Abstract
Ataxia telangiectasia and Rad3-related protein (ATR) kinase regulate a key cell regulatory node for maintaining genomic integrity by preventing replication fork collapse. ATR inhibition has been shown to increase replication stress resulting in DNA double-strand breaks (DSBs) and cancer cell death, and several inhibitors are under clinical investigation for cancer therapy. However, activation of cell-cycle checkpoints controlled by ataxia telangiectasia–mutated (ATM) kinase could minimize the lethal consequences of ATR inhibition and protect cancer cells. Here, we investigate ATR-ATM functional relationship and potential therapeutic implications. In cancer cells with functional ATM and p53 signaling, selective suppression of ATR catalytic activity by M6620 induced G1-phase arrest to prevent S-phase entry with unrepaired DSBs. The selective ATM inhibitors, M3541 and M4076, suppressed both ATM-dependent cell-cycle checkpoints, and DSB repair lowered the p53 protective barrier and extended the life of ATR inhibitor–induced DSBs. Combination treatment amplified the fraction of cells with structural chromosomal defects and enhanced cancer cell death. ATM inhibitor synergistically potentiated the ATR inhibitor efficacy in cancer cells in vitro and increased ATR inhibitor efficacy in vivo at doses that did not show overt toxicities. Furthermore, a combination study in 26 patient-derived xenograft models of triple-negative breast cancer with the newer generation ATR inhibitor M4344 and ATM inhibitor M4076 demonstrated substantial improvement in efficacy and survival compared with single-agent M4344, suggesting a novel and potentially broad combination approach to cancer therapy.
Introduction
In the face of continuous exposure to endogenous and exogenous DNA damage, mammalian cells have evolved complex molecular mechanisms to preserve their genomic integrity, referred to as the DNA damage response (DDR; refs. 1–3). Many chemotherapeutic agents act by inducing DNA damage and are often used to exploit genetic deficiencies in the DDR that confer synthetic lethality (1, 4, 5). Targeting different DDR components has emerged as a promising field for development of potential therapeutic agents, offering novel synergistic combination strategies for cancer therapy with DNA-damaging agents (4).
DNA double-strand breaks (DSBs) are the most lethal lesions, and mammalian cells have evolved two main mechanisms for their rapid and efficient repair: nonhomologous end-joining and homology-dependent repair (HDR). HDR begins with nuclease processing of the DNA ends at the site of the DSB, resulting in 3′ single-stranded DNA (ssDNA) tails (6). These ssDNA regions are protected and stabilized by the binding of replication protein-A (RPA); RPA also binds ssDNA at the sites of stalled replication forks (7). Ataxia telangiectasia and Rad3-related protein (ATR) is recruited to RPA-coated ssDNA segments of the genome to slow down and stabilize replication forks to prevent their collapse and the loss of genomic integrity (7). ATR also promotes DSB repair and protection of replication forks through phosphorylation of CHK1 and activation of further downstream signaling (8). Thus, ATR controls a checkpoint response to prevent replication stress by restricting replication origin firing during S-phase and premature entry into mitosis via G2 arrest (1, 9, 10), ultimately confining cells to a cell-cycle phase in which a homologous template is available for DSBs repair via high-fidelity HDR before mitosis.
Because ATR plays a critical role in maintaining genomic integrity in replicating cells, it has been considered an attractive target for cancer therapy (5, 10, 11). Many ATR inhibitors (ATRis) are being tested in clinical trials such as M6620 (berzosertib, VX-970), AZD6738 (ceralasertib), BAY 1895344 (elimusertib), RP-3500 (camonsertib), and ART0380 (12). The ATRis used in this study, M6620 and M4344 (Gartisertib; refs. 5, 13–16) have been shown to sensitize cancer cells to several chemotherapy agents, including platinum-based therapies (15, 17), ionizing radiation (17–19), gemcitabine (15, 18), and topoisomerase inhibitors (20), consistent with findings from other ATRis. Furthermore, it has been shown that ATR inhibition is synthetically lethal with ataxia telangiectasia mutated (ATM) or p53 deficiency using ATRis such as VE-821 (21), AZD6738 (22, 23), BAY 1895344 (24), and RP-3500 (25). Clinical responses have been observed with the ATRi BAY 1895344 as a monotherapy in a phase I trial in a subset of patients with ATM mutation or loss of protein expression (26).
ATM is a protein kinase that activates a signaling cascade to promote cell-cycle checkpoint activation and DSB repair by HDR. A critical component of this signaling process is ATM-dependent phosphorylation of the tumor suppressor p53, leading to disruption of the MDM2–p53 interaction and stabilization of the p53 protein (1). The p53 tumor suppressor acts as a guardian of the genome and activates G1–S or G2–M checkpoints to arrest cells and promote DNA repair (27). In cells unable to repair excessive DNA damage, p53 initiates a signaling cascade to induce apoptosis. TP53 is frequently mutated or lost in tumors, leading to disrupted cell-cycle regulation and increased replication stress (14, 28). ATM sustains loss-of-function mutations in approximately 5% of all cancers with more frequent alterations in cancers such as hepatocellular, colorectal, bladder, prostate, and stomach (29–31). ATM protein expression is also frequently lost in cancers such as prostate (32) and colorectal (33). The extent to which ATM function must be disabled through mutations or loss of expression to sensitize human tumors to ATRis is still debated, but it has recently been shown that clinical responses to the RP-3500 In the phase I/II TRESR trial, ATRi monotherapy was enriched in patients with the ATM mutation, particularly those with biallelic mutations (34). Classification of ATM loss-of-function cancers is complex and requires careful characterization of the genetic mutations, their allelic status, and ATM protein expression (35). The frequency of cancers with partial or complete loss of ATM function is poorly understood, and clinical exploitation of this genetic deficiency for ATRis remains challenging (31).
Here, we explore ATM functional deficiency achieved by selective small-molecule inhibitors and show that they offer synergistic combination partners to ATRis in a wide range of cellular and animal cancer models. Our findings provide mechanistic rationale to expand upon this paradigm. In ATM/TP53 wild-type cancer cells, the ATRi M6620 produced DSB damage and induced ATM-mediated G1 checkpoint activation, but in cells lacking intact ATM-p53 signaling, cells continue to progress through the cell cycle, ultimately resulting in cell death. Addition of M3541, a potent inhibitor of ATM (36), partially abrogates M6620-induced G1 arrest in ATM-p53 wild-type cancer cells, thereby lowering the barrier to mitotic entry and increasing the fraction of cycling cells with unrepaired DSBs. These cells showed elevated levels of chromosome defects, misalignment, and mis-segregation, leading to aberrant division and ultimately cell death. ATM inhibitors strongly potentiated ATRi-induced cancer cell death in multiple cancer cell lines and animal models at tolerated doses, suggesting that ATM inhibitors could offer an efficacious approach to enhancing the antitumor activity of ATRis currently under clinical investigation.
Materials and Methods
Cell lines and reagents
All cell lines were obtained, Mycoplasma free, from the Merck Tissue Culture Bank (Merck KGaA, Darmstadt, Germany). Cells were originally purchased from ATCC, European Collection of Animal Cell Cultures, or DSMZ. The CRISPR-generated A549 ATM-null clone and its parental line were provided by Alan D'Andrea (Dana-Farber Cancer Institute, Boston, MA). The A549 p53-null cells have been described previously (37). Short tandem repeats were analyzed to confirm the identity of all cell lines by PCR and electrophoretic fractionation. Mycoplasma infection was excluded by qPCR-based testing performed quarterly. All cell lines were serially passaged twice a week and used for experiments before reaching passage 10 from the original frozen stock. TP53 mutation status was obtained from the current version of the p53 Database (https://p53.fr/tp53-database). A375, A549, and HeLa cells were maintained in the DMEM from GIBCO. NCI-H460 cells were maintained in RPMI1640 (GIBCO). IMR90 and WI38 cells were maintained in Minimum Essential Medium from GIBCO. Culture medium was supplemented with 10% FBS (catalog No. 35-015-CV, Corning Life Science). M6620 (berzosertib), M4344 (gartisertib), M3541, and M4076 were synthesized in the department of Medicinal Chemistry at Merck KGaA, Darmstadt, Germany. The M3541 and M4076 chemical structures were published previously (36). The synthetic procedures for M3541, disclosed as compound 36 in U.S. patent applications (38), can be found in Examples 4a and 4b (p. 33–35). The synthetic procedure for M4076, disclosed as compound 1 in the patent application (39) can be found in Example 1 (p. 70–72). Alternative methods of obtaining compound 1 are described in Examples 2 and 3 (p. 73–77; ref. 39). All compounds were dissolved in DMSO to make 10 mmol/L stock solution and kept frozen at −80°C until use. The 34 cancer cell lines used for combination screening of ATR, ATM, and DNA-dependent protein kinase (DNA-PK) inhibitors performed at Oncolead are as follows: A204 (muscle), A375 (skin), A549 (lung), A673 (muscle), ASPC1 (pancreas), BXPC3 (pancreas), CALU6 (lung), COLO205 (colon), DU145 (prostate), GRANTA-519 (hematologic), HCT116 (colon), HEPG2 (liver), HL-60 (hematologic), HT1080 (connective tissue), HT29 (colon), IGROV1 (ovary), IMR90 (lung), LOVO (colon), MCF7 (breast), MDAMB231 (breast), MDAMB435 (skin), MDAMB436 (breast), MHHES1 (bone), MIAPACA2 (pancreas), MV4-11 (hematologic), NCIH460 (lung), PANC1 (pancreas), RAMOS (hematologic), RDES (bone), SAOS2 (bone), SW620 (colon), U2OS (bone), U87MG (brain), and WSU-NHL (hematologic; refs. 36, 40). The identity of these cell lines was tested and confirmed by short tandem repeat at Oncolead. Additional routine tests were performed to ensure that the cells were Mycoplasma free.
Immunofluorescence
Cells were seeded in chamber well slides (Millipore Millicell EZ Slides) and cultured overnight. The following day, cells were treated with compounds for various durations. Cells were then fixed and stained as described previously (41). Briefly, cells were fixed with 1% paraformaldehyde/2% sucrose for 15 minutes at room temperature, 100% methanol for 30 minutes at −20°C, and then 50% methanol/50% acetone for 20 minutes at −20°C. Cells were then incubated in permeabilization/blocking solution (Cell Signaling Technology #12411) at room temperature for 1 hour. Primary antibodies (Cell Signaling Technology #9718 or #80312 for γH2Ax, #2675 for phospho-53BP1, and #86298 for beta-tubulin) were diluted 1:250 in permeabilization/blocking solution and incubated with the cells at 4°C overnight. The secondary antibodies used were Alexa Fluor 488 (Invitrogen #A32766 donkey anti-mouse IgG or #A32790 donkey anti-rabbit IgG) and Alexa Fluor 555 (Invitrogen #A32773 donkey anti-Mouse IgG or #A32794 donkey anti-rabbit IgG). Three washes with PBS with Triton X-100 and four washes with PBS were performed after primary and secondary antibody. Cells were costained with DAPI (Invitrogen #D1306) to visualize the nuclei and micronuclei. Slides were imaged on a Zeiss MIC-074 microscope (Carl Zeiss Microscopy, LLC). Images were processed and analyzed using ImageJ software.
Western blot analysis
Cells (7.5 × 105) were seeded into 100-mm dishes. The next day, cells were treated with compounds of interest. Cells were harvested on ice in PBS, pelleted, and lysed in AZ lysis buffer [50 mmol/L Tris pH 8, 250 mmol/L NaCl, 1% NP-40, 0.1% SDS, 5 mmol/L ethylenediamine tetraacetic acid (EDTA), 10 mmol/L Na4P2O7, 10 mmol/L NaF, 1×cOmplete EDTA-free Protease Inhibitor Cocktail (Roche), 1× PhosSTOP (Roche)]. Protein concentration of each sample was determined using the Pierce BCA Protein Assay Kit (Thermo Fisher Scientific #23227). Samples were run on NuPAGE Bis-Tris gels (Thermo Fisher Scientific) and transferred to nitrocellulose. The primary antibodies used were: p-ATM S1981 (Abcam #ab81292), ATM (Santa Cruz Biotechnology #SC23921), p-KAP1 S824 (Abcam #ab133440), KAP1 (Abcam #ab22553), p-CHK2 T68 (Cell Signaling Technology #2197), CHK2 (Cell Signaling Technology #6334), p-CHK1 S317 (Cell Signaling Technology #12302), CHK1 (Cell Signaling Technology #2360), p-p53 S15 (Cell Signaling Technology #9284), p53 (Cell Signaling Technology #48818), MDM2 (Cell Signaling Technology #86934), p21 (Cell Signaling Technology #2947), and horseradish peroxidase–conjugated GAPDH (Cell Signaling Technology #3683). Western blots were analyzed using GE ImageQuant LAS 4000 with SuperSignal West Pico Chemiluminescent Substrate (Thermo Fisher Scientific #34080) and/or SuperSignal West Femto Maximum Sensitivity Substrate (Thermo Fisher Scientific #34094).
Cell-cycle analysis
Cell-cycle analysis was performed using the FITC bromodeoxyuridine (BrdU) Flow Kit from BD Biosciences (#559619) according to the manufacturer's protocol. Briefly, 7.5 × 105 of proliferating cells were seeded in T75 flasks. The following day, cells were treated with compounds of interest. The next day, cells were pulsed with BrdU for 1 hour and then washed, harvested, and stained with FITC-conjugated anti-BrdU antibody and 7-aminoactinomycin D (7-AAD). Cells were processed on a BD FACSCanto flow cytometer to determine cell-cycle profiles, and the percentages of cells in each phase was calculated using FlowJo v10 software (FlowJo, LLC).
IncuCyte live cell imaging
Cells were seeded at 500 cells per well in 96-well plates (Corning #353219) and cultured overnight. The next day, cells were treated with compounds of interest. Real-time cell death was monitored using the IncuCyte Live-Cell Imaging System (Essen Biosciences) by adding the mix-and-read IncuCyte CytoTox Reagent (Essen Biosciences #4632 or 4633). Relative cell death was calculated by dividing the number of positively CytoTox stained objects by percent cell confluence (both determined through the IncuCyte software).
Meso Scale Discovery sandwich ELISA for ATM activity
For abrogation of ATM activity by M6620 and ATM inhibitors, A549 (7.5 × 105) cells were seeded into 100-mm dishes and treated the following day with DMSO, M6620 (200 nmol/L), ATM inhibitor (1 μmol/L), or the combination thereof for 24 hours. Cells were harvested on ice in PBS, pelleted, and lysed in Meso Scale Discovery (MSD) lysis buffer (MSD #R60TX-3) supplemented with Roche cOmplete EDTA-free Protease Inhibitor Cocktail (Sigma-Aldrich #11836170001) and Roche phosSTOP phosphatase inhibitor (Sigma-Aldrich #4906837001). MSD MULTI-ARRAY 96-well plates (L15XA-3) were coated with capture antibodies against phosphorylated ATM (Abcam #ab208775) or total ATM (Santa Cruz Biotechnology #sc135663) and incubated at 4°C overnight. The next day, plates were washed with 1×TBST (Tris Buffered Saline + Tween 20, MSD #R61TX-2), blocked for nonspecific binding with Blocker A (MSD #R93BA-4) for 1 hour. Plates were incubated with equal microgram amounts of lysates for 2 hours, incubated with primary detection antibodies against total ATM (Santa Cruz Biotechnology #sc135663 or Abcam #ab199726) for 1 hour, and then incubated with secondary detection antibody (MSD #R32AC-5 for anti-mouse SULFO-Tag and MSD #R32AB-5 for anti-rabbit SULFO-Tag). Plates were washed after each incubation step. Reading buffer (MSD #R92TC-3) was added to each plate, and the plates were read by a Sector Imager 600 (MSD). Background levels were subtracted from experimental values, and phosphorylated ATM levels were normalized to total ATM levels. Normalized phosphorylated ATM levels were then normalized to DMSO.
CellTiter-Glo viability assay
Cells were seeded at 125 cells per well in 384-well plates (Corning #354663) and cultured overnight. The next day, cells were treated with compounds of interest. Cell viability was assessed using the CellTiter-Glo 2.0 cell viability assay (Promega #G9242) according to the manufacturer's protocol. Luminescence was detected using an EnVision plate reader (Perkin Elmer), and the IC50 values and Bliss synergy scores were determined using Combenefit software (42).
Bliss combination synergy analysis in cancer cell lines
Determination of Bliss combination synergy was performed at Oncolead (Karlsfeld, Germany) in a panel of 34 cancer cell lines using a sulforhodamine B staining assay as described previously (37, 40). The ATRis M6620 and M4344 were used at a fixed concentration of 200 and 40 nmol/L, respectively, either alone and together with increasing concentrations of a second DDR inhibitor (DNA-PK inhibitor peposertib, ATRis M6620 or M4344 or ATM inhibitor M4076).
Isolation of metaphase cells for spectral karyotyping
Cells were seeded in T75 flasks. The next day, cells were treated with DMSO, M6620 (200 nmol/L), M3541 (1 μmol/L), or M6620+M3541 combination for 24 hours. Colcemid (0.1 μg/mL) was added, and the cells were incubated for an additional 4 hours. Mitotic cells were collected by shake off and exposed to a hypotonic buffer (0.57% KCl) at 37°C for 30 minutes. Cells were fixed with fresh ice-cold methanol:acetic acid (3:1) and then processed and characterized at the Molecular Cytogenetic Core at Albert Einstein College of Medicine, Bronx, New York, as described previously (43). Ten randomly picked metaphase spreads were imaged and analyzed for each treatment condition.
Animal studies
To investigate the combination benefit of M4344 and M4076, two cell line–derived xenograft models [MV4.11, acute myeloid leukemia (AML), ATCC, CRL-9591, and MiaPaCa2, pancreatic cancer, ATCC, CRL-1420] and the panel of 26 triple-negative breast cancer (TNBC) patient-derived xenograft (PDX) models from XenTech (Evry, France) were used. Study design and animal use were approved by the local animal welfare authorities (Regierungspräsidium Darmstadt, Hesse, Germany, protocol registration number DA4/Anz.1014). Six- to 8-week-old female H2dRag2 (C;129P2-H2d-Rag2<tm1Fwa IL2rgtm1; Taconic Denmark) mice were used. They received subcutaneous injections in the right flank (3 × 106 cells in PBS/Matrigel). When tumor xenografts reached an average volume of about 100 mm3, mice (n = 10 per treatment arm, randomized from 15 mice per arm to obtain a similar mean and median within the treatment groups) received M4344 (suspended in 15% Captisol/0.1 mol/L hydrochloride acid, pH 3.5) as once-daily oral dose, M4076 (10 mg/kg, suspended in 0.5% Methocel/0.25% Tween20/50 mmol/L Citrate buffer, pH 3) at a once-daily oral dose of 50 or 100 mg/kg, or the combination thereof. For the combination, M4344 was applied 1 hour after M4076.
The PDX study was performed by XenTech (Evry, France) in accordance with French legislation concerning the protection of laboratory animals and in accordance with a license approved by the French Ministry of Higher Education, research and innovation (registration number APAFIS#11191v2). Twenty-six different TNBC models were tested using 5-week-old female athymic nude mice (Foxn1nu ENVIGO, France). Tumors were transplanted subcutaneously with tumor fragments of 20 mm3 under anesthesia (100 mg/kg ketamine hydrochloride and 10 mg/kg xylazine). In each of the 26 studies, mice with an established growing tumor volume between 60 and 290 mm3 were randomized according to their tumor volumes into the respective treatment groups to achieve homogenous mean and median tumor volume in each treatment arm, which consisted of three mice per treatment group and model. Inclusion rate (number of implanted vs. mice included into the study) was different for each individual model and ranged between 27% and 50%. Mice received M4344 at a daily oral dose of 10 mg/kg, M4076 at a twice-daily oral dose of 50 mg/kg, or the combination thereof. The control group was left untreated. M4344 was given 30 to 45 minutes after the first M4076 dose of each day. Tumor length (L) and width (W) were measured with calipers, and tumor volumes were calculated as L × W2/2. For the TNBC study, relative tumor volume (RTV) was used as an endpoint for analysis. RTV was calculated using the formula RTV (%) = ((TVend–TVstart)/TVstart)) × 100 was the day when the respective control group reached an absolute tumor volume of 800 mm3 (median). RECIST criteria were applied to evaluate response rates considering the relationship between change in diameter (response ≥30% decrease, progression ≥20% increase) and volume (response ≥65% decrease, progression ≥73% increase; ref. 44).
Statistical analysis
All statistical tests were performed using GraphPad PRISM software. For in vitro data, the mean values from at least three independent replicates were analyzed using unpaired t tests, and the graphed data are shown as mean ± SEM. Significance values are ****, P < 0.0001; ***, P < 0.001; **, P < 0.01; and *, P < 0.05. NS represents nonsignificant differences (P > 0.05). For analysis of in vivo xenograft data, a repeated measures two-way ANOVA with a Tukey multiple comparison test was applied on log-transformed tumor volume data. For transformation tumor volumes of 0 mm3 were set to 10 mm3. Significance was reported at the last day at which vehicle controls were still available. Significance values are *, P < 0.05 and NS represents nonsignificant differences (P > 0.05). An ordinary one-way ANOVA with multiple comparisons was used as a statistical test for the TNBC PDX study. The test was performed on median tumor volume change data from the respective treatment groups and models that consisted of three mice each.
Results
ATR inhibition activates key cell-cycle checkpoints
ATR plays an important role in maintaining genomic integrity in replicating cells by promoting replication fork stabilization under conditions of replication stress and controlling cell-cycle checkpoints to minimize lethal consequences of cycling with damaged DNA. Inhibition of ATR activity leads to an accumulation of DSBs, as shown by an increase in DNA repair foci formation (18, 45, 46). M6620 is a potent and selective small-molecule ATR kinase inhibitor shown to effectively suppress the catalytic activity of ATR and its functions in response to replication stress (47). We interrogated cellular events following suppression of the ATR pathway and the fate of cancer cells.
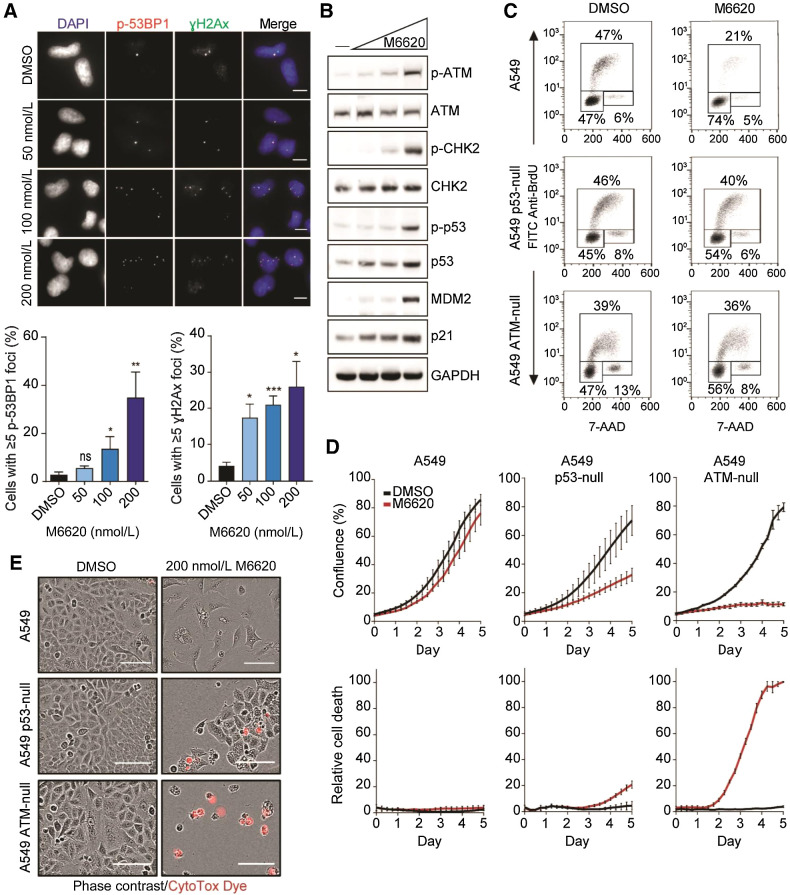
First, we assessed the effect of an ATRi on the number of γH2AX and phospho-53BP1 foci, two widely used markers of DNA DSBs (48, 49). M6620 dose-dependently increased the population of cells with five or more phospho-53PB1 and γH2AX foci, marking unrepaired DSBs, in A549 cancer cells (Fig. 1A), its p53-null clone, and the p53 dysfunctional HeLa cells (Supplementary Fig. S1). Because ATM is a sensor and controller of response to DSBs, we analyzed key proteins from the ATM-p53 signaling pathway in A549 cells exposed to M6620 by Western blotting (Fig. 1B). Concentration-dependent increase in the phosphorylation levels of ATM and CHK2 indicated that ATRi treatment triggers activation of ATM signaling. This led to an increased p53 phosphorylation at serine 15, stabilization of total p53, elevated expression of its transcriptional targets MDM2 and p21, a pan CDK kinase inhibitor and a key mediator of the p53-dependent cell-cycle checkpoint response (50). BrdU/7-AAD cell-cycle analyses demonstrated a substantial increase in the G1-phase population at the expense of S-phase, indicating a marked activation of the G1–S checkpoint (Fig. 1C). The abrogation of the G1 checkpoint was reversed in the engineered p53-null (37) and ATM-null isogenic A549 (Supplementary Fig. S1C) clones, suggesting that p53 regulated downstream targets are largely responsible for the G1 arrest (Fig. 1C). The G1 checkpoint activation by ATRi was maintained and even increased after an additional 24 hours of exposure to the ATRi, while no cell-cycle arrest was observed in the p53-null A549 cells (Supplementary Fig. S2A). G1 checkpoint activation was also observed in two additional p53 wild-type cancer cell lines, A375 and H460 (Supplementary Fig. S2B). However, HeLa cells, in which human papillomavirus E6 protein inhibits the phosphorylation of p53 and induces p53 degradation (51), failed to activate a G1 checkpoint (Supplementary Fig. S2B), further supporting the main role of p53 signaling in ATRi-induced G1 checkpoint activation.
Figure 1.
Inhibition of ATR by M6620 activates the ATM-p53 G1 checkpoint in response to DNA damage. A, Representative immunofluorescence images and analysis of phospho-53BP1 and γH2Ax foci in A549 cells after 24 hours exposure with M6620. Error bars, SEM; **, P < 0.01; and *, P < 0.05 by unpaired t test. Scale bars = 10 μm. B, Western blot analysis of components of the ATM-p53 signaling pathway in A549 cells after 24-hour exposure to DMSO or M6620 (50, 100, 200 nmol/L). C, BrdU/7-AAD cell-cycle analysis in A549 parental, p53-null, and ATM-null cells after 24-hour exposure to M6620 (200 nmol/L). D, Confluence (top row) and relative cell death (bottom row) from IncuCyte live imaging in A549 parental, p53-null, and ATM-null cells treated with DMSO or M6620 (200 nmol/L). Relative cell death was calculated from the number of cells positively stained with CytoTox dye normalized to confluence. Error bars, SEM. E, Representative images from IncuCyte live imaging (10× objective) of A549 parental, p53-null, and ATM-null cells after 6-day exposure with M6620 (200 nmol/L). Scale bars = 100 μmol/L.
Taken together, these results show that ATR inhibition leads to the accumulation of DSB damage and an ATM-p53–mediated G1 checkpoint response, protecting cells from entering replication. However, in cells lacking intact ATM-p53 signaling, ATR inhibition permits cell-cycle progression despite elevated DNA damage. Unchecked cell cycling could compromise chromosome integrity and lead to cell death. To assess this further, we examined the consequences of exposure to M6620 in the ATM-null and p53-null A549 isogenic cell lines by live cell imaging. Cell growth and viability were measured via cell confluence and cell death readouts (Fig. 1D). Proliferation of the parental A549 cells exposed to M6620 was only slightly impaired, and appearance of cells with a senescence-like morphology was evident, yet no induction of cell death was observed (Fig. 1D and E). These findings further support M6620-induced G1 checkpoint activation as a protective mechanism. A549 cells lacking p53 activity showed growth retardation in the presence of M6620, but also an increase in the population of dying cells. These results indicated that turning off p53-mediated checkpoints allows more cells to enter mitosis and undergo cell death. The ATM-null A549 cells exposed to M6620 showed nearly complete growth arrest and higher levels of cell death (Fig. 1D and E), suggesting that suppressing both p53-dependent and -independent ATM functions provides a most effective way for sensitizing cancer cells to M6620. Together, our data demonstrate that suppression of the ATM-p53 signaling axis in cancer cells strongly enhances cytotoxicity of ATR inhibitors.
ATM inhibition bypasses the ATRi-induced G1 checkpoint protection and enhances cancer cell death
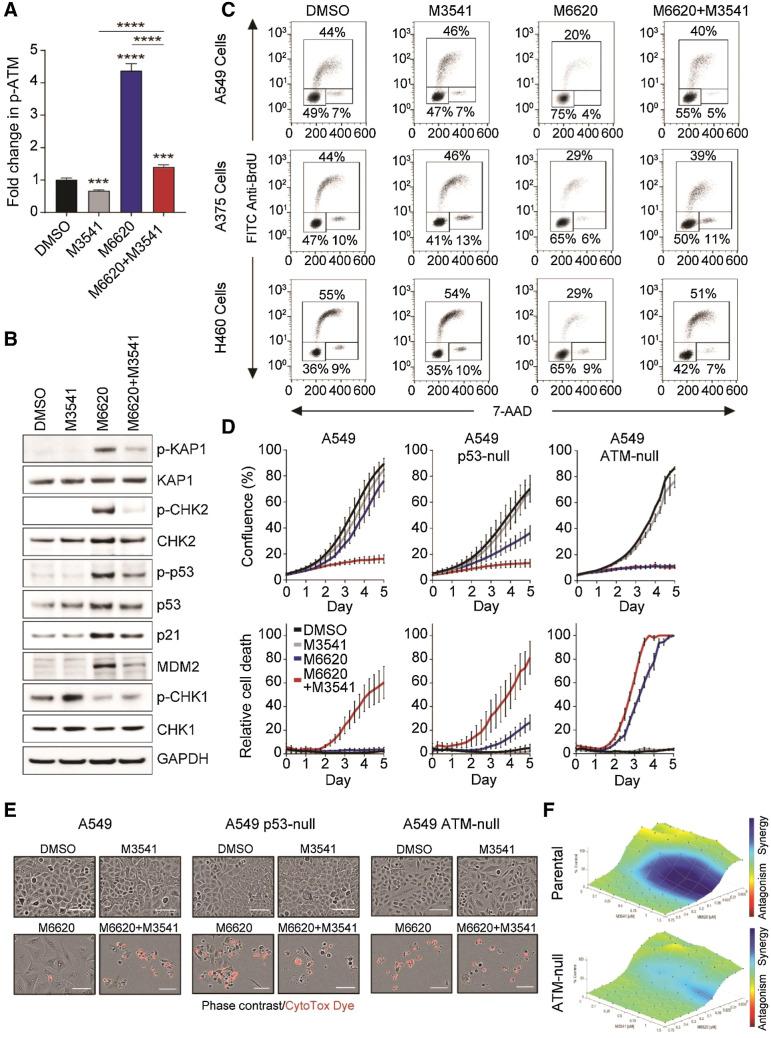
ATR inhibition has been shown to trigger compensatory activation of ATM-dependent HDR of DNA DSBs, as these two kinases cooperate to maintain genomic stability (52). We assessed the effect of the ATM inhibitor M3541 on M6620-induced phospho-ATM levels by sandwich ELISA. We chose 1 μmol/L concentration of M3541 because this suppresses radiation-induced phospho-ATM by over 90% in a variety of cancer cell lines (36). Treatment with M3541 for 24 hours decreased phospho-ATM levels slightly below baseline but dramatically reversed M6620-induced phospho-ATM levels (Fig. 2A). The effect of M3541 exposure on M6620-activated ATM signaling was further analyzed by Western blotting (Fig. 2B). The combination suppressed p-KAP1, p-CHK2, p-p53, and p21 levels elevated in response to M6620 alone. The combination also maintained p-CHK1 levels previously lowered by ATRi. The inability to effectively activate either ATR or ATM signaling in cells simultaneously exposed to M6620 and M3541 translated to a failure to trigger effective cell-cycle checkpoint response (Fig. 2C). While M3541 alone did not have any impact on the cell-cycle profiles of A549, A375, or H460 cells, addition of M3541 partially abrogated the M6620-induced G1 checkpoint and promoted cycling in all three cell lines (Fig. 2C). In cells with dysfunctional ATM-p53 signaling (A549 p53-null, A549 ATM-null, and HeLa), neither inhibitor alone nor their combination impacted cell-cycle distribution (Supplementary Fig. S3A).
Figure 2.
ATM inhibition by M3541 circumvents the M6620-induced G1 checkpoint, resulting in aberrant cell-cycle progression and cell death. A, Quantification of ATM autophosphorylation (p-ATMSer1981/total ATM) in A549 cells by MSD assay after 24 hours exposure to DMSO, M3541 (1 μmol/L), M6620 (200 nmol/L), or their combination. Error bars, SEM; ****, P < 0.0001 and ***, P < 0.001 by unpaired t test. B, Western blot analysis of components of the ATM/p53 signaling pathway in A549 cells after 24-hour exposure to DMSO, M3541 (1 μmol/L), M6620 (200 nmol/L), or their combination. C, BrdU/7-AAD cell-cycle analysis in A549, A375, and H460 cells after 24 hours after treatment with DMSO, M3541 (1 μmol/L), M6620 (200 nmol/L) or their combination. D, Confluence (top row) and relative cell death (bottom row) from IncuCyte live imaging in A549 parental, p53-null, and ATM-null cells exposed to DMSO, M3541 (1 μmol/L), M6620 (200 nmol/L) or their combination. Relative cell death was calculated from the number of cells positively stained with CytoTox dye normalized to confluence. Error bars, SEM. E, Representative images from IncuCyte live imaging (10× objective) of A549 parental, p53-null, and ATM-null cells after 6-day exposure to DMSO, M3541 (1 μmol/L), M6620 (200 nmol/L), or their combination. Scale bars = 100 μmol/L. F, Bliss synergy plots generated from CellTiter-Glo assay viability results. A549 paired isogenic cell lines (parental and ATM-null) were treated for 5 days with titrations of M3541 and M6620. Bliss synergy was determined using Combenefit software.
The consequences of dual ATR and ATM inhibition were further assessed by real-time cell imaging. While neither inhibitor alone impacted the parental A549 cells, the combined treatment with M3541 and M6620 hindered cell growth and induced cell death. The addition of M3541 further enhanced the effect on cell viability observed with M6620 alone in the A549 p53-null cells, while there was no additional effect in the A549 ATM-null cells (Fig. 2D and E). The enhanced cytotoxicity with dual ATR and ATM inhibition was also observed in three additional cell lines, A375, H460, and HeLa (Supplementary Fig. S3B and S3C). We next assessed whether dual ATR and ATM inhibition was synergistic using the isogenic parental and ATM-null A549 cancer cell lines (Fig. 2F). Cells were exposed to a concentration range of M6620 in the presence of increasing M3541 concentrations, and cell viability was assessed after 5 days. M3541 alone had minimal to no effect on the cell viability of either the parental or ATM-null A549 cells. However, although the parental cells were sensitive to M6620, especially at higher doses, there was an observable IC50 decrease in the ATM-null cells. Bliss excess analysis indicated synergism between M6620 and M3541 in the A549 parental cell line, but not in the ATM-null cell line.
ATM inhibition enhances cytotoxicity of ATRis by allowing mitotic entry of cancer cells with structurally damaged chromosomes
We further investigated the mechanism of cancer cell death following dual ATR/ATM inhibition. Halting the normal DNA damage response by combined ATR and ATM inhibition led to an enhanced accumulation of p-53BP1 foci (Fig. 3A and B; Supplementary Fig. S4), indicative of unrepaired DNA DSBs. As prolonged replication stress and suppression of the S-phase cell-cycle checkpoints promotes DNA damage that may carry over into mitosis, we examined spindle and chromosomal fidelity during the division of A549 cells exposed to vehicle, M6620, M3541, or M6620+M3541. Immunofluorescence analyses detected a high degree of abnormal mitotic events, primarily in the samples exposed to dual inhibitor treatment (Fig. 3C). These abnormal mitotic events included monopolar or multipolar spindle, chromosome bridges, improper chromosome condensation and alignment, and lagging chromosomes (Fig. 3D).
Figure 3.
Dual inhibition of ATR by M6620 and ATM by M3541 leads to chromosomal damage as cells progress through the cell cycle with unrepaired DSBs. A, Immunofluorescence analysis of phospho-53BP1 foci in A549 cells after 24-hour exposure to DMSO, M3541 (1 μmol/L), M6620 (50, 100, 200 nmol/L), or their combination. Error bars, SEM; ****, P < 0.0001; ***, P < 0.001; and **, P < 0.01 by unpaired t test. B, Representative immunofluorescence images of phospho-53BP1 foci in A549 cells after 1-day treatment with DMSO, M3541 (1 μmol/L), M6620 (50 nmol/L), or their combination. Scale bars = 10 μmol/L. C, Immunofluorescence analysis of abnormal mitotic events [chromosome bridging, lagging chromosome(s), misaligned tubulin, monopolar or multipolar spindles, or improper chromosome condensation and alignment] captured in unsynchronized A549 cells after 24 hours exposure to DMSO, M3541 (1 μmol/L), M6620 (50, 100, 200 nmol/L), or their combination. Error bars, SEM; ****, P < 0.0001; ***, P < 0.001; **, P < 0.01; and *, P < 0.05 by unpaired t test. D, Representative immunofluorescence images of abnormal mitotic events quantified in C. A549 cells were arrested in metaphase with colcemid (0.1 μg/mL) after 24-hour exposure to DMSO, M3541 (1 μmol/L), M6620 (200 nmol/L), or their combination. Ten randomly picked metaphase spreads were imaged and analyzed by spectral karyotyping for each treatment condition. E, Graph of the total number of chromosomal aberrations per metaphase spread and the occurrence of the specific type of chromosomal aberration (break, translocation, or deletion) across the 10 spreads for each treatment condition. Error bars, SD; ****, P < 0.0001; ***, P < 0.001; **, P < 0.01; and *, P < 0.05 by unpaired t test. F, Quantification of marker chromosomes per cell across the 10 spreads for each treatment condition. Error bars, SD; ****, P < 0.0001; ***, P < 0.001 by unpaired t test. G, Representative spectral karyotype images for each treatment condition.
We next investigated the extent of chromosome damage by spectral karyotyping. Proliferating A549 cells were treated with M3541, M6620, or the combination and then arrested in metaphase with colcemid. Treatment with M6620 alone led to a slight but significant increase in the number of chromosomal aberrations per cell, while dual M6620+M3541 treatment produced a twofold increase in the incidence of chromosomal aberrations compared with DMSO or single agents (Fig. 3E). Notably, M6620 alone or in combination with M3541 led to the generation of chromosomal breaks, which were not observed with DMSO or M3541 treatment (Fig. 3E). A large increase in the formation of marker chromosomes, unidentifiable chromosomes/fragments with gross structural damage (53, 54), was induced by exposure to the M6620+M3541 combination (Fig. 3F and G). Together, these series of experiments demonstrate that persistent DSBs resulting from simultaneous inhibition of ATR and ATM lead to structural chromosome alterations, compromising their ability to properly attach and segregate during mitosis, ultimately resulting in cell death. Because these mechanisms may also affect the viability of normal cells, we assessed the impact of dual ATM and ATR inhibition in two actively proliferating human embryonal fibroblast lines, IMR90 and WI38, by live cell imaging. While M6620 alone or in combination with M3541 caused growth inhibition in both cell lines, only a slight induction of cell death was observed (Supplementary Fig. S5A). However, treatment of proliferating IMR90 cells with staurosporine, an agent frequently used as an apoptosis inducer, led to a nearly complete ablation of the cell population (Supplementary Fig. S5B). Consistent with previous reports (21), these data suggest that, while the effect of M6620+M3541 combination is cytotoxic in most tested cancer cell lines, it is largely cytostatic in fibroblasts.
Newer generation of ATR and ATM inhibitors show synergistic combination activity
Although M6620 and M3541 are excellent tools for investigating cellular mechanisms, they have some limitations for clinical use. M6620 is only available for intravenous administration (55), and the low solubility of M3541 poses difficulties for achieving an optimal pharmacokinetic profile (56). However, newer optimized ATR (M4344) and ATM (M4076) inhibitors offer similar or better potency and selectivity, together with improved pharmacologic properties (16, 36). M4344 is an orally available potent and selective small-molecule inhibitor of ATR (14, 16). In comparative studies, M4344 alone increased the number of phospho-53BP1 and γH2AX foci, induced G1 checkpoint activation in cells with intact ATM/p53 signaling, and increased cell death in A549 p53-null and A549 ATM-null cells compared with the parental A549 cells (Supplementary Fig. S6), as previously demonstrated with M6620 (Fig. 1A; Supplementary Fig. S1A).
M4076 and M3541 are structurally related compounds with similar potency and selectivity, but M4076 has significantly improved water solubility and better pharmacokinetic profile in vivo (36, 57). As expected, experimental findings with M4076 in cellular studies were comparable with the M3541 results (36). The experimental results from the combination activity studies of M6620 and M4076 (Supplementary Fig. S7) were also comparable with the M6620+M3541 results (Fig. 2A, D, and E). Suppression of M6620-induced phosphorylation of ATM was effectively abrogated by the addition of M4076 (Supplementary Fig. S7A) as with M3541 (Fig. 2A). In addition, the results of real-time cell imaging showed that neither inhibitor alone affected the parental A549 cells, but the combined treatment with M4076 and M6620 hindered cell growth and induced cell death. As seen with M3541, the addition of M4076 further enhanced the effect on cell viability observed with M6620 alone in the A549 p53-null cells, while having no additional effect in the A549 ATM-null cells (Supplementary Fig. S7B and S7C). Dual inhibition of ATR (M4344) and ATM (M4076) increased the number of unrepaired DSBs (Supplementary Fig. S8) and enhanced cytotoxicity in several cancer cell lines (Supplementary Fig. S9). Taken together, these data confirmed that both ATM (M3541 or M4076) and ATR (M6620 or M4344) inhibitors have comparable combination potential that is derived from the same mechanism of action.
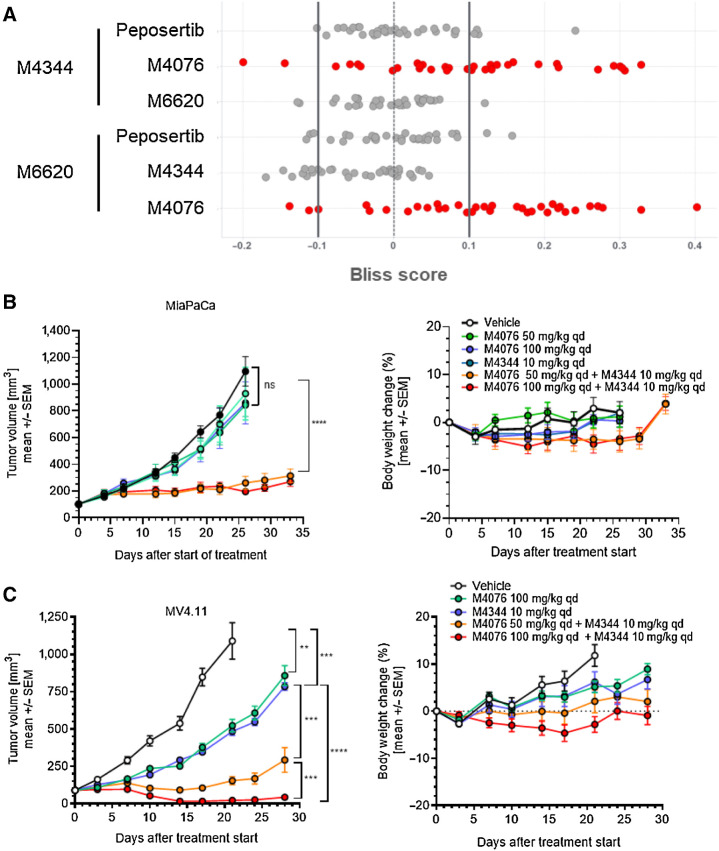
We then assessed the combination cytotoxicity of ATR (M6620 and M4344), ATM (M4076), and DNA-PK (peposertib, formerly M3814) inhibitors in 34 randomly selected cancer cell lines in vitro (Fig. 4A). Exponentially growing cells were exposed to ATRis used at single concentrations alone or in combination with M4076, peposertib, or M4344 used in a wide range of concentrations for 5 days. Cell growth and/or viability was determined by the sulforhodamin B assay as described previously (36, 40). The high Bliss excess scores measured in multiple cell lines indicated strong and comparable synergism between ATR and ATM inhibitors but not ATR and DNA-PK inhibitors. These results supported further exploration in vivo using the newer optimized ATR (M4344) and ATM (M4076) inhibitors.
Figure 4.
ATM inhibitor M4076 synergizes with ATRis M4344 or M6620 in cancer cells and mouse xenograft models. A, Pairwise analysis of the combination benefit of inhibitors of ATR (M4344, M6620), ATM (M4076), and DNA-PK (peposertib) in a panel of 34 randomly selected cancer cell lines. Cell lines were incubated with drug combinations for 5 days, and their growth and/or viability was determined by the sulforhodamin B assay as described previously (40). Bliss excess was calculated per drug and cell line and plotted (strong synergy >0.1; 0.1≥ weak to moderate additivity/antagonism ≥−0.1; strong antagonism <−0.1). B and C, The combination effect of M4344 and M4076 was evaluated in mice transplanted with MiaPaCa and MV4.11 cancer cells. Mice received subcutaneous injections in the right flank. M4344 (10 mg/kg), M4076 (50 or 100 mg/kg), or the combination thereof were applied orally, once a day throughout the study period, and M4344 was given 1 hour after M4076. Tumor volumes are shows as mean ± SEM (10 mice per group). For MiaPaCa2, P < 0.0001 for vehicle or any of the monotherapy treatment arms and M4076 100 mg/kg + M4344 10 mg/kg.
ATM inhibition enhances the antitumor efficacy of ATRis in vivo
We next aimed at assessing the antitumor potential of dual ATR and ATM inhibition in mouse models of human cancer using two xenograft models, MiaPaCa2 and MV4.11. In the MiaPaCa2 pancreatic cancer model, ATR (M4344) and ATM (M4076) inhibitors were inactive when given as single agents, but the combination resulted in complete tumor growth inhibition (Fig. 4B). The combination was further evaluated in the AML model MV4.11, which bears a mixed-lineage leukemia (MLL) fusion shown to mediate sensitivity to the ATRi AZ20 or the ATM inhibitor AZD0156 as monotherapies (58). In this model, M4344 and M4076 showed significant tumor growth inhibition as monotherapies, which was strongly enhanced in combination, leading to almost complete tumor regression (Fig. 4C). The combination was well tolerated in both studies based on body weight changes (Fig. 4B and C).
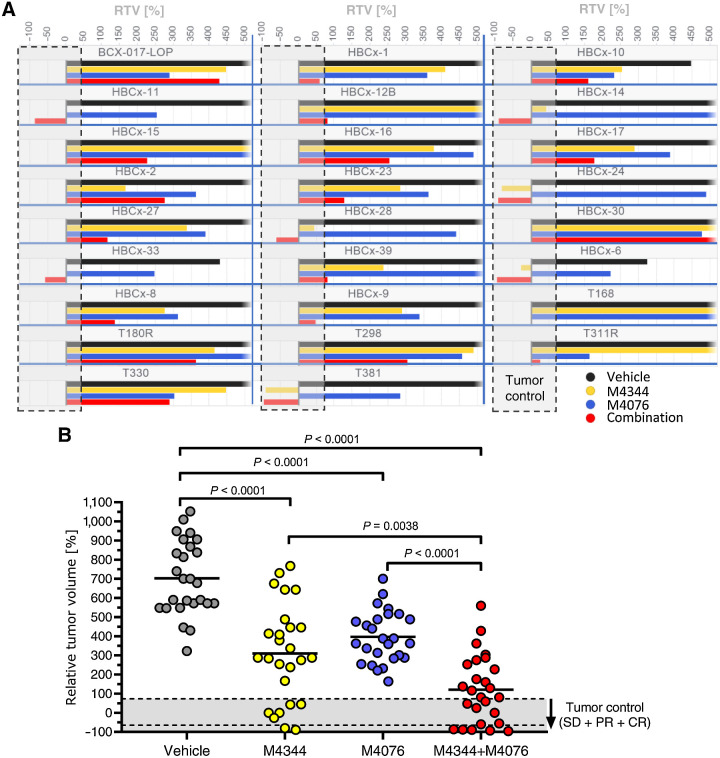
We then further explored the therapeutic potential of the M4344 and M4076 combination in a panel of 26 patient-derived TNBC xenograft (PDX) models. This indication was chosen because of the high medical need and prior clinical experience with DDR inhibitors. The ATM inhibitor M4076 improved the single-agent activity of the ATRi M4344 in most models by comparing RTV endpoints for the respective treatment groups (Fig. 5A). The mean RTV for all 26 TNBC models was 700% for the control group, 400% for the ATM inhibitor group, 310% for the ATRi group, and 120% for the combination (Fig. 5B). Single-agent ATM inhibitor was qualified as inactive after applying tumor volume adapted RECIST criteria. The ATRi M4344 showed a tumor control rate (including stable disease, partial response (PR) and complete response (CR) of approximately 27% (7/26) and an objective response rate (PR, CR) of approximately 8% (2/26). The combination treatment had a tumor control rate of approximately 42% (11/26) and an objective response rate of approximately 20% (5/26). These results indicated that ATM inhibitor potentiates the efficacy of ATRis in multiple TNBC PDX cancer models.
Figure 5.
ATM inhibitor M4076 enhances the efficacy of ATRi M4344 in PDX models of TNBC. The combination of M4344 and M4076 was tested in the 26 TNBC XenTech PDX panel. Mice were transplanted with tumor tissue and given a once-daily oral dose of M4344 (10 mg/kg) and a twice-daily oral dose of M4076 (50 mg/kg). M4344 was applied 30 to 45 minutes after the first M4076 dose of the day. RTVs calculated from the median of each treatment group (n = 3 mice per group) at the time when the controls reached 800 mm3 are plotted for each model and treatment group (A) and per treatment group for all 26 models (B).
Discussion
Targeting the DDR as a therapeutic strategy for cancer is of high interest in light of the success of PARP inhibitors (PARPis) in BRCA- and homologous recombination–deficient cancers (4). In particular, inhibition of the replication stress response with ATRis is a promising strategy because many ATRis have shown antitumor activities in preclinical models as monotherapies in biomarker-selected cancers and in combination with certain chemotherapies, radiation, and PARPis (11). However, selecting the appropriate clinical setting and combination partner for ATRis requires an improved understanding of the mechanistic basis of ATR inhibition in cancer cells and possible resistance mechanisms. Here, we show that ATR kinase inhibition activates the ATM signaling pathway and that dual inhibition of these two central DDR pathways produces synergistic cancer cell killing by abrogation of the ATM-p53 cell-cycle checkpoint and DSB repair processes.
The ATM kinase controls DSB-induced cell-cycle checkpoints and HDR to protect genomic integrity of proliferating mammalian cells. These functions are preserved in most cancers and are frequently associated with decreased response or resistance to DNA-damaging therapy. Therefore, the outcome of therapeutic strategies targeting DDR is likely to be affected by ATM functionality. This important role makes ATM a desirable target for inactivation in cancer (5). ATM inhibitors such as AZD0156, AZD1390, M3541, and M4076 have been shown to strongly potentiate DSB-inducing agents and offer combination partners for sensitizing cancer cells to radiation (36, 59), topoisomerase inhibitors (36, 60), and PARPis (36, 61). The ATM inhibitors AZD1390 and M4076 are currently in clinical development (12).
Genetically acquired ATM deficiency has been shown to increase sensitivity to ATRis and is currently being explored as a clinical biomarker for response (26). However, the frequency of this abnormality is relatively low. ATM mutations occur in approximately 5% of all human cancers, but the functional consequences of many of these mutations are not well understood (29–31, 35). It is becoming clearer that ATR-ATM synthetic lethality requires complete disabling of ATM function, and biallelic high-impact loss-of-function mutations are needed for full loss of ATM pathway activity. Clinical responses to the RP-3500 ATRi monotherapy were enriched in patients carrying the ATM mutation with biallelic mutations in the phase I/II TRESR trial (34). This requirement would limit the patient population, making patient selection a critical aspect of clinical trial design. Here, we explore the potential of selective ATM inhibition as an alternative strategy for imposing functional ATM deficiency in combination with ATRis.
We show that in cells with intact ATM signaling, ATR inhibition leads to increased DNA DSB damage, which activates the ATM-p53–driven G1 checkpoint. G1–S arrest sequesters a substantial portion of the cell population and prevents entry into DNA replication. This checkpoint response minimizes the potential of ATRis to generate DSBs during replication and the lethal consequences of dividing before their repair. Using two new ATM inhibitors (M3541 and M4076) and engineered p53-null and ATM-null isogenic clones of the p53/ATM wild-type A549 cells as molecular tools, we interrogated the importance of the canonical ATM functions. Deletion of TP53 largely abrogated ATM checkpoint function and increased the activity of ATRis M6620 and M4344. However, full disabling of the ATM pathway, which also suppressed ATM-dependent HDR was needed for maximal enhancement of their cytotoxicity. Inhibition of ATM by M3541 or M4076 was able to boost M6620 and M4344 activity in the parental A549 cells to the level observed in the A549 ATM-null cells, suggesting that it can effectively impose temporary functional ATM deficiency. Because the effect of ATM inhibitors is reversible, ATM activity could be controlled as needed.
Our experiments demonstrated that selective ATM inhibition strongly enhances the cytotoxicity of ATRis, and the main mechanism behind this effect is severe chromosomal damage arising from multiple unrepaired DSBs. Cancer cells entering mitosis with structurally impaired chromosomes are unable to properly attach and segregate at the mitotic spindle, leading to lagging chromosomes at anaphase, micronucleation, severe aneuploidy/polyploidy, and ultimately cell death. This mechanism is not unique to ATRis but rather a universal mode of cytotoxicity caused by DSB-inducing agents, including radiation and a variety of chemotherapeutic drugs. However, due to effective DSB repair and checkpoint controls, the cytotoxicity of these agents is limited by the fraction of the cell population with unrepaired breaks that enter mitosis. Pharmacologic DSB repair inhibitors can amplify the population of affected cancer cells, as we recently reported for the DNA-PK inhibitor peposertib in combination with radiation in p53-deficient cancers (37). The unique property of ATM inhibitors is that they can simultaneously suppress both DSB repair and the p53-driven checkpoint protection. Therefore, they can blunt p53 dependence of response and may not require patient selection based on TP53 status.
Our mechanistic and synergy findings using two ATR and two ATM inhibitors demonstrate that the combination effects are not specific to individual compounds and provide a clear rationale for dual inhibition of ATR and ATM as a therapeutic strategy. This combination approach could be tested clinically with many other ATR and ATM inhibitors in clinical development (12). The cancer-specific cytotoxicity is synergistic across a wide concentration range and diverse set of tumor-derived cell lines. These findings were extended beyond the in vitro setting with the use of newer pharmacologically optimized ATR (M4344) and ATM (M4076) inhibitors. Initial M4344 and M4076 combination testing in two xenograft models were encouraging and led to an in vivo screen in 26 TNBC patient-derived tumor models. We observed RECIST relevant antitumor activity with a tumor control rate (SD+PR+CR) of 42% in the combination arm. It is possible that optimization of dosing and scheduling of the combination would further improve the efficacy and RECIST response rates and that data reported herein are underestimating the therapeutic benefit.
As ATM plays a key cell protective role in the DDR, tolerability emerges as the main concern in targeting its activity for cancer therapy. ATM dysfunction observed in ataxia telangiectasia, a debilitating genetic disorder, can lead to enhanced sensitivity to radiation and cancer development (62). However, earlier studies have shown that temporary suppression of ATM activity is tolerated in irradiated cultured cells (63). Our in vitro experiments indicated that the combined ATR/ATM inhibition could arrest but not kill proliferating human fibroblasts cultures. The M4344/M4076 treatment was well tolerated in vivo, and the increased efficacy was not followed by proportionally increased toxicity. In fact, no overt toxicity was seen during treatment and the observation period in the 28 experimental mouse models. However, more elaborate in vivo toxicology studies and ultimately the clinical experience with the combination could inform us of the safety window for this novel combination. The tested TNBC PDX models showed encouraging activity in this difficult-to-treat cancer segment with high unmet medical need. Taken together, the in vitro and in vivo data presented here strongly support further clinical exploration of ATR/ATM inhibitor combination.
Supplementary Material
Supplementary figures
Acknowledgments
This research was supported by EMD Serono (CrossRef Funder ID:10.13039/100004755).
We thank Alan D'Andrea (Dana-Farber Cancer Institute, Boston, MA, USA) for the kind gift of ATM-deficient A549 cells and Florian Lorenz (the healthcare business of Merck KGaA, Darmstadt, Germany) for his editorial comments.
The publication costs of this article were defrayed in part by the payment of publication fees. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
This article is featured in Highlights of This Issue, p. 805
Footnotes
Note: Supplementary data for this article are available at Molecular Cancer Therapeutics Online (http://mct.aacrjournals.org/).
Authors' Disclosures
A. Zimmermann reports all authors are or have been employees of the healthcare business of Merck KGaA, Darmstadt, Germany, or EMD Serono, Billerica, MA, USA. Merck KGaA, Darmstadt, Germany, and/or its affiliates have certain rights in patents, patent applications pertaining to berzosertib/M6620 (WO10071837), gartisertib/M4344 (WO14089379), M4076 (WO20193660), M3541 (WO12028233), and peposertib (WO14183850). However, none of the authors are named inventors. A. Blaukat was an employee of the healthcare business of Merck KGaA, Darmstadt, Germany, when this article was prepared. No disclosures were reported by the other authors.
Authors' Contributions
A. Turchick: Conceptualization, data curation, formal analysis, investigation, methodology, writing–original draft, writing–review and editing. A. Zimmermann: Conceptualization, resources, data curation, formal analysis, supervision, validation, investigation, writing–original draft, writing–review and editing. L.-Y. Chiu: Conceptualization, data curation, formal analysis, methodology, writing–review and editing. H. Dahmen: Conceptualization, resources, formal analysis. B. Elenbaas: Supervision, writing–review and editing. F.T. Zenke: Resources, data curation, writing–review and editing. A. Blaukat: Resources, funding acquisition, writing–review and editing. L.T. Vassilev: Conceptualization, resources, formal analysis, supervision, writing–original draft, project administration, writing–review and editing.
References
- 1. Blackford AN, Jackson SP. ATM, ATR, and DNA-PK: the trinity at the heart of the DNA damage response. Mol Cell 2017;66:801–17. [DOI] [PubMed] [Google Scholar]
- 2. Majidinia M, Yousefi B. DNA repair and damage pathways in breast cancer development and therapy. DNA Repair 2017;54:22–9. [DOI] [PubMed] [Google Scholar]
- 3. Jackson SP, Bartek J. The DNA-damage response in human biology and disease. Nature 2009;461:1071–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. O'Connor MJ. Targeting the DNA damage response in cancer. Mol Cell 2015;60:547–60. [DOI] [PubMed] [Google Scholar]
- 5. Bradbury A, Hall S, Curtin N, Drew Y. Targeting ATR as cancer therapy: a new era for synthetic lethality and synergistic combinations? Pharmacol Ther 2020;207:107450. [DOI] [PubMed] [Google Scholar]
- 6. San Filippo J, Sung P, Klein H. Mechanism of eukaryotic homologous recombination. Annu Rev Biochem 2008;77:229–57. [DOI] [PubMed] [Google Scholar]
- 7. Zou L, Elledge SJ. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science 2003;300:1542–8. [DOI] [PubMed] [Google Scholar]
- 8. Saldivar JC, Cortez D, Cimprich KA. The essential kinase ATR: ensuring faithful duplication of a challenging genome. Nat Rev Mol Cell Biol 2017;18:622–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Costanzo V, Shechter D, Lupardus PJ, Cimprich KA, Gottesman M, Gautier J. An ATR- and Cdc7-dependent DNA damage checkpoint that inhibits initiation of DNA replication. Mol Cell 2003;11:203–13. [DOI] [PubMed] [Google Scholar]
- 10. Foote KM, Lau A, Nissink JW. Drugging ATR: progress in the development of specific inhibitors for the treatment of cancer. Future Med Chem 2015;7:873–91. [DOI] [PubMed] [Google Scholar]
- 11. Lecona E, Fernandez-Capetillo O. Targeting ATR in cancer. Nat Rev Cancer 2018;18:586–95. [DOI] [PubMed] [Google Scholar]
- 12. Ngoi NYL, Pham MM, Tan DSP, Yap TA. Targeting the replication stress response through synthetic lethal strategies in cancer medicine. Trends Cancer 2021;7:930–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Charrier JD, Durrant SJ, Golec JM, Kay DP, Knegtel RM, MacCormick S, et al. Discovery of potent and selective inhibitors of ataxia telangiectasia mutated and Rad3 related (ATR) protein kinase as potential anticancer agents. J Med Chem 2011;54:2320–30. [DOI] [PubMed] [Google Scholar]
- 14. Gorecki L, Andrs M, Rezacova M, Korabecny J. Discovery of ATR kinase inhibitor berzosertib (VX-970, M6620): clinical candidate for cancer therapy. Pharmacol Ther 2020;210:107518. [DOI] [PubMed] [Google Scholar]
- 15. Hall AB, Newsome D, Wang Y, Boucher DM, Eustace B, Gu Y, et al. Potentiation of tumor responses to DNA damaging therapy by the selective ATR inhibitor VX-970. Oncotarget 2014;5:5674–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Jo U, Senatorov IS, Zimmermann A, Saha LK, Murai Y, Kim SH, et al. Novel and highly potent ATR inhibitor M4344 kills cancer cells with replication stress, and enhances the chemotherapeutic activity of widely used DNA damaging agents. Mol Cancer Ther 2021;20:1431–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Leszczynska KB, Dobrynin G, Leslie RE, Ient J, Boumelha AJ, Senra JM, et al. Preclinical testing of an Atr inhibitor demonstrates improved response to standard therapies for esophageal cancer. Radiother Oncol 2016;121:232–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Fokas E, Prevo R, Pollard JR, Reaper PM, Charlton PA, Cornelissen B, et al. Targeting ATR in vivo using the novel inhibitor VE-822 results in selective sensitization of pancreatic tumors to radiation. Cell Death Dis 2012;3:e441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Tu X, Kahila MM, Zhou Q, Yu J, Kalari KR, Wang L, et al. ATR inhibition is a promising radiosensitizing strategy for triple-negative breast cancer. Mol Cancer Ther 2018;17:2462–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jossé R, Martin SE, Guha R, Ormanoglu P, Pfister TD, Reaper PM, et al. ATR inhibitors VE-821 and VX-970 sensitize cancer cells to topoisomerase i inhibitors by disabling DNA replication initiation and fork elongation responses. Cancer Res 2014;74:6968–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Reaper PM, Griffiths MR, Long JM, Charrier JD, Maccormick S, Charlton PA, et al. Selective killing of ATM- or p53-deficient cancer cells through inhibition of ATR. Nat Chem Biol 2011;7:428–30. [DOI] [PubMed] [Google Scholar]
- 22. Kwok M, Davies N, Agathanggelou A, Smith E, Petermann E, Yates E, et al. Synthetic lethality in chronic lymphocytic leukaemia with DNA damage response defects by targeting the ATR pathway. Lancet 2015;385:S58. [DOI] [PubMed] [Google Scholar]
- 23. Kwok M, Davies N, Agathanggelou A, Smith E, Oldreive C, Petermann E, et al. ATR inhibition induces synthetic lethality and overcomes chemoresistance in TP53- or ATM-defective chronic lymphocytic leukemia cells. Blood 2016;127:582–95. [DOI] [PubMed] [Google Scholar]
- 24. Wengner AM, Siemeister G, Lücking U, Lefranc J, Wortmann L, Lienau P, et al. The novel ATR inhibitor BAY 1895344 is efficacious as monotherapy and combined with DNA damage-inducing or repair-compromising therapies in preclinical cancer models. Mol Cancer Ther 2020;19:26–38. [DOI] [PubMed] [Google Scholar]
- 25. Roulston A, Zimmermann M, Papp R, Skeldon A, Pellerin C, Dumas-Bérube É, et al. RP-3500: a novel, potent, and selective ATR inhibitor that is effective in preclinical models as a monotherapy and in combination with PARP inhibitors. Mol Cancer Ther 2022;21:245–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Yap TA, Tan DSP, Terbuch A, Caldwell R, Guo C, Goh BC, et al. First-in-human trial of the oral ataxia telangiectasia and RAD3-related (ATR) inhibitor BAY 1895344 in patients with advanced solid tumors. Cancer Discov 2021;11:80–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Giono LE, Manfredi JJ. The p53 tumor suppressor participates in multiple cell cycle checkpoints. J Cell Physiol 2006;209:13–20. [DOI] [PubMed] [Google Scholar]
- 28. Gurpinar E, Vousden KH. Hitting cancers' weak spots: vulnerabilities imposed by p53 mutation. Trends Cell Biol 2015;25:486–95. [DOI] [PubMed] [Google Scholar]
- 29. Choi M, Kipps T, Kurzrock R. ATM mutations in cancer: therapeutic implications. Mol Cancer Ther 2016;15:1781–91. [DOI] [PubMed] [Google Scholar]
- 30. Knijnenburg TA, Wang L, Zimmermann MT, Chambwe N, Gao GF, Cherniack AD, et al. Genomic and molecular landscape of DNA damage repair deficiency across The Cancer Genome Atlas. Cell Rep 2018;23:239–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Jette NR, Kumar M, Radhamani S, Arthur G, Goutam S, Yip S, et al. ATM-deficient cancers provide new opportunities for precision oncology. Cancers 2020;12:687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kaur H, Salles DC, Murali S, Hicks JL, Nguyen M, Pritchard CC, et al. Genomic and clinicopathologic characterization of ATM-deficient prostate cancer. Clin Cancer Res 2020;26:4869–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sundar R, Miranda S, Rodrigues DN, Chénard-Poirier M, Dolling D, Clarke M, et al. Ataxia telangiectasia mutated protein loss and benefit from oxaliplatin-based chemotherapy in colorectal cancer. Clin Colorectal Cancer 2018;17:280–4. [DOI] [PubMed] [Google Scholar]
- 34. Yap TA, Silverman IM, Fontana E, Lee E, Spigel D, Højgaard M, et al. Genomic and pathologic determinants of response to RP-3500, an ataxia telangiectasia and Rad3-related inhibitor (ATRi), in patients (pts) with DNA damage repair (DDR) loss-of-function (LOF) mutant tumors in the Phase 1/2 TRESR trial [abstract]. In: Proceedings of the American Association for Cancer Research Annual Meeting 2022; 2022 Apr 8–13. Philadelphia (PA): AACR; Cancer Res 2022;82(suppl 12):Abstract nr CT030.
- 35. Neeb A, Herranz N, Arce-Gallego S, Miranda S, Buroni L, Yuan W, et al. Advanced prostate cancer with ATM loss: PARP and ATR inhibitors. Eur Urol 2021;79:200–11. [DOI] [PubMed] [Google Scholar]
- 36. Zimmermann A, Zenke FT, Chiu LY, Dahmen H, Pehl U, Fuchss T, et al. A new class of selective ATM inhibitors as combination partners of DNA double-strand break inducing cancer therapies. Mol Cancer Ther 2022;21:859–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sun Q, Guo Y, Liu X, Czauderna F, Carr MI, Zenke FT, et al. Therapeutic implications of p53 status on cancer cell fate following exposure to ionizing radiation and the DNA-PK inhibitor M3814. Mol Cancer Res 2019;17:2457–68. [DOI] [PubMed] [Google Scholar]
- 38. Fuchss T, Mederski W, Zenke F. Imidazo[4,5-C]quinolines as DNA-PK inhibitors patent US 2013/0172337; 2013.
- 39. Fuchss T, Becker A, Kubas H, Graedler U. Imidazolonylquinoline compounds and therapeutic uses thereof patent WO 2020/193660; 2020.
- 40. Zenke FT, Zimmermann A, Sirrenberg C, Dahmen H, Kirkin V, Pehl U, et al. Pharmacological inhibitor of DNA-PK, M3814, potentiates radiotherapy and regresses human tumors in mouse models. Mol Cancer Ther 2020;19:1091–101. [DOI] [PubMed] [Google Scholar]
- 41. Turchick A, Hegan DC, Jensen RB, Glazer PM. A cell-penetrating antibody inhibits human RAD51 via direct binding. Nucleic Acids Res 2017;45:11782–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Di Veroli GY, Fornari C, Wang D, Mollard S, Bramhall JL, Richards FM, et al. Combenefit: an interactive platform for the analysis and visualization of drug combinations. Bioinformatics 2016;32:2866–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ruf-Zamojski F, Ge Y, Pincas H, Shan J, Song Y, Hines N, et al. Cytogenetic, genomic, and functional characterization of pituitary gonadotrope cell lines. J Endocr Soc 2019;3:902–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Therasse P, Arbuck SG, Eisenhauer EA, Wanders J, Kaplan RS, Rubinstein L, et al. New guidelines to evaluate the response to treatment in solid tumors. European Organization for Research and Treatment of Cancer, National Cancer Institute of the United States, National Cancer Institute of Canada. J Natl Cancer Inst 2000;92:205–16. [DOI] [PubMed] [Google Scholar]
- 45. Min A, Im SA, Jang H, Kim S, Lee M, Kim DK, et al. AZD6738, a novel oral inhibitor of ATR, induces synthetic lethality with ATM deficiency in gastric cancer cells. Mol Cancer Ther 2017;16:566–77. [DOI] [PubMed] [Google Scholar]
- 46. Sanjiv K, Hagenkort A, Calderón-Montaño JM, Koolmeister T, Reaper PM, Mortusewicz O, et al. Cancer-specific synthetic lethality between ATR and CHK1 kinase activities. Cell Rep 2016;17:3407–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Knegtel R, Charrier JD, Durrant S, Davis C, O'Donnell M, Storck P, et al. Rational design of 5-(4-(Isopropylsulfonyl)phenyl)-3-(3-(4-((methylamino)methyl)phenyl)isoxazol-5-yl)pyrazin-2-amine (VX-970, M6620): optimization of intra- and intermolecular polar interactions of a New Ataxia telangiectasia mutated and Rad3-related (ATR) kinase inhibitor. J Med Chem 2019;62:5547–61. [DOI] [PubMed] [Google Scholar]
- 48. Burma S, Chen BP, Murphy M, Kurimasa A, Chen DJ. ATM phosphorylates histone H2AX in response to DNA double-strand breaks. J Biol Chem 2001;276:42462–7. [DOI] [PubMed] [Google Scholar]
- 49. Gupta A, Hunt CR, Chakraborty S, Pandita RK, Yordy J, Ramnarain DB, et al. Role of 53BP1 in the regulation of DNA double-strand break repair pathway choice. Radiat Res 2014;181:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. El-Deiry WS. p21(WAF1) mediates cell-cycle inhibition, relevant to cancer suppression and therapy. Cancer Res 2016;76:5189–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Ajay AK, Meena AS, Bhat MK. Human papillomavirus 18 E6 inhibits phosphorylation of p53 expressed in HeLa cells. Cell Biosci 2012;2:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Weber AM, Ryan AJ. ATM and ATR as therapeutic targets in cancer. Pharmacol Ther 2015;149:124–38. [DOI] [PubMed] [Google Scholar]
- 53. Jafari-Ghahfarokhi H, Moradi-Chaleshtori M, Liehr T, Hashemzadeh-Chaleshtori M, Teimori H, Ghasemi-Dehkordi P. Small supernumerary marker chromosomes and their correlation with specific syndromes. Adv Biomed Res 2015;4:140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Bochtler T, Granzow M, Stölzel F, Kunz C, Mohr B, Kartal-Kaess M, et al. Marker chromosomes can arise from chromothripsis and predict adverse prognosis in acute myeloid leukemia. Blood 2017;129:1333–42. [DOI] [PubMed] [Google Scholar]
- 55. Yap TA, O'Carrigan B, Penney MS, Lim JS, Brown JS, de Miguel Luken MJ, et al. Phase I trial of first-in-class ATR inhibitor M6620 (VX-970) as monotherapy or in combination with carboplatin in patients with advanced solid tumors. J Clin Oncol 2020;38:3195–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Waqar SN, Robinson C, Olszanski AJ, Spira A, Hackmaster M, Lucas L, et al. Phase I trial of ATM inhibitor M3541 in combination with palliative radiotherapy in patients with solid tumors. Invest New Drugs 2022;40:596–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Saal C, Becker A, Krier M, Fuchß T. Atropisomerism - a neglected way to escape out of solubility flatlands. J Pharm Sci 2021;111:206–13. [DOI] [PubMed] [Google Scholar]
- 58. Morgado-Palacin I, Day A, Murga M, Lafarga V, Anton ME, Tubbs A, et al. Targeting the kinase activities of ATR and ATM exhibits antitumoral activity in mouse models of MLL-rearranged AML. Sci Signal 2016;9:ra91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Durant ST, Zheng L, Wang Y, Chen K, Zhang L, Zhang T, et al. The brain-penetrant clinical ATM inhibitor AZD1390 radiosensitizes and improves survival of preclinical brain tumor models. Sci Adv 2018;4:eaat1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Davis SL, Hartman SJ, Bagby SM, Schlaepfer M, Yacob BW, Tse T, et al. ATM kinase inhibitor AZD0156 in combination with irinotecan and 5-fluorouracil in preclinical models of colorectal cancer. BMC Cancer 2022;22:1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Mak JPY, Ma HT, Poon RYC. Synergism between ATM and PARP1 inhibition involves DNA damage and abrogating the G(2) DNA damage checkpoint. Mol Cancer Ther 2020;19:123–34. [DOI] [PubMed] [Google Scholar]
- 62. Shiloh Y, Lederman HM. Ataxia-telangiectasia (A-T): an emerging dimension of premature ageing. Ageing Res Rev 2017;33:76–88. [DOI] [PubMed] [Google Scholar]
- 63. Rainey MD, C ME, Stanton RV, Kastan MB. Transient inhibition of ATM kinase is sufficient to enhance cellular sensitivity to ionizing radiation. Cancer Res 2008;68:7466–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary figures



![Figure 3. Dual inhibition of ATR by M6620 and ATM by M3541 leads to chromosomal damage as cells progress through the cell cycle with unrepaired DSBs. A, Immunofluorescence analysis of phospho-53BP1 foci in A549 cells after 24-hour exposure to DMSO, M3541 (1 μmol/L), M6620 (50, 100, 200 nmol/L), or their combination. Error bars, SEM; ****, P < 0.0001; ***, P < 0.001; and **, P < 0.01 by unpaired t test. B, Representative immunofluorescence images of phospho-53BP1 foci in A549 cells after 1-day treatment with DMSO, M3541 (1 μmol/L), M6620 (50 nmol/L), or their combination. Scale bars = 10 μmol/L. C, Immunofluorescence analysis of abnormal mitotic events [chromosome bridging, lagging chromosome(s), misaligned tubulin, monopolar or multipolar spindles, or improper chromosome condensation and alignment] captured in unsynchronized A549 cells after 24 hours exposure to DMSO, M3541 (1 μmol/L), M6620 (50, 100, 200 nmol/L), or their combination. Error bars, SEM; ****, P < 0.0001; ***, P < 0.001; **, P < 0.01; and *, P < 0.05 by unpaired t test. D, Representative immunofluorescence images of abnormal mitotic events quantified in C. A549 cells were arrested in metaphase with colcemid (0.1 μg/mL) after 24-hour exposure to DMSO, M3541 (1 μmol/L), M6620 (200 nmol/L), or their combination. Ten randomly picked metaphase spreads were imaged and analyzed by spectral karyotyping for each treatment condition. E, Graph of the total number of chromosomal aberrations per metaphase spread and the occurrence of the specific type of chromosomal aberration (break, translocation, or deletion) across the 10 spreads for each treatment condition. Error bars, SD; ****, P < 0.0001; ***, P < 0.001; **, P < 0.01; and *, P < 0.05 by unpaired t test. F, Quantification of marker chromosomes per cell across the 10 spreads for each treatment condition. Error bars, SD; ****, P < 0.0001; ***, P < 0.001 by unpaired t test. G, Representative spectral karyotype images for each treatment condition.](https://cdn.ncbi.nlm.nih.gov/pmc/blobs/df01/10320480/f6c4b4ff74e1/859fig3.jpg)