A collection of iPSC lines derived from AML patients capture leukemia genomes and phenotypes and produce xenografts nearly identical to the matched primary PDXs. iPSCs from different AML clones uniquely empower the study of subclonal mutations.
Abstract
The reprogramming of human acute myeloid leukemia (AML) cells into induced pluripotent stem cell (iPSC) lines could provide new faithful genetic models of AML, but is currently hindered by low success rates and uncertainty about whether iPSC-derived cells resemble their primary counterparts. Here we developed a reprogramming method tailored to cancer cells, with which we generated iPSCs from 15 patients representing all major genetic groups of AML. These AML-iPSCs retain genetic fidelity and produce transplantable hematopoietic cells with hallmark phenotypic leukemic features. Critically, single-cell transcriptomics reveal that, upon xenotransplantation, iPSC-derived leukemias faithfully mimic the primary patient-matched xenografts. Transplantation of iPSC-derived leukemias capturing a clone and subclone from the same patient allowed us to isolate the contribution of a FLT3-ITD mutation to the AML phenotype. The results and resources reported here can transform basic and preclinical cancer research of AML and other human cancers.
Significance:
We report the generation of patient-derived iPSC models of all major genetic groups of human AML. These exhibit phenotypic hallmarks of AML in vitro and in vivo, inform the clonal hierarchy and clonal dynamics of human AML, and exhibit striking similarity to patient-matched primary leukemias upon xenotransplantation.
See related commentary by Doulatov, p. 252.
This article is highlighted in the In This Issue feature, p. 247
INTRODUCTION
Since reprogramming of human somatic cells to pluripotency through transcription factors was made possible, a plethora of induced pluripotent stem cell (iPSC) lines have been generated as models of numerous inherited genetic diseases (1, 2). Cancers are fundamentally somatic genetic diseases, and thus iPSC technology could offer new opportunities for their study (3). iPSC models of cancer have distinct advantages, notably providing faithful human genetic models of driver genetic lesions in their endogenous genomic environment and in disease-relevant cell types and enabling functional studies through genetic perturbations and high-throughput assays, such as multiomics and screens. However, efforts to generate stable iPSC lines from human cancers—immortalized cell lines or primary tumors—have been mostly unsuccessful or yielded incompletely reprogrammed cells (4–7). These reports have led to speculation that inherent biological properties of malignant cells hinder their reprogramming to a pluripotent stem cell state (6, 7). In addition, how well iPSC-derived cells resemble their primary counterparts remains largely unknown, and very limited studies have been designed to address this fundamental question.
Acute myeloid leukemia (AML) is a hematologic malignancy with fulminant course and poor prognosis, characterized by excessive proliferation of hematopoietic stem/progenitor cells (HSPC), which are blocked in their differentiation. AML is a genetically heterogeneous disease with over 100 known genetic drivers, which include gene mutations, chromosomal translocations, generating fusion oncogenes, and large chromosomal deletions (8). Although the landscape of AML genetic drivers has now been extensively characterized, the mechanisms by which specific gene mutations drive leukemogenesis remain incompletely understood. The development of genetically engineered mouse models (GEMM) of some of these mutations has provided important insights, but GEMMs do not always provide accurate genetic models due to species differences in genome organization, synteny, the physiologic regulation of hematopoiesis, and the kinetics of leukemia development, among others. On the other hand, primary leukemia cells isolated from the bone marrow (BM) or peripheral blood (PB) of AML patients survive poorly and only short-term ex vivo. Although patient-derived xenograft (PDX) models can often be generated from these primary cells and enable drug testing and other preclinical studies, they impose limits to mechanistic interrogation due to clonal heterogeneity and, critically, due to the very limited—practically absent—ability of primary AML cells to be genetically modified to enable functional studies.
Here, we describe “Complete Capture of Mutational Burden (CCoMB)” reprogramming, a method tailored to the reprogramming of cancer cells. With this method, we derived a panel of AML-iPSCs from 15 different patients, with normal matched lines for 7 of them, capturing 8 genetic groups of AML [t(15;17) - PML-RARA; splicing factor-mutated; TP53/aneuploidy; t(8;21) - AML1-ETO; MLL-rearranged; NPM1-mutated; FLT3-ITD; post-MPN], in 21 genotypes with combinations of 24 distinct recurrent AML genetic lesions (mutations, translocations, and deletions). We thus demonstrate that most AML driver mutations do not impose absolute biological barriers to the reprogramming of AML cells, with a notable exception being the NPM1 gene mutation.
We demonstrate that hematopoietic cells derived from a range of genetically diverse AML-iPSC lines are engraftable in immunodeficient mice, in striking contrast to normal human pluripotent stem cell (hPSC)-derived hematopoietic cells that are invariably nonengraftable (9). Importantly, taking advantage of this engraftment ability, we generated patient-matched primary and iPSC-derived leukemias and corresponding mouse xenografts. Although leukemia cells derived through directed differentiation of AML-iPSCs in vitro show significant differences in their cellular composition and transcriptomes to the matched primary leukemia cells, these differences are largely eliminated upon transplantation and xenografted AML-iPSC–derived leukemias are strikingly similar to their primary counterparts. The results and resources that we report here can transform the modeling and study of human acute leukemia and, possibly, of other cancers.
RESULTS
A Panel of iPSC Lines Represent the Genetic Diversity of AML
We previously described the derivation of bona fide iPSC lines from 3 patients with AML harboring MLL translocations (10) and del7q (11, 12). To increase the efficiency and success rate of iPSC derivation from AML patient samples, we developed Complete Capture of Mutational Burden (CCoMB) reprogramming (Fig. 1A). We reasoned that specific genetic (and potentially epigenetic) alterations of cancer cells likely exert a sizable (positive or negative) impact on reprogramming efficiency (13–20), thus skewing the clonal representation of the heterogeneous starting cell population after reprogramming. With these restrictions in mind, in CCoMB reprogramming, we incorporated two key modifications to standard reprogramming protocols: comprehensive genetic characterization of the starting cell sample and inference of its clonal composition, to then guide saturating targeted genetic screening of all clonally reprogrammed cell colonies that can be derived.
Figure 1.
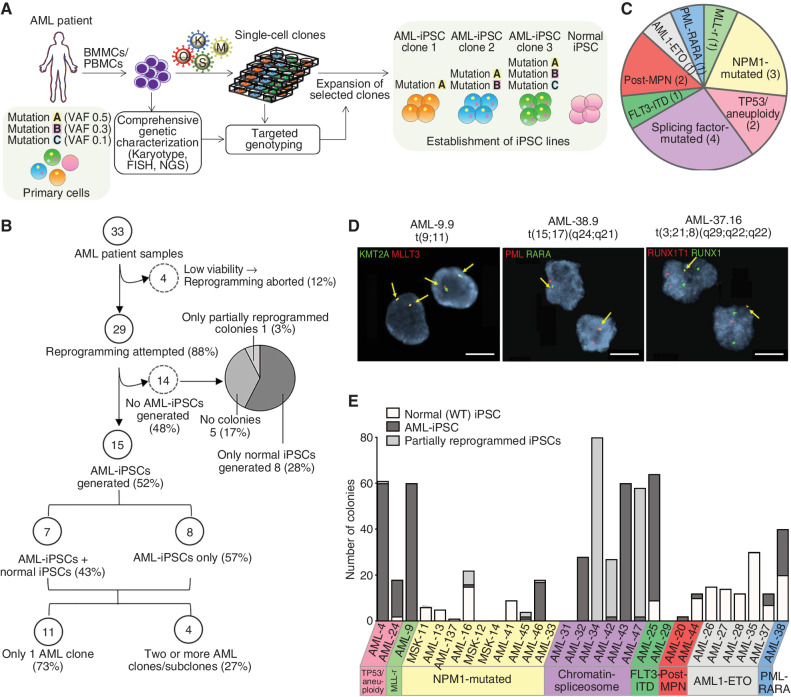
Generation of a panel of patient-derived iPSCs representative of the genetic diversity of human AML. A, Schematic overview of the reprogramming method. Bone marrow mononuclear cells (BMMC) or peripheral blood mononuclear cells (PBMC) from AML patients were first subjected to comprehensive genetic characterization (including karyotyping and FISH analysis for recurrent translocations and mutational analysis with gene panel sequencing) to infer the genetic and clonal/subclonal composition of the starting sample. Following transduction with Sendai vectors or a lentiviral vector expressing OCT4, SOX2, KLF4, and cMYC, the cells were plated in clonal density and all colonies were genotyped for all genetic alterations present in the primary sample using low-input methods (FISH and PCR with Sanger sequencing). Single-cell colonies corresponding to unique genotypes were selected and expanded to establish iPSC lines. B, Flowchart summarizing the reprogramming outcome of all 33 AML patient samples. C, Pie chart showing the distribution of the 15 AML patients from which AML-iPSCs could be established based on their genetic classification into AML genetic groups. Numbers in parentheses denote the number of patients within each genetic group. (Detailed information on patient and genetic characteristics is provided in Supplementary Tables S1 and S2.) D, Representative interphase FISH analyses for the indicated characteristic chromosomal translocations, as indicated. The respective iPSC line names are shown in the top. Scale bars, 20 μm. E, Bar plot showing the number of normal, AML-iPSC, as well as partially reprogrammed AML colonies obtained from each of the 29 patient samples with which reprogramming was attempted (from B, excluding the 4 samples with aborted reprogramming due to very low viability). Sample name and genetic groups are indicated. More detailed information is presented in Supplementary Table S2. Note that the term “AML-iPSC” here and throughout the manuscript refers to iPSC lines that harbor at least one myeloid malignancy driver mutation. Thus, preleukemic cells are also included under this umbrella term.
We obtained 33 AML patient samples (Supplementary Tables S1 and S2) and performed detailed genetic characterization of the starting cells, which included next-generation sequencing (NGS) of a comprehensive panel of myeloid malignancy driver genes and recurrent karyotypic abnormalities. Variant allele fraction (VAF) information was used to infer the clonal composition of the starting cell population and to inform the design of targeted PCR-based genotyping, suitable for rapid screening of tens to hundreds of individual iPSC colonies using low-input methods (21, 22). Clones with unique genotypes were selected and expanded to derive iPSC lines representative of as many distinct clones of the starting cell pool as possible, as well as normal [wild-type (WT)] cells, whenever possible.
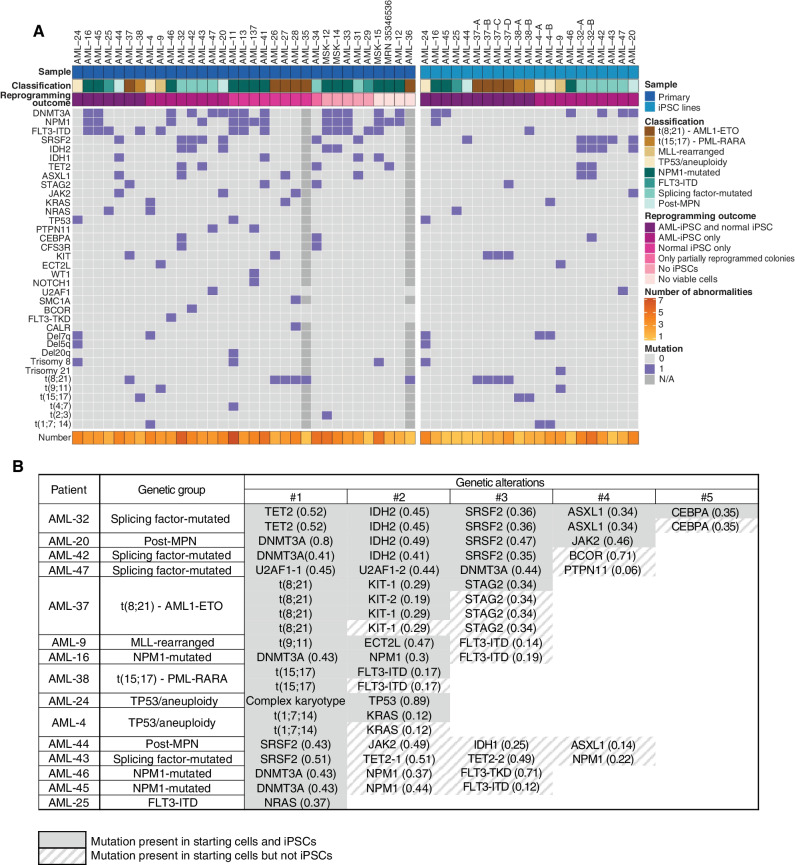
Four of the 33 samples did not contain viable cells after short-term culture, and reprogramming was thus aborted (Fig. 1B; Supplementary Table S2). For the remaining 29 patient samples, the reprogramming outcomes were: AML-iPSCs, i.e., iPSCs harboring at least one AML driver genetic lesion, were obtained from 15 of the samples (52%); 8 samples (28%) yielded only normal iPSCs, i.e., iPSCs without any driver mutations; 5 samples (17%) gave no colonies; and 1 sample gave only partially reprogrammed colonies, from which iPSC lines could not be established (Fig. 1B; Supplementary Table S2). The 15 AML cases from which AML-iPSCs could be derived included the 6 most common AML genetic groups, namely NPM1-mutated (patients AML-16, AML-45, and AML-46); t(15;17)-PML-RARA (patient AML-38); splicing factor-mutated (patients AML-32, AML-42, AML-43, and AML-47); TP53/aneuploidy (patients AML-4 and AML-24); t(8;21) RUNX1-RUNX1T1 (also known as AML1-ETO, patient AML-37); and t(9;11) X-KMT2A (MLL-rearranged, patient AML-9; Figs. 1C–E and 2A; Supplementary Fig. S1A and Supplementary Table S2). One additional sample (AML-25) had FLT3-ITD and NRAS mutations, but no other identified genetic lesions, and was assigned as “FLT3-ITD” and two more (AML-20 and AML-44) were from AML cases following a myeloproliferative neoplasm (post-MPN AML). Six of the 15 patients had clinically documented (AML-4, AML-24, and AML-32) or inferred—based on the presence of splicing factor mutations—(AML-42, AML-43, and AML-47), antecedent myelodysplastic syndrome (MDS) and one (AML-9) was a therapy-related case (Supplementary Table S1). We were able to derive matched normal (WT) iPSCs from 7 of the 15 patients (Supplementary Table S2). Furthermore, we were able to capture two or more distinct AML clones from 4 of the patients. Specifically, we obtained iPSCs from two AML clones/subclones from 3 of the samples: AML-32 (TET2, IDH2, SRSF2, ASXL1 with/without CEBPA mutations); AML-38 (PML-RARA with/without FLT3-ITD); and AML-4 (del7q with/without KRAS mutation; Fig. 1B; Supplementary Table S2). A fourth sample (AML-37) yielded 4 distinct clones: AML1-ETO alone; AML1-ETO with one of two distinct KIT mutations; and AML1-ETO with one KIT and one STAG2 mutation (Fig. 2A; Supplementary Table S2).
Figure 2.
AML-iPSCs capture both late and preleukemic clones arising during the clonal evolution of AML. A, Oncoplots showing all genetic lesions present in the AML patient samples (left) or the AML-iPSC lines generated (right). Letters (A, B, C, D) correspond to iPSC lines derived from different AML clones of the same patient. Classification refers to the genetic group classification of the patient AML. (Note that whereas AML-25 was classified as “FLT3-ITD,” all AML-iPSC lines obtained harbored NRAS and not FLT3 mutations. See also Supplementary Table S2.) B, Table showing the mutations captured in iPSCs from each of the 15 patients with respect to all mutations present in each starting patient sample. Numbers in parentheses show VAFs. The numbers on top represent the order by which mutations were acquired in each patient, inferred from VAF values and reprogramming outcomes (see also Fig. 3; Supplementary Fig. S2). U2AF1-1 and U2AF1-2 denote two different U2AF1 mutations—S34F and Q157R, respectively—present in patient AML-47 and in the derived iPSCs. TET2-1 and TET2-2 denote two different TET2 mutations—4044+1G>C and G1913D, respectively—present in patient AML-43. (See also Supplementary Table S2 for details.) KIT-1 and KIT-2 denote two different KIT mutations—N822K and D816V, respectively—present in patient AML-37 and in the derived iPSCs.
The reprogramming efficiency varied considerably across samples (Fig. 1E; Supplementary Fig. S1B and Supplementary Table S2).
The tumor burden of the sample, as determined by the allele fraction of the driver genetic lesions (highest or average of all allele fractions for each sample), did not appear to have a major impact on reprogramming success (Supplementary Fig. S1C and S1D). The age or sex of the patients did not influence the outcome either (Supplementary Fig. S1E and S1F). We also observed comparable efficiency of AML-iPSC generation from either BM or PB mononuclear cells, whereas BM was a better source for normal iPSCs (Supplementary Fig. S1G and S1H).
Although the relatively small number of samples prohibits deriving strong correlations between reprogramming efficiency and AML genetic groups, we made two notable observations.
First, reprogramming success was overall low for AML1-ETO cases (with only 1 out of 4 samples with viable cells yielding AML-iPSCs; Fig. 1E; Supplementary Table S2). Second, more strikingly, out of 11 NPM1-mutated cases with viable cells, none gave AML-iPSCs after one round of reprogramming (Supplementary Fig. S1I). All colonies generated from these samples were either WT (4 cases: AML-11, AML-13, AML-137, and AML-41) or had only DNMT3A without NPM1 mutations (AML-45). Three NPM1-mutated samples gave no colonies at all. NPM1-mutated colonies obtained from 3 samples were all partially reprogrammed and could not be established into bona fide iPSC lines with passaging. Finally, following a second round of reprogramming of an initially partially reprogrammed clone, we were able to derive one single iPSC line from one sample, AML-16 (Supplementary Fig. S1I; Supplementary Table S2). However, because of this multistep derivation history with presumed strong selection pressure, this NPM1-mutant iPSC line was not further used in this study. These results show strong negative selection against NPM1-mutated cells during reprogramming.
In summary, so far we showed that it is possible to reprogram all major genetic subtypes of AML into patient-derived iPSCs with a refined protocol, which we used to generate human genetic models of all major AML genotypes, encompassing 8 genetic groups (including post-MPN), 21 distinct genotypes, and 24 distinct AML driver genetic lesions (single-gene mutations, translocations, and aneuploidy; Fig. 2A; Supplementary Table S2).
CCoMB Reprogramming Aids Reconstruction of the Evolutionary Hierarchy of AML
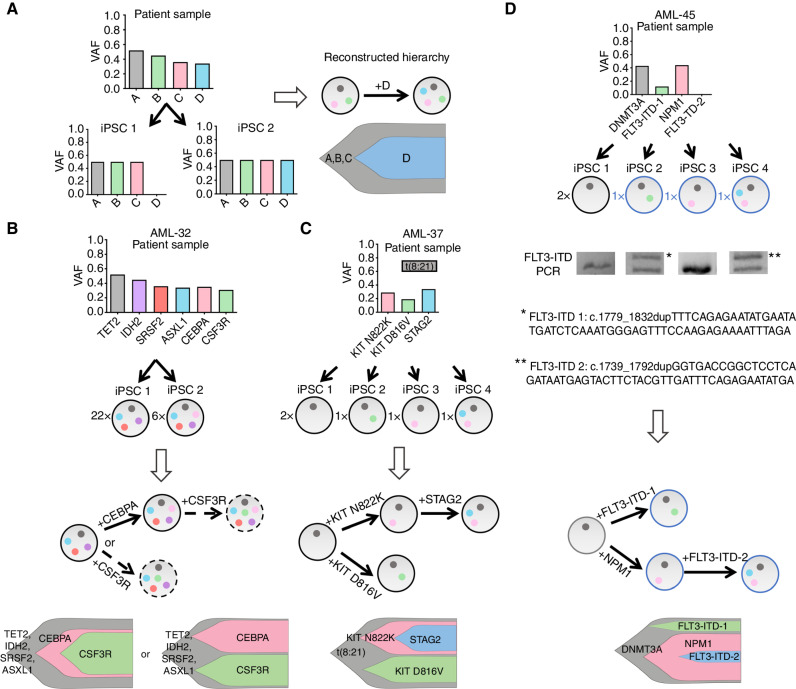
Although we and others have previously shown that patient cell reprogramming can capture distinct clones and subclones (10, 11, 23), whether reprogramming per se can inform cases of ambiguous clonal composition that bulk sequencing cannot resolve or uncover unsuspected clonal complexity has not been shown. In 7 of the 15 patients, reprogramming captured the most advanced disease clone, i.e., the clone harboring the complete set of mutations (Fig. 2B). In 4 of the remaining cases, the AML-iPSCs generated had all mutations except for the last subclonal mutation. In 4 more, the iPSCs captured only the preleukemic clone, which harbored single isolated mutations—specifically in DNMT3A (AML-45 and AML-46) or SRSF2 (AML-34 and AML-44; Fig. 2B). In 13 of the 15 cases from which we generated AML-iPSCs, reprogramming was informative with regard to clonal composition (Fig. 3A; Supplementary Fig. S2). (Partially reprogrammed clones were also included in these analyses to inform clonal composition.) In 7 of the cases, reprogramming confirmed a clonal hierarchy that could be relatively readily inferred from allele frequencies and past findings from population genetics studies (24–26). These included the occurrence of FLT3-ITD or PTPN11 mutations late in clonal progression (in cases AML-16, AML-9, AML-38, and AML-47; Supplementary Fig. S2A–S2D); the divergent acquisition of signaling activating mutations in independent subclones (AML-4 and AML-25; Supplementary Fig. S2E and S2F); and DNMT3A mutation being the initiating event in AML with DNMT3A, NPM1, and FLT3 mutations (AML-46; Supplementary Fig. S2G). In 4 cases, reprogramming helped resolve clonal relations that could not be inferred based on the bulk sequencing of the starting cells (AML-32, AML-42, AML-43, and AML-44; Fig. 3B; Supplementary Fig. S2H–S2J). More interestingly, in 2 additional cases, genotyping of the derivative iPSC lines yielded unexpected findings about the clonal composition of the starting sample. In patient AML-37, the reprogramming outcome revealed that a clone with the AML1-ETO t(8;21) translocation diverged into two clones, which then acquired a different KIT point mutation each, and one of them went on to acquire a subsequent STAG2 mutation (Fig. 3C). Reprogramming of the DNMT3A–NPM1–FLT3 AML-45 case revealed a more complex pattern of mutational acquisition than the expected linear DNMT3A→NPM1→FLT3 sequence. An initiating DNMT3A-mutant clone diverged into two lineages: one that evolved along the expected evolutionary path (with the acquisition of, first, an NPM1 mutation and, second, an FLT3-ITD mutation); and a second that acquired a different FLT3-ITD without an NPM1 mutation (Fig. 3D).
Figure 3.
Reprogramming illuminates the evolutionary history and clonal composition of AML. A, Schematic showing how reprogramming to pluripotency in clonal conditions can aid reconstruction of the clonal hierarchy of AML. As an example, the VAF values of 4 hypothetical mutations (A–D) in the patient cells (top) and in two derivative AML-iPSC lines (iPSC 1, iPSC 2) are shown. All mutations are clonal (VAF = 0.5 for heterozygous mutations) in the iPSCs, because each line is derived from a single starting cell. Because iPSC 1 contains mutations A–C and iPSC 2 contains mutations A–D, it can be concluded that mutation D was acquired after mutations A–C. The circles of different colors in the right represent the mutations A–D and the arrow denotes the order of mutational acquisition. Partially reprogrammed iPSC lines were also included in the clonal evolution analyses. B–D, Reconstruction of clonal evolution in 3 patients. The numbers to the left of each circle representing an iPSC clone denote numbers of colonies with the indicated genotype (see also Supplementary Table S2). In all fish plots each clone is represented by a different color and its height is proportional to the percentage of total cells that belong to a given clone (estimated from the VAF). Blue fonts and adjacent circles represent partially reprogrammed clones (which did not give iPSC lines but still informed clonal composition). B, Clonal evolution in patient AML-32. Two distinct clones were captured, one with 4 mutations (TET2, IDH2, SRSF2, and ASXL1) and one with 5 (the same 4 plus CEBPA). This result indicates that CEBPA was acquired after the other 4 mutations. Because no clones with CSF3R mutation were obtained, it cannot be determined if the CSF3R mutation (present in the patient sample) was acquired by the clone with 4 or 5 mutations. The two scenarios are thus represented with dashed arrows. C, Clonal evolution in patient AML-37. iPSC lines corresponding to 4 distinct clones were obtained. One clone only contained the t(8;21) translocation. Notably, two clones harbored each a different KIT mutation in addition to the translocation and a fourth clone harbored the t(8;21), one of the KIT mutations and, additionally, a STAG2 mutation. This result indicates the parallel evolution of two distance lineages with KIT mutations, one of which went on to subsequently also acquire a STAG2 mutation. (The cell fraction for the t(8;21) translocation in the starting sample was not available. Note that the STAG2 gene locus is on the X chromosome and the AML-37 patient is male; thus, the VAF for STAG2 is higher than that of the antecedent KIT mutations, which are heterozygous.) D, Clonal evolution in patient AML-45. Of the 4 clones obtained in iPSCs, one harbored an isolated DNMT3A mutation, which is thus unequivocally the initiating event. The other 3 clones all harbored the DNMT3A mutation and, in addition, an FLT3-ITD, an NPM1 mutation, or both an FLT3-ITD and an NPM1 mutation, respectively. Sequencing of the duplicated FLT3-ITD region in each clone revealed two different ITDs. These results allow us to conclude that, in this patient, an initial DNMT3A-mutant clone diverged into two lineages, one of which acquired an FLT3-ITD and the other both an NMP1 and an FLT3-ITD mutation.
These results highlight examples in which CCoMB reprogramming unveiled clonal hierarchies and show that reprogramming can provide new insights into the evolutionary process of AML.
AML-iPSCs Exhibit Phenotypic Hallmarks of AML
Reprogramming to pluripotency remodels the epigenome and resets a transcriptional and epigenetic state that sustains a pluripotency gene regulatory network (27). We previously showed that iPSCs derived from AML patients with MLL translocations and 7q deletions reset their epigenome at the pluripotent state, but reacquire a leukemic phenotype, transcriptome, and epigenome upon differentiation into HSPCs (10–12).
To test if this is a generalizable observation that extends to other genetic classes of AML, we phenotypically characterized our panel of genetically diverse AML-iPSCs following hematopoietic differentiation to HSPCs. We included in these analyses our previously derived AML-iPSC lines AML-4.10 and AML-4.24, harboring del7q (11) and the SU042.2 AML-iPSC line, harboring an MLL translocation (10). We excluded from these analyses 4 patients: AML-16, because of the strong selection pressure applied during two reprogramming rounds that were required for derivation of the only NPM1-mutated line from this patient; AML-45 and AML-46, because all derived lines captured only the preleukemic clone (with DNMT3A mutation only) and were thus not expected to exhibit leukemic features; and AML-37, because differentiation of multiple lines from this patient repeatedly yielded insufficient numbers of cells for meaningful phenotypic assessment or transplantation. We thus phenotypically assessed a total of 18 AML-iPSC lines, which encompassed 15 distinct genotypes (and 3 independent lines corresponding to the same clone), derived from 12 patients (Supplementary Table S3). For 3 of the patients (AML-4, AML-32, and AML-38), we characterized AML-iPSC lines capturing two distinct AML clones (Supplementary Table S3).
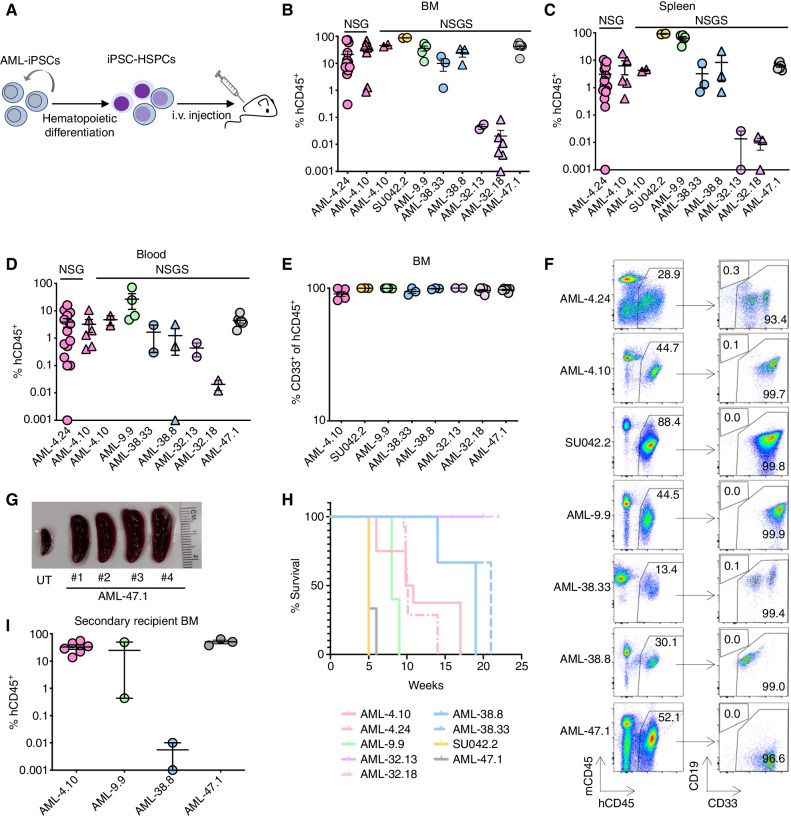
We previously showed that AML-iPSC–derived HSPCs (AML-iPSC-HSPC) are able to engraft into immunodeficient mice, in stark contrast to normal iPSC-HSPCs that are unable to engraft (10–12). We thus first assessed the engraftment ability of HSPCs from the new AML-iPSC panel, as the most stringent leukemic phenotype. We found that 6 AML-iPSC lines from 4 patients (AML-9, AML-38, AML-32, and AML-47), as well as the 3 previously characterized AML-iPSC lines (from patients AML-4 and SU042), gave rise to HSPCs that were able to engraft into NSG or NSGS mice. All mice had detectable engraftment of human cells, which were exclusively myeloid, in the BM, spleen, and PB (Fig. 4A–F). The mice showed signs of illness and splenomegaly, consistent with leukemia (Fig. 4G; Supplementary Fig. S3A and S3B). With the exception of mice transplanted with AML-iPSC–derived cells from patient AML-32, which had very low engraftment levels (0.1% or fewer hCD45+ cells), all mice succumbed to their disease (Fig. 4H). Transplantation of engrafted cells into secondary recipients resulted in robust engraftment and lethal leukemia in the case of AML-iPSCs from patients AML-4, AML-9, and AML-47 (Fig. 4I). Cells from the AML-38.8 line (patient AML-38) were not serially transplantable, despite robust engraftment in the primary recipients. We did not serially transplant engrafted iPSC-derived cells from patient AML-32 because of the low level of engraftment in the primary recipients. We have previously demonstrated the serial engraftment ability of the SU042.2 line (10).
Figure 4.
AML-iPSC–derived hematopoietic cells exhibit cardinal leukemia features. A, AML-iPSCs were differentiated into HSPCs and injected into immunodeficient mice (NSG or NSGS). Human cell engraftment was assessed after 13–15 weeks or earlier if signs of illness. B–D, Levels of human engraftment in the BM, spleen, and blood of NSG and NSGS mice, as indicated, 5 to 22 weeks after transplantation with 1 × 106 HSPCs derived from the indicated AML-iPSC lines. Each data point represents one mouse. Error bars show mean and SEM. E, Fraction of myeloid (CD33+) lineage cells within the hCD45+ population in the BM of mice transplanted with HSPCs derived from the indicated AML-iPSC lines. Each data point represents one mouse. Error bars show mean and SEM. F, Representative flow cytometry analyses from BM of recipient mice transplanted with HSPCs from the indicated AML-iPSC lines. G, Representative image showing marked splenomegaly in 4 mice transplanted with HSPCs from the AML-47.1 line. UT: untransplanted mouse. H, Kaplan–Meier curves showing survival of mice transplanted with HSPCs from the indicated AML-iPSC lines. I, Human engraftment levels in the BM of secondary recipient mice. Each data point represents a unique mouse. Mean and SEM are shown.
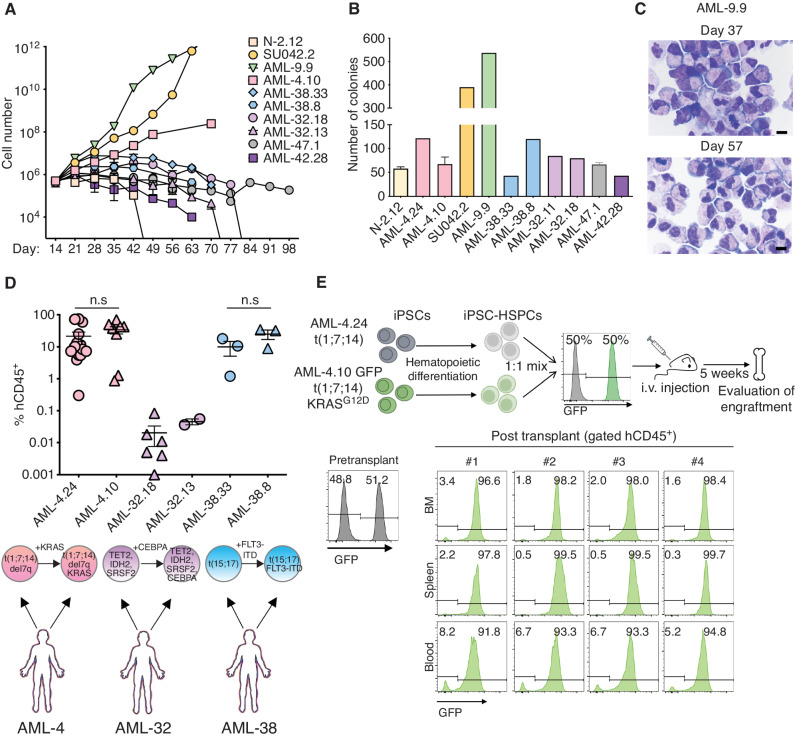
HSPCs from these AML-iPSC lines also exhibited in vitro leukemic features, specifically prolonged growth in culture, overall increased clonogenicity and markedly impaired differentiation along the monocytic and granulocytic lineages (Fig. 5A–C). These phenotypic findings are consistent with our previous observations of blocked differentiation and increased proliferation in vitro in other AML-iPSC models (11, 12, 28).
Figure 5.
AML-iPSC–derived hematopoietic cells mimic the in vivo clonal dynamics of patients. A, Cell counts of HSPCs derived from the indicated AML-iPSC lines at the indicated days of hematopoietic differentiation liquid culture. N-2.12: a normal iPSC line is shown for comparison. Mean of 3 independent differentiation experiments for each line is shown. B, Number of colonies obtained from 5,000 HSPCs derived from the indicated AML-iPSC lines seeded in methylcellulose assays on day 14 of hematopoietic differentiation. Error bars represent mean and SEM of 1–3 independent differentiation experiments. C, Wright–Giemsa staining of representative cytospin preparations of hematopoietic cells derived from the AML-9.9 line on days 37 and 57 days of hematopoietic differentiation liquid culture, showing predominantly cells with immature myeloid progenitor morphology and prominent mitotic figures. Scale bars, 10 μm. D, Levels of human engraftment (hCD45+) in the BM of NSGS mice transplanted with 1 × 106 HSPCs derived from the indicated 3 pairs of AML-iPSC lines. Each pair is derived from one AML patient and represents an earlier and a later clone, as indicated below the plot. n.s.: not significant (unpaired t test). E, Top panel: schematic overview of the experimental design. HSPCs derived from two lines representing an early (AML-24) and late (AML-4.10) clone from the same AML patient—the latter stably expressing GFP—were mixed 1:1 and intravenously injected into NSGS mice. Bottom: Left: flow cytometry assessment pre-transplant confirming equal mixing of the two clones. Right: flow cytometry assessment of 4 independent mice 5 weeks after transplantation.
We also compared the engraftment of iPSC-HSPCs from lines capturing two distinct AML clones from 3 patients (AML-4, AML-32, and AML-38). In all 3 cases, iPSC-HSPCs from the more advanced clone of the patient AML tended to engraft at higher levels (Fig. 5D; Supplementary Fig. S3C and S3D). To assess this in more defined conditions, we competitively transplanted two AML-iPSC lines, both derived from patient AML-4, harboring a t(1;7;14) translocation with del7q and subclonal RAS mutations (Supplementary Table S2): AML-4.24 harboring the translocation only; and AML-4.10, harboring, in addition, a KRAS mutation—the latter lentivirally marked with GFP. 5 weeks after transplantation with premixed equal numbers of HSPCs from each line, all 4 mice developed lethal leukemia that was almost exclusively derived from the more advanced AML-4.10 (GFP+) clone (Fig. 5E). These results show that AML-iPSC-HSPCs model the relative clonal fitness of the leukemia clones in the patients.
iPSC-HSPCs generated from preleukemic lines harboring isolated SRSF2 P95L mutations from patients AML-43 and AML-44 showed no detectable engraftment, as expected (Supplementary Fig. S3E; refs. 11, 28). AML-iPSC-HSPCs from two different lines from patient AML-42 (AML-42.19 and AML-42.28) did not show detectable engraftment (Supplementary Fig. S3F). Interestingly, matched primary leukemia cells from this patient did not engraft either (Supplementary Fig. S3F). This is consistent with the well-documented variability in the engraftment ability of primary human leukemias (29–31) and lends further support to the ability of the AML-iPSC model to mimic the behavior of the patient-specific leukemia. Finally, 5 AML-iPSC lines derived from 3 patients (AML-20, AML-24, and AML-25) repeatedly failed to produce CD45+ hematopoietic cells (Supplementary Fig. S4A). Upon hematopoietic differentiation, these lines produced KDR+ mesoderm and expressed CD34 and other endothelial markers, but failed to upregulate hematopoietic genes or markers of endothelial-to-hematopoietic transition (Supplementary Fig. S4B and S4C). These data are consistent with an early developmental block at a stage prior to the specification of the hematopoietic lineage, which prohibits the phenotypic assessment of the hematopoiesis derived from these lines.
AML-iPSC–Derived Xenografts Recapitulate the Cellular Architecture of Primary PDXs
The data presented so far here and our previous work (10–12, 28) establish that human leukemia cells can be reprogrammed to pluripotent stem cell lines and reproduce leukemic phenotypic features when differentiated back to hematopoietic cells. We have also previously shown that AML-iPSC-HSPCs recapitulate a phenotypic hierarchy, with cells with functional and genomic features of hematopoietic stem cells (HSC)/multipotent progenitor cells (MPP) on the apex, giving rise to more committed progenitors and mature cells, mimicking a leukemia stem cell hierarchy (12). However, how iPSC-derived leukemia cells compare to the primary patient leukemia remains unknown.
To investigate this, we first selected 4 patients from which we obtained AML-iPSCs—AML-4, AML-9, AML-32, and AML-47 (based on sample availability)—and created PDXs in NSGS mice. PDXs from all patients had very high levels of engraftment and developed lethal leukemia (Supplementary Fig. S5A and S5B). Comparison of immunophenotypic markers between human cells from these primary PDXs and the patient-matched AML-iPSC-derived PDXs revealed largely concordant immunophenotypes (Supplementary Fig. S5C). (AML-32 was excluded from this analysis because of very low-level engraftment of the AML-iPSC–derived cells; Fig. 4B–D.)
To further compare patient-specific iPSC-derived leukemias with the matched primary AML before and after transplantation, we performed single-cell transcriptome analyses of patient-matched primary and iPSC-derived leukemia cells ex vivo/in vitro or after transplantation and isolation from primary and matched iPSC-derived PDXs from 3 of the patients, AML-4, AML-9, and AML-47 (Figs. 6A–G, and 7A–E; Supplementary Figs. S6A–S6F and S7A–S7I). AML-iPSC–derived cells engrafted into secondary recipients were also analyzed in 2 of the 3 patients (AML-9 and AML-47). Two time points—7 and 13 weeks following transplantation—were analyzed in the case of AML-9 patient-derived iPSCs to evaluate potential changes in cellular composition over time. AML-32 was again excluded because engraftment of the AML-iPSC–derived cells was too low to yield sufficient cells for these analyses. DNA sequencing of cells isolated from PDXs from patient AML-4 determined that the vast majority of transplanted cells contained the subclonal NRAS mutation (VAF: 0.45). Thus for a more accurate comparison, we chose the AML-4.10 iPSC line, harboring a KRAS mutation (and not the AML-4.24 line that lacks a RAS mutation) as the comparator. In contrast, cells isolated from PDXs from patient AML-9 lacked the subclonal FLT3-ITD mutation of this patient and were thus compared with the AML-9.9 line that also lacks FLT3-ITD mutation.
Figure 6.
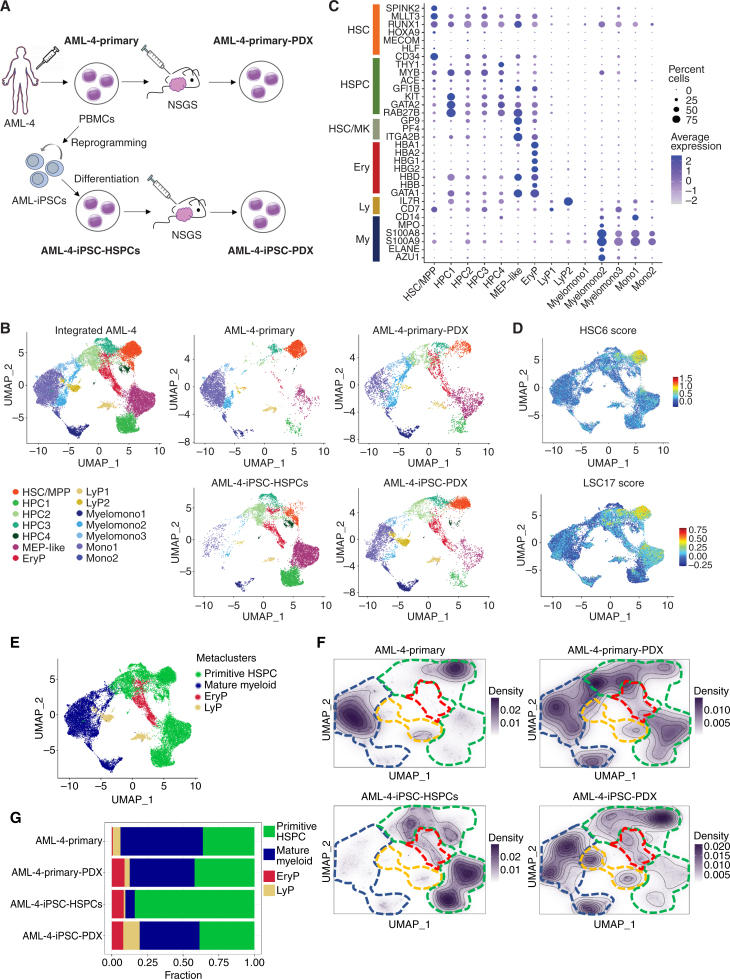
Single-cell RNA sequencing analyses of matched primary and iPSC-derived leukemia cells from patient AML-4 in vitro/ex vivo and after transplantation. A, Schematic representation of the experimental design. The samples analyzed by single-cell RNA sequencing are: (1) AML-4-Primary: peripheral blood mononuclear cells (PBMC) from AML patient AML-4; (2) AML-4-Primary-PDX: cells (sorted hCD45+) from a mouse xenograft of the AML-4-Primary cells; (3) AML-4-iPSC-HSPCs: cells obtained following in vitro differentiation of the iPSC line AML-4.10, which was derived from the AML-4-Primary cells; (4) AML-4-iPSC-PDX: cells (sorted hCD45+) from a mouse xenograft of the AML-4-iPSC-HSPCs. B, UMAP representation of single-cell transcriptome data colored by cluster. Left: Integrated analysis of all AML-4 subsets. Right: Individual samples, as indicated. Clusters were annotated using known lineage and stem cell marker genes found amongst the most differentially expressed genes in each cluster. HSC/MPP: hematopoietic stem cell/multipotent progenitor; HPC: hematopoietic progenitor cell; MEP: megakaryocyte/erythroid progenitor; EryP: erythroid progenitor; LyP: lymphoid progenitor; Myelomono: mature cells of myelomonocytic lineage; Mono: mature monocytic lineage cells. C, Dot plot showing the expression level of selected marker genes in each cluster. Dot size represents the percentage of cells expressing the marker gene, while dot color represents the scaled average expression of the gene across the various clusters (a negative value corresponds to expression level below the mean). D, HSC6 score based on the expression of the 6 genes RUNX1, HOXA9, MLLT3, MECOM, HLF, and SPINK2 (top) and LSC17 score (bottom), projected onto the integrated analysis UMAP from B. E, UMAP plot from B colored by metacluster. F, Cell density across the UMAP coordinates of each sample displayed as contours filled by a dark violet color gradient. The 4 metaclusters from E are indicated by dashed lines (green line: primitive HSPC; blue line: mature myeloid; red line: EryP; yellow line: LyP). G, Bar plots showing the proportion of cells in each metacluster for each sample.
Figure 7.
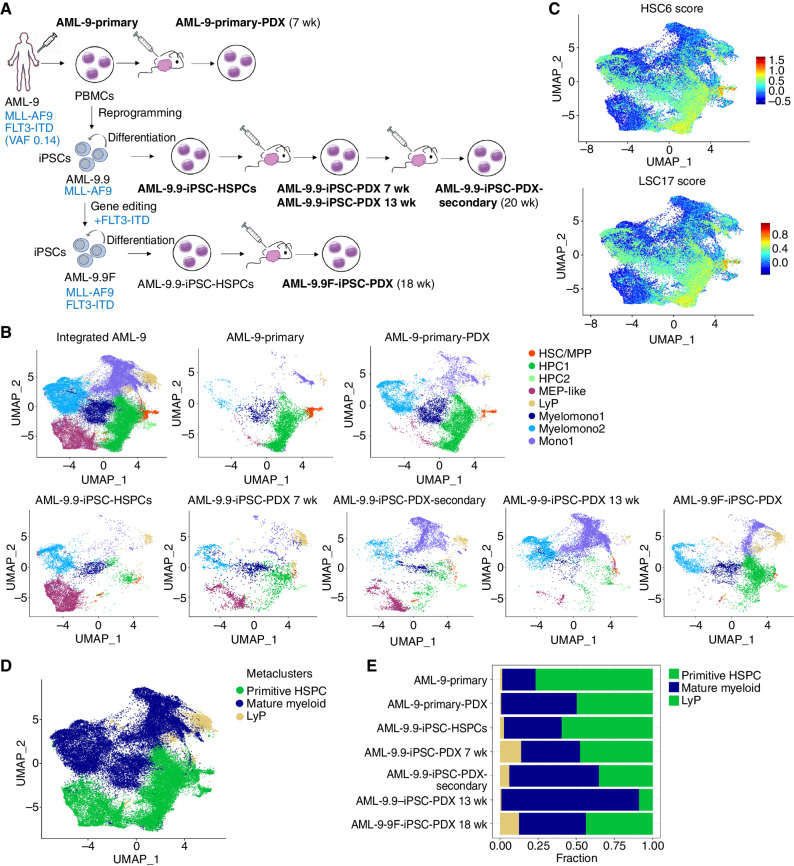
Single-cell RNA sequencing analyses of matched primary and iPSC-derived leukemia cells from patient AML-9 before and after xenotransplantation. A, Schematic representation of the experimental design. The samples analyzed by single-cell RNA sequencing are: (1) AML-9-Primary: peripheral blood mononuclear cells (PBMCs) from AML patient AML-9; (2) AML-9-Primary-PDX: cells from a mouse xenograft of the AML-9-Primary cells; (3) AML-9.9-iPSC-HSPCs: cells obtained following in vitro differentiation of the iPSC line AML-9.9, which was derived from the AML-9-Primary cells; (4) AML-9.9-iPSC-PDX 7 weeks and AML-9.9-iPSC-PDX 13 weeks: cells recovered from mouse xenografts of the AML-9.9-iPSC-HSPCs 7 and 13 weeks post transplant, respectively; (5) AML-9.9-iPSC-PDX-Secondary: cells obtained after serial transplantation of the AML-9-iPSC-PDX cells into secondary recipient mice; (6) AML-9.9F-iPSC-PDX: cells from a mouse xenograft of iPSC-HSPCs from the AML-9.9F line (see text for details). (The number of weeks post transplantation when the cells were recovered is indicated in parentheses next to the sample name.) B, UMAP representation of single-cell transcriptome data colored by cluster. Integrated analysis of all AML-9 datasets and the indicated individual samples are shown. Clusters were annotated using known lineage and stem cell marker genes found amongst the most differentially expressed genes in each cluster. HSC/MPP: hematopoietic stem cell/multipotent progenitor; HPC: hematopoietic progenitor cell; MEP: megakaryocyte/erythroid progenitor; LyP: lymphoid progenitor; My: mature myeloid lineage cells; Mono: mature monocytic lineage cells. C, HSC6 score based on the expression of the 6 genes RUNX1, HOXA9, MLLT3, MECOM, HLF, and SPINK2 (top) and LSC17 score (bottom), projected onto the integrated analysis UMAP from B. D, UMAP plot from B colored by metacluster. E, Bar plots showing the proportion of cells in each metacluster for each sample.
Each set of samples from each of the 3 patients was integrated and visualized with uniform manifold approximation and projection (UMAP). Clustering and manual assignment of clusters to cell types based on marker gene expression (Supplementary Table S4) revealed cells corresponding to HSC/MPPs, hematopoietic progenitor cells (HPC), and more mature cells of the myeloid lineage in all samples from all 3 patients at varying frequencies (Figs. 6A–C and 7A and B; Supplementary Fig. S6A and S6B). Identification of HSC/MPPs was further aided by the use of an “HSC6 score,” derived from the expression of 6 genes (RUNX1, HOXA9, MLLT3, MECOM, HLF, and SPINK2), recently identified as a signature that distinguishes HSCs from HPCs throughout developmental stages of human hematopoiesis in a comprehensive single-cell transcriptome study of human embryos (refs. 32; Figs. 6D and 7C; Supplementary Fig. S6C). Additionally, we used an established leukemia stem cell score, LSC17 (33). Overall cluster assignment was also informed by cell cycle and pseudotime analyses of the single-cell transcriptome data (Supplementary Fig. S7A–S7I). To facilitate comparisons across samples, we derived two metaclusters, primitive HSPC and mature myeloid, by merging the primitive (HSC/MPP and HPCs) and mature (myeloid, monocytic, and myelomonocytic) clusters, respectively (Figs. 6E and 7D; Supplementary Fig. S6D).
Leukemias derived through in vitro differentiation from iPSC lines exhibited both similarities and differences in their cellular composition and transcriptome, compared with the patient-matched ex vivo leukemias (Figs. 6B, F, and G, and 7B and E; Supplementary Figs. S6B and S6E, and S6F). However, the same primary and iPSC-derived cells, after transplantation, became strikingly more similar to each other (Figs. 6B, F, and G, and 7B and E; Supplementary Figs. S6B and S6E, and S6F). (The primary PDX sample from patient AML-47 did not pass quality control and was thus excluded from these analyses.) Integration and clustering of the single-cell RNA sequencing (scRNA-seq) data using orthogonal methods gave very similar results, reinforcing that the high similarity between primary and iPSC-derived leukemias after transplantation is not an artifact of the data integration approach (Supplementary Fig. S8A–S8C). In all 3 AML cases, leukemic cells derived in vitro from iPSCs contained a high proportion of cells within the primitive HSPC metacluster. This fraction decreased upon transplantation, gradually, as a function of time, in primary recipients sampled at different time points and in secondary recipients (Figs. 6F and G, and 7A, B, and E; Supplementary Fig. S6E and S6F). Conversely, the fraction of iPSC-derived cells within the mature myeloid metacluster increased progressively upon primary and secondary transplantation (Figs. 6F and G, and 7A, B, and E; Supplementary Fig. S6E and S6F). These results mimic the gradual loss of stemness upon serial transplantation, well-documented in PDX models.
To further investigate the similarities and differences between primary and iPSC-derived cells, we performed pseudobulk differential gene-expression (DGE) analyses (Supplementary Fig. S8D–S8F). Principal component analysis showed that primary and iPSC-derived cells are closer to each other after transplantation than before transplantation (Supplementary Fig. S8D). Comparison of primary to iPSC-derived cells prior to transplantation revealed 4,779 differentially expressed genes (DEGs), whereas this number dropped to 913 after transplantation (Supplementary Fig. S8E). To gain further insights into the sources of these differences and similarities, we performed DGE analysis specifically in the HSC/MPP cluster, which comprises the leukemia-initiating cells. These cells were essentially identical between primary and primary PDX samples (only 3 DEGs), and near-identical between primary PDX and iPSC-PDX (only 17 DEG), although they were more dissimilar between iPSC-PDX and iPSC-HSPCs (95 DEGs) or between primary cells and iPSC-HSPCs (604 DEGs). These results are consistent with a model whereby iPSCs give rise to a diverse repertoire of HSPC types in vitro and transplantation selects for leukemia-initiating cells that are transcriptionally very similar, if not indistinguishable, to the leukemia-initiating cells of the primary leukemias.
These results collectively demonstrate that patient-specific iPSC-derived myeloid leukemias recapitulate the cellular composition of the matched primary AML upon xenotransplantation, thus representing faithful models of human leukemias.
AML-iPSCs Provide Insights into the Contribution of Specific Mutations to the AML Phenotype
Currently, our ability to link genotypes to phenotypes of primary human AML is limited because no cell-surface markers exist to separate clones and subclones. Methods to combine the detection of mutations with transcriptomes or other genomic readouts at the single-cell level are increasingly improving, but the insights that can be derived from them remain limited. We therefore exploited the ability to capture distinct clones from the same patient in iPSC lines and thus isolate the effects of specific mutations in isogenic conditions, to interrogate the contribution of FLT3-ITD mutation to the AML phenotype. To this end, we used CRISPR/Cas9-mediated gene editing to introduce an FLT3-ITD mutation in line AML-9.9. Line AML-9.9 was derived from patient AML-9, who had an MLL-AF9 translocation and subclonal FLT3-ITD, and contained the MLL-AF9 translocation only (Fig. 2; Supplementary Table S2). We derived a CRISPR-edited iPSC line, AML-9.9F, with a clonal heterozygous FLT3-ITD mutation, which was differentiated and transplanted into NSGS mice.
Single-cell transcriptome analysis showed that the AML-9.9F clone generated more primitive HSPCs and, conversely, fewer mature myelomonocytic cells than the main clone without the FLT3-ITD mutation (AML-9.9; Fig. 7A, B, and E). Although the AML-9.9 line, representing the FLT3-WT clone, contained 48% primitive HSPCs after 7 weeks and only 9% primitive HSPCs at 13 weeks after transplant, the AML-9.9F line, representing the FLT3-ITD mutant subclone, contained 44% primitive cells at 18 weeks after transplant. These results suggest that the acquisition of an FLT3-ITD mutation skews the hierarchical structure of the mutant subclone toward a more primitive phenotype, which may account for the unfavorable prognosis and frequent emergence during disease relapse of FLT3-ITD mutations. More broadly, these experiments demonstrate the value of AML-iPSC models to reveal clonal and subclonal cellular hierarchies of AML.
DISCUSSION
Here we report the generation of a comprehensive panel of patient-derived AML-iPSC lines by tailoring a reprogramming protocol to genetically and clonally heterogeneous malignant cells. Our panel comprises iPSC models of all major genetic classes of human AML, including NPM1-mutated (approximately 30% of all AML); t(15;17) - PML-RARA (approximately 13% of all AML); splicing factor-mutated (approximately 13% of all AML); TP53/aneuploidy (approximately 10% of all AML); t(8;21) - AML1-ETO (approximately 7% of all AML); MLL-rearranged (approximately 4% of AML) and others. These provide new human genetic models of AML that can empower in vitro and in vivo functional studies into leukemia biology, as well as preclinical studies.
Human AMLs have characteristically very simple genomes, compared with most adult solid tumors, harboring only 2 to 5 genetic driver genetic lesions (8, 24). This has led to speculation that nongenetic lesions, such as heritable epigenetic lesions, or noncell autonomous factors, such as insults from the microenvironment, may also serve as drivers and contribute to the disease. Reprogramming to pluripotency erases any epigenetic marks of the starting cell and, as we show here, single leukemic cells reestablish the phenotypic and transcriptomic features of the original leukemia after reprogramming and differentiation back to hematopoietic cells. These results argue that, at least in many cases, the leukemia genome is largely sufficient to reproduce the leukemia epigenome and cellular phenotype. Additionally, the observation that the growth advantage of the more evolved subclone was recapitulated in cases where iPSCs from AML clones and subclones were available (Fig. 5D and E) argues that this advantage is largely cell autonomous.
Although the derivation of cellular models is the primary and most valuable output of reprogramming human AML samples, the reprogramming process per se allows the clonal deconvolution of the genetic composition of a given leukemia sample, much as was hitherto possible only by single-cell plating and assessment of colonies grown in methylcellulose media (34). Single-cell DNA sequencing approaches are increasingly informing the clonal architecture of AML, but the sensitivity of detection of different mutations remains highly variable (35). Although reprogramming to inform clonal relationships is neither a practical nor scalable approach to be useful in clinical practice, it could be of use to elucidate unclear cases of mutational order and comutation for research applications or even to aid minimal residual disease monitoring by NGS approaches in select cases.
Our results show that preleukemic clones—harboring isolated initiating preleukemic mutations—were more rarely captured, compared with fully leukemic clones, with the exception of NPM1-mutant cases (in which NPM1 mutations prevent reprogramming of the leukemic clone). This possibly reflects the rarity of preleukemic cells, compared with the fully evolved AML cells, in the blood or BM of patients with full-blown AML. At the same time, subclonal mutations were also less likely to be captured than clonal mutations, again likely reflecting the underrepresentation of the subclones, compared with the major clones, in the starting cells. In 11 of the 15 cases, only one leukemic or preleukemic clone per patient was captured, underscoring the complementarity of reprogramming with gene editing to add or correct specific mutations, in order to more comprehensively model clonal evolution (28, 36).
A subset of lines from 3 patients (AML-20, AML-24, and AML-25) presented a differentiation block prior to the specification of the hematopoietic lineage, akin to the embryonically lethal phenotype observed in some GEMMs with constitutive genetic lesions. Two distinct but genotypically identical lines from patients AML-24 (AML-24.14 and AML-24.15) and AML-25 (AML-25-14 and AML-25.16) showed identical phenotypes (Supplementary Fig. S4; Supplementary Table S3), arguing against spurious line-to-line variation as a potential source of this differentiation defect. Although underlying genetic lesions are the likely culprits, these are hard to pinpoint, as there appear to be no shared genetic lesions among the lines of these 3 patients. iPSCs from patient AML-24 had a complex karyotype and patient AML-25 likely harbored additional undetected mutations or larger-scale lesions, in addition to the characterized subclonal FLT3 and NRAS mutations.
Distinct but genetically identical lines derived from the same patients (AML-24, AML-25, and AML-42; Supplementary Table S3) showed identical phenotypes, in agreement with our previous observations (10–12, 28, 36–38). This consistent observation effectively eliminates random line-to-line variation as a discernible source of phenotypic differences in our study. AML-iPSC-HSPCs from different lines showed variable engraftment ability into immunodeficient mice, which, importantly, tracked with patient of origin. Even though lines from more advanced clones showed a relative advantage compared with the more ancestral ones, lines derived from the same patient exhibited comparable overall engraftment ability. Engraftment and survival of transplanted mice were also comparable between primary and iPSC-derived cells (Supplementary Fig. S5A and S5B). All MLLr lines generated from two patients showed high-level engraftment, as well as exceptionally high proliferation ability and clonogenic capacity ex vivo (Figs. 4 and 5A–C), mirroring the well-documented unusually high self-renewal capacity of this AML type in patients and other research models (39). Leukemia cells from lines derived from patient AML-42, whose primary leukemia was nonengraftable, also lacked engraftment ability themselves. In the case of AML-47, engraftment was slightly lower with iPSC-derived than with primary cells, but the primary xenografts survived longer than the iPSC-derived (Supplementary Fig. S5A and S5B), indicating that these differences are likely spurious. (It should also be noted that the endpoint for engraftment assessment was not fixed, but determined by the time of overt disease, and that the cohort size of the mice included in these analyses was limited by the number of primary cells.) Collectively, these observations support the ability of AML-iPSCs to accurately reproduce and model behaviors of primary human leukemia cells.
Although some differences revealed in our scRNA-seq analyses between the primary nontransplanted AML and the iPSC-derived cells could be due to comparison of a genetically heterogeneous primary AML sample to a clonal iPSC line, our DGE analyses suggest that a more likely source of these differences is the heterogeneity of the types of HSPCs generated in vitro from iPSCs through current directed differentiation protocols. Most of these differences are eliminated following the selection of a more rare subset of leukemia-initiating stem/progenitor cells that are transcriptionally very similar, if not identical, to the primary leukemia-initiating cells upon transplantation (Supplementary Fig. S8F). Both primary and iPSC-derived leukemia cells underwent maturation upon xenotransplantation in a time-dependent manner. This gradual maturation has been well documented in primary AML xenografts and is again mirrored by our iPSC models.
Differences in cellular composition and immunophenotype of human AML have long been recognized and reflected in classification systems used in the past, such as the French–American–British (FAB) classification. More recent studies have reinforced these diverse immunophenotypic features and correlated them with clinical outcomes, such as prognosis, patient survival, and responses to targeted treatments. A recent study, using computational approaches for the deconvolution of the cellular architecture from bulk transcriptomes of a large number of human AML, identified differences along a primitive-to-mature cell axis as a dominant prognostic factor (40). This and other recent studies also correlated certain mutations with differentiation state and hierarchy (35, 41). Specifically, RAS mutations were correlated with a more mature phenotype, whereas FLT3-ITD mutations were associated with a more primitive hierarchy (41–43). Although previous data from AML patient samples revealed these associations, they are correlative and noisy, due to significant heterogeneity inherent in these samples. In contrast, here we were able to exploit the strictly clonal and isogenic conditions afforded by our models to more unambiguously establish a causative link between the presence of FLT3-ITD and a more primitive AML phenotype, at least in one AML case. Thus, our data lend strong support to the observations in patients of a connection between subclonal mutations and leukemia maturation stage.
We observed a high overlap between the LSC17 and HSC6 scores, as both were high primarily in the HSC/MPP, MEP-like, and some HPC clusters (Figs. 6D and 7C; Supplementary Fig. S6C). Interestingly, in the case of AML-9, which is an MLLr AML, the cells with the highest LSC17 score were not the HSC/MPP (Fig. 7C), as in the two other patients (Fig. 6D; Supplementary Fig. S6C), but cells within an HPC cluster (HPC-2). This might reflect previous evidence of an origin of the leukemia-initiating cells in MLLr AML from committed myeloid progenitors rather than HSC/MPPs (44).
The ability to derive engraftable HSCs from hPSCs remains a highly desirable but still unattainable goal in the stem cell research and regenerative medicine fields, which, if realized, would revolutionize stem cell therapies. So far, AML-iPSCs are the only hPSCs able to support engraftable hematopoiesis of in vitro differentiated HSPCs lacking any transgenes. This remarkable property may inform not only the biology of human AML but also efforts to derive normal engraftable hematopoiesis from hPSC sources in the future. Our findings, showing that the mouse repopulation assay selects for iPSC-derived cells essentially identical to the primary repopulating cells—at least in an AML background—validate xenotransplantation as a relevant assay to aid these efforts toward protocols to generate engraftable iPSC-derived normal hematopoietic cells.
In summary, we have generated a diverse panel of AML-iPSCs and associated transcriptome data, as a valuable resource for the stem cell, leukemia, and cancer communities. Additionally, the results presented here demonstrate that xenotransplantation selects for effectively identical cells between primary and AML-iPSC-derived leukemia cells. Thus, iPSC reprogramming can faithfully and comprehensively model human AML and xenotransplantation of AML-iPSC–derived cells is essential to this end. The results presented here can pave the way toward the iPSC modeling of other human cancers.
METHODS
Generation of AML-iPSCs Through Patient Cell Reprogramming
Peripheral blood mononuclear cells (PBMC) or bone marrow mononuclear cells (BMMC) from AML patients were obtained with written informed consent under protocols approved by local Institutional Review Boards at the Icahn School of Medicine at Mount Sinai and Memorial Sloan Kettering Cancer Center. All studies were conducted in accordance with Declaration of Helsinki ethical guidelines. Cryopreserved PBMCs or BMMCs were thawed and cultured in X-VIVO 15 media with 1% nonessential amino acids (NEAA), 1 mmol/L L-glutamine, and 0.1 mmol/L β-mercaptoethanol (2ME) and supplemented with 100 ng/mL stem cell factor (SCF), 100 ng/mL Flt3 ligand (Flt3L), 100 ng/mL thrombopoietin (TPO), and 20 ng/mL IL3 for 1 to 4 days.
For induction of reprogramming, 10,000 to 300,000 cells were transduced with the excisable OKMS CMV-fSV2A lentiviral vector (37) or the viral cocktail CytoTune-iPS 2.0 Sendai reprogramming kit (Invitrogen), containing KLF4, OCT4, and SOX2 (KOS) virus, the c-MYC virus and the KLF4 virus. Twenty-four hours later, the cells were harvested and plated on mitotically inactivated MEFs in 6-well plates and the plates were centrifuged at 500 × g for 30 minutes at RT. The next day and every day thereof, half of the medium was changed to the hESC medium with 0.5 mmol/L valproic acid (VPA). Colonies with hPSC morphology were manually picked and expanded.
Mutational Analysis
An aliquot of the cells before culture was used for gene panel sequencing with a custom capture bait set including the coding regions of 163 myeloid malignancy genes and 1,118 genome-wide single-nucleotide polymorphism (SNP) probes for copy-number analysis, with on average one SNP probe every 3 Mb. Samples were sequenced with pair-end Illumina Hi-Seq at a median coverage of 600× per sample (range, 127–2,480×). Variants with VAF <2%, less than 20 total reads, or less than 5 mutant supporting reads, were excluded. After prefiltering of artifactual variants, likely germline SNPs were filtered out by considering the VAF density of variants, their presence in the Genome Aggregation Database (gnomAD), their annotation in the human variation database ClinVar and their recurrence in a panel of normal samples. From the list of likely somatic variants, putative oncogenic variants were distinguished from variants of unknown significance based on the mutational consequence and their recurrence in various databases of somatic mutations in cancer. The HeatmapAnnotation function of the “ComplexHeatmap” package was used to generate the oncoplots. iPSC colonies were genotyped by targeted PCR with primers listed in Supplementary Table S5 and Sanger sequencing.
Human iPSC Culture, Hematopoietic Differentiation, and In Vitro Phenotypic Characterization
Culture of human iPSCs on mitotically inactivated MEFs was performed as previously described (36). Hematopoietic differentiation was performed using a spin-EB protocol previously described (36). In the end of the differentiation culture, the cells were collected and dissociated with accutase into single cells and used for flow cytometry, cytological analyses, or clonogenic assays.
Cytological Analyses
Approximately 200,000 cells from liquid hematopoietic differentiation cultures were washed twice with PBS containing 2% FBS and resuspended in PBS. Cytospins were prepared on slides using a Shandon CytoSpin III cytocentrifuge (Thermo Electron Corporation). Slides were then air-dried for 30 minutes and stained with the Hema 3 staining kit (Fisher Scientific Company LLC). The slides were read on a Nikon Eclipse Ci microscope and digital images were taken with a Nikon DS-Ri2 camera and NIS-Elements D4.40.00 software.
Clonogenic Assays
For methylcellulose assays, the cells were resuspended in StemPro-34 SFM medium at a concentration of 3 × 104/mL. Cell suspension (500 μL) was mixed with 2.5 mL MethoCult GF+ (H4435, Stem Cell Technologies) and 1 mL was plated in duplicate 35-mm dishes. Colonies were scored after 14 days and averaged between the duplicate dishes.
Transplantation into NSG and NSGS Mice
NSG (NOD.Cg-PrkdcscidIl2rgtm1Wjl/SzJ) and NSGS (NOD.Cg-PrkdcscidIl2rgtm1WjlTg(CMV-IL3,CSF2,KITLG)1Eav/MloySzJ) mice were purchased from Jackson Laboratories and housed at the Center for Comparative Medicine and Surgery at Icahn School of Medicine at Mount Sinai. One day before transplantation, the mice were injected intraperitoneally with 30 mg/kg busulfan solution. Primary leukemia patient cells or AML-iPSC–derived hematopoietic cells from days 11 to 16 of hematopoietic differentiation were resuspended in StemPro-34 and injected via the tail vein using a 25-G needle at 1 × 106 cells per mouse in 100 μL. The mice were sacrificed when they showed signs of illness or at 22 weeks, if no signs of illness. BM was collected from the femurs and tibia. Total blood was collected by cardiac puncture, and spleens were harvested and their weight recorded. BM, blood, and spleen cells were hemolyzed with ACK lysing buffer and stained with anti-mCD45, anti-hCD45, anti-hCD33, anti-hCD19, anti-hCD34, and anti-hCD38. All mouse studies were performed in compliance with Icahn School of Medicine at Mount Sinai laboratory animal care regulations and approved by an Institutional Animal Care and Use Committee (IACUC). Secondary transplants were performed by the tail-vein injection of 1 × 106 fresh MACS- or FACS-sorted hCD45+ cells isolated from the BM of primary recipient mice in busulfan-treated NSGS mouse recipients.
Flow Cytometry and Cell Sorting
The following antibodies were used: CD34-PE (clone 563, BD Pharmingen), CD45-APC (clone HI30, BD PharMingen), CD33-BV421 (clone WM53, BD Horizon), mCD45-PE-Cy7 (clone 30-F11, BD Biosciences), CD19-PE (clone HIB19, BD Pharmingen), CD38-PE-CF594 (clone HIT2, BD Horizon), CD15-BV785 (clone W6D3, BioLegend), CD14-APC (clone M5E2, BD Horizon), and CD44-APC. Cell viability was assessed with DAPI (Life Technologies). Cells were assayed on a BD Fortessa and data were analyzed with FlowJo software (Tree Star). Cell sorting was performed on a BD FACSAria II.
qRT-PCR
RNA was isolated with TRIzol (Life Technologies). Reverse transcription was performed with Superscript III (Life Technologies) and qPCR was performed with the SsoFast EvaGreen Supermix (Bio-Rad) using primers listed in Supplementary Table S6. Reactions were performed in triplicate in a 7500 Fast Real-Time PCR System (Applied Biosystems).
CRISPR/Cas9-Mediated Gene Editing to Introduce FLT3-ITD Mutation
We used CRISPR/Cas9-mediated homology-directed repair to introduce the FLT3-ITD mutation in the AML-9.9 line, as previously described (28). Briefly, a gRNA targeting the FLT3 locus was designed and cloned under the U6 promoter in a plasmid also expressing Cas9 linked to mCitrine by a P2A driven by the CMV promoter. A donor template containing a 5′ homology arm (923 bp), the ITD sequence (102 bp), and a 3′ homology arm (714 bp), consisting, respectively, of nucleotides 28,608,318–28,609,240, 28,608,216–28,608,317 and 28,607,604–28,608,317 (hg19 human genome assembly), was assembled and cloned in a plasmid. The AML-9.9 iPSC line was nucleofected with 5 μg of the gRNA/Cas9 plasmid and 10 μg of the donor plasmid. mCitrine+ cells were FACS-sorted 48 hours after transfection and plated as single cells at clonal density. After 7 to 10 days, single colonies were picked and screened by PCR with primers F: CACTCTTTTGTTGCAGGCCC and R: CGGCAACCTGGATTGAGACT. Monoallelically targeted clones were selected, and the PCR products were cloned into the PCR-4 TOPO TA vector (Invitrogen) and sequenced. Clones with one FLT3-ITD allele and one WT intact allele without indels were selected.
scRNA-seq
Single-cell RNA-sequencing was performed with the Chromium 10x Genomics 3′ protocol (v3.0) on cryopreserved primary AML patient PBMCs, iPSC-HSPCs from day 11 (AML-9.9 and AML-47.1) or day 17 (AML-4.10) of differentiation and hCD45+ cells from primary and iPSC-derived xenografts sorted using Magnetic Activated Cell Sorting (MACS, Miltenyi Biotech Inc.).
scRNA-seq Data Quality Control and Preprocessing
Sequenced fastq files were aligned, filtered, barcoded, and unique molecular identifiers (UMI) counted using Cell Ranger Chromium Single-Cell RNA-seq version 6.1.0, by 10X Genomics with Cell Ranger, GRCh38 database (version 2020-A) as the human genome reference. Each data set was filtered to retain cells with ≥1,000 UMIs, ≥400 genes expressed, and <25% of the reads mapping to the mitochondrial genome. UMI counts were then normalized so that each cell had a total of 10,000 UMIs across all genes and these normalized counts were log-transformed with a pseudocount of 1 using the “LogNormalize” function in the Seurat package. The top 2,000 most highly variable genes were identified using the “vst” selection method of “FindVariableFeatures” function and counts were scaled using the “ScaleData” function. Data sets were processed using the Seurat package (version 4.0.3; ref. 45).
scRNA-seq Data Dimensionality Reduction and Integration
Principal component analysis was performed using the top 2,000 highly variable features (“RunPCA” function), and the top 30 principal components were used in the downstream analysis. Diffusion maps were generated as implemented in the destiny (version 3.4.0) R package (46) with default parameters and using 10,000 subsampled cells from each integrated data set. Data sets for each patient were integrated separately by using the “RunHarmony” function in the harmony package (version 0.1.0). K-Nearest eighbor graphs were obtained by using the “FindNeighbors” function, whereas the UMAPs were obtained by the “RunUMAP” function (47). The Louvain algorithm was used to cluster cells based on expression similarity. The resolution was set at 0.4 for the AML-4 integrated data set, and at 0.2 for AML-9 and AML-47 data sets for optimal clustering. Cell density estimations were performed using the stat_density_2d function of the ggplot2 (version 3.3.5) package.
Additionally, AML-4 data sets were also integrated with the Seurat R package. Individual sample data sets were normalized using the SCTransform normalization method. Standard Seurat integration workflow was followed with the identification of integration features with SelectIntegrationFeatures function where nfeatures was set to 3,000. The selected integration features were used with the PrepSCTIntegration and FindIntegrationAnchors methods to identify the integration anchors with canonical correlation analysis. These anchors were used to integrate the data with IntegrateData function using the SCT normalization method.
scRNA-seq Data Cell Type Annotation
Differential markers for each cluster were identified using the Wilcox test (“FindAllMarkers” function) with adjusted P < 0.05 and absolute log2 fold change >0.25. The top upregulated genes and curated genes from the literature were used to assign cell types to the clusters. Metaclusters were obtained by merging the manually annotated cell types into groups. The HSC6, LSC17, and cell-cycle score were generated using the “AddModuleScore” function from the Seurat package (version 4.0.3).
scRNA-seq Data Pseudotime Analysis
Pseudotime was computed on the diffusion map space as described (48). Diffusion pseudotime was implemented using the “DPT” function from the destiny (version 3.4.0) R package and using the HSC/MPP cell cluster of each patient data set as the root of the trajectories.
Chord Diagram Representation
A chord diagram was generated by merging the metadata between the Harmony-integrated samples and the Seurat-integrated samples, generating an adjacency matrix between the corresponding cell annotations, and utilizing the chordDiagram function from the circlize (version 0.4.13) R package to plot the matrix.
Pseudobulk Analysis
We identified DEGs using the muscat algorithm (ref. 49; version 1.4.0) with default parameters. Briefly, we first sum-collapsed the data, summing UMIs across cells for each patient and sample, to produce a bulk RNA-seq-style UMI profile for each sample. Aggregate raw counts for each gene and each biological sample were generated using the aggregateData function of the muscat package (v.1.2.1). The resulting matrix was used as input for DGE analysis with DESeq2 (v1.28.1) R package (50).
Statistical Analysis
Statistical analysis was performed with GraphPad Prism software. Pairwise comparisons between different groups were performed using a two-sided unpaired unequal variance t test. For all analyses, P < 0.05 was considered statistically significant. Investigators were not blinded to the different groups.
Data and Code Availability
Raw scRNA-seq data have been deposited in GEO with the accession number GSE210889.
Supplementary Material
Supplemental Figure 1. Generation of a panel of iPSCs from patients with AML. Supplemental Figure 2. Reprogramming aids reconstruction of the evolutionary history and clonal composition of AML. Supplemental Figure 3. Transplantation of AML-iPSCs into immunodeficient mice. Supplemental Figure 4. Developmental block in a subset of AML-iPSC lines. Supplemental Figure 5. Transplantation of primary AML cells and patient-matched AMLiPSC lines. Supplemental Figure 6. Single-cell RNA-sequencing analyses of matched primary and iPSC-derived leukemia cells from patient AML-47. Supplemental Figure 7. Cell cycle and pseudotime analyses. Supplemental Figure 8. Comparison of scRNA-Seq data integration and clustering methods and pseudobulk differential gene expression analyses.
Table S1. Patient characteristics. AML: acute myeloid leukemia; MDS: myelodysplastic syndrome; MPN: myeloproliferative neoplasm; ET: essential thrombocythemia; PBMCs: peripheral blood mononuclear cells; BMMCs: bone marrow mononuclear cells; PDX: patient-derived xenografts Table S2. All patient samples used in this study with genetic characterization and reprogramming outcomes. Blue font denotes partially reprogrammed (as opposed to bona fide iPSC) colonies and clones. Table S3. All AML-iPSC lines phenotypically characterized. Table S4. Top 50 upregulated genes (highest log2 fold change) in each cluster. Table S5. Primers used for genotyping. Table S6. Primers used for qRT-PCR analyses.
Acknowledgments
The authors thank Peter van Galen and Christopher Sturgeon for advice and helpful suggestions. This work was supported by NIH grants R01CA225231, R01CA260711, and R01CA271331, by the New York Stem Cell Board (NYSTEM), by a Leukemia and Lymphoma Society (LLS) Scholar Award, by an Edward P Evans Foundation Discovery Research Grant, by an LLS Blood Cancer Discoveries Grant, and by a 2021 AACR–MPM Oncology Charitable Foundation Transformative Cancer Research Grant (grant number 21-20-45-PAPA) to E.P. Papapetrou. M.G. Kharas is an LLS Scholar and supported by the US NIH National Institute of Diabetes Digestive and Kidney Diseases Career Development Award and grants R01DK101989, R01CA193842, R01HL135564, and R01CA225231, the LLS Translational Research Program, the Susan and Peter Solomon Fund, and the Tri-Institutional Stem Cell Initiative. This work was supported in part by the Bioinformatics for Next-Generation Sequencing (BiNGS) shared resource facility within the Tisch Cancer Institute at the Icahn School of Medicine at Mount Sinai, which is partially supported by NIH grant P30CA196521. This work was also supported in part by the computational resources and staff expertise provided by Scientific Computing at the Icahn School of Medicine at Mount Sinai, supported by the Clinical and Translational Science Awards (CTSA) grant UL1TR004419 from the National Center for Advancing Translational Sciences. Research reported in this paper was also supported by the NIH Office of Research Infrastructure award S10OD026880.
The publication costs of this article were defrayed in part by the payment of publication fees. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Footnotes
Note: Supplementary data for this article are available at Blood Cancer Discovery Online (https://bloodcancerdiscov.aacrjournals.org/).
Authors’ Disclosures
E. Bernard reports other support from Pfizer outside the submitted work. M.P. Chao reports other support from TenSixteen Bio, personal fees and other support from Gilead Sciences, other support from TigaTx, HepaTx, IconOVir, Leukemia and Lymphoma Society, Stanford Medical School Alumni Association, and Chimera Bioengineering outside the submitted work. R. Majeti reports personal fees and other support from Kodikaz Therapeutic Solutions, personal fees from TenSixteen Bio, Roche, Cullgen, 858 Therapeutics, grants and other support from Gilead, personal fees and other support from Pheast Therapeutics, Orbital Therapeutics, and other support from MyeloGene outside the submitted work. M.G. Kharas reports personal fees from 858 Therapeutics, Inc., Kumquat Biosciences, and AstraZeneca outside the submitted work. E.P. Papapetrou reports personal fees from Janssen and Regeneron outside the submitted work. No disclosures were reported by the other authors.
Authors’ Contributions
A.G. Kotini: Data curation, formal analysis, investigation, methodology. S. Carcamo: Formal analysis. N. Cruz-Rodriguez: Data curation, formal analysis, investigation. M. Olszewska: Investigation. T. Wang: Investigation. D. Demircioglu: Investigation. C. Chang: Investigation, methodology. E. Bernard: Software. M.P. Chao: Resources. R. Majeti: Resources. H. Luo: Resources. M.G. Kharas: Resources. D. Hasson: Formal analysis, supervision. E.P. Papapetrou: Conceptualization, resources, formal analysis, supervision, funding acquisition, investigation, methodology, writing–original draft, project administration, writing–review and editing.
References
- 1. Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 2007;131:861–72. [DOI] [PubMed] [Google Scholar]
- 2. Sterneckert JL, Reinhardt P, Scholer HR. Investigating human disease using stem cell models. Nat Rev Genet 2014;15:625–39. [DOI] [PubMed] [Google Scholar]
- 3. Papapetrou EP. Patient-derived induced pluripotent stem cells in cancer research and precision oncology. Nat Med 2016;22:1392–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kim J, Hoffman JP, Alpaugh RK, Rhim AD, Reichert M, Stanger BZ, et al. An iPSC line from human pancreatic ductal adenocarcinoma undergoes early to invasive stages of pancreatic cancer progression. Cell Rep 2013;3:2088–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Munoz-Lopez A, Romero-Moya D, Prieto C, Ramos-Mejia V, Agraz-Doblas A, Varela I, et al. Development refractoriness of MLL-rearranged human B cell acute leukemias to reprogramming into pluripotency. Stem Cell Reports 2016;7:602–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lee JH, Salci KR, Reid JC, Orlando L, Tanasijevic B, Shapovalova Z, et al. Brief report: human acute myeloid leukemia reprogramming to pluripotency is a rare event and selects for patient hematopoietic cells devoid of leukemic mutations. Stem Cells 2017;35:2095–102. [DOI] [PubMed] [Google Scholar]
- 7. Papapetrou EP. Modeling leukemia with human induced pluripotent stem cells. Cold Spring Harb Perspect Med 2019;9:a034868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cancer Genome Atlas Research N, Ley TJ, Miller C, Ding L, Raphael BJ, Mungall AJ, et al. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med 2013;368:2059–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wahlster L, Daley GQ. Progress towards generation of human haematopoietic stem cells. Nat Cell Biol 2016;18:1111–7. [DOI] [PubMed] [Google Scholar]
- 10. Chao MP, Gentles AJ, Chatterjee S, Lan F, Reinisch A, Corces MR, et al. Human AML-iPSCs reacquire leukemic properties after differentiation and model clonal variation of disease. Cell Stem Cell 2017;20:329–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kotini AG, Chang CJ, Chow A, Yuan H, Ho TC, Wang T, et al. Stage-specific human induced pluripotent stem cells map the progression of myeloid transformation to transplantable leukemia. Cell Stem Cell 2017;20:315–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wesely J, Kotini AG, Izzo F, Luo H, Yuan H, Sun J, et al. Acute myeloid leukemia iPSCs reveal a role for RUNX1 in the maintenance of human leukemia stem cells. Cell Rep 2020;31:107688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Utikal J, Polo JM, Stadtfeld M, Maherali N, Kulalert W, Walsh RM, et al. Immortalization eliminates a roadblock during cellular reprogramming into iPS cells. Nature 2009;460:1145–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Marion RM, Strati K, Li H, Murga M, Blanco R, Ortega S, et al. A p53-mediated DNA damage response limits reprogramming to ensure iPS cell genomic integrity. Nature 2009;460:1149–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Li H, Collado M, Villasante A, Strati K, Ortega S, Canamero M, et al. The Ink4/Arf locus is a barrier for iPS cell reprogramming. Nature 2009;460:1136–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kawamura T, Suzuki J, Wang YV, Menendez S, Morera LB, Raya A, et al. Linking the p53 tumour suppressor pathway to somatic cell reprogramming. Nature 2009;460:1140–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hong H, Takahashi K, Ichisaka T, Aoi T, Kanagawa O, Nakagawa M, et al. Suppression of induced pluripotent stem cell generation by the p53-p21 pathway. Nature 2009;460:1132–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Banito A, Rashid ST, Acosta JC, Li S, Pereira CF, Geti I, et al. Senescence impairs successful reprogramming to pluripotent stem cells. Genes Dev 2009;23:2134–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Muller LU, Milsom MD, Harris CE, Vyas R, Brumme KM, Parmar K, et al. Overcoming reprogramming resistance of Fanconi anemia cells. Blood 2012;119:5449–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Raya A, Rodriguez-Piza I, Guenechea G, Vassena R, Navarro S, Barrero MJ, et al. Disease-corrected haematopoietic progenitors from Fanconi anaemia induced pluripotent stem cells. Nature 2009;460:53–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Papapetrou EP, Lee G, Malani N, Setty M, Riviere I, Tirunagari LM, et al. Genomic safe harbors permit high beta-globin transgene expression in thalassemia induced pluripotent stem cells. Nat Biotechnol 2011;29:73–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Papapetrou EP, Sadelain M. Derivation of genetically modified human pluripotent stem cells with integrated transgenes at unique mapped genomic sites. Nat Protoc 2011;6:1274–89. [DOI] [PubMed] [Google Scholar]
- 23. Hsu J, Reilly A, Hayes BJ, Clough CA, Konnick EQ, Torok-Storb B, et al. Reprogramming identifies functionally distinct stages of clonal evolution in myelodysplastic syndromes. Blood 2019;134:186–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Papaemmanuil E, Gerstung M, Bullinger L, Gaidzik VI, Paschka P, Roberts ND, et al. Genomic classification and prognosis in acute myeloid leukemia. N Engl J Med 2016;374:2209–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Menssen AJ, Khanna A, Miller CA, Nonavinkere Srivatsan S, Chang GS, Shao J, et al. Convergent clonal evolution of signaling gene mutations is a hallmark of myelodysplastic syndrome progression. Blood Cancer Discov 2022;3:330–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ogawa S. Genetics of MDS. Blood 2019;133:1049–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Apostolou E, Stadtfeld M. Cellular trajectories and molecular mechanisms of iPSC reprogramming. Curr Opin Genet Dev 2018;52:77–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wang T, Pine AR, Kotini AG, Yuan H, Zamparo L, Starczynowski DT, et al. Sequential CRISPR gene editing in human iPSCs charts the clonal evolution of myeloid leukemia and identifies early disease targets. Cell Stem Cell 2021;28:1074–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Klco JM, Spencer DH, Miller CA, Griffith M, Lamprecht TL, O'Laughlin M, et al. Functional heterogeneity of genetically defined subclones in acute myeloid leukemia. Cancer Cell 2014;25:379–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lapidot T, Sirard C, Vormoor J, Murdoch B, Hoang T, Caceres-Cortes J, et al. A cell initiating human acute myeloid leukaemia after transplantation into SCID mice. Nature 1994;367:645–8. [DOI] [PubMed] [Google Scholar]
- 31. Bonnet D, Dick JE. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat Med 1997;3:730–7. [DOI] [PubMed] [Google Scholar]
- 32. Calvanese V, Capellera-Garcia S, Ma F, Fares I, Liebscher S, Ng ES, et al. Mapping human haematopoietic stem cells from haemogenic endothelium to birth. Nature 2022;604:534–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ng SW, Mitchell A, Kennedy JA, Chen WC, McLeod J, Ibrahimova N, et al. A 17-gene stemness score for rapid determination of risk in acute leukaemia. Nature 2016;540:433–7. [DOI] [PubMed] [Google Scholar]
- 34. Jan M, Snyder TM, Corces-Zimmerman MR, Vyas P, Weissman IL, Quake SR, et al. Clonal evolution of preleukemic hematopoietic stem cells precedes human acute myeloid leukemia. Sci Transl Med 2012;4:149ra18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Miles LA, Bowman RL, Merlinsky TR, Csete IS, Ooi AT, Durruthy-Durruthy R, et al. Single-cell mutation analysis of clonal evolution in myeloid malignancies. Nature 2020;587:477–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Chang CJ, Kotini AG, Olszewska M, Georgomanoli M, Teruya-Feldstein J, Sperber H, et al. Dissecting the contributions of cooperating gene mutations to cancer phenotypes and drug responses with patient-derived iPSCs. Stem Cell Reports 2018;10:1610–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kotini AG, Chang CJ, Boussaad I, Delrow JJ, Dolezal EK, Nagulapally AB, et al. Functional analysis of a chromosomal deletion associated with myelodysplastic syndromes using isogenic human induced pluripotent stem cells. Nat Biotechnol 2015;33:646–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Asimomitis G, Deslauriers AG, Kotini AG, Bernard E, Esposito D, Olszewska M, et al. Patient-specific MDS-RS iPSCs define the mis-spliced transcript repertoire and chromatin landscape of SF3B1-mutant HSPCs. Blood Adv 2022;6:2992–3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Yi G, Wierenga ATJ, Petraglia F, Narang P, Janssen-Megens EM, Mandoli A, et al. Chromatin-based classification of genetically heterogeneous AMLs into two distinct subtypes with diverse stemness phenotypes. Cell Rep 2019;26:1059–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zeng AGX, Bansal S, Jin L, Mitchell A, Chen WC, Abbas HA, et al. A cellular hierarchy framework for understanding heterogeneity and predicting drug response in acute myeloid leukemia. Nat Med 2022;28:1212–23. [DOI] [PubMed] [Google Scholar]
- 41. Zeng Z, Shi YX, Samudio IJ, Wang RY, Ling X, Frolova O, et al. Targeting the leukemia microenvironment by CXCR4 inhibition overcomes resistance to kinase inhibitors and chemotherapy in AML. Blood. 2009;113:6215–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Bottomly D, Long N, Schultz AR, Kurtz SE, Tognon CE, Johnson K, et al. Integrative analysis of drug response and clinical outcome in acute myeloid leukemia. Cancer Cell 2022;40:850–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Pei S, Pollyea DA, Gustafson A, Stevens BM, Minhajuddin M, Fu R, et al. Monocytic subclones confer resistance to venetoclax-based therapy in patients with acute myeloid leukemia. Cancer Discov 2020;10:536–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Krivtsov AV, Twomey D, Feng Z, Stubbs MC, Wang Y, Faber J, et al. Transformation from committed progenitor to leukaemia stem cell initiated by MLL-AF9. Nature 2006;442:818–22. [DOI] [PubMed] [Google Scholar]
- 45. Butler A, Hoffman P, Smibert P, Papalexi E, Satija R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol 2018;36:411–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Angerer P, Haghverdi L, Buttner M, Theis FJ, Marr C, Buettner F. destiny: diffusion maps for large-scale single-cell data in R. Bioinformatics 2016;32:1241–3. [DOI] [PubMed] [Google Scholar]
- 47. Korsunsky I, Millard N, Fan J, Slowikowski K, Zhang F, Wei K, et al. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat Methods 2019;16:1289–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Haghverdi L, Buttner M, Wolf FA, Buettner F, Theis FJ. Diffusion pseudotime robustly reconstructs lineage branching. Nat Methods 2016;13:845–8. [DOI] [PubMed] [Google Scholar]
- 49. Crowell HL, Soneson C, Germain PL, Calini D, Collin L, Raposo C, et al. muscat detects subpopulation-specific state transitions from multi-sample multi-condition single-cell transcriptomics data. Nat Commun 2020;11:6077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014;15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1. Generation of a panel of iPSCs from patients with AML. Supplemental Figure 2. Reprogramming aids reconstruction of the evolutionary history and clonal composition of AML. Supplemental Figure 3. Transplantation of AML-iPSCs into immunodeficient mice. Supplemental Figure 4. Developmental block in a subset of AML-iPSC lines. Supplemental Figure 5. Transplantation of primary AML cells and patient-matched AMLiPSC lines. Supplemental Figure 6. Single-cell RNA-sequencing analyses of matched primary and iPSC-derived leukemia cells from patient AML-47. Supplemental Figure 7. Cell cycle and pseudotime analyses. Supplemental Figure 8. Comparison of scRNA-Seq data integration and clustering methods and pseudobulk differential gene expression analyses.
Table S1. Patient characteristics. AML: acute myeloid leukemia; MDS: myelodysplastic syndrome; MPN: myeloproliferative neoplasm; ET: essential thrombocythemia; PBMCs: peripheral blood mononuclear cells; BMMCs: bone marrow mononuclear cells; PDX: patient-derived xenografts Table S2. All patient samples used in this study with genetic characterization and reprogramming outcomes. Blue font denotes partially reprogrammed (as opposed to bona fide iPSC) colonies and clones. Table S3. All AML-iPSC lines phenotypically characterized. Table S4. Top 50 upregulated genes (highest log2 fold change) in each cluster. Table S5. Primers used for genotyping. Table S6. Primers used for qRT-PCR analyses.
Data Availability Statement
Raw scRNA-seq data have been deposited in GEO with the accession number GSE210889.