Abstract
Caspases are very specific cell death proteases that are involved in apoptotic and non-apoptotic processes. While the role of caspases during apoptosis has been very well defined and many apoptotic proteolytic substrates of caspases have been identified and characterized, the role of caspases for non-apoptotic processes is not well understood. In particular, few non-apoptotic substrates of caspases have been identified thus far. Here, in order to facilitate the identification and characterization of potential caspase substrates, a protocol that allows the testing of candidate substrates in caspase cleavage assays in vitro is described. This protocol includes the production and purification of recombinant caspase proteins, the production of the candidate substrates either recombinantly or in a cell-free expression system, and the actual in vitro cleavage reaction followed by SDS-PAGE and immunoblotting. This protocol is tailored for the Drosophila caspases Dronc and Drice but can easily be adopted for caspases from other organisms including mammals.
SUMMARY:
Here we present a protocol to express and purify recombinant Drosophila caspases Dronc and Drice, and their use in in vitro cleavage assays.
INTRODUCTION:
Programmed cell death or apoptosis is executed by a class of highly specialized cell death proteases termed caspases (reviewed in reference1). Caspases are Cys proteases that contain a Cys residue in the catalytic site. They have defined consensus cleavage sites and cleave substrates proteolytically after Asp residues (although the Drosophila caspase Dronc has also been reported to cleave after Glu residues2). They are subdivided into initiator (also known as apical or upstream) and effector (executioner or downstream) caspases. Initiator caspases activate effector caspases. For example, in mammals, the initiator caspase Caspase-9 cleaves and activates the effector caspase Caspase-33. Likewise, in Drosophila melanogaster, the caspase-9-ortholog Dronc cleaves and activates the caspase-3-ortholog Drice2,4. During apoptosis, effector caspases cleave hundreds of substrates leading to the death of the cell5.
Caspases are synthesized as inactive proenzymes (zymogens) in the cell. In this form, they contain an N-terminal prodomain, a large subunit with the catalytic Cys in the central portion of the proenzyme, and a small subunit at the C-terminus1 (Figure 1). The mechanism of activation is different between initiator and effector caspases. Initiator caspases (Caspase-9, Dronc) require dimerization for activation which occurs by incorporation into a large protein complex, termed the apoptosome6. For the incorporation into the apoptosome, Caspase-9 and Dronc carry a caspase activation and recruitment domain (CARD) in the N-terminal prodomain (Figure 1). The apoptosome component Apaf-1 also contains a CARD and recruits Caspase-9 or Dronc via CARD/CARD interaction into the apoptosome3,6,7. While Caspase-9 and Dronc can be proteolytically processed in the apoptosome, this processing is not fully required for enzymatic activity8,9.
Figure 1: Domain structure of initiator caspases Caspase-9 and Dronc and effector caspases Caspase-3 and Drice.
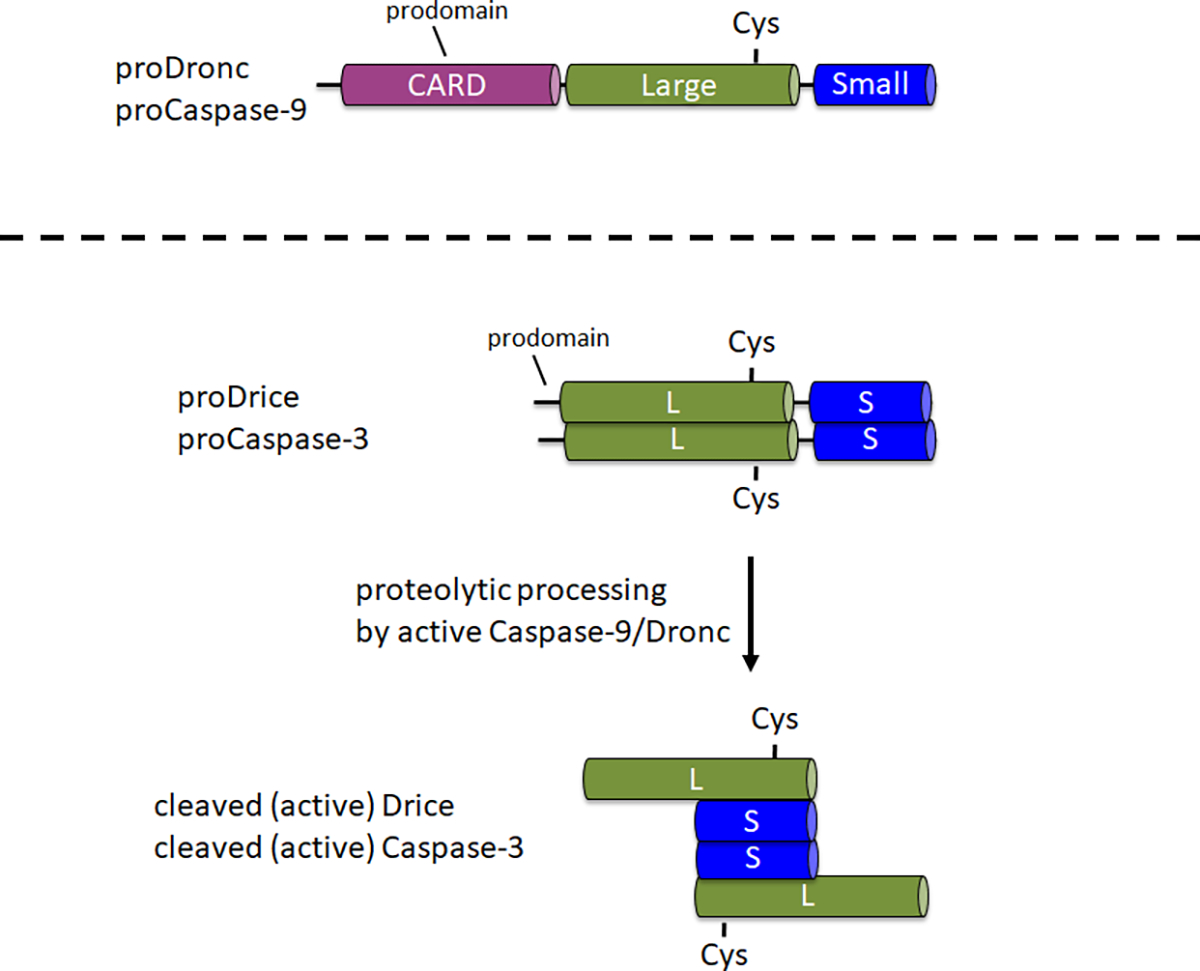
CARD – caspase activation and recruitment domain; Cys - relative location of the catalytic Cysteine residue; L – large subunit; S – small subunit. The location of the N-terminal prodomains is indicated.
In contrast, effector caspases (Caspase-3, Drice) do not carry a CARD in their prodomains and are not incorporated into large protein complexes for activation1. They are dependent on proteolytic cleavage by active Caspase-9 or Dronc1, respectively. The active effector caspase forms a tetramer composed of two large and two small subunits, and thus contains two catalytic sites (Figure 1). Importantly for this protocol, recombinant expression of caspases in E.coli causes auto-processing and activation of caspases including Drice10 and Dronc2,8,9,11,12, even in the absence of Apaf-1. This auto-processing allows to perform in vitro cleavage assays of candidate substrates with recombinant caspase protein.
Caspases are not only involved in apoptosis, but they can also have many non-apoptotic functions including proliferation, differentiation, cell migration, neuronal pruning, innate immunity, and others13–15. It is currently unknown how cells can survive despite containing active caspases during non-apoptotic processes. It is possible that these cells activate caspases only at sub-lethal levels16 or they sequester active caspases in non-apoptotic compartments of the cell such as the plasma membrane 17,18. Therefore, the identification and verification of non-apoptotic substrates will not only reveal how caspases mediate non-apoptotic processes but may also help to understand how cells can survive in the presence of active caspases.
Candidate proteins as caspase substrates can be identified using genetic and biochemical methods. Identified proteins can be checked for the presence of the consensus Dronc cleavage sites. This can be done by simply inspecting the protein sequence by hand or using more sophisticated online bioinformatic tools such as CasCleave (https://sunflower.kuicr.kyoto-u.ac.jp/~sjn/Cascleave/)19,20. These tools use the known consensus cleavage sites of caspases and structural considerations to predict novel targets of caspases. While CasCleave incorporates the information of verified substrates from human Caspases-1, -3, -6, -7, and -8, it may nevertheless also be useful for the purposes described here as these caspases and their consensus cleavage sites are well conserved. However, because the Dronc cleavage site is not well defined (two studies identified two different optimal cleavage sites, TATD/E2 and LALD9), the candidate substrates are also examined for the presence of other caspase cleavage site including Drice.
To validate the predicted substrates of caspases, additional assays are necessary. One of these assays is the demonstration that a given caspase can actually cleave the candidate protein in vitro. Here, we provide a convenient protocol for in vitro caspase cleavage assays. Using this protocol, candidate substrates are tested with Dronc as the caspase. They can also be tested as substrates of Drice. Although this protocol is written for the Drosophila caspases Dronc and Drice, it can also be adapted for caspases from other organisms.
The extraction and purification of Dronc and Drice along with the in vitro cleavage assay must be performed on the same day due to the loss of catalytic activity by these caspases. This protocol has been modified and optimized from previous publications8,9,11,12,21,22. In this protocol, four different caspase proteins are recombinantly expressed in the E.coli strain BL21(DE3)pLysS. These proteins are: 6xHis-Droncwt, 6xHis-DroncC318A, 6xHis-Dricewt and 6xHis-DriceC211A. Each of these proteins is tagged with 6- Histidine residues (6xHis) tag at the N-terminus for purification. Droncwt and Dricewt are the wild-type proteins and can auto-process into the active caspase upon recombinant expression. DroncC318A and DriceC211A encode mutant forms of Dronc and Drice that change the catalytic Cys residue to an Ala residue. These constructs are catalytically inactive and cannot auto-process (see also Figure 2A). They are used as controls in the cleavage assay. Because DriceC211A cannot auto-process, it is also used as the model substrate for Droncwt in the in vitro cleavage assay described here.
Figure 2. Representative results.
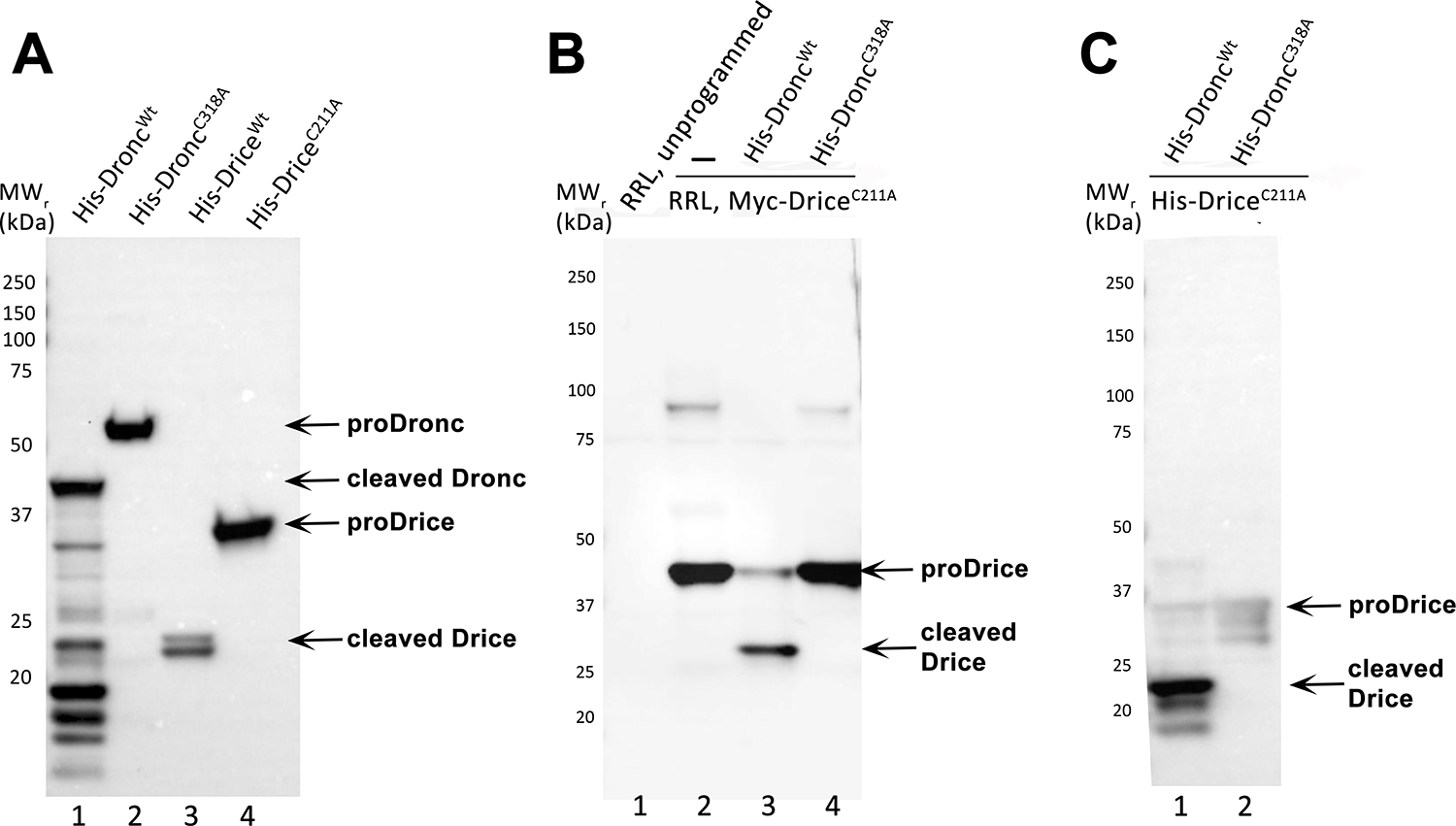
(A) Immunoblot analysis of purified recombinant 6xHis-Dronc and 6xHis-Drice preparations, probed with an anti-His antibody. Unprocessed (proDronc and proDrice) and cleaved 6xHis-Dronc and 6xHis-Drice are indicated by arrows. MW markers are indicated on the left.
(B) Immunoblot analysis of the in vitro cleavage reaction of RRL-generated N-Myc-Drice with caspases 6xHis-Droncwt (lane 3) or 6x-His-DroncC318A (lane 4) probed with an anti-Myc antibody. Unprogrammed and programmed (N-Myc-DriceC211A) RRL extracts are loaded and separated in lanes 1 and 2. Unprocessed (proDrice) and cleaved N-Myc-Drice are indicated by arrows. MW markers are indicated on the left.
(C) Immunoblot analysis of the in vitro cleavage reaction of bacterially expressed and purified recombinant 6xHis-DriceC211A with caspases 6xHis-Droncwt (lane 1) or 6x-His-DroncC318A (lane 2), probed with an anti-cleaved Drice antibody. Full-length (proDrice) and cleaved 6xHis-DriceC211A are indicated by arrows. MW markers are indicated on the left.
PROTOCOL:
1. Recombinant Caspase Expression in Bacteria
1.1. Clone the gene of interest (Caspase or putative substrate) into a bacterial expression vector with N- and/or C-terminal tag(s) using standard protocols23.
NOTE: Tags can increase the solubility of the recombinant proteins and are used for purification of the recombinant proteins. Here, Dronc, Drice and their catalytic mutants are cloned into the vector pET28a which provides an N-terminal 6xHis tag (pET28a-6xHis-Droncwt, pET28a-6xHis-DroncC318A, pET28a-6xHis-Dricewt, pET28a-6xHis-DriceC211A) (for primer information see Supplementary Table 2).
1.2. Transform the vectors with the genes of interest into competent BL21(DE3)pLysS E.coli cells using standard procedures23,24. Plate the transformation mixture on LB agar plates with the appropriate antibiotic (kanamycin in case of pET28a) to select transformed bacteria. (Refer to Supplementary Table 1).
1.3. Pick a colony from the plate and inoculate in 5 mL LB medium containing the appropriate antibiotic to grow overnight at 37 °C at 220 rpm on a shaking platform.
1.4. On the next day, prepare 30–50 mL (per sample) of LB medium with antibiotic and add 1 mL of the overnight grown culture. The optical density at 600 nm (OD600) using a bio photometer/spectrophotometer should be between 0.1 and 0.2.
1.5. Grow the culture at 37 °C at 220 rpm on a shaking platform until OD600 reaches 0.6. Check OD600 every hour until it reaches 0.6. This will take about 2 to 3 hours.
1.6. To induce protein (caspase) expression, add IPTG to a final concentration of 0.1–0.2 mM (dilute 1:1,000 – 1:500 from IPTG stock, see Supplementary Table 1).
1.7. Grow the cultures for 3 h at 30 °C at 220 rpm.
NOTE: Time and temperature are dependent on what kind of protein is being expressed and its solubility. These conditions may need to be adjusted.
1.6. After 3 hours, spin down the cultures in 50 mL centrifuge tubes for 20 min at 4 °C at 2000 × g. Discard the supernatant and proceed with the pellet.
NOTE: The protocol can be stopped at this point and continued later. Bacterial pellets can be frozen at −80 °C.
2. Small-scale Recombinant Caspases extraction
2.1. Remove the frozen tubes with the pelleted cultures from −80 °C storage and keep them on ice for 10 min to soften the pellet.
2.2. After 10 min, add 0.6 mL of bacterial cell lysis buffer (Supplementary Table 1) supplemented with freshly added protease inhibitors, 10 mg/mL of lysozyme and 50 units/mL benzonase into the pellet-containing tubes using a pipette controller with a 1 mL serological pipette.
2.3. With the same pipette tip, dissolve the pellet by pipetting up and down until a clear pale-yellow solution without any pellet particles is visible. Incubate for 30 min on ice.
2.4. Centrifuge the lysates for 40 min at 17,000 × g at 4 °C.
2.5. Transfer the supernatants into 1.5 mL microcentrifuge tube. This is the crude extract containing the caspases.
2.6. Keep the tubes on ice and proceed for purification with Ni-NTA agarose.
3. Small-scale His-tagged Caspases Purification.
3.1. Add 0.2 mL of the 50% slurry of Ni-NTA agarose to each tube of the caspase extracts from step 2.5. Rotate tubes on end-on-end rotator for 1 h at 4 °C.
3.2. After 1 h, add the extract with the Ni-NTA agarose to 1 mL polypropylene columns with the tip intact, placed in a rack. Let it stand for 5 min.
3.3. After 5 min, remove the cap of the columns and let the supernatant flow out by gravity flow.
3.4. Carefully add 1 mL wash buffer (Supplementary Table 1) to the columns without disturbing the packed resin of Ni-NTA agarose and wash it through gravity flow.
3.5. Perform washing step three times.
3.6. For elution of the caspases, place 1.5 mL microcentrifuge tubes under the collecting nozzle of the columns after complete draining of the wash buffer.
3.7. Add 0.5 mL of elution buffer (supplemented with 1x protease inhibitors immediately before use) to each column and collect the eluate in the 1.5 mL microcentrifuge tubes.
NOTE: The fine purified eluate will be transparent in color.
3.9. Keep the eluates on ice and measure the concentration of proteins by Bradford assay25. Confirm purity/homogeneity of purified caspases by SDS-PAGE followed by Coomassie Blue staining26.
NOTE: The yield of a 50 mL LB culture ranges between 0.5 and 1.5 mg of caspase protein. Given that 0.5 mL elution buffer is used for elution, the concentration ranges from 1 to 3 mg/mL. It is important to use the purified caspase eluates for in vitro cleavage assays on the same day of lysis and purification as these caspase preparations lose activity over night.
4. Expression of the putative caspase substrate in cell-free expression systems.
NOTE: In this protocol, Drice, the natural substrate of Dronc, is prepared by both recombinant expression in E.coli (see Section 3 above) and by expression in rabbit reticulocyte lysate (RRL), a mammalian cell-free expression system (this section, see below).
4.1. Clone the gene of the putative substrate into an expression vector containing either a T7, T3 or SP6 promoter using standard procedures23.
NOTE: In this protocol, DriceC211A is being used as the model substrate and was cloned into the vector pT7CFE1-N-Myc which carries a T7 promoter for gene expression and tags the putative protein substrate with an N-terminal Myc-tag for detection by immunoblotting.
4.2. In RRL, candidate substrates can be synthesized either radioactively or non-radioactively.
4.2.1. Non-radioactive synthesis: In a 0.5 mL microcentrifuge tube, add 25 μL of RRL lysate, 2 μL of reaction buffer, 0.5 μL of Amino Acid Mixture -Minus Leucine (1mM), 0.5 μL of Amino Acid Mixture-Minus Methionine (1mM), 1μL of Ribonuclease Inhibitor (40U/μL), 2μL of DNA Template (0.5 μg/μL) and 1 μL of T7-RNA Polymerase. Add nuclease-free water to adjust the final volume of the reaction to 50 μL. Mix the components by pipetting up and down five times.
NOTE: The RRL lysate contains 100–200 mg/ml of endogenous proteins. For in vitro translation reactions, provide the RRL at 50% concentration (here 25 μl/50μL reaction).
4.2.2. Radioactive synthesis: In a 0.5 mL microcentrifuge tube, add 25 μL of RRL lysate, 2 μL of reaction buffer, 0.5 μL of Amino Acid Mixture-Minus Methionine (1mM), 1 μL of Ribonuclease Inhibitor (40U/μL), 2 μL of DNA Template (0.5 μg/μL), 2 μL of S35-labeled Methionine (1000 Ci/mmol at 10 mCi/mL) and 1 μL of T7-RNA Polymerase. Add nuclease-free water to adjust the final volume of the reaction to 50 μL. Mix the components by pipetting up and down five times. Dispose of tips and tubes in a radioactive waste container.
NOTE: Do not add the Amino Acid Mixture without Leucine. The pT7CFE1 vector uses a T7 promoter for protein expression. Other vectors use T3 or SP6 promoters. In that case, T3 or SP6 RNA polymerases should be used instead of T7 RNA polymerase. Please make sure that the DNA template is of high purity.
4.3. Incubate the reaction at 30 °C for 90 min.
4.4. Spin briefly for 10 s and place tubes on ice. Check the expression level of the putative substrate in RRL by SDS-PAGE and immunoblotting/autoradiography.
NOTE: The amount of RRL extract added to the cleavage reaction should be based on the amount of protein that can be detected by the detection method (either immunoblotting or S35 autoradiography). Proceed to in vitro cleavage assay (next section) or store at − 80 °C.
5. In vitro Cleavage Assay with substrates generated with RRL
5.1. Take the putative substrate(s) generated, as described in Section 4. In this protocol, N-Myc-DriceC211A is used as model substrate.
5.2. Depending on the expression level of the putative substrate (to be determined separately by immunoblot or autoradiography analysis, see Step 4.4.), use 1–10 μl of the RRL lysate programmed with the protein of interest in the cleavage assay.
5.3. Add 10 μg of purified caspase protein generated in Sections 1, 2 and 3.
5.4. Bring the total reaction volume to 50 μL with caspase assay buffer.
5.5. Incubate the reactions in a water bath at 30 °C for 3 h. Make sure to include the appropriate controls in the cleavage assay. Here, the catalytic mutant DroncC318A is used as control.
5.6. After 3 h, stop the reactions by transferring the tubes on ice.
5.7. Add one volume of LDS sample buffer containing 50 mM DTT. The reaction is stopped completely with the addition of LDS sample buffer.
5.8. Incubate the samples at 75 °C in a heat block for 10 min.
5.9. Quickly spin, mix by flicking, load 24 μL per sample and run SDS-PAGE (see section 7) or store the samples at −20 °C.
6. In vitro cleavage assay with bacterially expressed recombinant substrate protein
6.1. Add 10 μg of purified candidate substrate (here 6xHis-DriceC211A, generated in Sections 1, 2 and 3) to a 1.5 mL microcentrifuge tube.
6.2. Add 10 μg of purified caspase proteins generated in Sections 1 and 2. Bring total volume to 50 μl using caspase assay buffer.
6.3. Follow steps 5.5. to 5.9.
7. SDS-PAGE and immunoblotting
7.1. Load the cleavage reactions (24 μL) onto 4–12% Tris-Glycine or Bis-Tris gradient gels (Table of Materials) and perform protein electrophoresis and immunoblotting (or autoradiography if using S35-labeled substrates) to visualize results using standard procedures27,28.
| Name of Material/Equipment | Company | Catalog Number | Comments/Description |
|---|---|---|---|
| 14 ml Polypropylene round bottom tubes | Fisher Scientific | 352029 | For growing plasmid cultures |
| 1 ml Serological Pipets, Sterile | celltreat | 229001B | For bacterial cell lysis in 50 ml |
| tubes | |||
| Ampicillin | Fisher | BP1760–25 | |
| Anti-His antibody | Sigma-Aldrich | MA1–21315 | |
| Anti-mouse IgG, HRP-linked Antibody | Cell signaling | 7076P2 | |
| Anti-Myc antibody | Santa Cruz Biotechnology | sc-40 | |
| Anti-rabbit IgG, HRP-linked Antibody | Cell signaling | 7074P2 | |
| Benchtop Centrifuge | Eppendorf | 5415 R | |
| Benzonase | Sigma-Aldrich | E1014–5KU | |
| BioPhotometer | Eppendorf | #6131 | |
| BL21(DE3)pLysS Competent Cells | Promega | L1195 | |
| Centrifuge rotor | Beckman Coulter | JA-25.50 | |
| CHAPS | Sigma-Aldrich | C3023–1G | |
| ChemiDoc with image software | Bio-Rad | Universal Hood II | For Chemiluminiscence imaging |
| Chemiluminiscence Substrate | Thermofisher Scientific | 34095 | For Chemiluminiscence imaging |
| Cleaved Drosophila ICE (drICE) (Asp230) Antibody | Cell Signaling Technology | 9478S | |
| Disposable cuvettes | Fisher Scientific | 14955128 | Used to measure bacterial |
| growth and protein concentration | |||
| Dithiothreitol (DTT) | Bio-Rad | #1610610 | |
| Erlenmeyer flasks, 1000 ml | Millipore sigma | CLS49801L | For LB agar media preparation |
| and autoclaving | |||
| Erlenmeyer flasks, 250 ml | Millipore sigma | CLS4980250 | For bacterial culture growth and |
| induction | |||
| Ethylene-diamine-tetra-acetic Acid (EDTA) | Sigma-Aldrich | E5134 | |
| Gel extraction kit | Qiagen | 28704 | |
| Gel tank SDS-PAGE system | Thermofisher Scientific | STM1001 | |
| Glycine | Sigma-Aldrich | G8898 | |
| Halt Protease Inhibitor Cocktail (100X) | Thermofisher scientific | 87786 | |
| HEPES | Sigma-Aldrich | H3375 | |
| His-Drice-pet28a | this study | N/A | Available from authors |
| His-DriceC211A-pet28a | this study | N/A | Available from authors |
| His-Dronc-pet28a | this study | N/A | Available from authors |
| His-DroncC318A-pet28a | this study | N/A | Available from authors |
| Imidazole | Sigma-Aldrich | I2399–100G | |
| Isopropyl-ß-D-thiogalactopyranoside (IPTG) | Thermofisher Scientific | FERR0392 | |
| Kanamycin | Fisher Scientific | BP906–5 | |
| LB Agar, Miller (Powder) | Fisher Scientific | BP1425–500 | |
| LB Broth, Miller | Fisher Scientific | BP1426–500 | |
| Lysozyme | Thermofisher Scientific | 90082 | |
| Microbiological plate incubator | Fisher Scientific | 11–690-650D | For colony growth after |
| transformation | |||
| Microcentrifuge tubes, 0.5mL | Eppendorf | 22363611 | |
| Microcentrifuge tubes, 1.5mL | Eppendorf | 22363204 | |
| Midiprep kit | Qiagen | 12243 | |
| Mini tube rotator | Fisher Scientific | 05–450-127 | for mixing bacterial lysates and |
| Ni-NTA agarose | |||
| Miniprep kit | Qiagen | 27106 | |
| Motorized Pipette Controller | Gilson | F110120 | For using serological pipettes |
| NaH2PO4 | Fisher Scientific | BP330–1 | |
| Ni-NTA Agarose | Qiagen | 30210 | |
| NuPAGE 4 to 12%, Bis-Tris, 1.0 mm, Midi Protein Gel, 20-well | Thermofisher Scientific | WG1402BOX | |
| NuPAGE LDS Sample Buffer (4X) | Thermofisher Scientific | NP0007 | |
| NuPAGE MOPS SDS Running Buffer (20X) | Thermofisher Scientific | NP0001 | |
| NuPAGE Transfer Buffer (20X) | Invitrogen | NP00061 | |
| Orbital shaking incubator with temperature control | New Brunswick Scientific | C25 incubator shaker | |
| Petridish 100mm × 15mm | Fisher Scientific | FB0875712 | |
| Plating beads | Zymo research | S1001 | For spreading culture on |
| AmpR/KanR plates | |||
| Polypropylene Columns (1 ml) | Qiagen | 34924 | For purification of |
| His-tagged proteins | |||
| Precision Plus Protein Standards | Bio-Rad | #161–0374 | |
| Protein Assay Dye Reagent Concentrate | Bio-Rad | #5000006 | |
| pT7CFE1-NMyc | Thermofisher Scientific | 88863 | For cloning substrates for RRL |
| expression | |||
| PVDF membrane | Invitrogen | LC2007 | |
| QiaRack | Qiagen | 19095 | For holding polypropylene |
| columns during purification | |||
| Refrigerated High speed Centrifuge | Beckman Coulter | Avanti J-25 | |
| rRNasin Ribonuclease Inhibitor | Promega | N251A | For RRL expression |
| Sodium chloride | Fisher Scientific | BP358–212 | |
| Sodium Hydroxide | Fisher Scientific | BP359–500 | |
| Sterile Falcon tubes, 15mL | Fisher Scientific | 05–527-90 | |
| Sterile Falcon tubes, 50ml | Fisher Scientific | 14–959-49A | |
| Sucrose | Sigma-Aldrich | S70903–250G | |
| TnT Coupled Reticulocyte Lysate -T7 | Promega | L4611 | |
| Tris-base | Fisher Scientific | BP154–1 | |
| Tween 20 | Sigma-Aldrich | P1379 | |
| Waterbath | Fisher Scientific | 2340 | |
| Western wet transferring cassette | Thermofisher Scientific | STM2001 |
NOTE: Here, in this protocol, Bis-Tris gradient gels with Mops running buffer and LDS loading buffer were used. In general, the commonly used PAGE protocols of Tris-Glycine gels run with Tris-Glycine-SDS running buffer and SDS-loading buffer do work well.
REPRESENTATIVE RESULTS:
This protocol provides step-by-step instructions for the caspase protein induction in E.coli, purification of the recombinant Drosophila caspases Dronc and Drice, the synthesis of candidate substrates and the in vitro cleavage reaction with candidate substrates (here DriceC211A) and the caspase Dronc. The catalytic mutant DriceC211A was used as model substrate in this assay because it does not have auto-processing activity (Figure 2A) and remains full-length until it is cleaved by Droncwt. DriceC211A should always be used as a positive control to validate that the Droncwt preparation has enzymatic activity.
Figure 2A provides a representative example of the expression and purification of recombinant caspases. Four different recombinant caspases were induced and purified: 6xHis-Droncwt, 6xHis-DroncC318A, 6xHis-Dricewt and 6xHis-DriceC211A. The purified caspases were run by SDS-PAGE, immunoblotted and the blot was probed with an anti-His antibody (diluted 1:5,000; followed by anti-mouse IgG, HRP-linked antibody (1:10,000)). Unprocessed 6xHis-Dronc (proDronc) runs at a relative molecular weight (MWr) of 55 kDa (lane 2), unprocessed 6xHis-Drice (proDrice) has a MWr of 35 kDa (lane 4). Auto-processing of the caspases is visible by the appearance of bands of smaller MWr which due to the presence of the His-tag at the N-terminus represent the large subunits of the caspases (Figure 1). In the case of 6x-Dronc, the large subunit has a MWr of 40 kDa (lane 1). The large subunit of 6x-Drice runs at 23 kDa (lane 3). The catalytic mutants 6xHis-DroncC318A and 6xHis-DriceC211A fail to auto-process and are only detectable as full-length proteins (lanes 2 and 4).
Figure 2B. To demonstrate that the bacterially-produced and purified 6xHis-Droncwt preparation has enzymatic activity, an in vitro cleavage assay as described in this protocol was performed. As negative control, the catalytic mutant 6xHis-DroncC318A was used. The substrate was RRL-generated N-Myc-DriceC211A which is tagged with a Myc-tag at the N-terminus. After the in vitro cleavage reaction, the proteins were separated by SDS-PAGE, immunoblotted and the blots were incubated with an anti-Myc antibody (diluted 1:1,000); followed by anti-mouse IgG, HRP-linked antibody (1:10,000)) to detect N-Myc-DriceC211A. Successful cleavage and thus enzymatic activity of the caspase can be demonstrated by the appearance of at least one band of smaller MWr compared to the full-length, unprocessed form of the substrate. Unprocessed, full-length N-Myc-DriceC211A has a MWr of 40 kDa (lanes 2 and 4), whereas the large subunit of processed N-Myc-DriceC211A runs at 30 kDa (lane 3). Lane 1 represents the unprogrammed RRL lysate (no plasmid/transcript added). Lane 2 demonstrates in vitro production of DriceC211A by RRL expression. Lane 3 contains the in vitro cleavage reaction with 6xHis-Droncwt. Lane 4 contains the in vitro cleavage reaction with 6xHis-DroncC318A.
Figure 2C. An in vitro cleavage reaction that uses both recombinant and purified caspase (6xHis-Droncwt and 6x-His-DroncC318A) and substrate (6xHis-DriceC211A) according to this protocol, was analyzed by SDS-PAGE and immunoblot. For analysis of the cleavage reaction, the anti-cleaved Drice antibody (diluted 1:5,000; followed by anti-rabbit IgG, HRP-linked antibody (1:10,000)) was used in this immunoblot. The anti-cleaved Drice antibody detects its neo-epitope in the large subunit (23 kDa) of 6xHis-DriceC211A only after processing by 6xHis-Droncwt (lane 1). The catalytic mutant 6xHis-DroncC318A is unable to process 6xHis-DriceC211A in this assay and full-length 6xHis-DriceC211A appears as faint unprocessed band of 35 kDa (lane 2).
DISCUSSION:
The bulk of our knowledge about caspases and caspase function has been derived from intense work in apoptosis during the last three decades. It is very well established that initiator caspases proteolytically process effector caspases, and hundreds of proteins have been identified as effector caspase substrates during apoptosis5,29. In contrast, much less is known about the function of caspases for non-apoptotic processes and which non-apoptotic substrates they are processing. It is conceivable that initiator caspases are key decision makers here. During apoptosis, they activate effector caspases causing cell death. However, to trigger non-apoptotic processes, they may activate different proteins (other than effector caspases) which control the non-apoptotic process. This protocol tests candidate proteins as substrates of the initiator caspase Dronc in Drosophila17,30.
The substrates to be tested in the cleavage assay can be produced either in an in vitro mammalian cell-free expression system such as RRL or by recombinant expression in E.coli. There are several advantages for in vitro expression using RRL over bacterial expression. The RRL expression protocol is simple and fast, allowing many different substrates to be prepared in parallel. In many cases, the RRL extract containing the protein of interest can be stored at −80 °C before use in the cleavage assay (although this needs to be determined separately for each substrate). The putative substrate can be labeled with S35-Met which allows easy analysis by autoradiography after SDS-PAGE. That is particularly useful, if no substrate-specific antibody is available. Alternatively, if S35-Met labeling is not desired, the putative substrate can be tagged with common tags such as Flag, HA or Myc tags which allows detection of caspase cleavage by immunoblotting.
It needs to be strongly emphasized that the success of this protocol depends on the careful and consistent purification of the recombinant Dronc and Drice proteins. Unfortunately, these proteins cannot be stored – not even short-term - neither in the fridge nor frozen. They lose enzymatic activity overnight in any stored form. Therefore, they need to be prepared freshly on the day of the cleavage assay. Regardless of the candidate protein(s) being tested, DriceC211A should always be used as a positive control to validate the enzymatic activity of the Droncwt preparation (see Figure 2B, C). Alternatively, the activity of the Dronc and Drice preparations can also be tested by in vitro cleavage of fluorogenic synthetic tetrapeptide substrates2,9,31.
If antibodies are used to detect the candidate substrates, they need to be validated by the user32,33. That applies also to commercially available antibodies that detect epitope tags such as Flag, HA, Myc or others. Poor antibody quality can obscure important results. Double-epitope-tagging of the candidate substrates on both the N- and C-terminus with different tags is also recommended34. If cleavage occurs, double tagging helps to trace both cleavage products and may help to elucidate if cleavage occurs at one or more sites.
While this protocol easily validates the known biological substrate of Dronc, Drice, there are also limitations. One limitation is that this is an in vitro protocol with recombinant proteins. In these assays, the caspases are present at an unphysiologically high concentration which is evident from the observation that they can spontaneously auto-process in E.coli. The spontaneous auto-processing usually does not occur under physiological conditions. This high caspase concentration may cause spurious activity resulting in false positives. False positives can be eliminated by lowering the caspase concentration in the in vitro cleavage reaction. Furthermore, as outlined in more detail below, additional assays are necessary to confirm genuine substrates and to eliminate false positives.
The recombinant caspases may not have the same specificity as they have in their normal cellular environment in vivo. For example, the activity of caspases can be modified by post-translational modifications. These are not present on recombinant proteins. Furthermore, in vivo, initiator caspases including Dronc are incorporated into large protein complexes such as the apoptosome. Under the conditions of this protocol, formation of the apoptosome is not achieved. That would require recombinant expression of Drosophila Apaf-1 (aka Dark or Hac-1)35–37 which has challenges of its own. Therefore, in vitro, Dronc may not have the same specificity that it has in vivo.
It is also conceivable that Dronc is incorporated into a different protein complex for non-apoptotic processes. This may confer a different cleavage specificity to Dronc which could also explain why Dronc does not induce apoptosis under non-apoptotic conditions. Related to this, CasCleave uses the known cleavage consensus sites to predict new caspase substrates. However, it is unknown if the same cleavage consensus sites are also used for non-apoptotic processes. In fact, recently it was shown that Caspase-3 changes its preferred consensus site during a non-apoptotic process in the developing auditory brainstem in the chick embryo38. Likewise, if initiator caspases are incorporated into different protein complexes, they may have a different specificity and thus may cleave at different consensus sequences.
These limitations illustrate that it is not sufficient to solely rely on the in vitro cleavage assays described in this protocol. Alternative approaches should be employed to further validate the results obtained using this protocol. Ideally, in vivo assays should be used to address the following questions: Is the candidate protein proteolytically processed during the non-apoptotic process in vivo? If so, is the same cleavage site used in vivo and in vitro? What is the consequence if the cleavage is blocked by mutagenesis of the cleavage site? Is the processing of the candidate substrate dependent on caspases, and if so, which one? What is the role of the cleavage fragments for the non-apoptotic process? These questions can be easily addressed in genetic model organisms such as C.elegans and Drosophila using standard genetic and transgenic methods.
In summary, this protocol describes a reliable and consistent method to produce enzymatically active caspases, specifically the Drosophila caspases Dronc and Drice. The ultimate goal of this protocol is to examine if Dronc can cleave candidate substrates in vitro which were identified by genetic, biochemical or bioinformatics approaches. As explained in the previous paragraph, additional assays are required to validate these proteins as caspase substrates in vivo. Given the degree of conservation of caspase genes across metazoa, it should be possible to adapt this protocol to caspases from other organisms as well.
Supplementary Material
ACKNOWLEDGMENTS:
We would like to thank Dr. Elif Kamber-Kaya for her help to establish the protocol in the lab. Dr. Guy Salvesen kindly provided the DriceC211A mutant9. This work was funded by a MIRA award from the National Institute of General Medical Sciences (NIGMS) of the National Institutes of Health (NIH) under grant number 2R35GM118330. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
A complete version of this article that includes the video component is available at http://dx.doi.org/10.3791/64392.
DISCLOSURES:
The authors declare no competing interests.
REFERENCES:
- 1.Kumar S Caspase function in programmed cell death. Cell Death and Differentiation. 14 (1), 32–43, (2007). [DOI] [PubMed] [Google Scholar]
- 2.Hawkins CJ et al. The Drosophila caspase DRONC cleaves following glutamate or aspartate and is regulated by DIAP1, HID, and GRIM. Journal of Biological Chemistry. 275 (35), 27084–27093, (2000). [DOI] [PubMed] [Google Scholar]
- 3.Li P et al. Cytochrome c and dATP-dependent formation of Apaf-1/caspase-9 complex initiates an apoptotic protease cascade. Cell. 91 (4), 479–489, (1997). [DOI] [PubMed] [Google Scholar]
- 4.Meier P, Silke J, Leevers SJ, Evan GI The Drosophila caspase DRONC is regulated by DIAP1. EMBO Journal. 19 (4), 598–611, (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Timmer JC, Salvesen GS Caspase substrates. Cell Death and Differentiation. 14 (1), 66–72, (2007). [DOI] [PubMed] [Google Scholar]
- 6.Zou H, Li Y, Liu X, Wang X An APAF-1.cytochrome c multimeric complex is a functional apoptosome that activates procaspase-9. Journal of Biological Chemistry. 274 (17), 11549–11556, (1999). [DOI] [PubMed] [Google Scholar]
- 7.Zou H, Henzel WJ, Liu X, Lutschg A, Wang X Apaf-1, a human protein homologous to C. elegans CED-4, participates in cytochrome c-dependent activation of caspase-3. Cell. 90 (3), 405–413, (1997). [DOI] [PubMed] [Google Scholar]
- 8.Stennicke HR et al. Caspase-9 can be activated without proteolytic processing. Journal of Biological Chemistry. 274 (13), 8359–8362, (1999). [DOI] [PubMed] [Google Scholar]
- 9.Snipas SJ, Drag M, Stennicke HR, Salvesen GS Activation mechanism and substrate specificity of the Drosophila initiator caspase DRONC. Cell Death and Differentiation. 15 (5), 938–945, (2008). [DOI] [PubMed] [Google Scholar]
- 10.Fraser AG, Evan GI Identification of a Drosophila melanogaster ICE/CED-3-related protease, drICE. EMBO Journal. 16 (10), 2805–2813, (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Denton D, Mills K, Kumar S Methods and protocols for studying cell death in Drosophila. Methods in Enzymology. 446 17–37, (2008). [DOI] [PubMed] [Google Scholar]
- 12.Dorstyn L, Kumar S A biochemical analysis of the activation of the Drosophila caspase DRONC. Cell Death and Differentiation. 15 (3), 461–470, (2008). [DOI] [PubMed] [Google Scholar]
- 13.Aram L, Yacobi-Sharon K, Arama E CDPs: caspase-dependent non-lethal cellular processes. Cell Death and Differentiation. 24 (8), 1307–1310, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Arama E, Baena-Lopez LA, Fearnhead HO Non-lethal message from the Holy Land: The first international conference on nonapoptotic roles of apoptotic proteins. FEBS Journal. 288 (7), 2166–2183, (2021). [DOI] [PubMed] [Google Scholar]
- 15.Baena-Lopez LA All about the caspase-dependent functions without cell death. Seminars in Cell and Developmental Biology. 82 77–78, (2018). [DOI] [PubMed] [Google Scholar]
- 16.Florentin A, Arama E Caspase levels and execution efficiencies determine the apoptotic potential of the cell. Journal of Cell Biology. 196 (4), 513–527, (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Amcheslavsky A et al. Plasma Membrane Localization of Apoptotic Caspases for Non-apoptotic Functions. Developmental Cell. 45 (4), 450–464 e453, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bergmann A Are membranes non-apoptotic compartments for apoptotic caspases? Oncotarget. 9 (60), 31566–31567, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Song J et al. Cascleave: towards more accurate prediction of caspase substrate cleavage sites. Bioinformatics. 26 (6), 752–760, (2010). [DOI] [PubMed] [Google Scholar]
- 20.Wang M et al. Cascleave 2.0, a new approach for predicting caspase and granzyme cleavage targets. Bioinformatics. 30 (1), 71–80, (2014). [DOI] [PubMed] [Google Scholar]
- 21.Kamber Kaya HE, Ditzel M, Meier P, Bergmann A An inhibitory mono-ubiquitylation of the Drosophila initiator caspase Dronc functions in both apoptotic and non-apoptotic pathways. PLoS Genetics. 13 (2), e1006438, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stennicke HR, Salvesen GS Caspases: preparation and characterization. Methods. 17 (4), 313–319, (1999). [DOI] [PubMed] [Google Scholar]
- 23.Green MR, Sambrook J Molecular Cloning: A Laboratory Manual (4th edition). (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press, 2012). [Google Scholar]
- 24.Froger A, Hall JE Transformation of plasmid DNA into E. coli using the heat shock method. Journal of Visualized Experiments. 10.3791/253 (6), 253, (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bradford MM A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Analytical Biochemistry. 72 248–254, (1976). [DOI] [PubMed] [Google Scholar]
- 26.Brunelle JL, Green R Coomassie blue staining. Methods in Enzymology. 541 161–167, (2014). [DOI] [PubMed] [Google Scholar]
- 27.Hirano S Western blot analysis. Methods in Molecular Biology. 926 87–97, (2012). [DOI] [PubMed] [Google Scholar]
- 28.Kim B Western Blot Techniques. Methods in Molecular Biology. 1606 133–139, (2017). [DOI] [PubMed] [Google Scholar]
- 29.Luthi AU, Martin SJ The CASBAH: a searchable database of caspase substrates. Cell Death and Differentiation. 14 (4), 641–650, (2007). [DOI] [PubMed] [Google Scholar]
- 30.Fogarty CE et al. Extracellular Reactive Oxygen Species Drive Apoptosis-Induced Proliferation via Drosophila Macrophages. Current Biology. 26 (5), 575–584, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Song Z et al. Biochemical and genetic interactions between Drosophila caspases and the proapoptotic genes rpr, hid, and grim. Molecular and Cellular Biology. 20 (8), 2907–2914, (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Edfors F et al. Enhanced validation of antibodies for research applications. Nature Communications. 9 (1), 4130, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Uhlen M et al. A proposal for validation of antibodies. Nature Methods. 13 (10), 823–827, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brizzard B Epitope tagging. Biotechniques. 44 (5), 693–695, (2008). [DOI] [PubMed] [Google Scholar]
- 35.Kanuka H et al. Control of the cell death pathway by Dapaf-1, a Drosophila Apaf-1/CED-4-related caspase activator. Molecular Cell. 4 (5), 757–769, (1999). [DOI] [PubMed] [Google Scholar]
- 36.Rodriguez A et al. Dark is a Drosophila homologue of Apaf-1/CED-4 and functions in an evolutionarily conserved death pathway. Nature Cell Biology. 1 (5), 272–279, (1999). [DOI] [PubMed] [Google Scholar]
- 37.Zhou L, Song Z, Tittel J, Steller H HAC-1, a Drosophila homolog of APAF-1 and CED-4 functions in developmental and radiation-induced apoptosis. Molecular Cell. 4 (5), 745–755, (1999). [DOI] [PubMed] [Google Scholar]
- 38.Weghorst F, Mirzakhanyan Y, Hernandez KL, Gershon PD, Cramer KS Non-Apoptotic Caspase Activity Preferentially Targets a Novel Consensus Sequence Associated With Cytoskeletal Proteins in the Developing Auditory Brainstem. Frontiers in Cell and Developmental Biology. 10 844844, (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


