Hypusination of the eIF5A translation factor is necessary for the development and maintenance of MYC-driven lymphoma, where hypusinated eIF5A controls the translation of factors that are essential for cell-cycle transit and DNA replication.
Abstract
The MYC oncoprotein is activated in a broad spectrum of human malignancies and transcriptionally reprograms the genome to drive cancer cell growth. Given this, it is unclear if targeting a single effector of MYC will have therapeutic benefit. MYC activates the polyamine–hypusine circuit, which posttranslationally modifies the eukaryotic translation factor eIF5A. The roles of this circuit in cancer are unclear. Here we report essential intrinsic roles for hypusinated eIF5A in the development and maintenance of MYC-driven lymphoma, where the loss of eIF5A hypusination abolishes malignant transformation of MYC-overexpressing B cells. Mechanistically, integrating RNA sequencing, ribosome sequencing, and proteomic analyses revealed that efficient translation of select targets is dependent upon eIF5A hypusination, including regulators of G1–S phase cell-cycle progression and DNA replication. This circuit thus controls MYC's proliferative response, and it is also activated across multiple malignancies. These findings suggest the hypusine circuit as a therapeutic target for several human tumor types.
Significance:
Elevated EIF5A and the polyamine–hypusine circuit are manifest in many malignancies, including MYC-driven tumors, and eIF5A hypusination is necessary for MYC proliferative signaling. Notably, this circuit controls an oncogenic translational program essential for the development and maintenance of MYC-driven lymphoma, supporting this axis as a target for cancer prevention and treatment.
See related commentary by Wilson and Klein, p. 248.
This article is highlighted in the In This Issue feature, p. 247
INTRODUCTION
MYC family oncoproteins are activated in a broad spectrum of human malignancies (1). However, MYC overexpression alone is not sufficient for the neoplastic transformation of normal human cells and additional cooperating events are necessary (e.g., loss of the tumor suppressor p53 or overexpression of the oncoprotein BCL-2). During cancer progression, MYC-overexpressing cells evolve by various means to bypass cell-cycle and apoptotic checkpoints and, as evidenced by the number and combination of mutations found in aggressive MYC-driven cancers (2), these tumors are particularly challenging to treat. Thus, finding a common MYC downstream effector that controls select factors/pathways (e.g., cell-cycle regulation, ribosome biogenesis, and metabolism) at a posttranscriptional level and that is required for MYC-induced tumorigenesis would be of great interest and should have broad therapeutic benefit.
One MYC-regulated metabolic pathway that appears nearly universally upregulated in cancer is polyamine biosynthesis, which directs the production of putrescine, spermidine, and spermine. Notably, polyamine levels are generally elevated in tumors (3, 4), and targeting the first enzyme of the pathway, ornithine decarboxylase (ODC), impairs the development of MYC-driven lymphoma and MYCN-driven neuroblastoma (5–7). Furthermore, the ODC suicide inhibitor α-difluoromethylornithine (DFMO) has shown activity as a chemoprevention agent in colorectal and prostate cancer trials (8, 9). However, DFMO has little activity as a single agent in cancer therapeutic trials (10), and this likely reflects the fact that polyamines are abundant in the diet and their intracellular levels can be restored by polyamine transport (11).
Interestingly, the 4-aminobutyl moiety of the polyamine spermidine is used as the sole substrate to posttranslationally modify a single lysine residue (Lys-50) of eIF5A (eIF5A1) and eIF5A2 in a process coined hypusination. This two-step conversion is directed by deoxyhypusine synthase (DHPS) and deoxyhypusine hydroxylase (DOHH), which covalently modify eIF5ALys50 with a unique amino acid, hypusine (12). eIF5A was initially identified as a translation initiation factor (13) and has been shown to play additional roles in translation elongation and termination in yeast (14, 15) as well as in start codon selection (16). Furthermore, hypusination of yeast eIF5A augments the translation of mRNAs containing consecutive (>3) polyproline (poly-P) codons (17). Finally, functional roles for hypusinated eIF5A (eIF5AHyp) have been linked to several physiologic processes, including preventing premature aging of the brain (18) and immune senescence (19), as well as in macrophage activation (20), and meta-inflammation (21).
The roles of the polyamine–hypusine circuit in cancer are currently controversial (22), where eIF5A has been suggested to play oncogenic roles in some contexts (23) and to function as a tumor suppressor in others (24, 25). To understand tumor cell–intrinsic effects of the hypusine circuit on tumorigenesis, we targeted the pathway specifically in the B cells of Eμ-Myc transgenic mice, a validated model for human B-cell lymphoma with MYC involvement (26). Here we report that hypusinated eIF5A plays critical roles in the maintenance of MYC-driven lymphoma and that DHPS has essential roles in malignant conversion that is necessary for lymphoma development. Further, unbiased global analyses of changes in gene expression, translation, and steady-state protein levels reveal that hypusination of eIF5A controls cell cycle and oncogenic programs that are necessary for lymphoma cell growth.
RESULTS
Activation of the Polyamine–Hypusine Circuit Is a Hallmark of Cancer
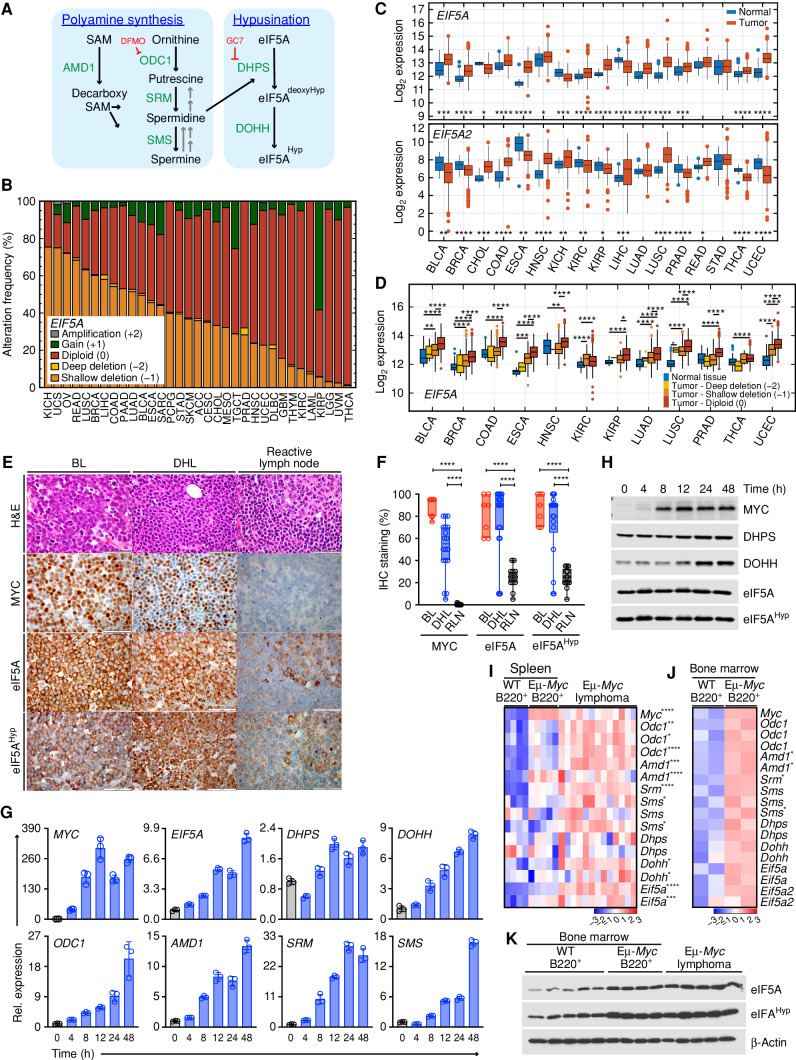
To assess the status of the hypusine circuit (Fig. 1A) in cancer in an unbiased fashion, genetic alterations and expression profiles of the genes involved in this process—EIF5A, EIF5A2, DHPS, and DOHH—were determined using publicly available data from The Cancer Genome Atlas (TCGA). Deep deletion (−2) or amplification (+2) is rare in EIF5A, DHPS, and DOHH (Supplementary Fig. S1A), although loss of heterozygosity (LOH) in the region of chromosome 17p that harbors the EIF5A and/or TP53 genes is frequent in many tumors (refs. 27–29; Fig. 1B, shallow deletion −1: orange). However, expression analyses across all cancer types revealed that EIF5A is significantly overexpressed in many cancers (12/17) compared with corresponding normal tissues, along with DOHH (16/17), and that there are modest increases in DHPS (8/17) expression in many tumor types (Fig. 1C; Supplementary Fig. S1B). Furthermore, analyses of EIF5A expression in the 12 EIF5A-overexpressing tumor types with respect to copy-number status revealed that EIF5A expression in tumors with shallow deletion is still higher than or equivalent to EIF5A expression in normal tissues (Fig. 1D). In contrast, due to a common amplification of chromosome 3q (30), EIF5A2 (at 3q26.2) is significantly amplified in some cancers including lung squamous (36%) and ovarian cancer (23%) (Supplementary Fig. S1A), and increased EIF5A2 expression is limited to cancers with 3q26 amplicons (Fig. 1C).
Figure 1.
Activation of the polyamine–hypusine circuit is a hallmark of MYC-driven cancers. A, Schematic of the polyamine–hypusine pathway. AMD1, adenosylmethionine decarboxylase-1; dcSAM, decarboxy SAM; DHPS, deoxyhypusine hydroxylase; DOHH, deoxyhypusine hydroxylase; DFMO, difluoromethylornithine; eIF5A, eukaryotic translation initiation factor 5A; GC7, N1-guanyl-1, 7-diamine-heptane; Hyp, hypusine; ODC, ornithine decarboxylase; SAM, S-adenosylmethionine; SMS, spermine synthase; SRM, spermidine synthase. B, Copy-number alterations (CNA) of EIF5A across different cancer types. The abbreviations for the tumor types are provided in Supplementary Fig. S1A. C,EIF5A and EIF5A2 levels in RNA sequencing datasets in 17 cancer types derived from TCGA PanCanAtlas. D,EIF5A transcript levels relative to normal tissue, classified based on EIF5A copy number (TCGA data sets). E, Representative hematoxylin and eosin (H&E) staining and IHC of MYC, eIF5A, and hypusinated eIF5A (eIF5AHyp) proteins in patients with either BL (n = 7) or DHL (n = 18), or reactive lymph node samples (n = 13; Moffitt Cancer Center; scale bar, 50 μm). F, Box plots showing IHC staining (%) of the indicated proteins for each patient (open circle). G, Expression of genes in the polyamine–hypusine circuit following induction of MYC in p493-6 human B-lymphoma cells, as analyzed by qRT-PCR (n = 3, fold change to expression at time 0). H, Immunoblot analyses of MYC, DHPS, DOHH, eIF5A, and eIF5AHyp levels in p493-6 human B-lymphoma cells following MYC induction. I and J, Gene-expression profiling of arrays showing the polyamine-hypusine genes in mouse splenic B220+ B cells from premalignant Eμ-Myc (n = 5) and wild-type (WT; n = 4) mice and Eμ-Myc lymphomas (n = 13) from GSE32239 (I), and bone marrow (BM) of premalignant Eμ-Myc (n = 2) and WT (n = 2) littermate mice from GSE33474 (J). Rows represent different probe sets. K, Representative immunoblots of eIF5A and eIF5AHyp in mouse B220+ B cells isolated from BM of 6-week-old WT mice (n = 5) and premalignant Eμ-Myc mice (n = 4), and in lymphomas isolated from Eμ-Myc mice (n = 5). Levels of significance determined by Mann–Whitney test (B), Dunnett multiple comparison (F) or unpaired t tests (J) are indicated as follows: *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
Previously, we and others have shown that the polyamine biosynthetic genes ODC1, AMD1, and SRM are direct transcription targets of MYC (Fig. 1A; ref. 5), and their expression correlates well with MYC expression across cancer types (Supplementary Fig. S1C). Notably, EIF5A and DHPS expression is highly elevated in MYC-overexpressing Burkitt lymphoma (BL; Supplementary Fig. S1D). Indeed, when compared with normal human B-cell subtypes, EIF5A is the second most significantly upregulated gene in BL (Supplementary Fig. S1E). Further, IHC analyses of BL and double-hit lymphoma (DHL) patients who bear MYC translocations (Moffitt Cancer Center) revealed elevated expression of MYC, eIF5A, and hypusinated eIF5A (eIF5AHyp) relative to levels expressed in normal lymph nodes (Fig. 1E and F; Supplementary Table S1).
The control of the hypusine circuit by MYC was evaluated using the p493-6 human B lymphoma cell line that harbors a tetracycline-repressible MYC transgene (31). Given EIF5A is overexpressed across many cancer types (Fig. 1C), it is evident that eIF5A is not regulated solely by MYC. In accord with this, eIF5A protein is expressed at basal levels even in the MYC-off state; however, MYC induction leads to significant increases in the levels of EIF5A, DHPS, and DOHH transcripts, and to increases in the expression of genes encoding the polyamine biosynthetic enzymes, ODC1, AMD1, SRM, and SMS (Fig. 1G). Finally, induction of MYC also leads to increases in the levels of DHPS and DOHH proteins (Fig. 1H).
Marked upregulation of the polyamine–hypusine circuit is also manifest in B220+ B cells isolated from the bone marrow (BM) and spleens of Eμ-Myc transgenic mice as well as in the lymphomas that arise in these mice (Fig. 1I and J). Furthermore, the hypusinated form of eIF5A (eIF5AHyp) is substantially elevated in BM premalignant and neoplastic Eμ-Myc B cells versus BM B cells from wild-type (WT) littermates (Fig. 1K). Thus, activation of the hypusine circuit is a hallmark of cancer and is especially elevated in MYC-driven lymphoma.
Hypusinated eIF5A Is Essential for Proliferation of MYC-Driven B-cell Lymphoma
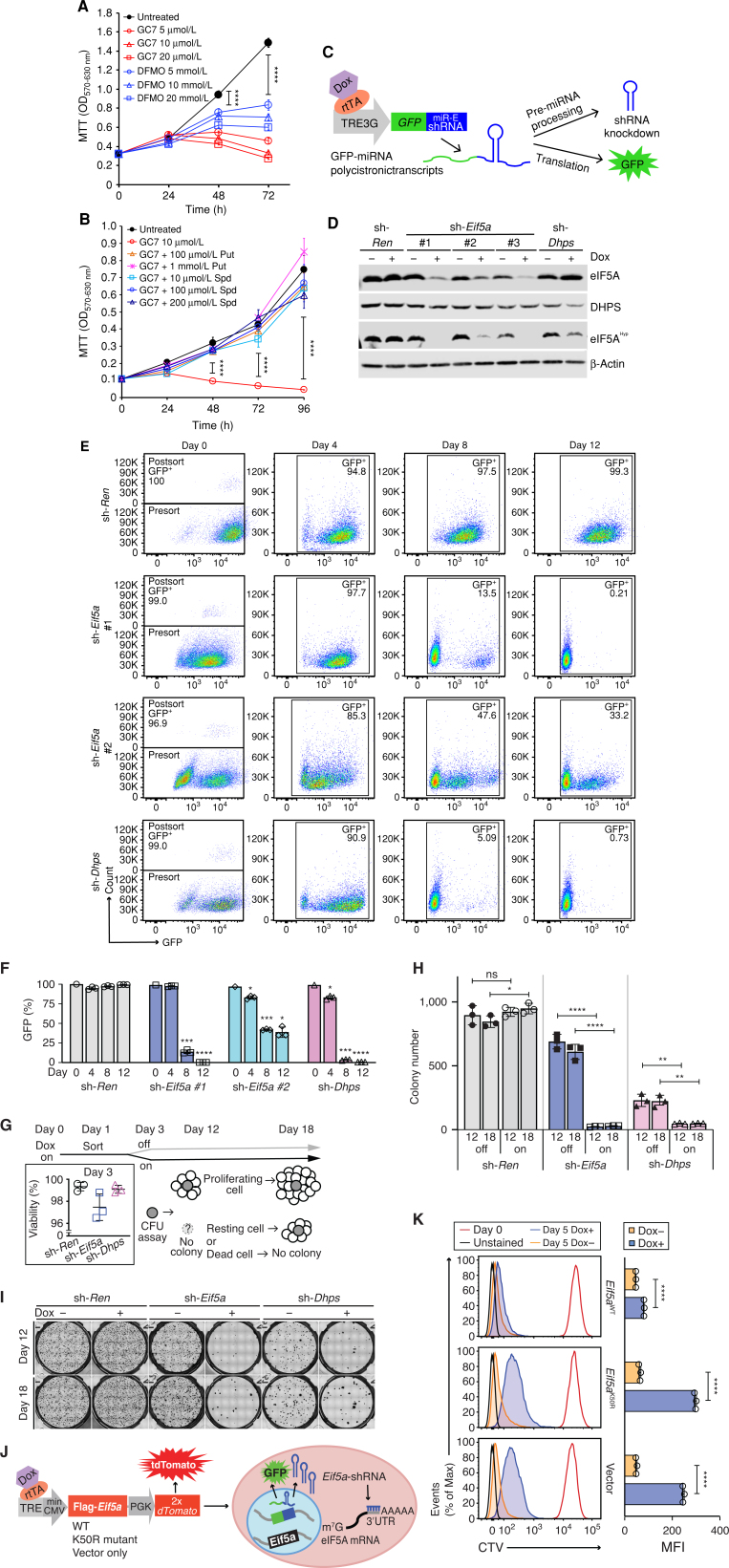
The spermidine analogue GC7 (N1-guanyl-1,7-diamineoheptane) competitively inhibits DHPS enzymatic activity (32) and, as expected, GC7 treatment of Eμ-Myc B lymphoma cells reduced levels of hypusinated eIF5A (Supplementary Fig. S2A). Moreover, in agreement with other studies (22), low concentrations of GC7 (5–20 μmol/L) effectively inhibited the growth of human Raji BL and mouse Eμ-Myc B lymphoma cells (Fig. 2A; Supplementary Fig. S2B), and this response was rescued by the addition of the polyamines putrescine (Put) or spermidine (Spd; Fig. 2B; Supplementary Fig. S2B). However, as previously noted (33), this spermidine analogue may affect other polyamine-engaged pathways independent of the hypusine axis.
Figure 2.
Depletion of eIF5A or DHPS suppresses the growth of human and mouse MYC-driven lymphoma. A and B, MTT assays showing growth inhibition of human Raji BL cells following treatment with GC7 or DFMO (A) and rescue of GC7-induced growth inhibition by addition of putrescine (Put) or spermidine (Spd; n = 4; B). C, Schematic illustrating the miRE shRNA system, which generates GFP-shRNA polycistronic transcripts. D, Immunoblots showing levels of indicated proteins in Eμ-Myc;Rosa26-rtTA cells inducibly expressing sh-Eif5a #1, sh-Eif5a #2, sh-Eif5a #3, sh-Dhps, or sh-Ren. E, Representative FACS profile showing retention of cells expressing indicated GFP-shRNAs following cell sort. F, Quantification of GFP+ cell populations at the indicated intervals based on results in E (n = 3). G–I, Methycellulose colony formation unit (CFU) assays evaluating long-term effects of depletion of eIF5A or DHPS on clonogenic potential. G, Schematic illustrating the experimental design; inset shows the viability of cells at day 3 as evaluated by trypan blue dye exclusion. H and I, Representative photos taken at days 12 and 18 showing colony size and number (H) and quantification of colony numbers (I; n = 3). J, Schematic of experimental design where sh-Eif5a-expressing cells also inducibly express either eIF5AWT or eIF5AK50R or the vector alone. K, Rescue of cell proliferation by induction of eIF5AWT in eIF5A-depleted cells, as determined by flow cytometry of CellTrace Violet (CTV) stained cells, and quantification of mean fluorescence intensity (MFI). Left, representative histograms; right, the quantified data of day 5 Dox+ (GFP+TdTomato+, blue) vs. Dox− cells (orange; n = 3). Levels of significance determined by unpaired t tests are indicated as follows: ns, not significant; *, P < 0.05; **, P < 0.01; ***, P< 0.001; ****, P < 0.0001. Error bar indicates mean ± SD. Max, maximum.
To directly examine the effects of loss of function of the hypusine axis, mouse lymphoma lines were derived from Eμ-Myc;Rosa26-rtTA2 tumors that express the reverse tetracycline transactivator rtTA2, and that were engineered to inducibly express short hairpin RNAs (shRNA) targeting Eif5a, using a doxycycline (Dox)-regulated system. Dox treatment provoked efficient Eif5a knockdown (Supplementary Fig. S2C) and impaired lymphoma cell growth (Supplementary Fig. S2D and S2E).
To precisely track the effects of silencing Eif5a or Dhps, we also generated Eμ-Myc;Rosa26-rtTA2 lymphoma cell lines that inducibly express GFP-shRNA polycistronic transcripts (Fig. 2C; ref. 34) and that effectively depleted eIF5A and DHPS proteins following Dox treatment, as well as levels of eIF5AHyp (Fig. 2D). These cells were then followed for GFP expression to monitor the retention (expression) of the shRNAs by flow cytometry. There was a remarkable selection against the growth of Eif5a or Dhps shRNA-expressing cells, but not against cells expressing a control shRNA targeting Renilla luciferase (sh-Ren), which maintained nearly 100% GFP-expressing cells (Fig. 2E and F); thus, Eif5a and Dhps expression is necessary for lymphoma cell growth. Finally, this response correlated with the efficiency of depletion of eIF5A protein, where lymphoma cells expressing sh-Eif5a#1 that was more efficient at knockdown had a more profound phenotype than lymphoma cells expressing the less efficient sh-Eif5a#2 (Fig. 2D and E).
Although the half-life of eIF5A protein is very long (>24 hours; ref. 35), tracking cell division with a tracing dye revealed that inhibition of cell proliferation in Eif5A- or Dhps-shRNA expressing cells was first evident 3 days after Dox treatment (Supplementary Fig. S2F). To assess the fate of Eif5A- or Dhps-shRNA expressing lymphoma cells, these Dox-induced knockdown cell lines (day 0) were sorted for GFP+ at day 1 and cultured in Dox-containing media (Fig. 2G). By day 3, there were no apparent effects on cell viability (Fig. 2G, inset), and an equal number of these cells cultured in Dox media were then plated in methocult media in either the presence or absence of Dox, and colony formation was monitored over time (days 12 and 18). When Dox was removed on day 3, a substantial number of DHPS- or eIF5A-depleted cells were still capable of forming colonies, indicating recovery from knockdown. In contrast, most eIF5A- or DHPS-depleted cells failed to form colonies when kept on Dox over time (day 12), and no newly formed colonies were detected by day 18 of culture (Fig. 2H and I; Supplementary Fig. S2G).
To test if eIF5A hypusination is specifically required for lymphoma cell proliferation, cells expressing Eif5a-shRNA that targets the 3′ untranslated region of Eif5a mRNA were engineered to also inducibly express comparable levels of FLAG-tagged WT eIF5A or mutant (K50R) eIF5A that cannot be hypusinated (or the vector control, Fig. 2J; Supplementary Fig. S2H). Importantly, forced expression of WT eIF5A, but not the eIF5A-K50R mutant, restored the proliferation of lymphoma cells depleted of endogenous eIF5A (Fig. 2K); thus, eIF5aHyp is necessary for lymphoma cell proliferation.
eIF5A Hypusination Contributes to Tumorigenic Potential of MYC-Driven Lymphoma
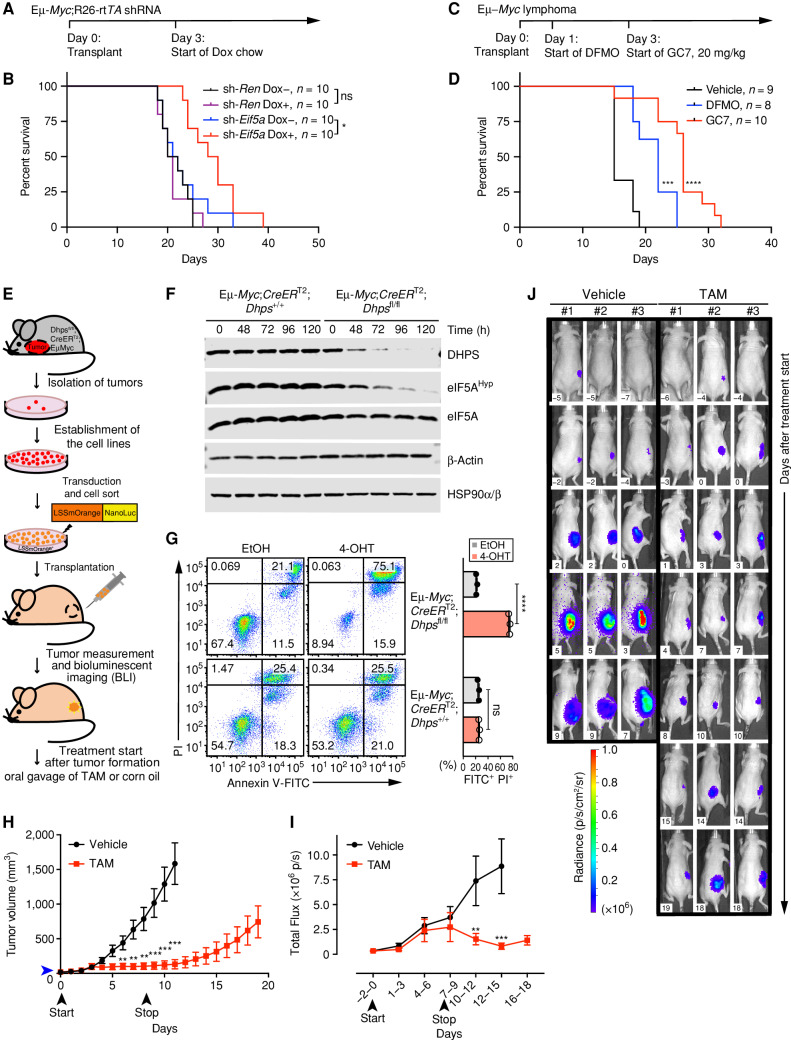
To examine the roles of the hypusine circuit on the tumorigenic potential of MYC-driven lymphoma, we tested the effects of eIF5A depletion in recipient mice bearing Eμ-Myc;Rosa26-rtTA2 lymphomas expressing Eif5a shRNA or the control Renilla shRNA. On day 3 following transplant, recipient mice were switched to Dox chow or kept on normal chow and were monitored daily (Fig. 3A). These lymphoma cells harbor E255K mutant p53, a DNA binding domain IV mutant whose equivalent site in human TP53, E258 (Supplementary Fig. S3A), is a recurrent hotspot in human cancer as revealed by cBioPortal analysis (36). Regardless, selective silencing of Eif5a, but not of Renilla, impaired lymphomagenic potential (Fig. 3B). Further, analyses of tumors at the endpoint in mice given the Dox-diet revealed that these lymphomas no longer expressed sh-Eif5a and reexpressed eIF5A, indicating a selection for tumor cells that escape silencing by the Eif5a shRNA (Supplementary Fig. S3B).
Figure 3.
Hypusinated eIF5A is necessary for the growth and maintenance of Myc-driven lymphoma. A, Experimental design of lymphoma transplant studies; B, Kaplan–Meier survival curves of mice transplanted i.v. with Eμ-Myc;Rosa26-rtTA2 lymphoma cells that inducibly express sh-Eif5a or sh-Ren. Median survival of cohorts: sh-Ren Dox− 21 days; sh-Ren Dox+ 20.5 days; sh-Eif5a Dox− 21.5 days; and sh-Eif5a Dox+ 29 days. C and D, Experimental design of Eμ-Myc lymphoma transplant studies testing the efficacy of DFMO (1% wt/vol, in drinking water) and GC7 (20 mg/kg, daily i.p.; C) versus vehicle and Kaplan–Meier survival curves of treated mice (D; vehicle, median survival 15 days; DFMO, median survival 22 days; GC7, median survival 26 days). DFMO- and GC7-treated mice showed no obvious toxicities. E, Experimental design of subcutaneous (sub-Q) transplant studies of lymphoma cells established from tumors of Eμ-Myc;CreERT2;Dhpsfl/fl or Eμ-Myc;CreERT2;Dhps+/+ mice. F, Immunoblots showing levels of indicated proteins in Eμ-Myc;Cre-ERT2;Dhpsfl/fl or Eμ-Myc;Cre-ERT2;Dhps+/+ lymphoma cells following treatment with 4-OHT. G, Left, representative FACS profiles showing apoptosis upon loss of Dhps as analyzed by Annexin V–FITC/PI labeling; right, quantification (n = 3). H, Lymphoma growth in transplanted mice following treatment with vehicle (corn oil) or TAM (tamoxifen). Blue arrowhead indicates the average tumor volume at the start of the TAM treatment: 21 mm3 (Vehicle group: 18 mm3). Black arrowheads indicate dates when treatment was started (day 0) and terminated (day 9; n = 13). I, Total flux analyzed by BLI of the same mice bearing Eμ-Myc;Cre-ERT2;Dhpsfl/fl lymphomas (n = 13). J, Representative BLI of mice that were treated with TAM or vehicle. Days after treatment start are indicated (white inset). Levels of significance determined by Mantel–Cox log-rank test (B) and (D), or unpaired t tests (G) are indicated as follows: *, P < 0.05; **, P < 0.01; ***, P< 0.001; ****, P < 0.0001. Error bar, mean ± SD (G) or mean ± SEM (H) and (I).
These findings were confirmed in pharmacologic studies, where GC7 treatment of Eμ-Myc lymphoma reduced eIF5AHyp levels (Supplementary Fig. S2A) and where GC7 treatment (4 or 20 mg/kg, daily i.p.) of mice transplanted with Eμ-Myc lymphoma significantly extended overall survival versus vehicle-treated mice, which rapidly succumbed to disease (Fig. 3C and D; Supplementary Fig. S3C and S3D). Indeed, GC7 treatment was more effective at impairing tumorigenic potential than the clinically used suicide inhibitor of ornithine decarboxylase, DFMO (given in drinking water 1% wt/v; Fig. 3D; Supplementary Fig. S3C and S3D). Thus, eIF5AHyp contributes to the tumorigenic potential of MYC-driven lymphoma.
Dhps Is Necessary for the Maintenance of MYC-Driven Lymphoma
Given these findings, we reasoned that eIF5A hypusination would be necessary for the maintenance of MYC-driven lymphoma. To establish a system where complete loss of hypusination is achievable, we used the Dhpsfl/fl conditional knockout mouse model (37), which we crossed to Eμ-Myc and Rosa26-CreERT2 (38) transgenic mice to generate Eμ-Myc;Rosa26-CreERT2;Dhpsfl/fl and Eμ-Myc;Rosa26-CreERT2;Dhps+/+ mice. Lymphomas arising in these mice were then harvested (Fig. 3E) and lymphoma cell lines were derived in which the floxed Dhps alleles can be deleted by ex vivo treatment with 4-hydroxytamoxifen (4-OHT) that activates Cre recombinase. Efficient deletion of the Dhps gene and loss of DHPS protein were confirmed by genomic PCR and immunoblot analyses (Supplementary Fig. S3E and S3F). Loss of DHPS protein led to marked reductions in levels of eIF5AHyp without affecting the total levels of eIF5A (Fig. 3F). Further, consistent with reduced clonogenic survival potential observed following shRNA silencing of Dhps or Eif5a (Fig. 2I; Supplementary Fig. S2G), Cre-directed loss of Dhps, and as a consequence loss of eIF5AHyp, ultimately led to cell death (Fig. 3G).
To assess the role of Dhps in tumor maintenance, Eμ-Myc;Rosa26-CreERT2;Dhpsfl/fl and Eμ-Myc;Rosa26-CreERT2;Dhps+/+ lymphoma cells engineered to express the bioluminescence reporter OgNLuc+ were injected subcutaneously into Nude mice and tumor growth was monitored daily by measurement of tumor volumes and twice a week by bioluminescence imaging (BLI; Fig. 3E and H–J). Once tumors were palpable, mice were randomized to vehicle or tamoxifen cohorts and were treated for 9 consecutive days. Importantly, Dhps loss significantly impaired tumor progression (Fig. 3H–J), whereas lymphoma growth continued unabated in vehicle-treated mice (Supplementary Fig. S3G). Remarkably, 5 out of 13 tamoxifen-treated mice bearing Eμ-Myc;Rosa26-CreERT2;Dhpsfl/fl lymphomas displayed nearly complete tumor regression. Nonetheless, once tamoxifen treatment was terminated (at day 9), following a lag period all tumors started to regrow (Fig. 3H), and these mice reexpressed DHPS (Supplementary Fig. S3H). Thus, there is a strong selection against loss of the hypusine circuit in the maintenance of MYC-driven lymphoma.
Depletion of eIF5A or DHPS Disrupts Select MYC-Dependent Transcription Programs
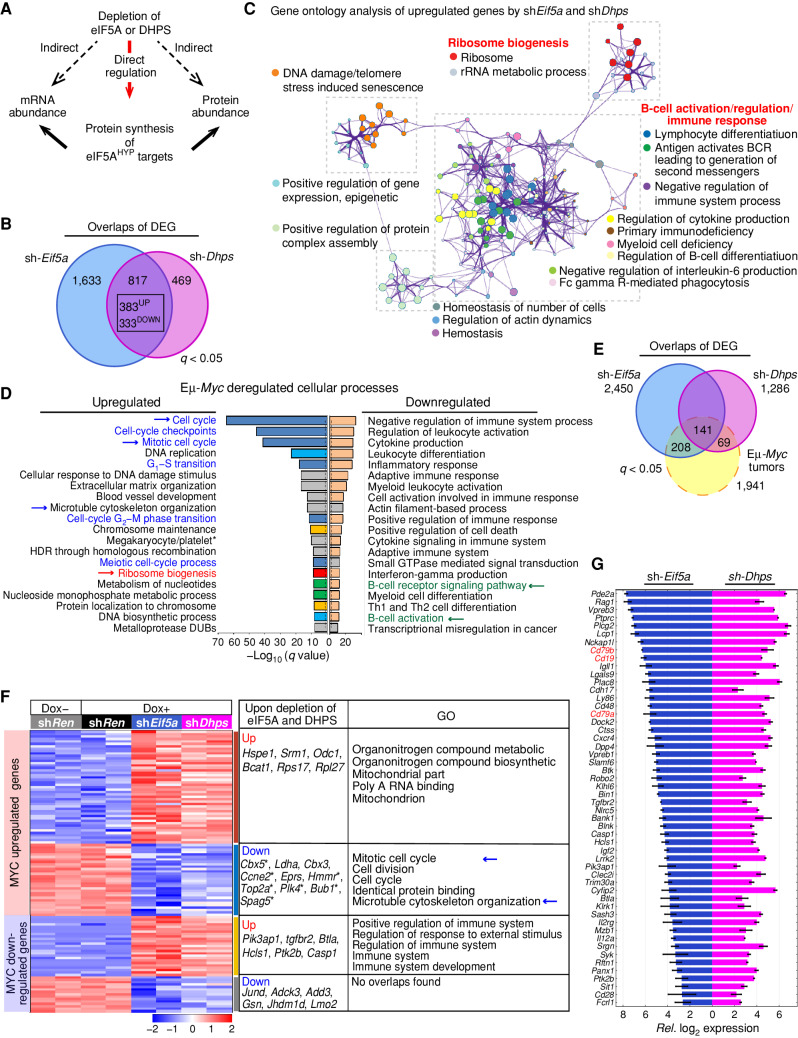
Hypusinated eIF5A likely affects diverse cellular pathways, for example, through direct regulation of its translational targets or via secondary effects on other biological processes (Fig. 4A). Therefore, we examined the global effects of knockdown of Eif5a and Dhps on the transcriptome, proteome, and translational landscape of MYC-induced lymphoma. For these studies, we used Eμ-Myc;Rosa26-rtTA2 lymphoma cells depleted of eIF5A or DHPS for 72 hours, an interval where the cells were still viable (Fig. 2G, inset). RNA sequencing (RNA-seq) revealed 2,450 differentially expressed genes (DEG) in Eif5a-depleted lymphoma cells, and 1,286 DEGs in Dhps-knockdown cells relative to control cells (sh-Ren). No DEGs were detected in control sh-Ren cells treated with and without Dox; thus, there are minimal effects of Dox on gene expression in this model (Supplementary Fig. S4A). Further, 383 significantly upregulated and 333 significantly downregulated genes were shared between Eif5a- and Dhps-depleted cells (Fig. 4B). Among these shared genes, Metascape gene set enrichment analysis (39) showed that ribosome biogenesis, B-cell regulation/activation, and the immune response are the major enriched functional categories represented by upregulated genes (Fig. 4C; Supplementary Fig. S4B), whereas enriched functional terms for shared downregulated genes include diverse central cellular processes, including cell division (Supplementary Fig. S4C). Further, genes in the polyamine–hypusine circuit, including the direct MYC transcription targets Odc1 and Srm1, were also upregulated (Supplementary Fig. S4D); thus, the initial cellular response to inhibiting the hypusine circuit includes a compensatory upregulation of this pathway.
Figure 4.
The hypusine circuit controls select transcriptional networks manifest in MYC-driven lymphoma. A, Schematic of possible direct or indirect effects provoked by silencing Eif5A or Dhps. B, Venn diagram of differentially expressed genes (DEG) in Eμ-Myc lymphoma cells following depletion of eIF5A (blue) or DHPS (pink; q < 0.05). C, Network enrichment analysis by Metascape (39) of the shared 383 genes significantly upregulated following depletion of eIF5A or DHPS in Eμ-Myc lymphoma cells. Each cluster is represented by different colors, and a circle node represents each enriched functional term. D, Metascape enrichment analysis of significantly up- and downregulated genes in B220+ splenic Eμ-Myc lymphoma cells versus WT B220+ splenic B cells (40). Cell-cycle regulation, ribosome biogenesis, and B-cell signaling/activation pathways are indicated in blue, red, and green, respectively, and the dotted line shows q = 0.05. E, Venn diagram showing overlap of DEGs present in Eμ-Myc lymphoma (dotted yellow line) with DEGs of eIF5A- or DHPS-depleted Eμ-Myc lymphoma cells (blue or pink, respectively, defined by comparing their expression to sh-Ren controls). 141 MYC-deregulated genes were differentially expressed in both eIF5A- and DHPS-depleted Eμ-Myc cells. F, Heat maps showing changes in mRNA levels manifest in MYC-dysregulated genes in Eμ-Myc lymphoma cells that are significantly and commonly altered by eIF5A or DHPS depletion (left), representative genes (middle), and top 5 pathways defined by ontology analysis (right). G, Relative log2 expression of genes of B-cell regulation/activation pathways (e.g., CD19, CD79a, CD79b, shown in red) whose expression is induced following depletion of eIF5A or DHPS. All genes are DEG (q < 0.05).
Next, we assessed how the depletion of eIF5A or Dhps affects MYC-controlled transcription programs. Previous studies established 1,941 MYC-dysregulated genes (q < 0.05) in Eμ-Myc lymphomas (40), and that upregulated genes represent pathways involved in the control of the cell cycle, ribosome biogenesis, and metabolism (Fig. 4D). In contrast, MYC-downregulated genes represent pathways controlling the immune response, and B-cell regulation/activation (ref. 40; Fig. 4D). Integrating our RNA-seq data with MYC-dysregulated genes (Fig. 4E) revealed that eIF5A or DHPS depletion has two major effects on MYC-regulated pathways. First, immune regulatory genes that are significantly downregulated in Eμ-Myc lymphoma are actually increased or reexpressed following silencing of Eif5a or Dhps (Fig. 4F; Supplementary Table S2). Interestingly, such immune genes encode cell-surface proteins such as CD19 that are targets for antibody and chimeric antigen receptor (CAR) T therapy, and that are occasionally silenced in treatment-naïve lymphoma and are often silenced in patients who fail CD19-targeted therapies (41, 42). Specifically, the Eμ-Myc;Rosa26-rtTA2 lymphoma line used in our studies is unusual in that it does not express several B-cell-specific receptors including CD19 and, quite strikingly, CD19 expression, as well as that of several other B cell–specific receptors (e.g., CD79a and CD79b), are induced following depletion of eIF5A or DHPS (Fig. 4G).
Second, expression of 20% and 10% of the 1,236 MYC-upregulated genes manifest in Eμ-Myc lymphoma was significantly altered following eIF5A or DHPS depletion (sh-Eif5a, 229; sh-Dhps, 132; 81 common genes, q < 0.05), respectively (Fig. 4F). Among these altered genes, the downregulated genes are enriched for cell-cycle control genes, including several E2F transcription targets (e.g., Ccne2 and Bub1, Fig. 4F; Supplementary Table S2). Thus, the initial proliferation inhibitory phenotype provoked by eIF5A or DHPS depletion (Fig. 2K; Supplementary Fig. S2F) is associated with the suppression of transcription targets that are necessary for cell-cycle progression.
The Hypusine-Dependent Translational Landscape of MYC-Driven Lymphoma
To identify eIF5AHyp-dependent translation targets that might contribute to lymphoma growth and survival, we determined the global effects of silencing Eif5a or Dhps on translation by performing ribosome profiling (43) in parallel with RNA-seq. Further, to assess potential effects on steady-state protein levels, tandem mass tag (TMT)-multilabeling mass spectrometry (44) was performed following Eif5a or Dhps knockdown. Translation efficiency (TE) was then determined as the ratio of the levels of ribosome-bound mRNAs (those being actively translated) to mRNA levels for each gene (45). Reproducibility for each transcript analyzed (RNA-seq, TE) was validated in biological duplicates of sh-Ren-, sh-Eif5a-, and sh-Dhps-expressing lymphoma cells (Supplementary Fig. S5A). Further, the 10-plex TMT proteomics time-course analyses identified over 6,400 proteins for the sh-Ren, sh-Eif5a, and sh-Dhps cohorts (Supplementary Fig. S5B).
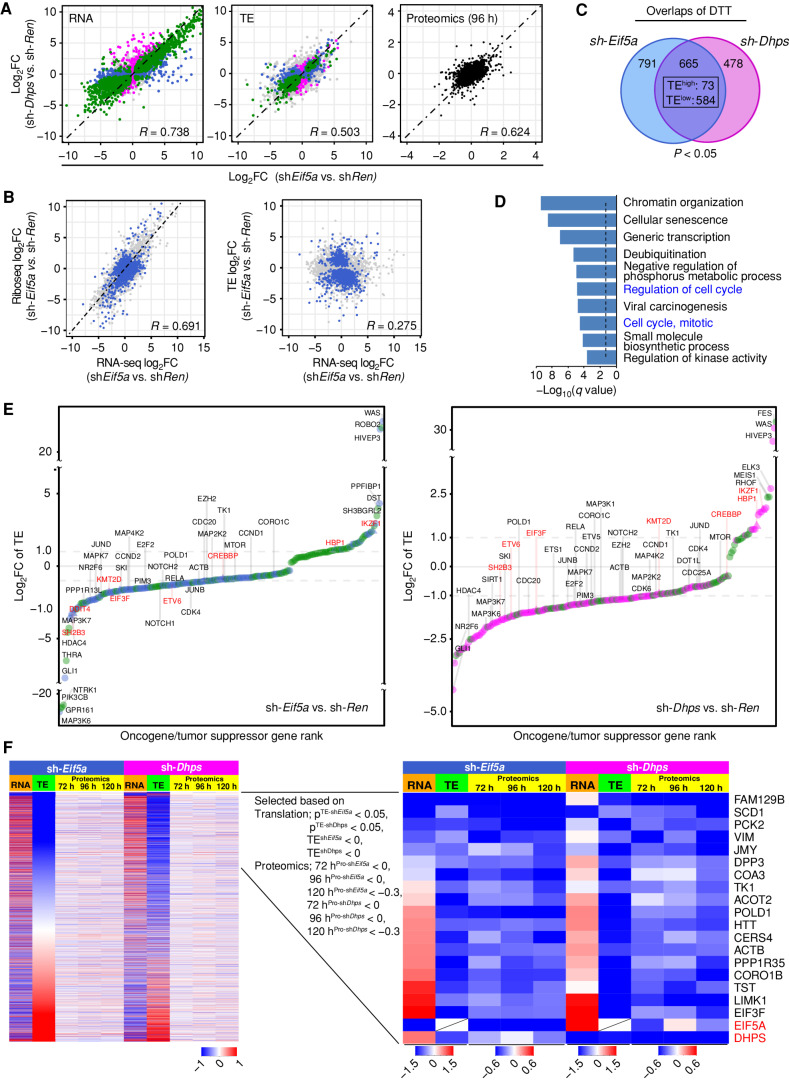
As expected given their functions in the hypusine axis, the directionality of log2-fold changes resulting from eIF5A depletion versus control shRen cells was highly concordant with those changes caused by depletion of DHPS in all three parameters: RNA-seq, TE, and steady-state protein levels (Fig. 5A). Furthermore, although there was an expected correlation of RNA-seq and ribosome sequencing (Ribo-seq), there were no overall correlations in TE and RNA-seq (Fig. 5B; Supplementary Fig. S5C). Thus, the changes in mRNA levels and their TE that are observed following depletion of eIF5A or DHPS are largely independent of each other, underscoring the key roles of hypusination in translational control.
Figure 5.
The eIF5AHyp-dependent translational landscape of Eμ-Myc lymphoma. A, Correlation plots showing concordant effects of Eif5A and Dhps knockdown on gene expression (left, R = 0.738), translation efficiency (middle, TE, R = 0.503), and steady-state protein levels (right, R = 0.624) in sh-Eif5a- and sh-Dhps- vs. sh-Ren-expressing Eμ-Myc lymphoma cells. Correlations are displayed as scatterplots where green, blue, and pink colors indicate significance for both sh-Eif5a- and sh-Dhps (green), for only sh-Eif5a (blue), or for only sh-Dhps (pink); gray, not significant. Black denotes proteomic samples at the 96 hours time point. q < 0.05 was used to determine the significance for RNA-seq and P < 0.05 was used for TE data sets. B, Scatter plots showing correlation among log2 fold changes in RNA-seq, Ribo-seq, and TE analyses following eIF5A depletion. Blue indicates differentially translated transcripts (DTT, P < 0.05); gray indicates nonsignificant transcripts. C, Venn diagram of DTTs in sh-Eif5a- (blue) or sh-Dhps- (pink) knockdown Eμ-Myc lymphoma cells (P < 0.05; 657 concordantly shared transcripts, TElow: 584 and TEhigh: 73). D, Metascape enrichment analysis of the 584 TElow genes showing the top 10 categories. The dotted line shows q = 0.05. E, A ranked list of log2-fold change of TE for 244 and 204 differentially translated oncogenes (circles) and tumor suppressors (triangles) in eIF5A- (left) or DHPS- (right) depleted cells. Dot colors indicate significance for both sh-Eif5a and sh-Dhps (green), for only sh-Eif5a (blue), or for only sh-Dhps (pink). Names of select oncogenes are in black; tumor suppressors are in red. F, Identification of shared TElow transcripts having reduced levels of their corresponding proteins following Eif5a or Dhps knockdown in Eμ-Myc lymphoma. Left, heat maps of all transcripts sorted based on low to high TE-log2 fold changes (green columns 2 and 7). Orange column (columns 1 and 6) indicates RNA-seq log2 fold changes of these transcripts (72 hours). Yellow columns indicate the time course of TMT proteomic analyses (72, 96, and 120 hours). Right, among the shared significant TElow transcripts (P < 0.05), a time-course analysis of concordantly reduced encoded proteins in both sh-Eif5a and sh-Dhps cells.
To assess the spectrum of lymphoma mRNAs whose translation is dependent on eIF5A or DHPS, global analyses of TE were performed. These analyses identified 665 shared differentially translated transcripts (DTT, P < 0.05) in sh-Eif5a- or sh-Dhps-expressing lymphoma cells (Supplementary Table S3). Importantly, 88% of these DDT had reduced TE (TElow: 584 mRNAs), indicating eIF5AHyp is indeed necessary for their efficient translation (Fig. 5C). Supporting previous studies showing that eIF5A promotes translation of mRNAs containing sequential polyproline (poly-P) codons (>3; ref. 28), analyses of the number of poly-P motifs within a given mRNA revealed that depletion of eIF5A or DHPS generally reduced the TE of mRNAs having multiple poly-P motifs (Supplementary Fig. S5D). Interestingly, however, the majority of mRNAs having TElow following depletion of eIF5A or DHPS lacked consecutive poly-P codons, indicating that the role of eIF5A in promoting efficient translation is not limited to transcripts containing poly-P codons. Finally, gene enrichment analysis of these hypusine-dependent translation targets revealed that TElow genes has roles in key cellular processes, including regulation of the cell cycle (Fig. 5D).
eIF5AHyp Controls Oncogenic Translation and Transcription Programs in Lymphoma
Notably, DTTs included a substantial number of genes that have established roles in cancer, including tumor suppressors and oncogenes. Specifically, from 2,579 potential oncogenic drivers and tumor suppressors, we identified 244 (TElow: 173) and 204 (TElow: 176) transcripts whose translation was significantly altered upon depletion of eIF5A or DHPS, respectively (129 shared mRNAs with 113 TElow, 15 TEhigh, and 1 discordant transcript; Supplementary Table S4). Ranking these DTTs based on fold changes in TE revealed that over 80% of mRNAs encoding potential oncogenic drivers had reduced translation efficiency following the knockdown of eIF5A or DHPS (Fig. 5E). Importantly, these TElow transcripts include genes known to play critical roles in cell growth across different lymphoma subtypes (e.g., E2f2, Pim3).
Because steady-state levels of proteins are a function of both their synthesis and turnover rates, not all TElow genes identified following the knockdown of Eif5a or Dhps showed an immediate reduction in their corresponding protein levels. We thus reasoned that TElow genes that contribute to phenotypes provoked by depletion of eIF5A or DHPS encode proteins whose steady-state levels are consistently reduced upon knockdown of Eif5a or Dhps. To assess this, we performed a time-course TMT proteomic analysis. These studies revealed that several TElow genes have consistent reductions in the levels of their corresponding proteins, including Huntingtin (HTT; Fig. 5F) that is widely known as the protein mutated in a neurologic disorder, Huntington's disease, but is also ubiquitously expressed in many cell types, including B cells (Supplementary Fig. S5E) and WT HTT possesses antiapoptotic function (46). In addition, there were reductions in POLD1 (Fig. 5F), which is the catalytic subunit of DNA polymerase-δ that directs the synthesis of the lagging strand during DNA replication and that, together with DNA polymerase-ε, directs DNA synthesis of over 90% of the genome (47). Interestingly, POLD1 is overexpressed in many cancer types, unlike POLD-2, -3, and -4 genes that encode accessory subunits (Supplementary Fig. S5F). Furthermore, there were reductions in the steady-state levels of the metabolic enzyme Stearoyl-CoA desaturase (SCD1; Fig. 5F), which generates unsaturated lipids required for the maintenance of endoplasmic reticulum (ER) membrane homeostasis and for preventing MYC-mediated ER stress. Notably, SCD1 contributes to the growth and survival of MYC-driven lymphoma (48).
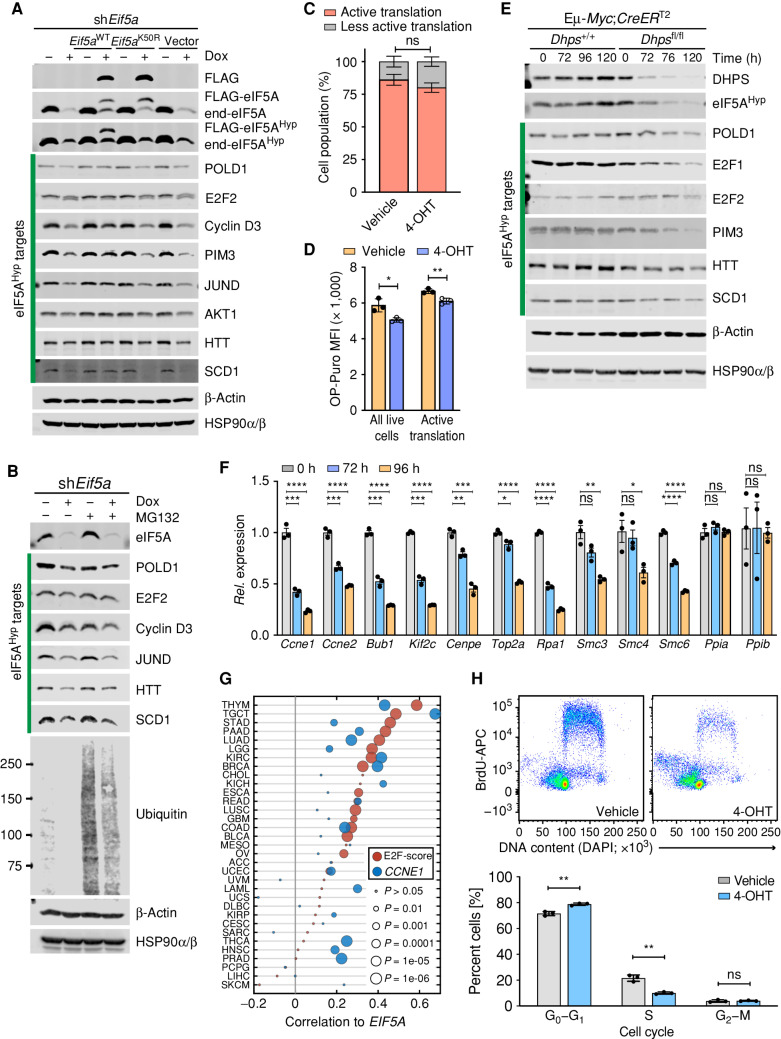
To validate these hypusine targets and to test if changes in their protein levels are associated with loss of hypusination, we evaluated their levels in eIF5A knockdown lymphoma cells that also inducibly express exogenous WT eIF5A or the hypusine-defective eIF5AK50R mutant. In accord with the profound defects in cell proliferation observed following depletion of eIF5AHyp (Fig. 2K; Supplementary Fig. S2F), immunoblot analyses confirmed that eIF5A depletion provoked reductions in protein levels of several hypusine targets known to contribute to cell-cycle progression, including POLD1, E2F2, PIM3, JUND, and Cyclin D3 (Fig. 6A and B, c.f., lanes 1 vs. lanes 2). Moreover, inducible expression of WT eIF5A but not eIF5AK50R at least partially restored the levels of these eIF5AHyp-dependent targets (Fig. 6A).
Figure 6.
eIF5AHyp controls translation of regulators of cell-cycle progression and DNA replication in lymphoma. A, Immunoblot analyses of the indicated proteins in Eμ-Myc;Rosa26-rtTA lymphoma cells inducibly expressing only sh-Eif5a or sh-Eif5a plus either Eif5aWT or Eif5aK50R, or the vector alone, following Dox treatment for 84 hours. Although β-Actin was identified as a TElow gene following eIF5A or DHPS knockdown, the levels of β-Actin are unaffected by immunoblot analysis. HSP90α/β served as a second control. B, Immunoblot analyses of the indicated eIF5AHyp target proteins in Eμ-Myc;Rosa26-rtTA2 cells expressing sh-Eif5a with or without treatment with the proteasome inhibitor MG132 (10 μmol/L for 4 hours). C, Nascent polypeptide synthesis evaluated by OP-puro incorporation showing cell population (%) undergoing active translation in Eμ-Myc;CreERT2;Dhpsfl/fl cells treated with 4-OHT (72 hours; n = 3). D, MFI showing OP-puro incorporation in actively translating Eμ-Myc;CreERT2;Dhpsfl/fl cells (n = 3). E, Time-course immunoblot analyses of the indicated proteins in Eμ-Myc;CreERT2;Dhpsfl/fl lymphoma before and following deletion of Dhps. F, Relative mRNA expression of E2F target genes at the indicated time points following Dhps deletion in Eμ-Myc;CreERT2;Dhpsfl/fl cells, as analyzed by qRT-PCR (n = 3). Housekeeping genes Ppia and Ppib served as controls. G, Correlation between EIF5A and HALLMARK E2F target gene sets (MsigDB, red circles) and CCNE1 expression (blue circles) across TCGA data sets. E2F score is the PC1 from a principal component analysis. H, BrdU incorporation assays showing effects of Dhps loss on DNA synthesis in Eμ-Myc;Cre-ERT2;Dhpsfl/fl lymphoma treated with 4-OHT (72 hours; top) and quantification of cells at different cell-cycle phases (bottom; n = 3). Levels of significance determined by unpaired t tests in C, D, F, and H are indicated as follows: ns, not significant; *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. Error bar indicates mean ± SD.
Loss of eIF5A could also conceivably affect proteasome degradation of putative eIF5AHyp targets. However, treatment with the proteasome inhibitor MG132 did not affect reduction of these targets observed following the knockdown of Eif5a (Fig. 6B), indicating that the effects of Eif5a knockdown are primarily at the level of translation of these targets.
To confirm these findings in an independent model, we used Eμ-Myc;Rosa26-CreERT2;Dhpsfl/fl lymphoma cells. First, we evaluated the effects of Dhps loss on protein synthesis by assessing O-Propargyl-puromycin (OP-puro) incorporation into nascent polypeptides in vehicle versus 4-OHT–treated cells. Consistent with selective effects on translation (rather than global effects), loss of Dhps was associated with modest reductions in OP-Puro incorporation versus control cells (Fig. 6C and D; Supplementary Fig. S6A). Furthermore, similar to the effects of eIF5A knockdown (Fig. 6A and B), immunoblot analyses confirmed that acute Dhps deletion provoked reductions in protein levels of most of the same targets, including E2F2, POLD1, PIM3, Cyclin D3, SCD1, and HTT (Fig. 6E; Supplementary Fig. S6B).
In accord with the antiproliferative effects of GC7 on BL and Eμ-Myc lymphoma cells (Fig. 2A; Supplementary Fig. S2B), pharmacologic inhibition of hypusination by GC7 also led to reductions in the steady-state levels of both E2F1 and E2F2, as well as to increased cleaved (c)-poly(ADP-ribose) polymerase (cPARP), and these effects were abolished by cotreatment with spermidine (Supplementary Fig. S6C and S6D).
These findings suggested the loss of eIF5AHyp should compromise the expression of E2F transcription targets that drive transit through the cell cycle. Indeed, analysis of our RNA-seq data revealed differential expression of 188 mouse homologs of the HALLMARK E2F gene set (as defined by MsigDB) upon depletion of eIF5A or DHPS (Supplementary Fig. S6E). This includes reductions in the expression of bona fide E2F targets required for cell-cycle progression and DNA replication (e.g., Ccne2, Bub1b, and Top2a; Supplementary Fig. S6F), whereas there were no consistent changes in the expression of E2f genes themselves (E2f1; log2FCsh-Eif5a = 0.31, q = 0.9, log2FCsh-Dhps = −0.71, q = 0.84, E2f2; log2FCsh-Eif5a = −1.62, q = 0.52, log2FCsh-Dhps = −0.35, q = 0.94). Further, qRT-PCR analyses established time-dependent reductions in these E2F transcription targets following knockout of Dhps in Eμ-Myc;Cre-ERT2;Dhpsfl/fl lymphoma cells (Fig. 6F). Interestingly, there are positive correlations between the expression of EIF5A and HALLMARK E2F targets across many human cancer types (Fig. 6G). Thus, in addition to previously reported MYC-E2F interplays (49), our study suggests that eIF5AHyp, at least in part, contributes to downstream E2F transcription programs by regulating the levels of E2F proteins across a broad spectrum of tumors.
Finally, these findings also suggested that eIF5AHyp regulates lymphoma cell-cycle progression through the G1–S transition and DNA replication, which we tested by 5-bromodeoxyuridine (BrdU) incorporation into DNA in Eμ-Myc;Rosa26-CreERT2;Dhpsfl/fl lymphoma cells treated with 4-OHT versus vehicle. Notably, these studies revealed that Dhps loss provoked marked reductions in S-phase and an accumulation of cells in G1 (Fig. 6H).
Dhps Is Essential for MYC-Driven Lymphomagenesis
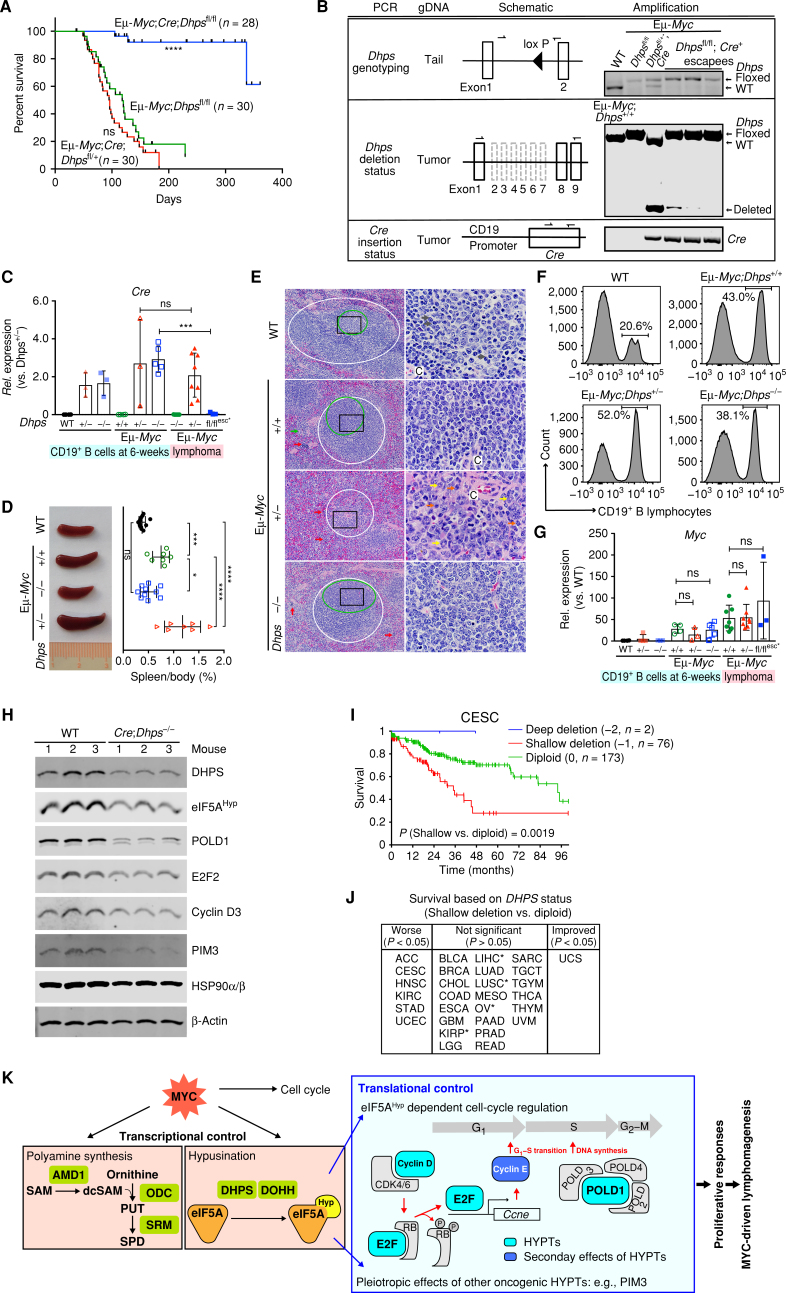
Eμ-Myc mice develop B-cell lymphoma with nearly 100% penetrance (26). The critical role of eIF5A hypusination in lymphoma maintenance indicated it might also be essential for lymphoma development. To test this hypothesis, we generated an Eμ-Myc;CD19-Cre;Dhpsfl/fl mouse, in which Dhps is selectively deleted in B lymphocytes via expression of Cre recombinase under the control of the B cell–specific CD19 promoter (Supplementary Fig. S7A), along with littermates lacking only one allele of Dhps (Eμ-Myc;CD19-Cre;Dhpsfl/+) or carrying the Dhps gene. Strikingly, long-term evaluation of Eμ-Myc;CD19-Cre;Dhpsfl/fl mice revealed that they did not develop lymphoma (Fig. 7A). Indeed, the three mice in this cohort that eventually succumbed to disease evaded Cre-mediated deletion of Dhps (Fig. 7B) by silencing Cre expression; i.e., these lymphomas retained the Cre recombinase transgene but failed to express the gene (Cre escapees; Fig. 7C). Consequently, Dhps is expressed in the Cre escapees (Supplementary Fig. S7B). Thus, there is strong selective pressure for retaining Dhps during lymphoma development.
Figure 7.
Dhps is essential for the development of MYC-driven lymphoma. A, Kaplan–Meier curves showing survival of Eμ-Myc;CD19-Cre;Dhpsfl/fl, Eμ-Myc;CD19-Cre;Dhpsfl/+, and Eμ-Myc;Dhpsfl/fl mice. B,Dhps deletion and Cre knock-in status, as determined by genomic PCR of the indicated tissues or tumor samples. C,Cre expression in CD19+ B cells from 6-week-old WT, CD19-Cre;Dhpsfl/+ (hereafter Dhps+/−), and CD19-Cre;Dhpsfl/fl (Dhps−/−); and from preneoplastic and malignant Eμ-Myc;CD19-Cre;Dhps cohorts (Eμ-Myc;Dhps+/+, Eμ-Myc;Dhps+/−, and Eμ-Myc;Dhps−/−) and the 3 Cre escapees (fl/flesc*), as determined by qRT-PCR (n ≥ 3 mice). Fold change was calculated relative to Cre levels in Dhps± B cells. D, Left, representative images; right, the weight of spleens (normalized to total body weight) from indicated 6-week-old mice (n > 7 for each cohort). E, Hematoxylin and eosin (H&E) staining of spleens from indicated 7-week-old mice. Periarteriolar lymphoid sheaths (PALS) and germinal centers are shown in white and green circles, respectively. Red, green, yellow, gray, and blue arrows indicate erythroid precursors, megakaryocytes, maturing granulocytic precursors, small lymphocytes, and large lymphocytes, respectively. Central arteriole is indicated as C. Black box in the left panels shows the area magnified in the right panels; scale bars, left and right, 50 μm and 10 μm, respectively. F, Representative CD19+ B-cell profiles of BM-derived lymphocytes of indicated 6-week-old mice. G,Myc expression in CD19+ B lymphocytes from indicated 6-week-old littermates, and from preneoplastic and malignant Eμ-Myc;CD19-Cre;Dhps cohorts (+/+, +/−, and −/−; n ≥ 3). H, Immunoblot analyses of identified eIF5AHyp targets in CD19+ B lymphocytes from CD19-Cre;Dhpsfl/fl and WT littermates (n = 3). I, Kaplan–Meier curves showing survival of cervical squamous cell carcinoma (CESC) based on DHPS copy-number status. J, Overall survival based on DHPS copy number (shallow deletion vs. diploid). TCGA cancer types in the middle panel that are associated with trends toward worse survival are marked as * (P < 0.1). K, Model for roles of eIF5AHyp in MYC-driven lymphoma. The requirement for eIF5AHyp for Eμ-Myc B-cell growth and malignant conversion is associated with MYC-induced hyperactivation of the polyamine–hypusine circuit and eIF5AHyp-dependent translation of targets such as E2F and POLD1 (turquoise boxes). Cyclin E (blue box) is the transcription target of E2F. HYPTs, eIF5AHyp targets; PUT, putrescine; SPD, spermidine; SAM, S-adenosylmethionine; dcSAM, decarboxy SAM. Levels of significance determined by Mantel–Cox log-rank test (A) and (I), unpaired t tests (C), or Dunnett multiple comparison (D) and (G) are indicated as follows: ns, not significant; *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. Error bar indicates mean ± SD.
In contrast, there were no statistically significant effects of Dhps heterozygosity on overall survival (median survival 95.5 days) versus WT Eμ-Myc littermates (median survival 118 days; Fig. 7A). Indeed, Eμ-Myc;CD19-Cre;Dhpsfl/+ mice tended to have a shorter latency period prior to disease onset, as judged by assessments of splenomegaly, a hallmark of disease in this model (Fig. 7D; Supplementary Fig. S7C). Furthermore, the splenic architecture in this cohort was severely disrupted (e.g., diminished periarteriolar lymphoid sheath), with many large lymphocytes infiltrating the red pulp and lymphoid follicles (Fig. 7E). Thus, Dhps heterozygosity appears to accelerate disease onset.
The failure of Eμ-Myc;CD19-Cre;Dhpsfl/fl mice to develop lymphoma was not due to effects of Dhps loss on B-cell homeostasis, as the proportion of CD19+ B cells in BM isolated from Eμ-Myc;CD19-Cre;Dhpsfl/fl mice actually exceeded those present in aged-matched (6-week) littermate WT mice (Fig. 7F). Dhps loss also had no effect on the elevated expression of the Myc mRNA or protein in preneoplastic Eμ-Myc B cells (Fig. 7G; Supplementary Fig. S7D).
Interestingly, morphologic analyses of BM, where disease arises in Eμ-Myc mice, revealed that Eμ-Myc;CD19-Cre;Dhpsfl/fl mice were similar to their premalignant Eμ-Myc littermates, where there were increased numbers of large lymphocytes (Supplementary Fig. S7E), likely due to proliferative activity induced by MYC. However, we observed that the percentage of CD19+ B cells in BM lymphocytes isolated from Eμ-Myc;CD19-Cre;Dhpsfl/fl mice was consistently lower than that of their premalignant Eμ-Myc littermates, whereas the BM of Dhps heterozygous Eμ-Myc;CD19-Cre;Dhpsfl/+ mice was even more dominated by B cells (Fig. 7F). Moreover, phenotypically, Eμ-Myc;CD19-Cre;Dhpsfl/fl mice failed to develop splenomegaly (Fig. 7D) and the spleens of these mice retained their normal architecture (Fig. 7E), indicating Dhps loss abolishes progression to the malignant state.
Finally, to confirm that key eIF5AHyp targets identified in Eμ-Myc lymphomas (Fig. 6) are dependent upon eIF5AHyp in B cells in vivo, we analyzed their expression in CD19+ B cells from 6-week-old CD19-Cre;Dhpsfl/fl mice versus WT mice. Here as well reductions in DHPS protein are associated with selective reductions in eIF5AHyp and its targets Cyclin D3, E2F2, POLD1, and PIM3 (Fig. 7H).
DHPS Status Is Associated with Outcomes of Some Cancer Types
The requirement for Dhps in MYC-driven lymphomagenesis suggested that DHPS deletion status might connote outcomes in human cancer. To test this hypothesis, survival analysis of subclassified patient groups based on DHPS copy-number alterations was performed across all available TCGA tumor types. As predicted for a gene that is essential for cancer, the number of patients with tumors having deep (−2) deletion of DHPS is exceedingly rare; indeed, only 16 cases exist in the entire TCGA. Moreover, although patient numbers are too small to assess statistical significance, in those rare cases where deep deletion of DHPS does occur there is improved patient survival (Fig. 7I; Supplementary Fig. S7F). Finally, similar to the accelerated disease phenotypes manifest in Dhps heterozygous Eμ-Myc mice (Fig. 7A, D and E), shallow deletion (−1) of DHPS was associated with worse patient outcomes in several tumor types, including cervical, head and neck and stomach cancers, and exhibited trends toward worse survival in several tumor types (Fig. 7I and J; Supplementary Table S5).
DISCUSSION
Here, we report the essential roles of the hypusine circuit in the development and maintenance of MYC-driven lymphoma. Specifically, pharmacologic inhibition, knockdown or knockout of DHPS, or knockdown of eIF5A, compromises the growth and survival of extant lymphoma both ex vivo and in vivo. Furthermore, Dhps is necessary for conversion to the fully malignant state in vivo, where the loss of Dhps abolishes the development of MYC-driven B-cell lymphoma. Finally, unbiased multiomics approaches revealed that hypusinated eIF5A is a translational regulator of key oncoproteins that drive the cell cycle and MYC's proliferative responses.
Notably, although Eif5a, Dhps, and Dohh are all essential for embryonic development (50–52), and as shown here Dhps is essential for MYC-driven lymphomagenesis, Dhps loss in Eμ-Myc;CD19-Cre;Dhpsfl/fl mice does not impair the percentage of BM B cells versus those found in WT mice. Collectively, the data support a model whereby increased levels of eIF5A and its DHPS/DOHH-modifying enzymes observed in lymphoma are necessary to sustain the highly anabolic state of a developing or extant tumor, and for the transition to the malignant state, as shown by analyses of the precancerous state of Eμ-Myc;CD19-Cre;Dhpsfl/fl mice.
These findings seemingly contrast with those of others who, using an Eμ-Myc hematopoietic stem/progenitor cell transplant model and shRNA screens, have suggested tumor suppressor roles of the polyamine–hypusine circuit in MYC-driven lymphomagenesis, by triggering apoptosis (24). There are two likely explanations for these discrepant findings. First, this could reflect developmental differences in the requirement for this circuit during tumorigenesis, where Dhps in our study was deleted specifically in B cells, whereas the aforementioned shRNA knockdown studies suppressed the expression of the circuit throughout the entire hematopoietic system. Second, given that Dhps heterozygosity appears to provoke a more aggressive course of disease in Eμ-Myc mice, it is possible that partial knockdown of this circuit using shRNA leads to one phenotype—accelerated disease—whereas inactivation of the pathway abolishes the conversion to, and maintenance of, the malignant state. Consistent with the required roles of Dhps in the conversion to the malignant state, Cre escapees of Dhps deletion via silencing Cre expression occur during lymphoma development in Eμ-Myc;CD19-Cre;Dhpsfl/fl mice. Thus, there is a clear intrinsic selection against tumor cells lacking the hypusine circuit.
Other evidence also argues for the necessary roles of hypusinated eIF5A in tumorigenesis. eIF5A is one of the most abundant proteins in eukaryotic cells (53), and our analyses establish that, regardless of MYC involvement, EIF5A levels are higher in most cancer types versus matched normal tissue, consistent with its role as an oncogene. Further, although there is LOH in the region of chromosome 17p that harbors EIF5A (and notably TP53), our analyses indicate that EIF5A expression from such tumors is generally equivalent to or higher than EIF5A expression in matched normal tissues.
One surprising finding is that shallow deletion of DHPS connotes worse outcomes in some human tumor types, and that lymphoma progression (e.g., splenomegaly onset and mean overall survival) appears more aggressive in Eμ-Myc;Dhps+/− mice versus Eμ-Myc mice. One possibility is that this could reflect compensatory upregulation that has been documented in Dhps+/− mice, where there are increased Ievels of Dhps mRNA in tissues of Dhps+/− mice versus their WT littermates (52). In support of this notion, we note that there are increased levels of Dhps transcripts in some lymphomas that arise in Eμ-Myc;Dhps+/− mice versus levels of Dhps mRNA expressed in lymphomas from Eμ-Myc;Dhps+/+ mice (Supplementary Fig. S7B).
Recent findings indicate that hypusinated eIF5A augments translation elongation, rather than functioning as a core translation factor. Furthermore, in accord with recent findings in S. cerevisiae (15), our analysis of TE suggests the requirement for hypusinated eIF5A is not limited to the translation of mRNAs containing polyproline codons. Important issues that remain to be resolved are: what features of mRNA (i.e., newly synthesized polypeptide structure) confer a requirement for eIF5AHyp for their efficient translation; and why eIF5AHyp-dependent targets are cell context specific, as distinct targets for eIF5AHyp have been reported in other models (19, 37).
Increasing evidence indicates deregulation of translation control is a hallmark of cancer (54). However, the repertoire of transcripts that are differentially translated in distinct cancer types is largely unknown, and such data could inform potential drug targets and combination strategies to treat cancer. As noted above, eIF5AHyp targets appear to vary among different cell types. For example, the translation of the autophagy transcription factor TFEB has been reported to be regulated by eIF5AHyp in normal B cells (19). However, TFEB expression is suppressed in MYC-overexpressing cancers (55, 56) and TFEB was not found as an eIF5AHyp target in our study.
Notably, at least in the context of MYC-driven lymphoma, eIF5AHyp directly or indirectly controls the levels of several key regulators of cell-cycle progression. First, targeting the hypusine circuit compromises the translation of E2F transcription factors that control expression of key cell-cycle genes, including Ccne (Cyclin E) that is essential for the G1–S phase transition of the cell cycle. Second, this circuit also appears to control DNA replication, as translation of POLD1, which appears to have roles in the synthesis of both the lagging and leading strands (47), is also impaired following Eif5a or Dhps knockdown, and by loss of Dhps. Collectively, these findings support a model whereby translational regulation by hypusinated eIF5A regulates MYC-driven proliferative responses at several levels (Fig. 7K).
Enforced MYC overexpression is sufficient to initiate a preneoplastic state, but disease progression requires the acquisition of additional alterations that contribute to neoplastic conversion (57–60). The myriad of mutations that are manifest in aggressive hematologic malignancies, including in MYC-driven lymphoma (2), make these tumors particularly challenging to treat, and they are highly prone to developing resistance. Underscoring this fact, although several downstream targets of MYC have been shown to contribute to lymphoma development in the Eμ-Myc model (5, 61–66), to date none of them are absolutely required for disease development as is the case for DHPS. Our findings suggest this profound phenotype is due to the pleiotropic effects of blocking eIF5A hypusination, which is required for the efficient translation of cooperating oncogenes and critical regulators of the cell cycle (Fig. 7K). As activation of the hypusine circuit is a hallmark of MYC-driven tumors, the development of improved small-molecule inhibitors of DHPS, or of agents that bind to and block eIF5AHyp function, are attractive therapeutic strategies for translating these findings into the clinic. Indeed, the recent discovery of the DHPS allosteric inhibitor bromobenzothiophene (67) provides a new avenue for targeting this circuit, which is needed given the off-target activities of spermidine mimetic compounds such as GC7 (33).
METHODS
Animals
All animal care and the treatment of animals were in strict accordance with Good Animal Practice as defined by the US Office of Laboratory Animal Welfare and were approved by the Moffitt/University South Florida (USF) Institutional Animal Care and Use Committee (IACUC).
Conditional knockout Dhpsfl/fl mice (37) were backcrossed for at least 10 generations onto a C57BL/6J (The Jackson Laboratory Stock #000664) background. These were crossed with hemizygous Eμ-Myc transgenic mice (26) and knock-in CD19-Cre mice to generate Eμ-Myc;CD19-Cre;Dhpsfl/fl mice. Eμ-Myc;Rosa26-CreERT2;Dhpsfl/fl mice were generated by crossing Dhpsfl/fl mice with Rosa26-CreERT2 (38) and Eμ-Myc transgenic mice (Supplementary Table S6).
Experiments were designed to include both males and females in all groups, and no sex-based bias was detected in the observations reported in this study. Unless otherwise stated in the text, all premalignant Eμ-Myc mice and Eμ-Myc derivatives were 6 to 7 weeks of age at the time of experimentation.
Cell Lines and Culture
Eμ-Myc lymphomas were harvested, homogenized in 1× HBSS buffer, filtered through a 100-μm strainer and cultured as a single-cell suspension in 45% DMEM, 45% IMDM, supplemented with 10% fetal bovine serum (FBS), penicillin/streptomycin (100 U/mL), 4 mmol/L GlutaMax, 25 μmol/L β-mercaptoethanol, 1 mmol/L sodium pyruvate, and 5 ng/mL mouse IL7.
Human Raji, Ramos, and Daudi BL cells were purchased from ATCC and were cultured in RPMI-1640 medium supplemented with 10% FBS and penicillin–streptomycin (100 U/mL) and 4 mmol/L L-glutamine at 37°C, 5% CO2.
P493-6 human B-lymphoma cells (31) were cultured in RPMI-1640 medium, 10% FBS, 4 mmol/L L-glutamine, and penicillin/streptomycin (100 U/mL) at 37°C, 5% CO2. To manipulate MYC transgene expression in these cells, they were cultured in the presence of tetracycline (Tet, 0.1 μg/mL) for 72 hours, resuspended in media without Tet following washing of cells in medium (3×) to induce MYC expression, and cultured for the times indicated.
EcoPack cells were maintained in DMEM with 10% FBS, 2 mmol/L L-glutamine, penicillin/streptomycin (100 U/mL) at 37°C, 5% CO2.
All cell lines (Supplementary Table S7) were routinely tested for mycoplasma contamination and confirmed to be negative using the Mycoplasma PCR detection kit (iNtRON) and/or MycoAlert mycoplasma detection kit (Lonza).
Bacteria Strains
TOP10 and Stbl3 E. coli cells (New England Biolabs) were grown in autoclave-sterilized Luria Bertani broth (Fisher Scientific), in a MAXQ6000 shaker (Thermo Scientific) set at 230 rpm and 37°C.
Human Tissue Specimen Studies
Human BL, DHL, and reactive lymph node patient biopsy samples were used for IHC and hematoxylin and eosin (H&E) staining. Formalin-fixed paraffin-embedded lymphoma (FFPE) biopsies from the patients were sectioned (4-μm) and incubated with anti-MYC, anti-eIF5A, and anti-Hypusine antibody (Supplementary Table S8). Slides were scanned using an OLYMPUS BX51 microscope and photographed using an OLYMPUS DP22 camera.
Ethics Reporting
We and our colleagues obtained written informed consent from the patients agreeing to the use of biopsy tissue for research studies. These studies were performed in accordance with recognized ethical guidelines (e.g., Declaration of Helsinki, CIOMS, Belmont Report, U.S. Common Rule), and this study was approved by an institutional review board (IRB).
Mouse Tissue Samples
Mouse spleens and femurs were collected following humane euthanasia. Spleens were formalin-fixed and paraffin-embedded. Femurs were treated with 70% EtOH and decalcified prior to embedding. Sections were stained with H&E, and the staining was visualized using a microscope with magnifications of 10×, 20×, or 40× (Version 3.0. Leica). Samples were processed, and the staining intensity of slides was evaluated at IDEXX BioAnalytics.
Primary B-cell Isolation
B lymphocytes were isolated from the BM of the indicated mice. BM samples were treated with Gey's solution to lyse red blood cells. B cells were isolated using magnetic beads conjugated with CD19 or B220 antibodies and Miltenyi magnetic separation columns per the manufacturer's instructions.
Immunoblotting
Cells were washed with cold Dulbecco's phosphate buffer saline (PBS), lysed in ARF buffer (50 mmol/L HEPES, pH7.5, 150 mmol/L NaCl, 1 mmol/L EDTA, 2.5 mmol/L EGTA, 0.1% Tween-20) supplemented with 1 mmol/L PMSF, 10 mmol/L β-glycerophosphate, 1 mmol/L NaF, 1 mmol/L NaVO4 phosphatase inhibitor and Complete mini protease inhibitor (Roche), and lysates were sonicated and cleared by centrifugation at 4°C. Total protein was quantified using the MicroBCA Protein Assay, and equal amounts of total proteins were loaded on 6%, 7%, 8%, 10%, 12%, or 15% SDS-polyacrylamide gels, transferred onto nitrocellulose membranes with a tank blotting system, and probed with the indicated primary antibodies specific for the proteins of interest (Supplementary Table S8). Membranes were then incubated with a corresponding fluorophore-conjugated secondary antibody and signals were then detected using Odyssey Fc imaging systems (LI-COR). Reagents used in this study are listed in Supplementary Table S9.
Genomic DNA Preparation
Genomic DNA from tails and lymphomas was extracted using the QIAamp DNA Mini Kit (QIAGEN) per the manufacturer's protocol.
Sanger Sequencing
Plasmid constructs generated in this study were confirmed using Sanger sequencing performed by Eurofins Genomics. To assess the mutation status of Tp53 in Eμ-Myc;Rosa26-rtTA2 lymphoma cells, the p53 coding region was PCR-amplified using cDNA prepared from these cells, and the fragments were sequenced.
Flow Cytometry Analysis and Cell Sorting
At least 10,000 events were acquired on BD Canto II or BD LSRII Flow Cytometer (BD Biosciences), and data were analyzed using the FlowJo software package (TreeStar). For time-course experiments, Rainbow calibration particles (BD Biosciences) were used to evaluate and calibrate the flow cytometer for each use. Additionally, cell sorting was performed using an Aria flow cytometer (BD Biosciences).
Expression of shRNAs targeting Renilla (Ren), Eif5a or Dhps expression in Eμ-Myc;Rosa26-rtTA2 lymphoma cells was accomplished by transducing these lymphoma cells with retroviruses expressing inducible miRE shRNA constructs (Supplementary Table S10), which was assessed by monitoring changes in GFP expression over time. GFP+ cells were isolated by sorting after Dox induction of these shRNAs (0.9 μg/mL) overnight.
Proliferation was assessed in Eμ-Myc;Rosa26-rtTA2 lymphoma cell lines transduced with miRE shRNA constructs using the CellTrace Violet (CTV) Cell Proliferation kit (Thermo Fisher Scientific) according to the manufacturer's protocol. For cell proliferation eIF5A rescue experiments, Eμ-Myc;Rosa26-rtTA2 lymphoma cells expressing tGFP-shEif5a#1 were engineered to also inducibly express either WT FLAG-tagged eIF5A (eIF5AWT) or FLAG-tagged K50R-eIF5A (eIF5AK50R), or the vector alone, which expresses tdTomato from an independent promoter. shEif5a #1 targets the 3′-UTR of the endogenous Eif5a gene; thus, expression of exogenous Eif5A is not affected by the shRNA. Once the stable cell lines were established, these cells were labeled with CTV as described above and cultured in the presence or absence of Dox. Cell division status was assessed by flow cytometry at the times indicated.
For analysis of nascent peptide synthesis, OP-Puro (20 μmol/L) was added for 1 hour to Eμ-Myc;Cre-ERT2;Dhpsfl/fl lymphoma cells that had been treated 4-OHT or ethanol for 3 days. Incorporation of OP-Puro into nascent peptides was assessed by conjugation of AlexaFluor 640 using the Click-iT technology (Thermo Fisher) and flow cytometry.
Analyses of early and late apoptosis of Eμ-Myc;Cre-ERT2;Dhpsfl/fl lymphoma cells following knockout of Dhps were performed by staining with FITC-conjugated Annexin V (BD Biosciences) and pro-pidium iodide.
For cell-cycle analyses of Eμ-Myc;Cre-ERT2;Dhpsfl/fl lymphoma cells, BrdU incorporation was performed using the BrdU assay kit (BD Biosciences) and DAPI staining according to the manufacturer's protocol.
To analyze CD19+ B-cell populations, following RBC removal, single-cell suspensions from mouse BM were stained using the PE-Cy7 anti-CD19 and Zombie cell viability dye (BioLegend).
Cell Growth Assays
Cell growth was measured by counting cells or by MTT assay (Millipore) daily. Human Raji cells were treated with the indicated concentrations of DFMO or GC7 and MTT assays were performed daily according to the manufacturer's instructions. For cell counting assays, Eμ-Myc;Rosa26-rtTA2 lymphoma cells inducibly expressing miR-30 shEif5a or a scrambled shRNA were seeded in 6-well plates in the absence or presence of Dox (0.9 μg/mL). Cells were counted at the indicated intervals using trypan blue stain (0.2%) and Cellometer Auto 2000 (Nexcelom Bioscience) per the manufacturer's instructions.
Methocult Colony-Forming Assays
Eμ-Myc;Rosa26-rtTA2 lymphoma cells transduced with retrovirus expressing inducible miRE shRNA targeting mouse Eif5a, Dhps, or Renilla were treated with Dox and sorted for GFP+ cells. On day 3, the cells were washed with media three times and 1.2 × 103 viable cells were resuspended in 1.1 mL MethoCult medium and cultured in 35-mm 6-well plates (SmartDish, STEMCELL Technology) in the presence or absence of Dox (1.0 μg/mL). The images were captured using an automated STEMvision system (STEMCELL Technology) at days 12 and 18 and colonies were counted manually.
Quantitative Real-time PCR Analysis
RNA was isolated from CD19+ B cells, tumors, or cultured cells using NucleoSpin RNA kit per the manufacturer's instructions. cDNA was generated using an iScript cDNA synthesis kit, and qRT-PCR was performed using iQ SYBR green mix (Bio-Rad) on a CFX96 Touch Real-Time PCR Detection System instrument (Bio-Rad). Data were analyzed using the ΔΔCt method, and data were normalized to the expression of Ubiquitin (Ubc). Primers are listed in the Sequence Information (Supplementary Table S11).
Generation of shRNA Knockdown and Eif5a-Expressing Eμ-Myc Lymphoma Cell Lines
Retroviruses that inducibly express shRNAs targeting mouse Eif5a or Dhps were constructed in the following manner. 97-mer oligos containing the shRNA sequences listed in Sequence Information (Supplementary Table S11) were synthesized (IDT), annealed, and cloned into either SIN-TREmiRE30-PIG or miRE shRNA (34) vectors. pRetroX-Tight-FLAG-tagged mouse Eif5a plasmid was generated by cloning the FLAG-Eif5a PCR fragment into the pRetroX-Tight-MCS-PGK-tdTomato vector. The K50R amino acid substitution mutant of eIF5A that is defective for hypusination was generated by site-directed mutagenesis using QuickChange II (Agilent).
EcoPack cells were transfected with the indicated plasmids using the ProFection transfection system (Promega). Retrovirus was collected at day 2, filtered through 0.45-μm filter, and added to mouse Eμ-Myc;Rosa26-rtTA2 lymphoma cells. Spin-infection was then performed at 2,500 rpm for 90 minutes in the presence of IL7 (5 ng/mL) and polybrene (8 ng/mL). After the expansion of cells in culture, transduced cells were isolated by selection in antibiotic (G418) or by cell sorting prior to experimental analyses.
Lymphoma Transplant Studies
To assess the effects of pharmacologic inhibition of ODC or DHPS on lymphoma tumorigenic potential, Eμ-Myc lymphoma cells were suspended in PBS, and 1 million cells were injected into the tail veins of 7-week-old C57BL/6J female mice and mice were randomly distributed into three groups (10 mice each). Three days after transplant, mice were treated with GC7 (4 or 20 mg/kg) or vehicle (PBS) via daily i.p. injection until animals reached the endpoint. DFMO was administered in drinking water (1% wt/vol) starting one day after the transplant.
To determine the effects of eIF5A depletion on lymphoma tumorigenic potential, Eμ-Myc;Rosa26-rtTA2 cells engineered to inducibly express shRNAs targeting Eif5a or Renilla were injected into the tail veins of 7-week-old nude female mice (3.5 × 105 cells/mouse) and mice were randomized into 2 groups. Three days after the transplant, half of these mice were shifted to a Dox chow diet (200 μg/kg), which was given daily until the endpoint.
To evaluate the effects of Dhps loss on tumor maintenance, 6-week-old athymic male nude recipient mice were transplanted with Eμ-Myc;Rosa26-CreERT2;Dhpsfl/fl; lymphoma cells that were transduced with pRetroX-Tight-MCS-PGK-OgNLuc, which expresses the LSSmOrange-NanoLuc reporter (68). Transduced cells were isolated by sorting for LSSmOrange and were suspended in a 1:1 mixture of PBS and Matrigel (Corning). Cell suspensions (1 million cells) were then subcutaneously implanted in the right flank of 6-week-old male nude mice (The Jackson Laboratory). The greatest longitudinal diameter (L), the transverse diameter (W), and the height (H) were measured daily using calipers, and the tumor volume was calculated using the formula, volume = π/6 (L × W × H; ref. 69). Once a palpable tumor formed, mice (bearing established tumors of ∼20 mm3) were randomly assigned to either tamoxifen or vehicle (corn oil) treatment and were dosed daily by oral gavage (145 mg/kg). 13 Eμ-Myc;Rosa26-CreERT2;Dhpsfl/fl mice were treated with tamoxifen or corn oil (for 9 consecutive days). As a control, 6 mice bearing Eμ-Myc;Dhps+/+;Rosa26-CreERT2 lymphomas were either treated with tamoxifen or corn oil. Luminescence signals were monitored twice a week using IVIS spectrum 200 (PerkinElmer) following i.p. injection of furimazine; 2-furanylmethyl-deoxy-coelenterazine (Promega) and were quantified using Living Image software (PerkinElmer).
Sample Preparation for RNA-seq, Ribosome Profiling, and TMT Multilabeling Mass Spectrometry
Eμ-Myc;Rosa26-rtTA2 lymphoma cells engineered to inducibly express shRNAs targeting Renilla, Eif5a, or Dhps were treated with Dox overnight and GFP+ cells were sorted by flow cytometry. Cells were incubated for a total of 72 hours in the presence of Dox, washed with PBS, and harvested. Total RNA for RNA-seq was isolated with the RNAeasy kit (QIAGEN) with in-column DNase treatment. cDNA libraries were generated using Epicentre ScriptSeq Complete Gold Library Prep (Epicentre Biotechnologies). RNA integrity was verified using Agilent 2100 Bioanalyzer (Agilent Technologies) prior to RNA-seq run (NextSeq 500, 2 × 75 High-Output Run). RNA-seq data shown in Fig. 4 and Supplementary Fig. S4 were analyzed at a gene level (see Analyses of RNA-seq data sets), although RNA-seq data used for TE calculation were analyzed at a transcript level (see Analyses of ribosome profiling data sets). For ribosome profiling, Eμ-Myc;Rosa26-rtTA2 lymphoma cells expressing shRNAs targeting Renilla, Eif5a, or Dhps were simultaneously prepared as described above, treated with cycloheximide for 15 minutes prior to harvest, and washed with PBS. Then, ribosome footprint cDNA libraries were prepared according to the protocol described by Ingolia and colleagues (45). Single-end 1 × 50 bp NextSeq runs were performed.
For time-course TMT proteomics analysis, Eμ-Myc;Rosa26-rtTA2 lymphoma cells that inducibly express shRNAs targeting Renilla, Eif5a, or Dhps were treated with Dox overnight and GFP+ cells were isolated by flow cytometry. Cells were then incubated for a total of 72, 96, or 120 hours in the presence of Dox, washed with PBS, and harvested. Cell pellets were then processed for 10 plex TMT-multilabeling mass spectrometry (44). sh-Ren cells without Dox treatment (0 hours) were also included as a control in the 10-plex.
Cells were lysed in denaturing buffer containing 8M urea, 20 mmol/L HEPES (pH 8), 1 mmol/L sodium orthovanadate, 2.5 mmol/L sodium pyrophosphate, and 1 mmol/L β-glycerophosphate. Following the determination of protein concentration using the Bradford assay, the proteins were reduced with 4.5 mmol/L dithiothreitol and alkylated with 10 mmol/L iodoacetamide. Trypsin digestion was carried out at room temperature overnight, and tryptic peptides were then acidified with 1% trifluoroacetic acid (TFA) and desalted with C18 Sep-Pak cartridges according to the manufacturer's procedure. An aliquot (100 μg) of tryptic peptide digest from each sample was labeled with TMT10-plex reagent using the following experimental design where the reporter ion channels are listed in parentheses: sh-Ren No-Dox (126), sh-Ren Dox-72hr (127N), sh-Ren Dox-96hr (127C), sh-Ren Dox-120hr (128N), sh-Eif5a Dox-72hr (128C), sh-Eif5a Dox-96hr (129N), sh-Eif5a Dox-120hr (129C), sh-Dhps Dox-72hr (130N), sh-Dhps Dox-96hr (130C), and sh-Dhps Dox-120hr (131). The label incorporation and correct label assignment were verified by LC-MS/MS; spectral counts were used to verify that at least 98% of the identified peptides were TMT labeled for each channel. The 10 samples were pooled and lyophilized.
After lyophilization, the peptides were redissolved in 250 μL of aqueous 20 mmol/L ammonium formate (pH 10.0), which was used as the bRPLC A solvent. The basic pH reversed phase separation was performed on a 4.6 mm ID × 100 mm long column packed with BEH C18 resin, 3.5 μm particle size, 130 Å pore size (XBridge, Waters). The peptides were eluted as follows: 5% B (10% aqueous 5 mmol/L ammonium formate, 90% acetonitrile, pH 10.0) for 10 minutes, 5%–15% B in 5 minutes, 15%–40% B in 47 minutes, 40%–100% B in 5 minutes and 100% B held for 10 minutes, followed by reequilibration at 5% B. The flow rate was 0.6 mL/minute, and 24 concatenated fractions were collected. Peptide fractions were dried by vacuum centrifugation (Speedvac, Thermo).
A nanoflow ultra-high performance liquid chromatograph interfaced with an electrospray benchtop orbitrap mass spectrometer (RSLCnano and Q-Exactive Plus, Thermo) was used for tandem mass spectrometry peptide sequencing experiments. The sample was first loaded onto a precolumn (2 cm ID × 100 μm length packed with C18 reversed phase resin, 5 μm particle size, 100 Å pore size (C18 PepMap100, Thermo) and washed for 8 minutes with aqueous 2% acetonitrile and 0.1% formic acid (LC-MS/MS solvent A). The trapped peptides were separated by a C18 reversed phase analytical column 75 μm ID × 25 cm, 2 μm particle size, and 100 Å pore size (C18 PepMap100, Thermo). The 120-minute gradient was programmed as: 95% solvent A (aqueous 2% acetonitrile + 0.1% formic acid) for 8 minutes, solvent B (aqueous 90% acetonitrile + 0.1% formic acid) from 5% to 38.5% in 90 minutes, then solvent B from 50% to 90% B in 7 minutes and held at 90% for 5 minutes, followed by solvent B from 90% to 5% in 1 minute and reequilibration for 10 minutes. The flow rate on the analytical column was 300 nL/minute. Twenty tandem mass spectra were collected in a data-dependent manner following each survey scan. The resolution settings were 60,000 and 45,000 for MS1 and MS/MS, respectively. The isolation window was 0.8 Th with 0.2 offset.
Analyses of Public Data Sets
Public data sets used in this study are listed in Supplementary Table S12.
Analyses of TCGA PanCanAtlas Data Sets.
To compare the expression of EIF5A, EIF5A2, DHPS, and DOHH in human malignant tumors (red) and related normal tissues (blue) across publicly available different cancer types, the TCGA PanCanAtlas data sets downloaded from the https://gdc.cancer.gov/about-data/publications/pancanatlas were log2 transformed. Samples listed in merged_sample_quality_annotations.tsv were removed from further analysis. Following QC, 17 cancer types were used throughout the study based on the availability of normal tissue sample (n > 5).
Analyses of copy-number alterations (CAN) were performed using the GISTIC data from the https://gdc.cancer.gov/about-data/publications/pancan-aneuploidy genes whitelisted tsv file. We observed significant overexpression of EIF5A in 12 of the aforementioned 17 TCGA cancer types. These 12 cancer types were classified based on EIF5A copy-number status; deep (−2), shallow (−1), and diploid (0, WT) and were compared with EIF5A log2 expression of these subgroups with the normal samples.
TCGA survival data were based on the TCGA Pan-Cancer Clinical Data resource (TCGA-CDR) as described by Liu and colleagues (70).
Analyses of GSE12453.
Data of BL patients and normal B-cell subsets were extracted from GSE12453 (71). The top 20 overexpressed genes were determined by comparing in BL and normal B-cell subsets using a two-sided t test to calculate each P value. Log2 fold change was calculated as mean expression for one group minus mean expression for the other group. A heat map showing expression profiling of genes of the hypusine circuit was generated to compare expression in BL and normal B-cell subsets genes. Significance of each probe was determined by an unpaired t test analysis.
Analyses of GSE32239 and GSE33474.
To compare the expression of genes involved in the polyamine–hypusine circuit in Eμ-Myc mice, we used our published WT B-cell and Eμ-Myc data sets: GSE32239, spleen (5) and GSE33474, BM (72). Expression values were first log2 transformed, and heat maps were generated by MATLAB R2020a. The fold change of expression of probe sets and their significance were determined as described above.
Analyses of GSE15907.
To analyze the expression of genes of the hypusine circuit during B-cell development, GSE15907 ImmGen data were downloaded from Gene-Expression Omnibus (GEO) as a Series Matrix File and log2 transformed (73).
Analyses of RNA-seq Data Sets
Processed raw reads were aligned to mouse genome mm10 using TopHat (v2.0.13; ref. 74). Gene expression was evaluated as read count at gene level with HTSeq (v0.6.1; ref. 75). The data were then normalized, and differential expression between experimental groups was analyzed using DEseq2 (76). Genes were considered to be differentially expressed if q < 0.05 (DEG). Expression data were log2 transformed, log2(x + 1), and the relative expression was calculated by removing the mean. Specific comparisons that are shown in Fig. 4 and Supplementary Fig. S4 represent averages calculated from two biological duplicates at the gene levels.
For expression analysis of mouse E2F target genes on the RNA-seq data, HALLMARK E2F target genes (GSEA, 200 human genes) were first mapped to 188 corresponding mouse genes and the first principal component was used to show differential expression between the controls and the Dhps or Eif5a knockdown samples.
Analyses of Ribosome Profiling Data Sets
Raw Sequence Read Alignment.
Single-end sequence reads from three independent runs were merged and clipped for the manually identified adapter: CTGTAGGCACCATCAAT. One base was then trimmed from the 5′ end and 3′ end using FASTX Toolkit (v0.0.13). Reads with 19 or fewer bases were discarded. Approximately 98% of the reads were retained afterward. Reads corresponding to ribosomal RNA (rRNA) sequence were removed by aligning to the mouse ribosomal RNA reference sequence using bowtie2. Resulting reads were aligned to the mouse genome assembly (GRCm38/mm10) using TopHat (v2.0.13; ref. 74). Aligned reads were ordered by name and the total number of reads for a given gene transcript were counted using HTSeq (v0.6.1; ref. 75).
TE Calculation.
Transcript expression levels from our RNA-seq experiment and ribosomal densities from the Ribo-seq experiment were normalized using the RPKM method. Pairwise scatter plot and Pearson correlation coefficients were used to assess the agreement and quality of RPKM data between biological replicates. The degree of TE for individual transcripts was then quantified by dividing Ribo-seq RPKM by RNA-seq RPKM. To avoid a zero denominator, we then added a very small number, 1e−10, to all RNA-seq RPKMs. The R LIMMA package (77) was used to perform differential TE analysis. False discovery rate was controlled using Benjamini–Hochberg method for a multiple testing correction in the differential TE analyses. DTTs (P < 0.05) that were identified from TE analysis are summarized in Supplementary Table S3.
TE Analysis of Oncogenes and Tumor Suppressor Genes.
The general human cancer-related gene list was obtained from the Bushman lab site (http://www.bushmanlab.org/links/genelists). Then, a literature search was performed to search for additional oncogenes and tumor suppressors, especially those that were related to lymphoma. Finally, among these combined human cancer genes, the mouse homologs were identified to generate our mouse cancer gene lists. Among those potential oncogene/tumor suppressor genes, DTTs were then ranked based on their log2 fold changes in TE (vs. sh-Ren cells). Note that genes whose roles have been reported to function as both an oncogene and a tumor suppressor were also included in the list. The oncogene/tumor suppressors that are differentially translated upon depletion of eIF5A or DHPS are listed in Supplementary Table S4.
Analyses of TMT Proteomics Data Sets
MaxQuant (version 1.5.2.8; ref. 78) was used to quantify the TMT reporter ion intensities. Database search was performed using trypsin with up to 2 missed cleavages, modifications included TMT labeling of lysines and peptide N-termini, carbamidomethylation of cysteine, and oxidation of methionine. Similar parameters were used for Mascot database searches to support data upload. Further, the intensity from all 10 samples was normalized using the Iterative rank-order normalization (IRON) algorithm (79).
Statistical Analyses of Experimental Data Sets
Statistical analyses were performed using GraphPad Prism 7 and MATLAB and a P value of less than 0.05 was considered statistically significant. The levels of significance were determined using an unpaired two-tailed Student t test or ANOVA, and P values are provided in the figures or the legends. Error bars presented in the figures indicate the mean ± SD unless otherwise specified. The statistical parameters are described in the individual figure legends. For survival analyses, overall survival was estimated with the Kaplan–Meier method and compared using a log-rank test.
Data Availability
This study did not generate any original code. Software and algorithms used in this study are listed in Supplementary Table S13. RNA-seq and Ribo-seq data sets were deposited in the GEO, GSE190669 and GSE190670, respectively. TMT multilabeling mass spectrometry data were deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the data set identifier Project accession: PXD031361 and Project DOI: 10.6019/PXD031361.
Supplementary Material
Supplementary Tables S1, S2, S5-S13 includes Supplementary Tables S1, S2, S5-S13. Supplementary Table S1 provides a summary of BL and DHL patient demographics for the immunohistochemistry study presented in Fig. 1, E and F. Supplementary Table S2 lists the MYC-dysregulated genes whose expression is significantly altered following depletion of eIF5A or DHPS, related to Fig. 4F. Supplementary Table S5 shows DHPS GISTIC count and survival of select TCGA PanCancer datasets, related to Fig. 7J. Supplementary Table S6 shows the genetic mouse models used in this study, related to Methods. Supplementary Table S7 shows the mouse and human cell lines used in this study, related to Methods. Supplementary Table S8 lists the antibodies used in this study, related to Methods. Supplementary Table S9 lists reagents used in this study, related to Methods. Supplementary Table S10 summarizes the plasmids used in this study, related to Methods. Supplementary Table S11 lists the sequences of the oligonucleotides used in this study, related to Methods. Supplementary Table S12 lists accession numbers and publicly deposited data from this study, related to Methods. Supplementary Table S13 lists software and algorithms used in this study, related to Methods.
Supplementary Table S3 lists 665 shared differentially translated transcripts (DTT) identified following depletion of eIF5A or DHPS, related to Fig. 5C.
Supplementary Table S4 lists oncogenes and tumor suppressor genes analyzed for changes in translation efficiency (TE) following depletion of eIF5A or DHPS, related to Fig. 5E.
Supplementary Figures S1-S7 includes Supplementary Figure S1-S7 and the figure legend for each figure. Supplementary Fig. S1 shows that the polyamine-hypusine circuit is activated in many human cancers including MYC-driven lymphoma. Supplementary Fig. S1 is related to Fig. 1. Supplementary Fig. S2 shows that inhibition of DHPS enzyme activity, or silencing eIF5A or DHPS, suppresses the growth of mouse MYC-driven lymphoma. Supplementary Fig. S2 is related to Fig. 2. Supplementary Fig. S3 shows that hypusinated eIF5A (eIF5AHyp) contributes to the tumorigenic potential and maintenance of MYC-driven lymphoma. Supplementary Fig. S3 is related to Fig. 3. Supplementary Fig. S4 shows the effects of eIF5A or DHPS depletion on the transcriptional landscape of MYC-driven lymphoma. Supplementary Fig. S4 is related to Fig. 4. Supplementary Fig. S5 shows that depletion of eIF5A or DHPS impairs the translation efficiency of subsets of mRNA in MYC-driven lymphoma. Supplementary Fig. S5 is related to Fig. 5. Supplementary Fig. S6 shows the validation of select eIF5AHyp translation targets identified by the multi-omics analyses, and that the translation of key regulatory cell cycle factors is controlled by hypusinated eIF5A. Supplementary Fig. S6 is related to Fig. 6. Supplementary Fig. S7 shows that hypusinated eIF5A is essential for the development of MYC-driven lymphoma. Supplementary Fig. S7 is related to Fig. 7.
Acknowledgments
We sincerely thank Dr. Nicholas Ingolia for technical advice on ribosome profiling analyses and critical review of this study. We also thank Dr. Srikumar Chellappan and Sathya Puthamveettil (Scripps Research) for critical review of this study. We also thank the Genomics Core of Scripps Research and the Molecular Genomics Core of Moffitt Cancer Center (MCC) for RNA-seq, the Proteomics and Metabolomics Core (MCC) for TMT analyses, the Tissue Core (MCC) for IHC analyses, the Flow Cytometry Core (MCC) for cell sorting and technical advice, and the Biostatistics and Bioinformatics Shared Resource for bioinformatics support. The miRE shRNA plasmid was a kind gift from Dr. Johannes Zuber (IMP, Austria). We also thank Dr. Maier Bernhard for providing the eIF5AHyp antibody and are grateful to Moffitt/USF Department of Comparative Medicine for exceptional animal care and mouse husbandry. This work was supported by R01 grants CA100603 and CA241713 (J.L. Cleveland), by NCI Comprehensive Cancer Center Grant P30 CA076292, by the Cortner-Couch Endowed Chair for Cancer Research from the University of South Florida School of Medicine (J.L. Cleveland), and by monies from the State of Florida to the H. Lee Moffitt Cancer Center and Research Institute. This work was also supported R01 grants DK124906 and DK060581 (R.G. Mirmira), and by the Diabetes Research Center Grant P30 DK020595 (R.G. Mirmira).
The publication costs of this article were defrayed in part by the payment of publication fees. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Footnotes
Note: Supplementary data for this article are available at Blood Cancer Discovery Online (https://bloodcancerdiscov.aacrjournals.org/).
Authors’ Disclosures
R.G. Mirmira reports grants from NIH during the conduct of the study. J.L. Cleveland reports grants from NCI/NIH and other support from Cortner-Couch Endowed Chair for Cancer Research from the University of South Florida during the conduct of the study. No disclosures were reported by the other authors.
Authors’ Contributions
S. Nakanishi: Conceptualization, resources, data curation, formal analysis, supervision, validation, investigation, visualization, methodology, writing–original draft, writing–review and editing. J. Li: Data curation, software, formal analysis, visualization, methodology, writing–original draft. A.E. Berglund: Data curation, software, formal analysis, visualization, methodology, writing–original draft, writing–review and editing. Y. Kim: Data curation, software, formal analysis, visualization, methodology, writing–original draft. Y. Zhang: Data curation, software, formal analysis, methodology, writing–original draft. L. Zhang: Data curation, formal analysis, methodology, writing–original draft. C. Yang: Data curation. J. Song: Resources. R.G. Mirmira: Resources. J.L. Cleveland: Conceptualization, resources, data curation, formal analysis, supervision, funding acquisition, validation, investigation, visualization, methodology, writing–original draft, project administration, writing–review and editing.
References
- 1. Gabay M, Li Y, Felsher DW. MYC activation is a hallmark of cancer initiation and maintenance. Cold Spring Harb Perspect Med 2014;4:a014241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Reddy A, Zhang J, Davis NS, Moffitt AB, Love CL, Waldrop A, et al. Genetic and functional drivers of diffuse large B cell lymphoma. Cell 2017;171:481–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Durie BG, Salmon SE, Russell DH. Polyamines as markers of response and disease activity in cancer chemotherapy. Cancer Res 1977;37:214–21. [PubMed] [Google Scholar]
- 4. Russell DH. Increased polyamine concentrations in the urine of human cancer patients. Nat New Biol 1971;233:144–5. [DOI] [PubMed] [Google Scholar]
- 5. Nilsson JA, Keller UB, Baudino TA, Yang C, Norton S, Old JA, et al. Targeting ornithine decarboxylase in Myc-induced lymphomagenesis prevents tumor formation. Cancer Cell 2005;7:433–44. [DOI] [PubMed] [Google Scholar]
- 6. Rounbehler RJ, Li W, Hall MA, Yang C, Fallahi M, Cleveland JL. Targeting ornithine decarboxylase impairs development of MYCN-amplified neuroblastoma. Cancer Res 2009;69:547–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hogarty MD, Norris MD, Davis K, Liu X, Evageliou NF, Hayes CS, et al. ODC1 is a critical determinant of MYCN oncogenesis and a therapeutic target in neuroblastoma. Cancer Res 2008;68:9735–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Simoneau AR, Gerner EW, Nagle R, Ziogas A, Fujikawa-Brooks S, Yerushalmi H, et al. The effect of difluoromethylornithine on decreasing prostate size and polyamines in men: results of a year-long phase IIb randomized placebo-controlled chemoprevention trial. Cancer Epidemiol Biomarkers Prev 2008;17:292–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Raj KP, Zell JA, Rock CL, McLaren CE, Zoumas-Morse C, Gerner EW, et al. Role of dietary polyamines in a phase III clinical trial of difluoromethylornithine (DFMO) and sulindac for prevention of sporadic colorectal adenomas. Br J Cancer 2013;108:512–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Casero RA Jr, Marton LJ. Targeting polyamine metabolism and function in cancer and other hyperproliferative diseases. Nat Rev Drug Discov 2007;6:373–90. [DOI] [PubMed] [Google Scholar]
- 11. Miller-Fleming L, Olin-Sandoval V, Campbell K, Ralser M. Remaining mysteries of molecular biology: the role of polyamines in the cell. J Mol Biol 2015;427:3389–406. [DOI] [PubMed] [Google Scholar]
- 12. Park MH, Cooper HL, Folk JE. The biosynthesis of protein-bound hypusine (N epsilon -(4-amino-2-hydroxybutyl)lysine). Lysine as the amino acid precursor and the intermediate role of deoxyhypusine (N epsilon -(4-aminobutyl)lysine). J Biol Chem 1982;257:7217–22. [PubMed] [Google Scholar]
- 13. Benne R, Brown-Luedi ML, Hershey JW. Purification and characterization of protein synthesis initiation factors eIF-1, eIF-4C, eIF-4D, and eIF-5 from rabbit reticulocytes. J Biol Chem 1978;253:3070–7. [PubMed] [Google Scholar]
- 14. Saini P, Eyler DE, Green R, Dever TE. Hypusine-containing protein eIF5A promotes translation elongation. Nature 2009;459:118–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Schuller AP, Wu CC, Dever TE, Buskirk AR, Green R. eIF5A functions globally in translation elongation and termination. Mol Cell 2017;66:194–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Manjunath H, Zhang H, Rehfeld F, Han J, Chang TC, Mendell JT. Suppression of ribosomal pausing by eIF5A Is necessary to maintain the fidelity of start codon selection. Cell Rep 2019;29:3134–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gutierrez E, Shin BS, Woolstenhulme CJ, Kim JR, Saini P, Buskirk AR, et al. eIF5A promotes translation of polyproline motifs. Mol Cell 2013;51:35–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Liang Y, Piao C, Beuschel CB, Toppe D, Kollipara L, Bogdanow B, et al. eIF5A hypusination, boosted by dietary spermidine, protects from premature brain aging and mitochondrial dysfunction. Cell Rep 2021;35:108941. [DOI] [PubMed] [Google Scholar]
- 19. Zhang H, Alsaleh G, Feltham J, Sun Y, Napolitano G, Riffelmacher T, et al. Polyamines control eIF5A hypusination, TFEB translation, and autophagy to reverse B cell senescence. Mol Cell 2019;76:110–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Puleston DJ, Buck MD, Klein Geltink RI, Kyle RL, Caputa G, O'Sullivan D, et al. Polyamines and eIF5A hypusination modulate mitochondrial respiration and macrophage activation. Cell Metab 2019;30:352–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Anderson-Baucum E, Pineros AR, Kulkarni A, Webb-Robertson BJ, Maier B, Anderson RM, et al. Deoxyhypusine synthase promotes a pro-inflammatory macrophage phenotype. Cell Metab 2021;33:1883–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Nakanishi S, Cleveland JL. Targeting the polyamine-hypusine circuit for the prevention and treatment of cancer. Amino Acids 2016;48:2353–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Fujimura K, Wright T, Strnadel J, Kaushal S, Metildi C, Lowy AM, et al. A hypusine-eIF5A-PEAK1 switch regulates the pathogenesis of pancreatic cancer. Cancer Res 2014;74:6671–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Scuoppo C, Miething C, Lindqvist L, Reyes J, Ruse C, Appelmann I, et al. A tumour suppressor network relying on the polyamine-hypusine axis. Nature 2012;487:244–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Liu Y, Chen C, Xu Z, Scuoppo C, Rillahan CD, Gao J, et al. Deletions linked to TP53 loss drive cancer through p53-independent mechanisms. Nature 2016;531:471–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Adams JM, Harris AW, Pinkert CA, Corcoran LM, Alexander WS, Cory S, et al. The c-myc oncogene driven by immunoglobulin enhancers induces lymphoid malignancy in transgenic mice. Nature 1985;318:533–8. [DOI] [PubMed] [Google Scholar]
- 27. El-Ghammaz AM, Abdelwahed E, Mostafa NN, Mansour DA. De novo deletion 17p13.1 as a predictor for disease progression in chronic lymphocytic leukemia. Clin Exp Med 2015;15:493–9. [DOI] [PubMed] [Google Scholar]
- 28. Seifert H, Mohr B, Thiede C, Oelschlagel U, Schakel U, Illmer T, et al. The prognostic impact of 17p (p53) deletion in 2272 adults with acute myeloid leukemia. Leukemia 2009;23:656–63. [DOI] [PubMed] [Google Scholar]
- 29. Baker SJ, Fearon ER, Nigro JM, Hamilton SR, Preisinger AC, Jessup JM, et al. Chromosome 17 deletions and p53 gene mutations in colorectal carcinomas. Science 1989;244:217–21. [DOI] [PubMed] [Google Scholar]
- 30. Fields AP, Justilien V, Murray NR. The chromosome 3q26 OncCassette: a multigenic driver of human cancer. Adv Biol Regul 2016;60:47–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Pajic A, Spitkovsky D, Christoph B, Kempkes B, Schuhmacher M, Staege MS, et al. Cell cycle activation by c-myc in a Burkitt lymphoma model cell line. Int J Cancer 2000;87:787–93. [DOI] [PubMed] [Google Scholar]
- 32. Jakus J, Wolff EC, Park MH, Folk JE. Features of the spermidine-binding site of deoxyhypusine synthase as derived from inhibition studies. Effective inhibition by bis- and mono-guanylated diamines and polyamines. J Biol Chem 1993;268:13151–9. [PubMed] [Google Scholar]
- 33. Oliverio S, Corazzari M, Sestito C, Piredda L, Ippolito G, Piacentini M. The spermidine analogue GC7 (N1-guanyl-1,7-diamineoheptane) induces autophagy through a mechanism not involving the hypusination of eIF5A. Amino Acids 2014;46:2767–76. [DOI] [PubMed] [Google Scholar]
- 34. Fellmann C, Hoffmann T, Sridhar V, Hopfgartner B, Muhar M, Roth M, et al. An optimized microRNA backbone for effective single-copy RNAi. Cell Rep 2013;5:1704–13. [DOI] [PubMed] [Google Scholar]
- 35. Torrelio BM, Paz MA, Gallop PM. The formation and stability of the hypusine containing protein in Chinese hamster ovary cells. Biochem Biophys Res Commun 1987;145:1335–41. [DOI] [PubMed] [Google Scholar]
- 36. Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 2013;6:pl1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Levasseur EM, Yamada K, Pineros AR, Wu W, Syed F, Orr KS, et al. Hypusine biosynthesis in beta cells links polyamine metabolism to facultative cellular proliferation to maintain glucose homeostasis. Sci Signal 2019;12:eaax0715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ventura A, Kirsch DG, McLaughlin ME, Tuveson DA, Grimm J, Lintault L, et al. Restoration of p53 function leads to tumour regression in vivo. Nature 2007;445:661–5. [DOI] [PubMed] [Google Scholar]
- 39. Tripathi S, Pohl MO, Zhou Y, Rodriguez-Frandsen A, Wang G, Stein DA, et al. Meta- and orthogonal integration of influenza “OMICs” data defines a role for UBR4 in virus budding. Cell Host Microbe 2015;18:723–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sabo A, Kress TR, Pelizzola M, de Pretis S, Gorski MM, Tesi A, et al. Selective transcriptional regulation by Myc in cellular growth control and lymphomagenesis. Nature 2014;511:488–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kelemen K, Braziel RM, Gatter K, Bakke TC, Olson S, Fan G. Immunophenotypic variations of Burkitt lymphoma. Am J Clin Pathol 2010;134:127–38. [DOI] [PubMed] [Google Scholar]
- 42. Masir N, Marafioti T, Jones M, Natkunam Y, Rudiger T, Hansmann ML, et al. Loss of CD19 expression in B-cell neoplasms. Histopathology 2006;48:239–46. [DOI] [PubMed] [Google Scholar]
- 43. Ingolia NT, Ghaemmaghami S, Newman JR, Weissman JS. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science 2009;324:218–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Thompson A, Schafer J, Kuhn K, Kienle S, Schwarz J, Schmidt G, et al. Tandem mass tags: a novel quantification strategy for comparative analysis of complex protein mixtures by MS/MS. Anal Chem 2003;75:1895–904. [DOI] [PubMed] [Google Scholar]
- 45. Ingolia NT, Brar GA, Rouskin S, McGeachy AM, Weissman JS. The ribosome profiling strategy for monitoring translation in vivo by deep sequencing of ribosome-protected mRNA fragments. Nat Protoc 2012;7:1534–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zheng Z, Diamond MI. Huntington disease and the huntingtin protein. Prog Mol Biol Transl Sci 2012;107:189–214. [DOI] [PubMed] [Google Scholar]
- 47. Nick McElhinny SA, Stith CM, Burgers PM, Kunkel TA. Inefficient proofreading and biased error rates during inaccurate DNA synthesis by a mutant derivative of Saccharomyces cerevisiae DNA polymerase delta. J Biol Chem 2007;282:2324–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Xie H, Tang CH, Song JH, Mancuso A, Del Valle JR, Cao J, et al. IRE1alpha RNase-dependent lipid homeostasis promotes survival in Myc-transformed cancers. J Clin Invest 2018;128:1300–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Leone G, Sears R, Huang E, Rempel R, Nuckolls F, Park CH, et al. Myc requires distinct E2F activities to induce S phase and apoptosis. Mol Cell 2001;8:105–13. [DOI] [PubMed] [Google Scholar]
- 50. Nishimura K, Lee SB, Park JH, Park MH. Essential role of eIF5A-1 and deoxyhypusine synthase in mouse embryonic development. Amino Acids 2012;42:703–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Sievert H, Pallmann N, Miller KK, Hermans-Borgmeyer I, Venz S, Sendoel A, et al. A novel mouse model for inhibition of DOHH-mediated hypusine modification reveals a crucial function in embryonic development, proliferation and oncogenic transformation. Dis Model Mech 2014;7:963–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Pallmann N, Braig M, Sievert H, Preukschas M, Hermans-Borgmeyer I, Schweizer M, et al. Biological relevance and therapeutic potential of the hypusine modification system. J Biol Chem 2015;290:18343–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Kulak NA, Pichler G, Paron I, Nagaraj N, Mann M. Minimal, encapsulated proteomic-sample processing applied to copy-number estimation in eukaryotic cells. Nat Methods 2014;11:319–24. [DOI] [PubMed] [Google Scholar]
- 54. Truitt ML, Ruggero D. New frontiers in translational control of the cancer genome. Nat Rev Cancer 2016;16:288–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Annunziata I, van de Vlekkert D, Wolf E, Finkelstein D, Neale G, Machado E, et al. MYC competes with MiT/TFE in regulating lysosomal biogenesis and autophagy through an epigenetic rheostat. Nat Commun 2019;10:3623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Yun S, Vincelette ND, Yu X, Watson GW, Fernandez MR, Yang C, et al. TFEB links MYC signaling to epigenetic control of myeloid differentiation and acute myeloid leukemia. Blood Cancer Discov 2021;2:162–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Lefebure M, Tothill RW, Kruse E, Hawkins ED, Shortt J, Matthews GM, et al. Genomic characterisation of Emu-Myc mouse lymphomas identifies Bcor as a Myc co-operative tumour-suppressor gene. Nat Commun 2017;8:14581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Eischen CM, Weber JD, Roussel MF, Sherr CJ, Cleveland JL. Disruption of the ARF-Mdm2-p53 tumor suppressor pathway in Myc-induced lymphomagenesis. Genes Dev 1999;13:2658–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Gorrini C, Squatrito M, Luise C, Syed N, Perna D, Wark L, et al. Tip60 is a haplo-insufficient tumour suppressor required for an oncogene-induced DNA damage response. Nature 2007;448:1063–7. [DOI] [PubMed] [Google Scholar]
- 60. Schmitt CA, McCurrach ME, de Stanchina E, Wallace-Brodeur RR, Lowe SW. INK4a/ARF mutations accelerate lymphomagenesis and promote chemoresistance by disabling p53. Genes Dev 1999;13:2670–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Rempel RE, Mori S, Gasparetto M, Glozak MA, Andrechek ER, Adler SB, et al. A role for E2F activities in determining the fate of Myc-induced lymphomagenesis. PLoS Genet 2009;5:e1000640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Baudino TA, Maclean KH, Brennan J, Parganas E, Yang C, Aslanian A, et al. Myc-mediated proliferation and lymphomagenesis, but not apoptosis, are compromised by E2f1 loss. Mol Cell 2003;11:905–14. [DOI] [PubMed] [Google Scholar]
- 63. Keller UB, Old JB, Dorsey FC, Nilsson JA, Nilsson L, MacLean KH, et al. Myc targets Cks1 to provoke the suppression of p27Kip1, proliferation and lymphomagenesis. EMBO J 2007;26:2562–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Lin CJ, Nasr Z, Premsrirut PK, Porco JA Jr, Hippo Y, Lowe SW, et al. Targeting synthetic lethal interactions between Myc and the eIF4F complex impedes tumorigenesis. Cell Rep 2012;1:325–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Farria AT, Plummer JB, Salinger AP, Shen J, Lin K, Lu Y, et al. Transcriptional activation of MYC-induced genes by GCN5 promotes B-cell lymphomagenesis. Cancer Res 2020;80:5543–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Zwolinska AK, Heagle Whiting A, Beekman C, Sedivy JM, Marine JC. Suppression of Myc oncogenic activity by nucleostemin haploinsufficiency. Oncogene 2012;31:3311–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Tanaka Y, Kurasawa O, Yokota A, Klein MG, Ono K, Saito B, et al. Discovery of novel allosteric inhibitors of deoxyhypusine synthase. J Med Chem 2020;63:3215–26. [DOI] [PubMed] [Google Scholar]
- 68. Schaub FX, Reza MS, Flaveny CA, Li W, Musicant AM, Hoxha S, et al. Fluorophore-NanoLuc BRET reporters enable sensitive in vivo optical imaging and flow cytometry for monitoring tumorigenesis. Cancer Res 2015;75:5023–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Tomayko MM, Reynolds CP. Determination of subcutaneous tumor size in athymic (nude) mice. Cancer Chemother Pharmacol 1989;24:148–54. [DOI] [PubMed] [Google Scholar]
- 70. Liu J, Lichtenberg T, Hoadley KA, Poisson LM, Lazar AJ, Cherniack AD, et al. An integrated TCGA pan-cancer clinical data resource to drive high-quality survival outcome analytics. Cell 2018;173:400–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Brune V, Tiacci E, Pfeil I, Doring C, Eckerle S, van Noesel CJ, et al. Origin and pathogenesis of nodular lymphocyte-predominant Hodgkin lymphoma as revealed by global gene expression analysis. J Exp Med 2008;205:2251–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Rounbehler RJ, Fallahi M, Yang C, Steeves MA, Li W, Doherty JR, et al. Tristetraprolin impairs myc-induced lymphoma and abolishes the malignant state. Cell 2012;150:563–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Heng TS, Painter MW; Immunological Genome Project C. The immunological genome project: networks of gene expression in immune cells. Nat Immunol 2008;9:1091–4. [DOI] [PubMed] [Google Scholar]
- 74. Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 2009;25:1105–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Anders S, Pyl PT, Huber W. HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics 2015;31:166–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014;15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 2015;43:e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol 2008;26:1367–72. [DOI] [PubMed] [Google Scholar]
- 79. Welsh EA, Eschrich SA, Berglund AE, Fenstermacher DA. Iterative rank-order normalization of gene expression microarray data. BMC Bioinformatics 2013;14:153. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Tables S1, S2, S5-S13 includes Supplementary Tables S1, S2, S5-S13. Supplementary Table S1 provides a summary of BL and DHL patient demographics for the immunohistochemistry study presented in Fig. 1, E and F. Supplementary Table S2 lists the MYC-dysregulated genes whose expression is significantly altered following depletion of eIF5A or DHPS, related to Fig. 4F. Supplementary Table S5 shows DHPS GISTIC count and survival of select TCGA PanCancer datasets, related to Fig. 7J. Supplementary Table S6 shows the genetic mouse models used in this study, related to Methods. Supplementary Table S7 shows the mouse and human cell lines used in this study, related to Methods. Supplementary Table S8 lists the antibodies used in this study, related to Methods. Supplementary Table S9 lists reagents used in this study, related to Methods. Supplementary Table S10 summarizes the plasmids used in this study, related to Methods. Supplementary Table S11 lists the sequences of the oligonucleotides used in this study, related to Methods. Supplementary Table S12 lists accession numbers and publicly deposited data from this study, related to Methods. Supplementary Table S13 lists software and algorithms used in this study, related to Methods.
Supplementary Table S3 lists 665 shared differentially translated transcripts (DTT) identified following depletion of eIF5A or DHPS, related to Fig. 5C.
Supplementary Table S4 lists oncogenes and tumor suppressor genes analyzed for changes in translation efficiency (TE) following depletion of eIF5A or DHPS, related to Fig. 5E.
Supplementary Figures S1-S7 includes Supplementary Figure S1-S7 and the figure legend for each figure. Supplementary Fig. S1 shows that the polyamine-hypusine circuit is activated in many human cancers including MYC-driven lymphoma. Supplementary Fig. S1 is related to Fig. 1. Supplementary Fig. S2 shows that inhibition of DHPS enzyme activity, or silencing eIF5A or DHPS, suppresses the growth of mouse MYC-driven lymphoma. Supplementary Fig. S2 is related to Fig. 2. Supplementary Fig. S3 shows that hypusinated eIF5A (eIF5AHyp) contributes to the tumorigenic potential and maintenance of MYC-driven lymphoma. Supplementary Fig. S3 is related to Fig. 3. Supplementary Fig. S4 shows the effects of eIF5A or DHPS depletion on the transcriptional landscape of MYC-driven lymphoma. Supplementary Fig. S4 is related to Fig. 4. Supplementary Fig. S5 shows that depletion of eIF5A or DHPS impairs the translation efficiency of subsets of mRNA in MYC-driven lymphoma. Supplementary Fig. S5 is related to Fig. 5. Supplementary Fig. S6 shows the validation of select eIF5AHyp translation targets identified by the multi-omics analyses, and that the translation of key regulatory cell cycle factors is controlled by hypusinated eIF5A. Supplementary Fig. S6 is related to Fig. 6. Supplementary Fig. S7 shows that hypusinated eIF5A is essential for the development of MYC-driven lymphoma. Supplementary Fig. S7 is related to Fig. 7.
Data Availability Statement
This study did not generate any original code. Software and algorithms used in this study are listed in Supplementary Table S13. RNA-seq and Ribo-seq data sets were deposited in the GEO, GSE190669 and GSE190670, respectively. TMT multilabeling mass spectrometry data were deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the data set identifier Project accession: PXD031361 and Project DOI: 10.6019/PXD031361.