Abstract
Background
There is an unmet medical need for biomarkers that capture host and environmental contributions in inflammatory bowel diseases (IBDs). This study aimed at testing the potential of circulating lipids as disease classifiers given their major roles in inflammation.
Methods
We applied a previously validated comprehensive high-resolution liquid chromatography-mass spectrometry–based untargeted lipidomic workflow covering 25 lipid subclasses to serum samples from 100 Crohn’s disease (CD) patients and 100 matched control subjects. Findings were replicated and expanded in another 200 CD patients and 200 control subjects. Key metabolites were tested for associations with disease behavior and location, and classification models were built and validated. Their association with disease activity was tested using an independent cohort of 42 CD patients.
Results
We identified >70 metabolites with strong association (P < 1 × 10-4, q < 5 × 10-4) to CD. Highly performing classification models (area under the curve > 0.84-0.97) could be built with as few as 5 to 9 different metabolites, representing 6 major correlated lipid clusters. These classifiers included a phosphatidylethanolamine ether (O-16:0/20:4), a sphingomyelin (d18:1/21:0) and a cholesterol ester (14:1), a very long-chain dicarboxylic acid [28:1(OH)] and sitosterol sulfate. These classifiers and correlated lipids indicate a dysregulated metabolism in host cells, notably in peroxisomes, as well as dysbiosis, oxidative stress, compromised inflammation resolution, or intestinal membrane integrity. A subset of these were associated with disease behavior or location.
Conclusions
Untargeted lipidomic analyses uncovered perturbations in the circulating human CD lipidome, likely resulting from multiple pathogenic mechanisms. Models using as few as 5 biomarkers had strong disease classifier characteristics, supporting their potential use in diagnosis or prognosis.
Keywords: comprehensive untargeted lipidomics, lipid biomarkers, subtype stratification, Crohn’s disease
Key messages.
What is already known?
It has previously been shown that Crohn’s disease has systemic impacts that can be detected in the circulation.
What is new here?
Testing of >1500 lipids, followed by mass spectrometry–based identification of Crohn’s disease–associated metabolites, enabled the discovery of biomarkers of dysregulated metabolism, dysbiosis, oxidative stress, compromised inflammation resolution, and intestinal membrane integrity that not only informed about the biology impacted by disease, but also formed the basis of powerful disease classification models.
How can this study help patient care?
This work provides the groundwork for the development of serum-based predictive tools to aid diagnosis and prognosis.
Introduction
Inflammatory bowel diseases (IBDs), namely Crohn’s disease (CD) and ulcerative colitis (UC), are becoming major public health concerns, due to their dramatic expansion worldwide, lack of a cure, and challenges in disease management. To date, more than 200 genomic loci reflecting a wide variety of biological functions have been found to be associated with susceptibility to these complex multifactorial diseases.1 However, these diseases exhibit heterogeneity in terms of disease onset, behavior, location, progression, and response to therapy. Currently, there is an unmet need to identify biomarkers that can capture both host and environmental contributions to IBD to improve diagnosis and patient stratification.
Given that circulating lipids play crucial roles in many biological mechanisms relevant to IBD pathophysiology2 and originate from metabolic processes occurring in specific host tissues and cells, gut microbiota, and diet, we hypothesized that the presence of specific circulating lipids could integrate multiple endogenous and exogenous contributions to IBD and therefore represent attractive candidate biomarkers. Lipids represent 75% of all circulating metabolites,3 and their analysis by mass spectrometry (MS)–based lipidomics, a subset of metabolomics, offers a great potential for biomarker discovery through untargeted data acquisition and mining, although the application to large clinical cohorts remains challenging.4 Notably, the ability to adequately resolve and identify the multiple existing lipid isomers is crucial for biological data interpretation, as different lipid subclasses and specific acyl side chains often reflect very different roles. For example, and of specific relevance to IBD pathophysiology, omega-6 vs omega-3 polyunsaturated fatty acids (PUFAs)—particularly in glycerophospholipids—distinguish between reservoirs of pro- and anti-inflammatory precursors,5 very long-chain fatty acids (VLCFAs), but not long-chain fatty acids (LCFAs), in sphingolipids impact colon barrier function and epithelial integrity,6,7 while even- vs odd-chain fatty acids (OCFAs) in all lipids may discriminate between host vs microbiome metabolism.8
Hence, we hypothesized that the use of a thoroughly validated comprehensive untargeted lipidomic workflow with large, well-powered cohorts of CD patients should enable the identification of subsets of various and functionally distinct circulating lipids associated with disease behavior or location. In this study, we applied a robust liquid chromatography-mass spectrometry (LC-MS)–based untargeted lipidomic workflow9 that enables measurement of >1500 high-quality MS signals, of which 509 structurally unique lipid species covering 25 lipid subclasses, including their acyl side chains, have been annotated to date by MS/MS analyses. Median inter- and intra-assay coefficients of variations (or relative standard deviation) in MS signal intensity for the resulting annotated lipids meet the criteria of the Food and Drug Administration (ie, >85% with relative standard deviation <20%), thereby enabling robust semi-quantification. First, we applied this workflow to 300 patients with CD patients and 300 healthy subjects, matched for sex, age, and ethnicity, collected by the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) IBD Genetics Consortium (IBDGC). Next we tested an independent set of 42 CD patients to explore the association of these key serum lipid metabolites with disease activity. Our analyses identified candidate circulating lipid biomarkers that are strongly associated with CD disease classifiers, with many being associated with disease location or disease behavior.
Methods
Human serum samples and study design
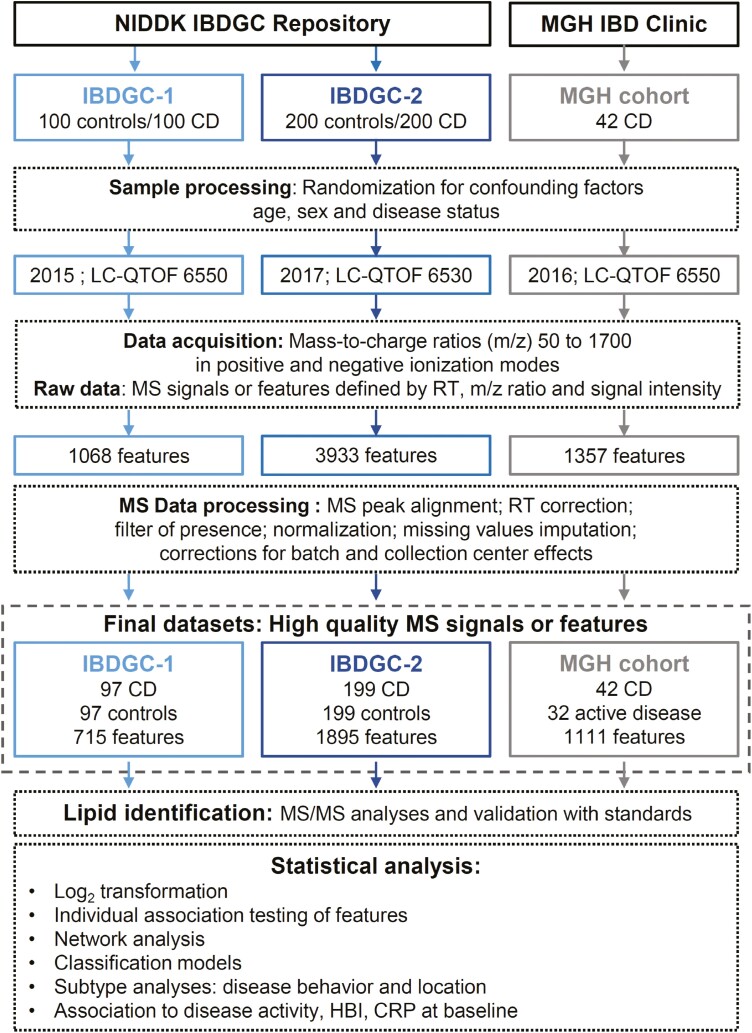
An overview of the current study is shown in Figure 1. First, serum samples from nonfasting CD patients and healthy donors matched for sex, age and ethnicity, collected by the 6 genetic research centers of the IBDGC (www.ibdgc.org) following a standardized protocol using one 10 mL nonadditive red top vacutainer tube (OR; Beckon Dickinson), allowing 30 minutes at room temperature for clotting prior to centrifugation, aliquoting of 125 µL of serum into 8 separate cryovials and storage at -80 °C within a maximum time interval between sample collection and freezing of 2-4 hours. Serum samples were processed in 2 separate phases, referred to as IBDGC-1 (100 CD patients and 100 control subjects) and IBDGC-2 (200 CD patients and 200 control subjects) as previously described.10 In the first phase (IBDGC-1), we selected patients with a more complicated disease behavior, namely stricturing (B2) and penetrating (B3) CD, according to the Montreal classification,11 while for the second phase (IBDGC-2), patients had a more representative proportion of disease behavior (including inflammatory CD; B1). Second, serum samples from 42 patients with moderate-to-severe CD recruited at the Massachusetts General Hospital were obtained from a previously described multicenter cohort.12 These serum samples were collected immediately prior to commencing vedolizumab therapy (week 0).
Figure 1.
Flow diagram of the study. The diagram depicts the analytical aspects of the study, in which the study design and the untargeted lipidomic screen are illustrated. First, lipid features were retrospectively measured in serum samples from 300 Crohn’s disease (CD) patients and 300 control subjects, on different liquid chromatography–quadrupole time-of-flight (LC-QTOF) instruments (6550 and 6530; Agilent Technologies Inc), in a 1.5-year interval (from October 7 to November 11, 2015, and from April 2 to May 31, 2017), as 2 independent phases referred as IBDGC-1 and -2. Mass spectrometry (MS) raw data were processed for peak picking and an in-house bioinformatic script encoded in both Perl and R languages for MS peak alignment, retention time (RT) correction, filter of presence, normalization of signal intensities using cyclic loess algorithm, imputation of missing values using k-nearest neighbors (setting k = 5) on scaled data and batch and collection center effect correction using Combat algorithm. The final National Institute of Diabetes and Digestive and Kidney Diseases IBD Genetics Consortium (IBDGC) datasets were analyzed using 3 approaches (individual testing, network analysis, and classification models), and MS/MS was performed on features associated with stricturing or penetrating CD patients vs control subjects with P < .05. Second, lipid features were measured in the serum of 42 CD subjects from July 19 to August 20, 2016, using the LC-QTOF 6550. Following processing of MS raw data, the MGH cohort final dataset was analyzed using individual testing. CRP, C-reactive protein; HBI, Harvey-Bradshaw Index; m/z, mass to charge.
Untargeted lipidomic screen using LC-MS
Three sets of serum samples were obtained as frozen aliquots (-80°C) that had not been previously thawed10 and analyzed using a previously validated semi-quantitative untargeted lipidomic workflow9 on different high-resolution LC-MS (LC–quadrupole-time-of-flight [LC-QTOF] 6550 and 6530; Agilent Technologies Inc) in a 1.5-year interval as 3 independent studies, hereafter referred as IBDGC-1, IBDGC-2, and MGH (Figure 1). Following recommended guidelines,4 stratified randomization of samples was achieved according to potential confounding factors, namely age, sex, and disease status, as well as treatment whenever applicable, in order to minimize batch-dependent bias. MS data were acquired in positive and negative modes. MS data quality-control analyses were performed by (1) injecting an “in-house” plasma pool quality-control sample at the beginning, at the end, and every 20 runs; (2) injecting blanks every 20 runs; and (3) monitoring 6 internal standards spiked in samples for signal intensity, mass-to-charge (m/z) ratios, and retention time (RT) accuracies.
Raw MS data were processed as previously described in detail9 using Mass Hunter Qualitative Analysis (version B.06 or B.07; Agilent Technologies Inc) for peak picking and using an in-house bioinformatic script encoded in both Perl and R languages that we developed for the following steps: (1) MS feature peak alignment and RT correction: MS features between chromatographic runs were aligned by selecting features that were present in all samples with no isobars (defined by mass 20 ppm) in a RT window of ±2 minutes, spread over the gradient as references for RT correction prior to MS features alignment; (2) filter of presence: features retained must be present in 80% of samples from at least one group, thereby setting a maximum value for the percentage of missing values for a given feature in any groups at 20%, and have coefficient of interindividual variation <80% among healthy donor samples; (3) normalization of signal intensities using cyclic loess algorithm; (4) imputation of missing values using k-nearest neighbors (KNN; with a setting at k = 5) on scaled data; and (5) batch and collection center effect correction using ComBat algorithm. The resulting final datasets listed high-quality MS signals, thereafter referred to as features, defined by their m/z ratios, RT, and signal intensity.
In a pilot study, the median interindividual MS signal intensity variations in lipid features for the analysis of serum samples from 4 nonfasting control subjects from the NIDDK Repository vs 4 nonfasting control subjects prepared on site using our standard optimal collection protocol was assessed using our lipidomic workflow and found to be similar, namely 44% vs 33% (data not shown), thereby suggesting the very good quality of the repository serum samples. The final IBDGC-1 and -2 datasets missed 6 (3 patients and 3 control subjects) and 2 (1 patient and 1 control) samples, respectively, lost during lipid extraction. The percentage of missing values for all MS features in the final datasets was similar for control subjects and CD patients and is reported in Supplementary Table 1 for IBDGC-1 and Supplementary Tables 2 and 4 for IBDGC-2, respectively; as an example, the median value for control subjects and cases was 1% and 2%, respectively, in the IBDGC-2 dataset (Supplementary Table 2).
Given the untargeted and discovery-based nature of our lipidomic screen, lipid features of interest were then annotated to unique lipids by MS/MS analysis, as previously described in detail.9 This step is crucial, given that about 50% of MS features are duplicate ions of the same unique lipid. For this study, the lipid annotation was focused on features that passed the selected threshold of significance for the group comparison of interest. This included all features associated with stricturing or internal penetrating (B2/B3) CD vs control with P values <.05 in IBDGC-1 or -2. The characteristics of MS/MS analyzed features, namely m/z ratios, RT, ionization mode, detected adducts, and MS/MS fragments considered for annotation, as well as their corresponding lipid IDs and category, are reported in Supplementary Tables 1 and 2 for IBDGC-1 and IBDGC-2, respectively. In addition, MS/MS spectra are reported for annotation in IBDGC-2 samples of cholesta-4,6-dien-3-one and the 5 annotated unique lipids that are part of our final classification model as well as for the validation using standards of cholesta-4,6-dien-3-one and sitosterol sulfate (SitS) IDs using available standards purchased from BOC Sciences and kindly provided by Pr Hubert Schaller, respectively (Supplementary Figure 1). All lipid IDs are mentioned in the text as annotated lipid molecular species, irrespective of validation with an analytical standard. Features without ID assumption were classified as unidentified and mentioned using their feature ID, defined as ionization mode:mass@RT.
Statistical analysis
MS data retained in the final IBDGC-1 and IBDGC-2 datasets were analyzed independently. First, individual testing identified features associated with B2/B3 vs control phenotype. Results were compared between the IBDGC phases to assess replicability. Second, the larger IBDGC-2 dataset was selected for correlation network analysis of B2/B3-associated features. Third, individual testing was used to assess association of lipid features with CD subtypes. Fourth, classification models validated the potential of circulating lipids to discriminate between CD patients and control phenotype.
Individual testing :
Independent testing was done on log2-transformed signal intensity for each feature using regression corrected for sex. For annotated features detected in both IBDGC datasets, evidence of association with B2/B3 vs control was compared and pooled using the z score. For this interstudy comparison, we used normalized log2-transformed signal intensity expressed as fold change (FC) for cases vs control subjects and corresponding P values from the final IBDGC-1 and IBDGC-2 datasets. A direct comparison of normalized feature MS absolute signal intensity values in the final datasets cannot be achieved between 2 projects given the semi-quantitative nature of our untargeted lipidomic screen and that MS raw data are normalized for each project independently. Analysis for disease behavior was performed as B2/B3 (stricturing/penetrating) vs inflammatory (B1), conditional on sex and age. Analysis for disease location was performed conditional on sex and age with ileocolonic (L3), considered statistically intermediate between colorectal (L2) and ileal (L1).
Significance thresholds:
The threshold for selecting associated features from individual testing was P <1 × 10-4 for both IBDGC datasets. Multiple testing was accounted for by evaluating false discovery rate using q values (R package qvalue).13
Network analysis:
Positive correlation (r > 0.4) between B2/B3-associated features (P < 1 × 10-4 and │log2(FC)│>0.3) was projected onto a 2-dimensional display using Fruchterman-Reingold layout algorithm (R package igraph).14 A starting point was given to the algorithm as the first 2 principal component analysis coordinates.
Classification models:
A classification model was built from the IBDGC-2 dataset using the 73 features associated with B2/B3 vs control (P < 1 × 10-4 and │log2(FC)│>0.3). The model was built using logistic regression, with a forward-backward parameter selection based on the Bayesian information criterion, as implemented in R (step function). More precisely, we started with the null model (no parameter) and proceeded to select a first regression model based on identified lipids. A second regression model was then built, again with a forward-backward approach but starting with the lipids included in the previous model and allowing any associated feature to enter (or leave) the model. This gave us the final model based on the 73 associated features, with prioritization of annotated lipids. Evaluation of model performances was represented using receiver-operating characteristic (ROC) curves and summarized with area under the curve (AUC). The 95% confidence intervals were computed using bootstrap (R package pROC).15 Given that these performances are expected to be overestimated when the model is built (trained) and validated within the same samples, estimates of out-of-sample AUC are provided in the Supplementary Materials.
Ethics approval and consent to participate
This study was approved by the Institutional Review Board of the Montreal Heart Institute known as the Comité d’éthique de la recherche et du développement des nouvelles technologies.
Results
An untargeted lipidomic screen identified circulating lipid features associated with stricturing or internal penetrating CD
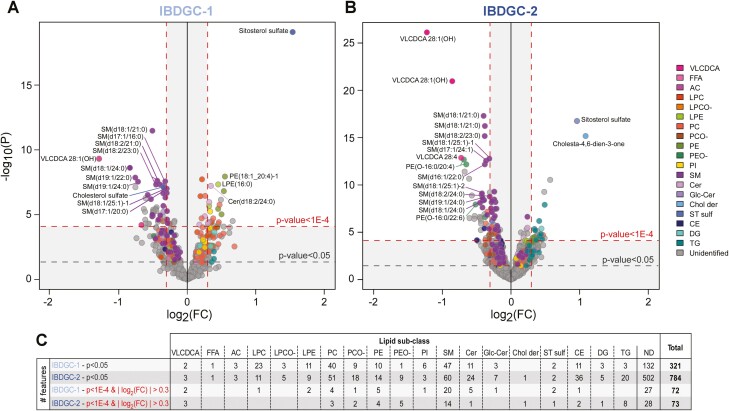
Serum samples from 300 CD patients and 300 healthy donors matched for sex, age, and ethnicity were analyzed in 2 independent phases (Figure 1). Demographic and clinical information for these subjects is presented in Table 1. In the first phase (IBDGC-1), we conducted an untargeted LC-MS–based lipidomic profiling of 100 CD patients and 100 control samples. Raw data consisting of 1068 MS signals were processed for peak alignment, RT correction, filters of presence, normalization of signal intensities, imputation of missing values, and corrections for batch and collection center effects (see Methods and Figure 1). The final dataset retained 715 lipid features, defined by their m/z ratios, RT, and signal intensity, which were tested for differences between patients with CD patients and control subjects. Applying liberal thresholds for significance (P < .05; corresponding to q < 0.03), 321 features were significantly different between CD patients and control subjects (Figure 2A and 2C). Given this large number, we focused our analyses on those of larger effect sizes (│log2(FC)│>0.3) and highest significance (P < 1 × 10-4, corresponding to q < 2 × 10-4). With these stringent thresholds, 72 features were significantly associated with CD.
Table 1.
Characteristics of the subjects in IBDGC-1 and IBDGC-2.
| IBDGC-1 | IBDGC-2 | |||
|---|---|---|---|---|
| Control Subjects (n = 97) | CD Patients (n = 97) | Control Subjects (n = 199) | CD Patients(n = 199) | |
| Age at recruitment, y | ||||
| Range | 19-70 | 19-70 | 16-76 | 16-87 |
| Median | 39 | 39 | 35 | 37 |
| IQR | 28-50 | 27-50 | 26-50 | 28-46 |
| Ethnicity | ||||
| White non-Jewish | 84 (86.6) | 83 (85.6) | 156 (78.4) | 156 (78.4) |
| Jewish | 8 (8.2) | 9 (9.3) | 43 (21.6) | 43 (21.6) |
| Black/African American | 5 (5.2) | 5 (5.1) | 0 (0) | 0 (0) |
| Female | 50 (51.5) | 52 (53.6) | 111 (55.8) | 111 (55.8) |
| Surgery history | ||||
| Yes | — | 86 (88.7) | — | 96 (48.2) |
| No | — | 10 (10.3) | — | 103 (51.8) |
| Missing | — | 1 (1) | — | 0 (0) |
| Tobacco use | ||||
| Smoker | 9 (9.3) | 21 (21.7) | 21 (10.5) | 47 (23.6) |
| Ex-smoker | 13 (13.4) | 8 (8.2) | 30 (15.1) | 20 (10.1) |
| Nonsmoker | 74 (76.3) | 67 (69.1) | 145 (72.9) | 129 (64.8) |
| Missing | 1 (1) | 1 (1) | 3 (1.5) | 3 (1.5) |
| Montreal classification | ||||
| Disease behavior | ||||
| Inflammatory (B1) | — | 0 (0) | — | 106 (53.3) |
| Stricturing (B2) | — | 51 (52.6) | — | 52 (26.1) |
| Penetrating (B3) | — | 46 (47.4) | — | 41 (20.6) |
| Disease location | ||||
| Ileal (L1) | — | 30 (31) | — | 53 (26.6) |
| Colorectal (L2) | — | 8 (8.2) | — | 47 (23.6) |
| Ileocolonic (L3) | — | 58 (59.8) | — | 99 (49.8) |
| Missing | — | 1 (1) | — | 0 (0) |
Values are n (%), unless otherwise indicated. This table presents the demographic and clinical phenotypes of the CD patients and healthy control subjects selected from the National Institute of Diabetes and Digestive and Kidney Diseases IBDGC Repository and retained into the final post–quality-control datasets for IBDGC-1 and IBDGC-2 phases of the current study.
Abbreviations: CD, Crohn’s disease; IBDGC, National Institute of Diabetes and Digestive and Kidney Diseases IBD Genetics Consortium; IQR, interquartile range.
Figure 2.
Circulating lipid features associated with stricturing or internal penetrating Crohn’s disease (CD) patients vs control subjects. A and B, Volcano plots show all 715 and 1894 lipid features retained in the final dataset following analysis of serum samples of CD patients and healthy control subjects in IBDGC-1 and IBDGC-2, respectively. Colors indicate lipid subclasses: very long-chain dicarboxylic acid (VLCDCA), free fatty acid (FFA), acylcarnitine (AC), lysophosphatidylcholine (LPC), LPC ether (LPCO-), lysophosphatidylethanolamine (LPE), phosphatidylcholine (PC), PC ether (PCO-), phosphatidylethanolamine (PE), PE ether (PEO-), phosphatidylinositol (PI), sphingomyelin (SM), ceramide (Cer), glucosylceramide (GlcCer), cholesterol derivative (Chol der), sterol sulfate (ST sulf), cholesterol ester (CE), diglyceride (DG), and triglyceride (TG) species. Lipid identification is shown for the 15 most significant B2/B3-associated features annotated by tandem mass spectrometry in each dataset. C, The number of B2/B3-associated features annotated by lipid subclasses. FC, fold change; ND, nondetermined.
Given the untargeted nature of this lipidomic screen, we then determined the identity of these lipid features by MS/MS analyses. This resulted in the annotation of 46 of these features (n = 46 of 72, 64%), corresponding to 37 unique lipid species once duplicate ions had been removed (Supplementary Table 1). Interestingly, these could be grouped into 4 lipid categories and 12 subclasses (Figure 2C), among which sphingomyelins (SMs) with VLCFAs (decreased in CD patients vs control subjects) are known to modulate inflammatory processes and intestinal epithelium barrier function.6,7
Given these positive results, we extended this lipidomic profiling to a larger set of samples consisting of 200 CD patients and 200 matched control subjects (IBDGC-2), that did not overlap with IBDGC-1. As opposed to the first set of samples that had a more complicated stricturing or internal penetrating disease behavior (B2/B3), IBDGC-2 had roughly equivalent numbers of cases in the inflammatory (B1) and stricturing (B2) or internal penetrating (B3) behavior categories (Table 1). Processing of raw MS data (total of 3933 features) retained 1894 features in the final IBDGC-2 dataset. The larger number of detected features, in comparison with IBDGC-1, was attributed to difference in sensitivity performances between the LC-QTOF instruments used for the 2 phases. For comparison with IBDGC-1, our initial analysis of IBDGC-2 focused on CD patients with the B2 or B3 phenotype. Using the same criteria as previously (P < 1 × 10-4 and │log2(FC)│>0.3), 73 features were significantly associated with the B2/B3 vs control phenotype (Figure 2B). MS/MS analyses enabled annotations of 45 of them (n = 45 of 73, 62%) (Supplementary Table 2), corresponding to 38 unique lipid species. Globally, the B2/B3 lipidomic profile observed in IBDGC-2 was remarkably similar to that in IBDGC-1 in terms of lipid categories and subclasses.
The high replicability between findings of the 2 IBDGC studies was also confirmed by interstudy alignment of results expressed as log2-transformed feature signal intensity expressed as FC for cases vs control subjects and corresponding P values given the semi-quantitative nature of our untargeted lipidomic screen (>85%) (Supplementary Table 3). However, given the greater sensitivity in IBDGC-2, we found more ether lipids of both phosphatidylcholine (ie, PCO-) and phosphatidylethanolamine (ie, PEO-) that were lower in CD patients, and 2 additional lipids that were associated to CD, but that were not detectable in the first phase, namely (1) cholesta-4,6-dien-3-one (higher in CD patients) and (2) cholesterol ester (CE) (14:1; lower).
Correlation structure identifies lipid subclasses, shared structures, and specific lipid entities perturbed in CD
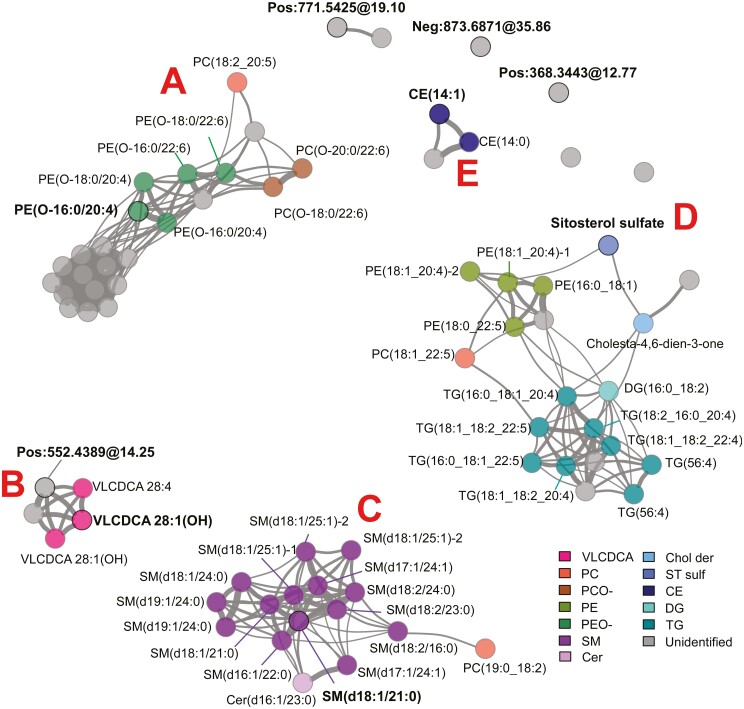
Next, because high-dimensional lipidomic data are expected to have a correlation structure,9 this was assessed in the 73 B2/B3-associated features from IBDGC-2. We found 6 correlation clusters and 4 individual features, with clusters primarily encompassing lipids of a same (sub)class or lipid entities from different subclasses but sharing similar structural characteristics with respect to their fatty acyl moieties (Figure 3, Supplementary Table 2). However, clustering could differ for lipids of the same (sub)class, as seen for the sterol species, namely SitS and cholesta-4,6-dien-3-one in cluster D and CE(14:1) in cluster E. A similar clustering was observed with the 72 B2/B3-associated lipids in IBDGC-1, albeit, as expected for this smaller dataset, correlations were less precisely defined (Supplementary Figure 2). Interestingly, lipids in cluster D were elevated in patient sera as compared with control subjects, whereas those in all other clusters were lower.
Figure 3.
Correlations between circulating lipid features associated with stricturing or internal penetrating Crohn’s disease patients vs control subjects. Positive correlations (r > 0.4) existing between the 73 B2/B3-associated features [P < 1 × 10-4 and │log2(fold change)│] in IBDGC-2 are illustrated using Fruchterman-Reingold algorithm, where the clusters with identified analytes are labeled A–E. Bond thickness corresponds to the strength of correlation between features. Circled features with bold ID are the 9 components of the classification model (see Figure 5). For expansions of the lipid subclasses, see Figure 2.
Circulating lipids are differently associated with CD disease behavior and location subtypes
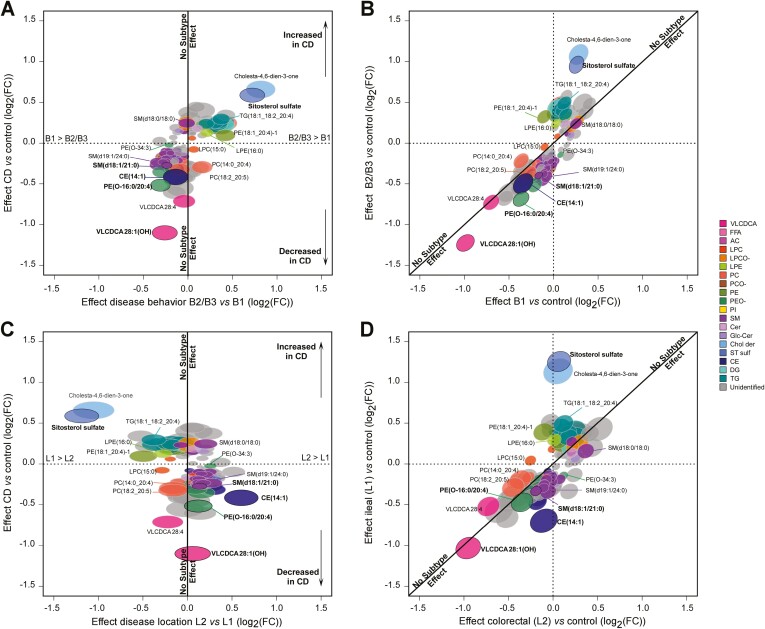
To explore the relationship between circulating lipids and CD subtypes, we further analyzed the entire IBDGC-2 dataset, which was larger and more representative in terms of disease behavior and location than IBDGC-1. First, we tested whether the inclusion of B1 impacted on our previous results that focused on B2 and B3 phenotypes and found 4 new features associated with CD (all patients) vs control subjects (P < 1 × 10-4 [q < 8 × 10-4] and │log2(FC)│>0.3), likely gained by the increase in power due to larger sample size (Supplementary Table 4). In contrast, half of the B2/B3-associated features failed to reach the significance threshold in the all CD patients vs control subjects analysis, suggesting an impact of disease subtype on association with some lipid features. To assess this directly, we tested each feature for association to a specific disease location or behavior and found 182 associations with P < 1 × 10-4 for at least 1 subtype (behavior [n = 138], location [n = 94]) (Supplementary Table 5), which we have plotted as effect sizes of these features with respect to disease status on separate axes (Figure 4). This representation highlights distinct patterns for multiple lipid features. Of note, in terms of disease behavior, the strongest effects were observed for PE(O-16:0/20:4), very long-chain dicarboxylic acid (VLCDCA) 28:1(OH), SitS, and cholesta-4,6-dien-3-one, all being more pronounced in B2/B3 than in B1 (Figure 4A and 4B). In terms of disease location, the strongest effects were observed for SitS, cholesta-4,6-dien-3-one, and CE(14:1) in ileal disease (L1) (Figure 4C and 4D). Taken together, these results suggest circulating levels of specific lipids are differentially perturbed among the phenotypic subgroups of CD, with SitS and cholesta-4,6-dien-3-one having the largest effect sizes.
Figure 4.
Comparative effect size of changes in circulating lipid features between Crohn’s disease (CD) subtypes: disease behaviors and locations. The panels show the 182 features associated with P < 1 × 10-4 for at least 1 CD subtype in IBDGC-2. Fold change (FC) (log2) in mass spectrometry signal intensity values for each feature with respect to disease subtypes is shown on separate axes. Disease behavior is illustrated in panels A and B, while disease location is illustrated in panels C and D. A subtype-dependent effect would fall outside the first diagonal in panels B and D or far from the vertical x = 0 axis in panels A and C. Ellipses correspond to 95% confidence intervals. Lipids with bold ID are components of the classification model (see Figure 5). For expansions of the lipid subclasses, see Figure 2.
Models using 9 or fewer lipid features are strong disease classifiers
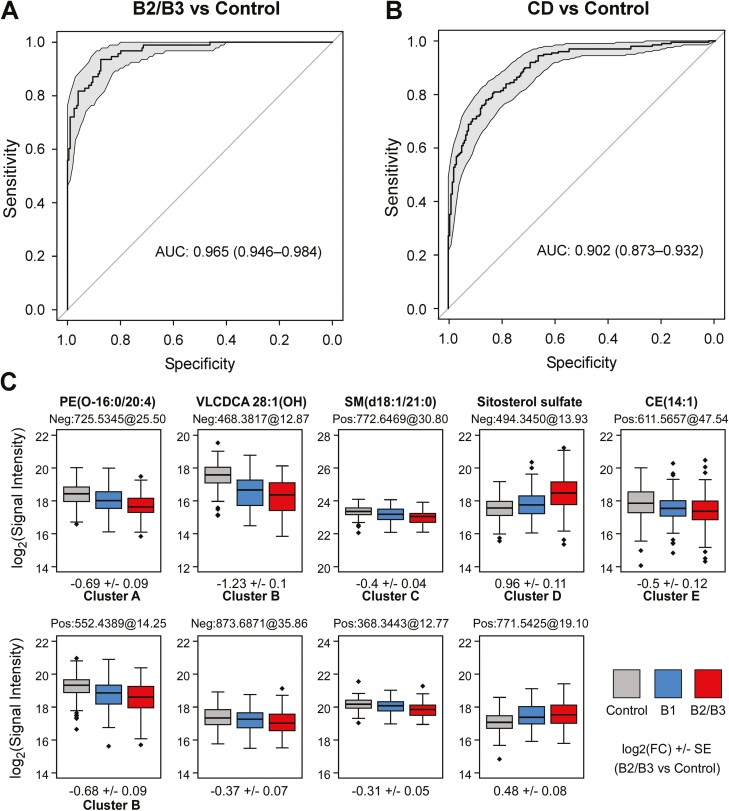
Given the strong effect sizes observed for many features, we assessed their ability in building a disease classifier. Specifically, we built a model using the 73 B2/B3-associated features from IBDGC-2, trained and tested using all B2/B3 and control samples in the IBDGC-2 dataset. The purpose of this model was to use a minimal number of features while reaching maximal classification performance. This model showed a very high performance, with an AUC of 0.97 for the ROC curve (Figure 5A), and also performed very well (AUC of 0.90) when tested against the entire IBDGC-2 dataset including B1 (Figure 5B). Discrimination from control subjects was however better (1) for ileal presentations (L1 and L3) than for colorectal-only presentations (L2) and (2) for stricturing or internal penetrating behavior (B2/B3) than for inflammatory behavior (B1) (Supplementary Figure 3). It is noteworthy that, despite the large number of B2/B3-associated features (Figure 2), the model was made from only 9 lipid features (Figure 5C) that were representative of 6 different correlation clusters and 2 individual features (classifiers are shown in bold in Figure 3), including the 5 annotated classifiers PE(O-16:0/20:4), VLCDCA [28:1(OH)], SM(d18:1/21:0), SitS, and CE(14:1) (cf. Supplementary Figure 1 for MS/MS spectra used for their annotation). Out-of-sample AUC estimates obtained from cross-validation (Supplementary Figure 4) and an alternate model (Supplementary Figure 5) demonstrated the great performance of the model, with an alternate model with as few as 5 lipid features producing high out-of-sample performances (ROC curves; AUCs of 0.79-0.9) (Supplementary Figure 5B). We next explored the effect of sex as a biological variable in our analyses and noted that while many of the CD-associated metabolites were associated with sex (Supplementary Table 7), male/female status itself was not associated with CD (P = .38), and consequently, performance of the model was not affected by the inclusion of sex information.
Figure 5.
Crohn’s disease (CD) classification model based on serum lipid features. A and B, The receiver-operating characteristic curve shows the performance of the classification model to discriminate case vs control phenotypes. Performance was tested (A) in the IBDGC-2 dataset consisting of all B2/B3 and control samples or (B) in the entire IBDGC-2 dataset (including all CD patients) The 95% confidence interval is shown in grandy and performances are expressed as the area under the curve (AUC). Estimations of out-of-sample performance are reported in Supplementary Figures 4 and 5. C, The boxplots represent the mass spectrometry signal intensity values (log2) with quartiles for the 9 classifiers, within control subjects (gray), B1 patients (blue), and B2/B3 patients (red) in IBDGC-2. For expansions of the lipid subclasses, see Figure 2. FC, fold change.
To explore whether any of these metabolites are associated with disease activity or with known biomarkers of disease activity, we performed lipidomic analyses of an independent set of 42 CD patients for whom this information was readily available at the time of serum collection (as this was not available for the IBDGC samples). Following data processing, 1111 lipid features were retained in the final dataset. Focusing on the metabolites with known identities that were strongly associated in the IBDGC cohort, we examined their association to clinically active disease (32 had active disease at serum collection), per clinical assessment and Harvey-Bradshaw Index (HBI) or HBI alone. Additionally, we tested for association between the CD-associated metabolites and serum C-reactive protein levels, and with steroid use. As can be seen in Supplementary Table 7, none of the metabolites in clusters A, B, or C were associated with disease activity and HBI at a P < .1, and only 1 metabolite [TG(18:2_16:0_20:4) in cluster D was associated with disease activity and another (TG(16:0_18:1_22:5)] with HBI score. In terms of association to C-reactive protein levels, while there was no significant association to most CD-associated metabolites, 2 metabolites in cluster C and 3 in cluster D were associated at P < .1 (Supplementary Table 7).
Discussion
IBD is a complex disease involving important contributions of multiple host cell types within the intestinal mucosa, notably immune cells, epithelial cells, and mesenchymal cells, as well as contributions from intestinal flora and mesenteric fat, known as creeping fat.16 Furthermore, alterations in epithelial permeability subsequent to colon inflammation may lead to extraintestinal manifestations, most notably in the liver.17 Importantly, well-powered genetic, transcriptomic, proteomic, and microbiome-based studies have increased our understanding of the biological pathways that contribute to IBD clinical heterogeneity, as well as provide candidate biomarkers of disease and clinical outcomes.10,18–21 More recently, metabolomic studies using patient stool or serum samples have also uncovered some metabolites and metabolic pathways associated with IBD pathophysiology.22–28 However, up to now, few if any studies have applied a comprehensive untargeted lipidomic profiling on serum samples, capturing both intestinal and extraintestinal contributions, from large cohorts of CD patients and healthy individuals.
In this study, we applied a validated comprehensive semi-quantitative untargeted lipidomic workflow, which has the potential to uncover key lipid subclasses and isomers with their acyl chains, originating from host cells, dietary intake, and intestinal flora. First, this workflow was applied to serum samples from a cohort of 100 patients with CD patients and 100 control subjects, matched for age, sex, and ethnicity. Findings were then replicated in an additional 200 CD patients and 200 control subjects, with the results being remarkably consistent between IBDGC-1 and IBDGC-2 despite being tested 18 months apart on 2 different instruments. This speaks to the robust nature of the standardized blood sampling protocol and storage of the NIDDK Repository in minimizing artifactual lipid variations, the platform and workflow, and the markers identified in the process.
Specifically, this led to the identification of >70 structurally unique lipids with strong association to CD (P < 1 × 10-4) and effect size [│log2(FC)│>0.3] in both datasets, of which over 60% were annotated by MS/MS. We observed that most associated lipid features fell into 5 major correlation clusters that included lipids predominantly of similar (sub)classes or sharing similar acyl side chains. However, a few other lipid features were independent of these major clusters.
Given the relatively strong effect sizes observed for the associated lipids, we were able to build a high-performance model with only 9 lipid features (ROC curve: AUC of 0.97), which was not affected by sex. Of note, the 9 lipid features were from the 6 correlation clusters plus 2 individual features, highlighting the nonredundant information provided by these different lipid clusters and likely distinct biological pathways. We were able to identity 5 of these classifiers, namely PE(O-16:0/20:4), VLCDCA 28:1(OH), SM(d18:1/21:0), SitS, and CE(14:1). Although this classifier model was tested in a retrospective fashion, the strong performance characteristics suggest that a small set of circulating lipids may be considered as promising candidate biomarkers to assist disease classification in CD. This will now need evaluating in a variety of prospective cohorts.
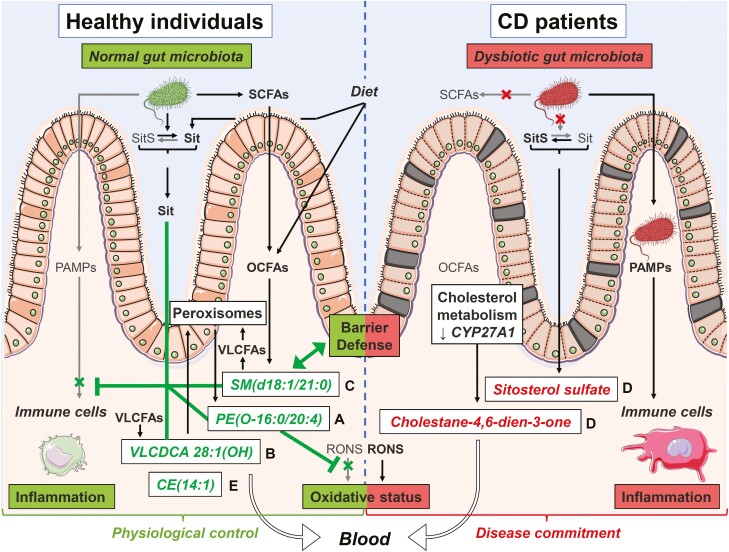
Given the many lipid entities identified in our study, this afforded us the opportunity to examine structural commonalities within each cluster to inform about their potential biological properties. Changes in serum levels of identified lipid classifiers or their correlated counterparts, as well as proposed alterations in their metabolism and potential impact on the (patho)physiology of IBD, are illustrated in Figure 6 and summarized as follows.
Figure 6.
Putative mechanisms underlying observed changes in lipid candidate biomarkers and their impact on Crohn’s disease (CD) pathophysiology. This figure illustrates the proposed mechanisms underlying the observed changes (lower in green, higher in red) in serum levels of candidate biomarker lipids and their correlated clusters (letters on the right of lipid names) in CD patients. We reason that these changes may restrict (┬) or stimulate (↑) various biological functions and mechanisms linked to the pathogenesis of CD. Lipids from clusters with lower serum levels in CD vs control (clusters A-C and E) will be unable to carry out their normal physiological roles, notably (1) restricting inflammation either by preventing its overtriggering by pathogen-associated molecular patterns (PAMPs) or by favoring its resolution through specialized proresolution mediator–like mechanisms (clusters A and B), (2) strengthening the defense barrier of the intestinal mucosa (cluster C), and/or (3) maintaining a physiological oxidative stress status by restricting reactive oxygen and nitrogen species (RONS) effects (cluster A). Furthermore, in CD, dysbiosis is expected to impair metabolism of (1) short-chain fatty acids (SCFAs) and ultimately that of odd-chain fatty acids (OCFAs), which are normally incorporated into sphingomyelins (SMs) (cluster C) and (2) noncholesterol sterols resulting in sitosterol (Sit) being converted into sitosterol sulfate (SitS) (cluster D). Dysregulated host metabolism involving peroxisomes or cytochrome P450 enzymes is also expected to impair formation of polyunsaturated fatty acid (PUFA)–containing ether lipids (cluster A), very long-chain dicarboxylic acids (VLCDCAs) (cluster B), very long-chain fatty acid (VLCFA)–containing SMs (cluster C), and cholesterol esters (CEs) (cluster E), while favoring the formation of a specific oxysterol, namely cholesta-4,6-dien-3-one (cluster D). Enhanced oxidative stress in CD may also contribute to lower serum levels of PUFA-containing ether lipids (cluster A), while the defective intestinal barrier function may result in serum over-representation of SitS and cholesta-4,6-dien-3-one (cluster D). Altogether, these changes and others may contribute to the progression from a physiological control of crucial intestinal processes toward a commitment of CD. Figure created by Servier Medical Art (SMART) images by Servier (http://smart.servier.com/), licensed under a Creative Commons Attribution 3.0 Unported License (CC BY 3.0).
Cluster A includes PE(O-16:0/20:4) and 5 other correlated ether lipids annotated by MS/MS analyses, which are lower in serum of CD patients vs control subjects. PE(O-16:0/20:4) was selected by the predictive models and was significantly associated with the B2/B3 phenotype but not with the disease location. Ether lipids are unusually abundant in neutrophil membranes, where they are essential for cell viability.29 They are intermediates in plasmalogen synthesis and also suggest a host dysmetabolism in CD of ether lipids in peroxisomes, which are specialized cellular organelles recently shown to function as hubs that coordinate responses to stress, metabolism, and immune signaling to maintain enteric health and the functionality of the gut-microbe interface.30 Lipids within this cluster also share a structural feature, namely that they have predominantly an omega-6 (C20:4) or omega-3 (C22:6) PUFA moieties in sn-2 position, which are susceptible to oxidative stress and likely contribute to their biological role via their metabolism to pro/anti-inflammatory as well as specialized proresolution mediators (SPMs).5 Hence, the lower circulating levels in PUFA-containing ether lipids may reflect an enhanced oxidative stress or dysregulated peroxisomal lipid metabolism and likely compromise inflammation resolution in CD.
Cluster B consists of VLCDCA 28:1(OH) and 4 correlated lipid features, including its nonhydroxylated and polyunsaturated relative VLCDCA 28:4, which were lower in sera of CD patients, associated with the B2/B3 phenotypes but not with disease location. While originally named gastrointestinal tract acids, VLCDCAs have been reported to have antiproliferative and anti-inflammatory properties in vitro31,32 and are reduced in patients with colorectal cancer,33–35 suggesting their categorization as bioactive lipids. Importantly, VLCDCAs are structurally and functionally related to SPMs,36 which are hydroxylated PUFAs that trigger the resolution phase of inflammation via signaling through G protein–coupled receptor–dependent pathways. To the best of our knowledge, this is the first report of lower circulating levels of VLCDCA 28:1(OH) and 28:4 in CD. This may reflect a metabolic dysregulation possibly via (1) reduced VLCFA metabolism via cytochrome P450 in the liver37 or (2) enhanced catabolism in peroxisomes.37 However, irrespective of the mechanism, this is likely impacting inflammatory processes possibly via SPM-like effects, which have not yet been explored in IBD.
Cluster C consists of SM(d18:1/21:0) and 11 correlated lipids that were lower in sera of CD patients vs control subjects and strongly associated with disease behavior but independent of location. Notably, all these sphingolipids bear OCFA or VLCFA moieties. OCFA levels likely reflect gut microbiota–mediated synthesis of its short-chain FA precursor, namely propionic acid, or host metabolism of branched-chain amino acids, which were reported to be both reduced in CD.22,38 As for VLCFAs, their incorporation into sphingolipids, which involves intestinal ceramide synthase 2, has been connected to intestinal defense in mice.6,7 In support of the current observation, a dysregulated intestinal metabolism of SMs was also observed in the feces from patients with CD.28 Hence, lower serum SMs bearing OCFAs or VLCFAs likely reflect dysbiosis or a defective synthesis and may impact on intestinal epithelial barrier function.
Cluster D consists of SitS and 15 other MS/MS-identified lipids, including cholesta-4,6-dien-3-one, which were all found to be elevated in sera of CD patients vs control subjects and were associated with disease behavior and location. Interestingly, 2 triglycerides containing PUFAs (20:4 or 22:5), which are precursors for bioactive molecules involved in inflammation and its resolution, were the only serum lipids found to be associated with disease activity and HBI in our independent cohort, suggesting rapid triglyceride acyl chain remodeling under these conditions. To the best of our knowledge, SitS and cholesta-4,6-dien-3-one are herein reported for the first time. The SitS precursor, sitosterol, is a dietary phytosterol with known antiproliferative and anti-inflammatory properties. By analogy with bile acids,39 one may speculate that dysbiosis favors elevated levels of its putative inactive 3-OH sulfated form (ie, SitS). Given that noncholesterol sterols are absorbed in the small intestine but their transfer into the lymph fluid is far less efficient, the elevated levels of SitS, which are more pronounced in B2/B3 and L1 presentation, may therefore reflect dysbiosis or intestinal epithelial barrier dysfunction. As for cholesta-4,6-dien-3-one, little is known about this oxysterol except for its accumulation in patients with cerebrotendinous xanthomatosis—50% of which suffer from chronic diarrhea—caused by mutations in the CYP27A1 gene.40 Several oxysterols play important roles in CD pathophysiology, acting through immune cell receptors such as GPR183 and liver X receptors, coded by IBD risk genes identified by genome-wide association studies.1 While the specific role of cholesta-4,6-dien-3-one in CD remains to be clarified, its higher serum levels concur with lower CYP27A1 messenger RNA levels in colon biopsies from CD patients41 and add to our knowledge on the dysregulated metabolism of cholesterol and bile acids in CD.
Cluster E consists of CE(14:1) and 2 other correlated lipids. CE(14:1) was found to be lower in CD patients vs control subjects and to be associated with ileal disease location. Just as SitS, CE(14:1) also belongs to the sterol lipid class but is likely to represent distinct biological information, given that they are in different clusters. Lower CE(14:1) in CD may result from impaired cholesterol metabolism, either its acylation by acyl-CoA cholesterol acyltransferase in the intestine or liver or by lecithin-cholesterol acyltransferase activity in high-density lipoproteins.42 This may also be linked to a cellular deficiency in plasmalogens, which is known to impact several steps of cholesterol homeostasis.43 Another interesting potential explanation is provided by the recent discovery of a microbial cholesterol dehydrogenase named ismA and that ismA+ species decreased fecal and serum cholesterol in humans.44 While this remains speculative, lower CE(14:1) in the sera of CD patients may reflect changes in the gut microbiome that increase the proportion of ismA+ species.
Taken together, these 5 annotated classifiers, as well as many of the correlated lipids within their clusters, appear to capture multiple different biologic mechanisms, which is reasonable to assume, and are associated with the etiology or pathology of CD. The strongest association of most of these metabolites was to B2/B3, suggesting that these capture biological pathways when there is a breakdown of intestinal homeostasis and barrier integrity.
Conclusions
In the current study, we identified circulating lipids associated with CD, disease location, and behavior and propose specific metabolic pathways that are perturbed in disease. While it remains to be determined how early in the disease process such metabolic imbalances are detectable in patients with CD, given the strong discriminating capacity that was observed with as few as 5 to 9 lipid features, it will be important to explore the potential of these candidate biomarkers to aid in the diagnosis of CD. It needs to be acknowledged that much work still needs to be done (eg, exploring potential confounders in large prospective cohorts, determining how early in disease course these lipid profiles are detected) in order to establish the clinical utility of these biomarkers. Nonetheless, this and recent studies demonstrate the power of MS-based metabolomics, particularly untargeted lipidomic approaches, to uncover mechanisms of CD pathophysiology and clinical outcomes.22–28
Supplementary Material
Acknowledgments
We thank Pr Hubert Schaller, at the Institut de Biologie Moléculaire des Plantes (Centre National de la Recherche Scientifique, Université de Strasbourg) for sharing analytical standards as well as Caroline Daneault and Isabelle Robillard Frayne for their assistance in lipidomic sample analysis.
Contributor Information
Romain Ferru-Clément, Research Center, Montreal Heart Institute, Montreal, QC, Canada; Laboratoire Histocompatibilité et Immunogénétique, Établissement français du sang–Nouvelle-Aquitaine, site de Poitiers, Poitiers, France.
Gabrielle Boucher, Research Center, Montreal Heart Institute, Montreal, QC, Canada.
Anik Forest, Research Center, Montreal Heart Institute, Montreal, QC, Canada.
Bertrand Bouchard, Research Center, Montreal Heart Institute, Montreal, QC, Canada.
Alain Bitton, Division of Gastroenterology, McGill University Health Centre, Montreal, QC, Canada.
Sylvie Lesage, Maisonneuve-Rosemont Hospital Research Center, Montreal, QC, Canada; Département de Microbiologie, Infectiologie et Immunologie, Université de Montréal, Montreal, QC, Canada.
Phil Schumm, Department of Public Health Sciences, University of Chicago, IL, USA.
Mark Lazarev, Harvey M. and Lyn P. Meyerhoff Inflammatory Bowel Disease Center, Department of Medicine, Johns Hopkins University School of Medicine, Baltimore, MD, USA.
Steve Brant, Department of Medicine, Rutgers Robert Wood Johnson Medical School, New Brunswick, NJ, USA; Department of Genetics and the Human Genetics Institute of New Jersey, Rutgers University, Piscataway, NJ, USA.
Richard H Duerr, Department of Medicine, University of Pittsburgh, Pennsylvania, PA, USA.
Dermot P B McGovern, F. Widjaja Foundation Inflammatory Bowel and Immunobiology Research Institute, Cedars-Sinai Medical Center, Los Angeles, CA, USA.
Mark Silverberg, Lunenfeld-Tanenbaum Research Institute, Mount Sinai Hospital, Toronto, ON, Canada.
Judy H Cho, Icahn School of Medicine at Mount Sinai, New York, NY, USA.
Ashwin Ananthakrishnan, Division of Gastroenterology, Massachusetts General Hospital, Boston, MA, USA; Harvard Medical School, Boston, MA, USA; Division of Gastroenterology, Brigham and Women’s Hospital, Boston, MA, USA.
Ramnik J Xavier, Division of Gastroenterology, Massachusetts General Hospital, Boston, MA, USA; Harvard Medical School, Boston, MA, USA; Division of Gastroenterology, Brigham and Women’s Hospital, Boston, MA, USA.
John D Rioux, Research Center, Montreal Heart Institute, Montreal, QC, Canada; Département de Médicine, Université de Montréal, Montreal, QC, Canada.
Christine Des Rosiers, Research Center, Montreal Heart Institute, Montreal, QC, Canada; Département de Nutrition, Université de Montréal, Montreal, QC, Canada.
Members of the iGenoMed Consortium: Alain Bitton, Gabrielle Boucher, Guy Charron, Christine Des Rosiers, Anik Forest, Philippe Goyette, Sabine Ivinson, Lawrence Joseph, Rita Kohen, Jean Lachaine, Sylvie Lesage, Megan Levings, John D. Rioux, Julie Thompson Legault, Luc Vachon, Sophie Veilleux, and Brian White-Guay.
Members of NIDDK IBD Genetics Consortium: Manisha Bajpai, Sondra Birch, Alain Bitton, Krzysztof Borowski, Gregory Botwin, Gabrielle Boucher, Steven R. Brant, Wei Chen, Judy H. Cho, Roberto Cordero, Justin Côté-Daigneault, Mark J. Daly, Lisa Datta, Richard H. Duerr, Melissa Filice, Philip Fleshner, Kyle Gettler, Mamta Giri, Philippe Goyette, Ke Hao, Talin Haritunians, Yuval Itan, Elyse Johnston, Liza Konnikova, Carol Landers, Mark Lazarev, Dalin Li, Dermot P. B. McGovern, Emebet Mengesha, Miriam Merad, Vessela Miladinova, Shadi Nayeri, Siobhan Proksell, Milgrom Raquel, John D. Rioux, Klaudia Rymaszewski, Ksenija Sabic, Bruce Sands, L. Philip Schumm, Marc B. Schwartz, Mark S. Silverberg, Claire L. Simpson, Joanne M. Stempak, Christine Stevens, Stephan R. Targan, and Ramnik Xavier.
Author Contribution
R.F.-C. (Data curation: Supporting; Writing—original draft: Equal; Visualization: Equal). G.B. (Formal analysis: Equal; Data curation: Equal; Writing—review & editing Equal; Visualization: Equal). A.F. (Formal analysis: Equal; Investigation: Equal; Data curation: Equal; Writing—review & editing: Equal; Visualization: Equal). B.B. (Investigation: Equal). A.B. (Resources: Equal). S.L. (Conceptualization: Supporting). P.S. (Resources: Equal). M.L. (Resources: Equal). S.B. (Resources: Equal). R.D. (Resources: Equal). D.M. (Resources: Equal). M.S. (Resources: Equal). J.C. (Resources: Equal; Funding acquisition: Supporting). A.A. (Resources: Equal). R.J.X. (Resources: Equal). J.D.R. (Conceptualization: Lead; Validation: Lead; Resources: Lead; Writing—original draft: Equal; Supervision: Lead; Project administration: Lead; Funding acquisition: Lead; Writing—review & editing: Lead). C.D.R. (Conceptualization: Lead; Validation: Lead; Resources: Lead, Writing—original draft: Equal; Visualization: Equal; Supervision: Lead; Project administration: Lead; Funding acquisition: Lead; Writing—review & editing: Lead).
Funding
The authors would like to acknowledge the financial support of Génome Québec; Genome Canada; the Government of Canada; the Ministère de l’enseignement supérieur, de la recherche, de la science et de la technologie du Québec; the Canadian Institutes of Health Research (with contributions from the Institute of Infection and Immunity, the Institute of Genetics, and the Institute of Nutrition, Metabolism and Diabetes); Genome BC; and Crohn’s Colitis Canada via the 2012 Large-Scale Applied Research Project competition (grant number GPH-129341). This work was also supported by grants from the National Institute of Diabetes and Digestive and Kidney Diseases (grant numbers DK062431 to S.R.B., DK062422 and DK062429 to J.H.C., DK062420 to R.H.D., DK062423 to M.S., DK062413 to D.P.B.M., and DK062432 to J.D.R.). J.D.R. holds a Canada Research Chair (#230625). This project also benefited from infrastructure supported by the Canada Foundation for Innovation (grant numbers 202695, 218944, 20415, and 36283 to J.D.R. and C.D.R.). The sponsors had no role in the study design and in the collection, analysis, and interpretation of data.
Conflicts of Interest
The authors have no potential conflicts (financial, professional, or personal) or competing interests relevant to the article.
Data Availability
Individual-level raw data will be made available via the controlled access NIDDK Central Repositories (https://repository.niddk.nih.gov/home/) and the NIDDK IBDGC Data Commons (https://ibdgc.datacommons.io).
References
- 1. Jostins L, Ripke S, Weersma RK, et al. . Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature. 2012;491(7422):119-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Chiurchiu V, Leuti A, Maccarrone M.. Bioactive lipids and chronic inflammation: managing the fire within. Front Immunol. 2018;9:38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Quehenberger O, Dennis EA.. The human plasma lipidome. N Engl J Med. 2011;365(19):1812-1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Burla B, Arita M, Arita M, et al. . MS-based lipidomics of human blood plasma: a community-initiated position paper to develop accepted guidelines. J Lipid Res. 2018;59(10):2001-2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Alhouayek M, Ameraoui H, Muccioli GG.. Bioactive lipids in inflammatory bowel diseases - From pathophysiological alterations to therapeutic opportunities. Biochim Biophys Acta Mol Cell Biol Lipids. 2021;1866(2):158854. [DOI] [PubMed] [Google Scholar]
- 6. Kim YR, Volpert G, Shin KO, et al. . Ablation of ceramide synthase 2 exacerbates dextran sodium sulphate-induced colitis in mice due to increased intestinal permeability. J Cell Mol Med. 2017;21(12):3565-3578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Oertel S, Scholich K, Weigert A, et al. . Ceramide synthase 2 deficiency aggravates AOM-DSS-induced colitis in mice: role of colon barrier integrity. Cell Mol Life Sci. 2017;74(16):3039-3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Jenkins B, West JA, Koulman A.. A review of odd-chain fatty acid metabolism and the role of pentadecanoic Acid (c15:0) and heptadecanoic Acid (c17:0) in health and disease. Molecules. 2015;20(2):2425-2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Forest A, Ruiz M, Bouchard B, et al. . Comprehensive and reproducible untargeted lipidomic workflow using LC-QTOF validated for human plasma analysis. J Proteome Res. 2018;17(11):3657-3670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Boucher G, Paradis A, Chabot-Roy G, et al. . Serum analyte profiles associated with Crohn’s disease and disease location. Inflamm Bowel Dis. 2022;28(1):9-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Silverberg MS, Satsangi J, Ahmad T, et al. . Toward an integrated clinical, molecular and serological classification of inflammatory bowel disease: report of a Working Party of the 2005 Montreal World Congress of Gastroenterology. Can J Gastroenterol. 2005;19Suppl A:5A-36A. [DOI] [PubMed] [Google Scholar]
- 12. Shelton E, Allegretti JR, Stevens B, et al. . Efficacy of vedolizumab as induction therapy in refractory IBD patients: a multicenter cohort. Inflamm Bowel Dis. 2015;21(12):2879-2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Storey JD, Bass A, Dabney A, Robinson D.. Q-value estimation for false discovery rate control. Release (3.14) ed. Accessed May 7, 2019. [Google Scholar]
- 14. Csárdi G, Nepusz T.. The igraph software package for complex network research. InterJournal, Complex Systems. 2006;16951704. [Google Scholar]
- 15. Robin X, Turck N, Hainard A, et al. . pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinformatics. 2011;12(1):77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Suau R, Pardina E, Domenech E, Loren V, Manye J.. The complex relationship between microbiota, immune response and creeping fat in Crohn’s disease. J Crohns Colitis. 2022;16(3):472-489. [DOI] [PubMed] [Google Scholar]
- 17. Rogler G, Singh A, Kavanaugh A, Rubin DT.. Extraintestinal manifestations of inflammatory bowel disease: current concepts, treatment, and implications for disease management. Gastroenterology. 2021;161(4):1118-1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Cleynen I, Boucher G, Jostins L, et al. . Inherited determinants of Crohn’s disease and ulcerative colitis phenotypes: a genetic association study. Lancet. 2016;387(10014):156-167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lee JC, Lyons PA, McKinney EF, et al. . Gene expression profiling of CD8+ T cells predicts prognosis in patients with Crohn disease and ulcerative colitis. J Clin Invest. 2011;121(10):4170-4179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. West NR, Hegazy AN, Owens BMJ, et al. . Oncostatin M drives intestinal inflammation and predicts response to tumor necrosis factor-neutralizing therapy in patients with inflammatory bowel disease. Nat Med. 2017;23(5):579-589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Pedamallu CS, Bhatt AS, Bullman S, et al. . Metagenomic characterization of microbial communities in situ within the deeper layers of the ileum in Crohn’s disease. Cell Mol Gastroenterol Hepatol. 2016;2(5):563-566.,e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Scoville EA, Allaman MM, Brown CT, et al. . Alterations in lipid, amino acid, and energy metabolism distinguish Crohn’s disease from ulcerative colitis and control subjects by serum metabolomic profiling. Metabolomics. 2018;14(1):17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Fan F, Mundra PA, Fang L, et al. . Lipidomic profiling in inflammatory bowel disease: comparison between ulcerative colitis and Crohn’s disease. Inflamm Bowel Dis. 2015;21(7):1511-1518. [DOI] [PubMed] [Google Scholar]
- 24. Murgia A, Hinz C, Liggi S, et al. . Italian cohort of patients affected by inflammatory bowel disease is characterised by variation in glycerophospholipid, free fatty acids and amino acid levels. Metabolomics. 2018;14(10):140. [DOI] [PubMed] [Google Scholar]
- 25. Guan S, Jia B, Chao K, et al. . UPLC-QTOF-MS-Based plasma lipidomic profiling reveals biomarkers for inflammatory bowel disease diagnosis. J Proteome Res. 2020;19(2):600-609. [DOI] [PubMed] [Google Scholar]
- 26. Iwatani S, Iijima H, Otake Y, et al. . Novel mass spectrometry-based comprehensive lipidomic analysis of plasma from patients with inflammatory bowel disease. J Gastroenterol Hepatol. 2020;35(8):1355-1364. [DOI] [PubMed] [Google Scholar]
- 27. Di Narzo AF, Houten SM, Kosoy R, et al. . Integrative analysis of the Inflammatory Bowel Disease serum metabolome improves our understanding of genetic etiology and points to novel putative therapeutic targets. Gastroenterology. 2022;162(3):828-843.e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Brown EM, Ke X, Hitchcock D, et al. . Bacteroides-derived sphingolipids are critical for maintaining intestinal homeostasis and symbiosis. Cell Host Microbe. 2019;25(5):668-680.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lodhi IJ, Wei X, Yin L, et al. . Peroxisomal lipid synthesis regulates inflammation by sustaining neutrophil membrane phospholipid composition and viability. Cell Metab. 2015;21(1):51-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Di Cara F. Peroxisomes in host defense. PLoS Pathog. 2020;16(7):e1008636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ritchie SA, Jayasinghe D, Davies GF, Ahiahonu P, Ma H, Goodenowe DB.. Human serum-derived hydroxy long-chain fatty acids exhibit anti-inflammatory and anti-proliferative activity. J Exp Clin Cancer Res. 2011;30:59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wood PL. Endogenous anti-inflammatory very-long-chain dicarboxylic acids: potential chemopreventive lipids. Metabolites. 2018;8(4):76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hata T, Takemasa I, Takahashi H, et al. . Downregulation of serum metabolite GTA-446 as a novel potential marker for early detection of colorectal cancer. Br J Cancer. 2017;117(2):227-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ritchie SA, Ahiahonu PW, Jayasinghe D, et al. . Reduced levels of hydroxylated, polyunsaturated ultra long-chain fatty acids in the serum of colorectal cancer patients: implications for early screening and detection. BMC Med. 2010;8:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wood PL, Donohue MM, Cebak JE, et al. . Reduced plasma levels of very-long-chain dicarboxylic acid 28:4 in Italian and Brazilian colorectal cancer patient cohorts. Metabolites. 2018;8(4):91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Serhan CN. Discovery of specialized pro-resolving mediators marks the dawn of resolution physiology and pharmacology. Mol Aspects Med. 2017;58:1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sanders RJ, Ofman R, Duran M, Kemp S, Wanders RJA.. Omega-oxidation of very long-chain fatty acids in human liver microsomes. Implications for X-linked adrenoleukodystrophy. J Biol Chem. 2006;281(19):13180-13187. [DOI] [PubMed] [Google Scholar]
- 38. Lloyd-Price J, Arze C, Ananthakrishnan AN, et al. . Multi-omics of the gut microbial ecosystem in inflammatory bowel diseases. Nature. 2019;569(7758):655-662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Duboc H, Rajca S, Rainteau D, et al. . Connecting dysbiosis, bile-acid dysmetabolism and gut inflammation in inflammatory bowel diseases. Gut. 2013;62(4):531-539. [DOI] [PubMed] [Google Scholar]
- 40. Duell PB, Salen G, Eichler FS, et al. . Diagnosis, treatment, and clinical outcomes in 43 cases with cerebrotendinous xanthomatosis. J Clin Lipidol. 2018;12(5):1169-1178. [DOI] [PubMed] [Google Scholar]
- 41. Guillemot-Legris O, Mutemberezi V, Buisseret B, et al. . Colitis alters oxysterol metabolism and is affected by 4beta-hydroxycholesterol administration. J Crohns Colitis. 2019;13(2):218-229. [DOI] [PubMed] [Google Scholar]
- 42. Gorshkova IN, Mei X, Atkinson D.. Arginine 123 of apolipoprotein A-I is essential for lecithin:cholesterol acyltransferase activity. J Lipid Res. 2018;59(2):348-356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Honsho M, Fujiki Y.. Plasmalogen homeostasis - regulation of plasmalogen biosynthesis and its physiological consequence in mammals. FEBS Lett. 2017;591(18):2720-2729. [DOI] [PubMed] [Google Scholar]
- 44. Kenny DJ, Plichta DR, Shungin D, et al. . Cholesterol metabolism by uncultured human gut bacteria influences host cholesterol level. Cell Host Microbe. 2020;28(2):245-257.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Individual-level raw data will be made available via the controlled access NIDDK Central Repositories (https://repository.niddk.nih.gov/home/) and the NIDDK IBDGC Data Commons (https://ibdgc.datacommons.io).