheung@ust.hk.
Materials availability
This study did not generate new unique reagents.
Data and code availability
This study did not generate any datasets.
Acknowledgments
We thank S. Tajbakhsh for providing Pax7-nGFP mice. We thank all members of Cheung Lab for the helpful discussion and optimization of the perfusion protocol. We thank T.W. Fung for the assistance with the graphical abstract. This work was supported by research grants from the Hong Kong Research Grant Council (GRF16102319, GRF16102420, C6018-19G, C6027-19G, AoE/M-604/16, and T13-605/18W), the Lee Hysan Foundation (LHF17SC01), and the Hong Kong Epigenome Project (Lo Ka Chung Charitable Foundation). This study was supported in part by the Innovation and Technology Commission (ITCPD/17-9). I.T.C.C. is a recipient of the Hong Kong Ph.D. Fellowship Scheme and 2022 Croucher Studentship. T.H.C. is the S. H. Ho Associate Professor of Life Science at the Hong Kong University of Science and Technology (HKUST). We also thank the Biosciences Central Research Facility at HKUST for their continuous technical support for this project.
Author contributions
Conceptualization, A.D., T.H.C.; investigation, A.D., I.T.C.C.; writing – original draft, A.D.; writing – review & editing, A.D., I.T.C.C., T.H.C.; funding acquisition, T.H.C.
Declaration of interests
The authors declare no competing interests.
Contributor Information
Anqi Dong, Email: adongaa@connect.ust.hk.
Tom H. Cheung, Email: tcheung@ust.hk.
References
- 1.Dong A., Liu J., Lin K., Zeng W., So W.-K., Hu S., Cheung T.H. Global chromatin accessibility profiling analysis reveals a chronic activation state in aged muscle stem cells. iScience. 2022;25:104954. doi: 10.1016/j.isci.2022.104954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Buenrostro J.D., Giresi P.G., Zaba L.C., Chang H.Y., Greenleaf W.J. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods. 2013;10:1213–1218. doi: 10.1038/nmeth.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Buenrostro J.D., Wu B., Chang H.Y., Greenleaf W.J. ATAC-seq: a method for assaying chromatin accessibility genome-wide. Curr. Protoc. Mol. Biol. 2015;109:21–29. doi: 10.1002/0471142727.mb2129s109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yue L., Wan R., Luan S., Zeng W., Cheung T.H. Dek modulates global intron retention during muscle stem cells quiescence exit. Dev. Cell. 2020;53:661–676.e6. doi: 10.1016/j.devcel.2020.05.006. [DOI] [PubMed] [Google Scholar]
- 5.Zeng W., Yue L., Lam K.S.W., Zhang W., So W.-K., Tse E.H.Y., Cheung T.H. CPEB1 directs muscle stem cell activation by reprogramming the translational landscape. Nat. Commun. 2022;13:947. doi: 10.1038/s41467-022-28612-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yue L., Cheung T.H. Protocol for isolation and characterization of in situ fixed quiescent muscle stem cells. STAR Protoc. 2020;1:100128. doi: 10.1016/j.xpro.2020.100128. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This study did not generate any datasets.