Abstract
Cancer remains elusive in many aspects, especially in its causes and control. After protein profiling, genetic screening, and mutation studies, scientists now have turned their attention to epigenetic modulation. This new arena has brought to light the world of noncoding RNA (ncRNA). Although very complicated and often confusing, ncRNA domains are now among the most attractive molecular markers for epigenetic control of cancer. Long ncRNA and microRNA (miRNA) have been studied best among the noncoding genome and huge data have accumulated regarding their inhibitory and promoting effects in cancer. Another sector of ncRNAs is the world of PIWI-interacting RNAs (piRNAs). Initially discovered with the asymmetric division of germline stem cells in the Drosophila ovary, piRNAs have a unique capability to associate with mammalian proteins analogous to P-element induced wimpy testis (PIWI) in Drosophila and are capable of silencing transposons. After a brief introduction to its discovery timelines, the present narrative review covers the biogenesis, function, and role of piRNAs in lung cancer. The effects on lung cancer are highlighted under sections of cell proliferation, stemness maintenance, metastasis, and overall survival, and the review concludes with a discussion of recent discoveries of another class of small ncRNAs, the piRNA-like RNAs (piR-Ls).
Keywords: PIWI protein, Drosophila, RNA, small interfering, Argonaute proteins, lung neoplasms, epigenetic repression
Lung cancer causes the highest mortality in cancer-related deaths worldwide [1]. One of the major causes of lung cancer is tobacco smoking, which may be associated with socioeconomic status [2, 3]. While raising awareness is necessary to control the growing incidence of lung cancer, understanding the cellular networks and discovering new targets for therapies are also important. In recent trends, scientists have examined the pool of noncoding RNAs (ncRNAs) and their specific roles in epigenetic regulation. So far, the most studied ncRNAs are microRNAs (miRNAs) and their regulatory roles in various cancers, including lung cancer, have been established [4].
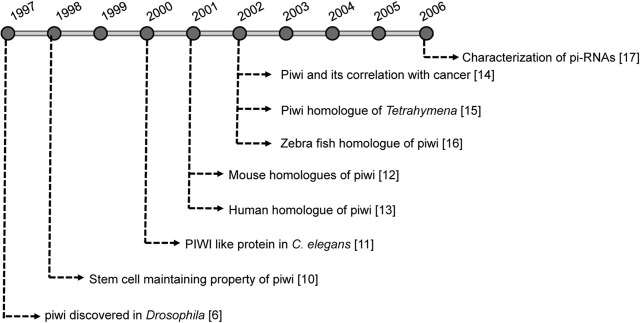
P-element induced wimpy testis (PIWI) proteins belong to the Argonaute family of proteins [5], which were first discovered in Drosophila melanogaster ovarian germ cells and follicular cells [6]. By interacting with Tudor domain-containing proteins (TDRDs), PIWI proteins mediate the biogenesis of PIWI-interacting RNAs (piRNAs), thereby silencing transposons [7]. Three types of PIWI subfamily proteins are found in Drosophila—piwi, which localizes in the nucleus of germ and gonadal somatic cells, Aubergine (aub), and Archipelago 3 or Argonaute 3 (ago3), which are both expressed in nuages, or germline granules, which are special cytoplasmic compartments in Drosophila melanogaster. All these proteins are involved in the biogenesis of piRNAs in Drosophila flies [8]. PIWI proteins use piRNAs as their guide to the specific DNA sequences for transposon silencing and gene regulation as well as playing major roles in their biogenesis [9]. Orthologs of Drosophila piwi proteins have been found in mice, zebrafish, C. elegans, and humans, in which they have similar roles in maintaining male and female fertility (Figure 1). In the human, genome 4 piwi orthologs (PIWIL1/HIWI, PIWIL2/HILI, PIWIL3, and PIWIL4/HIWI2) have been identified [5]. The first evidence of human PIWI association with cancer was studied in seminomas [14]. Aberrant expression of 1 or more of these 4 proteins is common, especially in breast, prostate, and colorectal carcinomas (Table 1). Limited findings have emerged in the case of lung cancer [4, 60,61,62]. Various studies have found interesting patterns of human PIWI-like protein expression and regulatory effects in lung cancers [63]. Human PIWIs and piRNAs may be important biomarkers and therapeutic targets for various cancers [64, 65].
Figure 1.
Early discoveries related to PIWI proteins and piRNA. piRNA, PIWI-interacting RNA; PIWI, P-element induced wimpy testis protein.
Table 1.
Important discoveries of PIWI/piRNA expression in various cancers†
| Year | PIWI/piRNA | Expression | Cancer type | Role |
|---|---|---|---|---|
| 2005 | PIWIL2 | Upregulated | Testicular seminoma | Inhibition of apoptosis and promotion of proliferation via Stat3/Bcl-XL signaling pathway [18] |
| 2006 | PIWIL1 | Upregulated | Human gastric cancer | Cell proliferation [19] |
| 2007 | PIWIL1 | Upregulated | Soft tissue sarcoma | Stem cell proliferation [20] |
| 2008 | PIWIL1 | Up/down-regulated | Adenocarcinoma | Poor prognosis and death [21] |
| 2009 | PIWIL1 | Presence in cytoplasm | Esophageal squamous cell carcinoma | Poor prognosis [22] |
| 2010 | PIWIL2 | Varied | Cervical neoplasia | Biomarker [23] |
| 2010 | PIWIL2 | Upregulated | Human breast cancers | Biomarker [24] |
| 2011 | PIWIL1 | Upregulated | Glioma | Tumor progression, poor outcome, and biomarker [25] |
| 2011 | piR-651 | Upregulated | Gastric, colon, lung, and breast cancer | Increases cell proliferation [26] |
| 2011 | PIWIL1 | Upregulated | Colorectal cancer | Leads to poor overall survival, biomarker [27] |
| 2012 | piR-823 | Downregulated | Gastric cancer | Increases cell proliferation [28] |
| 2012 | PIWIL2 | Upregulated | Colon cancer | Metastasis [29] |
| 2013 | PIWI | Upregulated | Stage III epithelial ovarian cancer | Promotes metastasis, biomarker [30] |
| 2013 | piR-932 | Upregulated | Breast cancer | Positive regulator of breast cancer stem cells [31] |
| 2014 | PIWIL1 | Upregulated | Human breast cancer | Cell proliferation [32] |
| 2014 | piRNA-823 | Upregulated | Multiple myeloma | Regulates angiogenesis [33] |
| 2014 | PIWIL1 and PIWIL4 | Varied | Renal cell carcinoma | Related to clinicopathological parameters [34] |
| 2014 | PIWIL1 | Upregulated | Cervical cancer | Promotes chemoresistance [35] |
| 2014 | PIWIL1 | Upregulated | Hepatocellular carcinoma | Reduces proliferation and migration [36] |
| 2015 | piRNA-DQ594040 | Downregulated | Bladder cancer | Promotes cell proliferation, colony formation and functions against apoptosis [37] |
| 2015 | piR-021285 | Upregulated | Breast cancer | Epigenetic remodeling [38] |
| 2015 | PIWIL2 | Upregulated | Prostate cancer | Metastasis [39] |
| 2015 | piR-017061 | Downregulated | Pancreatic cancer | Associated with diseased condition [40] |
| 2015 | piR-015551 | Downregulated | Colorectal cancer | Associated with long ncRNA expression [41] |
| 2015 | PIWIL1 | Downregulated | Chronic myeloid leukemia | Induces growth and metastasis [42] |
| 2015 | PIWIL2 | Upregulated | Cholangiocarcinoma | Involved in shorter survival span and metastasis [43] |
| 2015 | piR-57125 | Downregulated | Renal cell carcinoma | Associated with tumor recurrence and metastasis, prognostic biomarker [44] |
| piR-30924 | Upregulated | |||
| piR-38756 | Upregulated | |||
| 2015 | PIWIL1 | Upregulated | Type 1 endometrial cancer | Tumor progression by downregulating PTEN [45] |
| 2015 | PIWIL2 | Downregulated | Bladder cancer | Related to disease specific and progression free survival [46] |
| 2016 | PIWIL4 | Upregulated | Breast cancer | Metastasis, antiapoptotic activity and proliferation[47] |
| 2016 | FR140858 | Differentially expressed | Head and neck squamous cell carcinoma | Correlated to human papillomavirus infection [48] |
| 2016 | piR-598 | Upregulated | Glioma | Induces growth and proliferation [49] |
| 2017 | PIWIL3 | Upregulated | Melanoma | Induces metastasis [50] |
| 2017 | PIWIL4 | Upregulated | Retinoblastoma | Induces proliferation [51] |
| 2018 | piR-5937 | Varied | Colon cancer | Biomarker [52] |
| piR-28876 | ||||
| 2018 | piR-8041 | Downregulated | Glioblastoma | Related to tumor growth [53] |
| 2018 | PIWIL1 | Upregulated | Gastric cancer | Metastasis [54] |
| 2018 | PIWIL1 and PIWIL2 | Varied | Muscle invasive urothelial bladder cancer | Correlation with clinical factors, biomarkers [55] |
| 2018 | PIWIL4 | Upregulated | Human breast cancer | Cell motility [56] |
| 2019 | piR-823 | Upregulated | Multiple myeloma | Tumor progression [57] |
| 2019 | piR-39980 | Upregulated | Neuroblastoma | Tumor progression and drug resistance [58] |
| 2019 | piR-36712 | Downregulated | Breast cancer | Chemoresistance and tumor progression [59] |
| 2020 | piR-004987 | Upregulated | Lung cancer | Correlated with lung cancer from sputum as compared with normal [60] |
| piR-020809 | Upregulated | |||
| piR-023338 | Downregulated | |||
| piR-011186 | Downregulated | |||
| 2021 | piR-hsa-211106 | Downregulated | Lung cancer | Chemoresistance and tumor progression [61] |
| 2021 | piR-1008 | Upregulated | Lung cancer | |
| piR-28231 | Upregulated | Lung cancer | ||
| piR-11256 | Upregulated | Lung cancer | ||
| piR-30636 | Upregulated | Lung cancer | ||
| piR-24143 | Upregulated | Lung cancer | Biomarker lung cancer [62] | |
| piR-6842 | Upregulated | Lung cancer | ||
| piR-8757 | Upregulated | Lung cancer | ||
| piR-15572 | Upregulated | Lung cancer | ||
| piR-5444 | Upregulated | Lung cancer and serum exosome | ||
| piR-26925 | Upregulated |
Lung cancer until 2021 and other cancers until 2019. PIWI, PIWI protein (human); piRNA, Piwi protein-interacting RNA; PIWIL1, PIWI-like 1 protein (human) or HIWI protein (human); PIWIL2, HILI protein (human); PIWIL3, HIWI3 protein (human); and PIWIL4, HIWI2 protein (human); ncRNA, non-coding RNA.
The present narrative review aims to introduce the details of piRNA biogenesis and function and then detail the various discoveries as a timeline of events. The review aims to summarize the emerging roles of a relatively new group of sncRNAs, piRNAs, and their interacting PIWI family proteins in lung cancer. Finally, a correlation between the hallmarks of cancer and the piRNA/PIWI family of proteins has been attempted.
Search methodology
We used PubMed (MEDLINE inclusive), Google Scholar, Web of Science, and Scopus as principal online databases. To explore the literature related to PIWI RNA, the keyword “PIWI RNA” was used. To elaborate on its role in cancer the keywords “PIWI RNA” and “cancer” were used. Keywords “PIWI RNA” and “lung cancer” were used to narrow the specification to lung neoplasms. The genesis and logic for separation of dates to demark “PIWI RNA” and “cancer” and “PIWI RNA” and “lung cancer” as detailed in Table 1 was lung cancer until end 2021 and other cancers until 2019. This delimited the information and references related to PIWI RNA and cancers other than lung cancer to 2019. Preference was given to citation of references published in the past 5 years. The reference lists of identified articles were further examined for relevant publications. We have arranged our gathered information using the subtitles indicated in the review and further cross-verified each subheading content with appropriate keywords. We have assembled information for readers for ready reference and have added our personal comments summarizing information acquired along with an attempt to collate piRNA with the basics of cancer biology.
Biogenesis of piRNA
The pathway for piRNA generation is rather complicated and largely uncharted. However, studies conducted so far have confirmed 2 distinct pathways operate in the case of germline and gonadal somatic cells of Drosophila. Precursors of piRNAs are mainly transcribed from gene clusters flamenco and traffic jam. Flamenco is one of the major clusters for piRNA biogenesis in the somatic support cells of the Drosophila ovary and produces piRNA precursors. The cluster resides in the perichromatin region of the X chromosome in Drosophila. An approximately 180 kb stretch of the cluster transcribes into nascent piRNAs, which typically span about 150 kb. The transcripts are generated from a single strand of the DNA by unidirectional transcription orientation in the antisense direction. Partial or an entire loss of flamenco in Drosophila leads to malfunctioning in transposable element (TE) management [66, 67]. A similar, piRNA-producing locus in chromosome 2 in Drosophila is called traffic jam, whose primary function was identified as a crucial factor in gonadal morphogenesis in these flies. Loss of traffic jam leads to blockade in the differentiation of somatic cells into germ cells and ultimately the formation of follicular cells in Drosophila ovaries [68]. Although these 2 clusters are the widely studied and important sources of piRNA, other sources such as intergenic regions and transposons are also noted [69]. In these piRNA regions in the DNA, trimethylated lysine 9 of histone 3 (H3K9Me3) marks are abundant and contribute to piRNA expression [70]. In germ cells, piRNA clusters, to be transcribed, require a protein complex consisting of Rhino, Deadlock, and Cutoff (RDC) proteins [71]. TREX is another complex needed for the dual stranded cluster transcription recruited to the DNA in RDC dependent manner [8]. The length of mature piRNAs varies from 24 to 32 nucleotides [5].
Zucchini mediated pathway
In the gonadal somatic cells and follicular cells, after transcription, the precursors of piRNAs are transported from the nucleus to the Yb body (containing Piwi, Armitage (Armi), Tudor, Vreteno (Vret), RNA helicase Sister of Yb (SoYb)) in the cytosol [72]. Then premature piRNAs are processed at the 5′ end by a mitochondrial membrane endonuclease called Zucchini (Zuc) [73]. Subsequently, they are loaded onto Piwi by Shutdown and Hsp83 and their 3′ end is trimmed by a slicer enzyme [74]. Piwi has a bias for 5′ U. Aub and Ago3 proteins are not used in this pathway. This processed piRNA is further trimmed by a protein called Nibbler [75]. Ultimately, it is methylated at the 2′ O position by a methyltransferase Hen1 (HENMT1 in mice) [76]. This last step is believed to increase the stability of the piRNA. Processed and mature piRNAs are then transferred back to the nucleus.
Ping-pong pathway
The secondary amplification of piRNAs occurs in germ cells with the help of AUB and AGO3 proteins via the ping-pong pathway, which also leads to post-transcriptional gene silencing (PTGS). The ping-pong pathway starts with the loading of nascent piRNAs, transcribed from the clusters to Aub. Aub shows a 5′U bias for piRNAs and Ago3 shows a bias for adenine at the 10th position. The 5′ end is loaded on Aub with the help of the Shu and Hsp83 and then trimmed and methylated as described above for the Zuc mediated pathway [74]. piRNAs loaded onto Aub are antisense to specific TE mRNA and they eventually guide Aub to their complementary TE mRNA in the cytosol for targeted destruction. This process also generates the 5′ piRNA precursor. The primary piRNA precursor is loaded onto Ago3 and processed into secondary piRNA precursors. The secondary piRNA precursor is sense to TE and antisense to unprocessed piRNA; accordingly they cleave the newly attached piRNA precursors and continue the loop of transposon silencing and mature piRNA production [77].
Function
Transposon silencing
piRNAs save the germ cells from mutations by TE. The mechanism of piRNA biogenesis in Drosophila and many other species is itself a process of degrading TE at the post-transcriptional level. Mutations in Aub and Ago3 lead to an elevated level of transposons in the germ cells [78]. As discussed earlier for the ping-pong pathway, a piRNA targets a transposon with opposite orientation and degrades it into a new piRNA. Together these processes serve to slice TE and amplify piRNA. The transposon-containing regions of the DNA generate piRNAs, which in turn upon maturation, recruit Piwi and other proteins to silence the TE region [79, 80]. Other species, such as mice and zebrafish, also show a similar function for their PIWI family proteins. Knocking down of Piwi orthologs increased the abundance of transposons in these species [81].
piRNA mediated epigenetic regulation and transcriptional silencing
Piwi and Aub conduct position effect variegation in which a stretch of euchromatin is converted to heterochromatin to variable extents among cells within the same tissue [82]. Piwi interacts with heterochromatin protein 1a (HP1a) to promote heterochromatin formation [83]. It is shown by studies in Drosophila that Piwi helps in loading the H3K9Me3 mark on DNA and converts stretches of DNA into heterochromatin. So, the loss of Piwi makes TE available for pol II [84]. In mice, Mili and Miwi2 (mouse orthologs of PIWI) together promote retrotransposon silencing by CpG DNA methylation in male germ-line cells and piRNA alone can also regulate DNA methylation in these germ cells [81, 85].
Reproduction and development
PIWI proteins have stem cell maintaining properties for which their role in germline development and maintenance is well observed. Knockdown studies have shown anomalies in development. For instance, in Drosophila both male and female piwi protein-coding gene mutants fail to form primordial germ cells and renew germline stem cells, which leads to sterility [67]. Aub protein-coding gene mutation leads to compromised PTGS and DNA damage accumulation also resulting in sterility [86]. Females with mutant ago3 lay fewer eggs and most of the time are sterile [87]. Similar results were also obtained from studies on mice. PIWI orthologs, Mili, Miwi2, and Miwi mutant mice show defects in PTGS and spermatogenesis [88]. Zebrafish PIWI-like proteins Zili and Ziwi mutations decrease the number of germ cells and increase apoptosis of germ cells respectively [89]. Mutations in other piRNA biogenesis proteins such as Tudor, Vret, and Tej also result in DNA damage and defects in development [74, 79]. piRNA clusters from the chromosomes of Drosophila germ cells play a crucial role in maintaining the integrity of the telomeric regions. Loss of piRNAs in germ cells results in decreased levels of HP1a, Rhino and H3K9Me3 association with the telomeric region and disputed nuclear positioning of the telomere [90]. In silkworms and other insects, PIWI has a role in sex determination. Loss of Bmsiwi (insect PIWI) and histone methyltransferase BmAsh2 causes female-to-male sexual reversal [91].
Translational regulation by PIWI–piRNA pathway
piRNAs can be formed from the 3′UTR region of many protein-coding genes such as vas, traffic jam, and nanos and control the mRNA turnover and protein level expression of these genes. Drosophila flies with piwi mutant flies produce an excess of these gene products, which accumulate in the cell eventually damaging the DNA [70, 92, 93]. Mouse PIWIs interact with eIF4e, which forms a cluster of proteins to control translation [94].
Somatic functions
Although very little evidence has been found so far, studies are suggesting a greater somatic function of the PIWI/piRNA pathway. In the early stages of embryogenesis, Drosophila piwi is needed for chromatin structure maintenance and cell cycle progression [95]. piwi is also found on chromosomes of the salivary gland that mediate epigenetic regulation [96]. Loss of piwi proteins in Drosophila intestinal stem cells impairs the gut regenerative capacity of these flies [97]. In Aplysia, piRNA-mediated DNA methylation is needed for neuronal plasticity [98]. In various arthropods, the PIWI-piRNA pathway may perform as an antiviral defense mechanism in mosquitoes and silkworms [99]. In a rat model of diabetes, activities of pancreatic β-cells may be regulated by piRNAs. β-Cells express thousands of piRNAs and their expression changed when rat Piwi proteins were downregulated, resulting in defective insulin secretion [100]. Besides all these major and minor functions of the PIWI–piRNA pathway in various animals, aberrant expression in human cancers has been reported (Table 1), which will be discussed in detail in the following sections.
Role of PIWI–piRNA in lung cancer
Cell proliferation
Induced expression of PIWIL2 in A549 cells results in increased cell proliferation by elevated expression of CDK2 and cyclin A, both in vitro and in vivo. Similar results are obtained from H460 cells in vitro. By contrast, RNAi-mediated depletion of the protein results in cell cycle arrest (G2/M) and increased apoptosis [101]. An increase in PIWIL1 activity induces cell proliferation in A549 cells and decreases in H1299 cell proliferation when knocked down and increases the colony-forming capacity of tumor cells [63, 102].
A positive correlation of cell proliferation with aberrant expression of piRNAs can be postulated. piR-651 is one example of such piRNAs whose expression is altered significantly in many cancers including in lung cancer patient samples and cancer cell lines such as NCIH446 and 95-D [103]. piR-651 maintains the cell population by keeping proapoptotic proteins in check [103]. Inhibition of piR-651 decreases cell proliferation and increases apoptosis in A549 and HCC827 cells. piR-651 negatively regulates proapoptotic proteins while increasing the activities of antiapoptotic proteins [104]. piR-651 helps cyclin D1 and CDK-4 overexpression and upregulates proliferation of transfected A549 cells both in vitro and in vivo [105]. The expression of RASSF1C has been shown to regulate certain piRNA expression and cancer progression. Upregulation of this oncoprotein correlated with overexpression of piR-52200 and underexpression of piR-35127 inpatient samples while piR-34871 and piR-46545 were additionally up- and downregulated respectively in the non-small-cell lung cancer (NSCLC) cell line H1299 along with the previous 2 piRNAs. In vitro, overexpressing underexpressed and knocking down overexpressed piRNAs decreases cell proliferation. Specifically, knock down of piR-52200 in A549 cells, piR-34871 in HT520 cells decreases cell proliferation significantly. By contrast, H1299 responded in most knockdown and overexpression studies. A low level of colony formation was observed in normal lung tissues after manipulation of piRNA expression [106]. piR-55490 acted as an anticancer agent in vitro and in xenograft studies. In lung cancer cell lines such as A549, H460, and H1299, piR-55490 expression was originally suppressed and upon overexpression, these cell lines showed decreased proliferation. It is postulated that piR-55490 binds to mTOR and degrades it decreasing tumor cell proliferation [107]. Two piRNAs overlapping in the 15th chromosome and sharing a common single nucleotide polymorphism, rs11639347, piR-5247, and piR-5671, increase proliferation of A549 cells [108].
Stemness maintenance
Human PIWI proteins are now proven to maintain the stemness of certain cell populations when present in testis. Hiwi inhibition resulted in the loss of ALDH-1 (cancer cell marker) positive cells and decreased tumor mass in immunocompromised mice when injecting SSClo Aldebr stem cells isolated from an SPC-A1 cell line [109]. Overexpression of RASSF1C promotes CD133+ (stem cell marker) A549 cell tumor sphere formation. RASSF1C induces PIWIL1 expression, which maintains stem cell properties and regulates the wnt/β-catenin pathway. Coexpression of RASSF1C and IGFBP-5 reduces PIWIL1 expression [110].
Metastasis
The interplay between PIWIs and piRNAs aids more than one hallmark of cancer. Inhibition of piR-651 decreases migration of highly invasive cell lines 95-D, A549, and HCC827 cells [106, 107]. Inhibition of PIWIL1 interferes with metastatic activity in H1299 cells, while increased expression induces A549 cell migration [102].
Overall survival
Human case study databases like The Cancer Genome Atlas (TCGA) show a positive correlation between PIWIL1 expression and poor overall survival of patients [102]. Patient samples show similar results, increased PIWIL1 correlating to shorter time to relapse (TTR) and shorter overall survival. Whereas, decreased PIWIL4 correlated with shorter TTR and less overall survival [102]. Patients with higher piR-55490 expression have longer overall survival [107].
piRNA-like short noncoding RNA
Studies in NSCLC and lung squamous cell carcinoma (LSCC) have revealed another class of sncRNAs, the piRNA-Like RNAs (piR-Ls). These RNAs have similar as well as distinguishing features to piRNAs. Two variants have been discovered to date, piR-L-138 and piR-L-163, and both are similar to piRNAs in length. However, 2 major differences are that, unlike piRNAs, they are expressed in adult tissues, and they bind directly to phosphorylated protein targets (p-proteins) to regulate their functional efficacy. Therefore, they are designated as protein functional effector sncRNAs (pfeRNAs). piR-L-138 expression in LSCC increases after cisplatin-based chemotherapy, which eventually leads to chemoresistance by the tumor cells. By contrast, targeting piR-L-138 in LSCC cell lines such as H157 and SKMES-1 increases apoptosis. piR-L-138 regulates p60MDM2 to control cell proliferation [111, 58]. Another study showed piR-L-163 binds to Ezrin, Radixin, and Moesin (p-ERM), which in turn increases the binding capacity of p-ERM to EBP50 and F acting. Blocking piR-L-163 induces cell growth and invasion revealing it as a negative regulator of tumor progression [112].
piRNA biomarker and chemoresistance
Cancer prognosis is related to 2 important facets namely early detection and delayed chemoresistance. In a study with 20 pairs of malignant and nonmalignant tissues, piR-hsa-211106 was downregulated in all malignant tissues as it prevented metastasis and induced apoptosis in lung cancer cells [61]. This piRNA interacts with pyruvate carboxylase, which prevents cisplatin resistivity in lung cancer cells [61].
Lin et al. [60] and Li et al. [62] demonstrated that piRNAs can also be used for diagnosis of lung cancer. Sputum from 32 lung cancer patients was used as a source of epithelial cells from the bronchus and cRNA was profiled [60]. Lung cancer patients had upregulated piR-004987 and piR-020809 expression and downregulated expression of piR-023338 and piR-011186 [60]. Li et al. [62] examined 19 lung tissues from lung cancer patients and compared their piRNA profile with noncancerous lung tissues (from different sites in the same patients). They found 10 piRNAs unregulated in cancerous tissues compared with noncancerous tissue samples (see Table 1). Of these, 2 exosomal piRNAs, namely piR-hsa-26925 and piR-hsa-5444, were found in patient sera. Exosomes are double-layered lipid extracellular vesicles containing macromolecules like nucleic acids (in this case piRNA) used for cell-to-cell communication. Thus, such exosomes identifiable from sera can act as diagnostic markers for lung cancer.
Conclusion
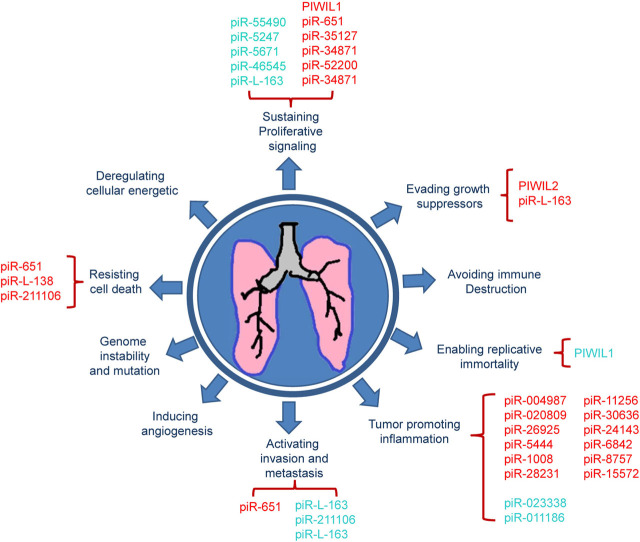
Accumulated data supports the importance of piRNAs in lung and other cancers. Presently we have emphasized lung cancer as it still reigns among all types of cancers in terms of the highest mortality in cancer-related deaths worldwide [1]. Lung cancer management, like any other cancer, revolves around both diagnoses and treatment. For both these factors, detailed molecular understanding of the disease is necessary for an effective outcome. piRNA contributes to cardinal features of cancer development, namely, cell proliferation, stemness maintenance, and metastasis; thus, also reflecting overall survival. Better understanding and clinical interpretation of these noncoding RNAs will not only aid in the understanding of the molecular perturbations, but may also provide insight into the selection of treatment modalities. More detailed screening and identification of anomalous expression of piRNAs may not only help in diagnosis, but also predict the prognosis of the disease. To collate the basics of cancer biology with the advances with knowledge of PIWI proteins or piRNAs, we merged our gathered information with the “Emerging hallmarks and enabling characteristics” as delineated by Hanahan and Weinberg [113, 105]. A diagrammatic representation of how the present finding of PIWI proteins or piRNA integrates with the hallmarks of cancer is depicted in Figure 2. Those PIWI proteins or piRNA that are positively regulated with the hallmarks may serve as a target for lung cancer therapy or as diagnostic or prognostic markers. By contrast, negatively regulated PIWI proteins or piRNA may serve as therapeutic options.
Figure 2.
Representation of how the present findings for PIWI proteins or piRNA integrate with the emerging hallmarks of cancer in the case of lung cancer. Turquoise represents positively regulated with the hallmark, while red represents negatively regulated with the hallmark. piRNA, PIWI-interacting RNA; P-element induced wimpy testis protein.
Acknowledgments
We did not receive any specific grant for this study from any funding agency in the public, commercial, or not-for-profit sectors.
Footnotes
Author contributions.
All authors contributed to the conception and design of the review. All authors contributed to the literature review, data extraction, and assessment of articles. PM and DM contributed to drafting the original manuscript and all authors critically revised it for important intellectual content. All authors approved the final version of the manuscript submitted for publication and agreed to be accountable for all aspects of the work and take responsibility for statements made in the published article.
Conflicts of interest statement.
Each of the authors has completed an ICMJE disclosure form. None of the authors declare any potential or actual relationship, activity, or interest related to the content of this article.
Data sharing statement.
The present review is based on the references cited. Further details, opinions, and interpretation are available from the corresponding authors on reasonable request.
Contributor Information
Shamee Bhattacharjee, Email: shamee.zoology@wbsu.ac.in.
Deba Prasad Mandal, Email: dpmandal.zoology@wbsu.ac.in.
References
- [1].Cai Z, Liu Q. Understanding the Global Cancer Statistics 2018: implications for cancer control. Sci China Life Sci. 2021;64:1017–20. doi: 10.1007/s11427-019-9816-1. [DOI] [PubMed] [Google Scholar]
- [2].Wong MCS, Lao XQ, Ho K-F, Goggins WB, Tse SLA. Incidence and mortality of lung cancer: global trends and association with socioeconomic status. Sci Rep. 2017;7:14300. doi: 10.1038/s41598-017-14513-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Nargis N, Yong H-H, Driezen P, Mbulo L, Zhao L, Fong GT. et al. Socioeconomic patterns of smoking cessation behavior in low and middle-income countries: emerging evidence from the Global Adult Tobacco Surveys and International Tobacco Control Surveys. PLoS One. 2019;14:e02202232019. doi: 10.1371/journal.pone.0220223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Sonea L, Buse M, Gulei D, Onaciu A, Simon I, Braicu C, Berindan-Neagoe I. Decoding the emerging patterns exhibited in non-coding RNAs characteristic of lung cancer with regard to their clinical significance. Curr Genomics. 2018;19:258–78. doi: 10.2174/1389202918666171005100124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Litwin M, Szczepańska-Buda A, Piotrowska A, Dzięgiel P, Witkiewicz W. The meaning of PIWI proteins in cancer development. Oncol Lett. 2017;13:3354–62. doi: 10.3892/ol.2017.5932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Lin H, Spradling AC. A novel group of pumilio mutations affects the asymmetric division of germline stem cells in the Drosophila ovary. Development. 1997;124:2463–76. doi: 10.1242/dev.124.12.2463. [DOI] [PubMed] [Google Scholar]
- [7].Gan B, Chen S, Liu H, Min J, Liu K. Structure and function of eTudor domain containing TDRD proteins. Crit Rev Biochem Mol Biol. 2019;54:119–32. doi: 10.1080/10409238.2019.1603199. [DOI] [PubMed] [Google Scholar]
- [8].Huang X, Tóth KF, Aravin AA. piRNA biogenesis in Drosophila melanogaster. Trends Genet. 2017;33:882–94. doi: 10.1016/j.tig.2017.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Yu T, Koppetsch BS, Pagliarani S, Johnston S, Silverstein NJ, Luban J. et al. The piRNA response to retroviral invasion of the koala genome. Cell. 2019;179:632–643.e12. doi: 10.1016/j.cell.2019.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Cox DN, Chao A, Baker J, Chang L, Qiao D, Lin H. A novel class of evolutionarily conserved genes defined by piwi are essential for stem cell self-renewal. Genes Dev. 1998;12:3715–27. doi: 10.1101/gad.12.23.3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Reinke V, Smith HE, Nance J, Wang J, Van Doren C, Begley R. et al. A global profile of germline gene expression in C. elegans. Mol Cell. 2000;6:605–16. doi: 10.1016/s1097-2765(00)00059-9. [DOI] [PubMed] [Google Scholar]
- [12].Kuramochi-Miyagawa S, Kimura T, Yomogida K, Kuroiwa A, Tadokoro Y, Fujita Y. et al. Two mouse piwi-related genes: miwi and mili. Mech Dev. 2001;108:121–33. doi: 10.1016/s0925-4773(01)00499-3. [DOI] [PubMed] [Google Scholar]
- [13].Sharma AK, Nelson MC, Brandt JE, Wessman M, Mahmud N, Weller KP, Hoffman R. Human CD34+ stem cells express the hiwi gene, a human homologue of the Drosophila gene piwi. Blood. 2001;97:426–34. doi: 10.1182/blood.v97.2.426. [DOI] [PubMed] [Google Scholar]
- [14].Qiao D, Zeeman A-M, Deng W, Looijenga LHJ, Lin H. Molecular characterization of hiwi, a human member of the piwi gene family whose overexpression is correlated to seminomas. Oncogene. 2002;21:3988–99. doi: 10.1038/sj.onc.1205505. [DOI] [PubMed] [Google Scholar]
- [15].Mochizuki K, Fine NA, Fujisawa T, Gorovsky MA. Analysis of a piwi-related gene implicates small RNAs in genome rearrangement in Tetrahymena. Cell. 2002;110:689–99. doi: 10.1016/s0092-8674(02)00909-1. [DOI] [PubMed] [Google Scholar]
- [16].Tan CH, Lee TC, Weeraratne SD, Korzh V, Lim TM, Gong Z. Ziwi, the zebrafish homologue of the Drosophila piwi: co-localization with vasa at the embryonic genital ridge and gonad-specific expression in the adults. Mech Dev. 2002;119(Suppl 1):S221–4. doi: 10.1016/s0925-4773(03)00120-5. [DOI] [PubMed] [Google Scholar]
- [17].Girard A, Sachidanandam R, Hannon GJ, Carmell MA. A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature. 2006;442(7099):199–202. doi: 10.1038/nature04917. [DOI] [PubMed] [Google Scholar]
- [18].Lee JH, Schütte D, Wulf G, Füzesi L, Radzun HJ, Schweyer S. et al. Stem-cell protein Piwil2 is widely expressed in tumors and inhibits apoptosis through activation of Stat3/Bcl-XL pathway. Human Mol Genet. 2005;15:201–11. doi: 10.1093/hmg/ddi430. [DOI] [PubMed] [Google Scholar]
- [19].Liu X, Sun Y, Guo J, Ma H, Li J, Dong B. et al. Expression of hiwi gene in human gastric cancer was associated with proliferation of cancer cells. Int J Cancer. 2006;118:1922–9. doi: 10.1002/ijc.21575. [DOI] [PubMed] [Google Scholar]
- [20].Taubert H, Greither T, Kaushal D, Würl P, Bache M, Bartel F. et al. Expression of the stem cell self-renewal gene Hiwi and risk of tumour-related death in patients with soft-tissue sarcoma. Oncogene. 2007;26:1098–100. doi: 10.1038/sj.onc.1209880. [DOI] [PubMed] [Google Scholar]
- [21].Grochola LF, Greither T, Taubert H, Möller P, Knippschild U, Udelnow A. et al. The stem cell-associated Hiwi gene in human adenocarcinoma of the pancreas: expression and risk of tumour-related death. Br J Cancer. 2008;99:1083–8. doi: 10.1038/sj.bjc.6604653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].He W, Wang Z, Wang Q, Fan Q, Shou C, Wang J. et al. Expression of HIWI in human esophageal squamous cell carcinoma is significantly associated with poorer prognosis. BMC Cancer. 2009;9:426. doi: 10.1186/1471-2407-9-426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].He G, Chen L, Ye Y, Xiao Y, Hua K, Jarjoura D. et al. Piwil2 expressed in various stages of cervical neoplasia is a potential complementary marker for p16INK4a. Am J Transl Res. 2010;2:156–69. [PMC free article] [PubMed] [Google Scholar]
- [24].Liu JJ, Shen R, Chen L, Ye Y, He G, Hua K. et al. Piwil2 is expressed in various stages of breast cancers and has the potential to be used as a novel biomarker. Int J Clin Exp Pathol. 2010;3:328–37. [PMC free article] [PubMed] [Google Scholar]
- [25].Sun G, Wang Y, Sun L, Luo H, Liu N, Fu Z, You Y. Clinical significance of Hiwi gene expression in gliomas. Brain Res. 2011;1373:183–8. doi: 10.1016/j.brainres.2010.11.097. [DOI] [PubMed] [Google Scholar]
- [26].Cheng J, Guo J-M, Xiao B-X, Miao Y, Jiang Z, Zhou H, Li Q-N. piRNA, the new non-coding RNA, is aberrantly expressed in human cancer cells. Clin Chim Acta. 2011;412:1621–5. doi: 10.1016/j.cca.2011.05.015. [DOI] [PubMed] [Google Scholar]
- [27].Yan ZE, Qu L-k, Lin M, Liu C-y, Bin D, Xing X-f. et al. HIWI expression profile in cancer cells and its prognostic value for patients with colorectal cancer. Chinese Med J (Engl) 2011;124:2144–9. [PubMed] [Google Scholar]
- [28].Cheng J, Deng H, Xiao B, Zhou H, Zhou F, Shen Z, Guo J. piR-823, a novel non-coding small RNA, demonstrates in vitro and in vivo tumor suppressive activity in human gastric cancer cells. Cancer Lett. 2012;315:12–7. doi: 10.1016/j.canlet.2011.10.004. [DOI] [PubMed] [Google Scholar]
- [29].Li D, Sun X, Yan D, Huang J, Luo Q, Tang H, Peng Z. Piwil2 modulates the proliferation and metastasis of colon cancer via regulation of matrix metallopeptidase 9 transcriptional activity. Exp Biol Med (Maywood) 2012;237:1231–40. doi: 10.1258/ebm.2012.011380. [DOI] [PubMed] [Google Scholar]
- [30].Chen C, Liu J, Xu G. Overexpression of PIWI proteins in human stage III epithelial ovarian cancer with lymph node metastasis. Cancer Biomark. 2013;13:315–21. doi: 10.3233/CBM-130360. [DOI] [PubMed] [Google Scholar]
- [31].Zhang H, Ren Y, Xu H, Pang D, Duan C, Liu C. The expression of stem cell protein Piwil2 and piR-932 in breast cancer. Surg Oncol. 2013;22:217–23. doi: 10.1016/j.suronc.2013.07.001. [DOI] [PubMed] [Google Scholar]
- [32].Wang D-W, Wang Z-H, Wang L-L, Song Y, Zhang G-Z. Overexpression of hiwi promotes growth of human breast cancer cells. Asian Pac J Cancer Prev. 2014;15:7553–8. doi: 10.7314/apjcp.2014.15.18.7553. [DOI] [PubMed] [Google Scholar]
- [33].Yan H, Wu Q-L, Sun C-Y, Ai L-S, Deng J, Zhang L. et al. piRNA-823 contributes to tumorigenesis by regulating de novo DNA methylation and angiogenesis in multiple myeloma. Leukemia. 2015;29:196–206. doi: 10.1038/leu.2014.135. [DOI] [PubMed] [Google Scholar]
- [34].Al-Janabi O, Wach S, Nolte E, Weigelt K, Rau TT, Stöhr C. et al. Piwi-like 1 and 4 gene transcript levels are associated with clinicopathological parameters in renal cell carcinomas. Biochim Biophys Acta. 2014;1842:686–90. doi: 10.1016/j.bbadis.2014.01.014. [DOI] [PubMed] [Google Scholar]
- [35].Liu W, Gao Q, Chen K, Xue X, Li M, Chen Q. et al. Hiwi facilitates chemoresistance as a cancer stem cell marker in cervical cancer. Oncol Rep. 2014;32:1853–60. doi: 10.3892/or.2014.3401. [DOI] [PubMed] [Google Scholar]
- [36].Xie Y, Yang Y, Ji D, Zhang D, Yao X, Zhang X. Hiwi downregulation, mediated by shRNA, reduces the proliferation and migration of human hepatocellular carcinoma cells. Mol Med Rep. 2014;11:1455–61. doi: 10.3892/mmr.2014.2847. [DOI] [PubMed] [Google Scholar]
- [37].Chu H, Hui G, Yuan L, Shi D, Wang Y, Du M. et al. Identification of novel piRNAs in bladder cancer. Cancer Lett. 2015;356:561–7. doi: 10.1016/j.canlet.2014.10.004. [DOI] [PubMed] [Google Scholar]
- [38].Fu A, Jacobs DI, Hoffman AE, Zheng T, Zhu Y. PIWI-interacting RNA 021285 is involved in breast tumorigenesis possibly by remodeling the cancer epigenome. Carcinogenesis. 2015;36:1094–102. doi: 10.1093/carcin/bgv105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Yang Y, Zhang X, Song D, Wei J. Piwil2 modulates the invasion and metastasis of prostate cancer by regulating the expression of matrix metalloproteinase-9 and epithelial-mesenchymal transitions. Oncol Lett. 2015;10:1735–40. doi: 10.3892/ol.2015.3392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Müller S, Raulefs S, Bruns P, Afonso-Grunz F, Plötner A, Thermann R. et al. Next-generation sequencing reveals novel differentially regulated mRNAs, lncRNAs, miRNAs, sdRNAs and a piRNA in pancreatic cancer. Mol Cancer. 2015;14:94. doi: 10.1186/s12943-015-0358-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Chu H, Xia L, Qiu X, Gu D, Zhu L, Jin J. et al. Genetic variants in noncoding PIWI-interacting RNA and colorectal cancer risk. Cancer. 2015;121:2044–52. doi: 10.1002/cncr.29314. [DOI] [PubMed] [Google Scholar]
- [42].Wang Y, Jiang Y, Ma N, Sang B, Hu X, Cong X, Liu Z. Overexpression of Hiwi inhibits the growth and migration of chronic myeloid leukemia cells. Cell Biochem Biophys. 2015;73:117–24. doi: 10.1007/s12013-015-0651-3. [DOI] [PubMed] [Google Scholar]
- [43].Chen YJ, Xiong XF, Wen SQ, Tian L, Cheng WL, Qi YQ. Expression and clinical significance of PIWIL2 in hilar cholangiocarcinoma tissues and cell lines. Genet Mol Res. 2015;14:7053–61. doi: 10.4238/2015.June.26.15. [DOI] [PubMed] [Google Scholar]
- [44].Busch J, Ralla B, Jung M, Wotschofsky Z, Trujillo-Arribas E, Schwabe P. et al. Piwi-interacting RNAs as novel prognostic markers in clear cell renal cell carcinomas. J Exp Clin Cancer Res. 2015;34:61. doi: 10.1186/s13046-015-0180-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Chen Z, Che Q, Jiang F-Z, Wang H-H, Wang F-Y, Liao Y, Wan X-P. Piwil1 causes epigenetic alteration of PTEN gene via upregulation of DNA methyltransferase in type I endometrial cancer. Biochem Biophys Res Commun. 2015;463:876–80. doi: 10.1016/j.bbrc.2015.06.028. [DOI] [PubMed] [Google Scholar]
- [46].Taubert H, Wach S, Jung R, Pugia M, Keck B, Bertz S. et al. Piwil 2 expression is correlated with disease-specific and progression-free survival of chemotherapy-treated bladder cancer patients. Mol Med. 2015;21:371–80. doi: 10.2119/molmed.2014.00250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Wang Z, Liu N, Shi S, Liu S, Lin H. The role of PIWIL4, an Argonaute family protein, in breast cancer. J Biol Chem. 2016;291:10646–58. doi: 10.1074/jbc.M116.723239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Firmino N, Martinez VD, Rowbotham DA, Enfield KS, Bennewith KL, Lam WL. HPV status is associated with altered PIWI-interacting RNA expression pattern in head and neck cancer. Oral Oncol. 2016;55:43–8. doi: 10.1016/j.oraloncology.2016.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Jacobs DI, Qin Q, Lerro MC, Fu A, Dubrow R, Claus EB. et al. PIWI-interacting RNAs in gliomagenesis: evidence from post-GWAS and functional analyses. Cancer Epidemiol Biomarkers Prev. 2016;25:1073–80. doi: 10.1158/1055-9965.EPI-16-0047. [DOI] [PubMed] [Google Scholar]
- [50].Gambichler T, Kohsik C, Höh AK, Lang K, Käfferlein HU, Brüning T. et al. Expression of PIWIL3 in primary and metastatic melanoma. J Cancer Res Clin Oncol. 2017;143:433–7. doi: 10.1007/s00432-016-2305-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Sivagurunathan S, Arunachalam JP, Chidambaram S. PIWI-like protein, HIWI2 is aberrantly expressed in retinoblastoma cells and affects cell-cycle potentially through OTX2. Cell Mol Biol Lett. 2017;22:17. doi: 10.1186/s11658-017-0048-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Vychytilova-Faltejskova P, Stitkovcova K, Radova L, Sachlova M, Kosarova Z, Slaba K. et al. Circulating PIWI-interacting RNAs piR-5937 and piR-28876 are promising diagnostic biomarkers of colon cancer. Cancer Epidemiol Biomarkers Prev. 2018;27:1019–28. doi: 10.1158/1055-9965.EPI-18-0318. [DOI] [PubMed] [Google Scholar]
- [53].Jacobs DI, Qin Q, Fu A, Chen Z, Zhou J, Zhu Y. piRNA-8041 is downregulated in human glioblastoma and suppresses tumor growth in vitro and in vivo. Oncotarget. 2018;9:37616–26. doi: 10.18632/oncotarget.26331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Gao C-L, Sun R, Li D-H, Gong F. PIWI-like protein 1 upregulation promotes gastric cancer invasion and metastasis. Onco Targets Ther. 2018;11:8783–89. doi: 10.2147/OTT.S186827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Eckstein M, Jung R, Weigelt K, Sikic D, Stöhr R, Geppert C. et al. Piwi-like 1 and-2 protein expression levels are prognostic factors for muscle invasive urothelial bladder cancer patients. Sci Rep. 2018;8:17693. doi: 10.1038/s41598-018-35637-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Heng ZS, Lee JY, Subhramanyam CS, Wang C, Thanga LZ, Hu Q. The role of 17β-estradiol-induced upregulation of Piwi-like 4 in modulating gene expression and motility in breast cancer cells. Oncol Rep. 2018;40:2525–35. doi: 10.3892/or.2018.6676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Li B, Hong J, Hong M, Wang Y, Yu T, Zang S, Wu Q. piRNA-823 delivered by multiple myeloma-derived extracellular vesicles promoted tumorigenesis through re-educating endothelial cells in the tumor environment. Oncogene. 2019;38:5227–38. doi: 10.1038/s41388-019-0788-4. [DOI] [PubMed] [Google Scholar]
- [58].Roy J, Das B, Jain N, Mallick B. PIWI-interacting RNA 39980 promotes tumor progression and reduces drug sensitivity in neuroblastoma cells. J Cell Physiol. 2020;235:2286–99. doi: 10.1002/jcp.29136. [DOI] [PubMed] [Google Scholar]
- [59].Tan L, Mai D, Zhang B, Jiang X, Zhang J, Bai R. et al. PIWI-interacting RNA-36712 restrains breast cancer progression and chemoresistance by interaction with SEPW1 pseudogene SEPW1P RNA. Mol Cancer. 2019;18:9. doi: 10.1186/s12943-019-0940-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Lin Y, Holden V, Dhilipkannah P, Deepak J, Todd NW, Jiang F. A non-coding RNA landscape of bronchial epitheliums of lung cancer patients. Biomedicines. 2020;8:88. doi: 10.3390/biomedicines8040088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Liu Y, Dong Y, He X, Gong A, Gao J, Hao X. et al. piR-hsa-211106 inhibits the progression of lung adenocarcinoma through pyruvate carboxylase and enhances chemotherapy sensitivity. Front Oncol. 2021;11 doi: 10.3389/fonc.2021.651915. 651915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Li J, Wang N, Zhang F, Jin S, Dong Y, Dong X. et al. PIWI-interacting RNAs are aberrantly expressed and may serve as novel biomarkers for diagnosis of lung adenocarcinoma. Thorac Cancer. 2021;12:2468–77. doi: 10.1111/1759-7714.14094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Fathizadeh H, Asemi Z. Epigenetic roles of PIWI proteins and piRNAs in lung cancer. Cell Biosci. 2019;9:102. doi: 10.1186/s13578-019-0368-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Dana PM, Mansournia MA, Mirhashemi SM. PIWI-interacting RNAs: new biomarkers for diagnosis and treatment of breast cancer. Cell Biosci. 2020;10:44. doi: 10.1186/s13578-020-00403-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Yu Y, Xiao J, Hann SS. The emerging roles of PIWI-interacting RNA in human cancers. Cancer Manag Res. 2019;11:5895–909. doi: 10.2147/CMAR.S209300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Sokolova OA, Ilyin AA, Poltavets AS, Nenasheva VV, Mikhaleva EA, Shevelyov YY, Klenov MS. Yb body assembly on the flamenco piRNA precursor transcripts reduces genic piRNA production. Mol Biol Cell. 2019;30:1544–54. doi: 10.1091/mbc.E17-10-0591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Ishizu H, Kinoshita T, Hirakata S, Komatsuzaki C, Siomi MC. Distinct and collaborative functions of Yb and Armitage in transposon-targeting piRNA biogenesis. Cell Rep. 2019;27:1822–35.e8. doi: 10.1016/j.celrep.2019.04.029. [DOI] [PubMed] [Google Scholar]
- [68].Saito K, Inagaki S, Mituyama T, Kawamura Y, Ono Y, Sakota E. et al. A regulatory circuit for piwi by the large Maf gene traffic jam in Drosophila. Nature. 2009;461:1296–9. doi: 10.1038/nature08501. [DOI] [PubMed] [Google Scholar]
- [69].Aguiar ERGR, de Almeida JPP, Queiroz LR, Oliveira LS, Olmo RP, de Faria IJDS. et al. A single unidirectional piRNA cluster similar to the flamenco locus is the major source of EVE-derived transcription and small RNAs in Aedes aegypti mosquitoes. RNA. 2020;26:581–94. doi: 10.1261/rna.073965.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Le Thomas A, Rogers AK, Webster A, Marinov GK, Liao SE, Perkins EM. et al. Piwi induces piRNA-guided transcriptional silencing and establishment of a repressive chromatin state. Genes Dev. 2013;27:390–9. doi: 10.1101/gad.209841.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Chen P, Luo Y, Aravin AA. RDC complex executes a dynamic piRNA program during Drosophila spermatogenesis to safeguard male fertility. PLoS Genet. 2021;17:e1009591. doi: 10.1371/journal.pgen.1009591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Qi H, Watanabe T, Ku H-Y, Liu N, Zhong M, Lin H. The Yb body, a major site for Piwi-associated RNA biogenesis and a gateway for Piwi expression and transport to the nucleus in somatic cells. J Biol Chem. 2011;286:3789–97. doi: 10.1074/jbc.M110.193888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Pane A, Wehr K, Schüpbach T. zucchini and squash encode two putative nucleases required for rasiRNA production in the Drosophila germline. Dev Cell. 2007;12:851–62. doi: 10.1016/j.devcel.2007.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Olivieri D, Senti K-A, Subramanian S, Sachidanandam R, Brennecke J. The cochaperone shutdown defines a group of biogenesis factors essential for all piRNA populations in Drosophila. Mol Cell. 2012;47:954–69. doi: 10.1016/j.molcel.2012.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Xie W, Sowemimo I, Hayashi R, Wang J, Burkard TR, Brennecke J. et al. Structure-function analysis of microRNA 3′-end trimming by Nibbler. Proc Natl Acad Sci U S A. 2020;117:30370–79. doi: 10.1073/pnas.2018156117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Ding D, Chen C. Zucchini: the key ingredient to unveil piRNA precursor processing. Biol Reprod. 2020;103:452–54. doi: 10.1093/biolre/ioaa090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Pippadpally S, Venkatesh T. Deciphering piRNA biogenesis through cytoplasmic granules, mitochondria and exosomes. Arch Biochem Biophys. 2020;695 doi: 10.1016/j.abb.2020.108597. 108597. [DOI] [PubMed] [Google Scholar]
- [78].Sokolova OA, Iakushev EIu, Stoliarenko AD, Mikhaleva EA, Gvozdev VA, Klenov MS. [The interplay of transposon silencing genes in the Drosophila melanogaster germline] Mol Biol (Mosk) 2011;45:633–41. [in Russian, English abstract] [PubMed] [Google Scholar]
- [79].Ozata DM, Gainetdinov I, Zoch A, O’Carroll D, Zamore PD. PIWI-interacting RNAs: small RNAs with big functions. Nat Rev Genet. 2019;20:89–108. doi: 10.1038/s41576-018-0073-3. [DOI] [PubMed] [Google Scholar]
- [80].Russell SJ, LaMarre J. Transposons and the PIWI pathway: genome defense in gametes and embryos. Reproduction. 2018;156:R111–24. doi: 10.1530/REP-18-0218. [DOI] [PubMed] [Google Scholar]
- [81].Aravin AA, Sachidanandam R, Girard A, Fejes-Toth K, Hannon GJ. Developmentally regulated piRNA clusters implicate MILI in transposon control. Science. 2007;316:744–7. doi: 10.1126/science.1142612. [DOI] [PubMed] [Google Scholar]
- [82].Pal-Bhadra M, Leibovitch BA, Gandhi SG, Chikka MR, Bhadra U, Birchler JA, Elgin SCR. Heterochromatic silencing and HP1 localization in Drosophila are dependent on the RNAi machinery. Science. 2004;303:669–72. doi: 10.1126/science.1092653. [DOI] [PubMed] [Google Scholar]
- [83].Teo RYW, Anand A, Sridhar V, Okamura K, Kai T. Heterochromatin protein 1a functions for piRNA biogenesis predominantly from pericentric and telomeric regions in Drosophila. Nat Commun. 2018;9:1735. doi: 10.1038/s41467-018-03908-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Zhao K, Cheng S, Miao N, Xu P, Lu X, Zhang Y. et al. A Pandas complex adapted for piRNA-guided transcriptional silencing and heterochromatin formation. Nat Cell Biol. 2019;21:1261–72. doi: 10.1038/s41556-019-0396-0. [DOI] [PubMed] [Google Scholar]
- [85].Watanabe T, Tomizawa SI, Mitsuya K, Totoki Y, Yamamoto Y, Kuramochi-Miyagawa S. et al. Role for piRNAs and noncoding RNA in de novo DNA methylation of the imprinted mouse Rasgrf1 locus. Science. 2011;332:848–52. doi: 10.1126/science.1203919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Théron E, Maupetit-Mehouas S, Pouchin P, Baudet L, Brasset E, Vaury C. The interplay between the Argonaute proteins Piwi and Aub within Drosophila germarium is critical for oogenesis, piRNA biogenesis and TE silencing. Nucleic Acids Res. 2018;46:10052–65. doi: 10.1093/nar/gky695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Zhang Y, Liu W, Li R, Gu J, Wu P, Peng C. et al. Structural insights into the sequence-specific recognition of Piwi by Drosophila Papi. Proc Natl Acad Sci U S A. 2018;115:3374–79. doi: 10.1073/pnas.1717116115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Wenda JM, Homolka D, Yang Z, Spinelli P, Sachidanandam R, Pandey RR, Pillai RS. Distinct roles of RNA helicases MVH and TDRD9 in PIWI slicing-triggered mammalian piRNA biogenesis and function. Dev Cell. 2017;41:623–637.e9. doi: 10.1016/j.devcel.2017.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Zhu J, Zhang D, Liu X, Yu G, Cai X, Xu C. et al. Zebrafish prmt5 arginine methyltransferase is essential for germ cell development. Development. 2019;146 doi: 10.1242/dev.179572. dev179572. [DOI] [PubMed] [Google Scholar]
- [90].Radion E, Morgunova V, Ryazansky S, Akulenko N, Lavrov S, Abramov Y. et al. Key role of piRNAs in telomeric chromatin maintenance and telomere nuclear positioning in Drosophila germline. Epigenetics Chromatin. 2018;11:40. doi: 10.1186/s13072-018-0210-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Li Z, You L, Yan D, James AA, Huang Y, Tan A. Bombyx mori histone methyltransferase BmAsh2 is essential for silkworm piRNA-mediated sex determination. PLoS Genet. 2018;14:e1007245. doi: 10.1371/journal.pgen.1007245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Robine N, Lau NC, Balla S, Jin Z, Okamura K, Kuramochi-Miyagawa S. et al. A broadly conserved pathway generates 3′UTR-directed primary piRNAs. Curr Biol. 2009;19:2066–76. doi: 10.1016/j.cub.2009.11.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Rouget C, Papin C, Boureux A, Meunier A-C, Franco B, Robine N. et al. Maternal mRNA deadenylation and decay by the piRNA pathway in the early Drosophila embryo. Nature. 2010;467(7319):1128–32. doi: 10.1038/nature09465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Dai P, Wang X, Gou L-T, Li Z-T, Wen Z, Chen Z-G. et al. A translation-activating function of MIWI/piRNA during mouse spermiogenesis. Cell. 2019;179:1566–81.e16. doi: 10.1016/j.cell.2019.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Mani SR, Megosh H, Lin H. PIWI proteins are essential for early Drosophila embryogenesis. Dev Biol. 2014;385:340–9. doi: 10.1016/j.ydbio.2013.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Bahn JH, Zhang Q, Li F, Chan T-M, Lin X, Kim Y. et al. The landscape of microRNA, Piwi-interacting RNA, and circular RNA in human saliva. Clin Chem. 2015;61:221–30. doi: 10.1373/clinchem.2014.230433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Lenart P, Novak J, Bienertova-Vasku J. PIWI-piRNA pathway: setting the pace of aging by reducing DNA damage. Mech Ageing Dev. 2018;173:29–38. doi: 10.1016/j.mad.2018.03.009. [DOI] [PubMed] [Google Scholar]
- [98].Rajasethupathy P, Antonov I, Sheridan R, Frey S, Sander C, Tuschl T, Kandel ER. A role for neuronal piRNAs in the epigenetic control of memory-related synaptic plasticity. Cell. 2012;149:693–707. doi: 10.1016/j.cell.2012.02.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Kolliopoulou A, Santos D, Taning CNT, Wynant N, Vanden Broeck J, Smagghe G, Swevers L. PIWI pathway against viruses in insects. Wiley Interdiscip Rev RNA. 2019;10:e1555. doi: 10.1002/wrna.1555. [DOI] [PubMed] [Google Scholar]
- [100].Henaoui IS, Jacovetti C, Mollet IG, Guay C, Sobel J, Eliasson L, Regazzi R. PIWI-interacting RNAs as novel regulators of pancreatic beta cell function. Diabetologia. 2017;60:1977–86. doi: 10.1007/s00125-017-4368-2. [DOI] [PubMed] [Google Scholar]
- [101].Zhou S, Yang S, Li F, Hou J, Chang H. P-element Induced WImpy protein-like RNA-mediated gene silencing 2 regulates tumor cell progression, apoptosis, and metastasis in oral squamous cell carcinoma. J Int Med Res. 2021;49 doi: 10.1177/03000605211053158. 3000605211053158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Xie K, Zhang K, Kong J, Wang C, Gu Y, Liang C. et al. Cancer-testis gene PIWIL1 promotes cell proliferation, migration, and invasion in lung adenocarcinoma. Cancer Med. 2018;7:157–66. doi: 10.1002/cam4.1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Yao J, Wang YW, Fang BB, Zhang SJ, Cheng BL. piR-651 and its function in 95-D lung cancer cells. Biomed Rep. 2016;4:546–50. doi: 10.3892/br.2016.628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Zhang S-J, Yao J, Shen B-Z, Li G-B, Kong S-S, Bi D-D. et al. Role of piwi-interacting RNA-651 in the carcinogenesis of non-small cell lung cancer. Oncol Lett. 2018;15:940–6. doi: 10.3892/ol.2017.7406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Li D, Luo Y, Gao Y, Yang Y, Wang Y, Xu Y. et al. piR-651 promotes tumor formation in non-small cell lung carcinoma through the upregulation of cyclin D1 and CDK4. Int J Mol Med. 2016;38:927–36. doi: 10.3892/ijmm.2016.2671. [DOI] [PubMed] [Google Scholar]
- [106].Reeves ME, Firek M, Jliedi A, Amaar YG. Identification and characterization of RASSF1C piRNA target genes in lung cancer cells. Oncotarget. 2017;8:34268–82. doi: 10.18632/oncotarget.15965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Chen S, Ben S, Xin J, Li S, Zheng R, Wang H. et al. The biogenesis and biological function of PIWI-interacting RNA in cancer. J Hematol Oncol. 2021;14:93. doi: 10.1186/s13045-021-01104-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Rui Y. piRNAs variants and lung cancer risk: a post-GWAS study. New Haven (CI): Yale Univ; 2016. [Masters Public Health thesis] Available from: https://elischolar.library.yale.edu/ysphtdl/1335 . [Google Scholar]
- [109].Liang D, Dong M, Hu L-J, Fang Z-H, Xu X, Shi E-H, Yang Y-J. Hiwi knockdown inhibits the growth of lung cancer in nude mice. Asian Pac J Cancer Prev. 2013;14:1067–72. doi: 10.7314/apjcp.2013.14.2.1067. [DOI] [PubMed] [Google Scholar]
- [110].Reeves ME, Firek M, Chen ST, Amaar YG. Evidence that RASSF1C stimulation of lung cancer cell proliferation depends on IGFBP-5 and PIWIL1 expression levels. PloS One. 2014;9:e101679. doi: 10.1371/journal.pone.0101679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Wang Y, Gable T, Ma MZ, Clark D, Zhao J, Zhang Y. et al. A piRNA-like small RNA induces chemoresistance to cisplatin-based therapy by inhibiting apoptosis in lung squamous cell carcinoma. Mol Ther Nucleic Acids. 2017;6:269–78. doi: 10.1016/j.omtn.2017.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Brock M, Mei Y. Protein functional effector sncRNAs (pfeRNAs) in lung cancer. Cancer Lett. 2017;403:138–43. doi: 10.1016/j.canlet.2017.06.013. [DOI] [PubMed] [Google Scholar]
- [113].Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–74. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]