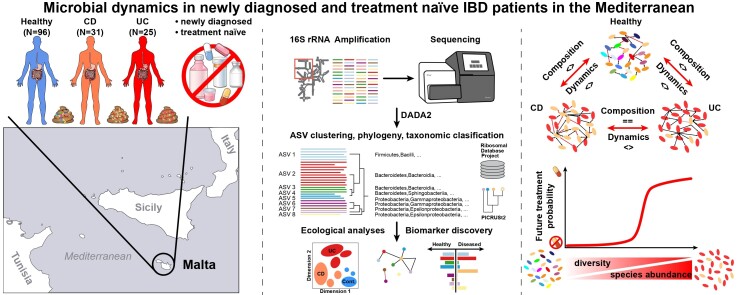
Abstract
Background
Microbial communities have long been suspected to influence inflammatory processes in the gastrointestinal tract of patients with inflammatory bowel disease. However, these effects are often influenced by treatments and can rarely be analyzed in treatment-naïve onset cases. Specifically, microbial differences between IBD pathologies in new onset cases have rarely been investigated and can provide novel insight into the dynamics of the microbiota in Crohn’s disease (CD) and ulcerative colitis (UC).
Methods
Fifty-six treatment-naïve IBD onset patients (67.3% CD, 32.7% UC) and 97 healthy controls were recruited from the Maltese population. Stool samples were collected after diagnosis but before administration of anti-inflammatory treatments. Fecal microbial communities were assessed via 16S rRNA gene sequencing and subjected to ecological analyses to determine disease-specific differences between pathologies and disease subtypes or to predict future treatment options.
Results
We identified significant differences in community composition, variability, and diversity between healthy and diseased individuals—but only small to no differences between the newly diagnosed, treatment-naïve UC and CD cohorts. Network analyses revealed massive turnover of bacterial interactions between healthy and diseased communities, as well as between CD and UC communities, as signs of disease-specific changes of community dynamics. Furthermore, we identified taxa and community characteristics serving as predictors for prospective treatments.
Conclusion
Untreated and newly diagnosed IBD shows clear differences from healthy microbial communities and an elevated level of disturbance, but only the network perspective revealed differences between pathologies. Furthermore, future IBD treatment is to some extent predictable by microbial community characteristics.
Keywords: inflammatory bowel disease, Crohn’s disease, ulcerative colitis, microbiome, treatment naïve, treatment prediction, biomarker
Graphical abstract
Graphical Abstract.
Key Messages.
What is already known?
The microbiota has been implicated in the pathology of CD and UC.
Pharmaceutical treatments, particularly anti-inflammatory treatments, influence microbial communities in many ways.
Investigations of new onset- and treatment-naïve individuals are rare and allow a pristine insight into the microbiome of IBD patients.
What is new here?
Inflammatory bowel disease fecal microbial communities of treatment-naïve patients differ significantly in diversity and structure from healthy individuals and show higher community variability.
Crohn’s disease and UC communities do not differ in composition or diversity in the early phases of inflammation but show very disparate community dynamics and bacterial interactions.
Community diversity, single community members, and functional characteristics are indicative of future medical treatments.
How can this study help patient care?
Community interactions and not necessarily community composition may help to distinguish and treat CD and UC in early stages of IBD.
Single taxa and community patterns can be indicators for disease development and may guide specific treatment regiments.
Introduction
The disease complex of inflammatory bowel disease (IBD) consists primarily of Crohn’s disease (CD) and ulcerative colitis (UC), which differ in their phenotypic and immunologic characteristics. Crohn’s disease often involves granulomas, discontinuous bowel involvement, and transmural inflammation, which can be present throughout the gastrointestinal tract. Ulcerative colitis is mainly restricted to the mucosa and submucosa of the large intestine and rectum. However, both pathologies overlap genetically in a considerable way.1 Genetic predispositions and heritability1,2 account only for a limited amount of disease development, and its incidence seems to be influenced by different environmental factors.3 Globally, the prevalence of IBD has increased over the recent years, potentially due to a multitude of lifestyle factors.3
However, it has become increasingly evident that the bodies’ microbial communities play an undeniably important role in the pathogenesis of IBD. Numerous animal experiments and population studies identified a multitude of ways the microbiome varies and interacts with its host, eventually leading to inflammation.4,5 Current evidence suggests that inflammation may not be caused by single pathogens, but rather driven by dysbiosis such as a shift in the balance of a beneficial microbial community as a whole in favor of a pro-inflammatory community, which eventually leads to and sustains intestinal inflammation.4,5 These shifts can be the result of disturbances introduced through environmental and lifestyle factors like diet, hygiene, medication, or even the host’s genetic makeup.6,7 Also, prior infections and their treatment can trigger and sustain IBD in previously healthy individuals.8 Thus, analyzing early changes in the microbial communities and reducing the influence of treatment-related disturbances offer immense opportunities to trace the true pathological dynamics in the microbiota that drive chronicity and inflammation.
The Maltese population is comparably small (approximately 550 000) and has not been investigated on the microbial level, let alone its association to IBD. However, small scale genetic studies identified several shared IBD risk genes with other cohorts but also population-specific patterns.9,10 The Maltese people have a long history of settlement and admixture, resulting in a distinct cultural and genetic makeup.11 Studies of populations with particular environmental and genetic background have the power to reveal new and validate already established patterns, adding to the overall understanding of host-microbiome interactions and its involvement in IBD. Thus, CD and UC may display not only patterns already shown in published population studies but may also reveal local dynamics in relation to IBD.12 In this study, we analyze the fecal microbial communities in the early stages of IBD development in a newly diagnosed and treatment-naïve cohort of patients suffering from CD and UC, which is an advantageous setup to investigate these complex pathologies.
Materials and Methods
Patient Recruitment and Sampling
Ethical approval was obtained from the University of Malta Research Ethics Committee (Ref 32/2017), and patients were recruited from the gastroenterology outpatient clinic at Mater Dei Hospital, Malta, between January 2018 and September 2019 (Table 1). Fecal samples were collected from patients diagnosed with IBD for the first time and randomly from healthy controls above the age of 18 years. Exclusion criteria included,
Table 1.
Summary of cohort characteristics of healthy controls, CD, and UC patients, summarizing subject numbers for general anthropometric characteristics and continuous measurements of general subject characteristics and severity measures (average values ± SD). Further, for diseased individuals we summarize the distribution among CD and UC subtypes, as well current and future treatments.
| Category | Variable | Levels | Healthy | CD | UC |
|---|---|---|---|---|---|
| Anthropometrics | Gender | male | 50 | 19 | 14 |
| female | 46 | 12 | 11 | ||
| Smoking | Yes | 6 | 3 | 7 | |
| No | 80 | 26 | 16 | ||
| Ex | 10 | 2 | 2 | ||
| Continuous | Age (years) | mean ± SD | 44.71±16.00 | 37.81±16.62 | 47.36±16.60 |
| measures | Weight (kg) | 74.06±12.92 | 67.24±10.53 | 70.01±8.77 | |
| Height (cm) | 166.88±7.91 | 163.52±6.96 | 162.24±7.52 | ||
| BMI (kg/m2) | 26.53±3.90 | 25.095±3.00 | 26.66±3.52 | ||
| Sunlight (hours) | 3.87±1.15 | 4.08±1.47 | |||
| CDAI | 23.10±21.33 | ||||
| SCCAI | 0.87±1.14; NA:2 | ||||
| Godin leisure time index | 6.94±9.82 | 3.84±4.59 | |||
| Harvey-Bradshaw Index | 1.45±1.73 | ||||
| Disease subtypes | Subtype A | A1 | 4 | ||
| (age of onset) | A2 | 20 | |||
| A3 | 7 | ||||
| Subtype B/P | B1 | 28 (P:3) | |||
| (disease behavior) | B2 | 0 | |||
| B3 | 3 (P:1) | ||||
| Subtype L | L1 | 16 | |||
| (disease location) | L2 | 2 | |||
| L3 | 13 | ||||
| Subtype E | E1 | 7 | |||
| (disease behavior) | E2 | 8 | |||
| E3 | 10 | ||||
| Current medication | Non-IBD medication | Yes/1 | 6 | 6 | |
| No/0 | 25 | 19 | |||
| Antibiotics | Yes/1 | 8 | 9 | ||
| (6 weeks prior) | No/0 | 23 | 16 | ||
| Antibiotics | Yes/1 | 13 | 11 | ||
| (6 months prior) | No/0 | 18 | 14 | ||
| Future IBD | Mesalazine | Yes/1 | 9 | 13 | |
| treatment | No/0 | 22 | 12 | ||
| (52 week follow-up) | Azathioprine | Yes/1 | 9 | 3 | |
| No/0 | 22 | 22 | |||
| Anti-TNF | Yes/1 | 7 | 0 | ||
| No/0 | 24 | 25 | |||
| Prednisolone | Yes/1 | 4 | 2 | ||
| No/0 | 27 | 23 | |||
| general IBD | Yes/1 | 19 | 14 | ||
| medication (combined) | No/0 | 12 | 11 |
antibiotics, probiotics, bowel-cleansing medications, systemic corticosteroids (3 months prior);
previous anti-inflammatory treatments;
IBD, acute illness gastrointestinal disease, chronic autoimmune/inflammatory conditions, history of malignancy, gastrointestinal surgery;
and pregnancy, breastfeeding.
DNA Extraction and 16S rRNA Sequencing
DNA from stool samples was extracted using the QIAamp DNA fast stool mini kit automated on the QIAcube (Qiagen, Hilden, Germany) and sequenced as described elsewhere.13 Data processing was performed using DADA2 1.10,14 and Amplicon Sequence Variants (ASVs) tables were classified via the naïve Bayesian classifier (RDP16, see supplemental material).15 Unstratified EC categories metagenome predictions were generated using the native ASV abundances and sequences in PICRUSt 2.5.0 using the default workflows.16 Raw sequence data and relevant meta-data can be accessed online under the accession number PRJEB47161 (IBD cases) and PRJEB47162 (controls) at the European Nucleotide Archive (https://www.ebi.ac.uk/ena/).
Statistical Methods for Microbiome Analyses
The microbiome data were rarefied to 11 800 reads/sample to ensure comparable and sufficient coverage across samples (average Good’s coverage 99.88% ± 0.001 SD). Species richness (Chao1), Simpson diversity (1-D), and phylogenetic alpha diversity (Nearest Taxon Index [NTI], Net Relatedness Index [NRI]) were calculated and analyzed in R 3.5.3.17–19 Beta diversity analyses were conducted via distance-based (conditional) Redundancy analyses and permutative ANOVA, as well as with a multivariate test for homogeneity of variances (10 000 permutations),20,21 using Jaccard distance (presence/absence) and Bray-Curtis dissimilarity (differential abundance).
Indicator species analysis was used to detect predictive taxa for the respective patient characteristics (present in >10% of samples),22 and differential abundance analyses were performed with DESeq2 (bacteria present >5% of samples) including age and body mass index (BMI) as covariates.23 Continuous variables were not transformed if used in a statistical model (ie, SCCAI, CDAI, BMI, age, HBI, Godin Leisure-Time Exercise index, time spent outside) and samples with missing values were excluded from the respective analyzes. Binomial generalized linear models (GLMs) were employed to predict future treatments (yes/1, no/0) by alpha diversity, bacterial abundances, and functions. All P values were adjusted via false discovery rate (FDR). Similar methods were applied to investigate functional characteristics of the bacterial communities, as based on imputed bacterial functions and pathways via PICRUSt2, with the exception of using the Wald test based on DESeq2 analyses and filtering.16 Weighted SparCC co-abundance networks were generated based on ASV abundances (present >10% of samples, PFDR ≤ .050, |R| ≥ 0.250).24 Networks were generated individually for the UC and CD cohorts, as well as for the controls (complete cohort, 3 subsets, N=31). Node-based (degree, betweenness, PageRank-index, eigenvalue-centrality) and network-wide measures (diameter, radius, assortativity) were calculated in igraph 1.2.4.1.25–27 To assess whether bacteria were more important than expected by chance, observed centralities (mean of control subsamples) were compared against a permuted set of networks (10 000 times, combined for control subsamples) via one-sided Z tests.
Results
Description of a Newly Diagnosed, Treatment-naïve IBD Cohort and Accompanying Healthy Subjects From Malta
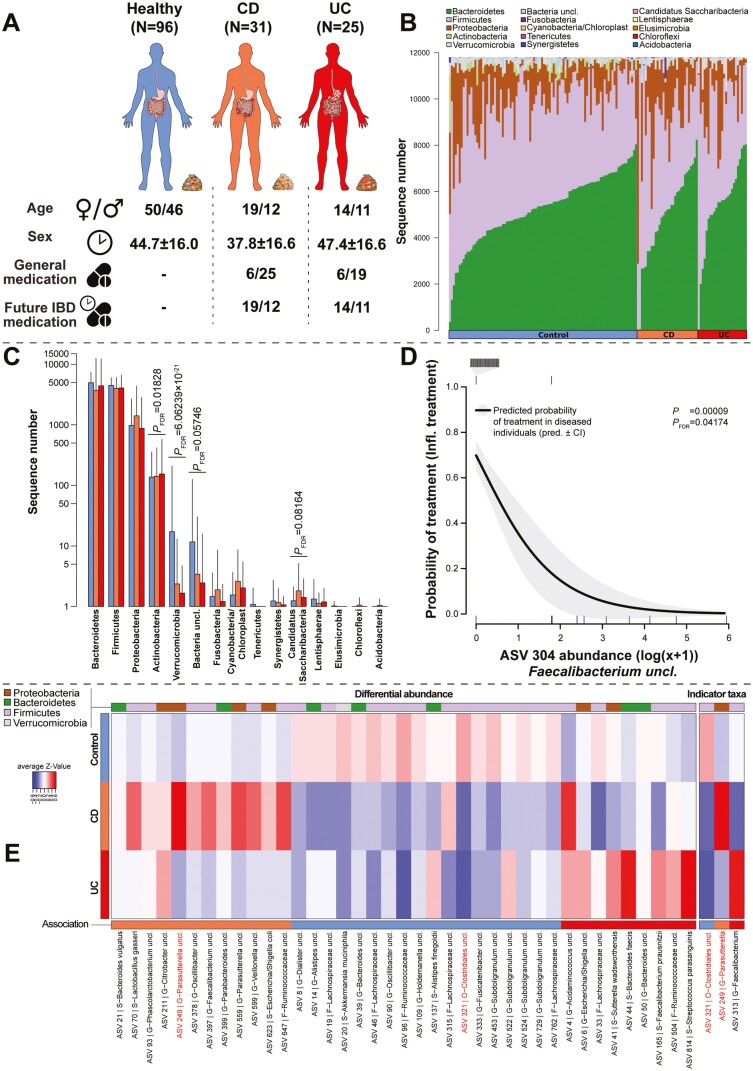
Our cohort consists of 56 patients with histologically confirmed IBD (CD, n = 31; UC, n = 25), as well as 96 healthy controls (Table 1). The average age was 44.7 years among the healthy individuals, 47.4 in UC patients, and 37.8 years in CD patients (χ2 = 5.3143, P > 0.050; Kruskal-Wallis test). The mean BMI was 26.5 kg/m2 in the healthy cohort, 26.7 kg/m2 in UC patients, and 25.1 kg/m2 in CD patients (χ2 = 3.9658, P > 0.050; Kruskal-Wallis test). A more detailed classification of CD and UC patients into subtypes was performed following the Montreal classification. Additional information was analyzed, as well, including medication (IBD specific, other medication), disease severity (Simple Clinical Colitis Activity Index [SCCAI], Crohn’s Disease Activity Index [CDAI], Harvey-Bradshaw Index [HBI]), and life quality (Godin Leisure-Time Exercise index, sunlight hours; Figure 1A and Table 1).
Novel and Established Bacterial Associations with IBD, Disease Subtypes, and Prospective IBD Treatments
The most abundant community members belonged to the phylum Firmicutes, followed by Bacteroidetes, Proteobacteria, Actinobacteria, Verrucomicrobia, and other low abundant bacterial phyla. The distribution of the major groups is fairly comparable between healthy and diseased individuals (Figure 1B). However, the abundance of Actinobacteria is increased in diseased patients, similar to members of the Candidatus saccharibacteria. In contrast, verrucomicrobia and yet unclassified bacteria are more abundant among the healthy cohort in our sample population (Figure 1C). On a more fine-grained analytical level, we identified 39 ASVs differentially abundant between the different health conditions. Eighteen taxa were more abundant in healthy individuals (eg, ASV 14- Alistipes uncl., ASV 20- Akkermansia muciniphila), whereas 12 taxa were more abundant in CD patients (eg, ASV 70- Lactobacillus gasseri, ASV 249- Parasutterella uncl.). We also identified 9 ASVs that show higher abundances among UC patients (eg, ASV 6-Escherichia/Shigella uncl., ASV 41-Sutterella wadsworthensis, ASV 44-Bacteroides faecis) compared with the other conditions (Figure 1E; Table S1). The ASV 249 belonging to the Parasutterella was found to be overabundant in CD and also detected to be indicative of a CD-associated microbiome via indicator species analysis. This approach determines taxa, which are predictive for a certain environment, based on its abundance and frequency like the Firmicutes Flavonifractor plautii (ASV 87).22 Crohn’s disease patients were further characterized through the associations with L. gasseri (ASV 70), which is also overabundant in CD (Table S2). Interestingly, UC patients showed a strong association to the usually probiotic Faecalibacteria (ASV 313). However, ASV 321 (Clostridia uncl.) and ASV 96 (Ruminococcaceae uncl.), 2 known butyrate producers, were strongly indicative of a healthy microbiota (Figure 1E). Other known probiotic bacteria were associated with healthy controls as well, like A. muciniphila (ASV 20),28,29Fusicatenibacter uncl. (ASV 333),30 and Subdoligranulum uncl. (ASV 453). However, these additional yet interesting associations did not reach the adjusted P value cutoff (PFDR ≤ .100) but overlapped strongly with the differentially abundant bacteria (10 ASVs, Table S2).
Figure 1.
A, Schematic overview of cohort characteristics (see Table 1 for further information). B, The stacked barplots visualize the individual community composition at the phylum level sorted by the relative phylum abundances within each cohort. C, The barplots visualize phylum abundances with respect to IBD status and highlight significant differentially abundant phyla (PFDR < 0.100), as based on DESeq2.23 Figure (D) displays the association between ASV 304 (Faecalibacterium uncl.) abundance in diseased individuals (CD and UC) and the probability of receiving future anti-inflammatory treatment (eg, 5-ASA, AZA, anti-TNF, or prednisolone) within one year after sampling (DF = 1,54, Deviance = 15.15736, P = .00009, PFDR = .04174, binomial GLM). Ticks on the bottom of the graph indicate samples without treatment, whereas ticks on the top indicate samples with future treatment. E, The heatmap displays in the left panel significant differentially abundant ASVs (DESeq2) and on the right panel significant indicator species22 with respect to IBD status/health condition (PFDR≤ .05). Overlapping associations between both methods are highlighted in red. Phylum membership of the respective ASVs is visualized in the upper color bar and the direction of association (association, maximum abundance) is indicated by the color bar at the bottom.
Disease subtypes, which categorize disease severity by the age of onset (subtype A), disease behavior (subtype B/P), and disease location (subtype L) in CD and disease magnitude/location in UC (subtype E), significantly differed in abundance of several bacterial taxa (228 unique ASVs; Table S3, Figure S1-S4). Several of these bacteria displayed repeated associations among the subtype categories (85 ASVs), which were detected in the indicator analyses to some extent (similar direction, 19 ASVs; different direction, 66), which itself detected 80 unique ASVs associated with disease subtypes in CD and UC patients (Table S4).
Because this is an inception cohort, the stool samples were taken when these patients were still not receiving any IBD-related medications. However, this cohort received non-IBD specific medication and in some cases antibiotics approximately 6 weeks to 6 months prior to sampling. We investigated the influence of non-IBD-related medications on the abundance and occurrence of bacteria in CD and UC patients. Several taxa appeared to associate in similar ways with the same treatment in UC and CD, as well as among the different detection methods (Table S5 and S6; Figure S5 and S6). In addition, we attempted to associate future IBD-specific treatments in UC and CD individuals with bacterial abundances. By investigating all diseased individuals combined, we identified only Faecalibacterium (ASV 304) to be associated with future treatments, as its abundance correlates with a reduced probability of IBD-related medication usage (P = 9.8912 × 10-5, PFDR = .0417; binomial GLM; Figure 1D).
Diversity of Fecal Communities in Newly Diagnosed IBD Cases Is Lower Than in Healthy Individuals and Is Associated with Prospective IBD Treatments
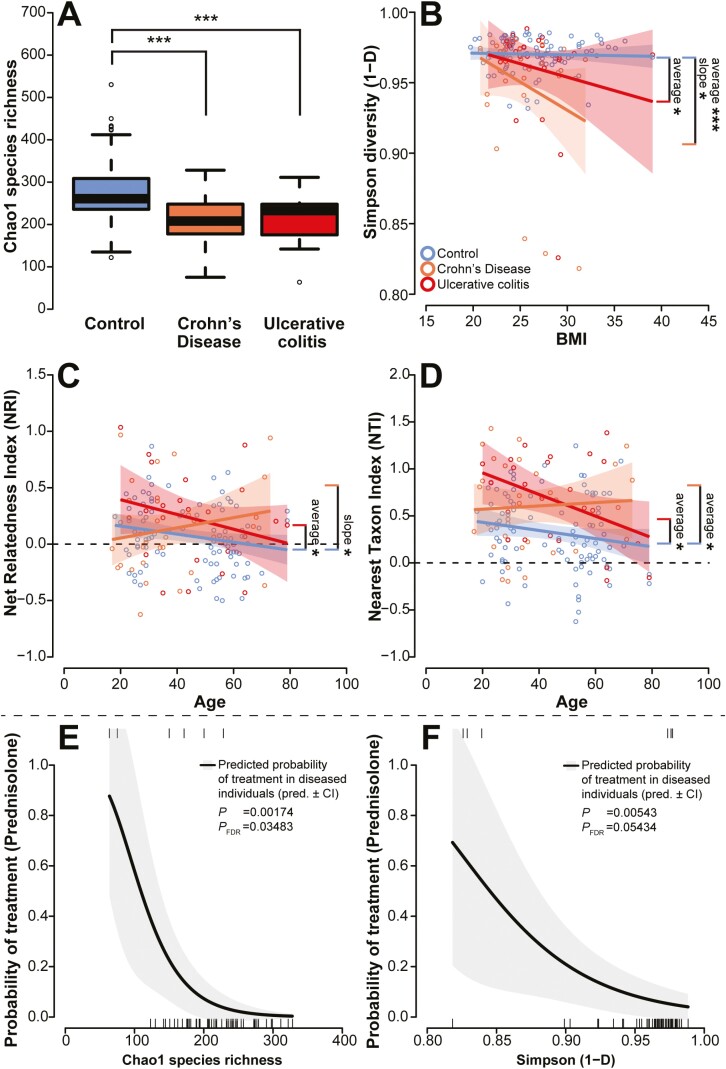
A central characteristic of microbial communities is the distribution of species within communities, termed alpha diversity, which is associated with productivity and stability of the respective communities.31 The strongest factor influencing the complexity of the fecal microbiota was the health condition of the individuals (healthy controls, CD, UC). Patients with IBD displayed a reduced average species richness and community evenness compared with healthy controls, as described by the metrics Chao1 species richness and Simpson diversity, respectively (Figure 2A and 2B; Table 2). With respect to Simpson diversity, we also observed an overall decrease in relation to BMI, a pattern that was strongest among CD patients (Figure 2B). Also in the phylogenetic diversity, as expressed by phylogenetic clustering ( > 0, low diversity) or overdispersion ( < 0, high diversity) of species over the phylogenetic tree, we observed phylogenetic dispersion with increasing age over the whole phylogenetic tree (NRI) and among more closely related bacteria (NTI), particularly in healthy individuals (Figure 2C and 2D). However, this pattern changed slightly in UC patients (clustering: NRI > 0, NTI > 0), whereas CD patients showed an increased phylogenetic clustering in older CD patients (strongest in NRI; Figure 2C and 2D; Table 2). Overall, we did not identify significant differences in alpha diversity between CD and UC patients.
Figure 2.
A, Alpha diversity is expressed through the Chao1 species richness among the different cohorts, which shows lower alpha diversity in patients with IBD compared with controls, while Simpson diversity (B) shows an average decrease of evenness in CD and UC patients compared with healthy subjects, as well as a negative correlation of evenness with BMI. Figures (C) and (D) display the correlation of the phylogenetic diversity measures Net Relatedness Index (NRI) and Nearest Taxon Index (NTI) with age for the different disease cohorts. The dashed zero line marks the cutoff between phylogenetic overdispersion (NRI < 0, NTI < 0) and phylogenetic clustering (NRI > 0, NTI > 0). Figures (E) and (F) depict the probability of future anti-inflammatory treatments (5-ASA, AZA, anti-TNF, or prednisolone) based on the current species richness (E) and evenness (F) in IBD patients. Ticks on the bottom of the graph indicate samples without treatment, while ticks on the top indicate samples with future treatment.
Table 2.
Linear model results of alpha diversity analyses focusing on Chao1 species richness, Simpson diversity (1-D), phylogenetic clustering of distantly (NRI) and closely related bacteria (NTI), after model selection (covariates: health condition, age, BMI, medication).
| Pairwise comparison | ||||||
|---|---|---|---|---|---|---|
| Diversity metric | Model | DF | F | P | Comparison | P |
| Chao1 | ~IBD | 2,149 | 15.46929 | 0.000001 | CD—Contr. | 0.00001 |
| UC—Contr. | 0.00024 | |||||
| CD—UC | 0.67680 | |||||
| Simpson | ~BMI | 1,146 | 1.06818 | 0.303066 | BMI | 0.55512 |
| +IBD | 2,146 | 8.417464 | 0.000347 | IBD(Contr.,CD) | 0.00003 | |
| +IBD:BMI | 2,146 | 2.943861 | 0.055796 | IBD(Contr.,CD):BMI | 0.01271 | |
| BMI | 0.33708 | |||||
| IBD(Contr.,UC) | 0.02192 | |||||
| IBD(Contr.,UC):BMI | 0.13964 | |||||
| BMI | 0.14430 | |||||
| IBD(UC,CD) | 0.18330 | |||||
| IBD(UC,CD):BMI | 0.52740 | |||||
| NRI | ~IBD | 2,146 | 2.02453 | 0.135746 | Age | 0.30089 |
| +Age | 1,146 | 2.04422 | 0.154922 | IBD(Contr.,CD) | 0.42558 | |
| +IBD:Age | 2,146 | 2.53737 | 0.082557 | IBD(Contr.,CD):Age | 0.04372 | |
| Age | 0.02469 | |||||
| IBD(Contr.,UC) | 0.02672 | |||||
| IBD(Contr.,UC):Age | 0.48271 | |||||
| Age | 0.92660 | |||||
| IBD(UC,CD) | 0.45790 | |||||
| IBD(UC,CD):Age | 0.10060 | |||||
| NTI | ~IBD | 2,148 | 21.48979 | 0.000008 | Age | 0.07742 |
| +Age | 1,148 | 4.23903 | 0.041246 | IBD(Contr.,CD) | 0.00365 | |
| Age | 0.02140 | |||||
| IBD(Contr.,UC) | 0.00033 | |||||
| Age | 0.35450 | |||||
| IBD(UC,CD) | 0.54700 | |||||
Inflammatory bowel disease subtypes, following the Montreal classification for CD and UC,32 had only minor effects on community diversity in UC patients and no significant influence on the community of CD patients. In UC, we observed a decrease of NTI (less phylogenetically clustered) in patients with subtype E3 (pancolitis) compared with the less severe subtypes E1 and E2 (F2,22 = 3.5799, P = .0451). When we focus on medications, neither non-IBD-related medication nor previous antibiotic use (at least 6 weeks or 6 months prior) affected the complexity of CD and UC microbiota (Table S7). We also analyzed data on IBD-related pharmaceutical treatments required within 12 months postdiagnosis. A combined analysis of CD and UC patients revealed that patients with a less diverse community were more likely to receive prednisolone treatment (Chao1: P = .00174, PFDR = .03483; Simpson: P = .00543, PFDR = .05434; binomial GLM, Figure 2E and 2F). However, nonspecific treatments and other anthropogenic factors influence disease trajectories, as well. Thus, this relatively small study can only be suggestive and will need further validation.
Diseased Fecal Communities Are Strongly Differentiated From Healthy Communities, Accompanied by Higher Community Variability
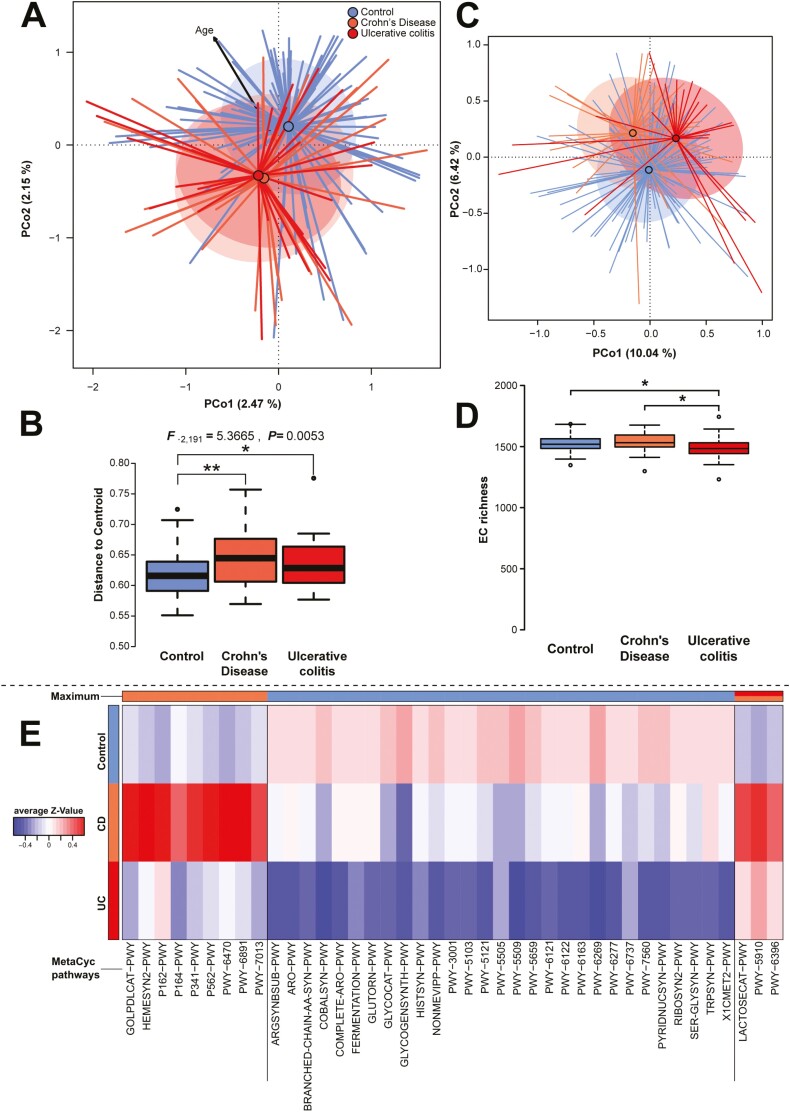
Disease condition not only changes the diversity within individuals but also influences differences in community composition between individuals in a systematic way.12,33 Highly significant differences in taxonomic community composition and structure (Bray-Curtis, Jaccard) with respect to health conditions were present, although the separation between the different health conditions was relatively weak (R2Bray-Curtis = 0.01651, R2Jaccard = 0.01631; Figure 3A, S8A; Table 3). In particular, differences between microbial communities of healthy individuals to either disease were comparably high, although we cannot observe a significant separation between CD and UC communities (Table 3). Correcting for potential confounding factors, like age or BMI, did not influence the effect of health condition on community differences (PBray-Curtis = .0001, PJaccard = .0001; conditional distance based redundancy analysis [dbRDA]). Patient gender but not smoking status influenced community structure, as well. However, compared with other patient characteristics, disease type was the most prominent factor influencing the microbial community composition and significantly increased community variability of diseased communities (Figure 3D, Figure S8D). This pattern has been called the “Anna Karenina principle,” by which the microbial communities of dysbiotic individuals vary more than communities of healthy individuals.36 These patterns are also present in the functional capacities of the different communities, which differentiate the cohorts in a highly significant way (F2,149 = 2.1353, P = .0002, R2 = 0.02784, adj. R2 = 0.01479; P = .0001 [conditional dbRDA]; Figure 3C), but functional diversity is decreased in UC patients (Figure 3D). Disease subtypes of CD and UC patients did not show any significant influence on the taxonomic or functional community composition, with the exception of weak differences in community composition associated with disease location in CD (subtype L; Table S8 and S9). Neither UC nor CD subtypes influenced the microbial community variability in any significant manner (Table S10). Noteably, functional and taxonomic community differences correlated to disease severity (SCCAI) in UC. Among individuals suffering from CD, we only observed nominally significant correlations with subject age (Bray-Curtis) and the average time spent outside (sunlight hours; Jaccard and Jaccard PICRUSt; Table S8 and S9), but no effect of other patient characteristics. In communities of UC patients, as well as in CD and UC patients combined, we could discern weak community differences between patients that will or will not receive IBD-specific medication, particularly prednisolone (CD, Bray-Curtis: P = .0395, PFDR = 0.1975; Jaccard: P = .0266, PFDR = 0.1330; IBD, Bray-Curtis: P = .0121, PFDR = 0.0968; Jaccard: P = .0041, PFDR = 0.0328). However in CD patients, no such associations were detected.
Table 3.
Beta diversity analyses of fecal microbial communities focusing on differential abundance (Bray-Curtis) or differential presence (Jaccard) of bacterial ASVs, as well as differential presence of imputed functions (Jaccard, EC categories based on PICRUSt2), with respect to the main anthropometric characteristics (IBD condition, gender, smoking status).
| Distance | Main factor | Pairwise | DF | F | P | P FDR | R 2 | adj. R2 |
|---|---|---|---|---|---|---|---|---|
| Bray-Curtis | IBD | 2,149 | 1.25099 | 0.00010 | 0.00024 | 0.01651 | 0.00331 | |
| (ASV based) | Contr.-CD | 1,125 | 1.36280 | 0.00010 | 0.00024 | 0.01078 | 0.00287 | |
| Contr.-UC | 1,119 | 1.25171 | 0.00040 | 0.00080 | 0.01041 | 0.00209 | ||
| CD-UC | 1,54 | 1.00161 | 0.43886 | 0.45955 | 0.01821 | 0.00003 | ||
| Sex | 1,150 | 1.00042 | 0.45955 | 0.45955 | 0.00663 | 0.00000 | ||
| Smoking | 2,149 | 1.04960 | 0.10829 | 0.14439 | 0.01389 | 0.00066 | ||
| Jaccard | IBD | 2,149 | 1.23488 | 0.00010 | 0.00024 | 0.01631 | 0.00310 | |
| (ASV based) | Contr.-CD | 1,125 | 1.33672 | 0.00010 | 0.00024 | 0.01058 | 0.00267 | |
| Contr.-UC | 1,119 | 1.22098 | 0.00010 | 0.00024 | 0.01016 | 0.00184 | ||
| CD-UC | 1,54 | 1.03744 | 0.07189 | 0.10784 | 0.01885 | 0.00068 | ||
| Sex | 1,150 | 1.05555 | 0.02330 | 0.03994 | 0.00699 | 0.00037 | ||
| Smoking | 2,149 | 1.01414 | 0.18588 | 0.22306 | 0.01343 | 0.00019 | ||
| Jaccard | IBD | 2,149 | 2.13380 | 0.00010 | 0.00030 | 0.02784 | 0.01479 | |
| (function based) | Contr.-CD | 1,125 | 2.14675 | 0.00050 | 0.00100 | 0.01688 | 0.00902 | |
| Contr.-UC | 1,119 | 2.50453 | 0.00010 | 0.00030 | 0.02061 | 0.01238 | ||
| CD-UC | 1,54 | 1.41523 | 0.05239 | 0.07859 | 0.02554 | 0.00749 | ||
| Sex | 1,150 | 1.03485 | 0.36386 | 0.36386 | 0.00685 | 0.00023 | ||
| Smoking | 2,149 | 1.04512 | 0.33287 | 0.36386 | 0.01383 | 0.00060 |
Figure 3.
A, Principal Coordinate Analysis (PCoA) of Bray-Curtis dissimilarity with respect to IBD status including the nominal significant correlation of subject ages with community dissimilarity. The control cohort is significantly separated from the CD and UC cohorts, whereas CD and UC patient groups do not differ significantly (Table 3). B, The boxplot visualizes the community variability within each group measured as the distance to the centroid within each group, revealing significantly higher community variability among IBD individuals than among control individuals (see Table S8; *P ≤ .05, **P ≤ .01, ***P ≤ .001). C, The PCoA displays functional community differences based on differential presence (Jaccard distance) of PICRUSt2 imputed functions among individuals (F1,149 = 2.1338, P = .0009, adj. R2 = 0.0148, PERMANOVA; EC/Enzyme Commission numbers). D, Functional alpha diversity (number of ECs) is most significantly decreased in UC patients as compared with healthy individuals and CD patients (F2,149 = 4.6156, P = .01135). E, The heatmap displays significant (PFDR ≤ .05), differentially abundant MetaCyc34 pathways among cohorts as detected via DESeq2 (Table S12). Columns are ordered by the direction of association/maximum abundance of the pathways (top color bar).35.
Interestingly, investigating the communities for individual functional differences, we could identify several functions and pathways involved in amino acid metabolism, oxidative stress, and other metabolic processes associated with a less anaerobic environment and associated with the diseased cohorts (Figure S9, Table S11 and S12). We found several oxidoreductases (eg, succinate-semialdehyde dehydrogenase, [EC 1.2.1.16]) and other enzymes involved in aerobic metabolism (eg, malonate-semialdehyde dehydrogenase [acetylating], [EC 1.2.1.18]), amino acid degradation (eg, 3-hydroxyisobutyrate dehydrogenase, [EC 1.1.1.31]), and detoxification of reactive oxygen species (eg, glutathione reductase, [EC 1.8.1.7]) to be significantly more abundant in CD patients (Table S11). Functions involved in butyrate degradation and nucleotide degradation appear more abundant in CD patients (EC 2.8.3.12, EC 3.1.21.2). Individuals suffering from ulcerative colitis have a higher abundance of enzymes involved in nucleotide (EC 2.7.1.113, EC 2.7.1.74) and sugar/glycan digestion (EC 2.7.1.66, EC 2.7.1.144, EC 2.7.1.144, EC 3.2.1.85). However, we also observe a higher abundance of several enzymes involved in amino acid biosynthesis, particularly in CD such as glutamate dehydrogenase (EC 1.4.1.3), glycine dehydrogenase (EC 1.4.1.10), or methionine γ-lyase (EC 4.4.1.11). In contrast, healthy individuals display higher abundances of enzymes cobalamin synthesis (precorrin-8X methylmutase/cobalt-precorrin-8 methylmutase EC 5.4.99.60/EC 5.4.99.61) and amino acid synthesis (eg, tryptophan synthase [EC 4.2.1.20], cysteine synthase [EC 2.5.1.47], branched-chain-amino-acid transaminase [EC 2.6.1.42], glutamine synthetase [EC 6.3.1.2], histidine biosynthesis [EC 3.6.1.31, EC 4.2.1.19]). The signals at the single enzyme level are reflected, as well, at the pathway abundance level (Table S12), with amino acid biosynthesis pathways abundant in healthy individuals (eg, L-tryptophan biosynthesis, superpathway of branched chain amino acid biosynthesis/superpathway of aromatic amino acid biosynthesis), and nucleotide and amino acid degradation more abundant among IBD patients (P164-PWY, P162-PWY). Interestingly, we detected an overabundance of a glycerol degradation pathway (GOLPDLCAT-PWY) in CD patients, which is particularly associated with Enterobacteriaceae, which are strong IBD indicators in this study. In association with IBD-related treatments, we could identify an imputed function involved in peptidoglycan synthesis, which can confer resistance to vancomycin (EC 6.1.2.1, D-alanine-[R]-lactate ligase; Deviance1,54 = 18.45282, P = 1.7400 × 10-5, PFDR = .03121).
Network Analyses Reveal Disease-specific Taxon Importance and Strong Network Differences Between Pathologies
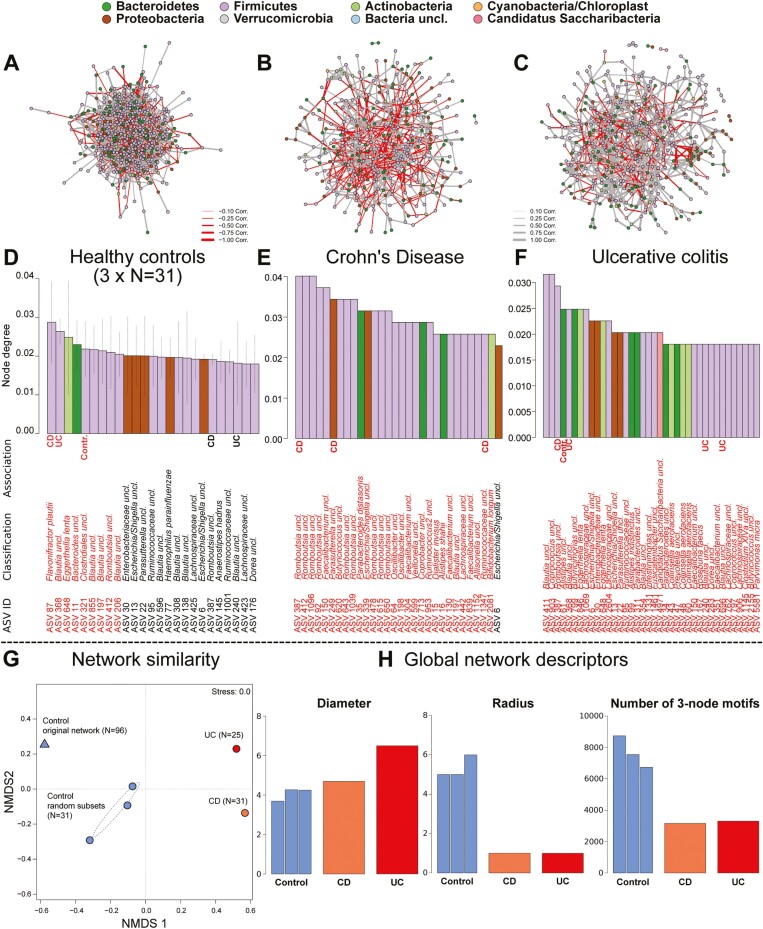
To investigate the bacterial communities as a complex system of interacting agents, whose interactions may change as a cause or consequence of IBD, we employed network analyses of bacterial co-abundance/correlation networks split by the different health conditions (Figure 4A). As the control cohort had a higher sample number, we took 3 equally sized random community subsets and generated networks in parallel (3 × N = 31). Based on the correlation of different small, connected, simple network components across networks, we can estimate their similarity.37,38 It became clear that bacterial correlation networks of diseased and healthy individuals, but also CD and UC patients, differed strongly in composition and topology (Figure 4G). This result implies a large change in community dynamics and interactions in cases of IBD, as well as distinct patterns for the different IBD pathologies in the early stages of disease.
Figure 4.
Displayed are ASV level correlation networks based on SPARCC correlation performed separately in (A) healthy individuals (3 times subsampled to N = 31, for comparability to IBD cohorts), (B) CD patients and (C) UC patients. The node colors indicate phylum membership, and edge color indicates the direction of correlation. Barplots below the networks visualize the relative node importance (node degree) in (D) healthy controls (average of 3 subsets), CD (E), and UC (F). ASVs highlighted in red have a higher than expected importance, based on 10’000 network permutations (Table S13, Figure S11). Additional markups signify the association of a taxon with a health condition. Bacteria marked by CD, UC, or Contr. are indicator species22 for the respective health state and thus highlight the overlap between structurally or ecologically important taxa and their association to healthy or diseased individuals. (G) The Nonmetric Multidimensional Scaling plot (NMDS) visualizes the similarity between the different community networks based on the different health conditions (stress value <0.01). The NMDS shows the healthy network based on the complete healthy cohort (N = 96) and its similarity to the 3 subset healthy networks (N = 31), which themselves are fairly similar to each other. Furthermore networks based on CD (N = 31) and UC (N = 25) communities are structurally as different to each other as they are to networks based on healthy individuals. H, The barplots display global network characteristics for this single networks generated for each health condition, which describe the network diameter (shortest path between most distant nodes), radius (minimum longest paths between any nodes), and diversity and connectedness (number of 3-node motifs) of the networks (additional metrics Figure S10).
Global network characteristics like network diameter, radius, or the number of node-triplets (corresponds to the connectedness and network density) showed large differences between health conditions (Figure 4H). The co-abundance networks in UC also display the highest network diameter but kept a small network radius. This pattern is an indicator of sparsity and strong differentiation of the network, as the average “shortest paths” between nodes become longer, which leads to increased clustering of nodes and subgraphs (eg, transitivity and degree-assortativity increases in UC and CD; Figure 4H, Figure S9A, and S9B). The connectedness in healthy networks was higher, and centralization was lower compared with the pathological states (average node degree constant, betweenness increases), which means that the relative number of connections is constant, whereas their importance/centrality increases in a pathological state (Figure S9C and S9D). This may make communities of individuals with IBD more prone to failure or dysbiosis after community disturbance, as the loss of central players does affect network structure more severely in clustered/centralized networks.39,40
Direct testing of single nodes revealed several ASVs in important and central positions in the different networks, which in part were also identified as indicator- and differentially abundant species as described previously (Figure 4D-4F, Figure S10, Table S13). In total, only 3 ASVs were in significantly prominent positions among all 3 health conditions (ASV 6- Escherichia/Shigella uncl., ASV 30- Enterobacteriaceae uncl., and ASV 387- Romboutsia uncl.). Due to the central roles of these bacteria, they may represent central pillars of the microbial communities irrespective of the disease state. The ASV 6 (Escherichia/Shigella uncl.) was further influenced by a multitude of factors like medication (future and present) and disease subtype (CD, B1, P, L3; UC, E3), thus further implying a dynamic and important role in the diseased gut and an apparent association to inflammatory processes. Romboutsia (eg, ASV 387) is a member of Peptostreptococcaceae (Firmicutes) and was the most frequently detected significant member across networks. Several ASVs classified as Romboutsia (Firmicutes) appeared important specifically and more frequently in CD networks, for which it was also a direct indicator (ASV 387; Figure S10, Table S13). Interestingly, only CD indicators were found among the most important bacteria in CD networks, whereas networks of UC and control individuals did include any type of indicator taxon. Thus, bacteria with a “prominent role” in CD communities may be more disease-specific than bacteria important in a healthy gut community or in cases of UC. Networks of CD and UC communities in general have a higher number and proportion of private significantly important bacteria (CD, n = 48 [13.71%]; UC, n = 31 [6.98%]; Figure S11) than the healthy networks have (NControl = 11 [3.37%]). Only 5 ASVs are repeatedly and exclusively central in networks derived from healthy individuals, like Bacteroides uncl. (ASV 11), Blautia uncl. (ASV 206, ASV 855), Haemophilus parainfluenzae (ASV 77), and Clostridiales uncl. (ASV 321, indicator of health condition), which also overlap with health-associated bacteria reported in Gevers et al.33 Several taxa were almost exclusively important in the disease-specific networks (eg, CD, Parasutterella uncl. [ASV 249] Veillonella uncl. [ASV 599]; UC, Alistipes [ASV 99, ASV 61], Candidatus Saccharibacteria uncl. [ASV 4971]; Table S13).
These strong disparities of taxon importance between networks likely emerge through shifts from previously unimportant positions in a healthy network topology to a central position in the diseased networks. By comparing the positions of bacteria among the networks, we could observe a clear shift of node importance among communities, in particular the significantly important bacteria in the respective networks (Figure S12). The majority of significantly important network members in a healthy network became less important in the case of IBD and vice versa (Figure S12).
Discussion
The etiology of IBD is suspected to be caused by an interaction of genetic susceptibilities and environmental triggers, like increasing urbanization/westernization, in particular dietary alterations, pharmaceutical use, pollution, and other factors.41 However, the microbiota has been identified as one of the central pillars of several diseases, in particular IBD. In this cohort of newly diagnosed and yet treatment-naïve Maltese individuals, we had the opportunity to investigate community dynamics in the early stages of the main IBD pathologies in an otherwise pharmaceutically undisturbed condition.
In the early stages of disease, we only detected small community differences between the disease conditions, which may be the result of the early diagnosis and stages of transition in disease development. However, also in a large pediatric cohort of newly diagnosed and treatment-naïve individuals, only minor community differences between healthy controls and CD patients were detected—particularly among the fecal microbial communities.33 Smaller cohorts do show significant differences in community diversity and composition between healthy and treatment-naïve IBD patients, similar to the patterns we have detected.42,43 However, microbial community differences between IBD pathologies have rarely been investigated in untreated individuals in the early stages of disease development. Disturbances induced by active inflammatory processes and the flare-like nature of IBD may not manifest immediately and consistently in the microbial community early in the diseases’ progression and may yet be too similar between UC and CD in the early stages of the diseases. The high community variation we detected in CD and UC patients compared with healthy controls further implies ongoing community turnover of the fecal microbial communities in the diseased cohorts. A similar pattern has recently been shown in a cohort of healthy and diseased twins, which showed increasing taxonomic and functional variability from healthy, UC, and CD individuals.44 Patterns like these have been identified as signs of chaotic community behavior induced by environmental stressors, the so-called “Anna Karenina” pattern, in which “all happy families look alike; each unhappy family is unhappy in its own way”.36 In other words, this observation implies that at least in these early stages of intestinal inflammation, communities react in a stochastic way, making diseased communities more variable than communities present in healthy individuals. These dynamics increase community differences between individuals in an unstructured way, which in turn reduces the distinctiveness of the communities between CD and UC due to stochastic changes. In the early stages of inflammation, “chaotic” dynamics are displayed in the co-abundance networks, as well. Furthermore, the diseased networks are as dissimilar to each other as they are to the healthy controls. This implies large changes in community dynamics and associations, while community composition changes less systematically between disease conditions. Network characteristics (eg, sparsity and network differentiation in UC/CD, increased clustering in UC/CD) are indicators for communities more prone to failure or “dysbiosis” in the IBD specific networks, as the loss of central players will affect community integrity more strongly and leads to a faster community collapse.39,40 In addition to those topological differences, we can observe a radical turnover of the positional importance of community members between healthy and diseased communities. Thus, bacteria less important in healthy communities (ie, less central) increase in importance and change their associations in the diseased state, potentially due to the emergence of new niches in the gut environment due to inflammation. This dynamic also appears to be present when we compare bacterial network importance between CD and UC.
Geographic or population differences can influence community signatures in general but also in the context of IBD and, therefore, may explain the deviation of the Maltese population from previous studies.12,45 However, the study of distinct human populations also reveals novel patterns of diversity and community dynamics in health46 and disease.12 The population of Malta further adds to the diversity of the global human-associated microbiome with its unique combination of high development, geographic, cultural, and genetic distinctness, and its current underrepresentation in microbiome studies.11 Thus, some single taxon associations are not in correspondence with previous findings42,43 and may be attributable to a variety of factors (eg, geography, recruitment, sampling procedure, technical aspects). For example, Romboutsia spp. has not yet been described in association with IBD outside of our study but was shown to be less abundant in cancerous gut mucosa47 or associated with renal disease.48 This genus is well adapted to the intestinal ecosystem and possesses bile acid–modifying and host glycan–associated pathways while being dependent on external sources of amino acids, and thus being in a central position among health conditions.49
The low abundance of Akkermansia in our CD and UC patients supports previous observations and may further underline the universal importance of A. muciniphilia for host wellbeing.29Akkermansia has been documented to be decreased in IBD patients in other studies.50 Amplicon sequence variants belonging to the Faecalibacteria, a well-known beneficial taxon,51 were mainly central in the CD and UC communities and are indicators for healthy controls and UC patients in our study. This variation with respect to its IBD associations has also been reported in other studies43 and attributed to different F. prausnitzii phylotypes able to temporarily multiply in an inflamed, micro-aerobic gut environment.52Faecalibacterium prausnitzii has been associated with an increased response rate to anti-TNF treatment,53 as well as a reduced risk of disease relapse after cessation of anti-inflammatory treatments (anti-TNF),54 just as other taxa influence IBD treatment efficacy.55 However, Faecalibacteria (ie, ASV 304) are associated with a reduced risk of anti-inflammatory treatments in diseased individuals, which may be the positive effect of butyrate production by this taxon, particularly as we could find a higher abundance of functions involved in butyrate consumption in CD patients.51 In contrast, we observed a strong association of Escherichia/Shigella abundance and anti-TNF treatment in CD patients, which speaks for their potentially negative role IBD. Despite their central position across health conditions in this study, Escherichia/Shigella or Enterobacteriaceae are often found to be pathological/detrimental community members and associated with inflammation.5,33,56 This is further underlined by an increased abundance of functions involved in glycerol degradation (GOLPDLCAT-PWY, PWY-7013), often found in Proteobacteria, particularly Enterobacteriacea.57
In general, we found strong overlap with prominent IBD-associated functions. In congruence with prominent findings by Franzosa et al 2019,58 we identified several functions more abundant in IBD patients, for example glutathione reductase (EC 1.8.1.7), an enzyme involved in oxidative stress resistance, or ethanolamine ammonia-lyase (EC 4.3.1.7), an enzyme responsible for crucial steps in glycerophospholipid synthesis. Also, the IBD-specific signal of Mg2+-importing ATPase (EC 3.6.3.2) could be replicated in this CD cohort,58 which might facilitate magnesium deficiency in IBD patients.59 An additional prominent feature we could identify is a pattern of functions involved in amino acid degradation, althouth we saw an increase of functions associated with amino acid biosynthesis in healthy individuals as shown before.60 Particularly interesting is the comparative lack of tryptophan biosynthesis in CD patients, which may lead to lower uptake by the gut epithelia. Crohn’s disease patients and animal models of colitis were shown to have lowered levels of tryptophan and its metabolites in the fecal and serum metabolome.61,62 Furthermore, the microbiome can directly influence tryptophan uptake and increase the propensity of intestinal inflammation.63
In our treatment-naïve IBD cohort, we identified an association of increasing community diversity with decreasing probability of future anti-inflammatory treatments. In general, declining bacterial diversity has been linked previously to active IBD, and factors decreasing diversity are strong risk factors for IBD development and disease aggravation or escalating dysbiosis.64,65 In addition, diversity-restoring procedures like fecal matter transplants (FMTs) appear to have some success in treating IBD.65 Low community diversity appears to predetermine treatment with anti-inflammatory medication, as observed in other cohorts before64,65 also in our study. Thus, diversity or even single bacterial taxa can be indicative of disease progression and to some extent treatment options like dietary or pharmaceutical interventions (eg, tryptophan supplementation)62,66 and may support already well-established or experimental sets of physiological and genetic markers (eg, C-reactive protein, fecal calprotectin, NOD2).67 However, previous studies showed only limited benefit of microbial variables for disease progress prediction compared with other biomarkers and will demand larger cohorts to validate and evaluate their efficacy.68
Supplementary Material
Acknowledgments
Authors would like to thank Ilona Urbach, Ines Wulf, and Tonio Hauptmann of the IKMB microbiome laboratory and the staff of the IKMB sequencing facilities for their excellent technical support. This study was funded by the University of Malta and supported by the Deutsche Forschungsgemeinschaft (DFG) Research Unit 5042 (“miTarget-The Microbiome as a Therapeutic Target in Inflammatory Bowel Diseases”).
Contributor Information
Philipp Rausch, Institute of Clinical Molecular Biology, Christian-Albrechts-University of Kiel, Kiel, Germany; Laboratory of Genomics and Molecular Biomedicine, Department of Biology, University of Copenhagen, Copenhagen, Denmark.
Sarah Ellul, Division of Pediatric Surgery, Department of Surgery, Mater Dei Hospital, Malta.
Anthea Pisani, Division of Gastroenterology, Department of Medicine, Mater Dei Hospital, Malta.
Corinna Bang, Institute of Clinical Molecular Biology, Christian-Albrechts-University of Kiel, Kiel, Germany.
Trevor Tabone, Division of Gastroenterology, Department of Medicine, Mater Dei Hospital, Malta.
Claire Marantidis Cordina, Department of Microbiology, Mater Dei Hospital, Malta.
Graziella Zahra, Molecular Diagnostics, Department of Pathology, Mater Dei Hospital, Malta.
Andre Franke, Institute of Clinical Molecular Biology, Christian-Albrechts-University of Kiel, Kiel, Germany.
Pierre Ellul, Division of Gastroenterology, Department of Medicine, Mater Dei Hospital, Malta.
Author Contributions
S.E., P.E., and A.F. designed the research. S.E., P.R., P.E., C.B., A.P., T.T., C.C.M., and G.Z. performed research and generated data. P.R. analyzed the data. P.R. wrote the manuscript. P.R., C.B., and P.E. edited the drafts.
Funding
This study was funded by the University of Malta and supported by the Deutsche Forschungsgemeinschaft (DFG) Research Unit 5042 (“miTarget-The Microbiome as a Therapeutic Target in Inflammatory Bowel Diseases”).
Conflicts of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
- 1. Jostins L, Ripke S, Weersma RK, et al. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease Nature. 2012;491(7422):119-124. doi: 10.1038/nature11582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Gordon H, Trier Moller F, Andersen V, Harbord M.. Heritability in inflammatory bowel disease: from the first twin study to genome-wide association studies. Inflamm Bowel Dis. 2015;21(6):1428-1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Thia KT, Loftus JEV, Sandborn WJ, Yang S-K.. An update on the epidemiology of inflammatory bowel disease in Asia. Am J Gastroenterol. Comparative Study Review. 2008;103(12):3167-3182. [DOI] [PubMed] [Google Scholar]
- 4. Lloyd-Price J, Arze C, Ananthakrishnan AN, et al. Multi-omics of the gut microbial ecosystem in inflammatory bowel diseases. Nature. 2019;569(7758):655-662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Mancabelli L, Milani C, Lugli GA, et al. Identification of universal gut microbial biomarkers of common human intestinal diseases by meta-analysis. FEMS Microbiol Ecol. 2017;93(12):1-10. [DOI] [PubMed] [Google Scholar]
- 6. Rausch P, Rehman A, Künzel S, et al. Colonic mucosa-associated microbiota is influenced by an interaction of Crohn disease and FUT2 (Secretor) genotype. Proc Natl Acad Sci. Comparative Study Research Support, Non-U.S. Gov’t. 2011;108(47):19030-19035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lewis James D, Chen Eric Z, Baldassano Robert N, et al. Inflammation, antibiotics, and diet as environmental stressors of the gut microbiome in pediatric Crohn’s disease. Cell Host and Microbe. 2015;18(4):489-500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Rodríguez LAG, Ruigómez A, Panés J.. Acute gastroenteritis is followed by an increased risk of inflammatory bowel disease. Gastroenterology. 2006;130(6):1588-1594. [DOI] [PubMed] [Google Scholar]
- 9. Schembri J, Pace N, Vella S, et al. P822 genome-wide association study (GWAS) of a Maltese inflammatory bowel disease cohort. J Crohn’s Colitis. 2019;13(Supplement_1):S534-S534. [Google Scholar]
- 10. Schembri J, Pace N, Degenhardt F, Franke A, Ellul P.. P825 Low-prevalence of NOD2 polymorphisms in a Maltese IBD cohort. J Crohn’s Colitis. 2019;13(Supplement_1):S536-S536. [Google Scholar]
- 11. Capelli C, Redhead N, Romano V, et al. Population structure in the Mediterranean basin: a Y chromosome perspective. Ann Hum Genet. 2006;70(Pt 2):207-225. [DOI] [PubMed] [Google Scholar]
- 12. Rehman A, Rausch P, Wang J, et al. Geographical patterns of the standing and active human gut microbiome in health and IBD. Gut. 2015;65(2):238-248. [DOI] [PubMed] [Google Scholar]
- 13. Trautmann T, Bang C, Franke A, Vincent D, Reinshagen K, Boettcher M.. The impact of oral sodium chloride supplementation on thrive and the intestinal microbiome in neonates with small bowel ostomies: a prospective cohort study. Front Immunol. 2020;11:1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJA, Holmes SP.. DADA2: High-resolution sample inference from Illumina amplicon data. Nat Meth. Brief Communication. 2016;13(7):581-583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Cole JR, Wang Q, Cardenas E, et al. The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucl Acids Res. 2009;37(suppl_1):D141-D145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Douglas GM, Maffei VJ, Zaneveld JR, et al. PICRUSt2 for prediction of metagenome functions. Nat Biotechnol. 2020;38(6):685-688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Oksanen J, Blanchet FG, Kindt R, et al. vegan: Community Ecology Package. http://CRAN.R-project.org, 2011
- 18. Kembel SW, Cowan PD, Helmus MR, et al. Picante: R tools for integrating phylogenies and ecology. Bioinformatics. Research Support, Non-U.S. Gov’t Research Support, U.S. Gov’t, Non-P.H.S. 2010;26(11):1463-1464. [DOI] [PubMed] [Google Scholar]
- 19. Turley P, Walters RK, Maghzian O, et al. Multi-trait analysis of genome-wide association summary statistics using MTAG. Nat Genet. 2018;50(2):229-237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Legendre P, Anderson MJ.. Distance-based redundancy analysis: testing multispecies responses in multifactorial ecological experiments. Ecol Monogr. 1999;69(1):1-24. [Google Scholar]
- 21. Anderson MJ, Ellingsen KE, McArdle BH.. Multivariate dispersion as a measure of beta diversity. Ecology Lett. 2006;9(6):683-693. [DOI] [PubMed] [Google Scholar]
- 22. De Cáceres M, Legendre P.. Associations between species and groups of sites: indices and statistical inference. Ecology. 2009;90(12):3566-3574. [DOI] [PubMed] [Google Scholar]
- 23. Love MI, Huber W, Anders S.. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Friedman J, Alm EJ.. Inferring Correlation Networks from Genomic Survey Data. PLoS Comput Biol. 2012;8(9):e1002687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Brin S, Page L.. The anatomy of a large-scale hypertextual web search engine. Comput Netw ISDN Syst. 1998;30(1-7):107-117. [Google Scholar]
- 26. Freeman LC. Centrality in social networks conceptual clarification. Social Networks. 1979;1(3):215-239. [Google Scholar]
- 27. Csardi G, Nepusz T.. The igraph software package for complex network research. InterJournal. 2006;Complex Systems:1695. [Google Scholar]
- 28. Derrien M, Vaughan EE, Plugge CM, de Vos WM.. Akkermansia muciniphila gen. nov., sp. nov., a human intestinal mucin-degrading bacterium. Int J Syst Evol Microbiol. 2004;54(5):1469-1476. [DOI] [PubMed] [Google Scholar]
- 29. Cani PD, de Vos WM.. Next-generation beneficial microbes: the case of akkermansia muciniphila. Front Microbiol. 2017;8:1765-1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Parker BJ, Wearsch PA, Veloo ACM, Rodriguez-Palacios A.. The Genus Alistipes: Gut Bacteria With Emerging Implications to Inflammation, Cancer, and Mental Health. Front Immunol. Review. 2020;11(906). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Liang J, Zhou M, Tobin PC, McGuire AD, Reich PB.. Biodiversity influences plant productivity through niche–efficiency. Proc Natl Acad Sci USA. 2015;112(18):5738-5743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Silverberg MS, Satsangi J, Ahmad T, et al. Toward an integrated clinical, molecular and serological classification of inflammatory bowel disease: report of a working party of the 2005 Montreal world congress of gastroenterology. Can J Gastroenterol. 2005;19(Suppl A):5A269076-5A26936A. [DOI] [PubMed] [Google Scholar]
- 33. Gevers D, Kugathasan S, Denson Lee A, et al. The treatment-naive microbiome in new-onset Crohn s disease. Cell Host and Microbe. 2014;15(3):382-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Karp PD, Riley M, Paley SM, Pellegrini-Toole A.. The MetaCyc Database. Nucleic Acids Res. 2002;30(1):59-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ward JH. Hierarchical grouping to optimize an objective function. J Am Stat Assoc. 1963;58(301):236-244. [Google Scholar]
- 36. Zaneveld JR, McMinds R, Vega Thurber R.. Stress and stability: applying the Anna Karenina principle to animal microbiomes. Nat Microbiol. 2017;2(9):17121. [DOI] [PubMed] [Google Scholar]
- 37. Hayes W, Sun K, Pržulj N.. Graphlet-based measures are suitable for biological network comparison. Bioinformatics. 2013;29(4):483-491. [DOI] [PubMed] [Google Scholar]
- 38. Hočevar T, Demšar J.. Computation of Graphlet Orbits for Nodes and Edges in Sparse Graphs. J Stat Softw. network analysis; graphlets; data mining; bioinformatics. 2016;71(10):24. [Google Scholar]
- 39. Albert R, Jeong H, Barabasi A-L.. Error and attack tolerance of complex networks. Nature. 2000;406(6794):378-382. doi: 10.1038/35019019 [DOI] [PubMed] [Google Scholar]
- 40. Jeong H, Mason SP, Barabasi AL, Oltvai ZN.. Lethality and centrality in protein networks. Nature. 2001;411(6833):41-42. doi: 10.1038/35075138 [DOI] [PubMed] [Google Scholar]
- 41. Bach J-F. The Effect of infections on susceptibility to autoimmune and allergic diseases. N Engl J Med. Research Support, Non-U.S. Gov’t Review. 2002;347(12):911-920. [DOI] [PubMed] [Google Scholar]
- 42. Kowalska-Duplaga K, Gosiewski T, Kapusta P, et al. Differences in the intestinal microbiome of healthy children and patients with newly diagnosed Crohn’s disease. Sci Rep. 2019;9(1):18880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Assa A, Butcher J, Li J, et al. Mucosa-Associated ileal microbiota in new-onset pediatric Crohn’s disease. Inflamm Bowel Dis. 2016;22(7):1533-1539. [DOI] [PubMed] [Google Scholar]
- 44. Brand EC, Klaassen MAY, Gacesa R, et al. Healthy cotwins share gut microbiome signatures with their inflammatory bowel disease twins and unrelated patients. Gastroenterology. 2021;160(6):1970-1985. [DOI] [PubMed] [Google Scholar]
- 45. Lazaridis I, Patterson N, Mittnik A, et al. Ancient human genomes suggest three ancestral populations for present-day Europeans. Nature. 2014;513(7518):409-413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Smits SA, Leach J, Sonnenburg ED, et al. Seasonal cycling in the gut microbiome of the Hadza hunter-gatherers of Tanzania. Science. 2017;357(6353):802-806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Mangifesta M, Mancabelli L, Milani C, et al. Mucosal microbiota of intestinal polyps reveals putative biomarkers of colorectal cancer. Sci Rep. 2018;8(1):13974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kang Y, Feng D, Law HK-w, et al. Compositional alterations of gut microbiota in children with primary nephrotic syndrome after initial therapy. BMC Nephrol. 2019;20(1):434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Gerritsen J, Hornung B, Renckens B, et al. Genomic and functional analysis of Romboutsia ilealis CRIB(T) reveals adaptation to the small intestine. PeerJ. 2017;5(9):1-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Rajilić-Stojanović M, Shanahan F, Guarner F, de Vos WM.. Phylogenetic analysis of dysbiosis in ulcerative colitis during remission. Inflamm Bowel Dis. 2013;19(3):481-488. [DOI] [PubMed] [Google Scholar]
- 51. Sokol H, Pigneur B, Watterlot L, et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc Natl Acad Sci. Randomized Controlled Trial Research Support, Non-U.S. Gov’t. 2008;105(43):16731-16736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Lopez-Siles M, Martinez-Medina M, Surís-Valls R, et al. Changes in the abundance of Faecalibacterium prausnitzii phylogroups i and ii in the intestinal mucosa of inflammatory bowel disease and patients with colorectal cancer. Inflamm Bowel Dis. 2016;22(1):28-41. [DOI] [PubMed] [Google Scholar]
- 53. Magnusson MK, Strid H, Sapnara M, et al. Anti-TNF therapy response in patients with ulcerative colitis is associated with colonic antimicrobial peptide expression and microbiota composition. J Crohn’s Colitis. 2016;10(8):943-952. [DOI] [PubMed] [Google Scholar]
- 54. Rajca S, Grondin V, Louis E, et al. Alterations in the intestinal microbiome (dysbiosis) as a predictor of relapse after infliximab withdrawal in Crohn’s disease. Inflamm Bowel Dis. 2014;20(6):978-986. [DOI] [PubMed] [Google Scholar]
- 55. Zhou Y, Xu ZZ, He Y, et al. Gut microbiota offers universal biomarkers across ethnicity in inflammatory bowel disease diagnosis and infliximab response prediction. mSystems. 2018;3(1):e00188-e00117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Darfeuille-Michaud A, Neut C, Barnich N, et al. Presence of adherent Escherichia coli strains in ileal mucosa of patients with Crohn’s disease. Gastroenterology. 1998;115(6):1405-1413. [DOI] [PubMed] [Google Scholar]
- 57. Bobik TA, Ailion M, Roth JR.. A single regulatory gene integrates control of vitamin B12 synthesis and propanediol degradation. J Bacteriol. 1992;174(7):2253-2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Franzosa EA, Sirota-Madi A, Avila-Pacheco J, et al. Gut microbiome structure and metabolic activity in inflammatory bowel disease. Nat Microbiol. 2019;4(2):898293-898898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Galland L. Magnesium and inflammatory bowel disease. Magnesium. 1988;7(2):78-83. [PubMed] [Google Scholar]
- 60. Morgan XC, Tickle TL, Sokol H, et al. Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment. Genome Biol. 2012;13(9):R79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Kolho KL, Pessia A, Jaakkola T, de Vos WM, Velagapudi V.. Faecal and serum metabolomics in paediatric inflammatory bowel disease. J Crohn’s Colitis. 2017;11(3):321-334. [DOI] [PubMed] [Google Scholar]
- 62. Nikolaus S, Schulte B, Al-Massad N, et al. Increased tryptophan metabolism is associated with activity of inflammatory bowel diseases. Gastroenterology. 2017;153(6):1504-1516.e2. [DOI] [PubMed] [Google Scholar]
- 63. Hashimoto T, Perlot T, Rehman A, et al. ACE2 links amino acid malnutrition to microbial ecology and intestinal inflammation. Nature . 2012;487(7408):477-481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Clooney AG, Eckenberger J, Laserna-Mendieta E, et al. Ranking microbiome variance in inflammatory bowel disease: a large longitudinal intercontinental study. Gut. 2021;70(3):499-510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Gong D, Gong X, Wang L, Yu X, Dong Q.. Involvement of reduced microbial diversity in inflammatory bowel disease. Gastroenterol Res Pract. 2016;2016:6951091-6951091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Makki K, Deehan EC, Walter J, Bäckhed F.. The impact of dietary fiber on gut microbiota in host health and disease. Cell Host and Microbe. 2018;23(6):705-715. [DOI] [PubMed] [Google Scholar]
- 67. Argmann C, Hou R, Ungaro RC, et al. Biopsy and blood-based molecular biomarker of inflammation in IBD. Gut. 2022:1-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Kugathasan S, Denson LA, Walters TD, et al. Prediction of complicated disease course for children newly diagnosed with Crohn’s disease: a multicentre inception cohort study. Lancet. 2017;389(10080):1710-1718. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.