ABSTRACT
Inflammatory bowel disease (IBD) is a multifactorial disease with increasing incidence in the U.S. suggesting that environmental factors, including diet, are involved. It has been suggested that excessive consumption of linoleic acid (LA, C18:2 omega-6), which must be obtained from the diet, may promote the development of IBD in humans. To demonstrate a causal link between LA and IBD, we show that a high fat diet (HFD) based on soybean oil (SO), which is comprised of ~55% LA, increases susceptibility to colitis in several models, including IBD-susceptible IL10 knockout mice. This effect was not observed with low-LA HFDs derived from genetically modified soybean oil or olive oil. The conventional SO HFD causes classical IBD symptoms including immune dysfunction, increased intestinal epithelial barrier permeability, and disruption of the balance of isoforms from the IBD susceptibility gene Hepatocyte Nuclear Factor 4α (HNF4α). The SO HFD causes gut dysbiosis, including increased abundance of an endogenous adherent invasive Escherichia coli (AIEC), which can use LA as a carbon source. Metabolomic analysis shows that in the mouse gut, even in the absence of bacteria, the presence of soybean oil increases levels of LA, oxylipins and prostaglandins. Many compounds in the endocannabinoid system, which are protective against IBD, are decreased by SO both in vivo and in vitro. These results indicate that a high LA diet increases susceptibility to colitis via microbial and host-initiated pathways involving alterations in the balance of bioactive metabolites of omega-6 and omega-3 polyunsaturated fatty acids, as well as HNF4α isoforms.
KEYWORDS: IBD, PUFAs, soybean oil, olive oil, oxylipins, adherent invasive E.Coli, HNF4α, gut microbiome, metabolomics, epithelial barrier function
Introduction
Inflammatory bowel disease (IBD) is a multifactorial disorder, the pathogenesis of which can be influenced by host genetics, immune dysfunction, intestinal microbiota, and a variety of environmental factors, including diet.1 Over the last century there has been a shift in the composition of the American diet with soybean oil (SO) being the component that has increased the most,2 paralleling the increased incidence of IBD in the U.S.3 While soybean oil is comprised of ~55% linoleic acid (LA, C18:2 ω6), an essential fatty acid, only 1–2% kcal of this polyunsaturated fatty acid (PUFA) is required in the human diet.4 Hence, with the increased use of soybean oil, Americans are now consuming at least five times the minimally required amount of LA.2 These trends are not limited to the U.S. as there are reports indicating increasing “westernization” of global dietary patterns as well as the emergence of IBD as a global disease.3,5 While high dietary LA has been positively correlated with the development of Ulcerative Colitis and Crohn’s disease6,7 and while previous studies have demonstrated the role of diet, especially the high-fat/low-fiber Western diet, in IBD pathogenesis,8,9 there are conflicting reports on whether dietary soybean oil promotes or protects against IBD.10–13 Additionally, LA, and its downstream metabolite arachidonic acid (AA, C20:4 ω6), are precursors to bioactive lipids such as oxylipins and endocannabinoids,14,15 which have been linked to IBD.16–18 However, dietary LA as a causal factor in IBD has not been established, and the impact of the combined dysregulation of the oxylipin and endocannabinoid systems on colitis is not well defined.
LA is the endogenous ligand for HNF4α,19 an IBD susceptibility gene,20 and a member of the nuclear receptor superfamily of ligand-dependent transcription factors.21 The human and mouse HNF4A genes are highly conserved and contain two promoters (P1 and P2) that drive the expression of P1 isoforms (HNF4α1–6) and P2 isoforms (HNF4α7–12) which have distinct first exons.22 While the whole-body knockout of HNF4α is embryonic lethal,23 exon swap mice that express exclusively P1- or P2-derived HNF4α24 can be used to study the role of HNF4α isoforms in various tissues, including the intestines. Both HNF4α promoters are active in the small intestines and colon, albeit in different parts of the crypt: P1-HNF4α is expressed in the differentiated portion at the top of the colonic crypt and P2-HNF4α in the proliferative compartment in the bottom half of the crypt25. While both α1HMZ (expressing only P1-HNF4α) and α7HMZ (expressing only P2-HNF4α) exon-swap mice are healthy under unstressed conditions, we have shown previously that on a low-fat, high-fiber diet, α1HMZ males are less susceptible than α7HMZ males to developing dextran sulfate sodium (DSS)-induced colitis.25
Intestinal microbiota plays an important role in the regulation of gut homeostasis and perturbation in the gut microbiome composition (dysbiosis) can lead to intestinal inflammation and various intestinal pathologies, including IBD.26–28 Notably, increased incidence of an adherent, invasive Escherichia coli (AIEC) has been reported in patients with IBD,29,30 while a Western diet leads to increased intestinal AIEC colonization in genetically susceptible mice.31 We recently isolated a novel mouse AIEC (mAIEC) with 90% DNA sequence homology to the human AIEC and showed that it can cause colitis and exacerbate intestinal inflammation in mice.32
In a previous study, we showed that a diet high in soybean oil similar to the current American diet can shorten intestinal colonic crypt length33 and that oxylipin metabolites of LA and alpha-linolenic acid (ALA, C18–3 ω3) in the liver positively correlate with SO-induced obesity in mice.34 In the current study, we determine the impact of a soybean oil-based HFD on the development of IBD using three different models of colitis – DSS-induced, IL10 knockout mice, and HNF4α exon swap mice. In all three models we observed increased susceptibility to colitis in adult males fed the SO diet high in LA. In contrast, HFDs consisting of a genetically modified soybean oil that is low in LA (Plenish) or olive oil (which is naturally low in LA) did not increase susceptibility to colitis in wild-type (WT) or IL10 knockout mice, respectively. We examined potential underlying mechanisms including the effect of the high-LA SO diet on HNF4α protein levels as well as the gut microbiome and both host and bacterial metabolomes. The results show that while the SO diet, by itself, does not lead to colitis in WT mice, it does decrease intestinal epithelial barrier function, which could contribute to increased susceptibility to colitis. It also leads to gut dysbiosis, including an expansion of mAIEC in the intestines of SO-fed mice. We show a disruption of the balance between HNF4α isoform protein levels with an elevation of P2-HNF4α which is associated with colitis and colon cancer.25,35,36 Finally, metabolomic analysis of both host intestinal cells and SO-mAIEC grown in vitro suggests a role for elevated levels of LA and oxylipins, as well as reduced levels of metabolites in the endocannabinoid system and the omega-3 eicosapentaenoic acid (EPA), in colitis susceptibility. Taken together, our results indicate that a diet high in soybean oil (and hence LA), analogous to the current American diet, may indeed be a predisposing environmental factor in the development of IBD and that intestinal barrier dysfunction, HNF4α isoform imbalance, gut bacterial dysbiosis, as well as disruption of the ratio of omega 6 to omega 3 bioactive metabolites, singly or in combination, may contribute to this effect.
Results
To examine the hypothesis that a diet high in LA can cause susceptibility to IBD and to identify potentially relevant factors, we performed experiments on four in vivo models using a total of five diets as well as three in vitro experiments with cultured bacteria (Figure 1).
Figure 1.
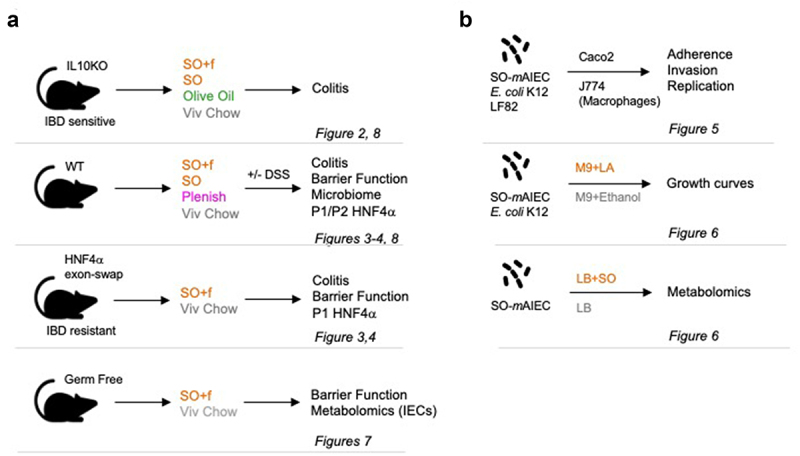
Experimental design.
Notes: (a) In vivo experiments using four different models (C57BL6 adult males) and the indicated treatments with high fat diets (SO+f, soybean oil plus fiber; SO, soybean oil; Plenish, low LA soybean oil and olive oil), DSS, and/or gavage with linoleic acid (LA). (b) In vitro experiments with the indicated bacterial strains and treatment of mammalian cells (CaCO2, J774) or growth condition. The assays used in each model are indicated as well as the figures presenting the results.
A diet high in LA accelerates onset of colitis in IL-10 deficient mice
To determine if there is an interaction between the IBD susceptibility loci IL-10 and LA, we fed the B6 strain of IL-10 deficient mice (IL-10−/−, also referred to as IL-10 KO, develops a mild form of colitis) either a low-fat vivarium (Viv) chow (13 kcal% fat, 4.5 kcal% LA) or a high fat diet (HFD) based on soybean oil (35 kcal% fat, 18.6 kcal% LA) for 10 weeks. The amount of fat in the SO HFD is comparable to the amount in the current American diet (~40 kcal%) but less than the amount typically used in high-fat rodent diets (~55–60 kcal%).37,38 Since a high fiber diet is considered to be protective against IBD,39 this soybean oil diet (SO+f) also contained 19% fiber analogous to the 23% fiber in the Viv chow (Supplementary Table S1).
The IL-10−/− mice on the SO+f diet showed accelerated development of disease indices for colitis – i.e., body weight loss starting at 7 weeks and appearance of blood in the stool after 3 weeks on the diet. In contrast, blood was apparent in the stool of Viv chow-fed animals after only 9 weeks and there was no significant drop in body weight even at 10 weeks (Figure 2a,b). Even though both groups of mice had similar weights at the beginning of the dietary treatments, the SO+f diet mice had significantly lower body weights than the Viv chow group at the time of sacrifice (Figure 2a). This is notable given that the SO+f diet is a high-fat, high-caloric diet. Colon lengths in the two groups were similar, and there were no obvious differences in gross colonic histology although mice in the SO+f group had significantly shorter crypt lengths compared to the Viv chow control (9.8% decrease) (Figure 2c and Supplementary Figure S1A). Compared to WT mice, the crypt length was elongated in IL-10−/− mice on both diets (Figures 2c and 3f), which is consistent with previous studies.40,41 No effect was observed on overall small intestine length by the SO+f diet in IL-10−/− mice (or spleen weight). The SO+f diet decreased the liver as a percent of body weight in these mice (Supplementary Figure S1B) similar to what we have observed with other soybean oil diets in WT mice (unpublished data).
Figure 2.
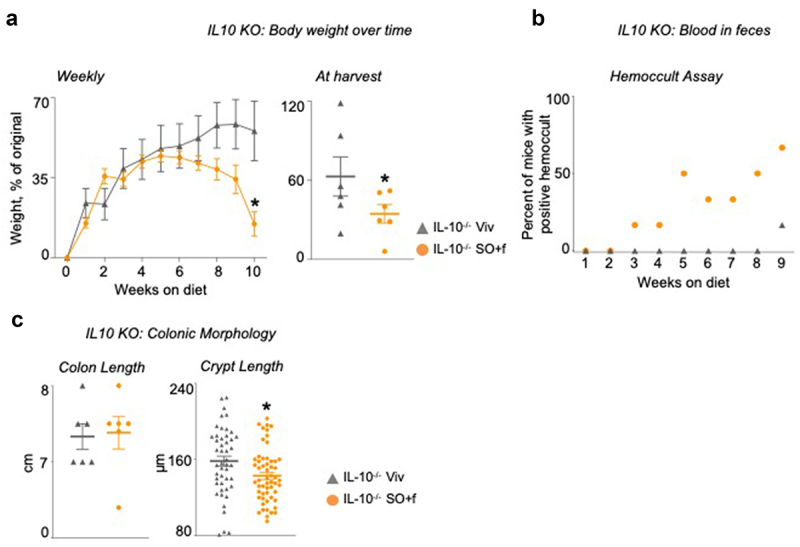
A diet high in LA increases susceptibility to colitis in IL10−/− mice. IL10−/− mice fed SO+f or Viv diet for 10 weeks.
Notes: (s) Average weekly body weights and at harvest; the body weight at harvest is a few days after the last data point in the weekly body weight graph. (b) hemoccult assay. (c) colon length and crypt length. Scale bar is 400 microns. Colon sections are shown in Supplementary Figure 1. * vs Viv. T-test, P < .05 N = 5–12 per group.
Figure 3.
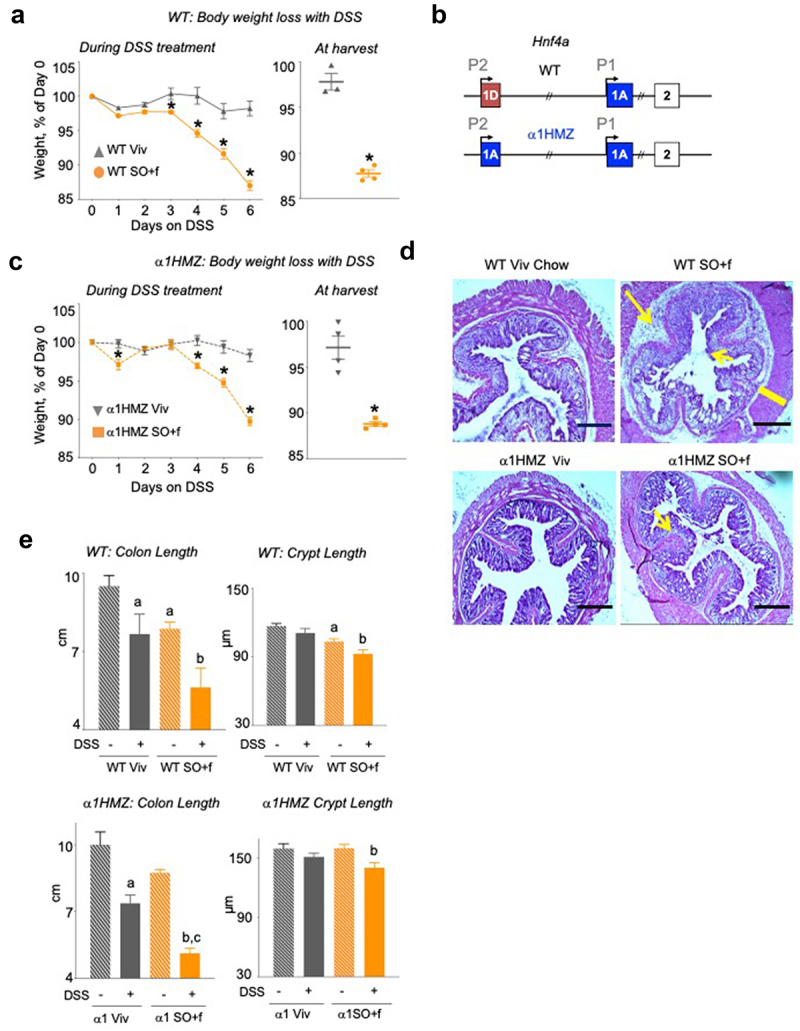
A diet high in LA increases colitis susceptibility and decreases barrier function in WT and α1HMZ mice.
Notes: (a) WT mice on Viv chow or SO+f diet for 15 weeks were treated with 2.5% DSS in their drinking water for 6 d: % weight loss of DSS-treated mice, body weight at harvest; the body weight at harvest is a few days after the last data point in the weekly body weight graph. * vs Viv P < .05, T-test N = 3–4 per group (b) Schematic of the mouse Hnf4a gene showing the two promoters (P1 and P2) (top), and the exon-swap (1D to 1A) that drives the expression of only P1-HNF4α in α1HMZ mice. α1HMZ mice on Viv chow or SO+f diet for 15 weeks were treated with 2.5% DSS for 6 d. (c) % body weight loss, weight at harvest, * vs α1HMZ Viv P < .05, T-test N = 3–4 per group. (d) Representative colonic histology. Big arrow, immune infiltrate; small arrow, loss of crypt structure; line, thickening of muscularis in SO+f. Scale bar is 400 microns. (e) Colon length and crypt length (at least 10 crypts were measured per mouse) Additional sections are shown in Supplementary Figures S2 and 3. a vs untreated WT, b vs untreated α1HMZ Viv, c vs untreated α1HMZ SO+f. One-way ANOVA, Tukey’s post-hoc. N = 5–12 per group.
A diet high in LA increases susceptibility to DSS-induced colitis in WT mice and overcomes resistance to DSS-induced colitis in HNF4α exon swap mice
To determine whether excessive amounts of dietary L.A. impact susceptibility to colitis in wild-type (WT) animals, male C57BL6/N mice were fed either the SO+f diet or the Viv chow for 15 weeks, followed by treatment with DSS. Consistent with our previous studies using a diet high in soybean oil but low in fiber,33,34 after 15 weeks on the SO+f diet the mice gained significantly more weight than the Viv chow controls (Supplementary Figure S2A). When challenged with 2.5% DSS in the drinking water after 15 weeks on the diet, we observed more loss in body weight in the SO+f group compared to the Viv chow control starting with day 3 of the DSS treatment (Figure 3a). The percentage of body weight at harvest was also significantly lower in the SO+f diet mice (88% vs 98% in Viv chow) (Figure 3a).
To determine whether soybean oil can impact susceptibility to colitis in a genetic model of IBD resistance, just as it does in the IBD-sensitive IL-10−/− model, we examined the effect of the SO+f diet on the IBD susceptibility loci HNF4α using the HNF4α exon swap mice which express only the P1 isoform of HNF4α (α1HMZ) (Figure 3b). We have previously shown that α1HMZ male mice exhibit resistance to DSS-induced colitis on Viv chow while mice expressing only the P2 isoforms of HNF4α are highly sensitive to DSS treatment.25 Interestingly, the α1HMZ male mice fed the SO+f diet did not show significant weight gain compared to the Viv chow group until 19 weeks on the diet versus 10 weeks for the WT mice (Supplementary Figure S3A vs Supplementary Figure S2A). When treated with 2.5% DSS for 6 d, the α1HMZ SO+f diet mice lost significantly more weight (10.3%) than the Viv chow controls (1.7%) (Figure 3c), although the body weight loss after DSS treatment in SO+f-fed mice was somewhat less in α1HMZ mice than in the WT mice (10.3 vs 13% for WT) (Supplementary Figure S3B). These results are consistent with the less susceptible phenotype of α1HMZ mice that we had noted previously on Viv chow.25
Histological analysis of the distal colon from DSS-treated WT mice fed Viv chow or the SO+f diet shows that the latter have extensive loss of crypt structure, increased submucosal inflammation and thickened muscularis (Figure 3d left and Supplementary Figure S2E). As in the WT mice, DSS treatment of α1HMZ mice caused greater loss of crypt structure and more immune cell infiltration in the SO+f diet group compared to the Viv chow control (Figure 3d, right and Supplementary Figure S3E).
As we have seen previously with another SO HFD,33 the SO+f diet decreased both the colon length and the crypt length in the WT mice; DSS treatment caused colon lengths to shorten further in both the Viv chow and SO+f groups, with a larger effect in the latter (29% vs 19% decrease) (Figure 3e, left). Crypt length measurements revealed a similar pattern as colon length, with DSS treatment shortening the crypts in both groups but with only the SO+f mice showing a significant difference (10.5% decrease) (Figure 3e, left). At 15 weeks on the diet, the colon length in α1HMZ SO+f fed mice was not significantly different compared to the Viv chow control (8.8 vs 10 cm, P = 0.06), and there was no difference in crypt lengths between the two diets (Figure 3e, right). After DSS treatment, similar to WT mice, colon length in the SO+f α1HMZ mice decreased significantly (from 8.8 to 5.1 cm) and was significantly shorter than the DSS-treated Viv chow group (7.4 cm). DSS treatment also significantly decreased crypt length in SO+f α1HMZ mice compared to the untreated SO+f group (12.5% decrease). However, in α1HMZ mice, there was no significant difference in crypt length between Viv chow and SO+f diet without the DSS treatment (Figure 3e, right) as there was in WT mice (Figure 3e, left).
Since immune dysfunction is an important manifestation of IBD, we characterized immune cell populations in intestinal intraepithelial leukocytes (IELs) and lamina propria leukocytes (LPLs) from both untreated (naive) and DSS-treated Viv chow and SO+f diet mice, both WT and α1HMZ. Compared to Viv chow-fed mice, a significant increase in the eosinophil population in the intestinal LPLs was observed in the WT SO+f mice after DSS treatment (Figure 4a) but not in untreated mice or in IELs from treated or untreated mice (Supplementary Figure S2C). In contrast, no significant difference was observed between SO+f or Viv chow fed mice for monocytes or neutrophils in the LPLs nor in the IELs or in any of the immune cell populations peripheral blood mononuclear cells (PBMCs) (Figures 4a and 2d). Taken together, these results indicate that DSS treatment causes greater immune dysregulation in mice fed SO+f than in control mice fed Viv chow. Immune cell analysis also revealed an increase in immune cells after DSS treatment in the α1HMZ mice on the SO+f diet although the profile was different from that observed in WT mice (Figure 4b). While the SO+f diet did not alter the percentage of eosinophils in the lamina propria in the α1HMZ mice as it did in WT mice (Figure 4a), there was a significant increase in neutrophils in the LPL population and monocytes in the IEL population with the SO+f diet (Figure 4b). Like the WT mice, there were no significant differences observed in α1HMZ mice in the PBMCs on the two different diets, indicating that the immune dysregulation was localized to the intestine (Supplementary Figure S3C). Taken together, these results indicate that the SO+f diet increased susceptibility to DSS-induced colitis in WT mice, including immune cell dysregulation and decreased colon and crypt lengths, all of which are correlated with increased colonic inflammation.40 The SO+f diet had a similar effect even in the α1HMZ that are less susceptible to DSS-induced colitis. The observation that the same diet can induce different immune responses in different genotypes suggests that the underlying mechanism for the recruitment and activity of the immune cells is complex and warrants further investigation.
Figure 4.
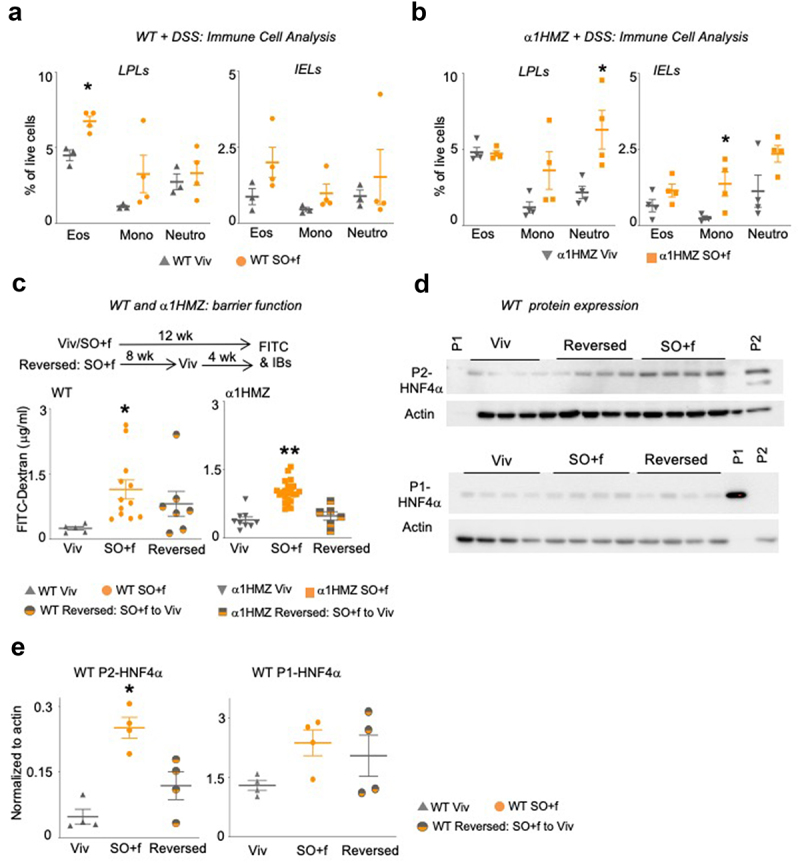
A diet high in LA increases immune dysfunction and decreases barrier function in WT and α1HMZ mice.
Notes: Immune cell analysis (Eos, eosinophils; Mono, monocytes; Neutro, neutrophils) in DSS-treated WT (a) or α1HMZ (b) mice (see Supplementary Figure S2D and S3D for data from untreated mice); * vs WT or α1 HMZ Viv P < .05, T-test was performed between Viv and SO+f for each cell type, N = 3–4 per group. (c) Epithelial barrier permeability in WT and α1HMZ mice fed either Viv or SO+f diets for 12 weeks or SO+f for 8 weeks, followed by 4 weeks of Viv (reversed) (one outlier removed from WT reversed group). * vs Viv, ** vs Viv and Reversed. P < .05, one-way ANOVA, Sidak’s post hoc comparison. N = 6–22 per group. (d) HNF4α immunoblots of whole cell extracts (WCE, 30 µg) from distal colon of WT mice fed Viv chow or SO+f diet for 12 weeks or SO+f for 8 weeks followed by Viv chow for 4 weeks (reversed). Each lane contains WCE from a different mouse. P1 control-nuclear extract from HCT116 cells expressing P1-HNF4α; P2 control-nuclear extract from α7HMZ mouse (see Supplementary Figure S4 for entire blots for WT mice and P1 blot for α1HMZ mice). (e) Quantification of the P1-and P2-HNF4α immunoblot signals (shown in Figure 4d) normalized to total protein, as determined by actin staining of the same blot. * P < .05, one-way ANOVA, Tukey’s post hoc comparison. N = 3–4 per group.
A diet high in LA increases intestinal barrier dysfunction: role for colonic HNF4α isoforms
Having established that the SO+f diet increases susceptibility to colitis in three different models – IL-10−/− (colitis-sensitive), DSS in WT (chemically induced colitis) and α1HMZ (less susceptible to colitis) mice – we next investigated the underlying mechanisms by examining the intestinal epithelial barrier function using the FITC-Dextran assay. We found that after 12 weeks, the SO+f diet significantly increased epithelial permeability (as determined by increased FITC-Dextran in the serum) in both WT and α1HMZ mice (Figure 4c). To determine whether this barrier defect could be reversed by changing the diet, we fed another group of mice the SO+f diet for 8 weeks, followed by 4 weeks of Viv chow. We found that while the barrier permeability in WT mice trended lower after the diet reversal, it was not significant. In contrast, in α1HMZ mice, replacing the SO+f diet with Viv chow significantly improved the barrier function (Figure 4c).
Immunoblot (IB) analysis of distal colon whole cell extracts showed an increase in the P2-HNF4α isoform in the colons of WT mice fed the SO+f diet (Figure 4d,e and Supplementary Figure S4A). Reverting the diet back to Viv chow for 4 weeks did not significantly decrease the P2-HNF4α levels, suggesting that the SO+f diet may cause a persistent imbalance in the intestinal expression of the HNF4α isoforms (Figure 4d, e, Supplementary Figure S4A). This could explain the lack of rescue in barrier function by Viv chow in WT mice (Figure 4c): we have reported previously that mice expressing only the P2-HNF4α isoform have reduced intestinal epithelial barrier function.25 In contrast, the level of P1-HNF4α protein in the distal colon of WT (and α1HMZ) mice was not significantly altered by the SO+f diet (Figure 4d,e, Supplementary Figure S4B, C, D). Taken together, these results suggest that a diet high in LA, such as that found in soybean oil, may increase susceptibility to colitis in part by increasing the expression of the P2-HNF4α isoform in the intestines, thereby compromising epithelial barrier function.
A diet high in LA causes an increase in intestinal mAIEC
Another potential mechanism by which the SO diet may increase susceptibility to colitis is via alteration of the gut microbiome. Examination of the intestinal microbiota revealed that a diet high in soybean oil (with no added fiber, referred to as SO, see Supplementary Table S1) causes dysbiosis of the bacteria associated with the intestinal epithelial cells, with a notable increase in the relative abundance of a specific Escherichia coli phylotype (Figure 5a, top orange segment). Since a portion of the rRNA ITS region of this E. coli phylotype had 100% sequence identity with an adherent, invasive Escherichia coli (AIEC), we recently characterized in another genetic mouse model of IBD susceptibility,32 and we conducted a phenotypic characterization of the isolate (Figure 5b). In Caco2-brush border epithelial cells (Caco-2BBE), the E. coli isolate (designated SO-mAIEC) demonstrated increased adherence and invasion compared to nonpathogenic E. coli K-12. It also had values similar to those of the well characterized AIEC LF82, which is associated with IBD in humans.42 SO-mAIEC also showed greater replication in murine macrophages (J774A.1) than both E. coli K-12 and AIEC LF82: given that LF82 is a human isolate, and it may not invade and replicate in mouse macrophages as well as the mouse AIEC. These results confirm that the E. coli phylotype and isolate, which increased in abundance in the intestines of mice fed the SO diet, is indeed an AIEC, hence the designation SO-mAIEC, the full strain designation being UCR-SoS5.
Figure 5.
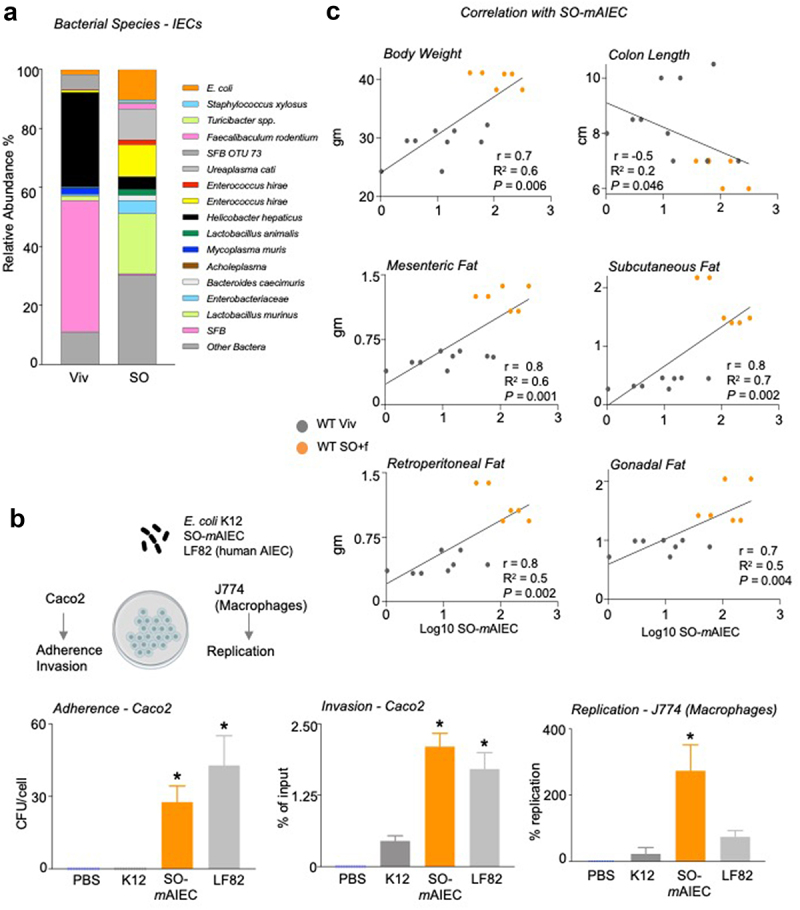
A diet high in LA increases the abundance of SO-mAIEC in WT mouse intestines.
Notes: (a) Bacterial species plots of intestinal epithelial cells (IECs) from the small and large intestines (N = 4–5) of mice fed Viv chow or an SO diet, with no added fiber. (b) Correlations between indicated mouse metadata and log10 of SO-mAIEC relative abundance in the IECs of mice fed Viv or SO+f diets. Spearman’s correlation coefficient (r) (for the body weight and adipose tissue) and Pearson correlation coefficient (for colon length) (r) goodness of fit or R2 values for linear regression and P value of the correlations (P) are indicated on the graphs. (c) Phenotypic characterization of the E. coli isolate enriched by SO (SO-mAIEC), compared with the human AIEC LF82 and the nonpathogenic E. coli K12. Assessments were made for bacterial adherence to Caco-2BBe cells, intracellular invasion of CaCo-2BBe cells, and replication in J774A.1 murine macrophages as indicated. * P < .05, one-way ANOVA, Tukey’s post hoc comparison. N = 12 across four experiments.
Interestingly, the intestinal abundance of the SO-mAIEC positively correlates with body weight and adipose tissue weight in a significant fashion (Figure 5c), suggesting it may play a role in the obesogenic properties of the SO diet.33,34 Conversely, colon length showed a modest negative correlation with SO-mAIEC abundance (Figure 5c), raising the possibility that this bacterium could also play a role in the reduction of colon length in mice fed the SO+f diet (Figure 3e).
SO-Maiec can accumulate, utilize, and metabolize LA
To further investigate the link between the SO diet and the SO-mAIEC, we conducted a targeted quantitative metabolomic analysis of SO-mAIEC grown in vitro in media without (LB) or with soybean oil (LB+SO) (Figure 6a). Since soybean oil is high in PUFAs (~55% LA and 7% alpha-linolenic acid, ALA, C18:3 ω3), we used a platform that measures bioactive metabolites of PUFAs, such as oxylipins, endocannabinoids, and N-acylethanolamines (NAEs). Principal component analysis (PCA) of the results shows a notable difference in the profiles obtained for SO-mAIEC grown in the absence or presence of soybean oil for five of six samples (Figure 6a). This difference was driven mainly by the levels of the omega-6 LA and the omega-3 ALA and their epoxy-derivatives, which coalesced into a single cluster component (Cluster 10), and the omega-3 EPA, which is derived from ALA, and its metabolites (Cluster 12) (Supplementary Figure S5A and Supplementary Table S3). The levels of LA and ALA were increased significantly in SO-mAIEC grown in the presence of soybean oil compared to control media (Figure 6b), as were the levels of three oxylipin metabolites of LA (9-KODE, 9,10-EpOME and 12,13-EpOME) (Figure 6c). Given that none of these LA-derived compounds showed a significant difference in levels in the growth media lacking bacteria (Supplementary Figure S5C), these results suggest that the SO-mAIEC isolate can accumulate and metabolize LA from soybean oil.
Figure 6.
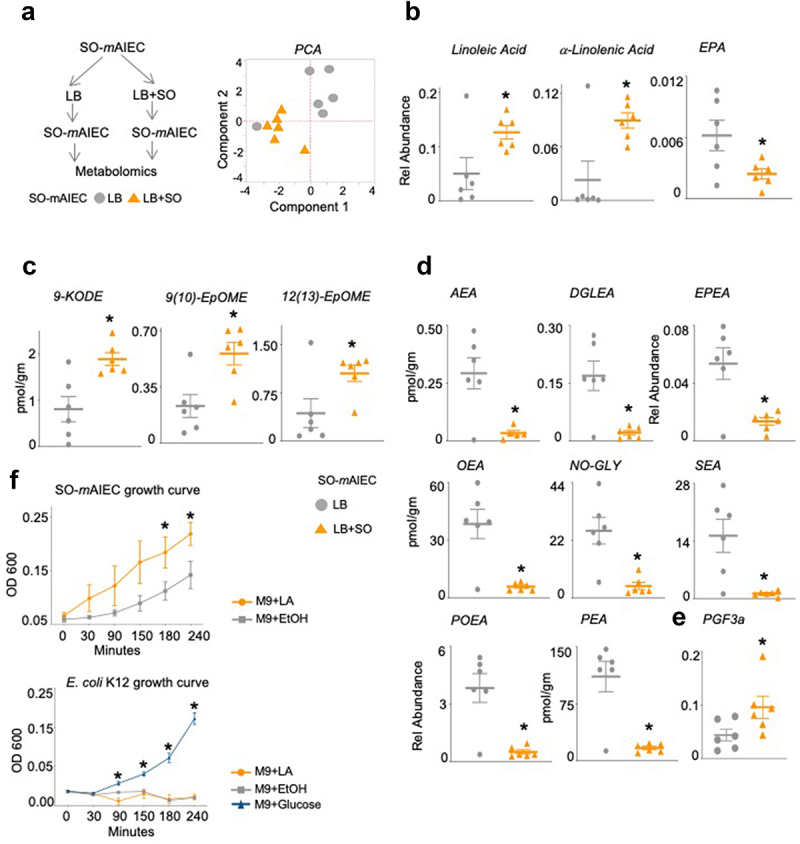
Soybean oil increases oxylipins and decreases endocannabinoid system metabolites in SO-mAIEC cultured in vitro.
Notes: (a) Experimental workflow and Principal Components Analysis (PCA) of oxylipin and endocannabinoid/NAE metabolites in SO-mAIEC grown with or without SO in the media. Absolute levels of fatty acids (b), oxylipin metabolites (c), endocannabinoid system metabolites (d) and (e) prostaglandins measured in SO-mAIEC grown in vitro in the presence or absence of SO in the media. *P < .05, T-test. N = 6 per group. (f) Growth curves for SO-mAIEC and E. coli K-12 grown in Minimum Essential Medium (M9) with LA, ethanol, or glucose as the carbon source. *P < .05, T-test. N = 3 replicates per culture. See Supplementary Figure S6.
Levels of an NAE derived directly from LA – (linoleoylethanolamide, LEA), ALA (alpha-linolenoylethanolamide, aLEA), and DHA (docosahexaenoylethanolamide, DHEA) – were not significantly different in SO-mAIEC grown in the absence or presence of soybean oil (Supplementary Table S4). However, the endocannabinoid anandamide (arachidonoyl ethanolamide, AEA) and closely related NAEs – dihomo-γ-linolenoylethanolamide (DGLEA), eicosapentaenoylethanolamide (EPEA), oleoylethanolamide (OEA), stearoylethanolamide (SEA), palmitoylethanolamide (PEA), and palmitoleoylethanolamide (POEA), as well as N-oleyl glycine (NOGLY) – are all significantly decreased in SO-mAIEC grown in the presence of soybean oil (Figure 6d). In contrast, the prostaglandin PGF3a is increased in the SO-mAIEC (Figure 6e).
To determine whether the SO-mAIEC can utilize LA as an energy source, we grew SO-mAIEC in a minimal medium (M9) supplemented with either ethanol (vehicle) or LA as the sole source of carbon. The SO-mAIEC grew significantly faster in the presence of LA, while the nonpathogenic E. coli K12 did not grow at all (Figure 6f). The viability of the initial K12 culture was confirmed by adding glucose to a parallel culture as well as to the LA (and ethanol) cultures after 4 h (Figure 6f and Supplementary Figure S5D). These results suggest that LA may be impairing the growth of E. coli K12, consistent with published results indicating that LA is bacteriostatic to a number of microorganisms, including the probiotic Lactobacillus species.43 Indeed, a daily LA gavage for 3 d in mice resulted in a change in the beta diversity of the fecal microbiome composition, including a decrease in the abundance of Lactobacillus murinus (Supplementary Figure S5E). Interestingly, the pathogenic human AIEC, LF82 also did not grow in the presence of LA (Supplementary Figure S5D). This could be explained by the fact that the LF82, unlike the SO-mAIEC, was not isolated from an LA-rich environment and thus may not possess the ability to use LA as a carbon source as does SO-mAIEC.
Intestinal epithelial barrier function in germ-free mice is affected by a diet high in LA
The results thus far suggest that a diet high in soybean oil may induce susceptibility to colitis by providing an environment that is conducive for the outgrowth of the pathobiont SO-mAIEC which has increased levels of LA oxylipins and decreased levels of NAEs and the endocannabinoid AEA, at least in vitro. To determine whether the host can also contribute to SO-induced susceptibility to colitis, we employed germ-free (GF) mice. Since these animals do not have any gut bacteria, any effects that are observed in response to the high soybean oil diet can be attributed to the host. GF animals are known to have a reduced mucus layer due to the absence of bacteria43 and hence are extremely sensitive to epithelial injury during DSS treatment.44 Therefore, we examined the intestinal epithelial barrier function in both GF and conventionally raised (Conv) mice fed the SO+f diet (but no DSS treatment) using the FITC-Dextran assay as an indicator of IBD susceptibility.45
As shown in Figure 4c, the SO+f diet significantly increased barrier permeability in Conv mice fed SO+f for 12 weeks versus the Viv chow control. While the difference in barrier permeability between GF mice on Viv chow versus SO+f diet was not significant (Figure 7a), FITC levels in the serum of GF SO+f mice were significantly greater than in the Conv SO+f mice (P < 0.005) (Supplementary Figure S6D). Taken together, these results suggest that while the presence of gut bacteria in Conv mice is important for decreased barrier function induced by the SO+f diet, some components, or host-derived metabolites, of the SO+f diet may also exacerbate barrier dysfunction in GF mice.
Figure 7.
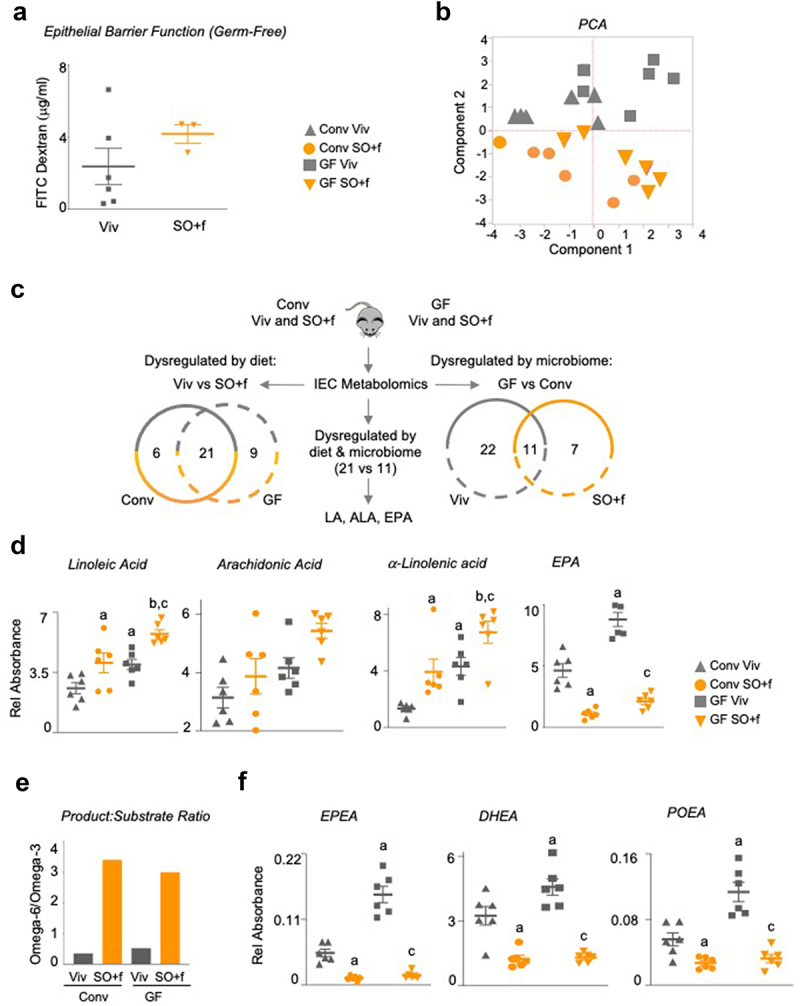
A diet high in LA decreases barrier function and alters the metabolome in the intestines of conventionally raised and germ-free mice.
Notes: (a) Epithelial barrier permeability as measured by FITC-Dextran in the blood in GF mice fed either Viv chow or SO+f diet for 12 weeks. T-test. N = 3–5 per group. See Supplementary Figure S6D for comparison of Conv and GF FITC. (b) PCA of oxylipin and endocannabinoid/NAE metabolites in IECs isolated from Conv and GF mice fed either Viv chow or SO+f diet for 8 weeks. N = 6 per group. (c) Schematic and Venn analysis of metabolomics data in B. Absolute levels of fatty acids (d), graph showing ratio of product-substrate for metabolism of LA to AA (omega-6) and ALA to EPA (omega-3) (e) and absolute levels of endocannabinoids/NAEs (f) measured in IECs from Conv and GF mice fed either Viv chow or SO+f diets for 8 weeks. a vs Conv Viv, b vs Conv SO+f, c vs GF Viv P < .05, P = .07 for AA Conv Viv vs GF Viv, One-way ANOVA (Holm-Sidak posthoc). N = 6 per group. See Supplementary Figures S7 and S8.
Impact of a diet high in LA on the gut metabolome
To determine whether the gut metabolome is affected by the SO diet and whether the gut microbiota plays a role, we applied the metabolomic analysis in Figure 6 to the IECs isolated from Conv and GF mice fed either Viv chow or the SO+f diet. We chose a shorter time on the diet (8 weeks) to identify compounds that might cause barrier dysfunction without the confounding factor of obesity seen at later time points. At 8 weeks on the diet, Conv mice on the SO+f diet were just beginning to show increases in body weight and serum FITC; in contrast, there was no difference in body weight between the GF Viv chow and SO+f mice (Supplementary Figure S6A vs Supplementary Figure S6B). This is consistent with previous reports showing that GF mice are resistant to HFD-induced obesity.46,47 PCA analysis of the metabolites shows that the two dietary groups (SO+f and Viv chow) are separated from each other regardless of the microbiome status (Conv or GF) of the mice, reinforcing the notion of host-dependent effects (Figure 7b). Variables with the largest contribution to this separation were EPA and docosahexaenoic acid (DHA) and their lipoxygenase and soluble epoxide hydrolase derivatives (Cluster 1) (Supplementary Figure S6C and Supplementary Table S3).
To better decipher the contributions of diet and the microbiota to the gut metabolome in vivo, we conducted a comparative analysis of the metabolites that were significantly dysregulated in IECs harvested from the four conditions: Conv and GF mice fed Viv chow or SO+f. Analysis of metabolites altered by diet identified a total of 27 compounds that were significantly dysregulated in Conv mice fed Viv chow versus the SO+f diet; in the GF comparison there were 30 compounds (Figure 7c, Supplementary Figure S7). A total of 21 compounds were dysregulated in both the Conv and GF comparisons (and in the same direction), suggesting that diet and/or host cells rather than bacteria play a role in their accumulation/metabolism.
Analysis of metabolites whose levels are significantly altered by the microbiome (Conv versus GF comparison for both Viv chow and SO+f diet) identified 33 metabolites that differ in the Viv chow comparison and 18 in the SO+f comparison (Figure 7c and Supplementary Figure S8). Of these, 11 are common to both Viv chow and SO+f diet, suggesting that the presence or absence of gut microbiota is the determining factor for these metabolites rather than diet. Notably, LA, ALA, and eicosapentaenoic acid (EPA) were the only three compounds dysregulated by both diet (21 common metabolites in Viv chow versus SO+f) and the microbiome (11 common metabolites in GF versus Conv mice) (Figure 7c).
In both GF and Conv mice, LA and ALA were increased by SO+f while EPA was decreased (Figure 7d). Intriguingly, all three compounds were increased in GF Viv chow mice compared to Conv mice, suggesting that the gut bacteria may sequester these compounds. Arachidonic acid (AA), the pro-inflammatory metabolite of LA, had a similar profile to LA and ALA in the Conv and GF mice, although none of the differences were significant (Figure 7d). The product:substrate ratio for omega-6 (AA:LA) was similar for Viv and SO diets in both Conv and GF mice. However, the product:substrate ratio for omega-3 (EPA:ALA) was reduced almost 10-fold by SO HFD in both Conv and GF mice (Supplementary Figure S7C). Consequently, the ratio of the omega-6:omega-3 product:substrate was greatly increased by the SO diet in both Conv and GF mice (Figure 7e), suggesting that ALA metabolism to EPA was severely impaired by the SO diet.
Other NAEs were similarly dysregulated by both diet and gut microbiota. For example, EPEA (derived from EPA), DHEA (derived from EPA via DHA), and POEA (derived from palmitoleic acid, C16:1) are all decreased by the SO+f diet in both GF and Conv mice, and in Conv versus GF mice (Figure 7f, Supplementary Figures S7, S8). This is analogous to the striking decrease observed for many NAEs in the LB+SO bacterial culture (Figure 6d).
Role of LA in diet-induced susceptibility to colitis and gut dysbiosis
To determine whether the enhanced susceptibility to colitis is indeed due to the high LA content of traditional soybean oil, we first examined the effect of an isocaloric HFD made with a genetically modified soybean oil low in LA in WT mice treated with 2.5% DSS (Plenish: 7.42% LA in the oil, 2.6 kcal% LA in the HFD vs 19 kcal% LA in the SO HFD, Supplementary Table S1). The results show that Plenish-fed mice lost less weight and had fewer morphological changes in the colon including less immune infiltrate and crypt cell damage than the SO-fed mice (Figure 8(a,b)). These results are consistent with a reduced disease activity index (Supplementary Table S2) of the IL-10 deficient mice fed a HFD comprised olive oil compared to the SO HFD. Notably, the diet high in olive oil, which has a fatty acid composition similar to that of Plenish with 4.5 kcal% LA versus 3.3 kcal% in Viv chow (Supplementary Table S1) yields a disease activity index essentially identical to that of the low-fat Viv chow (Figure 8c).
Figure 8.
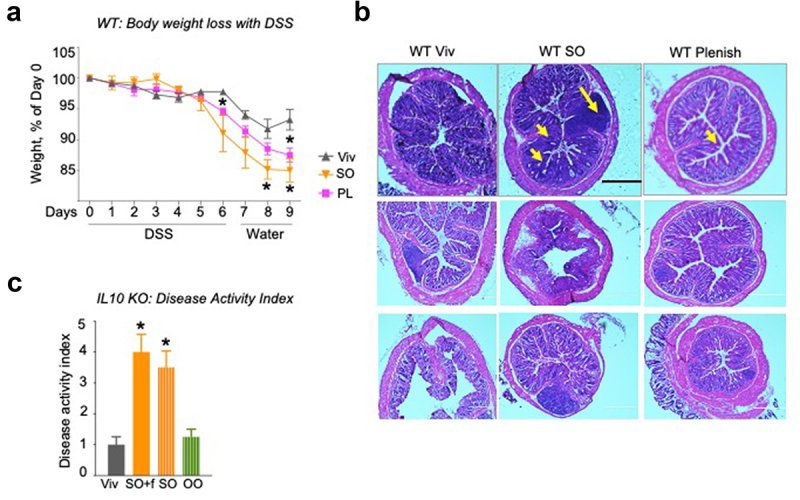
Impact of LA on gut microbiome.
Notes: WT mice on Viv chow, SO or PL diets for 12 weeks were treated with 2.5% DSS for 6 d, followed by 3 d recovery: % body weight loss (a) and colonic histology (b) of DSS-treated mice. Big arrow, immune infiltrate; small arrow, loss of crypt structure; Scale bar is 400 microns. Additional sections are shown in Supplementary Figure 9. (c) Disease activity index (DAI) after 6 weeks on an indicated diet. See Supplementary Table 2 for details on DAI scores. * vs Viv, a vs untreated WT Viv, b vs untreated WT SO+f. T-test, P < .05 N = 5–12 per group.
Since some studies have shown ambiguous results regarding the impact of inulin, the fiber in the SO+f diet, on IBD,48–51 we also fed the IL-10−/− mice an isocaloric SO diet (35 kcal% fat) lacking fiber for 6 weeks followed by an assessment of disease activity index (Supplementary Table S2). The results show nearly identical disease indices between the two SO diets, with and without fiber, both of which were significantly greater than the index of the Viv chow-fed mice (Figure 8c).
Discussion
Soybean oil, which consists of more than 50% LA (C18:2 omega-6), is currently the most highly consumed cooking oil in the U.S. and the second most produced edible oil in the world,2,52 and its increased consumption parallels the rise in IBD incidence in humans3. While there is an increasing awareness that diets high in saturated as well as unsaturated fats, such as LA, are implicated in the pathophysiology of IBD,1,6,7 there is relatively little known about the underlying mechanism. Here, we show that a HFD based on soybean oil, with amounts of the essential fatty acid LA far exceeding the minimum daily requirement, increases susceptibility to colitis in mice via a complex mechanism which impacts the microbiome, the balance of omega-6 to omega-3 bioactive metabolites and the IBD susceptibility gene HNF4α.
Using several different in vivo models of colitis and in vitro cell-based assays, we demonstrate for the first time that ingestion of a HFD based on soybean oil can: (i) increase the abundance of an adherent, invasive E. coli (AEIC), a known pathobiont associated with colitis, and decrease the abundance of probiotic Lactobacillus (ii) increase pro-inflammatory LA-derived oxylipins in both host and bacterial cells; and other LA metabolites such as arachidonic acid (AA) and prostaglandins in the host; (iii) decrease levels of omega-3 EPA and anti-inflammatory metabolites found in the endocannabinoid system; (iv) cause dysregulation of immune cells; (v) increase the level of P2-HNF4α in the colon; (vi) increase intestinal epithelial barrier permeability; and (vii) cause other phenotypic changes typical of colitis such as loss of body weight, altered colon and crypt length, and disruption of colon morphology. Importantly, isocaloric HFDs low in LA – olive oil and a genetically modified soybean oil (Plenish) – did not increase susceptibility to colitis, suggesting that the high LA content in soybean oil could be the determining factor. Finally, given that the effects of the soybean oil diet were similar with or without fiber (Figure 8), this suggests that any potential beneficial effect that might have been derived from the addition of dietary fiber was evidently overshadowed by the high LA content of the SO diet.
Based on these findings, we propose a bipartite mechanism for LA/SO-induced colitis susceptibility involving both gut bacteria and host cells (Figure 9a). On the one hand, the high LA content of SO provides an endogenous AIEC normally present in the gut in very low amounts with a growth advantage over other bacteria, including beneficial ones like Lactobacillus species. The SO-mAIEC can not only use LA as a carbon source but also metabolize LA into bioreactive oxylipins and, in the presence of SO, decrease its production of anandamide and other NAEs. On the other hand, experiments in GF mice show that the SO diet can directly affect host cells, resulting in elevated oxylipins from LA as well as pro-inflammatory arachidonic acid (AA, derived from LA) and prostaglandins derived from AA. Interestingly, the levels of the other essential fatty acids in soybean oil, ALA (C18:3 omega-3), were also elevated in the mouse gut and the in vitro SO-mAIEC culture in the presence of soybean oil. However, unlike LA, which appears to be metabolized by both SO-mAIEC and the host IECs, oxylipins derived from ALA as well as EPA and its NAE metabolites were all decreased, except one (Figure 9b, Supplementary Figure S7). The net result is a shift in the balance of anti-inflammatory omega-3 ALA-derived metabolites to pro-inflammatory omega-6 LA-derived metabolites. This shift, along with the increase in the pathogen SO-mAIEC, could tip the balance even further to a proinflammatory state (Figure 9c), a condition commonly found in IBD patients.53 The intestinal immune dysregulation noted in WT mice fed the SO HFD could also be linked to a pro-inflammatory state caused by an elevated omega-6/omega-3 ratio.54,55 Finally, the balance of HNF4α isoforms in the colonocytes is altered, resulting in sustained levels of P2-HNF4α, which is associated with decreased barrier function and sensitivity to colitis (Figures 9a,c).
Figure 9.
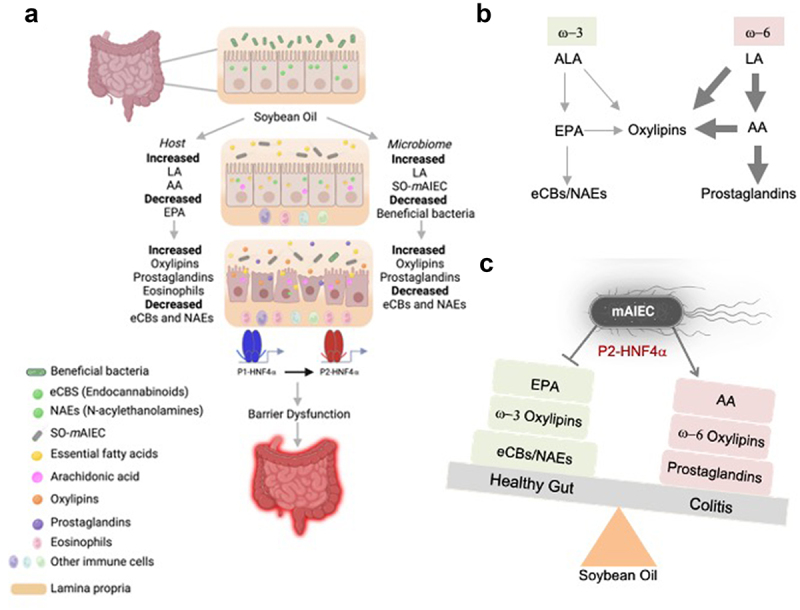
Proposed model by which a soybean oil-based HFD increases susceptibility to colitis.
Notes: (a) Impact of a diet high in soybean oil on the intestinal epithelium showing specific effects on host (left) and microbial cells (right) which ultimately leads to disruption of barrier function and IBD. (b) Simplified metabolic pathway of linoleic (LA) and alpha-linolenic acid (ALA). (c) Disrupted balance between ALA- and LA-derived anti- and pro-inflammatory metabolites caused by a diet high in soybean oil, which is tipped even further by the pathobiont SO-mAIEC. See Discussion for additional details.
Diet-induced adherent invasive E. coli (AIEC) in susceptibility to colitis
The pathobiont AIEC has recently been shown to play an important role in IBD in humans30,42 and mouse AIEC.32 There are also reports indicating that diet can impact the abundance or colonization efficiency of exogenous AIECs added to live animals.56–58 However, to our knowledge, this is the first study to demonstrate that a specific diet can result in a selection of an endogenous AIEC in an in vivo system. We also show that a key component of that diet (LA) can confer a nutritional and survival advantage for this pathogen over other gut microbes. The net result is dysbiosis in mice similar to the disruption of the balance between beneficial (e.g., Lactobacillus) and detrimental bacteria (AIEC) observed in human IBD.42,59–61
Increased levels of oxylipins and prostaglandins in susceptibility to colitis
LA oxylipins are bioactive, pro-inflammatory molecules that have been recently linked to IBD.17,62 Our in vitro results suggest that once established in the gut, SO-mAIEC can maintain high levels of LA-derived oxylipins. It remains to be determined whether the SO-mAIEC can also endogenously produce these compounds from the LA in soybean oil. Similarly, metabolomic analysis of Conv versus GF mice fed the SO HFD indicates that the IECs of the host also have high levels of LA-derived oxylipins, even in the absence of bacteria. Indeed, IECs from GF mice had higher levels of LA than those from Conv mice, providing in vivo evidence that the gut microbiota (such as AIEC) can take up and metabolize LA. Increased levels of oxylipins in the intestines of SO-fed mice are reminiscent of our previous findings showing that LA (and ALA) oxylipin levels in the liver correlate with obesity, which is also linked to chronic inflammation.34,63 While not all changes in oxylipin metabolites in the bacterial culture in vitro were the same as those identified in vivo (Supplementary Figures S5, S7 and S8), a complex pattern of some oxylipins being increased while others are decreased has been reported in colon biopsies from UC patients.17
Decreased endocannabinoid system metabolites and EPA in susceptibility to colitis
The endocannabinoid AEA and numerous other NAEs – derived from LA, ALA, and other fatty acids – were downregulated in both the bacterial and mouse metabolomic analyses. Particularly intriguing was the fact that, compared to the media alone, SO-mAIEC grown in the control media had significant levels of these compounds which were markedly decreased in the presence of SO (AEA, DGLEA, EPEA, NO-GLY, OEA, PEA, POEA, SEA). This suggests not only that these bacteria can produce endocannabinoids and NAEs but also that the presence of SO in the media can either inhibit that production and/or cause increased degradation or secretion of these bioactive compounds. The finding that bacteria can produce endocannabinoids is a relatively new one;16,64 there are only a couple of published works on soybean oil impacting endocannabinoid levels but none in bacteria or the gut.
There were also decreased levels of NAEs (aLEA, DGLEA, DHEA, EPEA, NA-Gly, PEA, and POEA) in the mice on the SO diet (Conv and/or GF), suggesting that host IECs also have decreased production/increased degradation of endocannabinoid system components in the presence of soybean oil. For example, AEA, which is derived from LA and considered to be a ‘gate opener’ in the intestinal epithelium, is upregulated in GF mice by the SO diet; in contrast, PEA, a protector of barrier function, is downregulated in SO-mAIEC and GF mice in the presence of SO.16 These results are consistent with decreased endocannabinoid levels being associated with increased colitis in humans, and relevant therapeutically in that endocannabinoids (and cannabis) are increasingly being explored for potential therapeutic activity against IBD.16,17,65
One of the best-studied omega-3 PUFAs in terms of beneficial health effects, EPA, was significantly decreased in the in vivo metabolome in the presence of soybean oil. This is despite the fact that its parent fatty acid, ALA, is enriched roughly 30-fold in the soybean oil diet (Supplemental Table S1) suggesting that some aspects of the ALA to EPA metabolic pathway in the host are compromised by dietary soybean oil and/or that EPA is selectively degraded in the presence of SO. One possible explanation for the observed imbalance could be that the excessive amount of LA in the SO diet outcompetes ALA in the utilization of the shared set of enzymes involved in their metabolism.66 This is supported by the fact that while the product/substrate ratio for AA:LA is similar between Viv and SO+f diet fed mice, the ratio for EPA:ALA is decreased 12-fold in mice fed the SO HFD compared to those on Viv chow, leading to a remarkable enhancement of the omega-6 pathway over the omega-3 pathway (Figure 7e). The higher levels of EPA observed in GF compared to Conv mice also suggests that the presence of one or more gut bacterium may lead to lower levels of EPA in the mouse IECs. Many EPA metabolites, which are considered to be anti-inflammatory – e.g., DiHETEs, HEPEs, PGF3a15,67 – were also decreased by the SO+f diet in both GF and Conv mice, suggesting a host effect. In contrast, the PGF3a levels were increased in the SO-mAIEC culture. It remains to be determined whether this prostaglandin is also increased in bacterial cells in the mouse gut and if so whether it is related to the decrease observed in the host IECs.
Role of HNF4α isoforms in soybean oil-induced susceptibility to colitis
We have previously shown that the increased intestinal expression of P2-HNF4α leads to decreased barrier function and increased DSS-induced colitis.25 Here, we show that the soybean oil diet increased P2-HNF4α protein as well as epithelial permeability (Figure 4), suggesting that an imbalance of the HNF4α isoforms in the gut may play a role in SO-induced colitis susceptibility (Figure 9). While the cause of the increase in the expression of P2-HNF4α remains to be determined, bacterial dysbiosis caused by high dietary LA could be involved as HNF4α expression has been shown to be modulated by the microbiome,68,69 although different isoforms were not examined in those studies. Another possibility is that LA itself alters the balance of the HNF4α isoforms. We previously identified LA as the endogenous ligand for HNF4α and showed that it can decrease its transcriptional activity and protein stability.19 However, since both P1- and P2-HNF4α contain identical ligand-binding domains, any potential impact of LA on the HNF4α isoform balance would likely be a complex one. Finally, others have recently noted that HNF4α shapes the intestinal epithelial lymphocyte compartment via direct regulation of immune signaling molecules,70 consistent with the SO HFD increasing monocytes in the IEL compartment in the α1HMZ exon swap mice.
Conclusion
Our results in the GF mice suggest that increased levels of omega-6 oxylipins in conjunction with decreased levels of omega-3 and endocannabinoid system metabolites can decrease barrier function. A similar mechanism appears to be at play in conventional mice with the added feature of selection of the SO-mAEIC pathobiont by the high LA content of SO and tipping the balance even further in the direction of colitis. The fact that the same types of compounds are altered in both host and bacteria underscores the importance of determining exactly how these bioactive lipids can alter microbial and host intestinal barrier function.
Finally, our results suggest that the naturally high LA content of soybean oil may be contributing to increased IBD in the U.S. by multiple mechanisms involving both the gut microbiota and the host cells and creating an immunoactive environment typical of IBD by both increasing pro-inflammatory and decreasing anti-inflammatory molecules. The specifics of this mechanism remain to be determined in future studies.
Methods
Animals and diets
Care and treatment of animals was in accordance with guidelines from and approved by the University of California, Riverside Institutional Animal Care and Use Committee, and followed NIH guidelines. Young adult male mice were maintained on a 12:12 h light–dark cycle in either a conventional, nonspecific-pathogen free vivarium or in a gnotobiotic facility, as indicated. Conventionally raised, wild-type (WT) C57BL/6N (Charles River), exon swap HNF4α (α1HMZ)24 and IL-10−/− mice71 (Jackson Labs, Stock#: 002251) were used. The IL-10−/− mice were in the B6 strain, which is known to develop a milder form of spontaneous colitis (JAX Labs, catalog # 002251). Germ-free mice (C57BL/6N, Taconic) were raised under gnotobiotic conditions, as described previously.72 All the mice were fed a standard vivarium chow from Newco until they were weaned 21 d after birth (autoclaved LabDiet 5K52 for the germ-free mice and non-autoclaved LabDiet 5001 for the conventionally raised mice). Post-weaning, the mice were either continued on the low-fat vivarium chow or fed one of four isocaloric high fat diets for up to 24 weeks: a soybean oil-based high fat diet with (SO+f) or without added fiber (SO), a low LA soybean oil diet (Plenish) or an olive oil diet. We decided to use inulin as a source of fiber (along with cellulose) because we did not want to introduce a dietary component that could potentially compound the effect of soybean oil. The fiber composition of Viv chow is an ill-defined mix of cellulose, hemicellulose and lignin. For the SO+f diet inulin was used as a source of fiber (along with cellulose) to avoid compounding the potential effects of soybean oil due to either a deficiency of fiber or addition of a less-well characterized source of fiber. All four high-fat diets have 35 kcal% fat and were formulated by Research Diets Inc. See Supplementary Table S1 for detailed composition of diets. All mice had ad libitum access to food and water. At the end of the study, mice were euthanized by carbon dioxide inhalation, in accordance with NIH guidelines.
Dextran sulfate sodium (DSS) treatment
WT or α1HMZ mice that had been on the diets for 8–15 weeks were treated with 2.5% dextran sodium sulfate salt (DSS) (reagent grade, MW 3.6–5 kDa, MP Biomedicals, #160110, Santa Ana, CA) in water ad libitum for 6 d and sacrificed immediately or allowed to recover up to 3 d with tap water. Mice were continued on the same diet before, during and after DSS treatment and monitored daily for changes in body weight, stool consistency, ruffled fur and activity level. The presence of blood in the stool was checked every other day using Hemoccult Dispensapak Plus (catalog no. 61130, Beckman Coulter).
Disease activity index
A composite disease activity index (DAI) score was calculated based on the following criteria:73,74 (1) weight loss, (2) colonic length-to-weight ratio, (3) hemoccult reading, and (4) gross morphological changes in colon. See Supplementary Table S2 for details on scoring criteria.
Linoleic acid gavage
WT mice on vivarium chow were gavaged with either pharma grade linoleic acid (catalog no. 39269-10 G, Millipore Sigma) or water for 3 d using disposable animal feeding needles (catalog no. 01-208-87, Thermo Fisher Scientific) and syringes (catalog no. 14-823-434, BD Slip tip, Thermo Fisher Scientific). The daily dose of LA gavage was 0.26 mg/kg body weight per day which corresponds to 2.33 kcal of energy from LA per day which makes up 17.38% of the daily caloric intake of a mouse (which is 13.44 kcal, based on average consumption of 4 g of food day/mouse and 3.36 kcal being provided by 1 gm of Viv chow). The Viv chow diet already provides 1.22 kcal % of energy from LA. Thus, the total kcals being derived from LA for a Viv chow fed mouse that is gavaged with 0.26 mg/kg LA is 17.38 + 1.22 which equals 18.6, which is equal to the kcal% LA in the SO+f and SO diets and is similar to what has been used in previous studies.75 The fecal samples were collected and stored at −80°C prior to the first gavage and at the end of the experiment (i.e., 24 h after the third gavage). These samples were used for bacterial rRNA internal transcribed spacer (ITS) library construction.
Tissue collection
Tissues were collected and snap-frozen in liquid nitrogen prior to storage at −80°C for immunoblotting and metabolomic analysis or fixed in 10% neutral-buffered formalin for 24 h before storing in 30% sucrose plus PBS solution at 4°C for subsequent histological analysis. Liver and adipose tissue (mesenteric, perirenal, gonadal, and flank subcutaneous) were excised and weighed.
Tissue embedding, sectioning, and staining
The entire large intestine was excised, and its length measured. A 1.0 to 1.5-cm piece adjacent to the rectum was cut and fixed in 10% neutral-buffered formalin as above, embedded in optimal cooling temperature (OCT) compound, and sectioned at 5-µm thickness on a Microm Cryostat and stored at −20°C. All slides were rehydrated in 95% ethanol for 7 min, tap water for 7 min, ddH20 for 2 min, and stained in hematoxylin (Ricca Chemical) for 40 s. Slides were then dipped in tap water for 30 s, running tap water for 90 s, 95% ethanol for 15 s and subsequently counterstained in eosin (Sigma-Aldrich) for 3 s and dipped in 95% and 100% ethanol two and three times, respectively, for 20 s each time. Slides were left in Citrisolve (Fisher Scientific) for at least 40 s. This staining process was completed in succession in a single session. Slides were fixed and preserved with Permount (Fisher Chemicals). Histology images were captured on an Evos Microscope (Life Technologies). Crypt length and submucosal thickness were measured using SPOT Imaging software (Sterling, MI).
Immunoblot analysis
Whole cell extracts were prepared from tissues stored in liquid nitrogen and analyzed by immuno-chemiluminescence after determination of protein concentration by the Bradford Assay, as described previously.25,76 The protein extracts were separated by 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to Immobilon membrane (EMD Millipore, Billerica, MA). The membrane was blocked with 5% nonfat milk for 30 min, incubated in primary antibodies (mouse monoclonal anti-HNF4α P1 and P2; catalog no. PP-K9218-00 and PP-H6939-00, respectively, R&D Systems) in 1% milk, overnight at 4°C. After several washes in TBST (Tris-buffered saline, 0.1% Tween 20), the blots were incubated in horseradish peroxidase (HRP)-conjugated goat anti-mouse (GαM-HRP) secondary antibody (Jackson ImmunoResearch Laboratories) for 40 min at room temperature followed by three 5-min washes in TBST and two 5-min washes in TBS (Tris-buffered saline). Blots were developed using SuperSignal™ West Pico PLUS Chemiluminescent Substrate (Thermo Fisher Scientific) and imaged in a Chemi-Doc imaging system (Bio-Rad). Coomassie staining of blots verified equal protein loading. The blots were re-probed for beta-actin (rabbit anti-actin; catalog no. A2066, Sigma) by washing twice in TBST before incubating in a stripping buffer (0.5 M NaOH solution) at room temperature for 5 min with shaking, washed twice with TBST and once with TBS (3 min each). Blots were blocked in 5% milk, and the immunoblot procedure described above was followed.
In-vivo permeability assay
Mice were fasted overnight on wood chip bedding (Newco Specialty, catalog # 91100). After 15 h, mice were weighed and gavaged with FITC-Dextran (FD-4 Sigma) diluted in water at a dose of 600 µg/gm body weight. The gavage was performed under yellow lights and staggered for 2 minbetween animals. Mice were sacrificed 4 h after gavage and blood was collected via cardiac puncture (BD 3-ml Luer-Lok Syringes, catalog no. 14-823-435, Fisher and BD 26 G ⅝ inch hypodermic needles, catalog no. 14–826-6A, Fisher) and transferred to 1.5-ml Eppendorf tubes. Samples were placed on ice for 45 min and then centrifuged at 9.3 rcf for 15 min after which serum was collected into a fresh tube. Samples were loaded into black 96-well plates (Corning, catalog no. 3991) in triplicate, at a dilution of 1:5 in water. Serum FITC-Dextran concentration was determined on a Veritas Microplate Luminometer (Turner Biosystems, Sunnyvale, CA), GloMax software (Promega, Madison, WI), using excitation/emission wavelengths of 490/520 nm.32 The relative fluorescence units obtained for the samples were compared to the values obtained from a standard curve generated by diluting the fluorophore stock in water.
Intestinal immune cell profile
IEL and LPL isolation
Intraepithelial (IELs) and lamina propria leukocytes (LPLs) were isolated from the mouse's small intestines as described previously.77 Briefly, the entire small intestine was excised and gently flushed with cold PBS and mesenteric fat and Peyer’s Patches were dissected away. The intestine was cut into 3 to 4-inch segments; each segment was rolled on a paper towel moistened with Gibco RPMI 1640 Media to remove any residual fat tissue. The segments were inverted using curved forceps and placed in 30 ml extraction medium – RPMI plus 93 µl 5% (w/v) dithiothreitol (DTT), 60 µl 0.5 M EDTA, and 500 µl fetal bovine serum per small intestine, ~40 cm long – and stirred at 500 rpm for 15 min at 37°C. A steel strainer was used to separate tissue pieces from the IEL-rich supernatant, which was placed on ice. Residual mucus from the tissue segments was removed by blotting on a dry paper towel. These fragments were then put in a 1.5-ml Eppendorf tube with 600 µl of digestion medium: 25 ml RPMI plus 12.5 mg dispase, 37.5 mg collagenase II, and 300 µl FBS. Dispase (Gibco, catalog no. 17105041) and collagenase (Gibco, catalog no. 17101015) were added immediately before use. The tissue was minced inside the tube using scissors, put in a cup containing 25 ml of digestion media, and stirred at 500 rpm for 15 min. Any large chunks of tissue were broken up by pipetting up and down with a serological pipette and stirring at 37°C was continued for an additional 15 min. Digested tissue and the IEL containing supernatant were passed through a 100-µm cell strainer into a 50-ml tube followed by a rinse with 20 ml of RPMI containing 10% FBS. The filtered solution was centrifuged at 500 × g for 10 min at 4°C; the supernatant was carefully decanted, and the pellet resuspended in 1 ml of RPMI containing 10% FBS. The resuspended cells were filtered through a 40-µm cell strainer into a 50-ml tube followed by a rinse with 20 ml of RPMI containing 10% FBS. The filtered solution was centrifuged at 500 × g for 10 min at 4°C; the supernatant was carefully decanted, and the pellet resuspended in 1 ml of RPMI containing 2% FBS. This suspension was then used for flow cytometry as described below.
PBMC isolation
Mice were euthanized, and 1 ml of blood was collected and immediately mixed with 1 mL of 4% sodium citrate. Next, 1 mL of wash media (RPMI with 5% heat-inactivated Fetal Calf Serum, FCS) was added and mixed, followed by the slow underlay of 1 mL Histopaque-1077 (catalog no: 10771, Sigma-Aldrich). This mix was centrifuged at 400 g for 30 min at room temperature. PBMCs were carefully aspirated from the interphase and washed with culture medium.
Flow cytometry
Isolated PBMC, IELs and LPLs were washed in fluorescence activated cell sorting buffer (FACS) buffer, incubated with Fc block (25 µg/mL αCD16/32 and 10 µg/ml rat IgG) and stained for 30 min with flow antibodies: F4/80 (Cl:A3–1, Bio Rad MCA497FB), SiglecF (E50–2440, BD Biosciences 562,757), CD4 (RM4–5, BD Biosciences 550,954), Ly6C (HK 1.4, Biolegend 128,018), CD11b (M1/70, Biolegend 101,226), CD11c (N418, Biolegend 117,310), Ly6G (1A8, Biolegend 127,628), MHCII (M5/114.15.2, Biolegend 107,622), CD19 (1D3, BD Biosciences 562,956), CD115 (AFS98, Biolegend 135,517) and CD8 (53–6.7, Biolegend 100,742). All cells were acquired on the BD LSRII (BD Biosciences) and analyzed using FlowJo (FlowJoTM v10). Cell populations were identified as follows: macrophage (CD11b+ F4/80+), eosinophils (CD11b+ SiglecF+), monocytes (CD11b+ Ly6C+), neutrophils (CD11b+ Ly6G+), CD4+ T cells (CD4+ CD8−), CD8+ T cells (CD8+ CD4−) and B cells (CD4− CD19+).
Isolation of intestinal epithelial cells (IECs) for metabolome and microbiome analysis
Intestinal epithelial cells (IECs) were isolated for microbiome and metabolomic analysis as previously described.32 Bacterial DNA was isolated from the IECs using the DNeasy PowerSoil Kit (Qiagen, Valencia, CA) with a 30-s bead-beating step using a Mini-Beadbeater-16 (BioSpec, Bartlesville, OK). For metabolomics analysis, the cells were flash frozen in liquid nitrogen and stored at −80°C until processing.
Bacterial rRNA internal transcribed spacer (ITS) library construction and sequencing
Illumina bacterial rRNA ITS gene libraries were constructed as follows: PCR was performed in an MJ Research PTC-200 thermal cycler (Bio-Rad Inc., Hercules, CA) in 25-μl reactions containing 50 mM Tris (pH 8.3), bovine serum albumin (BSA) at 500 μg/ml, 2.5 mM MgCl2, 250 μM of each deoxynucleotide triphosphate (dNTP), 400 nM of the forward PCR primer, 200 nM of each reverse PCR primer, 2.5 μl of DNA template and 0.625 units JumpStart Taq DNA polymerase (Sigma-Aldrich, St. Louis, MO). PCR primers targeted a portion of the small-subunit (ITS-1507F, GGTGAAGTCGTAACAAGGTA) and large-subunit (ITS-23SR, GGGTTBCCCCATTCRG) rRNA genes and the hypervariable ITS region,78 with the reverse primers including a 12-bp barcode and both primers including the sequences needed for Illumina cluster formation; primer binding sites were the reverse and complement of the commonly used small-subunit rRNA gene primer 1492 R79 and the large-subunit rRNA gene primer 129F.80 PCR primers were only frozen and thawed once. Thermal cycling parameters were 94°C for 5 min; 35 cycles of 94°C for 20 s, 56°C for 20 s, and 72°C for 40 seconds, followed by 72°C for 10 min. PCR products were purified using a Qiagen QIAquick PCR Purification Kit (Qiagen) according to the manufacturer’s instructions. DNA sequencing (single-end 150 base) was performed using an Illumina MiSeq (Illumina, Inc., San Diego, CA).
Bacterial rRNA ITS sequence processing and analysis
The UPARSE pipeline was used for de-multiplexing, length trimming, quality filtering, and amplicon sequence variant (ASV) picking using default parameters or recommended guidelines81 updated at https://www.drive5.com/usearch/manual10/uparse_pipeline.html. Briefly, after demultiplexing and using the recommended 1.0 expected error threshold, sequences were trimmed to a uniform length of 145 bp and then de-replicated. De-replicated sequences were subjected to error correction (denoised) and chimera filtering to generate zero radius operational taxonomic units (ZOTUs) using UNOISE3.82 An ASV table was generated using the otutab command. ASVs having non-bacterial DNA were identified by performing a local BLAST search83 of their seed sequences against the nucleotide database. ASVs were removed if any of the highest scoring BLAST hits contained taxonomic IDs within the rodent family, the kingdoms Fungi or Viridiplantae kingdoms, or PhiX. Taxonomic assignments of the bacterial ASVs were made by finding the lowest common taxonomic level of the highest BLAST hits excluding unclassified designations. Data were normalized within each sample by dividing the number of reads in each ASV by the total number of reads in that sample. The bacterial rRNA ITS sequences have been deposited in the National Center for Biotechnology Information (NCBI)’s Sequence Read Archive (SRA) under the SRA BioProject Accession PRJNA622821.
Bacterial rRNA ITS sequence analysis
QIIME84 was used to create the abundance tables for the phylotypes at various taxonomic levels. Correlation analyses and plots, as well as bacteria species plots, were performed using Prism (GraphPad, La Jolla, CA).
Beta diversity plot
QIIME84 was used to calculate a Hellinger beta diversity distance matrix (for the LA gavage experiment), which was depicted using principal coordinates analysis and statistically assessed using Adonis PERMANOVA tests.
Isolation of mAIEC
The mAIEC strain used in this study (UCR-SoS5, referred to as SO mAIEC in the Results) was isolated from subcutaneous fat collected from mice fed the high soybean oil diet with fiber (SO+f) using a selective medium, E. coli ChromoSelect Agar B, as described by the manufacturer (Sigma-Aldrich, St. Louis, MO). The strain was purified by selecting a colony from the selective media and then performing two successive streak plating procedures on LB agar to obtain single colonies. The strain was confirmed to have the identical rRNA ITS nucleotide sequence as the E. coli phylotype identified by the Illumina sequence analysis by PCR amplifying the rRNA ITS region of the strain and sequencing the amplicons using the Sanger method.
mAIEC phenotypic tests
Adherence and invasion ability of mAIEC to Caco-2 brush border epithelial (Caco2-BBe) cells was examined using previously described methods.32 Intracellular replication of mAIEC was performed on J774A.1 cells (ATCC TIB-67), murine macrophages maintained in RPMI plus 10% FBS, penicillin (100 U/ml), and streptomycin (100 µg/ml) until the day before the experiment, at which point they were maintained in the same media without antibiotics. Intracellular replication of mAIEC in J774A.1 cells was determined by a gentamicin survival assay using 24-well plates with 2 ml of media per well. J774A.1 monolayers were infected at an MOI of 20 bacteria/macrophage and incubated for 2 h at 37°C with 5% CO2. Infected macrophages were washed twice with PBS and incubated for 1 h in RPMI plus 10% FBS and 150 mg/ml gentamicin to kill extracellular bacteria. The J774A.1 cells were washed once with PBS and then lysed by incubation with 1 ml of 1% Triton-X 100 for 30 min; using two extra uninoculated wells, an average J774A.1 cell count per well was obtained at this point. The lysed cell solutions (1 ml) were collected, centrifuged at 14,000 × g for 10 s, the supernatants were decanted, and the cells were resuspended in 120 µl PBS. The lysed cell solutions were serially diluted, spread-plated on LB agar, incubated overnight at 37°C, and the bacteria were enumerated. This process was repeated for another plate but after the 1-h 150 mg/ml gentamicin incubation described above, the cells were washed once with PBS, and then RPMI plus 10% FBS and 20 mg/ml gentamicin was added and the cells were allowed to incubate for another 23 h at 37°C. Intracellular replication of the mAIEC and control bacteria was expressed as the mean percentage of the number of bacteria recovered 24 h post-infection divided by the number of bacteria 1 h post-infection, with the two J774A.1 cell counts being used to normalize the bacterial counts. The control bacteria were the LF82 human AIEC (kindly provided by the late Dr. Arlette Darfeuille-Michaud) and K12 (a noninvasive E. coli, ATCC 25,404).
Culture of mAIEC for metabolomic analysis
The mAIEC was grown aerobically in LB broth with 10% soybean oil for 20 h at 37°C with shaking at 350 rpm. The bacteria were collected by centrifugation at 14,000 × g for 30 s and the supernatant removed; this procedure was repeated two more times to remove as much of the soybean oil as possible. The bacterial pellets were flash-frozen in liquid nitrogen and stored at −80°C until they were analyzed. Controls were the mAIEC grown in LB broth without soybean oil as well as uninoculated LB with and without soybean oil that was subjected to incubation for 20 h at 37°C with shaking at 350 rpm.
Sole carbon source experiments
mAIEC or E. coli K-12 were grown aerobically in LB media for 20 h at 37°C at 300 rpm and then diluted (1:50) in minimum essential medium (standard M9 medium but without glucose), supplemented with vitamins (ATCC, catalog no. MD-VS) and trace minerals (ATCC, catalog no. MD-TMS). This medium was supplemented with either 2 mM linoleic acid dissolved in ethanol (Cayman Chemicals, catalog no. 9015050) or an equal volume of ethanol. A third treatment was included for E. coli K-12 in which the M9 medium was supplemented with 0.4% glucose. The OD600 measurements were taken on a Synergy HTX Microplate reader at 0, 30, 90, 150, 180 and 240 min after inoculation. To show the viability of the K-12 bacteria after LA and ethanol treatment, glucose was added at a concentration of 0.4% after 240 min and two additional OD readings, at 360 and 1440 min were taken. There were three replicate cultures per condition: results from one representative experiment out of at least two independent experiments are shown.
Metabolomic analysis
Cell pellet: non-esterified oxylipins, endocannabinoids, N-acylethanolamines, and polyunsaturated fatty acids extraction
Cell pellets (of IECs and bacteria) were weighed prior to extraction. Oxylipins, endocannabinoids, N-acylethanolamines (NAEs) and polyunsaturated fatty acids (PUFAs) were isolated by liquid extraction protocol using acetonitrile/isopropanol/water mixture [3:3:2 v/v] from approximately 80 mg of cell pellet and quantified by UPLC-MS/MS using internal standard methods. Briefly, ~80 mg of cell pellet was mixed with 20 µL BHT/EDTA (1:1 MeOH:water), 20 µL of 1250 nM deuterated oxylipins and endocannabinoids surrogates in methanol and 20 µL of 1-cyclohexyl ureido, 3-dodecanoic acid (CUDA) and 1-phenyl ureido 3-hexanoic acid (PUHA) at 5 µM in 1:1 methanol:acetonitrile. The internal standards CUDA and PUHA were added to samples after extraction and before analysis to allow estimation of isotopically labeled internal standards used for analyte quantification (see Supplementary Table S4 for complete list of internal standards). Samples were then homogenized using Geno/Grinder 2010 after addition of 0.5 mL of acetonitrile/isopropanol/water (3:3:2), together with three 3-mm stainless steel beads. Homogenate was centrifuged at 15,000 rcf for 10 min and filtered through a 0.1 µm PVDF spin filter and collected for mass spectrometry analysis described below.
Bacterial culture media: non-esterified oxylipins, endocannabinoids, N-acylethanolamines, and polyunsaturated fatty acids extraction
Non-esterified oxylipins, endocannabinoids, NAEs, and polyunsaturated fatty acids were isolated using solid-phase extraction with 60 mg Hydrophilic-Lipophilic-Balanced columns (Oasis, Waters Corporation, Milford, MA). Briefly, the columns were washed with one column volume of ethyl acetate followed by two column volumes of methanol and further conditioned with two column volume of 5% methanol, 0.1% acetic acid in water. Next, the columns were spiked with 5 µL BHT/EDTA (1:1 MeOH:water) and 5 µL of 250 nM deuterated oxylipins and endocannabinoid surrogates in methanol. Samples (200 µL) were mixed with 800 µL of 5% methanol, 0.1% acetic acid in water, transferred onto the column and extracted under the gravity. Columns were washed with 1 column volume of 30% methanol and 0.1% acetic acid in water. Analytical targets were eluted with 0.5 mL methanol followed by 1.5 mL ethyl acetate. Eluents were dried under the vacuum, reconstituted in 50 µL of 1-cyclohexyl ureido, 3-dodecanoic acid (CUDA) and 1-phenyl ureido 3-hexanoic acid (PUHA) at 5 µM in 1:1 methanol:acetonitrile, filtered through 0.1 µm PVDF spin filter and collected for mass spectrometry analysis described below.
Mass spectrometry analysis
The residues in the extracts were separated on a 2.1 mm × 150 mm, 1.7 μm BEH C18 column (Waters, Milford, MA) and detected by electrospray ionization with multi-reaction monitoring on an API 6500 QTRAP (Sciex; Redwood City, CA) and quantified against 7- to 9-point calibration curves of authentic standards as previously reported.85 Levels are reported in absolute amounts for all compounds for which the standards could be run simultaneously on the LC/MS. The exceptions are LA, ALA, EPEA, and POEA for which we report relative abundance since their respective standards are prone to oxidation. We have previously determined that the relative abundance of these compounds are consistent with their independent quantitative measurements on a GC/MS platform.
Statistical analysis
Data are presented as the mean ± standard error of the mean (SEM) using GraphPad Prism 6. One-way ANOVA or repeated measures (RM) ANOVA followed by post-hoc testing (Tukey’s for multiple comparisons or Sidak for pairwise comparisons, as specified in the figure legends) or Student’s T-test were used as appropriate and are indicated in the figure legends. Data were tested for normality using the Shapiro–Wilk normality test (see supplementary Table S3 for details). Statistical significance was set at an alpha level of 0.05 with a P-value less than or equal to 0.05 being considered significant. Linear regression analysis was performed between body weight, adipose tissue weight, liver as percent body weight, colon length, and relative abundance of mAIEC in intestinal epithelial cells. The following cutoffs were used to determine significance: Pearson’s or Spearman’s coefficient r > 0.5 with P ≤ 0.05. For the metabolomics data, outliers were first removed using the robust Huber M test, and missing data were imputed using multivariate normal imputation. Further, variables were clustered separately for each cell type using the JMP variable clustering algorithm, an implementation of the SAS VARCLUS procedure, and converted into cluster components for data reduction. Curated metabolomics data are presented in Supplementary Table S4. Cluster components were used for PCA analysis of experimental samples to provide an overview of metabolic changes.
Supplementary Material
Acknowledgments
We thank DuPont for Plenish oil. Model in Figure 9 was created with BioRender.com.
Funding Statement
NIH R01 DK053892, DK127082 (FMS); Crohn’s Colitis Foundation Career Development Award 454808 (PD); UCR Metabolomics Core Seed Grant (FMS, PD); FY17-18 P&F Grant from the NIH/UC Davis WCMC DK097154 (JB, PD). NIH R35 GM124724, R01 AI157106 (AH); Crohn’s and Colitis Foundation Senior Research Award, NIH R01 DK091281, American Gastroenterological Association IBD Research Award (DFM). NIH R01 AI153195 (MGN). Additional support was provided by the USDA Intramural project 2032-51530-022-00D and 2032-51530-025-00D (JWN). The USDA is an equal opportunity provider and employer. USDA National Institute of Food and Agriculture Hatch project CA-R-NEU-5680 (FMS).
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The bacterial rRNA ITS sequences have been deposited in the National Center for Biotechnology Information (NCBI)’s Sequence Read Archive (SRA) under the SRA BioProject Accession PRJNA622821 (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA622821/). The metabolomics data supporting the findings of this study are available within the article and the supplementary materials.
Supplementary material
Supplemental data for this article can be accessed online at https://doi.org/10.1080/19490976.2023.2229945
References
- 1.Knight-Sepulveda K, Kais S, Santaolalla R, Abreu MT.. Diet and inflammatory bowel disease. Gastroenterol Hepatol. 2015;11:511–28. [PMC free article] [PubMed] [Google Scholar]
- 2.Blasbalg TL, Hibbeln JR, Ramsden CE, Majchrzak SF, Rawlings RR. Changes in consumption of omega-3 and omega-6 fatty acids in the United States during the 20th century. Am J Clin Nutr. 2011;93(5):950–962. doi: 10.3945/ajcn.110.006643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Molodecky NA, Soon IS, Rabi DM, Ghali WA, Ferris M, Chernoff G, Benchimol EI, Panaccione R, Ghosh S, Barkema HW, et al. Increasing incidence and prevalence of the inflammatory bowel diseases with time, based on systematic review. Gastroenterology. 2012;142(1):46–54.e42. quiz e30. doi: 10.1053/j.gastro.2011.10.001. [DOI] [PubMed] [Google Scholar]
- 4.Barr LH, Dunn GD, Brennan MF. Essential fatty acid deficiency during total parenteral nutrition. Ann Surg. 1981;193(3):304–311. doi: 10.1097/00000658-198103000-00009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.da Costa GG, da Conceição Nepomuceno G, da Silva Pereira A, Simões BFT, da Costa GG, da Conceição Nepomuceno G, da Silva Pereira A. Worldwide dietary patterns and their association with socioeconomic data: an ecological exploratory study. Global Health. 2022;18(1):31. doi: 10.1186/s12992-022-00820-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tjonneland A, Olsen A, Overvad K, Bergmann MM, Boeing H, Nagel G, Linseisen J, Hallmans G, Palmqvist R, Sjodin H, et al. IBD in EPIC study investigators: linoleic acid, a dietary n-6 polyunsaturated fatty acid, and the aetiology of ulcerative colitis: a nested case-control study within a European prospective cohort study. Gut. 2009;58:1606–1611. [DOI] [PubMed] [Google Scholar]
- 7.Alzoghaibi MA, Walsh SW, Willey A, Iii FA, Graham MF. Linoleic acid, but not oleic acid, upregulates the production of interleukin-8 by human intestinal smooth muscle cells isolated from patients with Crohn’s disease. Clin Nutr. 2003;22:529–535. doi: 10.1016/S0261-56140300083-9. [DOI] [PubMed] [Google Scholar]
- 8.Hintze KJ, Benninghoff AD, Cho CE, Ward RE. Modeling the Western Diet for Preclinical Investigations. Adv Nutr. 2018;9(3):263–271. doi: 10.1093/advances/nmy002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marion-Letellier R, Savoye G, Ghosh S. IBD: in food we trust. J Crohns Colitis. 2016;10(11):1351–1361. doi: 10.1093/ecco-jcc/jjw106. [DOI] [PubMed] [Google Scholar]
- 10.Rashvand S, Somi MH, Rashidkhani B, Hekmatdoost A. Dietary fatty acid intakes are related to the risk of ulcerative colitis: a case–control study. Int J Colorectal Dis. 2015;30(9):1255–1260. doi: 10.1007/s00384-015-2232-8. [DOI] [PubMed] [Google Scholar]
- 11.Barros KV, Xavier RAN, Abreu GG, Martinez CAR, Ribeiro ML, Gambero A, Carvalho PO, Nascimento CMO, Silveira VLF. Soybean and fish oil mixture increases IL-10, protects against DNA damage and decreases colonic inflammation in rats with dextran sulfate sodium (DSS) colitis. Lipids Health Dis. 2010;9(1):68. doi: 10.1186/1476-511X-9-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wiese DM, Horst SN, Brown CT, Allaman MM, Hodges ME, Slaughter JC, Druce JP, Beaulieu DB, Schwartz DA, Wilson KT, et al. Serum fatty acids are correlated with inflammatory cytokines in ulcerative colitis. PLoS One. 2016;11(5):e0156387. doi: 10.1371/journal.pone.0156387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moldal T, Løkka G, Wiik-Nielsen J, Austbø L, Torstensen BE, Rosenlund G, Dale OB, Kaldhusdal M, Koppang EO. Substitution of dietary fish oil with plant oils is associated with shortened mid intestinal folds in Atlantic salmon (Salmo salar). BMC Vet Res. 2014;10(1):60. doi: 10.1186/1746-6148-10-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alvheim AR, Torstensen BE, Lin YH, Lillefosse HH, Lock EJ, Madsen L, Hibbeln JR, Malde MK. Dietary linoleic acid elevates endogenous 2-arachidonoylglycerol and anandamide in Atlantic salmon (Salmo salar L.) and mice, and induces weight gain and inflammation in mice. Br J Nutr. 2013;109(8):1508–1517. doi: 10.1017/S0007114512003364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gabbs M, Leng S, Devassy JG, Monirujjaman M, Aukema HM. Advances in our understanding of oxylipins derived from dietary PUFAs. Adv Nutr. 2015;6(5):513–540. doi: 10.3945/an.114.007732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cani PD, Plovier H, Van Hul M, Geurts L, Delzenne NM, Druart C, Everard A. Endocannabinoids — at the crossroads between the gut microbiota and host metabolism [internet]. Nat Rev Endocrinol. 2016;12(3):133–143. doi: 10.1038/nrendo.2015.211. [DOI] [PubMed] [Google Scholar]
- 17.Diab J, Al-Mahdi R, Gouveia-Figueira S, Hansen T, Jensen E, Goll R, Moritz T, Florholmen J, Forsdahl G. A quantitative analysis of colonic mucosal oxylipins and endocannabinoids in treatment-naïve and deep remission ulcerative colitis patients and the potential link with cytokine gene expression [internet]. Inflammatory Bowel Diseases 2019; 25:490–497. 10.1093/ibd/izy349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang W, Li H, Dong H, Liao J, Hammock BD, Yang G-Y. Soluble epoxide hydrolase deficiency inhibits dextran sulfate sodium-induced colitis and carcinogenesis in mice. Anticancer Res. 2013;33(S1):5261–5271. doi: 10.1096/fasebj.27.1_supplement.1104.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yuan X, Ta TC, Lin M, Evans JR, Dong Y, Bolotin E, Sherman MA, Forman BM, Sladek FM, Laudet V. Identification of an endogenous ligand bound to a native orphan nuclear receptor. PLoS One. 2009;4(5):e5609. doi: 10.1371/journal.pone.0005609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Genetics Consortium UI, Barrett JC, Lee JC, Lees CW, Prescott NJ, Anderson CA, Phillips A, Wesley E, Parnell K, Zhang H, et al. Genome-wide association study of ulcerative colitis identifies three new susceptibility loci, including the HNF4A region. Nat Genet. 2009;41:1330–1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sladek FM, Zhong WM, Lai E, JE D Jr.. Liver-enriched transcription factor HNF-4 is a novel member of the steroid hormone receptor superfamily. Genes Dev. 1990;4(12b):2353–2365. doi: 10.1101/gad.4.12b.2353. [DOI] [PubMed] [Google Scholar]
- 22.Ko HL, Zhuo Z, Ren EC. HNF4α combinatorial isoform heterodimers activate distinct gene targets that differ from their corresponding homodimers. Cell Rep. 2019;26(10):2549–57.e3. doi: 10.1016/j.celrep.2019.02.033. [DOI] [PubMed] [Google Scholar]
- 23.Chen WS, Manova K, Weinstein DC, Duncan SA, Plump AS, Prezioso VR, Bachvarova RF, JE D Jr.. Disruption of the HNF-4 gene, expressed in visceral endoderm, leads to cell death in embryonic ectoderm and impaired gastrulation of mouse embryos. Genes Dev. 1994;8(20):2466–2477. doi: 10.1101/gad.8.20.2466. [DOI] [PubMed] [Google Scholar]
- 24.Briancon N, Weiss MC. In vivo role of the HNF4α AF-1 activation domain revealed by exon swapping. Embo J. 2006;25(6):1253–1262. doi: 10.1038/sj.emboj.7601021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chellappa K, Deol P, Evans JR, Vuong LM, Chen G, Briançon N, Bolotin E, Lytle C, Nair MG, Sladek FM. Opposing roles of nuclear receptor HNF4α isoforms in colitis and colitis-associated colon cancer. Elife [Internet]. 2016;5 doi: 10.7554/eLife.10903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Buttó LF, Haller D. Dysbiosis in intestinal inflammation: cause or consequence. Int J Med Microbiol. 2016;306(5):302–309. doi: 10.1016/j.ijmm.2016.02.010. [DOI] [PubMed] [Google Scholar]
- 27.Ohno H. Impact of commensal microbiota on the host pathophysiology: focusing on immunity and inflammation [internet]. Semin Immunopathol. 2015;37(1):1–3. doi: 10.1007/s00281-014-0472-2. [DOI] [PubMed] [Google Scholar]
- 28.Tamboli CP, Neut C, Desreumaux P, Colombel JF. Dysbiosis in inflammatory bowel disease. Gut. 2004;53(1):1–4. doi: 10.1136/gut.53.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Darfeuille-Michaud A, Neut C, Barnich N, Lederman E, Di Martino P, Desreumaux P, Gambiez L, Joly B, Cortot A, Colombel JF. Presence of adherent Escherichia coli strains in ileal mucosa of patients with Crohn’s disease. Gastroenterology. 1998;115:1405–1413. doi: 10.1016/S0016-50859870019-8. [DOI] [PubMed] [Google Scholar]
- 30.Palmela C, Chevarin C, Xu Z, Torres J, Sevrin G, Hirten R, Barnich N, Ng SC, Colombel J-F. Adherent-invasive Escherichia coli in inflammatory bowel disease [internet]. Gut 2018; 67:574–587. doi: 10.1136/gutjnl-2017-314903 [DOI] [PubMed] [Google Scholar]
- 31.Martinez-Medina M, Denizot J, Dreux N, Robin F, Billard E, Bonnet R, Darfeuille-Michaud A, Barnich N. Western diet induces dysbiosis with increased E coli in CEABAC10 mice, alters host barrier function favouring AIEC colonisation. Gut. 2014;63(1):116–124. doi: 10.1136/gutjnl-2012-304119. [DOI] [PubMed] [Google Scholar]
- 32.Shawki A, Ramirez R, Spalinger MR, Ruegger PM, Sayoc-Becerra A, Santos AN, Chatterjee P, Canale V, Mitchell JD, Macbeth JC, et al. The autoimmune susceptibility gene, PTPN2, restricts expansion of a novel mouse adherent-invasive E. coli. Gut Microbes. 2020;11:1547–1566. doi: 10.1080/19490976.2020.1775538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Deol P, Evans JR, Dhahbi J, Chellappa K, Han DS, Spindler S, Sladek FM. Soybean oil is more obesogenic and diabetogenic than coconut oil and fructose in mouse: potential role for the liver. PLoS One. 2015;10:e0132672. doi: 10.1371/journal.pone.0132672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Deol P, Fahrmann J, Yang J, Evans JR, Rizo A, Grapov D, Salemi M, Wanichthanarak K, Fiehn O, Phinney B, et al. Omega-6 and omega-3 oxylipins are implicated in soybean oil-induced obesity in mice. Sci Rep. 2017;7:12488. doi: 10.1038/s41598-017-12624-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chellappa K, Jankova L, Schnabl JM, Pan S, Brelivet Y, Fung CL, Chan C, Dent OF, Clarke SJ, Robertson GR, et al. Src tyrosine kinase phosphorylation of nuclear receptor HNF4α correlates with isoform-specific loss of HNF4α in human colon cancer. Proc Natl Acad Sci U S A. 2012;109(7):2302–2307. doi: 10.1073/pnas.1106799109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vuong LM, Chellappa K, Dhahbi JM, Deans JR, Fang B, Bolotin E, Titova NV, Hoverter NP, Spindler SR, Waterman ML, et al. Differential effects of hepatocyte nuclear factor 4α isoforms on tumor growth and t-cell factor 4/AP-1 interactions in human colorectal cancer cells. Mol Cell Biol. 2015;35:3471–3490. doi: 10.1128/MCB.00030-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shan Z, Rehm CD, Rogers G, Ruan M, Wang DD, Hu FB, Mozaffarian D, Zhang FF, Bhupathiraju SN. Trends in dietary carbohydrate, protein, and fat intake and diet quality among US adults, 1999-2016. JAMA. 2019;322:1178–1187. doi: 10.1001/jama.2019.13771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Speakman JR. Use of high-fat diets to study rodent obesity as a model of human obesity. Int J Obes. 2019;43:1491–1492. doi: 10.1038/s41366-019-0363-7. [DOI] [PubMed] [Google Scholar]
- 39.Yusuf K, Saha S, Umar S. Health benefits of dietary fiber for the management of inflammatory bowel disease. Biomedicines Int. 2022;10 doi: 10.3390/biomedicines10061242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee JW, Bajwa PJ, Carson MJ, Jeske DR, Cong Y, Elson CO, Lytle C, Straus DS. Fenofibrate represses interleukin-17 and interferon-γ expression and improves colitis in interleukin-10–deficient Mice. Gastroenterology. 2007;133:108–123. doi: 10.1053/j.gastro.2007.03.113. [DOI] [PubMed] [Google Scholar]
- 41.Berg DJ, Davidson N, Kühn R, Müller W, Menon S, Holland G, Thompson-Snipes L, Leach MW, Rennick D. Enterocolitis and colon cancer in interleukin-10-deficient mice are associated with aberrant cytokine production and CD4(+) TH1-like responses. J Clin Invest. 1996;98:1010–1020. doi: 10.1172/JCI118861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Darfeuille-Michaud A, Boudeau J, Bulois P, Neut C, Glasser A-L, Barnich N, Bringer M-A, Swidsinski A, Beaugerie L, Colombel J-F. High prevalence of adherent-invasive Escherichia coli associated with ileal mucosa in Crohn’s disease. Gastroenterology. 2004;127:412–421. doi: 10.1053/j.gastro.2004.04.061. [DOI] [PubMed] [Google Scholar]
- 43.Nieman C. Influence of trace amounts of fatty acids on the growth of microorganisms. Bacteriol Rev. 1954;18:147–163. doi: 10.1128/br.18.2.147-163.1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hernández-Chirlaque C, Aranda CJ, Ocón B, Capitán-Cañadas F, Ortega-González M, Carrero JJ, Suárez MD, Zarzuelo A, Sánchez de Medina F, Martínez-Augustin O. Germ-free and antibiotic-treated Mice are highly susceptible to epithelial injury in DSS colitis. J Crohns Colitis. 2016;10:1324–1335. doi: 10.1093/ecco-jcc/jjw096. [DOI] [PubMed] [Google Scholar]
- 45.Martini E, Krug SM, Siegmund B, Neurath MF, Becker C. Mend your fences: the Epithelial Barrier and its relationship with mucosal immunity in inflammatory bowel disease. Cell Mol Gastroenterol Hepatol. 2017;4(1):33–46. doi: 10.1016/j.jcmgh.2017.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bäckhed F, Manchester JK, Semenkovich CF, Gordon JI. Mechanisms underlying the resistance to diet-induced obesity in germ-free mice. Proc Natl Acad Sci U S A. 2007;104:979–984. doi: 10.1073/pnas.0605374104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Turnbaugh PJ, Backhed F, Fulton L, Gordon JI. Diet-induced obesity is linked to marked but reversible alterations in the mouse distal gut microbiome. Cell Host & Microbe. 2008;3:213–223. doi: 10.1016/j.chom.2008.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Miles JP, Zou J, Kumar M-V, Pellizzon M, Ulman E, Ricci M, Gewirtz AT, Chassaing B. Supplementation of low- and high-fat diets with fermentable fiber exacerbates severity of DSS-induced acute colitis. Inflamm Bowel Dis. 2017;23:1133–1143. doi: 10.1097/MIB.0000000000001155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Singh V, Yeoh BS, Walker RE, Xiao X, Saha P, Golonka RM, Cai J, Bretin ACA, Cheng X, Liu Q, et al. Microbiota fermentation-NLRP3 axis shapes the impact of dietary fibres on intestinal inflammation. Gut. 2019;68:1801–1812. doi: 10.1136/gutjnl-2018-316250. [DOI] [PubMed] [Google Scholar]
- 50.Videla S, Vilaseca J, Antolín M, García-Lafuente A, Guarner F, Crespo E, Casalots J, Salas A, Malagelada JR. Dietary inulin improves distal colitis induced by dextran sodium sulfate in the rat. Am J Gastroenterol. 2001;96:1486–1493. doi: 10.1111/j.1572-0241.2001.03802.x. [DOI] [PubMed] [Google Scholar]
- 51.Schultz M, Munro K, Tannock GW, Melchner I, Gottl C, Schwietz H, Scholmerich J, Rath HC. Effects of feeding a probiotic preparation (SIM) containing inulin on the severity of colitis and on the composition of the intestinal microflora in HLA-B27 transgenic rats. Clin Diagn Lab Immunol. 2004;11:581–587. doi: 10.1128/CDLI.11.3.581-587.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Food and Agriculture Organization . World food and agriculture - statistical yearbook 2022. Rome: FAO; 2022. 10.4060/cc2211en. [DOI] [Google Scholar]
- 53.Lee SH, Kwon JE, Cho M-L. Immunological pathogenesis of inflammatory bowel disease. Intest Res. 2018;16:26–42. doi: 10.5217/ir.2018.16.1.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Calder PC. Polyunsaturated fatty acids and inflammation. Biochem Soc Trans. 2005;33:423–427. doi: 10.1042/BST0330423. [DOI] [PubMed] [Google Scholar]
- 55.Chapkin RS, Davidson LA, Ly L, Weeks BR, Lupton JR, McMurray DN. Immunomodulatory effects of (n-3) fatty acids: putative link to inflammation and colon cancer. J Nutr. 2007;137:200S–204S. doi: 10.1093/jn/137.1.200s. [DOI] [PubMed] [Google Scholar]
- 56.Agus A, Denizot J, Thévenot J, Martinez-Medina M, Massier S, Sauvanet P, Bernalier-Donadille A, Denis S, Hofman P, Bonnet R, et al. Western diet induces a shift in microbiota composition enhancing susceptibility to adherent-invasive E. coli infection and intestinal inflammation. Sci Rep. 2016;6:19032. doi: 10.1038/srep19032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Viennois E, Bretin A, Dubé PE, Maue AC, Dauriat CJG, Barnich N, Gewirtz AT, Chassaing B. Dietary emulsifiers directly impact adherent-invasive E. coli Gene expression to drive chronic intestinal inflammation. Cell Rep. 2020;33:108229. doi: 10.1016/j.celrep.2020.108229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gimier E, Chervy M, Agus A, Sivignon A, Billard E, Privat M, Viala S, Minet-Quinard R, Buisson A, Vazeille E, et al. Methyl-donor supplementation prevents intestinal colonization by Adherent-Invasive E. coli in a mouse model of Crohn’s disease. Sci Rep. 2020;10:12922. doi: 10.1038/s41598-020-69472-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kaur N, Chen C-C, Luther J, Kao JY. Intestinal dysbiosis in inflammatory bowel disease. Gut Microbes. 2011;2:211–216. doi: 10.4161/gmic.2.4.17863. [DOI] [PubMed] [Google Scholar]
- 60.Gkouskou KK, Deligianni C, Tsatsanis C, Eliopoulos AG. The gut microbiota in mouse models of inflammatory bowel disease. Front Cell Infect Microbiol. 2014;4:28. doi: 10.3389/fcimb.2014.00028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Alam MT, Amos GCA, Murphy ARJ, Murch S, Wellington EMH, Arasaradnam RP. Microbial imbalance in inflammatory bowel disease patients at different taxonomic levels. Gut Pathog. 2020;12:1. doi: 10.1186/s13099-019-0341-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Marton LT, de A GR, de CA, Barbalho SM. Omega fatty acids and inflammatory bowel diseases: an overview. Int J Mol Sci Int. 2019;20(19):4851. doi: 10.3390/ijms20194851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ellulu MS, Patimah I, Khaza’ai H, Rahmat A, Abed Y. Obesity and inflammation: the linking mechanism and the complications. Arch Med Sci. 2017;13:851–863. doi: 10.5114/aoms.2016.58928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.de Vos WM, Tilg H, Van Hul M, Cani PD, de Vos WM. Gut microbiome and health: mechanistic insights. Gut. 2022;71(5):1020–1032. doi: 10.1136/gutjnl-2021-326789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Naftali T. Is cannabis of potential value as a therapeutic for inflammatory bowel disease? dig. Dis Sci. 2019;64(10):2696–2698. doi: 10.1007/s10620-019-05763-8. [DOI] [PubMed] [Google Scholar]
- 66.Schmitz G, Ecker J. The opposing effects of n−3 and n−6 fatty acids. Prog Lipid Res. 2008;47(2):147–155. doi: 10.1016/j.plipres.2007.12.004. [DOI] [PubMed] [Google Scholar]
- 67.Ramstedt U, Serhan CN, Lundberg U, Wigzell H, Samuelsson B. Inhibition of human natural killer cell activity by (14R,15S)-14,15-dihydroxy-5Z,8Z,10E,12E- icosatetraenoic acid. Proc Natl Acad Sci USA. 1984;81(22):6914–6918. doi: 10.1073/pnas.81.22.6914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Davison JM, Lickwar CR, Song L, Breton G, Crawford GE, Rawls JF. Microbiota regulate intestinal epithelial gene expression by suppressing the transcription factor Hepatocyte nuclear factor 4 alpha. Genome Res Int. 2017;27(7):1195–1206. doi: 10.1101/gr.220111.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lickwar CR, Davison JM, Kelly C, Mercado GP, Wen J, Davis BR, Tillman MC, Semova I, Andres SF, Vale G, et al. Transcriptional integration of distinct microbial and nutritional signals by the small intestinal Epithelium. Cell Mol Gastroenterol Hepatol. 2022;14(2):465–493. doi: 10.1016/j.jcmgh.2022.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lei X, Ketelut-Carneiro N, Shmuel-Galia L, Xu W, Wilson R, Vierbuchen T, Chen Y, Reboldi A, Kang J, Edelblum KL, et al. Epithelial HNF4A shapes the intraepithelial lymphocyte compartment via direct regulation of immune signaling molecules. J Exp Med. 2022;219:e20212563. doi: 10.1084/jem.20212563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kühn R, Löhler J, Rennick D, Rajewsky K, Müller W. Interleukin-10-deficient mice develop chronic enterocolitis. Cell. 1993;75:263–274. doi: 10.1016/0092-86749380068-P. [DOI] [PubMed] [Google Scholar]
- 72.Alavi S, Mitchell JD, Cho JY, Liu R, Macbeth JC, Hsiao A. Interpersonal gut microbiome variation drives susceptibility and resistance to Cholera Infection. Cell. 2020;181:1533–46.e13. doi: 10.1016/j.cell.2020.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lo Sasso G, Phillips BW, Sewer A, Battey JND, Kondylis A, Talikka M, Titz B, Guedj E, Peric D, Bornand D, et al. The reduction of DSS-induced colitis severity in mice exposed to cigarette smoke is linked to immune modulation and microbial shifts. Sci Rep. 2020;10:3829. doi: 10.1038/s41598-020-60175-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Montbarbon M, Pichavant M, Langlois A, Erdual E, Maggiotto F, Neut C, Mallevaey T, Dharancy S, Dubuquoy L, Trottein F, et al. Colonic inflammation in mice is improved by cigarette smoke through iNKT cells recruitment. PLoS One. 2013;8:e62208. doi: 10.1371/journal.pone.0062208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Rodrigues HG, Vinolo MAR, Magdalon J, Vitzel K, Nachbar RT, Pessoa AFM, dos Santos MF, Hatanaka E, Calder PC, Curi R, et al. Oral administration of oleic or linoleic acid accelerates the inflammatory phase of wound healing. J Invest Dermatol. 2012;132(1):208–215. doi: 10.1038/jid.2011.265. [DOI] [PubMed] [Google Scholar]
- 76.Maeda Y, Hwang-Verslues WW, Wei G, Fukazawa T, Durbin ML, Owen LB, Liu X, Sladek FM. Tumour suppressor p53 down-regulates the expression of the human hepatocyte nuclear factor 4α (HNF4α) gene. Biochem J. 2006;400:303–313. doi: 10.1042/BJ20060614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Couter CJ, Surana NK. Isolation and flow cytometric characterization of murine small intestinal lymphocytes. J Vis Exp Int. 2016;111. doi: 10.3791/54114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ruegger PM, Clark RT, Weger JR, Braun J, Borneman J. Improved resolution of bacteria by high throughput sequence analysis of the rRNA internal transcribed spacer. J Microbiol Methods. 2014;105:82–87. doi: 10.1016/j.mimet.2014.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Frank JA, Reich CI, Sharma S, Weisbaum JS, Wilson BA, Olsen GJ. Critical evaluation of two primers commonly used for amplification of bacterial 16S rRNA genes. Appl Environ Microbiol. 2008;74:2461–2470. doi: 10.1128/AEM.02272-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hunt DE, Klepac-Ceraj V, Acinas SG, Gautier C, Bertilsson S, Polz MF. Evaluation of 23S rRNA PCR primers for use in phylogenetic studies of bacterial diversity. Appl Environ Microbiol. 2006;72:2221–2225. doi: 10.1128/AEM.72.3.2221-2225.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Edgar RC. UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat Methods. 2013;10:996–998. doi: 10.1038/nmeth.2604. [DOI] [PubMed] [Google Scholar]
- 82.Edgar RC. UNOISE2: Improved error-correction for Illumina 16S and ITS amplicon sequencing[Internet]. 10.1101/081257. [DOI] [Google Scholar]
- 83.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 84.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Pedersen TL, Gray IJ, Newman JW. Plasma and serum oxylipin, endocannabinoid, bile acid, steroid, fatty acid and nonsteroidal anti-inflammatory drug quantification in a 96-well plate format. Anal Chim Acta. 2021;1143:189–200. doi: 10.1016/j.aca.2020.11.019. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The bacterial rRNA ITS sequences have been deposited in the National Center for Biotechnology Information (NCBI)’s Sequence Read Archive (SRA) under the SRA BioProject Accession PRJNA622821 (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA622821/). The metabolomics data supporting the findings of this study are available within the article and the supplementary materials.


