Summary:
In this issue of Blood Cancer Discovery, Nakanishi et al. uncover a critical role for the elevated activity of the translation initiation factor eIF5A in the malignant growth of MYC-driven lymphoma. eIF5A is posttranslationally modified by hypusination through MYC oncoprotein-mediated hyperactivation of the polyamine–hypusine circuit, which may represent a promising therapeutic target because an enzyme of this circuit that is required for hypusinating eIF5A proved to be essential for lymphoma development.
Aberrant expression or overexpression of MYC oncoproteins promotes cell growth and survival of a range of malignancies of different cellular origins (1). MYC-driven lymphomas, which include Burkitt lymphoma and subtypes of diffuse large B-cell lymphoma, are typically aggressive and their treatment is challenging (2, 3). Dissecting the downstream consequences of oncogenic MYC activity is a major objective in the quest to identify new Achilles’ heels for the treatment of MYC-driven tumors. Nakanishi and colleagues focused on the MYC-regulated metabolic pathway of polyamine biosynthesis, which appears to be elevated in many cancers (4). Inhibitors of this pathway exist but are deemed inefficient as single agents in cancer therapy presumably because polyamines in the diet are readily taken up by cells (5). Nakanishi and colleagues posited that a target of the polyamine pathway, specifically the translation initiation factor eIF5A that is posttranslationally modified by a process called hypusination, may represent a critical downstream mediator of oncogenic MYC activity.
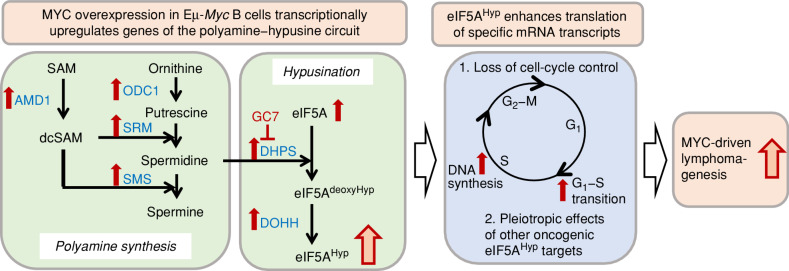
First to the main players that are at the center of this story: In the eIF5A hypusination reaction, the enzymes deoxyhypusine synthase (DHPS) and deoxyhypusine hydroxylase (DOHH) covalently modify a lysine residue in the eIF5A protein with the unique amino acid hypusine that derives from the polyamine spermidine (Fig. 1).
Figure 1.
Model: Proposed transcriptional and translational mechanisms involved in mediating MYC-driven lymphomagenesis in Eμ-Myc mice as the results of hyperactivation of the polyamine–hypusine circuit and eIF5AHyp-dependent translation of specific target mRNAs. Left, MYC overexpression in Eμ-Myc B cells transcriptionally upregulates genes encoding enzymes of the polyamine–hypusine circuit, in turn generating increased amounts of hypusinated eIF5A. Red arrows indicate the upregulation of gene expression. Hyp, hypusine; SAM, S-adenosylmethionine; dcSAM, decarboxy SAM; AMD1, adenosylmethionine decarboxylase-1; ODC1, ornithine decarboxylase-1; SRM, spermidine synthase; SMS, spermine synthase; DHPS, deoxyhypusine synthase; DOHH, deoxyhypusine hydroxylase; GC7, N1-guanyl-1, 7-diamine-heptane. GC7 is an inhibitor of DHPS. Right, Hypusinated eIF5A enhances translation of mRNA transcripts with roles in the cell cycle and known oncogenic effects in cancer. Red arrows indicate faster progression through the G1–S transition and DNA replication. Nakanishi and colleagues propose that these mechanisms substantially contribute to MYC-driven lymphomagenesis observed in Eμ-Myc mice, in addition to other MYC-controlled (eIF5AHyp-independent) biological programs.
Alerted by their earlier findings that several polyamine biosynthetic genes are MYC targets, Nakanishi and colleagues performed a meta-analysis to determine genetic alterations and expression profiles of genes involved in the polyamine-hypusine circuit in a range of cancers. The results suggested that EIF5A, DOHH, and to a lesser extent, DHPS are overexpressed in many cancer types. Most notably, they found that EIF5A and DHPS expression is highly elevated in MYC-overexpressing Burkitt lymphoma. This finding was corroborated by experimental evidence from an MYC-on/off system demonstrating that MYC induction caused marked increases in the expression levels of EIF5A, DHPS, and DOHH in addition to genes encoding polyamine biosynthetic enzymes. The same observations were made in precancerous B cells and lymphomas derived from Eμ-Myc transgenic mice, an informative animal model for mechanistic studies of MYC-driven B-cell malignant transformation (6).
Nakanishi and colleagues then used Eμ-Myc mice and cell lines established from lymphomas of those mice, as well as human Burkitt lymphoma cell lines, to test their prediction that ablation of the polyamine–hypusine circuit—leading to impaired hypusination of eIF5A—affects the growth of tumor cells with MYC overexpression. The observed growth inhibition of Burkitt lymphoma and Eμ-Myc lymphoma cell lines upon using a competitive inhibitor of DHPS, the spermidine analogue GC7, was confirmed by elegant genetic experiments in which DHPS or eIF5A expression was ablated by short hairpin (sh)RNAs in an inducible fashion. A rescue experiment in which vectors expressing either wild-type or mutant eIF5A that cannot be hypusinated were introduced into eIF5A knockdown lymphoma cells conclusively demonstrated that lymphoma growth was specifically linked to the hypusination of eIF5A. eIF5A hypusination contributed to the growth of Eμ-Myc lymphoma cell lines transplanted into recipient mice, as demonstrated by shRNA-mediated ablation of eIF5A and GC7-mediated inhibition of DHPS in mice bearing Eμ-Myc lymphomas.
Nakanishi and colleagues doubled down on these promising findings by showing with a suitable transgenic animal model that DHPS is required for the maintenance of MYC-driven lymphomas in vivo. Lymphomas were generated in Eμ-Myc mice carrying two loxP-flanked Dhps alleles, allowing Cre recombinase-mediated inducible deletion of Dhps in the cells. Upon subcutaneous injection of the lymphoma cells into Nude mice, homozygous Dhps deletion markedly impaired tumor progression compared with the uninduced controls. Overall, the dramatic phenotypes observed upon genetic and pharmacologic inhibition of the polyamine–hypusine circuit in MYC-driven lymphomas raised the question: What are the cellular pathways controlled by eIF5AHyp that are essential for MYC-induced lymphoma growth? Nakanishi and colleagues addressed this question by determining how the ablation of eIF5AHyp or DHPS in lymphoma cells affects the transcriptome, proteome, and translational landscape.
The transcriptomic analysis revealed that interfering with the polyamine–hypusine circuit resulted in a compensatory upregulation of genes acting in this pathway, further underscoring the functional importance of this circuit in MYC-driven lymphomas. Moreover, a focused analysis of the effects of eIF5A or DHPS depletion on the MYC-controlled transcriptional program uncovered major disruptions of this program. Most notably, proliferation-associated genes were downregulated, providing a mechanistic link to the tumor-cell growth inhibition observed upon gene ablation.
Two parallel experimental strategies, comparing eIF5A- or DHPS-depleted versus wild-type control cells, identified putative eIF5A-dependent translation targets with roles in the lymphoma growth phenotype: ribosome profiling combined with RNA sequencing, and tandem mass tag multilabeling mass spectrometry, which can assess effects on steady-state protein levels. Integration of these approaches demonstrated that hypusination of eIF5A affected the translation efficiency of a subset of mRNA transcripts. eIF5AHyp-dependent translation targets included genes involved in the progression of the cell cycle and many known cancer-promoting genes. Once more, elegant rescue experiments could verify the roles of eIF5AHyp-dependent targets critically involved in cell-cycle control. Similar findings in independent experimental systems further supported Nakanishi and colleagues’ conclusion that eIF5AHyp drives cell-cycle progression in MYC-driven lymphomas partly by controlling the G1–S transition and DNA replication (Fig. 1).
Finally, Nakanishi and colleagues asked whether hypusination of eIF5A is essential in Eμ-Myc mice not only for lymphoma maintenance but also for the progression of the precancerous MYC-expressing B cells to frank lymphoma. To address this question, they generated Eμ-Myc mice with one or two loxP-flanked Dhps alleles and a CD19-Cre allele to achieve B cell–specific deletion of Dhps. Intriguingly, compared with the 100% lymphoma incidence in Eμ-Myc control mice, the rare lymphomas that could be detected in a small fraction of Eμ-Myc mice with two floxed Dhps alleles all lacked deletion of the Dhps gene due to silencing of Cre expression, indicating that the precancerous B cells that progressed to lymphomas were Cre escapees. This finding demonstrates that, by facilitating hypusination of eIF5A, Dhps is required for the development of MYC-driven lymphomas, at least in this transgenic model. Moreover, a series of well-conceived experiments suggested that Dhps deletion impaired the progression of precancerous B cells to form a malignant tumor rather than abolish a particular B-cell developmental stage, which could potentially result in the depletion of the tumor-precursor cell population.
Surprisingly, Eμ-Myc mice with heterozygous deletion of Dhps, which developed lymphomas with 100% penetrance, appeared to have an accelerated disease onset and a markedly distorted splenic architecture compared with the DHPS-proficient Eμ-Myc mice. Support for this notion comes from an independent study in which constitutional heterozygous deletion of Dhps showed marked upregulation of Dhps, Dohh, and eIF5a mRNA levels compared with wild-type animals (7). This phenotype was suggested to be linked to a compensatory feedback mechanism leading to the upregulation of the polyamine–hypusine circuit, which would protect against the loss of physiologic levels of DHPS activity (7).
Although the focus of this study is on lymphoma, an MYC-driven enhancement of the polyamine–hypusine circuit is likely to be involved in other human malignancies. An in silico analysis of genomic data from cases published in The Cancer Genome Atlas suggested that only very few patients show deep deletion of DHPS (possibly biallelic deletion), and these patients were noted to have improved survival. Interestingly, cases with shallow deletion of DHPS (possibly reflecting monoallelic deletion) were associated with unfavorable outcomes in several tumor types, seemingly recapitulating the aggravated disease phenotype observed in Eμ-Myc mice with heterozygous deletion of Dhps. These initial findings provide a strong rationale for exploring the potential role of an MYC-driven polyamine–hypusine circuit in different cancer types. The work by Nakanishi and colleagues constitutes both a conceptual framework and an exemplary blueprint regarding the experimental methodologies for such studies.
Overall, Nakanishi and colleagues propose a model in which elevated eIF5AHyp levels, facilitated by enhanced DHPS and DOHH action in MYC-driven lymphoma, drive the transition of the precancerous tumor-precursor cells to the malignant state and sustain the high anabolic requirements of the lymphoma cells (4). This model contrasts with the findings made in a previous work that similarly aimed at understanding MYC-driven lymphomagenesis using an Eμ-Myc hematopoietic stem and progenitor cell (HSPC) transplant model (8). In this study, shRNA-mediated knockdown of several polyamine–hypusine circuit genes, including Dhps, promoted lymphomagenesis of premalignant HSPCs from Eμ-Myc mice, suggesting a tumor suppressor role rather than an oncogenic function for the polyamine–hypusine circuit in disease progression of MYC-driven lymphoma (8). Nakanishi and colleagues discuss two possibilities that could explain the discrepant observations. First, the polyamine–hypusine circuit may have distinct developmental stage-specific roles in the different tumor-precursor cells, representing B cells versus HSPCs. The second explanation relates to their observation that heterozygous deletion of Dhps in an Eμ-Myc genetic background appears to cause a more aggressive disease phenotype compared with DHPS-proficient Eμ-Myc mice. In this scenario, if shRNA-mediated knockdown of DHPS expression resulted in incomplete ablation of the enzyme, compensatory feedback mechanisms may enhance the activity of the polyamine–hypusine circuit and promote disease progression. Clearly, it will be important to resolve these discrepancies. At the very least, the contradictory findings and the corresponding alternative interpretations proposed in these works teach us the lesson that any future studies into the role of the polyamine–hypusine circuit in MYC-driven tumorigenesis must consider the possibility of cell type and context-dependent biological activities of the circuit. In this regard, Nakanishi and colleagues point out critical issues to be clarified: What makes an mRNA a target for eIF5AHyp to be translated efficiently, and what determines the apparent cell-context specificity of eIF5AHyp-dependent targets.
How about the translational potential of the new findings? The remarkable effects on the inhibition of lymphoma growth observed upon deletion of Dhps in Eμ-Myc mice raise hopes that more effective treatments against human MYC-driven lymphomas are on the horizon. Future work will need to find out to what extent the malignant growth of those lymphomas relies on the enhancement of the polyamine–hypusine circuit and eIF5A hypusination. Also, because the circuit is highly conserved, eIF5AHyp is likely to have essential functions associated with the activity of MYC—and likely not only MYC—in the physiology of healthy body cells, which implies that inhibiting the circuit may be toxic. Indeed, induced Cre-mediated homozygous deletion of Dhps in adult mice results in lethality (7). Moreover, it is paramount to elucidate the role of the polyamine–hypusine circuit in MYC-dependent B-cell activation and the germinal center reaction that establish humoral immunity and include the putative cells of origin of human MYC-driven lymphomas (3, 9). Overall, it will be crucial to determine the consequences of inhibiting the polyamine–hypusine circuit on the physiology of healthy cells, because this may predict potential systemic side effects of inhibitory drugs.
A challenge for strategies aimed at pharmacologically targeting DHPS is the aforementioned observation that, whereas complete deletion of Dhps in B cells abolished lymphoma development in Eμ-Myc mice, heterozygous deletion of Dhps was associated with a more severe disease phenotype. Because DHPS depletion in healthy cells results in lethality, for any pharmacologic strategies with candidate DHPS inhibitors (such as bromobenzothiophene; ref. 10), it will be critical to identify the therapeutic window in which DHPS inhibition impairs lymphoma progression while retaining sufficient functional enzyme to satisfy physiologic demands in healthy cells without risking lymphoma progression through compensatory feedback mechanisms. An alternative strategy would involve agents that specifically block eIF5AHyp. Moreover, the dissection of the eIF5AHyp-dependent translation targets that mediate pleiotropic oncogenic effects may identify targetable oncogenic pathways. Although the road may yet be long and winding, the intriguing results by Nakanishi and colleagues provide all the right reasons to fully explore the role of the polyamine–hypusine circuit in MYC-driven cancers, as this may uncover previously unanticipated tumor-cell vulnerabilities.
Acknowledgments
E.B. Wilson and U. Klein are supported by Blood Cancer UK and the Kay Kendall Leukaemia Fund.
Authors’ Disclosures
No disclosures were reported.
References
- 1. Dhanasekaran R, Deutzmann A, Mahauad-Fernandez WD, Hansen AS, Gouw AM, Felsher DW. The MYC oncogene: the grand orchestrator of cancer growth and immune invasion. Nat Rev Clin Oncol 2022;19:23–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Roschewski M, Staudt LM, Wilson WH. Burkitt's lymphoma. N Engl J Med 2022;387:1111–22. [DOI] [PubMed] [Google Scholar]
- 3. Ennishi ED, His ED, Steidl C, Scott D. Toward a new molecular taxonomy of diffuse large B-cell lymphoma. Cancer Discov 2020;10:1267–81. [DOI] [PubMed] [Google Scholar]
- 4. Nakanishi S, Li J, Berglund AE, Kim Y, Zhang Y, Zhang L, et al. The polyamine-hypusine circuit controls an oncogenic translational program essential for malignant conversion in MYC-driven lymphoma. Blood Cancer Discov 2023;4:294–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Miller-Fleming L, Olin-Sandoval V, Campbell K, Ralser M. Remaining mysteries of molecular biology: the role of polyamines in the cell. J Mol Biol 2015;427:3389–406. [DOI] [PubMed] [Google Scholar]
- 6. Adams JM, Harris AW, Pinkert CA, Corcoran LM, Alexander WS, Cory S, et al. The cmyc oncogene driven by immunoglobulin enhancers induces lymphoid malignancy in transgenic mice. Nature 1985;318:533–8. [DOI] [PubMed] [Google Scholar]
- 7. Pällmann N, Braig M, Sievert H, Preukschas M, Hermans-Borgmeyer I, Schweizer M, et al. Biological relevance and therapeutic potential of the hypusine modification system. J Biol Chem 2015;290:18343–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Scuoppo C, Miething C, Lindqvist L, Reyes J, Ruse C, Appelmann I, et al. A tumour suppressor network relying on the polyamine-hypusine axis. Nature 2012;487:244–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dominguez-Sola D, Victora GD, Ying CY, Phan RT, Saito M, Nussenzweig MC, et al. The proto-oncogene MYC is required for selection in the germinal center and cyclic reentry. Nat Immunol 2012;13:1083–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Tanaka Y, Kurasawa O, Yokota A, Klein MG, Ono K, Saito B, et al. Discovery of novel allosteric inhibitors of deoxyhypusine synthase. J Med Chem 2020;63:3215–26. [DOI] [PubMed] [Google Scholar]