Abstract
Purpose
Cardiac radioablation is an emerging therapy for recurrent ventricular tachycardia. Electrophysiology (EP) data, including electroanatomic maps (EAM) and electrocardiographic imaging (ECGI), provide crucial information for defining the arrhythmogenic target volume. The absence of standardized workflows and software tools to integrate the EP maps into a radiation planning system limits their use. This study developed a comprehensive software tool to enable efficient utilization of the mapping for cardiac radioablation treatment planning.
Methods and Materials
The tool, HeaRTmap, is a Python-scripted plug-in module on the open-source 3D Slicer software platform. HeaRTmap is able to import EAM and ECGI data and visualize the maps in 3D Slicer. The EAM is translated into a 3D space by registration with cardiac magnetic resonance images (MRI) or computed tomography (CT). After the scar area is outlined on the mapping surface, the tool extracts and extends the annotated patch into a closed surface and converts it into a structure set associated with the anatomic images. The tool then exports the structure set and the images as The Digital Imaging and Communications in Medicine Standard in Radiotherapy for a radiation treatment planning system to import. Overlapping the scar structure on simulation CT, a transmural target volume is delineated for treatment planning.
Results
The tool has been used to transfer Ensite NavX EAM data into the Varian Eclipse treatment planning system in radioablation on 2 patients with ventricular tachycardia. The ECGI data from CardioInsight was retrospectively evaluated using the tool to derive the target volume for a patient with left ventricular assist device, showing volumetric matching with the clinically used target with a Dice coefficient of 0.71.
Conclusions
HeaRTmap smoothly fuses EP information from different mapping systems with simulation CT for accurate definition of radiation target volume. The efficient integration of EP data into treatment planning potentially facilitates the study and adoption of the technique.
Introduction
Ventricular tachycardia (VT) is the most common cause of sudden cardiac death in the United States.1 VT often arises from local structural damage in the ventricular muscle resulting from myocardial infraction or other heart diseases. The scar creates a reentry circuit or self-activated myocardial abnormality, leading to cardiac arrhythmia. Scar related ventricular arrhythmia can be challenging to treat and may persist despite pharmacologic and catheter-based therapy. Cardiac radioablation (CR), also referred as cardiac stereotactic body radiation therapy, is a novel emerging treatment option for VT.2 A number of studies have demonstrated the efficacy of the treatment in reducing VT burden in patients refractory to drugs and standard catheter ablation3, 4, 5 and in patients with advanced heart failure.6
CR delivers ablative radiation dose precisely to the arrhythmogenic scar while minimizing damage to surrounding tissue. Stereotactic body radiation therapy planning is performed on target volumes and organs at risk contoured on 3-dimensional (3D) simulation computed tomography (CT). The location within what may be extensive myocardial scar is often most precisely determined during a catheter-based procedure in which the local electrical properties of the myocardium are registered in 3D space to create an electroanatomic mapping (EAM). Patients referred for CR typically have ventricular arrhythmia refractory to catheter ablation, and thus the EAM can provide critical information for determination of CR target volume. However, integration of surface-based electrophysiology (EP) data poses a technical barrier for enabling a modern radiation treatment planning system (TPS) to use the information.
A current clinically used solution to integrate EAM is limited to side-by-side visual comparison of 3D simulation images with the surface mapping. Recently, Brett et al implemented a workflow that converted the maps into binary volumes and fused the volumes with simulation CT using anatomic landmarks.7 Hohmann et al developed a plugin module on the open-source software, 3D Slicer, to read and use EAM obtained on 3 commonly used mapping systems.8 Both studies were performed for specific EAM data and required physicians to outline the scars on 2-dimensional image slices based on the landmarks defined on the EAM surfaces. To improve the workflow, we incorporated the 2 methods and developed a software tool, named as HeaRTmap, a 3D Slicer module that enables integration of EP information for CR treatment planning with minimal user operations. The tool assembled a number of functionalities in 3D Slicer into a comprehensive solution to facilitate clinical implementation of the emerging treatment.
Methods and Materials
EAM and electrocardiographic imaging (ECGI) have been regularly used for CR target definition.9 ECGI builds patient-specific heart-torso geometries from anatomic MR or CT images and reconstructs cardiac activation time and/or voltage maps using the potentials recorded by skin electrodes during ventricular arrhythmia induced in a noninvasive program stimulation procedure. Tracking the mapping catheters in an invasive EP procedure, EAM creates electrical property maps of the myocardial surface using the signals recorded with catheter electrodes. In comparison, EAM data are in their native space, while ECGI maps are spatially associated with 3D anatomic images.
As the mapping surface data are incompatible with a radiation TPS, we designed the tool to enable outlining a scar on EP maps and then converting the contours into The Digital Imaging and Communications in Medicine Standard in Radiotherapy (DICOM RT) that a TPS can read and process. A modern TPS typically has no capabilities to directly register surfaces to simulation CT but has rich functions for 3D volume registration. Multimodal image registration is routinely performed in the treatment planning process. Therefore, our workflow enforces landmark and surface registrations to translate EAM into 3D anatomic image space. Subsequent registration of the 3D images to simulation CT in a TPS simultaneously fuses the scar for delineating target volume on the CT.
EAM
HeaRTmap integrated the open-source 3D Slicer plugin EAMapReader,8 which can import EAM from currently available mapping systems, including CARTO 3 (Biosense Webster), Ensite (Abbott), and RHYTHMIA HDx (Boston Scientific). The maps represent electrical properties measured on the catheter-sampling points (ie, vertices), forming a triangular mesh of the endocardial surface. In 3D Slicer, dedicated color tables (Fig. 1) were built for Ensite EAM that correspond to the ones that we are currently using, so the activation time and voltage maps display in the 3D view in similar color scales as in the vendor-supplied mapping software.
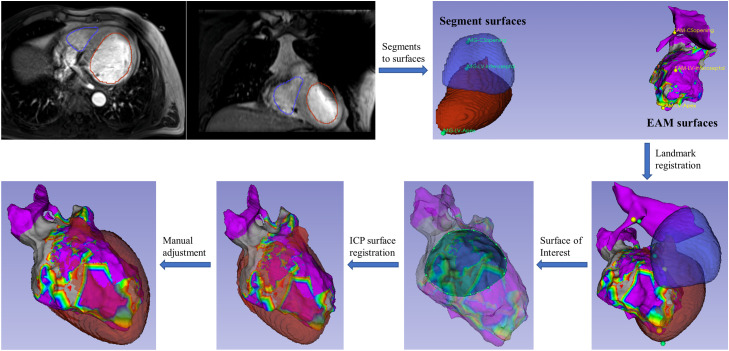
Figure 1.
Generation of arrhythmogenic scar from electrophysiology maps. (A) Manually outline scar area. (B) Create scar closed surface. (C) Overlap scar structure on anatomic images. In the images, I and II indicate electroanatomic map and electrocardiographic imaging, respectively. Curves on the images of (C): blue is left ventricle segmented on the images; cyan is left ventricle endocardium converted from electroanatomic map surface; and red is the scar structure.
The tool uses the markup editor module to enable manually drawing a closed curve on the EAM surface to outline scar area. Physicians can manipulate the surface 3D view and the mapping color tables to assist the process. As Fig. 1 shows, the software subsequently cuts the annotated patch from the EP surface, then perpendicularly extends the open patch outward by 2 mm to generate a closed surface. The closed surface represents the outlined scar, which can be converted to a scar structure into a structure set associated with 3D image space of the EAM. The scar structure overlaps on the image slices, illustrating the arrhythmogenic endocardial area that is derived from EP information by a multidisciplinary physician team.
The EAM data are translated to 3D image space by registering it with a cardiac MR or CT volume before scar contouring, which is necessary for the scar structure to spatially associate with the image data. Figure 2 shows the registration flowchart. After cardiac MR/CT data are imported into 3D Slicer, the endocardial chambers and other structures are contoured and converted to cardiac surface models to enable surface-based registration with EAM. The tool aggregates several registration functionalities in 3D Slicer to carry out the task. A typical procedure starts with manually placing landmark pairs on the EAM and cardiac surfaces and performing landmark registration for initial alignment. A closed curve can be manually drawn on the EAM to extract a partial surface of interest that is a high-quality mesh derived from densely catheter sampling in the EP study. Automatic iterative closest point registration of the partial surface with the cardiac surfaces is then conducted. Lastly, the surface alignment is manually reviewed and refined in the transformation module to finalize the registration of EAM to 3D cardiac images.
Figure 2.
Registration of EAM surface to cardiac magnetic resonance imaging. It starts from segmentation of left atrial and left ventricular endocardium on the images and follows a series of surface registrations. Abbreviations: EAM = electroanatomic map; ICP = iterative closest point.
The closed surfaces of the scar and the mapped anatomies are converted to structures in a structure set for the cardiac MR/CT. Functions for conversions between structures and surface models are included in the tool. In addition, an interface to export the 3D images and structure set as DICOM RT files is built. DICOM RT is the standard protocol for radiation therapy to use and transfer patient data.10 Subsequently, a TPS reads the files and integrates the EP structures for delineating a transmural target volume for irradiation.
ECGI
ECGI cardioelectrical maps are created in association with their 3D anatomic images. The same workflow, except registering the maps to additional cardiac images, is used to integrate ECGI maps in a TPS. The CardioInsight System (Medtronic Inc) combines ECG signals from the skin mapping vest and CT scan of a patient to construct EP maps on the epicardial surfaces that are contoured from the CT.11 The system exports CT DICOM data, a triangulated surface mesh, anatomic landmarks, and electric scalar data on the mesh vertices. A reading function is designed in HeaRTmap to import the CT and EP maps into 3D Slicer. The same color tables that are used in the ECGI mapping system are built to display the maps. Other anatomy surfaces that have been contoured in the mapping process are also imported using the tool. The scar area is then outlined on the electrical mapping and converted into a closed surface. The surfaces are converted to structures as a structure set for the ECGI CT. Finally, the CT and structure set are exported as DICOM RT data that can be imported into a TPS to aid in target volume definition.
Software implementation
The tool was written using Python language as a plug-in module for 3D Slicer. It is seamlessly integrated into the platform's main user interface.12 The various functionalities included in the system can be invoked to process and visualize 3D image and surfaces. However, these functions are provided across different modules, making it difficult to use for a specific application. HeaRTmap aggregates these functions as a comprehensive tool, by either combining the source codes or invoking the modules. These include the EAMapReader to read EAM maps,8 DICOM RT to read and export anatomic images and RT structures,13 iterative closest point registration, and Insight Segmentation and Registration Tookit/Visualization Toolkit processing of surface meshes.
Results
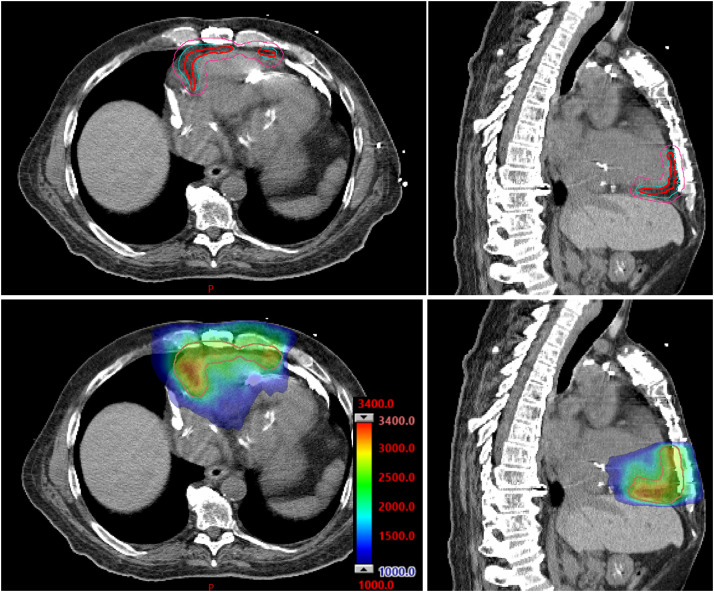
The tool has been successfully used in cardiac radioablation on 2 patients who had EAM acquired with an EnSite NavX system. The patients had prior catheter ablations, then radiation was recommended for treating the residual arrhythmogenic substrate. The first patient was treated for the VT in the left ventricle (LV). The integration of EAM data exactly followed the proposed workflow. The EAM was registered to the most recent cardiac MR on which the endocardial chambers were segmented as reference surfaces. The second patient was treated for right ventricular tachycardia. The clinical team registered the EAM to patient's cardiac CT in the mapping system, then exported the registered maps and anatomy surfaces that were segmented on the CT. Using the tool, the physician teams outlined the scar area on the EAM maps. Importing into the Eclipse TPS (Varian, Siemens Healthineers), the cardiac images were registered to simulation CT, and the scar structure was simultaneously overlapped on the simulation CT and showed the arrhythmogenic area around the CT endocardial surface (Fig. 3). Accordingly, radiation oncologists contoured the transmural target volume on the CT, referred to as gross target volume for radiation planning. The internal target volume (ITV) and planning target volume (PTV) were then created (Fig. 3) to account for the patient's internal motion and treatment setup uncertainties. Radiation plans with a prescription dose of 25 Gy to the PTV were designed and delivered using the volumetric modulated arc therapy technique (Fig. 3). The 2 patients were treated for PTVs of 142 cm3 on the LV and 156 cm3 on the right ventricle, respectively.
Figure 3.
Treatment planning based on a scar structure from electrophysiology data. Top row: physician creates internal target volume (cyan) and planning target volume (pink) on simulation computed tomography from registered scar (red) that is generated and exported from 3D Slicer. Bottom row: dose distribution with the color bar in centigray.
The third patient who was implanted with a left ventricular assist device received radioablation for late-stage VT burden. Catheter ablation and EAM acquisition were considered clinically infeasible for the patient. The clinical team worked with inHeart (inHeart Medical), whose 3D cardiac modeling software solution was used to postprocessed a contrast-enhanced cardiac CT of the patient and create the wall thinning map. The scar target was contoured on the map and exported with the cardiac CT in DICOM RT for treatment planning. On the day of CT simulation, ECGI of the patient was performed using a CardioInsight system. The EP maps were used by “side-by-side” visual comparison to assist the scar delineation on the wall thinning maps with the inHeart team. In a retrospective evaluation, we used HeaRTmap to generate a scar structure from the ECGI data. After the clinical workflow in the TPS, the ECGI CT and the scar structure were registered to the simulation CT, and a transmural target volume and then PTV were created. The Dice coefficient (2 times the volume of overlap divided by the total volumes of both structures), measuring the similarity of the volumetric overlap between this and the clinically used PTV (92 cm3), was 0.71.
Discussion
This work developed a software tool to efficiently integrate both ECGI and EAM data into a TPS for CR target volume definition. The tool was built upon previously published work. Brett et al first reported a workflow using 3D Slicer to convert CARTO3 EAM to DICOM images for CR.7 3D Slicer displayed the maps and enabled users to place fiducial marks to annotate scar boundaries. Converting the map into a binary volume, the users then manually delineated the scar structure to encompass the fiducial markers on the binary image slices. Registering cardiac CT with CARTO3 EAM in 3D Slicer, Hohman et al contoured scar structure on the CT to enclose the points that were labeled on the maps and projected on the image slices.8 However, the delineation on image slice views is prone to user-variability and can be labor-intensive. In comparison, our workflow only requires one-time annotation of the scar on the maps. The tool automatically converts the outlined surface patch into a structure associated with 3D anatomic images. Thus, HeaRTmap allows the clinical team to focus on defining the arrhythmogenic scar based on EP information, minimizing the physicians’ time and efforts.
The registration of EAM surfaces with 3D anatomic images determines the accuracy of using EP information for target definition. The workflow7 performed the fusion in a TPS based on manual alignment of anatomic landmark structures identified on both the EAM data and the CT. Instead, our workflow follows the procedure,8 which registered EAM to cardiac images in 3D Slicer using not only the anatomic landmarks but also the surfaces of the EAM and the corresponding structures segmented on the images. The many methods of surface registration provided in 3D Slicer can therefore be used. Our procedure is first landmark registration for global matching, then automatic surface registration for local alignment, and finally manual refining and confirmation. The invasive EAM procedure likely samples the endocardial surface nonuniformly, such as densely sampling the regions surrounding VT substrate but sparsely on distant areas. By providing a function to extract a portion of the EAM surface mesh, the tool enables region-of-interest-based surface registration. Nevertheless, due to the differences of cardiac cycles and respiratory phases between the mapping and cardiac imaging, thorough evaluation and further development of the fusion process are warranted.
Thanks to Hohman et al,8 our tool integrates their open-source EAMapReader to import EAM maps from 3 widely used mapping systems. Additionally, we developed the function to read ECGI mapping from the CardioInsight system. The tool can also handle ECGI data in the conventional VTK format because 3D Slicer can read and process them. Volumetric registration of ECGI anatomic images and simulation CT in a TPS readily fuses the scar structure for radiation target volume definition. However, studies have demonstrated there are discrepancies between the scars measured by ECGI and EAM.14 We currently use the ECGI to cognitively assess the scar location derived from EAM or contrast-enhanced scar imaging. In all, HeaRTmap provides a comprehensive solution to use EP data for target definition of CR.
It is important to recognize that registering an EAM map with the patient's anatomic images poses the biggest challenge for the workflow. First, the geometric accuracy of the EAM surface suffers from the respiratory and cardiac motions during the procedure, besides the limited catheter-sampling points of the mapping. Second, the mapping and imaging are likely performed at different phases of respiration and cardiac cycle, for example, ECG-gating EAM versus 4-dimensional free-breathing CT in our cases. Due to the phase differences, the morphologic changes of the heart may make the registration problematic. Third, the 2 data types have incomparable formats and different resolutions and require specific preprocessing. With various sources of uncertainties, we acknowledge that while the present cases demonstrate the workflow's efficacies, this study lacks quantitative assessment of the registration. Our future work will focus on evaluating and improving the accuracy and reliability of the data fusion.
As an emerging treatment modality, CR currently has no established guidance for target definition. Our method contours the arrhythmogenic area identified on EAM maps as “gross” disease. Another approach is to first divide the heart chamber contoured on CT into 17 segments, following the American Heart Association 17-segment model,15 and then select the segments including the critical substrate as target volume.16 The 17-segment model is the current standard in the cardiologic specialties in describing the heart among different imaging modalities.15 Although the 17-segment method addresses some of the uncertainties in the registration of EAM maps by targeting the whole segments, it could substantially increase radiation to healthy cardiac tissue and nearby organs at risk. In the current workflow, the clinical team visually assess the alignment of the maps with CT and cognitively verify that the EAM scar is located on the specific segments. A potential approach for quantitative evaluation is to delineate the 17 segments on simulation CT and assess the matching of the segments enclosing the fused scar with those identified by cardiologists during the EP procedure. With the development of the 17-segment atlas for radiation oncologists17,18 and automatic segmentation,19,20 the current workflow incorporating the 17-segment evaluation would improve the reliability and reproducibility of CR target delineation. The current software tool will be extended to develop this aspect in future work.
HeaRTmap was developed as a plugin for 3D Slicer which hosts numerous functions for medical image processing, distributed across different modules. Using Python scripting, the tool has a unified user interface to invoke the functions and modules to build a custom workflow for the application. This open-source tool is freely available at https://github.com/NYU-RadOnc/HEaRTmap as a 3D Slicer module.
Conclusion
Cardiac radioablation is a promising, noninvasive treatment option for refractory VT. Incorporating electrophysiological mapping in the treatment planning is critical for accurate definition of the target volume. This work built a software tool that enables smooth integration of EP data with minimal physician effort. It could facilitate further development and adoption of the emerging therapy for VT, and potentially other cardiac arrhythmias.
Disclosures
Chirag R. Barbhaiya reports consulting fees from Abbott and Biosense Webster. Larry A. Chinitz reports personal fees from Medtronik, Biotronik, Abbott, Biosense Webster, Phillips, Sanofi, Pfizer, and EPD solutions. David Barbee reports travel support from Sun Nuclear outside the submitted work. Ye Yuan is an employee of Atropos Heath outside the submitted work.
Acknowledgments
The authors thank the staff from Abbott for help in collecting NavX electroanatomic maps data, Qingguo Zeng from Medtronic for assisting in acquiring and processing CardioInsight data, and Hubert Cochet from inHeart Medical for myocardial wall thinning mapping and target definition. The work is partially under a research agreement with Medtronic for use of the CardioInsight data.
Footnotes
Sources of support: This work had no specific funding.
All data generated and analyzed during this study are included in this published article.
References
- 1.Tang PT, Shenasa M, Boyle NG. Ventricular arrhythmias and sudden cardiac death. Card Electrophysiol Clin. 2017;9:693–708. doi: 10.1016/j.ccep.2017.08.004. [DOI] [PubMed] [Google Scholar]
- 2.van der Ree MH, Blanck O, Limpens J, et al. Cardiac radioablation: A systematic review. Heart Rhythm. 2020;17:1381–1392. doi: 10.1016/j.hrthm.2020.03.013. [DOI] [PubMed] [Google Scholar]
- 3.Cuculich PS, Schill MR, Kashani R, et al. Noninvasive cardiac radiation for ablation of ventricular tachycardia. N Engl J Med. 2017;377:2325–2336. doi: 10.1056/NEJMoa1613773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Robinson CG, Samson PP, Moore KMS, et al. Phase I/II trial of electrophysiology-guided noninvasive cardiac radioablation for ventricular tachycardia. Circulation. 2019;139:313–321. doi: 10.1161/CIRCULATIONAHA.118.038261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chin R, Hayase J, Hu P, et al. Non-invasive stereotactic body radiation therapy for refractory ventricular arrhythmias: An institutional experience. J Interv Card Electrophysiol. 2021;61:535–543. doi: 10.1007/s10840-020-00849-0. [DOI] [PubMed] [Google Scholar]
- 6.Lloyd MS, Wight J, Schneider F, et al. Clinical experience of stereotactic body radiation for refractory ventricular tachycardia in advanced heart failure patients. Heart Rhythm. 2020;17:415–422. doi: 10.1016/j.hrthm.2019.09.028. [DOI] [PubMed] [Google Scholar]
- 7.Brett CL, Cook JA, Aboud AA, Karim R, Shinohara ET, Stevenson WG. Novel workflow for conversion of catheter-based electroanatomic mapping to DICOM Imaging for noninvasive radioablation of ventricular tachycardia. Pract Radiat Oncol. 2021;11:84–88. doi: 10.1016/j.prro.2020.04.006. [DOI] [PubMed] [Google Scholar]
- 8.Hohmann S, Henkenberens C, Zormpas C, et al. A novel open-source software-based high-precision workflow for target definition in cardiac radioablation. J Cardiovasc Electrophysiol. 2020;31:2689–2695. doi: 10.1111/jce.14660. [DOI] [PubMed] [Google Scholar]
- 9.Siedow M, Brownstein J, Prasad RN, et al. Cardiac radioablation in the treatment of ventricular tachycardia. Clin Transl Radiat Oncol. 2021;31:71–79. doi: 10.1016/j.ctro.2021.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Law MY, Liu B. Informatics in radiology: DICOM-RT and its utilization in radiation therapy. Radiographics. 2009;29:655–667. doi: 10.1148/rg.293075172. [DOI] [PubMed] [Google Scholar]
- 11.Cheniti G, Puyo S, Martin CA, et al. Noninvasive mapping and electrocardiographic imaging in atrial and ventricular arrhythmias (CardioInsight) Card Electrophysiol Clin. 2019;11:459–471. doi: 10.1016/j.ccep.2019.05.004. [DOI] [PubMed] [Google Scholar]
- 12.Fedorov A, Beichel R, Kalpathy-Cramer J, et al. 3D Slicer as an image computing platform for the quantitative imaging network. Magn Reson Imaging. 2012;30:1323–1341. doi: 10.1016/j.mri.2012.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pinter C, Lasso A, Wang A, Jaffray D, Fichtinger G. SlicerRT: Radiation therapy research toolkit for 3D Slicer. Med Phys. 2012;39:6332–6338. doi: 10.1118/1.4754659. [DOI] [PubMed] [Google Scholar]
- 14.Sapp JL, Zhou S, Wang L. Mapping ventricular tachycardia with electrocardiographic imaging. Circ Arrhythm Electrophysiol. 2020;13 doi: 10.1161/CIRCEP.120.008255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cerqueira MD, Weissman NJ, Dilsizian V, et al. Standardized myocardial segmentation and nomenclature for tomographic imaging of the heart. A statement for healthcare professionals from the Cardiac Imaging Committee of the Council on Clinical Cardiology of the American Heart Association. Circulation. 2002;105:539–542. doi: 10.1161/hc0402.102975. [DOI] [PubMed] [Google Scholar]
- 16.Lydiard PS, Blanck O, Hugo G, O'Brien R, Keall P. A review of cardiac radioablation (CR) for arrhythmias: Procedures, technology, and future opportunities. Int J Radiat Oncol Biol Phys. 2021;109:783–800. doi: 10.1016/j.ijrobp.2020.10.036. [DOI] [PubMed] [Google Scholar]
- 17.Duane F, Aznar MC, Bartlett F, et al. A cardiac contouring atlas for radiotherapy. Radiother Oncol. 2017;122:416–422. doi: 10.1016/j.radonc.2017.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brownstein J, Afzal M, Okabe T, et al. Method and atlas to enable targeting for cardiac radioablation employing the American Heart Association segmented model. Int J Radiat Oncol Biol Phys. 2021;111:178–185. doi: 10.1016/j.ijrobp.2021.03.051. [DOI] [PubMed] [Google Scholar]
- 19.Liang X, Garnavi R, Wail S, Sisi L, Prasanna P. Automatic segmentation of the left ventricle into 17 anatomical regions in cardiac MR imaging. Annu Int Conf IEEE Eng Med Biol Soc. 2015;2015:6531–6535. doi: 10.1109/EMBC.2015.7319889. [DOI] [PubMed] [Google Scholar]
- 20.van der Ree MH, Visser J, Planken RN, et al. Standardizing the cardiac radioablation targeting workflow: Enabling semi-automated angulation and segmentation of the heart according to the American Heart Association segmented model. Adv Radiat Oncol. 2022;7 doi: 10.1016/j.adro.2022.100928. [DOI] [PMC free article] [PubMed] [Google Scholar]