Graphical abstract
Keywords: Thanatin, LptA, Antimicrobial peptide, Genome mining, Lpt pathway inhibitor
Abstract
Podisus maculiventris thanatin has been reported as a potent antimicrobial peptide with antibacterial and antifungal activity. Its antibiotic activity has been most thoroughly characterized against E. coli and shown to interfere with multiple pathways, such as the lipopolysaccharide transport (LPT) pathway comprised of seven different Lpt proteins. Thanatin binds to E. coli LptA and LptD, thus disrupting the LPT complex formation and inhibiting cell wall synthesis and microbial growth. Here, we performed a genomic database search to uncover novel thanatin orthologs, characterized their binding to E. coli LptA using bio-layer interferometry, and assessed their antimicrobial activity against E. coli. We found that thanatins from Chinavia ubica and Murgantia histrionica bound tighter (by 3.6- and 2.2-fold respectively) to LptA and exhibited more potent antibiotic activity (by 2.1- and 2.8-fold respectively) than the canonical thanatin from P. maculiventris. We crystallized and determined the LptA-bound complex structures of thanatins from C. ubica (1.90 Å resolution), M. histrionica (1.80 Å resolution), and P. maculiventris (2.43 Å resolution) to better understand their mechanism of action. Our structural analysis revealed that residues A10 and I21 in C. ubica and M. histrionica thanatin are important for improving the binding interface with LptA, thus overall improving the potency of thanatin against E. coli. We also designed a stapled variant of thanatin that removes the need for a disulfide bond but retains the ability to bind LptA and antibiotic activity. Our discovery presents a library of novel thanatin sequences to serve as starting scaffolds for designing more potent antimicrobial therapeutics.
Introduction
Antimicrobial peptides (AMPs) are important components of the host innate immune defense system against pathogens. Despite their small sizes, AMPs can possess potent antiviral, antibacterial, and antifungal activity. Podisus maculiventris thanatin, an AMP isolated from the spined soldier bug, is a 21-residue peptide with broad inhibitory properties towards bacteria and fungi (Fehlbaum, 1996). Although the mechanism of action for P. maculiventris thanatin towards Gram-positive bacteria and fungi remains to be elucidated, recent studies suggest that P. maculiventris thanatin can interfere with multiple pathways in Gram-negative bacteria. In E. coli, binding of P. maculiventris thanatin to lipopolysaccharide (LPS) on the outer layer of the outer membrane dislodges Ca2+ and Mg2+ ions, causing membrane instability and bacterial agglutination for host clearance (Sinha et al., 2017; Ma, 2019). Likewise, P. maculiventris thanatin has been reported to disrupt the LPS translocation from the inner membrane to the outer membrane by binding to components of the lipopolysaccharide transport system (Okuda et al., 2016), LptA and LptD (Vetterli et al., 2018, Moura et al., 2020), ultimately disrupting the assembly of the bacterial outer membrane (Fig. S1A).
Apo P. maculiventris thanatin has been shown to adopt an architecture of β-hairpin held together by a disulfide bond in solution (Mandard et al., 1998). Using a truncated E. coli LptA protein that removes a C-terminal β-strand of LptA (Fig. S1B-D) to maintain a monomeric state in solution (LptAm), Vetterli and colleagues reported that P. maculiventris thanatin pairs its own β-hairpin with the N-terminal edge of the LptAm β-sheet, thus disrupting the head-to-tail LptA oligomer required for LPS transport (Fig. S1A) (Vetterli et al., 2018, Laguri, 2017). In contrast to the conflicting reports about the requirement of a cysteine disulfide bond for thanatin activity (Ma, 2016, Taguchi et al., 2000, Lee et al., 2002, Imamura et al., 2008); truncation and mutagenesis studies in general support the observations that the N-terminal loop of thanatin is dispensable (Fehlbaum, 1996) and the cationic loop between the two β-strands (Sinha et al., 2017, Taguchi et al., 2000) and the C-terminal region are required for in vivo inhibition (Fehlbaum, 1996).
In this study, we report the discovery of novel thanatin orthologs from insect and bacterial species by mining publicly available genomic databases. We show that thanatin orthologs from Chinavia ubica and Murgantia histrionica bind E. coli LptAm more tightly in vitro and display stronger antibiotic activity than the widely-studied thanatin from P. maculiventris (Fehlbaum, 1996, Ma, 2019, Vetterli et al., 2018, Ma, 2016). Our analyses of high-resolution crystal structures of thanatin orthologs from P. maculiventris; C. ubica and M. histrionica in complex with E. coli LptAm reveal key residues in M. histrionica thanatin that contribute to the tighter LptA binding affinity and superior antibiotic activity towards E. coli.
Results
Novel thanatin orthologs reveal a consensus sequence
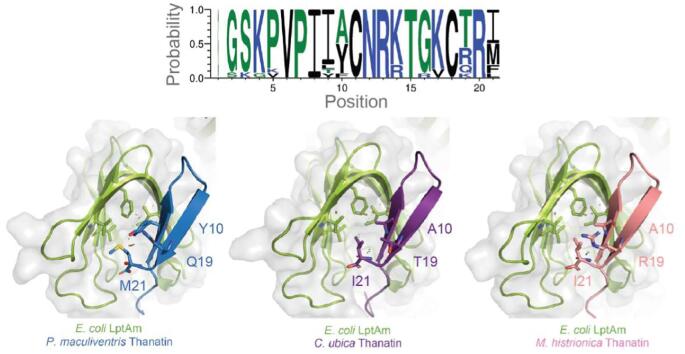
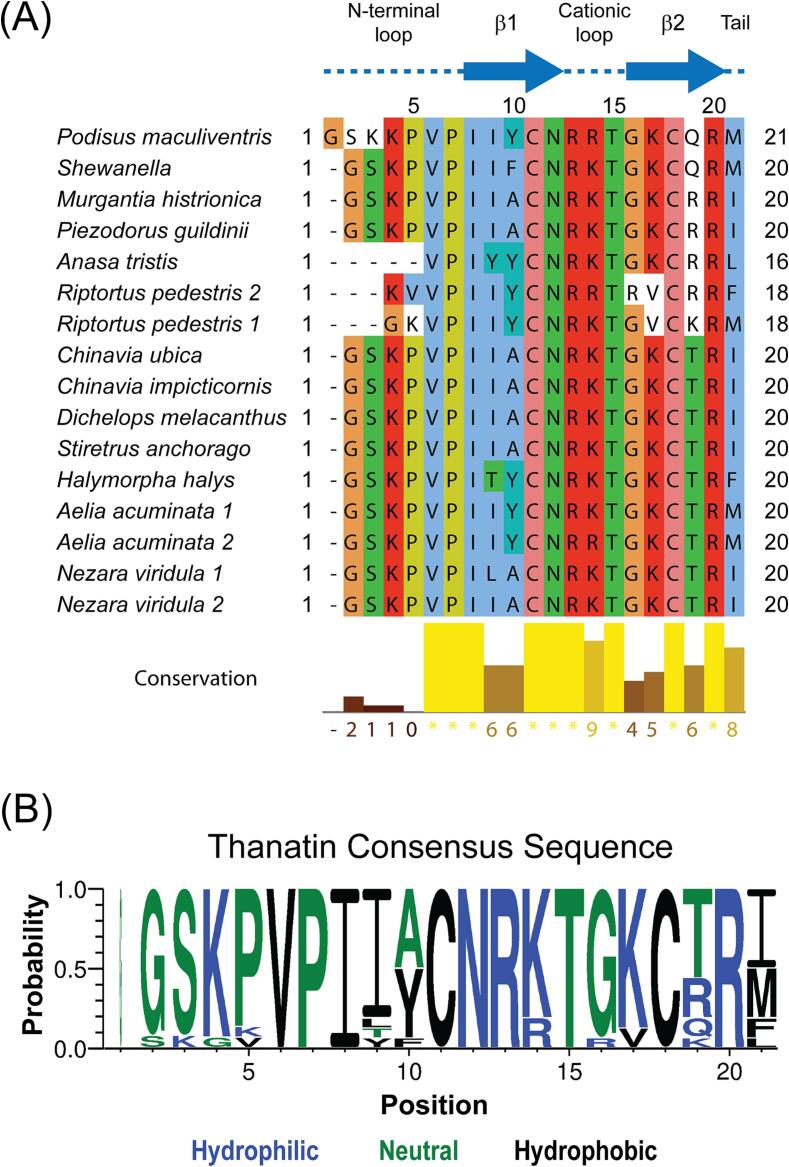
Thanatins from P. maculiventris and R. pedestris have previously been reported with potent inhibitory properties against Gram-negative bacteria (Fehlbaum, 1996, Park et al., 2018). To search for additional thanatin orthologs, we used the amino acid sequence of thanatin from P. maculiventris to query genomic databases deposited at the National Center for Biotechnology. A total of 16 thanatin-like sequences from 13 unique species were discovered (Fig. 1A), which can be aligned and grouped into 11 unique sequences. Analysis of the consensus sequence logo shows that thanatin peptides share a pattern of G2S3K4P5V6P7I8I9(A/Y)10C11N12R13K14T15G16K17C18(T/R/Q/K)19R20(I/M/F/L)21 (Fig. 1B). It is important to note that A10 only occurs when position 21 is an isoleucine (I21), whereas Y10 is observed with multiple types of hydrophobic residues at position 21, such as methionine, phenylalanine, and leucine.
Fig. 1.
Sequence alignment of thanatin orthologs reveals a consensus sequence. A sequence alignment of 16 thanatin sequences identified through genomic databases, with sequence conservation shown as a bar graph underneath the sequences. There are 11 unique sequences from 13 distinct species. B, A consensus logo generated from the thanatin sequences shows overall high conservation except for N-terminal loop (positions 1–5) and positions 9, 10, 19 and 21.
Thanatin orthologs bind more tightly to E. coli LptA and display stronger antibiotic activity than P. maculiventris thanatin
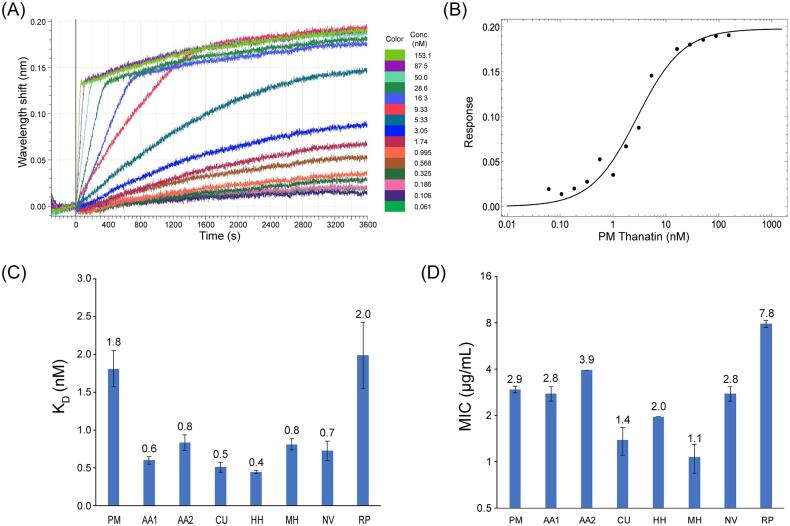
In order to assess the interaction of these thanatin orthologs towards E. coli LptA, we developed a bio-layer interferometry (BLI) binding assay. Eight thanatin orthologs from Aelia acuminata (two isoforms), Chinavia ubica, Halyomorpha halys, Murgantia histrionica, Nezara viridula, Podisus maculiventris, and Riptortus pedestris together with E. coli LptAm (Laguri, 2017) were cloned and purified. LptAm was biotinylated and immobilized onto streptavidin biosensors, which were then immersed into solutions containing different concentrations of target thanatin peptides. Representative titration and binding curves of P. maculiventris thanatin with LptAm are shown in Fig. 2A and 2B. The steady-state binding association was fit to a 1:1 binding model, yielding a KD value of 1.8 ± 0.2 nM. Representative titration curves and steady-state binding analyses of individual thanatin orthologs are summarized in Fig. 2C (raw data shown in Fig. S2). Surprisingly, with the exception of R. pedestris thanatin, which binds similarly to P. maculiventris thanatin (KD = 2.0 ± 0.4 nM versus KD = 1.8 ± 0.2 nM), all of the remaining thanatin peptides bind LptAm at least 2-fold more tightly (KD = 0.4 – 0.8 nM).
Fig. 2.
Characterization of novel thanatin orthologs. A, Representative BLI binding curves of the P. maculiventris thanatin titration with LptAm immobilized on the BLI biosensor tip. B, Steady-state binding curve of P. maculiventris thanatin with LptAm. C, Comparison of binding affinities of thanatin orthologs with LptAm. Error bars indicate SEM (minimum of at least n = 3). D, MIC values of thanatin orthologs against E. coli ATCC 25922. MIC values are calculated as geometric means, and the error bars represent SEM (minimum of at least n = 3).
Two of these thanatin orthologs displayed stronger antibiotic activity against E. coli by at least 2-fold (Fig. 2D). P. maculiventris thanatin had an MIC of 2.9 ± 0.2 µg/mL against the E. coli ATCC 25922 strain, whereas thanatin from C. ubica had a MIC value of 1.4 ± 0.3 µg/mL. Thanatin from M. histrionica appeared to be the most potent antibiotic, showing an MIC of 1.1 ± 0.2 µg/mL, improving the potency of P. maculiventris thanatin by approximately 2.8-fold.
Crystal structure of P. maculiventris thanatin bound to E. coli LptAm reveals a domain-swapped dimer of heterodimers
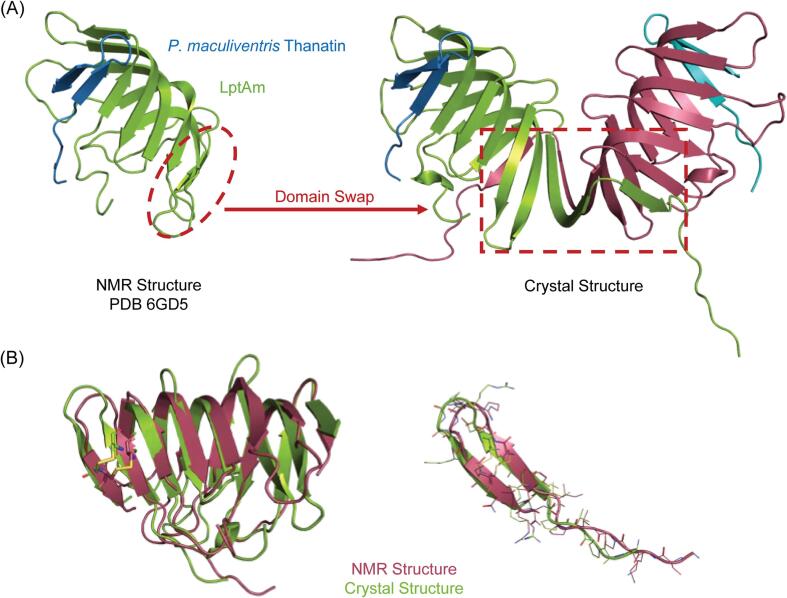
In order to obtain a molecular understanding of the P. maculiventris thanatin interaction with E. coli LptA, we crystallized the LptAm – P. maculiventris thanatin complex in the P212121 space group, which diffracted to 2.43 Å resolution. Unexpectedly, the crystal structure revealed a domain-swapped dimer of LptAm in the shape of a butterfly, with the terminal two β-strands on each protomer pairing up with the edge of the central β-sheet in the adjacent protomer (Fig. 3A). P. maculiventris thanatin forms a β-hairpin held together by a disulfide bond and appends to LptAm via parallel β-strand interactions through the N-terminal β-strand of LptAm, similar to the previously reported solution NMR structure (PDB 6GD5). As LptAm and its complex with P. maculiventris thanatin are primarily monomeric in solution with a small dimeric population, shown via SEC-MALS (Fig. S3), the dimeric state of LptAm likely results from the high protein concentrations under crystallization conditions.
Fig. 3.
Structural comparison of LptAm in complex with P. maculiventris thanatin by NMR and crystallography. A, Previously published solution NMR structure (PDB ID: 6GD5) showed a monomeric LptAm-thanatin complex. Crystal structure reveals a domain-swapped dimer of the heterodimeric LptAm-thanatin complex. B, Overlay of the NMR structure and crystal structure showed good backbone agreement for the β-strands, but poor agreement in the loop regions and side chain rotamers.
The overall features of the P. maculiventris thanatin interaction with LptAm in the crystal structure are similar to that of the NMR structure (PDB 6GD5). When disregarding the terminal two β-strands of LptA, the backbones are in good agreement with a RMSD of ∼1.9 Å (Fig. 3B). The areas of poor agreement are largely in the loop regions, along with great variance in the side chain rotamers (overall total RMSD of ∼2.4 Å), which is expected due to dynamic movement in solution (Fig. 3B). Furthermore, as the domain-swapping occurs at the C-terminus of LptAm (Fig. 3A and Fig. S1D), which is far removed from the binding interface between LptAm and thanatin, the thanatin binding interface is entirely unperturbed.
Shared features of LptAm-thanatin ortholog complexes
We next determined the crystal structures of C. ubica thanatin and M. histrionica thanatin in complex with E. coli LptAm to probe how these thanatin orthologs bind more tightly to E. coli LptA than P. maculiventris thanatin. Both C. ubica thanatin and M. histrionica thanatin were co-crystallized with E. coli LptAm in the same P212121 space group as P. maculiventris thanatin in complex with E. coli LptAm, and their crystal structures were resolved to 1.90 Å and 1.80 Å, respectively. Each unit cell similarly contains two LptAm – thanatin complexes as seen for the LptAm – P. maculiventris thanatin complex, with LptAm forming a domain-swapped dimer (Fig. S4; refinement statistics for all three structures can be found in the supplementary Table S1).
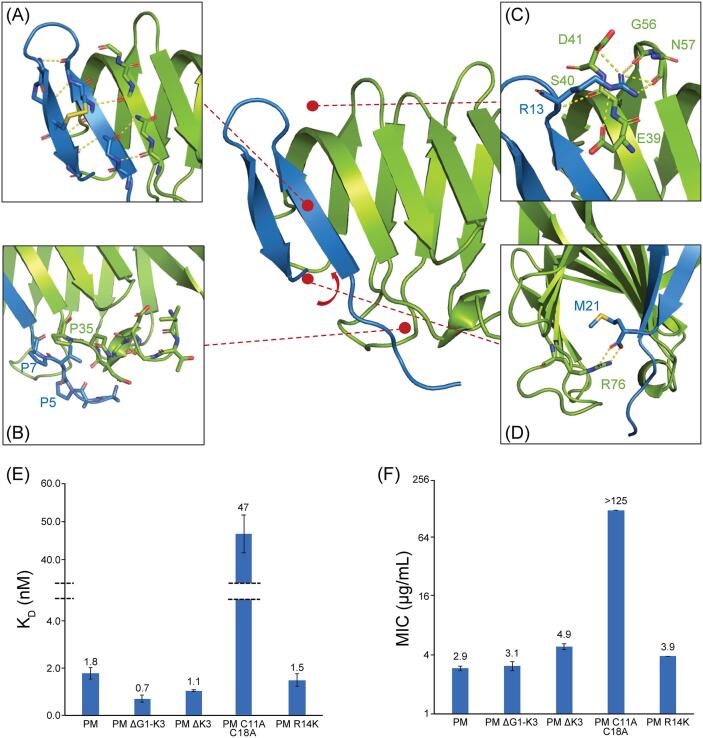
In all three structures, LptAm forms a β-jellyroll domain and thanatin forms a β-hairpin held together by an invariant disulfide bond (Fig. 4A). Removal of the disulfide bond in P. maculiventris thanatin via a C11A/C18A mutation reduced the LptAm binding affinity by 26-fold (KD: 47 ± 5 nM versus WT KD: 1.8 ± 0.2 nM) (Fig. 4E) and completely abolished the antibiotic activity (MIC > 125 (µg/mL) (Fig. 4F), suggesting that the disulfide bond stabilized β-hairpin structure is an important feature for maintaining the activity of thanatin.
Fig. 4.
Characteristics of P. maculiventris thanatin binding to E. coli LptAm. A, Thanatin binds to the N-terminal β-strand of LptA in a parallel orientation mediated through backbone interactions. Thanatin forms a β-hairpin held together by a disulfide bond and provides a β-strand edge to the complex that disfavors further oligomerization. B, Thanatin contains a flexible N-terminal tail not well defined in the electron density map, thus missing the first several residues and side chains. C, Thanatin R13 of the cationic loop is extensively involved in intermolecular interactions. D, LptA R76 stabilizes the C-terminus of thanatin through bidentate hydrogen bonds. E, Probing importance of shared structural regions by binding. Error bars indicate SEM (minimum of at least n = 3). F, MIC values of thanatin mutants against E. coli ATCC 25922. MIC values are calculated as geometric means, and the error bars represent SEM (minimum of at least n = 3).
The thanatin peptides bind to the N-terminus of LptAm by aligning their own N-terminal β-strands at the interface parallel to the N-terminal β -strand of LptAm and forming backbone interactions (Fig. 4A). As a result, thanatin disrupts the LptA head-to-tail connected β-jellyroll “bridge” in lipid A transport by binding to the N-terminus of LptAm.
All thanatin peptides contain a dispensable and flexible N-terminal tail that is poorly defined in the crystal structure (Fig. 4B). Deletion of first three residues (ΔG1-K3) in P. maculiventris thanatin did not affect its interaction with LptAm (KD: 0.7 ± 0.1 nM) (Fig. 4E) nor its antibiotic activity (MIC: 3.1 ± 0.3 µg/mL) (Fig. 4F) suggesting that this region is dispensable for efficient binding. This is in agreement with previously reported results of minimal effect observed between full-length and truncated (ΔG1-K3) P. maculiventris thanatin on E. coli inhibition (Fehlbaum, 1996). Similarly, deletion of K3 (ΔK3) in P. maculiventris thanatin, which mimics the shorter peptide length observed in several orthologs, has no major consequence due to its location in the dispensable N-terminal tail (KD: 1.1 ± 0.03 nM, MIC: 4.9 ± 0.3 µg/mL) (Fig. 4E, F).
The cationic loop, specifically R13, in all three thanatin peptides is involved in extensive intermolecular interactions with LptAm (Fig. 4C), suggesting that it is an important residue for LptAm binding. Mutations of R13 that break contact to LptAm, such as R13C or R13H, were reported to abolish inhibition of E. coli growth (Taguchi et al., 2000). In contrast to R13, the residue at position 14 has no obvious contact with LptAm, though these two residues are invariantly conserved as basic residues (Arg/Lys). R14K has no significant changes in LptAm binding and antibiotic activity (KD: 1.5 ± 0.3 nM, MIC: 3.9 ± 0.0 µg/mL) (Fig. 4E, F). However, removal of positively charged residues in the cationic loop via P. maculiventris thanatin R13A/R14A, was reported to abolish inhibition of E. coli growth (Sinha et al., 2017), showing that it plays a critical role in the efficacy of thanatin presumably due to enhanced membrane permeability.
The C-terminal carboxyl group in all three thanatin peptides forms bidentate hydrogen bonds with the sidechain of LptAm R76 to position thanatin with the hydrophobic core of LptAm (Fig. 4D). C-amidation of P. maculiventris thanatin was reported to increase the MIC value by 2-fold, most likely due to the loss of the charged interaction with LptA R76 (Fehlbaum, 1996). Deletions of residues 19 – 21, 20 – 21, or 21 in P. maculiventris thanatin were also reported to abolish inhibition of E. coli growth, highlighting the importance of the C-terminal region (Fehlbaum, 1996).
Molecular basis of the enhanced LptAm binding and antibiotic activity by thanatin orthologs from C. ubica and M. histrionica
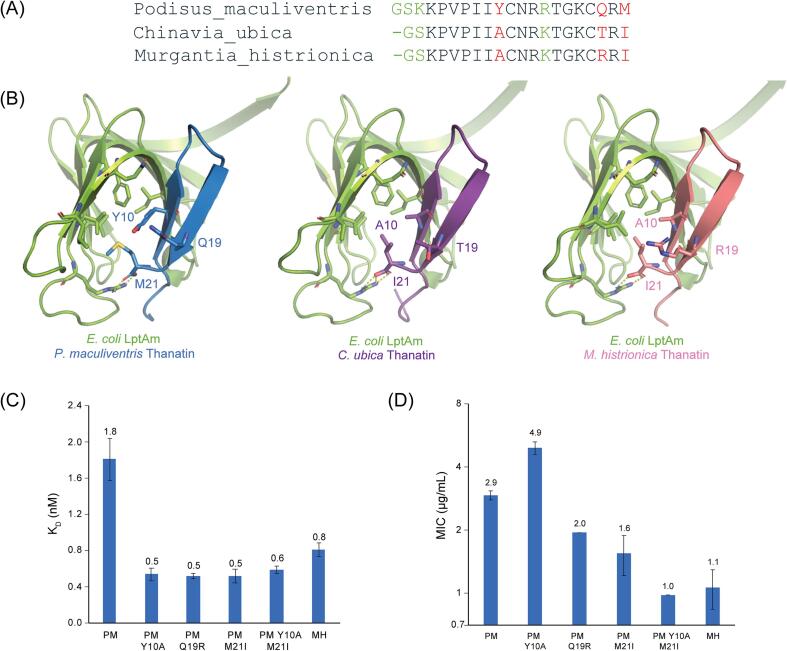
When comparing the sequences of thanatin from P. maculiventris, C. ubica, and M. histrionica, Y10A, Q19R, Q19T, and M21I highlight the major differences at the interface of thanatin with the hydrophobic core of LptAm (Fig. 5A, B). The hydrophobic surface of LptAm consists of I38, L45, V52, and F54, which is the main interface that residues 10 and 21 of thanatin pack against. P. maculiventris thanatin contains Y10 and M21, while C. ubica thanatin and M. histrionica thanatin contain A10 and I21. Residues A10 and I21 provide a better interface, as reflected in tighter binding to LptAm (C. ubica KD of 0.5 ± 0.1 nM and M. histrionica KD of 0.8 ± 0.1 nM vs P. maculiventris KD of 1.8 ± 0.2 nM) (Fig. 2C). The tighter binding of C. ubica thanatin and M. histrionica thanatin to LptAm also contributes to lowering the MIC values (Fig. 2D).
Fig. 5.
Differences among P. maculiventris, C. ubica, and M. histrionica thanatin. A, Sequence alignment of thanatin peptides highlights neutral differences (ΔK3 and R14K) in green and major differences (Y10A, Q19T, Q19R, and M21I) in red. B, Major differences are found at the interface of thanatin with the hydrophobic core of LptA. C, Assessing effects on binding to E. coli LptAm of single- and double-point mutations. Values given are averages of KD (nM) with SEM as error bars (minimum of at least n = 3). D, Assessing effects on inhibition to E. coli ATCC 25922, a clinical strain, of single- and double-point mutations. Values given are geometric mean of MIC (µg/mL) with SEM as error bars (minimum of at least n = 3). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
To evaluate the contributions of the variant surface residues of thanatin orthologs to LptAm binding, we generated single point mutations of Y10A, Q19R, and M21I and double mutant Y10A/M21I on the P. maculiventris backbone. Binding for all mutants to LptAm improved by roughly 3-fold (Fig. 5C). However, a more gradual improvement of antibiotic activity was observed (Fig. 5D). None of the single point mutants Y10A, Q19R, and M21I significantly lowered the MIC value greater than 2-fold, but a double point mutant Y10A/M21I reduced the MIC by 3-fold, suggesting a synergistic relationship between the two positions. The lack of strong correlation between the binding data and antibiotic activity, in particular the Y10A mutant, indicate that thanatin may additionally affect other mechanisms of actions, such as binding to LPS to perturb the outer membrane or binding to LptD to disrupt the Lpt pathway (Sinha et al., 2017, Vetterli et al., 2018), thus resulting in the varied profile. Further experiments will be needed to explore these other paths.
Redesigning M. histrionica thanatin
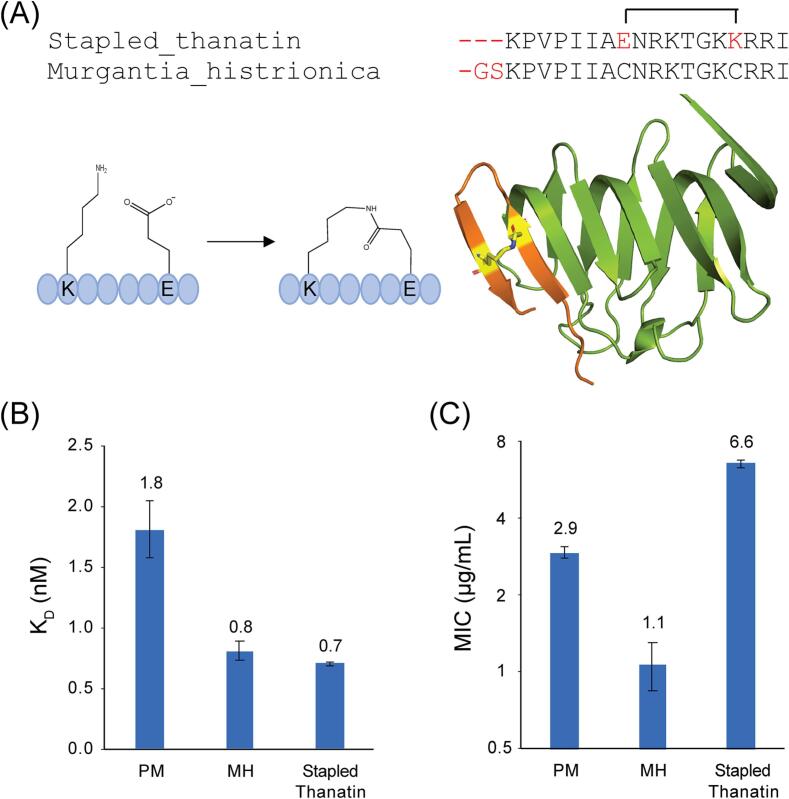
Next, we sought to improve M. histrionica thanatin, the most potent thanatin ortholog identified from our studies. We showed that the N-terminal region of P. maculiventris thanatin is dispensable (Fig. 4E, F) and poorly defined in any of the crystal structures, suggesting that the N-terminal flexibility applied to the other thanatin orthologs as well. We showed that the disulfide bond was necessary for binding and inhibition (Fig. 4E, F). Our redesign approach focused on truncating M. histrionica thanatin and removing the need of the disulfide bond, which are prone to environmental redox conditions. We mutated the disulfide bond to C11E and C18K and created a lactam bridge across the peptide (peptide synthesized by Genscript) (Fig. 6A). This created a staple across thanatin that fully retained binding to E. coli LptAm (KD: 0.70 ± 0.01 nM vs. 0.8 ± 0.1 nM for M. histrionica thanatin) (Fig. 6B), However, potency against E. coli was 6-fold worse (MIC: 6.6 ± 0.2 µg/mL vs. 1.1 ± 0.2 µg/mL for M. histrionica thanatin) (Fig. 6C), possibly reflecting a compromised ability of the stapled peptide to penetrate the bacterial membrane.
Fig. 6.
Redesigning M. histrionica thanatin into a novel scaffold. A, The sequence from M. histrionica thanatin was truncated on the N-terminus and the disulfide bond was replaced with a lactam bridge. Modeling predicts stapled thanatin can retain its structure with the modifications. B, Assessment of stapled thanatin binding to E. coli LptAm (minimum of at least n = 3). C, Assessment of stapled thanatin potency against E. coli ATCC 25922. Values given are geometric mean of MIC (µg/mL) with SEM as error bars (minimum of at least n = 3).
Discussion
Through genomic databases mining for thanatin orthologs, we have discovered more effective thanatin peptides that potently target E. coli. We have developed a BLI assay to measure their binding to E. coli LptA and a MIC assay to evaluate their antibiotic activity against E. coli. We have crystallized and determined the structures of P. maculiventris thanatin and two of the best thanatin orthologs, from C. ubica and M. histrionica, bound to E. coli LptAm for structural characterization. We have conducted mutagenesis studies to better understand the mechanism of improved binding and antibiotic activity. Overall, Y10A, Q19R, and M21I increased binding affinity, and the improvement in antibiotic activity was synergistically observed through Y10A and M21I. Furthermore, we have demonstrated that it is possible to redesign a scaffold from M. histrionica thanatin, the most potent thanatin ortholog, by shortening the peptide and removing the need for a disulfide bond.
The discovery of novel thanatin orthologs adds to the growing field of microproteomics and cryptic peptides. Recent efforts have been made to mine the human genome for microproteins (around 100 amino acids or less) that have been largely ignored until lately. Microproteins have been found to play various roles in regulating stress signaling (PIGBOS), muscle and fat metabolism (MOTS-c), and mitochondrial activity (SHMOOSE) (Chu et al., 2019, Miller et al., 2023, Lee et al., 2015). They have also been linked to various diseases such as cancer, diabetes, and Alzheimer’s disease (Chu et al., 2019, Miller et al., 2023, Lee et al., 2015). In addition, recent efforts have been made to mine the human genome for cryptic peptides that serve as antibiotics (Torres et al., 2021, Pane et al., 2017). Cryptic peptides are active peptides created from the degradation of or cleavage from a larger protein. P. maculiventris thanatin is expressed within a precursor of 109 residues (Accession number ATG84180.1). After cleavage from the precursor, P. maculiventris thanatin becomes an active antimicrobial peptide of 21 residues. Thanatin sequences found in our genomic search were mostly from whole genome sequences and their corresponding precursors were not identified. However, the precursor for R. pedestris thanatin has been reported, suggesting similar processing and maturation of the peptides (Accession number BAN67668). Our studies show that mining non-human genomes, such as insects in our case, could identify a valuable library of unexplored microproteins with antimicrobial activity.
Variance in the thanatin peptides can reveal what role these peptides play in their host by elucidating their microbial targets. The search for thanatin orthologs yielded 16 thanatin-like sequences, which can be grouped into 11 unique sequences, from 13 species. The majority of these orthologs are from stink bugs and other insects. It has been reported that thanatin in P. maculiventris (spined soldier bug) and R. pedestris (bean bug) are found in the insect’s midgut region and help regulate the microbiome to promote symbiosis (Fehlbaum, 1996, Park et al., 2018). It is plausible that the newly reported thanatins in other insects play a similar role in their respective hosts. Different thanatin sequences could target different pathogens to various degrees, which may reflect the microbiome population of each host and helps build the broadly inhibitory profile of thanatin. Interestingly, one thanatin ortholog was found in Shewanella, an anaerobic Gram-negative bacteria found in marine sponges. It is plausible that Shewanella produces thanatin as a toxin and secretes it into the environment to suppress the growth of competing bacteria or fungi. Further studies will be needed to elucidate the role of thanatin in these other species.
Our early design of a stapled thanatin retains binding but slightly reduces potency. It would be worthwhile to apply computational tools, such as OSPREY, to our stapled thanatin to increase binding affinity and regain potency (Hallen, 2018). Another design approach would be to create hybrid peptides to combine our improved thanatins with other antimicrobial peptides to synergistically attack pathogens.
Overall, we have identified a more potent thanatin peptide from M. histrionica that targets E. coli through strong binding to LptA to disrupt the lipopolysaccharide transport system. High-resolution crystal structures allowed us to better understand the mechanism of action between thanatin and LptA. We identified that the key to improved binding and potency lies in the cooperativity between thanatin A10 and I21. We also redesigned M. histrionica thanatin to remove the need for a disulfide bond and presented a scaffold for the next generation of peptide design. Progress in computational protein design may help improving the affinity and potency of thanatin and enhance its antibiotic activity to other Gram-negative bacteria beyond E. coli studied here.
Materials and methods
Sequence analysis
Identification of novel thanatin orthologs was performed with the tblastn and blastp suites (https://blast.ncbi.nlm.nih.gov) (AltschuP et al., May 1990). The amino acid sequence from P. maculiventris thanatin was used as the query sequence. Sequence identities and positive substitutions were used to determine sequences of interest. A multiple sequence alignment was generated with Clustal Omega (https://www.ebi.ac.uk/Tools/msa/clustalo/) (Sievers et al., 2011). A consensus sequence was generated with WebLogo (https://weblogo.berkeley.edu) (Crooks et al., 2004). The accession numbers corresponding to all thanatin orthologs mentioned in this publication are: Aelia acuminata (CAJVQN010000565), Anasa tristis (GEII01018028), Chinavia impicticornis (GIVF01040023), Chinavia ubica (GBFA01023333), Dichelops melacanthus (GBES01007078), Halyomorpha halys (XP_014293651), Murgantia histrionica (GECQ01170005), Nezara viridula (GIBW01225186 and GGPJ01242699), Piezodorus guildinii (CM041296), Podisus maculiventris (AAB36066), Riptortus pedestris (BAN67668 and JAAEAN010010806), Shewanella (WP_238898582), and Stiretrus anchorago (WUAS01076668).
Cloning, Expression, and purification of E. coli LptA
A monomeric mutant (Δ159-185) of LptA (Laguri, 2017) without the signal peptide sequence (Δ1-27) was inserted into a modified pET24b vector through Takara Bio In-Fusion cloning to yield the final construct of His6-SUMO-LptA. For BLI binding studies, a C-terminal flexible linker and AviTag were incorporated to yield the final construct of His6-SUMO-LptA-GSGGSGSG-AviTag. After confirmation through DNA sequencing, the plasmid was transformed into BL21 (DE3) competent cells on LB plates containing kanamycin. Cells were grown in Luria-Bertani (LB) media with kanamycin at 37 °C until OD600 ≈0.6 with shaking and then induced with 1 mM IPTG at 18 °C for 18 h. The cells were harvested by centrifugation and frozen at −80 °C until purification.
The cell pellet was thawed, resuspended in Buffer A (20 mM Tris pH 8.0, 250 mM NaCl) with a protease inhibitor tablet, and lysed through a microfluidizer. The lysate was centrifuged at 20,000g for 20 mins to remove cellular debris. The supernatant was passed through a Ni-NTA column, washed with Buffer A + 25 mM imidazole, eluted with Buffer A + 250 mM imidazole, and further eluted with Buffer A + 500 mM imidazole. Elution fractions containing protein were pooled and digested with SENP1 protease (1:100 M protease:protein) while being dialyzed (MWCO 3.5 kDa) against 20 mM Tris pH 8.0, 150 mM NaCl, 1 mM DTT for 20 h at 4 °C. The dialyzed and digested solution was passed through a Ni-NTA column, washed with Buffer A + 25 mM imidazole, eluted with Buffer A + 250 mM imidazole, and further eluted with Buffer A + 500 mM imidazole. The flow through and wash fractions containing protein were concentrated (MWCO 3 K) and passed through a Superdex200 column in Buffer B (50 mM Tris pH 8.5, 200 mM NaCl). The fractions containing purified LptA were concentrated (MWCO 3 kDa) and flash frozen in liquid nitrogen for storage at −80 °C. For BLI binding studies, the protein was biotinylated with the BirA biotin-protein ligase standard reaction kit from Avidity and passed through a S75 column in Buffer C (1X PBS) before being flash frozen for storage at −80 °C.
Cloning, expression, and purification of thanatin peptides
The thanatin sequence was inserted into a pET15b vector through Takara Bio In-Fusion cloning to yield the final construct of His10-GB1-TEV cleavage site-Thanatin. Single point and double point mutations were made with a Q5 Site-Directed Mutagenesis Kit from New England BioLabs. After confirmation through DNA sequencing, the plasmid was transformed into BL21 (DE3) competent cells on LB plates containing ampicillin. Cells were grown in LB media with ampicillin at 37 °C until OD600 ≈0.6 with shaking and then induced with 1 mM IPTG at 18 °C for 18 h. The cells were harvested by centrifugation and frozen at −80 °C until purification.
The cell pellet was thawed, resuspended in Buffer D (50 mM Tris pH 8.0, 200 mM NaCl) with a protease inhibitor tablet, and lysed through a microfluidizer. The lysate was centrifuged at 20,000 g for 20 mins to remove cellular debris. The supernatant was passed through a Ni-NTA column, washed with Buffer D + 25 mM imidazole, eluted with Buffer D + 250 mM imidazole, and further eluted with Buffer D + 500 mM imidazole. Elution fractions containing protein were pooled, concentrated (MWCO 3 kDa), and passed through a Superdex200 column in Buffer B. The fractions containing protein were pooled and digested with TEV protease (1:100 M protease:protein) for 20 h at 4 °C. The digested solution was passed through a Ni-NTA column, washed with Buffer B + 25 mM imidazole, eluted with Buffer B + 250 mM imidazole, and further eluted with Buffer B + 500 mM imidazole. The flow through and wash fractions containing purified peptide were concentrated (MWCO 3 K), buffer exchanged into Buffer B, and frozen for storage at −20 °C.
Synthesis of the stapled thanatin peptide was carried out by Genscript. Peptide was resuspended in Buffer B for experiments.
Bio-layer interferometry (BLI) binding assay
A BLI binding assay was developed on an Octet Red96e System. All assays were performed at 25 °C, at 700 r.p.m., and in buffer (1X Octet Kinetics Buffer: 1X PBS, 1% BSA, 0.1% Tween-20, 0.1% Kathon). Octet Streptavidin (SA) Biosensors were pre-wet for 15 min in buffer. The order of steps for each assay were as follows: (1) baseline: immersion in buffer for 300 s, (2) loading: immersion in biotinylated E. coli LptA-Avi at 1 µg/mL for 300 s, (3) baseline: immersion in buffer for 300 s, (4) association: immersion in peptide for 3600 s. A 15-point titration of peptide from 153.1 nM to 0.6 nM was used. Controls for double-referencing each point were the following: (1) reference sample well: SA biosensor with LptA-Avi loaded immersed into buffer, (2) reference sensor: SA biosensor with no LptA-Avi loaded immersed into various peptide concentrations. Curve fitting and data analysis for reporting dissociation constants (KD) was performed using the following steady-state 1:1 binding model: . The SA Biosensors loaded with LptA-Avi were regenerated for multiple uses as follows: (1) baseline: immersion in buffer for 100 s, (2) regeneration: immersion in 10 mM glycine pH 1.7 for 10 s, (3) neutralization: immersion in buffer for 10 s, repeat steps (2) and (3) for a total of 3 times, (4) baseline: immersion in buffer for 100 s.
Minimal inhibition concentration (MIC) assay
A MIC assay was developed with E. coli ATCC 25922 based on the Clinical & Laboratory Standards Institute (CLSI) standard protocol (Clinical and Laboratory Standards Institute, 2018). E. coli was streaked onto an antibiotic free LB plate from a cryo stock and incubated at 37 °C overnight. A single colony from this plate was streaked onto an antibiotic free LB plate and incubated at 37 °C overnight. A single colony was added to LB media (10 mL) and grown at 37 °C to OD600 ≈0.1 (corresponding to ≈1 – 2 × 108 CFU/mL) with shaking. Cells were added to a final concentration of ≈5 × 105 CFU/mL in Cation-Adjusted Mueller-Hinton Broth (CAMHB) over a titration of peptide inhibitor in clear 96 well plates. The plate was incubated at 37 °C for 20 h. MTT (3-(4,5-Dimethylthiazol-2-yl)-2,5-Diphenyltetrazolium Bromide) (10 µL of 5 mg/mL) was added to each well and incubated at 37 °C for 3 h. Isopropanol + 0.1 M HCl (100 µL) were added to each well. The plate was imaged and read at OD690 and OD5750 for analysis. The MIC was determined to be the maximum peptide concentration where no growth was observed.
Crystallization and structure determination of E. coli LptA with P. maculiventris thanatin
Purified E. coli LptA and P. maculiventris thanatin were incubated together and co-eluted through a Superdex200 column in Buffer B. The fractions containing the complex were concentrated (MWCO 3 kDa) to 5 and 10 mg/mL. Crystal trays were set up at each concentration using the sitting drop method with 1 µL protein and 1 uL of crystallization solution, along with seeding from E. coli LptA with P. maculiventris M21W crystals using the scratch method (structure not released). Crystallization solutions that yielded crystals ranged from 60 to 70% MPD at 0.1 M MES pH 5.8. The crystallization condition of the reported dataset was a mix of 10 mg/mL LptA:thanatin and 0.1 M MES pH 5.8, 60% MPD in 1:1 ratio.
Diffraction data were collected at NE-CAT 24-ID-C beamline. The 3D model was constructed using molecular replacement from the PHASER module in PHENIX with an AlphaFold2 model (generated through online hosted notebook (Evans et al., 2021, Jumper et al., 2021)) of E. coli LptAm and P. maculiventris thanatin as the input model. Coot (Emsley and Cowtan, 2004) and PHENIX (Afonine et al., 2018) were used for iterative model building and refinement.
Crystallization and structure determination of E. coli LptA with C. ubica thanatin
Purified E. coli LptA and C. ubica thanatin were incubated together and co-eluted through a Superdex200 column in Buffer B. The fractions containing the complex were concentrated (MWCO 3 kDa) to 6, 8, and 15 mg/mL. Crystal trays were set up at each concentration using the sitting drop method with 1 µL protein and 1 uL of crystallization solution. Crystallization solutions that yielded crystals ranged from 60 to 77.5% MPD and pH 5.8–8.5 (0.1 M MES, HEPES, or Tris). The crystallization condition of the reported dataset was a mix of 6 mg/mL LptA:thanatin and 0.1 M MES pH 5.8, 70% MPD in a 1:1 ratio.
Diffraction data were collected at NE-CAT 24-ID-E beamline. The 3D model was constructed using molecular replacement from the PHASER module in PHENIX with an AlphaFold2 model (generated through online hosted notebook (Evans et al., 2021, Jumper et al., 2021)) of E. coli LptA and C. ubica thanatin as the input model. Coot (Emsley and Cowtan, 2004) and PHENIX (Afonine et al., 2018) were used for iterative model building and refinement.
Crystallization and structure determination of E. coli LptA with M. histrionica thanatin
Purified E. coli LptA and M. histrionica thanatin were incubated together and co-eluted through a Superdex200 column in Buffer B. The fractions containing the complex were concentrated (MWCO 3 kDa) to 10.5 mg/mL. Crystal trays were set up at each concentration using the sitting drop method with 1 µL protein and 1 uL of crystallization solution. Crystallization solutions that yielded crystals ranged from 0.095 to 0.1 M sodium citrate pH 5.6, 19–20% isopropanol, 19–20% PEG 4000, and 0–5% glycerol. The crystallization condition of the reported dataset was a mix of 10.5 mg/mL LptA:thanatin and 0.095 M sodium citrate pH 5.6, 19% isopropanol, 19% PEG 4000, and 5% glycerol in a 1:1 ratio.
Diffraction data were collected at NE-CAT 24-ID beamline. The 3D model was constructed using molecular replacement from the PHASER module in PHENIX with an AlphaFold2 model (generated through online hosted notebook (Evans et al., 2021, Jumper et al., 2021)) of E. coli LptA and M. histrionica thanatin as the input model. Coot (Emsley and Cowtan, 2004) and PHENIX (Afonine et al., 2018) were used for iterative model building and refinement.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Acknowledgements
We thank all members of the Zhou laboratory, Donald laboratory, Dr. Abhishek Chhetri, and Dr. Terrence Oas for helpful discussions.
Author disclosure statement
BRD is a founder of Ten63 Therapeutics, Inc. K.H., B.R.D., and P.Z. are co-inventors of a provisional patent on designed thanatin peptides.
Funding information
This work is in part supported by grants from NIH (AI139216 to P.Z. and GM144042 to B.R.D.).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.yjsbx.2023.100091.
Contributor Information
Bruce R. Donald, Email: brd+jsb23@cs.duke.edu.
Pei Zhou, Email: peizhou@biochem.duke.edu.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Data availability
The coordinates of the E. coli LptAm complexes with thanatins from P. maculiventris, C. ubica, and M. histrionica have been deposited to Protein Data Bank (www.pdb.org) with PDB access codes of 8GAJ, 8GAK, and 8GAL, respectively. Other data are available upon request.
References
- Afonine P.V., et al. Real-space refinement in PHENIX for cryo-EM and crystallography. Acta Crystallogr. D Struct. Biol. 2018;74:531–544. doi: 10.1107/S2059798318006551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- AltschuP S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. JMB. 1990;215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Chu Q., Martinez T.F., Novak S.W., Donaldson C.J., Tan D., Vaughan J.M., Chang T., Diedrich J.K., Andrade L., Kim A., Zhang T., Manor U., Saghatelian A. Regulation of the ER stress response by a mitochondrial microprotein. Nat. Commun. 2019;10(1) doi: 10.1038/s41467-019-12816-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clinical and Laboratory Standards Institute, 2018. Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically, 11th ed., vol. 2, 38 vols. 950 West Valley Road, Suite 2500, Wayne, Pennsylvania 19087, USA: Clinical and Laboratory Standards Institute, 2018.
- Crooks G.E., Hon G., Chandonia J.-M., Brenner S.E. WebLogo: A sequence logo generator. Genome Res. 2004;14(6):1188–1190. doi: 10.1101/gr.849004. https://10.1101/gr.849004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P., Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Evans R., et al. Protein complex prediction with AlphaFold-Multimer. Bioinformatics. 2021 doi: 10.1101/2021.10.04.463034. preprint. [DOI] [Google Scholar]
- Fehlbaum P., et al. Structure-activity analysis of thanatin, a 21-residue inducible insect defense peptide with sequence homology to frog skin antimicrobial peptides. PNAS. 1996;93(3):1221–1225. doi: 10.1073/pnas.93.3.1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallen M.A., et al. OSPREY 3.0: Open-source protein redesign for you, with powerful new features. J. Comput. Chem. 2018;39(30):2494–2507. doi: 10.1002/jcc.25522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imamura T., Yamamoto N., Tamura A., Murabayashi S., Hashimoto S., Shimada H., Taguchi S. NMR based structure–activity relationship analysis of an antimicrobial peptide, thanatin, engineered by site-specific chemical modification: Activity improvement and spectrum alteration. Biochem. Biophys. Res. Commun. 2008;369(2):609–615. doi: 10.1016/j.bbrc.2008.02.057. [DOI] [PubMed] [Google Scholar]
- Jumper J., et al. Highly accurate protein structure prediction with AlphaFold. Nature. 2021;596(7873):583–589. doi: 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laguri C., et al. Interaction of lipopolysaccharides at intermolecular sites of the periplasmic Lpt transport assembly. Sci. Rep. 2017;7(9715):13. doi: 10.1038/s41598-017-10136-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M.-K., Cha L.-N., Lee S.-H., Hahm K.-S. Role of amino acid residues within the disulfide loop of thanatin, a potent antibiotic peptide. BMB Rep. 2002;35(3):291–296. doi: 10.5483/BMBRep.2002.35.3.291. [DOI] [PubMed] [Google Scholar]
- Lee C., Zeng J., Drew B., Sallam T., Martin-Montalvo A., Wan J., Kim S.-J., Mehta H., Hevener A., de Cabo R., Cohen P. The mitochondrial-derived peptide MOTS-c promotes metabolic homeostasis and reduces obesity and insulin resistance. Cell Metab. 2015;21(3):443–454. doi: 10.1016/j.cmet.2015.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma B., et al. The disulfide bond of the peptide thanatin is dispensible for its antimicrobial activity in vivo and in vitro. Antimicrob. Agents Chemother. 2016;60(7):4283–4289. doi: 10.1128/AAC.00041-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma B., et al. The antimicrobial peptide thanatin disrupts the bacterial outer membrane and inactivates the NDM-1 metallo-β-lactamase. Nat. Commun. 2019;10(1):3517. doi: 10.1038/s41467-019-11503-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandard N., Sodano P., Labbe H., Bonmatin J.-M., Bulet P., Hetru C., Ptak M., Vovelle F. Solution structure of thanatin, a potent bactericidal and fungicidal insect peptide, determined from proton two-dimensional nuclear magnetic resonance data. Eur. J. Biochem. 1998;256(2):404–410. doi: 10.1046/j.1432-1327.1998.2560404.x. [DOI] [PubMed] [Google Scholar]
- Miller B., Kim S.-J., Mehta H.H., Cao K., Kumagai H., Thumaty N., Leelaprachakul N., Braniff R.G., Jiao H., Vaughan J., Diedrich J., Saghatelian A., Arpawong T.E., Crimmins E.M., Ertekin-Taner N., Tubi M.A., Hare E.T., Braskie M.N., Décarie-Spain L., Kanoski S.E., Grodstein F., Bennett D.A., Zhao L., Toga A.W., Wan J., Yen K., Cohen P. Mitochondrial DNA variation in Alzheimer’s disease reveals a unique microprotein called SHMOOSE. Mol. Psychiatry. 2023;28(4):1813–1826. doi: 10.1038/s41380-022-01769-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moura E.C.C.M., Baeta T., Romanelli A., Laguri C., Martorana A.M., Erba E., Simorre J.-P., Sperandeo P., Polissi A. Thanatin impairs lipopolysaccharide transport complex assembly by targeting LptC–LptA interaction and decreasing LptA stability. Front. Microbiol. 2020;11 doi: 10.3389/fmicb.2020.00909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuda S., Sherman D.J., Silhavy T.J., Ruiz N., Kahne D. Lipopolysaccharide transport and assembly at the outer membrane: the PEZ model. Nat. Rev. Microbiol. 2016;14(6):337–345. doi: 10.1038/nrmicro.2016.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pane K., Durante L., Crescenzi O., Cafaro V., Pizzo E., Varcamonti M., Zanfardino A., Izzo V., Di Donato A., Notomista E. Antimicrobial potency of cationic antimicrobial peptides can be predicted from their amino acid composition: application to the detection of ‘cryptic’ antimicrobial peptides. J. Theor. Biol. 2017;419:254–265. doi: 10.1016/j.jtbi.2017.02.012. [DOI] [PubMed] [Google Scholar]
- Park K.-E., Jang S.H., Lee J., Lee S.A., Kikuchi Y., Seo Y.-s., Lee B.L. The roles of antimicrobial peptide, rip-thanatin, in the midgut of Riptortus pedestris. Dev. Comp. Immunol. 2018;78:83–90. doi: 10.1016/j.dci.2017.09.009. [DOI] [PubMed] [Google Scholar]
- Sievers F., Wilm A., Dineen D., Gibson T.J., Karplus K., Li W., Lopez R., McWilliam H., Remmert M., Söding J., Thompson J.D., Higgins D.G. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011;7(1) doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha S., Zheng L., Mu Y., Ng W.J., Bhattacharjya S. Structure and interactions of a host defense antimicrobial peptide thanatin in lipopolysaccharide micelles reveal mechanism of bacterial cell agglutination. Sci. Rep. 2017;7(1):17795. doi: 10.1038/s41598-017-18102-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taguchi S., Kuwasako K., Suenaga A., Okada M., Momose H. Functional mapping against Escherichia coli for the broad-spectrum antimicrobial peptide, thanatin, based on an in vivo monitoring assay system. J. Biochem. 2000;128(5):745–754. doi: 10.1093/oxfordjournals.jbchem.a022811. [DOI] [PubMed] [Google Scholar]
- Torres M.D.T., Melo M.C.R., Crescenzi O., Notomista E., de la Fuente-Nunez C. Mining for encrypted peptide antibiotics in the human proteome. Nat. Biomed. Eng. 2021;6(1):67–75. doi: 10.1038/s41551-021-00801-1. [DOI] [PubMed] [Google Scholar]
- Vetterli S.U., Zerbe K., Müller M., Urfer M., Mondal M., Wang S.-Y., Moehle K., Zerbe O., Vitale A., Pessi G., Eberl L., Wollscheid B., Robinson J.A. Thanatin targets the intermembrane protein complex required for lipopolysaccharide transport in Escherichia coli. Sci. Adv. 2018;4(11) doi: 10.1126/sciadv.aau2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The coordinates of the E. coli LptAm complexes with thanatins from P. maculiventris, C. ubica, and M. histrionica have been deposited to Protein Data Bank (www.pdb.org) with PDB access codes of 8GAJ, 8GAK, and 8GAL, respectively. Other data are available upon request.