Abstract
The first week after birth is a critical time for the establishment of microbial communities for infants. Preterm infants face unique environmental impacts on their newly-acquired microbiomes, including increased incidence of Cesarean section delivery and exposure to antibiotics as well as delayed enteral feeding and reduced human interaction during their intensive care unit stay. Using contextualized paired metabolomics and 16S sequencing data we profiled the development of the gut, skin, and oral microbiomes of infants daily for the first week after birth and found that the skin microbiome appears robust to early life perturbation, while direct exposure of infants to antibiotics, rather than presumed maternal transmission, delayed microbiome development and prevented the early differentiation based on body site regardless of delivery mode. Metabolomic analyses identified development of all gut metabolomes of preterm infants towards full-term infant profiles, but a significant increase of primary bile acid metabolism only in the non-antibiotic treated vaginally birthed late preterm infants. This study provides a framework for future multi-omic, multi-body site analyses on these high-risk preterm infant populations and suggests opportunities for monitoring and intervention, with infant antibiotic exposure as the primary driver of delays in microbiome development.
Keywords: preterm infant, microbiome, delivery mode, antibiotics, early neonatal development
Daily, multi-body-site microbiome sampling for the first week of life revealed that preterm infant antibiotic exposure delays the differentiation of body-site specific microbiomes. Primary bile acid metabolism in the gut metabolome was also present only in the non-antibiotic-treated, vaginally-birthed preterm infants, further highlighting the disruptive impact of antibiotics. This study provides a framework for future studies of infant microbiome development.
Introduction
It is now well-established that the human microbiome, the community of microbes that inhabit nearly every organ in the human body[1,2] but particularly the skin, mouth, and gut[3,4], is linked to numerous facets of human health and disease including inflammatory bowel disease, asthma, obesity, dental caries, atopic dermatitis, type 1 and 2 diabetes, cancer, and even mental health or neurological conditions such as schizophrenia and Parkinson’s disease[3,5,6]. Diet, geography, medication, hygiene, housing, and physical activity each shape the structure and diversity of the community, but the earliest effects that create our baseline microbiome begin at birth[3,7,8].
Birthmode, antibiotic-exposure, gestational age, and feeding are each known to impact the development of the infant gut microbiome through extensive characterization by 16S rRNA gene amplicon sequencing[9–15]. Infants born via Cesarean section are initially colonized by skin microbiota from their mothers and other adults, whereas infants born via vaginal delivery primarily receive a mix of vaginal and stool microbes[12,16,17]. This difference is reported to delay gut colonization by common adult-associated microbes[12,18,19], and is associated with adverse health effects[20–22], but the precise mechanisms remain unclear. Gestational age at birth also profoundly impacts development of the infant gastrointestinal tract, and is associated with additional short and long term health outcomes[23,24], perhaps partly attributable to altered gut microbiome development[21,25]. However, studies of gestational age are often confounded by birthmode and antibiotic-exposure because lower gestational age is associated with birth by Cesarean section and high antibiotic use[26–28]. Studies of the impact of antibiotics rarely examine proximal effects, and there is likely considerable under-reporting of antibiotic exposure in infants born via vaginal delivery[29,30], and over-reporting of antibiotic exposure in infants born via Cesarean section, because studies typically assume infants born via Cesarean section are either exposed to antibiotics via their universally antibiotic-treated mothers or otherwise affected by antibiotic effects (e.g. alteration in breast milk microbiota or metabolite composition of breast milk)[31,32].
After birth, infants are exposed to their first forms of food: colostrum, breast milk (either from their mother or a donor), and/or formula[18,33]. Breast milk is a complex mixture including micronutrients, macronutrients, and bioactives, such as human milk oligosaccharides (HMOs), along with immune factors and breast milk microbiota, which change in composition with infant age and differ between mothers based on maternal characteristics.[34–36] HMOs, a group of diverse compounds unique to each mother[37], cannot be metabolized by human cells but are converted into other metabolites including short-chain fatty acids (SCFA), a primary energy source, by gut microbes[38]. Although formulas contain HMOs, these HMOs are less complex than in breast milk; formula also lacks diverse immune factors found in breast milk[39]. Different types of infant diet and feeding practices, including differences between direct and indirect breastfeeding[40], have been shown to correlate with the gut microbiome and later health implications[33,41–44]. However, the reported relationships between the type of milk (breast milk vs formula) an infant receives and their microbiomes vary among studies. Over larger time scales, taxonomic groups change based on diet such that gut samples in a longitudinal study were more similar to each other in the first sample than the final sample collected at age 2.5 years[45], with developmental milestones such as the introduction of solid foods or the weaning off of breast milk resulting in a specific decrease in Bifidobacteria, the bacteria that reduce the conversion of HMOs to SCFA[46].
Premature birth correlates with disparities in the metabolomic and microbial compositions in infant microbiomes[47–49], with delayed colonization of “traditional” bacteria in very-low birth weight preterm (VLBW) infants[50]. Establishment of the microbiome in the first days to weeks after birth can have important implications later in life, due to the far-reaching health associations. Infants born late preterm (LP) and with very-low birthweight (VLBW) are at higher risk for infant mortality, sepsis, hypoglycemia, feeding difficulties, and long-term neurodevelopmental impairment[47,48,51,52] compared to infants born at full-term (FT), yet the mechanisms for these risks are poorly understood. Their higher incidence of delivery by Cesarean section, routine administration of antibiotics to both mothers and preterm infants and frequent use of formula places them at high risk for developing an abnormal or delayed gut microbial community[19,33,45]. Furthermore, although differences in the microbiome have been observed in the gut communities of FT compared to those of preterm or VLBW infants[17,53,54], few studies have examined the development and diversification of distinct, body-site specific (e.g. skin, oral, nares, and stool) infant microbiomes, and only on the order of weeks and months[12,15,54]. Distinct oral and gut communities were not observed until 15 days after birth in a small cohort of 6 preterm infants (5 without antibiotic exposure), or as late as six weeks in a larger cohort of 162 FT infants[15], while disparities between the gut, oral, and skin microbial communities and metabolomes between preterm and VLBW infants in the neonatal intensive care unit (NICU) remains largely unexamined.
In order to provide additional resolution and further this understanding, we conducted 16S rRNA gene amplicon sequencing and untargeted metabolomic analyses on gut, oral, and skin samples collected daily for the first week after birth from LP infants born either via Cesarean section or vaginally in San Diego and who were fed mother’s milk supplemented with either donor human milk (DHM) or formula. We also examined the impact of birthweight in shaping the development of the microbial community in VLBW infants born via Cesarean section who were housed in the NICU during this same period of collection. Together these cohorts enabled us to characterize the effect of early life exposures, including delivery mode, antibiotics and type of milk on the initial colonization and differentiation of their gut, oral, and skin microbiota and metabolomes during this critical window of holobiont development. We show that although the microbiome at each body site changed over the first week of life, robust differentiation among these microbiomes was only observed in non-antibiotic exposed late preterm infants regardless of delivery mode. In addition, we find that the metabolomic profile of each body site is dominated initially by host chemistry, but microbial influence begins to appear within the first week after birth, with a stronger impact in non-antibiotic exposed infants.
Results
Study groups and exclusion criteria
Eighty-five preterm infants (60 LP and 25 VLBW) were recruited from the UC San Diego NICU for approximately daily sampling of the oral, skin, and feces during the duration of their stay, resulting in a total of 1799 samples (Figure 1A). Untargeted metabolomics data were generated for 1682 samples and 16S rRNA gene amplicon sequencing was performed on 1654 samples, with paired data generated for 1547 samples. For this investigation, we focused on changes in the microbial community and metabolome as a function of birth weight, delivery mode and antibiotic use in the first 7 days after birth, resulting in the exclusion of 482 16S and 495 metabolomic samples. Insufficient samples were available for examining VLBW infants born via Cesarean section without antibiotic administration, or born vaginally over time resulting in the exclusion of an additional 100 16S and 107 metabolomic samples. Finally, 130 samples with fewer than 500 16S sequencing reads were excluded (Figure 1A, Table 1). A total of 1080 samples remained for metabolomic analyses and 942 samples remained for 16S analyses from 75 infants, consisting of three cohorts: 29 late preterm infants born via Cesarean section (LP-C), 28 late preterm infants born vaginally (LP-V), and 18 very low birth weight preterm infants born via Cesarean section (VLBW-C) (Figure 1B) with samples available from at least 30 infants across all three cohorts for each body site for each day of sampling. (Extended Data Figure 1).
Figure 1: Sample counts and workflow diagram for 16S rRNA gene amplicon and untargeted metabolomics data.

a) Sample filtering steps displaying the remaining samples after each filtering criterion. b) Sample data density for each infant over the first week after birth for stool, oral, and skin. Each column represents one infant and fill color indicates presence of 16S and/or metabolomics data. Infants divided by birth mode and birth weight.
Table 1: Demographics of preterm infant cohorts.
Group significance determined by Chi-square test when appropriate.
| LP-Vaginal | LP-C-section | VLBW-C-section | Group Significance | |
|---|---|---|---|---|
| Number of infants | 28 | 29 | 18 | |
| Birth gestational age (weeks) | 35.0 (0.86) | 35.0 (0.85) | 28.4 (2.06) | 1.45E-29 |
| Birth weight (grams) | 2548.8 (471.49) | 2555.2 (557.49) | 1079.2 (280.3) | 4.22E-17 |
| Maternal age (years | 32.9 (5.27) | 32.8 (5.69) | 31.5 (7.78) | n.s. |
| % Female | 39.3 | 41.4 | 55.6 | n.s. |
| Ethnicity | ||||
| White | 28.6 | 44.8 | 44.4 | n.s. |
| Hispanic | 42.9 | 31 | 33.3 | n.s. |
| African American | 3.6 | 17.2 | 22.2 | n.s. |
| Other | 25 | 6.9 | 0 | 0.013124 |
| Randomization | ||||
| Donor breast milk | 15 | 15 | n/a | |
| Formula | 13 | 14 | n/a | |
| 16S Samples | ||||
| Stool n | 101 | 119 | 85 | |
| % antibiotics exposed | 31.7 | 20.2 | 100 | |
| Oral n | 117 | 126 | 87 | |
| % antibiotics exposed | 29.9 | 20.6 | 100 | |
| Skin n | 116 | 136 | 102 | |
| % antibiotics exposed | 25 | 20.6 | 100 | |
| Metabolomics Samples | ||||
| Stool n | 131 | 140 | 89 | |
| % antibiotics exposed | 29 | 20 | 100 | |
| Oral n | 114 | 142 | 109 | |
| % antibiotics exposed | 29.8 | 21.1 | 100 | |
| Skin n | 108 | 139 | 108 | |
| % antibiotics exposed | 32.4 | 20.1 | 100 |
Determination of significant drivers of microbial community separation
Stepwise redundancy analysis (RDA)[55] of robust Aitchison distances between all samples from the 16S sequencing data, which account for the compositional nature of these data[56], revealed that as expected, individual had the strongest impact on microbial community composition (stepwise ANOVA, R2=0.196, F=4.8, p=0.002)(Extended Data Table 1). To better parse out the more subtle effects on composition, we removed the individual from the model and re-ran the stepwise redundancy analysis. Body site had the next strongest impact on the microbial community (stepwise ANOVA, R2=0.170, F=97.14, p=0.002), followed by antibiotic exposure (R2=0.095, F=121.59, p=0.002), age (day after birth) (R2=0.043, F=59.16, p=0.002), birth weight (R2=0.016, F=23.04, p=0.002), and delivery mode (R2=0.014, F=20.51, p=0.002). Employing stepwise RDA within body sites (again removing individual from the formula) revealed significant effects for all primary variables of interest. In stool, the strongest effect on the microbial community was delivery mode (R2=0.141, F=50.05, p=0.002) followed by significant effects from antibiotic exposure, age, and delivery weight (Extended Data Table 1). LP infants were also randomized into two groups that received either supplemental donor breast milk or supplemental formula (Table 1, Methods). This randomization group had a small, but significant effect on the stool microbial community (R2=0.013, F=3.69, p=0.002). Within the oral community, antibiotic exposure had the strongest effect (R2=0.249, F=104.35, p=0.002) followed by age, birth weight, and delivery mode (Extended Data Table 1). For skin, age had the strongest effect (R2=0.114, F=43.11, p=0.002), followed by birth weight and delivery mode (Extended Data Table 1).
Alpha and beta diversity changes over the first week after birth
The microbiome of each infant is influenced by their initial exposures outside the womb, and our multi-body-site analyses enabled us to assess the differentiation of each body site over time in our cohorts. As previous reports have detailed the impact of antibiotics and delivery mode on the infant gut community[12,54,57], we subdivided individuals into five groups for comparison: LP-V/Abx(+/−), LP-C/Abx(+/−), and VLBW-C/Abx(+); too few VLBW-C subjects were not administered antibiotics to enable adequate comparisons in the VLBW cohort. No differences in alpha diversity of the microbes (Shannon index) were found across any of the groups over time, with a contraction in alpha diversity of all microbiomes to a similarly-restricted community over the first week after birth (Extended Data Figure 2). We then compared changes within each body site by comparing robust Aitchison distances over time to the earliest available samples (day 0 for LP infants and day 1 for VLBW infants) within each infant. Skin microbiomes changed significantly in all cohorts over time (linear mixed effects models, p=0.001 for LP-V; p=0.003 for LP-C and p=0.032 for VLBW-C). Oral microbial communities significantly changed in all cohorts (linear mixed effects models p=0.001 for LP-V; p=0.002 for LP-C; p=0.002 for VLBW-C). The stool microbiome was only significantly changed in the LP-C and VLBW-C groups (linear mixed effects models p=0.0001 for LP-C; p=0.025 for VLBW-C), however, the LP-V infant stool at day 1 already had relatively high distance from day 0 samples.
Changes in infant microbiome differentiation
As clear changes occurred in most body sites and groups relative to their origins, we tested whether these changes were reflective of niche differentiation by body site. Robust Aitchison principal components analysis (PCA) revealed a pattern wherein the microbiomes were significantly differentiated at 1 day since birth in both LP/Abx(−) groups (V and C), but not in the VLBW-C/Abx(+) group (Figure 2a,b). Over the first week after birth, differences emerged in the degree of separation among the three body sites, as assessed by PERMANOVA pseudo-F score. Increased body site separation was seen only in the Abx(−) groups while the Abx(+) groups were not significantly differentiated or were significantly differentiated at fewer time points with a low pseudo-F score (Figure 2b). Classifying samples by maternal exposure to antibiotics showed a stark difference between infants receiving antibiotics compared to those assumed to be exposed via maternal antibiotic administration perinatally (Extended Data Figure 4). However, stratification of the data after applying the principle of metabolome-informed microbiome analyses[58] to provide empirical evidence of maternal antibiotic transfer demonstrated a pattern most similar to the classification of direct infant exposure only (Extended Data Figure 5).
Figure 2. Impact of antibiotics exposure on the differentiation of the microbial community of preterm infants over the first week after birth.
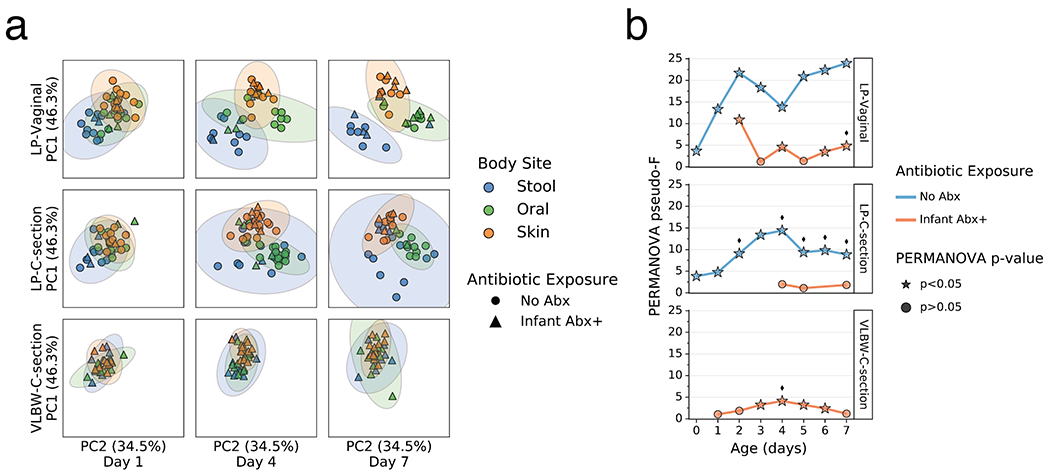
a. Robust Aitchison Principal Components Analysis (PCA) based on 16S V4 amplicon sequence variants (ASVs) of the gut (blue), oral (green), and skin (orange) microbiomes collected at day 1, 4 and 7 after birth from late preterm infants (LPs) born vaginally (LP-Vaginal) or by Cesarean section (LP-C-section) and very-low birth weight (VLBW) preterm infants (VLBW-C-section), with samples from children exposed to antibiotics as reported by clinical metadata (Infant Abx(+); triangles) and those without exposure to antibiotics (No Abx(−); circles) differentiated. b. PERMANOVA for differences in sample type (stool, oral, skin) based on Aitchison distances between all within-group samples for LP-Vaginal, LP-C-section, and VLBW-C-section preterm infants daily over the first week after birth, separated by Abx exposure. Diamonds above points indicate significant Permdisp tests (p<0.05) which indicate significantly different dispersion among body sites. Refer to Extended Data Table 3 for sample sizes in each comparison.
Comparison of infant microbiomes to adult microbiomes
As clear differentiation occurred over the first week after birth in the Abx(−) groups, we evaluated the proportion of each microbiome in our analytical groups that could be attributed to their more mature equivalents and potential sources. We trained a Bayesian microbial-source tracking algorithm on 16S data from the gut (n=4,434), oral (n=2,550), skin (n=1,975), and vaginal (n=427) communities from 11 public studies of adults age 20-80, stool samples from a cohort of FT infants age 0.5-4 months (n=87) (list of studies and details in Extended Data Table 2 [3,59–63]) and determined the percentages of each community that could be assigned to the expected body site or source of origin. Approximately half of the skin microbial community in the LP-V and VLBW-C groups was attributable to adult skin from the first day after birth, while this fraction exceeded 75% in the LP-C groups (Figure 3, right columns). By 6 days after birth, this proportion increased to nearly 100%, after an initial period of attribution to the vaginal microbiome in LP-V groups. The oral microbiome of all infants except LP-V/Abx(−) was primarily attributable to skin for all infant samples for the first three days since birth, however by day 4, Abx(−) infant samples were almost entirely attributable to the adult oral community while the Abx(+) infant samples remained primarily attributable to skin (Figure 3, center columns) with a trend towards increased oral attribution near the end of sampling for the LP-V/Abx(+) group. The brief dominance of oral attribution at day 4 in LP-C/Abx(+) was driven by a large relative abundance of Streptococcus in a minority of samples at that time point (Figure 3b, center panel). Attribution to adult stool was highest in the gut microbiome of LP-V/Abx(−) samples with a slight increase over time, though never exceeding 25% attribution (Figure 3a). Instead, the gut microbiome of these infants rapidly increased to resemble the gut of FT non-antibiotic exposed infants born vaginally (FT-V/Abx(−)) with delayed and reduced pattern of increasing attribution observed in LP-C/Abx(−) samples (Figure 3a, left panels). In contrast, while the gut microbiome of LP-V/Abx(+) samples initially resembled FT-V/Abx(−) stool, it quickly declined and the microbiome of all three groups of Abx(+) samples was <50% attributable to FT-V/Abx(−) stool at day 7 (Figure 3b, upper left panel). Grouping samples based on maternal, rather than infant antibiotic administration, obscured these differences and a universal pattern of increasing attribution to the expected body site for all infant groups (Extended Data Figure 6).
Figure 3. Antibiotics and delivery mode alter maturation of microbial communities over the first week after birth.
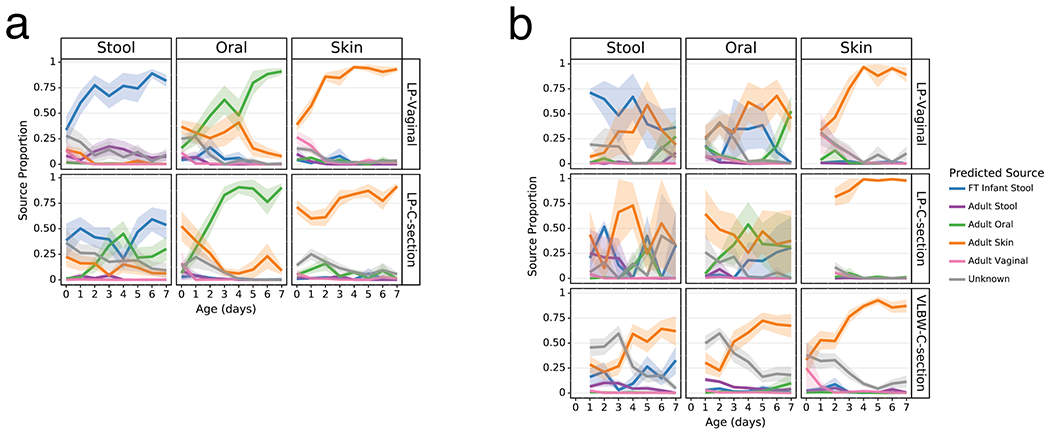
SourceTracker2 analysis showing proportion of LP and VLBW infant microbial communities attributed to adult or full term (FT) infant microbiomes based on 11 public studies in Qiita and a cohort of 87 FT infants not exposed to antibiotics aged 0.5-4 months (Methods). Lines represent the mean (+/− SEM) proportion of the microbial profile attributed to a specific source for all samples from preterm infants within each cohort displayed by birth-mode (rows) and body site (column). a) Preterm infant samples not exposed to antibiotics. b) Preterm infant samples after exposure to antibiotics. Refer to Extended Data Table 3 for sample sizes of the lP and VLBW infants at each time point.
Differentially abundant microbes due to age, antibiotics, and breast milk supplementation
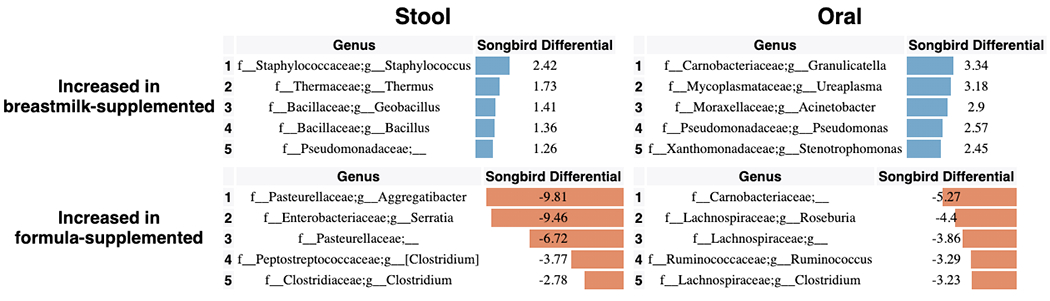
To identify specific changes in microbial abundances associated with differentiation and development, we used Songbird[64], a tool that overcomes the compositionality of microbiome data by ranking features based on log-fold changes with respect to variables of interest. We constructed models for each body site separately using only non-antibiotic exposed LP infants using age, delivery mode, and randomization (i.e. breast milk or formula supplementation) as explanatory variables (Figure 4). Each of these models resulted in a Q2>0, indicating that the model including the variable attained higher predictive accuracy than the baseline model not including the variable. Bifidobacteria in stool and Rothia in oral samples were most highly associated with increased age, while the common urinary tract genus Ureaplasma, was strongly associated with oral and stool samples from younger infants. The genera Aggregatibacter and Serratia were most associated with formula supplementation and Staphylococcus was most associated with donor human milk supplementation (Extended Data Figure 7) in stool. In contrast, no microbial taxa were found to be highly associated with these factors in skin (Songbird Q2>0).
Figure 4. Top differentially abundant microbes based on Songbird differentials.
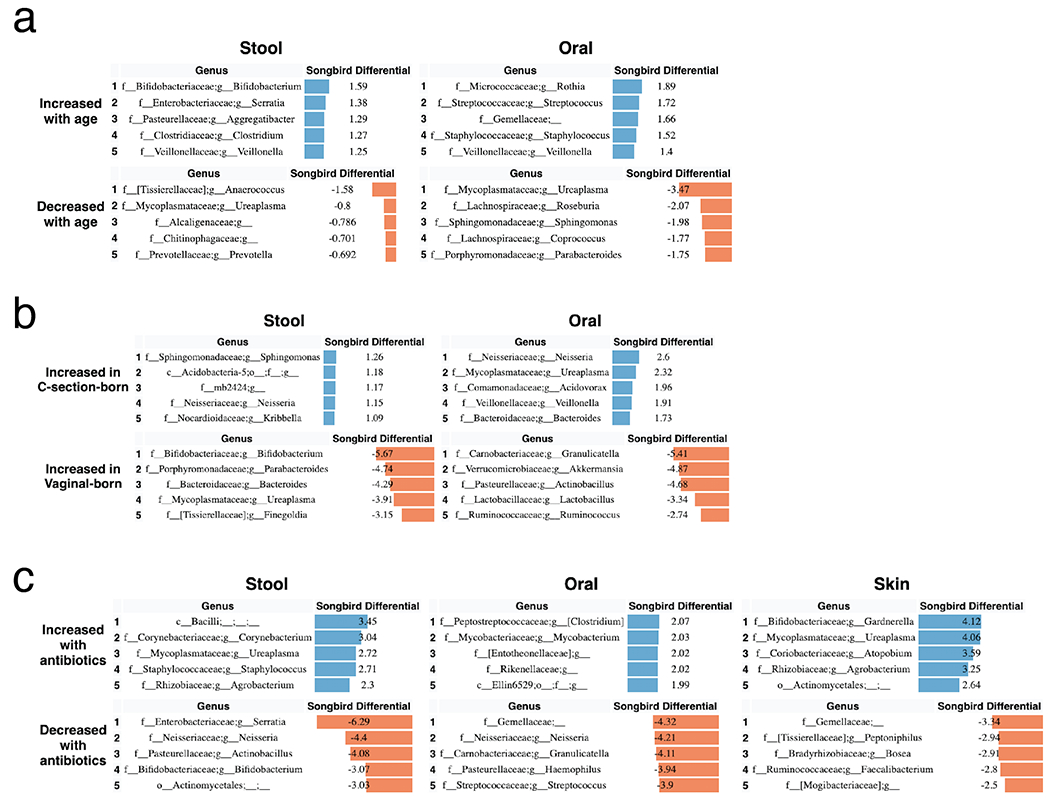
Top 5 genera positively and negatively associated with a) age and b) delivery mode in non-antibiotic-exposed samples from LP infants in stool and oral microbial communities based on 16S sequencing. The songbird model fit for stool (Q2=0.072) and oral (Q2=0.014). Top 5 genera positively and negatively associated with c) antibiotics based on stool, oral, and skin samples from LP and VLBW infants based on 16S sequencing. The Songbird model fit for stool (Q2=0.222), oral (Q2= 0.316), and skin (Q2=0.038). Results for skin are not shown in a and b because the Songbird model was unable to fit to data. Songbird differential values describe the log-fold change of genera with respect to the metadata category given.
To identify differentially abundant microbes associated with infant antibiotic use, separate models were constructed by body site including both the antibiotic-exposed LP and VLBW infants with antibiotic exposure as the explanatory variable (Figure 4c). In stool, Corynebacterium and an unidentified genus in the class Bacilli were most associated with antibiotic exposure, while the genera Serratia, Neisseria, Actinobacullus, and Bifidobacterium were most associated with non-antibiotic exposed samples. In skin samples, Ureaplasma and the common vaginal microbe Gardnerella were most associated with antibiotic exposure.
Metabolomic changes in the first week after birth
Given the major microbiome changes observed during the first week after birth, we sought to examine whether there was evidence of these changes in the metabolomic profiles of each body site within each infant for our cohorts. PCoA of the composition of unique metabolites, assessed by Jaccard distance, revealed that each body site within each cohort was significantly different throughout the first week after birth as expected for unique host compartments (Extended Data Figure 8). However, when comparing Jaccard distances over time to the earliest available samples (day 0 for LP infants and day 1 for VLBW infants) within each infant, a different pattern emerged. While changes were not significantly different in any cohort for the skin profile (linear mixed model p>0.05) the profile of oral samples was significantly different with time for the LP-C cohort (linear mixed model p=0.001; Figure 5a). In contrast, the stool metabolome of each infant changed significantly over the first week after birth in all cohorts (linear mixed model p=0.003 for LP-V, p=0.018 for LP-C and p=0.001 for VLBW-C; Figure 4a). Differential abundance analysis via Songbird revealed that several metabolites increased with time including diet-related metabolites such as oleic acid and conjugated linoleic acid, while several primary conjugated bile acids, such as taurocholic acid and glycocholic acid, decreased over time (Figure 5b).
Figure 5. Metabolomics profiles change over the first week after birth.
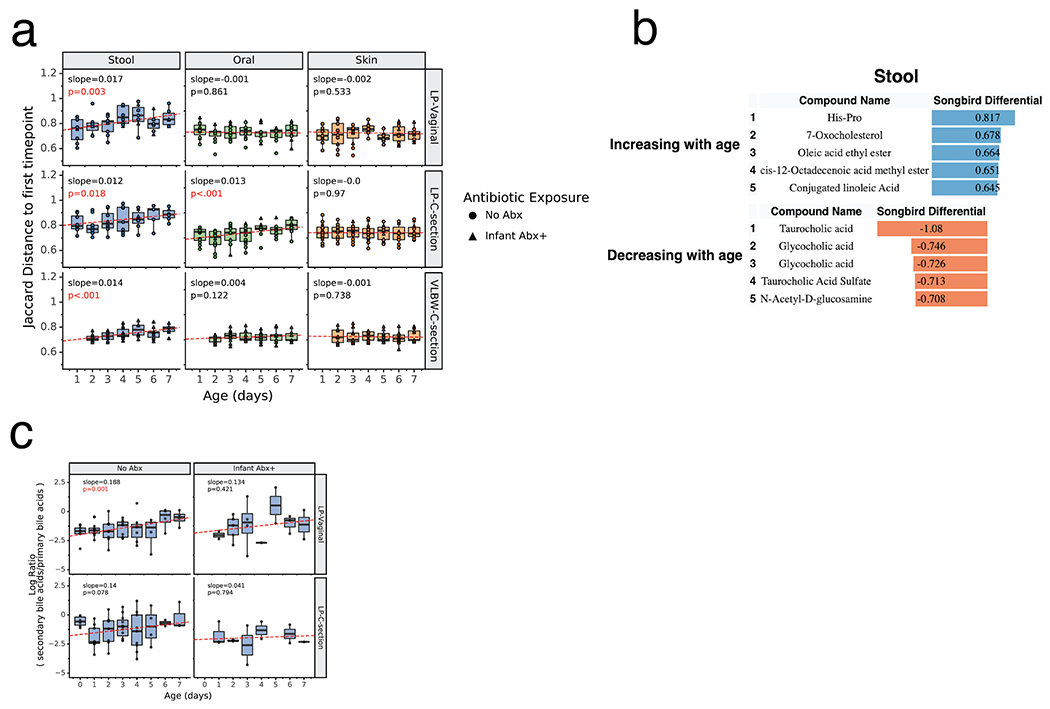
a) Within-infant metabolome distance to first sample over time. For each infant, the Jaccard distance to the first sample (day 0 for LP infants, day 1 for VLBW infants) is plotted over time for each body site. Boxplots show median and interquartile range with whiskers extending to the furthest value within 1.5 times the edge of the interquartile range. Red lines show the fitted linear mixed effect model with individual as a random effect and age as a fixed effect. P-value and slope of the age variable shown. Stool samples for all cohorts (LP-Vaginal, LP-C-section, VLBW C-section) and oral samples for LP-C-section infants have a significant increasing slope, corresponding to increased distance from the first time point over the first week after birth. All data points are shown with shape indicating infant antibiotic exposure. b) Top 5 identified metabolites positively and negatively associated with age based on Songbird model including LP infants. Songbird model fit Q2=0.017. c) Log ratios of secondary bile acids over primary bile acids, identified using GNPS annotations to structural family matches. Linear mixed effects models were run in the same way as panel a. Refer to Extended Data Table 3 for sample sizes in each comparison.
Bile acid changes in stool metabolome
We further investigated the changes in the log ratio of the relative abundances of non-microbially modified primary bile acids vs secondary, i.e. microbially-modified, bile acids in infant stool samples over time. As primary bile acids are transformed into secondary bile acids through microbial metabolism, the ratio of these should give a measure of changes in microbial metabolism in our groups. We observed a significant change in the ratio of secondary to primary bile acids in stool samples over the first week after birth only in the LP-V/Abx(−) group (linear mixed effects, p=0.001, slope=0.188; Figure 5c). The LP-C infants show a similar increasing trend, but had a high ratio at day 0 for unknown reasons. The log-ratio for LP-C/Abx(+) infants mostly remained constant, while the LP-V/Abx(+) infants showed an increasing trend, but the model was not significant due to low sampling density and higher variance. This data supports the idea that in the absence of antibiotic exposure, the gut microbiome of LP infants differentiates and gut-specific microbial metabolism occurs within the first week after birth.
Microbe-associated changes in stool metabolome
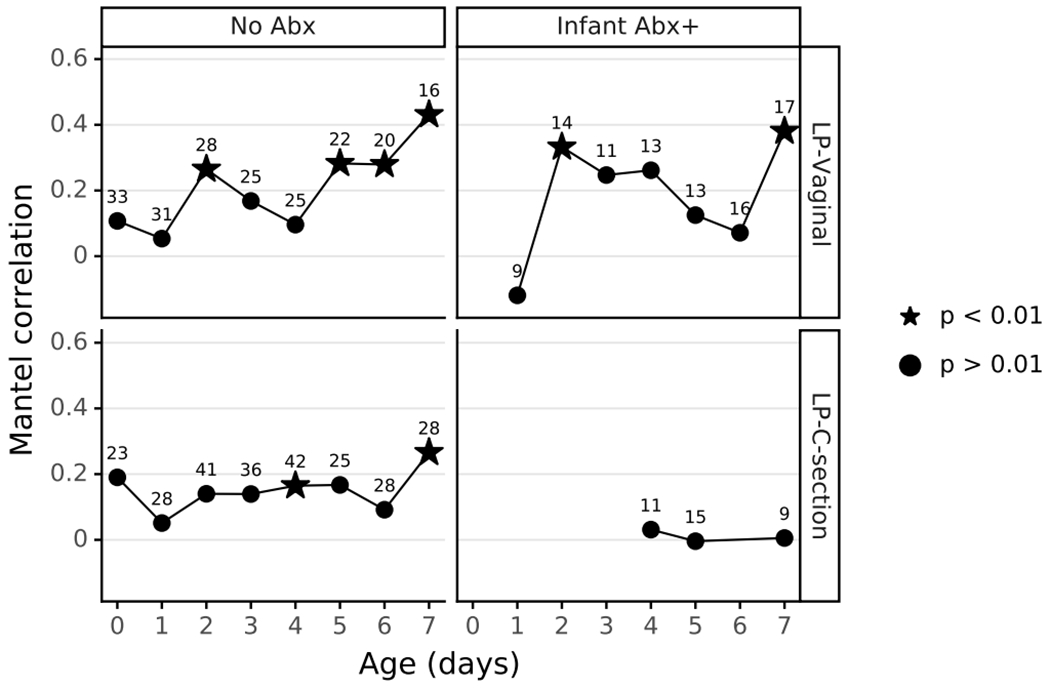
Given our observation of increasing microbial differentiation by body site over the first week after birth, we hypothesized that microbes would have an increasing effect on the infant metabolome over that time. We compared the composition of matched 16S and metabolomic samples from LP infants using Mantel tests with data divided by infant day after birth, antibiotic exposure, and delivery mode, each observed to be major factors influencing the microbial profile. Mantel tests for the first four days after birth were largely non-significant, however days 5, 6, and 7 in LP-V/Abx(−) infants were significant with an increasing trend in the Mantel correlation. Furthermore, the microbial community and metabolome were significantly correlated in all LP groups except LP-C/Abx(+) infants (Extended Data Figure 9).
Conclusion
Research regarding the development of the human microbiome has increased dramatically in the last decade, and will increase as researchers and clinicians identify new methods of intervention and establish best practices during critical early windows of microbial community niche differentiation. LP and VLBW infants represent two of the most at-risk populations for adverse impacts of early life exposures, and even a brief NICU stay may increase these risks via inadequate exposure to the necessary milieu of microbes, exposure to antibiotics, and/or delays in feeding[65]. Early life provides a unique opportunity for potential microbiome-directed interventions and for observing microbiomes with few exposures compared to adults[8,21,45], although key factors in early development such as antibiotic use and diet play a significant role for the gut microbiome regardless of age[3].
Here, by implementing daily sampling, and a multi-omic, multi-body site analysis in the context of publicly available human data from infants and adults in Qiita[66] and publicly available information in GNPS[67] , we were able to examine the impact of these factors on the developing microbiomes of preterm infants in the first week after birth. The skin microbiome in all infants appeared to develop a resemblance to adults regardless of exposure or delivery mode, suggesting that at least one niche may be resilient to these effects. The lack of oral vs stool microbiome differentiation and development in the VLBW infants, who are typically both born via Cesarean section and administered antibiotics, is of concern, and suggests that these infants may be able to benefit most from any interventions, e.g. vaginal-seeding[68], or compensatory actions. Crucially, by separating the effects of presumed intrapartum antibiotic exposure from clinically-reported direct antibiotic administration, we demonstrate that only the latter form of exposure appears to have a significant impact on the differentiation of the microbial communities and the establishment of the normal stool and oral microbial communities. While we saw empirical evidence that antibiotic exposure was occurring in infants from maternal use and/or other sources, reclassifying antibiotic-exposed samples per body site did not impact these results. Together these data suggest that future studies should separate these groups to better elucidate differential impacts of antibiotics on the infant microbiome via maternal vs direct exposure when examining short or long-term impacts of delivery mode.
Using compositionally-aware methods enabled us to identify specific taxa enriched in infants supplemented with formula or DHM. Our observation that members of the Staphylococcus genus were associated with DHM supplementation is perhaps unsurprising given the source. Staphylococcus infections remain a leading cause of infections in the NICU, and it is poorly understood what promotes this vulnerability, though removal of S. aureus in particular appears to reduce the risk of infection. This suggests the need to consider treating upstream sources of milk[69,70] (the human milk donor, or screening of human milk for S. aureus), which may mitigate the need for this vulnerable preterm population to directly receive antibiotic treatment that would disrupt microbiome development. In addition, the increased ratio of Aggregatibacter in formula-supplemented infants, observed previously[33], may indicate an early risk exposure for the later development of caries[71,72] as well as localized aggressive periodontitis[73–75] which may require treatment with antibiotics. Limiting antibiotics to a local application may ameliorate the global dysfunctional differentiation observed with systemic antibiotic exposure in our cohorts. Together, our results suggest that there is opportunity for treatment options specific to the type of feeding supplementation for preterm infants in the NICU.
An organ-specific metabolomic profile dominated in oral, stool, and skin samples in the first week after birth. However, the increasing correlation between the microbial community and metabolome of stool samples over the first week after birth, especially in the LP-V/Abx(−) group, suggests that as the microbial community expands and establishes itself, a greater degree of influence is exerted on the metabolomic profile, as also described in Bittinger et al[76]. The late preterm infant cohort reflects a low microbial biomass host, mirroring conditions explored in Quinn et al.[77], where germ-free and specific pathogen free mice were used to demonstrate that microbes profoundly reshaped the metabolomic profile of each organ. In addition, an increasing trend in the ratio of secondary to primary bile acids, again primarily in the LP-V/Abx(−) group provides further evidence that the microbial community is actively engaged in modifying this axis early in human development. The lack of this trend is most evident in the LP-C/Abx(+) group, suggesting that the combination of antibiotics and birth by Cesarean section may result in not just delayed microbiome development, but metabolomic development as well. Microbes enriched for producing secondary bile acids may therefore be a key component of successful seeding efforts to ameliorate the impact of early antibiotic exposure and birth mode.
Ultimately, the microbes living both on and in humans and their corresponding metabolites are influenced by and, in turn, influence the holobiont’s collective health. Future studies into the causes of health disparities and potential interventions should ensure that this critical window of development is captured for the oral and stool microbiome, including the metabolome, rather than solely focusing on the gut microbial community, to ensure robust clinical interpretation. As the identification of metabolites continues to improve through the public sharing, curation, and aggregation of data using tools such as ReDU[78] and MASST[79], the data in this study will remain available for providing context for future studies. Our study adds to the body of knowledge of preterm infant development and provides a model for future investigations as we continue to modify and improve our standard of care of preterm infants to optimize their long term health.
Methods
Cohorts
The data presented in this study originate from three independent cohorts: late preterm infants (LP), very low birth weight infants (VLBW) and full-term infants (FT, included for comparison), including birth by both vaginal delivery and Cesarean section. For all analyses, the gut, oral, and skin microbiome samples were separated by sample type as well as by delivery mode, in order to prevent possible confounding effects. Due to independent collection efforts, some methods are data set specific, as described below.
Recruitment:
For each study participant, a parent provided a written informed consent, and the study was approved by the Institutional Review Board at the University of California, San Diego [LP cohort: IRB approval number 151713; VLBW cohort: IRB approval number 151689] or the University of Michigan Institutional Review Board [FT: University of Michigan IRB HUM00103575].
LP:
Pregnant English and Spanish speaking women at 34-36 6/7 weeks gestation who were expected to, or had delivered, prior to 37 weeks gestation were approached by a member of the research team. Infants outside of the gestational age criteria were excluded, as well as infants with major congenital anomalies including gastrointestinal anomalies, known or suspected metabolic disorder requiring a specialized diet, and infants who had received any formula prior to randomization.
77 families representing 86 babies (7 sets of twins, 1 set of triplets) were approached regarding the study. Of those approached, 57 of the families consented, resulting in enrollment of 60 infants including 1 set of triplets, and 4 sets of twins. There were 2 infants consented but not randomized in error and 1 infant consented who was not randomized due to delivery after 37 weeks gestation.
VLBW:
English or Spanish speaking mothers who were likely to deliver an infant prematurely were approached by a member of the research team from June 2016 until May 2017. Consent and enrollment in the study were finalized if the infant birth weight was 500g-1500 g and between 23-34 weeks gestational age. Infants with medical problems incompatible with life or birth weight <500 g were excluded as the outcomes for these infants are poor. Most consents with the mother were initialized before the birth of their infant, however occasionally they were consented shortly after the delivery if the birth was imminent and unexpected. The first 25 patients from Hillcrest Medical Center were enrolled prior to establishment of a consistent oral colostrum care policy and the latter 25 patients were enrolled from the Jacobs Medical Center after the oral colostrum care policy was put into place.
FT:
Infant-mother dyads were recruited from the community when infants were between 2 weeks and 2 months of age. Mothers provided written informed consent for themselves and their infants. Inclusion criteria were: (1) Child was born at 37.0 – 42.0 weeks gestation, with weight appropriate for gestational age, and no significant perinatal or neonatal complications. Exclusions were: (1) non-fluency in English in the parent; (2) foster child; (3) mother < 18 years old; (4) medical problems or known diagnosis affecting current or future eating, growth or development; (5) child protective services involvement in the neonatal period; (6) infant does not consume at least 2 ounces in one feeding from an artificial nipple and bottle at least once per week.
Sample collection
All samples were collected using sterile dual-tip swabs (BD Swube®; (Becton, Dickinson and Company, Sparks, MD). One swab was used for 16S sequence analysis and the other for untargeted metabolomics analysis.
LP:
Gloves and a mask were worn while collecting the samples to try to prevent contamination. 3 double-tip swabs were used to gently swab the inside of the cheek, the skin of the axilla, and stool from the diaper. Once the collection was completed, the swabs were placed in the freezer, typically within 15-30 minute of collection, and stored at −80 degree C until analysis. Swabs were collected once daily until the infant was discharged or had reached 21 days after birth. Mother’s breast milk and donor milk from the LP cohort were collected into 2 mL cryovials and stored at −80 C until analysis.
VLBW:
For sample collection a research team member or RN working with the infant used 1 double-tip swab to gently swab the inside of the cheek, the skin of the axilla, and stool from the diaper each day for the first 7 days after birth. Once the collection was completed, the swabs were stored at −80 degree C until analysis.
16S rRNA gene sequencing
DNA was extracted using the MO BIO PowerSoil DNA extraction kit according to Earth Microbiome Project (EMP) standard protocols [80](http://www.earthmicrobiome.org/emp-standard-protocols/). PCR targeting the V4 region of the 16S rRNA bacterial gene was performed with the 515F/806R primers, utilizing the protocol described in [81]. Amplicons were barcoded and pooled in equal concentrations for sequencing. The amplicon pool was purified with the MO BIO UltraClean PCR Clean-up kit and 2 x 150 bp sequencing was performed on the MiSeq sequencing platform at the Institute for Genomic Medicine at UCSD.
16S amplicon sequencing data processing and analysis
Raw sequencing data was deposited in Qiita[66] and processed using the default parameters. Briefly, forward reads were demultiplexed and quality control filtered, followed by trimming to 150 bp and then processed with deblur[82] and filtered to remove amplicon sequence variants (ASVs) with a total count below 10 across all samples. The resulting ASVs were inserted into the Greengenes tree[83] via SEPP (SATé-Enabled Phylogenetic Placement using q2-fragment-insertion in QIIME2[84,85]. The feature table was collapsed to the genus level for use in differential abundance testing.
Metabolomics data acquisition
Swab tips were transferred to 96 well deep well plates, cutting or breaking the wooden swab handle slightly above the cotton swab. Breast milk samples were extracted in a final concentration of 80% MeOH:20% water (Optima LC-MS grade; Fisher Scientific, Fair Lawn, NJ, USA).
Untargeted mass spectrometry data for VLBW sample sets were collected using a ultra-high performance liquid chromatography system (Vanquish, Thermo) coupled to an orbitrap mass spectrometer (Q-Exactive, Thermo). Reverse chromatographic separation was accomplished using a Kinetic C18 column. The column compartment was held at 40 °C with a flow rate of 500 μL/min. Mobile phase composition was (A) 0.1% formic acid in water and (B) 0.1% formic acid in acetonitrile. The elution gradient used was: 0-1.0 min isocratic at 5% B, 1.0-9.0 min 100% B, 9.0-11.0 min isocratic at 100% B, 11.0-11.5 min 5% B, and 11.5-12.5 min isocratic at 5% B. Data dependent acquisition for Full MS: Resolution set at 35,000, AGC target 5e4, scan range 100 to 1500 m/z. For dd-MS/MS: Resolution at 17,500, AGC target 2e4, loop count 5, and (N)CE/stepped (N)CE 20, 30, 40. Data Dependent acquisition settings were set at: minimum AGC target 2.00e4, and Apex trigger 3 to 15 sec.
LP data were collected using the data-dependent acquisition method outlined in Gauglitz et al.[58]. Extracts were dried down using centrifugal evaporation (Labconco) and resuspended in 50% MeOH:50% water (Optima LC-MS grade; Fisher Scientific, Fair Lawn, NJ, USA), with 1uM sulfadimethoxine. Untargeted metabolomics was carried out using an ultra-high-performance liquid chromatography system (UltiMate 3000, Thermo Scientific, Waltham, MA) coupled to a Maxis Q-TOF (Bruker Daltonics, Bremen, Germany) mass spectrometer with a Kinetex C18 column (Phenomenex Torrance, CA, USA). A linear gradient was applied: 0-0.5 min isocratic at 5% B, 0.5-8.5 min 100% B, 8.5-11 min isocratic at 100% B, 11-11.5 min 5% B, 11.5-12 min 5% B; where mobile phase A is water with 0.1% formic acid (v/v) and phase B is acetonitrile 0.1% formic acid (v/v) (LC-MS grade solvents, Fisher Chemical). Electrospray ionization in positive mode was used.
MS1 Feature Finding and Data Processing
qToF files (.d) were exported using DataAnalysis (Bruker) as .mzXML files after lock mass correction using hexakis (1H, 1H, 2H-difluoroethoxy)phosphazene (Synquest Laboratories, Alachua, FL), with m/z 622.029509. Data quality was assessed by qualitatively evaluating the m/z error and retention time of the LC-MS standard solution (i.e. mixture of 6 compounds), which was analyzed at least once in every 96-well plate.
MS1 feature finding was performed on the .mzXML files in MZmine2 (version MZmine-2.37.corr16.4)[86]. The mzMINE parameters used for feature finding are as follows: mass detection (centroid; ms1: 1.5E3; MS2: 90); ADAP Chromatogram builder (min group size in # of scans: 4; group intensity threshold: 5E3; min highest intensity: 2E3; m/z tolerance: 0.001 m/z to 20 ppm); chromatogram deconvolution (LMS: chromatographic threshold of 96%, search minimum in RT range (min) of 0.03, minimum relative height of 5%, minimum absolute height of 2E3, min ratio of peak top/edge of 1 and peak duration range (min) of 0 - 2; m/z center calculation set to auto; m/z range for MS2 scan pairing (Da) of 0.02 and RT range for MS2 scan pairing (min) of 0.15); isotope peaks grouper (m/z tolerance set to 0.0015 m/z or 10 ppm; retention time tolerance of 0.05, maximum charge of 3 and representative isotope set to most intense); order peak lists; join aligner (m/z tolerance set at 0.0015 m/z or 15 ppm; weight for m/z of 2; retention time tolerance of 0.2 min; weight for RT of 1. A filter was used such that only features present in at least 2 samples were included. The output was a data matrix of variables (i.e. MS1 features that triggered MS2 scans) by samples, exported for GNPS (.mgf and .csv quant table). Tables were normalized by sample wherein metabolite relative abundances were determined by dividing the integrated intensity for each feature by the sum of the intensities across all features for that sample.
MS2 molecular networking
The resulting .mgf and .csv quantification table produced by MZmine2 were imported into the GNPS interface for use in the feature based molecular networking workflow[67,87]. The parameters were set at: Precursor Ion Mass Tolerance 0.02 Da, Fragment Ion Mass Tolerance 0.02 Da, Minimum Pairs Cosine 0.7, Minimum Matched Fragment Ions 6, Maximum shi between precursors 500 Da, Network TopK 10, Maximum Connected Component Size (Beta) 100, Library Search Min Matched Peaks 6, Score Threshold 0.7. The resulting annotations fall under the Metabolomics Standards Initiative metabolite identification levels 1-3[88].
16S Diversity Analyses
Alpha diversity was measured by Shannon diversity after rarefying the ASV table to a depth of 1250 sequences[89]. Beta diversities for ASVs were calculated using robust Aitchison distances[90,91]. Principal components analysis (PCA) of these distances were plotted and distances were used to calculate body site differentiation by PERMANOVA as implemented by skbio. To determine the overall contribution of metadata factors to microbial diversity, stepwise redundancy analysis was run using the ordistep function from the vegan package in R. The data used in the analysis were the robust Aitchison distances on samples from each body site separately. The metadata factors included in the ordistep formula were antibiotic use, age, and delivery mode.
Metabolomics analysis
Beta diversity of normalized metabolite intensities was calculated with Jaccard distances. Metabolite profile change over time was determined by calculating the Jaccard distance of each infant sample to the sample from the same body site at day 0 for LP infants and day 1 for VLBW infants. These distances to the earliest time point over time were split by birth-weight, delivery-mode, and antibiotic exposure and used to calculate linear mixed models using individual as a random effect and age as a fixed effect. Slopes and p-values of the age variable in linear mixed models were reported.
Differential abundance testing
Differential abundances of microbes and metabolites were determined by Songbird. Songbird takes in a feature table, sample metadata, and a formula to calculate differential scores for each feature as a measure of differential abundance depending on metadata variables specified in the formula. Songbird also supplies a Q2 that behaves similar to an R2. Following advice from the Songbird creators, only models with positive Q2 values were included in results. The genus collapsed table was used for differential abundance testing of microbes. Songbird was run separately for each body site. To determine differential abundances due to delivery-mode, age, and formula/donor breast milk supplementation, only non-antibiotic exposed LPI infant samples were included. The formula in this case was “age + delivery-mode + milk_supplementation”. To determine the effect of antibiotics, all LP and VLBW infant samples were included and the formula “age + delivery-mode + milk_supplementation + antibiotics” was used. Songbird differentials for metabolite abundances were calculated using normalized metabolite abundances with all LP infant samples using the formula“age + delivery-mode + milk_supplementation”. Log ratios of secondary to primary bile acids were calculated using Qurro[92]. Linear mixed effects models (age as fixed variable and individual as random variable) were run on these log ratios split by delivery mode and antibiotic exposure over time and the slope, and p-value of the age variable in these models were reported.
SourceTracker 2
To compare infant microbiomes to healthy adult microbiomes, 16S-V4 sequencing data from healthy adults, including 4434 stool, 2550 oral, 1975 skin samples, and 427 vaginal samples were downloaded (https://msystems.asm.org/content/5/1/e00630-19). We excluded all samples that indicated exposure to antibiotics within the last year. We compared the LP and VLBW infants to healthy FT infants, using the cohort of 100 FT vaginally-born infants from a publicly available dataset in Qiita. Only stool samples were available from FT infants, not skin or oral samples. All data was processed in the same manner as for preterm infants described above, with an additional filtering to remove all ASVs present in less than 1% of samples to reduce complexity. All adult samples from all three body sites and stool samples from FT infants were merged into a single database that was used as a source for Sourcetracker2[93] analysis on each preterm infant sample. The relative contribution of each source (adult stool, adult oral, adult skin, and FT infant stool) was determined for each preterm infant sample. Preterm infant samples were grouped by delivery mode, birth weight (LP vs VLBW), and antibiotic status, and the mean and standard error was calculated for each group over time and plotted.
Mantel test
Correlation between microbe abundances and metabolite intensities were calculated using a Mantel test as implemented by skbio. The genus collapsed feature table and normalized metabolite profile were subsampled to only include samples present in both datasets. Samples from all three body sites were included in this analysis. Samples were divided by age, delivery-mode, and antibiotic exposure, and then the Mantel test was run on each dataset independently. The Mantel correlation and p-value were plotted over time for each sample set.
Statistical Analysis
The methods used for each specific component of the analyses outlined above are detailed in those respective sections, but a general summary follows.
Pre-processing:
Amplicon sequencing data were quality and noise-filtered within Qiita using recommended settings, removing low abundance ASVs (<10 across the dataset). Sequences were further rarefied to 1250 reads per sample for calculation of alpha diversity, but no other analyses utilized a rarefied dataset. Metabolomic feature relative abundances determined by MS1 feature finding using mzMINE were calculated using per-sample normalization.
Data presentation:
Tabular data were summarized in Microsoft Excel, manipulated using the Pandas package in Python, and plotted using the Python packages matplotlib and plotnine, a Python implementation of the R package ggplot2. For within-individual distances, boxplots were generated showing the median and interquartile range with whiskers extending to the furthest value within 1.5 times the edge of the interquartile range. For PCA or PCoA plots, confidence ellipses were calculated in matplotlib showing the boundary of 3 standard deviations from the centroid. For alpha diversity and SourceTracker 2 analysis results, line graphs were generated showing the mean with shaded areas indicating the standard error of the mean.
Sample size:
From 85 infants enrolled in the study, 1799 samples were obtained, 942 16S amplicon sequencing tables (from 61 infants) and 1080 metabolomic feature tables (from 75 infants) across 3 body sites and 7 days of life were analyzed. The exact sample sizes per body site, per day varied considerably and a detailed breakdown of subgroups compared for clinical characteristics can be found in Table 1, while the subgroups analyzed for metagenomic and metabolomic attributes are listed in Extended Data Table 3 and shown in the flowchart and heatmaps in Figure 1.
Statistical methods:
Permutational multivariate analysis of variance (PERMANOVA), stepwise redundancy analysis (RDA), linear mixed models, and Mantel tests were all performed with a testing value of p = 0.05 to determine significance (https://github.com/ucsd-cmi/preterm_infant/tree/master/results/tables). Multiple test correction was not required for the analyses conducted.
Software:
PERMANOVA, linear mixed models, and Mantel tests were performed using the scikit-learn and skbio Python libraries. Stepwise redundancy analysis was run in R using the ordistep function in the vegan package.
Extended Data
Extended Data Figure 1. Sample counts for preterm infants by day after birth.
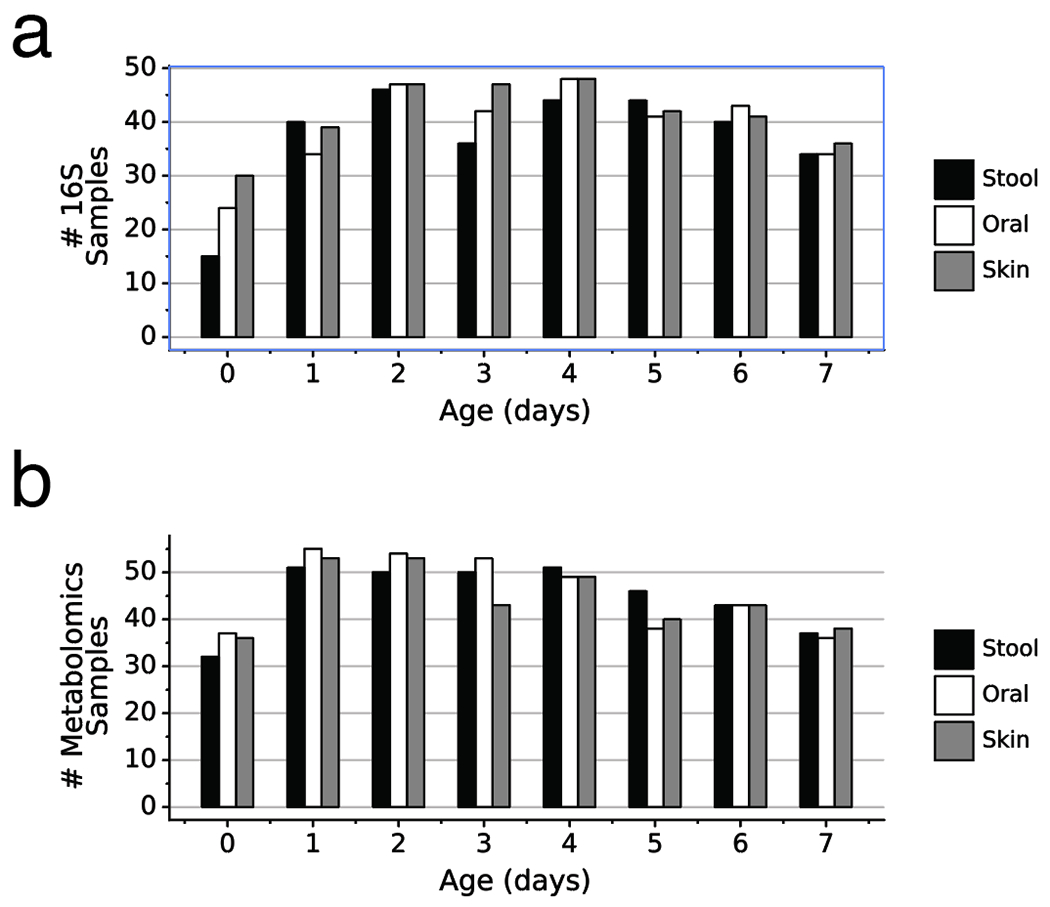
a) Number of 16S samples for each body site by infant day after birth. b) Number of metabolomics samples for each body site by infant day after birth. Stool (black), Oral (white), Skin (grey).
Extended Data Figure 2. Alpha diversity in preterm infants over the first week after birth.
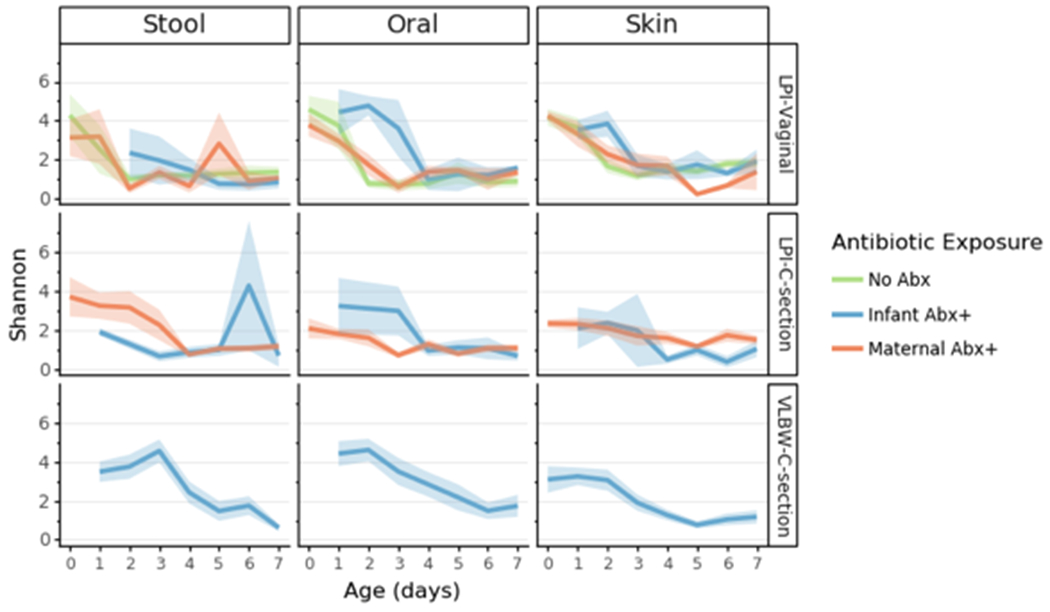
Lines represent the mean (+/− SEM) alpha diversity as measured by Shannon index for all samples from preterm infants within each cohort: late preterm (LP) or very low birth weight (VLBW) displayed by birth-mode (rows) and body site (column). Lines are colored based on reported exposure to antibiotics, none (green), direct exposure (blue) or potential exposure due to maternal transfer (orange). Refer to Extended Data Table 3 for sample sizes in each comparison.
Extended Data Figure 3. 16S profile changes over the first week after birth in LP and VLBW infants.
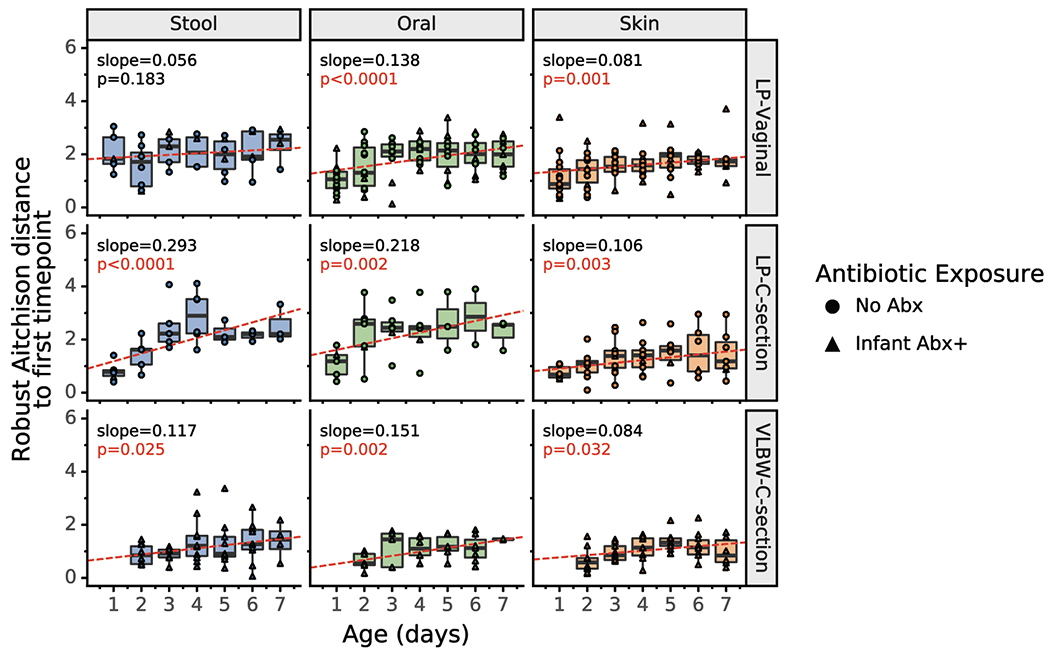
Within-infant 16S Robust Aitchison distance to first sample over time. For each infant, the Robust Aitchison distance to the first sample (day 0 for LPI infants, day 1 for VLBW infants) is plotted over time for each body site. Red lines show the fitted linear mixed effects model (individual as a random effect and age as a fixed effect); the corresponding p-value and slope is shown. All body sites show a trend of increasing distance over time, indicating that microbial profiles change over the first week after birth. Boxplots show median and interquartile range with whiskers extending to the furthest value within 1.5 times the edge of the interquartile range. Refer to Extended Data Table 3 for sample sizes in each comparison.
Extended Data Figure 4. Impact of maternal antibiotics exposure on the differentiation of the microbial community of preterm infants over the first week after birth.
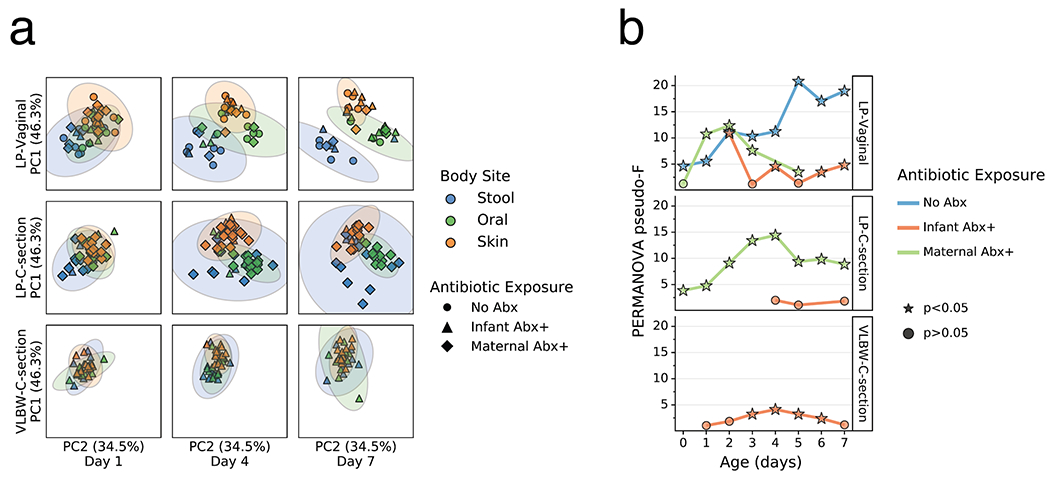
a) Robust Aitchison Principal Components Analysis (PCA) based on 16S V4 amplicon sequence variants (ASVs) of the gut (blue), oral (green), and skin (orange) microbiomes collected at day 1, 4 and 7 after birth from late preterm infants (LPs) born vaginally (LP-Vaginal) or by Cesarean section (LP-C-section) and very-low birth weight (VLBW) preterm infants (VLBW-C-section), with samples from children exposed to antibiotics as reported by clinical metadata (Infant Abx(+); triangles), from children whose mothers were exposed to antibiotics (Maternal Abx; diamonds), and those without exposure to antibiotics (No Abx(−); circles) differentiated. b) PERMANOVA for differences in sample type (stool, oral, skin) based on Aitchison distances between all within-group samples for LP-Vaginal, LP-C-section, and VLBW-C-section preterm infants daily over the first week after birth, separated by maternal Abx exposure based on clinical metadata. Refer to Extended Data Table 3 for sample sizes in each comparison.
Extended Data Figure 5. 16S profile stratified by metabolome-informed antibiotic exposure.
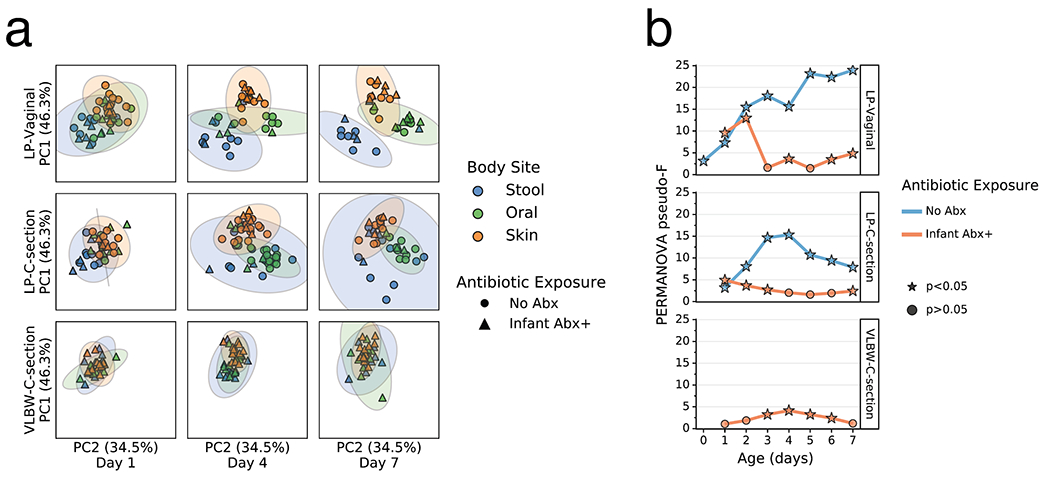
a) Robust Aitchison Principal Components Analysis (PCA) based on 16S V4 amplicon sequence variants (ASVs) of the gut (blue), oral (green), skin (orange) microbiomes collected at day 1, 4 and 7 after birth from late preterm infants (LPs) born vaginally (LP-Vaginal) or by Cesarean section (LPI-C-section) and very-low birth weight (VLBW) preterm infants (VLBW-C-section), with samples from children separated by Abx exposure based on metabolically-informed metadata. b) PERMANOVA for differences in sample type (stool, oral, skin) based on Aitchison distances between all within-group samples for LP-Vaginal, LP-C-section, and VLBW-C-section preterm infants daily over the first week after birth, separated by Abx exposure based on metabolically-informed metadata. Refer to Extended Data Table 3 for sample sizes in each comparison.
Extended Data Figure 6. Sourcetracker2 analysis stratified by maternal antibiotic exposure.
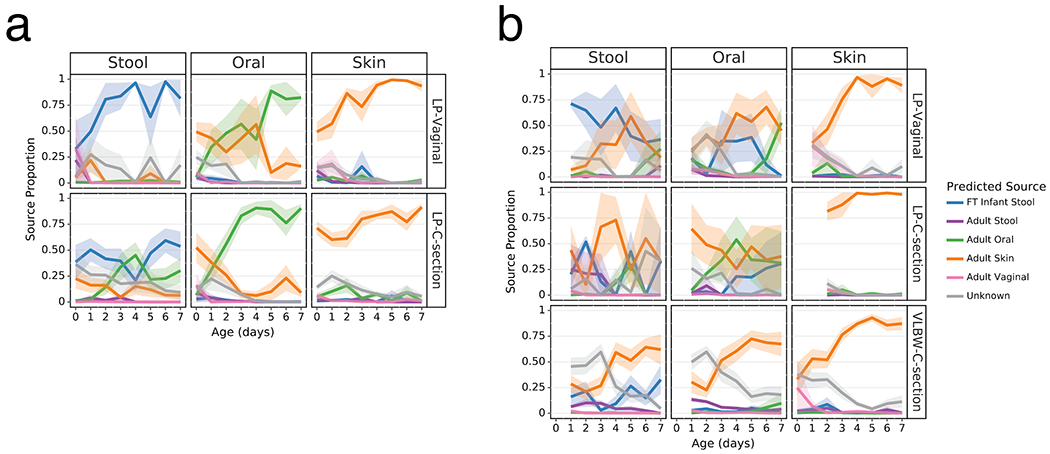
SourceTracker2 analysis showing proportion of LP infant microbial communities attributed to adult or full term (FT) infant microbiomes based on 11 public studies in Qiita and a cohort of 87 FT infants not exposed to antibiotics aged 0.5-4 months (Methods). Lines represent the mean (+/− SEM) proportion of the microbial profile attributed to a specific source for all samples from preterm infants within each cohort displayed by birth-mode (rows) and body site (column). a) LP infant samples with no infant or maternal antibiotic exposure. b) LP infant samples with maternal antibiotic exposure but without infant antibiotic exposure. Refer to Extended Data Table 3 for sample sizes at each time point.
Extended Data Figure 7. Sourcetracker2 analysis stratified by metabolomics-informed antibiotic exposure.
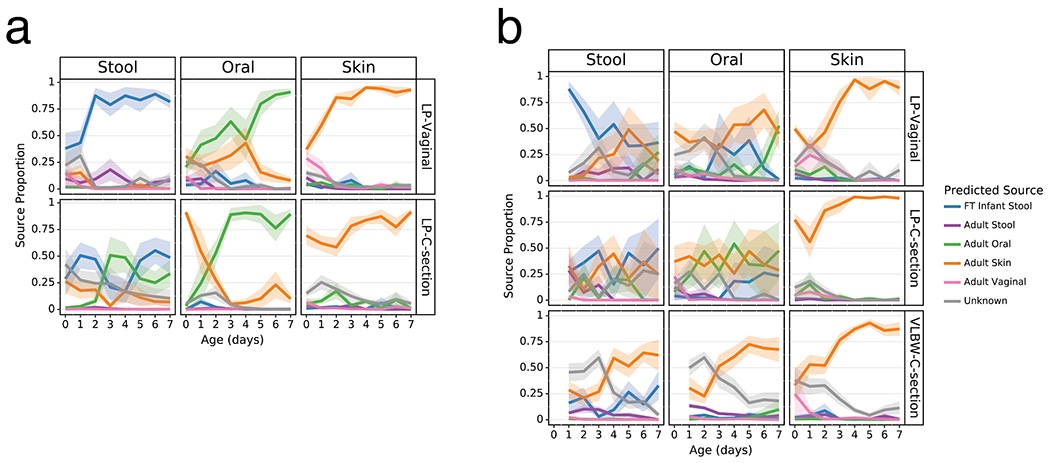
SourceTracker2 analysis showing proportion of LP infant microbial communities attributed to adult or full term (FT) infant microbiomes based on 11 public studies in Qiita and a cohort of 87 FT infants not exposed to antibiotics aged 0.5-4 months (Methods). Antibiotic exposure is defined by infants exposed to antibiotics by clinical metadata or antibiotics detected by metabolomics. Lines represent the mean (+/− SEM) proportion of the microbial profile attributed to a specific source for all samples from preterm infants within each cohort displayed by birth-mode (rows) and body site (column). a) LP infant samples with no antibiotic exposure. b) LP infant samples with antibiotic exposure. Refer to Extended Data Table 3 for sample sizes at each time point.
Extended Data Figure 8. Impact of antibiotics exposure on the differentiation of the metabolome of preterm infants over the first week after birth.
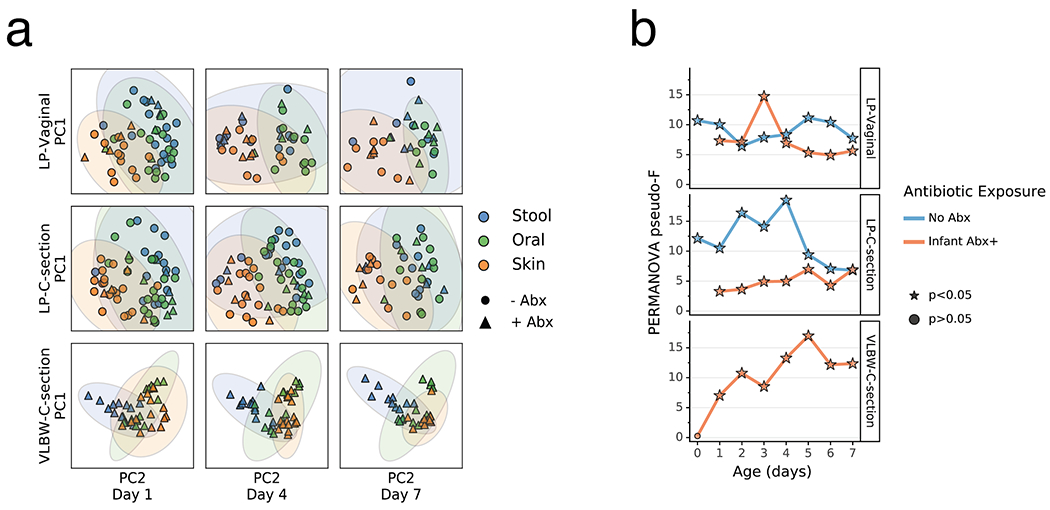
a) Principal Coordinates Analysis (PCoA) based on Jaccard distances of metabolomics profile of the gut (blue), oral (green), and skin (orange) at day 1, 4, and 7 after birth from late preterm infants (LPs) born vaginally (LP-Vaginal) or by Cesarean section (LP-C-section) and very-low birth weight (VLBW) preterm infants (VLBW-C-section), with samples from children exposed to antibiotics as reported by clinical metadata (Infant Abx(+); triangles) and those without exposure to antibiotics (No Abx(−); circles) differentiated. Jaccard distances and PCoA for VLBW infants calculated separately from LP-infants because samples were run on different instruments. Principal Components (PCs) differ for LP-infants and VLBW-infants (LP-infant PC1 variance= 54.5%, PC2 variance= 30.6%; VLBW PC1 variance= 70.3%, PC2 variance= 25.1%). b) PERMANOVA for differences in sample type (stool, oral, skin) based on Jaccard distances between all within-group samples for LP-Vaginal, LP-C-section, and VLBW-C-section preterm infants daily over the first week after birth, separated by Abx exposure. Refer to Extended Data Table 3 for sample sizes in each comparison.
Extended Data Figure 9. Microbial genera associated with supplemental formula or breastmilk.
Songbird differentials for top 5 genera positively and negatively associated with supplemental breastmilk or supplemental formula in the stool and oral samples from LP infants. The songbird model fit for stool (Q2=0.072) and oral (Q2=0.014).
Extended Data Figure 10. Mantel correlation between metabolome and 16S profile over time.
Stool, oral, and skin samples with matched 16S and metabolomics data were divided by delivery mode, and antibiotic exposure, then Mantel tests were run for each day to measure the relation between the two data types over time. Significant Mantel correlations are depicted as stars. Sample size for each comparison shows as the number above each point.
Extended Data Table 1.
Stepwise RDA of metadata variables influencing microbial composition of all samples. Separate analyses run for samples for all body sites together and for each body site individually.
| All body sites (including individual) | |||||
|---|---|---|---|---|---|
|
| |||||
| Factor | R2.adj | Df | AIC | F | Pr(>F) |
|
| |||||
| host_subject_id | 0.196 | 60 | 5979.54 | 4.80 | 0.002 |
| sample_type | 0.169 | 2 | 5759.22 | 118.18 | 0.002 |
| age | 0.038 | 1 | 5702.12 | 56.85 | 0.002 |
| <All variables> | 0.001 | ||||
| All body sites (excluding individual) | |||||
|
| |||||
| Factor | R2.adj | Df | AIC | F | Pr(>F) |
|
| |||||
| body_site | 0.170 | 2 | 5953.00 | 97.14 | 0.002 |
| metadata_abx | 0.095 | 1 | 5840.19 | 121.59 | 0.002 |
| age | 0.043 | 1 | 5784.52 | 59.16 | 0.002 |
| birth_weight | 0.016 | 1 | 5763.61 | 23.04 | 0.002 |
| delivery_mode | 0.014 | 1 | 5745.17 | 20.51 | 0.002 |
| <All variables> | 0.001 | ||||
| Stool (including individual) | |||||
|
| |||||
| Factor | R2.adj | Df | AIC | F | Pr(>F) |
|
| |||||
| host_subject_id | 0.451 | 58 | 1512.02 | 5.21 | 0.002 |
| <All variables> | 0.042 | ||||
| Stool (excluding individual) | |||||
|
| |||||
| Factor | R2.adj | Df | AIC | F | Pr(>F) |
|
| |||||
| delivery_mode | 0.141 | 1 | 1595.21 | 50.05 | 0.002 |
| metadata_abx | 0.078 | 1 | 1567.77 | 30.62 | 0.002 |
| age | 0.042 | 1 | 1552.43 | 17.62 | 0.002 |
| randomization | 0.013 | 2 | 1548.98 | 3.69 | 0.002 |
| delivery_weight | 0.006 | 1 | 1547.43 | 3.49 | 0.022 |
| <All variables> | 0.000 | ||||
| Oral (including individual) | |||||
|
| |||||
| Factor | R2.adj | Df | AIC | F | Pr(>F) |
|
| |||||
| host_subject_id | 0.441 | 59 | 1565.52 | 5.15 | 0.002 |
| age | 0.080 | 1 | 1518.29 | 42.90 | 0.002 |
| <All variables> | 0.004 | ||||
| Oral (excluding individual) | |||||
|
| |||||
| Factor | R2.adj | Df | AIC | F | Pr(>F) |
|
| |||||
| metadata_abx | 0.249 | 1 | 1605.94 | 104.35 | 0.002 |
| age | 0.100 | 1 | 1562.50 | 48.44 | 0.002 |
| birth_weight | 0.044 | 1 | 1541.63 | 23.42 | 0.002 |
| delivery_mode | 0.005 | 1 | 1540.16 | 3.43 | 0.024 |
| <All variables> | 0.003 | ||||
| Skin (including individual) | |||||
|
| |||||
| Factor | R2.adj | Df | AIC | F | Pr(>F) |
|
| |||||
| host_subject_id | 0.146 | 58 | 1485.42 | 1.96 | 0.002 |
| age | 0.119 | 1 | 1436.70 | 44.84 | 0.002 |
| <All variables> | 0.001 | ||||
| Skin (excluding individual) | |||||
|
| |||||
| Factor | R2.adj | Df | AIC | F | Pr(>F) |
|
| |||||
| age | 0.114 | 1 | 1446.51 | 43.11 | 0.002 |
| birth_weight | 0.050 | 1 | 1428.25 | 20.71 | 0.002 |
| delivery_mode | 0.008 | 1 | 1426.00 | 4.22 | 0.006 |
| <All variables> | 0.007 | ||||
Extended Data Table 2.
List of studies from which 16S data was used for Sourcetracker analysis.
| Qiita ID | Study Name | N | Publication |
|---|---|---|---|
| 550 | Moving pictures of the human microbiome | Oral: 508 | Caporaso et al (2011) |
| 1774 | Puerto Rico and Plantanal | Oral: 48 | None |
| 1841 | controlCUforehead | Skin: 1293 Oral: 642 |
None |
| 2010 | Partial restoration of the microbiota of cesarean-born infants via vaginal microbial transfer | Oral: 72 Skin: 65 |
Dominguez-Bello et al (2016) |
| 2024 | The microbiota at multiple body sites during pregnancy in a rural Tanzanian population and the effects of Moringa supplemented probiotic yogurt | Oral: 254 | Bisanz et al (2016) |
| 2202 | Host lifestyle affects human microbiota on daily timescales | Oral: 285 | David et al (2016) |
| 10052 | The microbiome of uncontacted Amerindians | Oral: 16 | Clemente et al (2015) |
| 10317 | American Gut Project | Stool: 2825 Oral: 547 Skin: 440 |
McDonald et al (2018) |
| 10894 | MG_baby timeseries | vaginal mucus: 427 | None |
| 10918 | Eating behavior development in infants | Stool: 96 | None |
| 11052 | Knight_ABTX | Oral: 178 Skin: 177 |
None |
Extended Data Table 3.
Sample counts for 16S and metabolomics data.
| cohort | antibiotics | body_site | age | 16S_sample_count | metabolomics_sample_count |
|---|---|---|---|---|---|
| LP-C-section | no | Oral | 0 | 7 | 7 |
| LP-C-section | no | Oral | 2 | 8 | 10 |
| LP-C-section | no | Oral | 16 | 19 | |
| LP-C-section | no | Oral | 3 | 14 | 18 |
| LP-C-section | no | Oral | 4 | 16 | 18 |
| LP-C-section | no | Oral | 5 | 11 | 15 |
| LP-C-section | no | Oral | 6 | 10 | 15 |
| LP-C-section | no | Oral | 7 | 9 | 10 |
| LP-C-section | no | Skin | 0 | 12 | 15 |
| LP-C-section | no | Skin | 1 | 11 | 13 |
| LP-C-section | no | Skin | 2 | 14 | 18 |
| LP-C-section | no | Skin | 3 | 13 | 17 |
| LP-C-section | no | Skin | 4 | 15 | 18 |
| LP-C-section | no | Skin | 5 | 10 | 11 |
| LP-C-section | no | Skin | 6 | 9 | 9 |
| LP-C-section | no | Skin | 7 | 10 | 10 |
| LP-C-section | no | Stool | 0 | 7 | 9 |
| LP-C-section | no | Stool | 1 | 12 | 17 |
| LP-C-section | no | Stool | 2 | 14 | 17 |
| LP-C-section | no | Stool | 3 | 11 | 16 |
| LP-C-section | no | Stool | 4 | 15 | 18 |
| LP-C-section | no | Stool | 5 | 11 | 14 |
| LP-C-section | no | Stool | 6 | 11 | 11 |
| LP-C-section | no | Stool | 7 | 10 | 10 |
| LP-C-section | yes | Oral | 1 | 3 | 4 |
| LP-C-section | yes | Oral | 2 | 2 | 3 |
| LP-C-section | yes | Oral | 3 | 4 | 5 |
| LP-C-section | yes | Oral | 4 | 5 | 5 |
| LP-C-section | yes | Oral | 5 | 5 | 5 |
| LP-C-section | yes | Oral | 6 | 3 | 4 |
| LP-C-section | yes | Oral | 7 | 3 | 4 |
| LP-C-section | yes | Skin | 1 | 1 | 3 |
| LP-C-section | yes | Skin | 2 | 3 | 4 |
| LP-C-section | yes | Skin | 3 | 2 | 3 |
| LP-C-section | yes | Skin | 4 | 5 | 5 |
| LP-C-section | yes | Skin | 5 | 5 | 5 |
| LP-C-section | yes | Skin | 6 | 3 | 4 |
| LP-C-section | yes | Skin | 7 | 3 | 4 |
| LP-C-section | yes | Stool | 1 | 3 | 4 |
| LP-C-section | yes | Stool | 2 | 2 | 4 |
| LP-C-section | yes | Stool | 3 | 3 | 4 |
| LP-C-section | yes | Stool | 4 | 4 | 4 |
| LP-C-section | yes | Stool | 5 | 5 | 5 |
| LP-C-section | yes | Stool | 6 | 2 | 3 |
| LP-C-section | yes | Stool | 7 | 3 | 4 |
| LP-Vaginal | no | Oral | 0 | 16 | 15 |
| LP-Vaginal | no | Oral | 1 | 11 | 12 |
| LP-Vaginal | no | Oral | 2 | 12 | 14 |
| LP-Vaginal | no | Oral | 3 | 9 | 11 |
| LP-Vaginal | no | Oral | 4 | 9 | 9 |
| LP-Vaginal | no | Oral | 5 | 8 | 7 |
| LP-Vaginal | no | Oral | 6 | 7 | 7 |
| LP-Vaginal | no | Oral | 7 | 6 | 5 |
| LP-Vaginal | no | Skin | 0 | 15 | 15 |
| LP-Vaginal | no | Skin | 1 | 13 | 12 |
| LP-Vaginal | no | Skin | 2 | 12 | 11 |
| LP-Vaginal | no | Skin | 3 | 12 | 8 |
| LP-Vaginal | no | Skin | 4 | 9 | 8 |
| LP-Vaginal | no | Skin | 5 | 9 | 7 |
| LP-Vaginal | no | Skin | 6 | 7 | 6 |
| LP-Vaginal | no | Skin | 7 | 6 | 6 |
| LP-Vaginal | no | Stool | 0 | 8 | 11 |
| LP-Vaginal | no | Stool | 1 | 10 | 16 |
| LP-Vaginal | no | Stool | 2 | 12 | 16 |
| LP-Vaginal | no | Stool | 3 | 9 | 15 |
| LP-Vaginal | no | Stool | 4 | 8 | 10 |
| LP-Vaginal | no | Stool | 5 | 9 | 11 |
| LP-Vaginal | no | Stool | 6 | 7 | 8 |
| LP-Vaginal | no | Stool | 7 | 6 | 6 |
| LP-Vaginal | yes | Oral | 1 | 4 | 4 |
| LP-Vaginal | yes | Oral | 2 | 5 | 5 |
| LP-Vaginal | yes | Oral | 3 | 3 | 4 |
| LP-Vaginal | yes | Oral | 4 | 5 | 5 |
| LP-Vaginal | yes | Oral | 5 | 5 | 5 |
| LP-Vaginal | yes | Oral | 6 | 6 | 6 |
| LP-Vaginal | yes | Oral | 7 | 6 | 5 |
| LP-Vaginal | yes | Skin | 1 | 3 | 4 |
| LP-Vaginal | yes | Skin | 2 | 4 | 5 |
| LP-Vaginal | yes | Skin | 3 | 4 | 5 |
| LP-Vaginal | yes | Skin | 4 | 4 | 5 |
| LP-Vaginal | yes | Skin | 5 | 4 | 4 |
| LP-Vaginal | yes | Skin | 6 | 5 | 6 |
| LP-Vaginal | yes | Skin | 7 | 5 | 6 |
| LP-Vaginal | yes | Stool | 1 | 2 | 3 |
| LP-Vaginal | yes | Stool | 2 | 5 | 6 |
| LP-Vaginal | yes | Stool | 3 | 4 | 5 |
| LP-Vaginal | yes | Stool | 4 | 4 | 4 |
| LP-Vaginal | yes | Stool | 5 | 5 | 6 |
| LP-Vaginal | yes | Stool | 6 | 6 | 8 |
| LP-Vaginal | yes | Stool | 7 | 6 | 6 |
| VLBW-C-section | yes | Oral | 0 | 1 | 4 |
| VLBW-C-section | yes | Oral | 1 | 8 | 11 |
| VLBW-C-section | yes | Oral | 2 | 12 | 17 |
| VLBW-C-section | yes | Oral | 3 | 12 | 15 |
| VLBW-C-section | yes | Oral | 4 | 13 | 17 |
| VLBW-C-section | yes | Oral | 5 | 12 | 15 |
| VLBW-C-section | yes | Oral | 6 | 17 | 17 |
| VLBW-C-section | yes | Oral | 7 | 10 | 13 |
| VLBW-C-section | yes | Skin | 0 | 3 | 4 |
| VLBW-C-section | yes | Skin | 1 | 11 | 12 |
| VLBW-C-section | yes | Skin | 2 | 14 | 15 |
| VLBW-C-section | yes | Skin | 3 | 16 | 16 |
| VLBW-C-section | yes | Skin | 4 | 15 | 16 |
| VLBW-C-section | yes | Skin | 5 | 14 | 15 |
| VLBW-C-section | yes | Skin | 6 | 17 | 16 |
| VLBW-C-section | yes | Skin | 7 | 12 | 14 |
| VLBW-C-section | yes | Stool | 1 | 13 | 14 |
| VLBW-C-section | yes | Stool | 2 | 13 | 14 |
| VLBW-C-section | yes | Stool | 3 | 9 | 11 |
| VLBW-C-section | yes | Stool | 4 | 13 | 13 |
| VLBW-C-section | yes | Stool | 5 | 14 | 14 |
| VLBW-C-section | yes | Stool | 6 | 14 | 14 |
| VLBW-C-section | yes | Stool | 7 | 9 | 9 |
Acknowledgements
This work was supported by a Seed Grant from the Center for Microbiome Innovation. We would like to thank Gregory Humphrey, Christine Aceves, Caitriona Brennan, Lindsay DeRight-Goldasich, A. Cole Heale, Morgan Panitchpakdi, Karenina Sanders, and Tara Schwartz for sample processing, Gail Ackermann for assistance with metadata curation, and Jeff DeReus for data handling and processing. We gratefully acknowledge the following funding sources: NIH R01HD084163, EIA14660045, an American Heart Association Established Investigator Award to Julie Lumeng. We gratefully acknowledge conversations with C. Martino, Y. Vasquez-Baeza, and S. Miller-Montgomery. This work was supported in part by an Emerald Foundation Distinguished Investigator Award (R.K.). This work was supported in part by the Chancellor’s Initiative in the Microbiome and Microbial Sciences (R.K., A.D.S.) and by Illumina, Inc. through reagent donation in partnership with the Center for Microbiome Innovation at UC San Diego.
Footnotes
Data and code availability
All raw and processed sequencing data is publically available in Qiita study 11712 (https://qiita.ucsd.edu/study/description/11712) and EBI project ERP122936 for the LP cohort, Qiita study 11713 (https://qiita.ucsd.edu/study/description/11713) and EBI project ERP122952 for the VLBW infant cohort. Mass spectral data files are available on MassIVE (http://massive.ucsd.edu) under the following IDs: MSV000083559 (VLBW infant stool, oral, skin), MSV000083462 (LP stool, oral, skin), MSV000083463 (LP mother breast milk / donor breast milk). All code, feature tables, QIIME2 artifacts and tables containing results used to generate figures and visualizations used in these analyses are available at https://github.com/ucsd-cmi/preterm_infant.
Conflict of Interest
J.H.K. is a medical advisor for Evolve BioSystems and a consultant for Nutricia. R.K., A.D.S., and S.J.S. are directors at the Center for Microbiome Innovation at UC San Diego, which receives industry research funding for various microbiome initiatives, but no industry funding was provided for this study.
References
- [1].Poore GD, Kopylova E, Zhu Q, Carpenter C, Fraraccio S, Wandro S, Kosciolek T, Janssen S, Metcalf J, Song SJ, Kanbar J, Miller-Montgomery S, Heaton R, Mckay R, Patel SP, Swafford AD, Knight R, Nature 2020, 579, 567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Nejman D, Livyatan I, Fuks G, Gavert N, Zwang Y, Geller LT, Rotter-Maskowitz A, Weiser R, Mallel G, Gigi E, Meltser A, Douglas GM, Kamer I, Gopalakrishnan V, Dadosh T, Levin-Zaidman S, Avnet S, Atlan T, Cooper ZA, Arora R, Cogdill AP, Khan MAW, Ologun G, Bussi Y, Weinberger A, Lotan-Pompan M, Golani O, Perry G, Rokah M, Bahar-Shany K, Rozeman EA, Blank CU, Ronai A, Shaoul R, Amit A, Dorfman T, Kremer R, Cohen ZR, Harnof S, Siegal T, Yehuda-Shnaidman E, Gal-Yam EN, Shapira H, Baldini N, Langille MGI, Ben-Nun A, Kaufman B, Nissan A, Golan T, Dadiani M, Levanon K, Bar J, Yust-Katz S, Barshack I, Peeper DS, Raz DJ, Segal E, Wargo JA, Sandbank J, Shental N, Straussman R, Science 2020, 368, 973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].McDonald D, Hyde E, Debelius JW, Morton JT, Gonzalez A, Ackermann G, Aksenov AA, Behsaz B, Brennan C, Chen Y, DeRight Goldasich L, Dorrestein PC, Dunn RR, Fahimipour AK, Gaffney J, Gilbert JA, Gogul G, Green JL, Hugenholtz P, Humphrey G, Huttenhower C, Jackson MA, Janssen S, Jeste DV, Jiang L, Kelley ST, Knights D, Kosciolek T, Ladau J, Leach J, Marotz C, Meleshko D, Melnik AV, Metcalf JL, Mohimani H, Montassier E, Navas-Molina J, Nguyen TT, Peddada S, Pevzner P, Pollard KS, Rahnavard G, Robbins-Pianka A, Sangwan N, Shorenstein J, Smarr L, Song SJ, Spector T, Swafford AD, Thackray VG, Thompson LR, Tripathi A, Vázquez-Baeza Y, Vrbanac A, Wischmeyer P, Wolfe E, Zhu Q, American Gut Consortium, , Knight R, mSystems 2018, 3, DOI 10.1128/mSystems.00031-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Proctor LM, Creasy HH, Fettweis JM, Lloyd-Price J, Mahurkar A, Zhou W, Buck GA, Snyder MP, Strauss JF, Weinstock GM, White O, Huttenhower C, Nature 2019, 569, 641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Paun A, Danska JS, Pediatr. Diabetes 2016, 17, 469. [DOI] [PubMed] [Google Scholar]
- [6].Baumgart DC, Carding SR, Lancet 2007, 369, 1627. [DOI] [PubMed] [Google Scholar]
- [7].He Y, Wu W, Zheng H-M, Li P, McDonald D, Sheng H-F, Chen M-X, Chen Z-H, Ji G-Y, Zheng Z-D-X, Mujagond P, Chen X-J, Rong Z-H, Chen P, Lyu L-Y, Wang X, Wu C-B, Yu N, Xu Y-J, Yin J, Raes J, Knight R, Ma W-J, Zhou H-W, Nat. Med 2018, 24, 1532. [DOI] [PubMed] [Google Scholar]
- [8].Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, Magris M, Hidalgo G, Baldassano RN, Anokhin AP, Heath AC, Warner B, Reeder J, Kuczynski J, Caporaso JG, Lozupone CA, Lauber C, Clemente JC, Knights D, Knight R, Gordon JI, Nature 2012, 486, 222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Salava A, Aho V, Lybeck E, Pereira P, Paulin L, Nupponen I, Ranki A, Auvinen P, Andersson S, Lauerma A, Exp. Dermatol 2017, 26, 861. [DOI] [PubMed] [Google Scholar]
- [10].Ubeja RG, Bhat C, Int J Clin Pediatr Dent 2016, 9, 326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Adlerberth I, Lindberg E, Aberg N, Hesselmar B, Saalman R, Strannegård I-L, Wold AE, Pediatr. Res 2006, 59, 96. [DOI] [PubMed] [Google Scholar]
- [12].Dominguez-Bello MG, Costello EK, Contreras M, Magris M, Hidalgo G, Fierer N, Knight R, Proc. Natl. Acad. Sci. U. S. A 2010, 107, 11971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Grölund M-M, Lehtonen O-P, Eerola E, Kero P, J. Pediatr. Gastroenterol. Nutr 1999, 28, 19. [DOI] [PubMed] [Google Scholar]
- [14].Grönlund M-M, Grześkowiak Ł, Isolauri E, Salminen S, Gut Microbes 2011, 2, 227. [DOI] [PubMed] [Google Scholar]
- [15].Chu DM, Ma J, Prince AL, Antony KM, Seferovic MD, Aagaard KM, Nat. Med 2017, 23, 314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Asnicar F, Manara S, Zolfo M, Truong DT, Scholz M, Armanini F, Ferretti P, Gorfer V, Pedrotti A, Tett A, Segata N, mSystems 2017, 2, DOI 10.1128/mSystems.00164-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Ferretti P, Pasolli E, Tett A, Asnicar F, Gorfer V, Fedi S, Armanini F, Truong DT, Manara S, Zolfo M, Beghini F, Bertorelli R, De Sanctis V, Bariletti I, Canto R, Clementi R, Cologna M, Crifò T, Cusumano G, Gottardi S, Innamorati C, Masè C, Postai D, Savoi D, Duranti S, Lugli GA, Mancabelli L, Turroni F, Ferrario C, Milani C, Mangifesta M, Anzalone R, Viappiani A, Yassour M, Vlamakis H, Xavier R, Collado CM, Koren O, Tateo S, Soffiati M, Pedrotti A, Ventura M, Huttenhower C, Bork P, Segata N, Cell Host Microbe 2018, 24, 133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Hall MA, Cole CB, Smith SL, Fuller R, Rolles CJ, Arch. Dis. Child 1990, 65, 185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Penders J, Thijs C, Vink C, Stelma FF, Snijders B, Kummeling I, van den Brandt PA, Stobberingh EE, Pediatrics 2006, 118, 511. [DOI] [PubMed] [Google Scholar]
- [20].Kristensen K, Henriksen L, J. Allergy Clin. Immunol 2016, 137, 587. [DOI] [PubMed] [Google Scholar]
- [21].Mueller NT, Whyatt R, Hoepner L, Oberfield S, Dominguez-Bello MG, Widen EM, Hassoun A, Perera F, Rundle A, Int. J. Obes 2015, 39, 665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Walker WA, Nestle Nutr. Inst. Workshop Ser 2017, 88, 23. [DOI] [PubMed] [Google Scholar]
- [23].Shah P, Kaciroti N, Richards B, Oh W, Lumeng JC, Pediatrics 2016, 138, e20153496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Fouhy F, Watkins C, Hill CJ, O’Shea C-A, Nagle B, Dempsey EM, O’Toole PW, Ross RP, Ryan CA, Stanton C, Nat. Commun 2019, 10, 1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Francis AP, Dominguez-Bello MG, Trends Microbiol. 2019, 27, 567. [DOI] [PubMed] [Google Scholar]
- [26].Chernikova DA, Madan JC, Housman ML, Zain-Ul-Abideen M, Lundgren SN, Morrison HG, Sogin ML, Williams SM, Moore JH, Karagas MR, Hoen AG, Pediatr. Res 2018, 84, 71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Puopolo KM, Benitz WE, Zaoutis TE, COMMITTEE ON FETUS AND NEWBORN, COMMITTEE ON INFECTIOUS DISEASES, Pediatrics 2018, 142, DOI 10.1542/peds.2018-2896. [DOI] [Google Scholar]
- [28].Salvatore S, Baldassarre ME, Di Mauro A, Laforgia N, Tafuri S, Bianchi FP, Dattoli E, Morando L, Pensabene L, Meneghin F, Dilillo D, Mancini V, Talarico V, Tandoi F, Zuccotti G, Agosti M, J. Pediatr 2019, 212, 44. [DOI] [PubMed] [Google Scholar]
- [29].Demoré B, Le Govic D, Thilly N, Boivin J-M, Pulcini C, Clin. Microbiol. Infect 2017, 23, 486.e7. [DOI] [PubMed] [Google Scholar]
- [30].Zhao Y, Zhou Y, Zhu Q, Xia B, Ma W, Xiao X, Shi H, Zhang Y, Environ. Int 2019, 123, 70. [DOI] [PubMed] [Google Scholar]
- [31].Dunn AB, Jordan S, Baker BJ, Carlson NS, MCN Am. J. Matern. Child Nurs 2017, 42, 318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Hermansson H, Kumar H, Collado MC, Salminen S, Isolauri E, Rautava S, Front Nutr 2019, 6, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Parra-Llorca A, Gormaz M, Alcántara C, Cernada M, Nuñez-Ramiro A, Vento M, Collado MC, Front. Microbiol 2018, 9, 1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Perrone S, Longini M, Zollino I, Bazzini F, Tassini M, Vivi A, Bracciali C, Calderisi M, Buonocore G, Nutrition 2019, 62, 158. [DOI] [PubMed] [Google Scholar]
- [35].Andreas NJ, Kampmann B, Mehring Le-Doare K, Early Hum. Dev 2015, 91, 629. [DOI] [PubMed] [Google Scholar]
- [36].Asbury MR, Butcher J, Copeland JK, Unger S, Bando N, Comelli EM, Forte V, Kiss A, LeMay-Nedjelski L, Sherman PM, Stintzi A, Tomlinson C, Wang PW, O’Connor DL, Cell Host Microbe 2020, 28, 669. [DOI] [PubMed] [Google Scholar]
- [37].Bode L, Glycobiology 2012, 22, 1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Sela DA, Chapman J, Adeuya A, Kim JH, Chen F, Whitehead TR, Lapidus A, Rokhsar DS, Lebrilla CB, German JB, Price NP, Richardson PM, Mills DA, Proc. Natl. Acad. Sci. U. S. A 2008, 105, 18964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Martin CR, Ling P-R, Blackburn GL, Nutrients 2016, 8, DOI 10.3390/nu8050279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Moossavi S, Sepehri S, Robertson B, Bode L, Goruk S, Field CJ, Lix LM, de Souza RJ, Becker AB, Mandhane PJ, Turvey SE, Subbarao P, Moraes TJ, Lefebvre DL, Sears MR, Khafipour E, Azad MB, Cell Host Microbe 2019, 25, 324. [DOI] [PubMed] [Google Scholar]
- [41].Cai C, Zhang Z, Morales M, Wang Y, Khafipour E, Friel J, Free Radic. Biol. Med 2019, 142, 146. [DOI] [PubMed] [Google Scholar]
- [42].Shen RL, Thymann T, Østergaard MV, Støy AC, Krych Ł, Nielsen DS, Lauridsen C, Hartmann B, Holst JJ, Burrin DG, Sangild PT, Am. J. Physiol. Gastrointest. Liver Physiol 2015, 309, G310. [DOI] [PubMed] [Google Scholar]
- [43].Rossiter MD, Colapinto CK, Khan MKA, McIsaac J-LD, Williams PL, Kirk SFL, Veugelers PJ, Matern. Child Health J 2015, 19, 2048. [DOI] [PubMed] [Google Scholar]
- [44].Li B, Wu RY, Horne RG, Ahmed A, Lee D, Robinson SC, Zhu H, Lee C, Cadete M, Johnson-Henry KC, Landberg E, Alganabi M, Abrahamsson T, Delgado-Olguin P, Pierro A, Sherman PM, Mol. Nutr. Food Res 2020, 64, e2000519. [DOI] [PubMed] [Google Scholar]
- [45].Koenig JE, Spor A, Scalfone N, Fricker AD, Stombaugh J, Knight R, Angenent LT, Ley RE, Proc. Natl. Acad. Sci. U. S. A 2011, 108 Suppl 1, 4578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].de Muinck EJ, Trosvik P, Nat. Commun 2018, 9, 2233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Younge NE, Newgard CB, Cotten CM, Goldberg RN, Muehlbauer MJ, Bain JR, Stevens RD, O’Connell TM, Rawls JF, Seed PC, Ashley PL, Sci. Rep 2019, 9, 8167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Stewart CJ, Embleton ND, Marrs ECL, Smith DP, Fofanova T, Nelson A, Skeath T, Perry JD, Petrosino JF, Berrington JE, Cummings SP, Microbiome 2017, 5, 75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Hill CJ, Lynch DB, Murphy K, Ulaszewska M, Jeffery IB, O’Shea CA, Watkins C, Dempsey E, Mattivi F, Tuohy K, Paul Ross R, Anthony Ryan C, O’Toole PW, Stanton C, Microbiome 2017, 5, DOI 10.1186/s40168-017-0240-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Yoshioka H, Fujita K, Sakata H, Murono K, Iseki K-I, Bifidobact. Microflora 1991, 10, 11. [Google Scholar]
- [51].Fanaroff AA, in Neonatology: A Practical Approach to Neonatal Diseases (Eds.: Buonocore G, Bracci R, Weindling M), Springer Milan, Milano, 2012, pp. 94–101. [Google Scholar]
- [52].Wandro S, Osborne S, Enriquez C, Bixby C, Arrieta A, Whiteson K, mSphere 2018, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Sakata H, Yoshioka H, Fujita K, Eur. J. Pediatr 1985, 144, 186. [DOI] [PubMed] [Google Scholar]
- [54].Costello EK, Carlisle EM, Bik EM, Morowitz MJ, Relman DA, MBio 2013, 4, e00782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Falony G, Joossens M, Vieira-Silva S, Wang J, Darzi Y, Faust K, Kurilshikov A, Bonder MJ, Valles-Colomer M, Vandeputte D, Tito RY, Chaffron S, Rymenans L, Verspecht C, De Sutter L, Lima-Mendez G, D’hoe K, Jonckheere K, Homola D, Garcia R, Tigchelaar EF, Eeckhaudt L, Fu J, Henckaerts L, Zhernakova A, Wijmenga C, Raes J, Science 2016, 352, 560. [DOI] [PubMed] [Google Scholar]
- [56].Martino C, Morton JT, Marotz CA, Thompson LR, Tripathi A, Knight R, Zengler K, mSystems 2019, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Bokulich NA, Chung J, Battaglia T, Henderson N, Jay M, Li H, Lieber AD, Wu F, Perez-Perez GI, Chen Y, Schweizer W, Zheng X, Contreras M, Dominguez-Bello MG, Blaser MJ, Science Translational Medicine 2016, 8, 343ra82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Gauglitz JM, Morton JT, Tripathi A, Hansen S, Gaffney M, Carpenter C, Weldon KC, Shah R, Parampil A, Fidgett AL, Swafford AD, Knight R, Dorrestein PC, mSystems 2020, 5, DOI 10.1128/mSystems.00635-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Bisanz JE, Enos MK, PrayGod G, Seney S, Macklaim JM, Chilton S, Willner D, Knight R, Fusch C, Fusch G, Gloor GB, Burton JP, Reid G, Applied and Environmental Microbiology 2015, 81, 4965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].David LA, Materna AC, Friedman J, Campos-Baptista MI, Blackburn MC, Perrotta A, Erdman SE, Alm EJ, Genome Biol 2014, 15, R89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Clemente JC, Pehrsson EC, Blaser MJ, Sandhu K, Gao Z, Wang B, Magris M, Hidalgo G, Contreras M, Noya-Alarcón Ó, Lander O, McDonald J, Cox M, Walter J, Oh PL, Ruiz JF, Rodriguez S, Shen N, Song SJ, Metcalf J, Knight R, Dantas G, Dominguez-Bello MG, Sci Adv 2015, 1, DOI 10.1126/sciadv.1500183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Caporaso JG, Lauber CL, Costello EK, Berg-Lyons D, Gonzalez A, Stombaugh J, Knights D, Gajer P, Ravel J, Fierer N, Gordon JI, Knight R, Genome Biol 2011, 12, R50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Dominguez-Bello MG, De Jesus-Laboy KM, Shen N, Cox LM, Amir A, Gonzalez A, Bokulich NA, Song SJ, Hoashi M, Rivera-Vinas JI, Mendez K, Knight R, Clemente JC, Nat. Med 2016, 22, 250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Morton JT, Marotz C, Washburne A, Silverman J, Zaramela LS, Edlund A, Zengler K, Knight R, Nat. Commun 2019, 10, 2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Rougé C, Goldenberg O, Ferraris L, Berger B, Rochat F, Legrand A, Göbel UB, Vodovar M, Voyer M, Rozé J-C, Darmaun D, Piloquet H, Butel M-J, de La Cochetière M-F, Anaerobe 2010, 16, 362. [DOI] [PubMed] [Google Scholar]
- [66].Gonzalez A, Navas-Molina JA, Kosciolek T, McDonald D, Vázquez-Baeza Y, Ackermann G, DeReus J, Janssen S, Swafford AD, Orchanian SB, Sanders JG, Shorenstein J, Holste H, Petrus S, Robbins-Pianka A, Brislawn CJ, Wang M, Rideout JR, Bolyen E, Dillon M, Caporaso JG, Dorrestein PC, Knight R, Nat. Methods 2018, 15, 796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Wang M, Carver JJ, Phelan VV, Sanchez LM, Garg N, Peng Y, Nguyen DD, Watrous J, Kapono CA, Luzzatto-Knaan T, Porto C, Bouslimani A, Melnik AV, Meehan MJ, Liu W-T, Crüsemann M, Boudreau PD, Esquenazi E, Sandoval-Calderón M, Kersten RD, Pace LA, Quinn RA, Duncan KR, Hsu C-C, Floros DJ, Gavilan RG, Kleigrewe K, Northen T, Dutton RJ, Parrot D, Carlson EE, Aigle B, Michelsen CF, Jelsbak L, Sohlenkamp C, Pevzner P, Edlund A, McLean J, Piel J, Murphy BT, Gerwick L, Liaw C-C, Yang Y-L, Humpf H-U, Maansson M, Keyzers RA, Sims AC, Johnson AR, Sidebottom AM, Sedio BE, Klitgaard A, Larson CB, C. A. B. P, Torres-Mendoza D, Gonzalez DJ, Silva DB, Marques LM, Demarque DP, Pociute E, O’Neill EC, Briand E, Helfrich EJN, Granatosky EA, Glukhov E, Ryffel F, Houson H, Mohimani H, Kharbush JJ, Zeng Y, Vorholt JA, Kurita KL, Charusanti P, McPhail KL, Nielsen KF, Vuong L, Elfeki M, Traxler MF, Engene N, Koyama N, Vining OB, Baric R, Silva RR, Mascuch SJ, Tomasi S, Jenkins S, Macherla V, Hoffman T, Agarwal V, Williams PG, Dai J, Neupane R, Gurr J, Rodríguez AMC, Lamsa A, Zhang C, Dorrestein K, Duggan BM, Almaliti J, Allard P-M, Phapale P, Nothias L-F, Alexandrov T, Litaudon M, Wolfender J-L, Kyle JE, Metz TO, Peryea T, Nguyen D-T, VanLeer D, Shinn P, Jadhav A, Müller R, Waters KM, Shi W, Liu X, Zhang L, Knight R, Jensen PR, Palsson BO, Pogliano K, Linington RG, Gutiérrez M, Lopes NP, Gerwick WH, Moore BS, Dorrestein PC, Bandeira N, Nat. Biotechnol 2016, 34, 828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Mueller NT, Dominguez-Bello MG, Appel LJ, Hourigan SK, BJOG 2020, 127, 301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Milstone AM, Voskertchian A, Koontz DW, Khamash DF, Ross T, Aucott SW, Gilmore MM, Cosgrove SE, Carroll KC, Colantuoni E, JAMA 2019, DOI 10.1001/jama.2019.20785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Barbosa-Cesnik C, Schwartz K, Foxman B, JAMA 2003, 289, 1609. [DOI] [PubMed] [Google Scholar]
- [71].Avila WM, Pordeus IA, Paiva SM, Martins CC, PLoS One 2015, 10, e0142922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Kriswandini IL, Tantiana DI, N. P, B. T, P. Ia, Pnbn T, Infect. Dis. Rep 2020, 12, 8722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Oettinger-Barak O, Dashper SG, Catmull DV, Adams GG, Sela MN, Machtei EE, Reynolds EC, J. Oral Microbiol 2013, 5, DOI 10.3402/jom.v5i0.20320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Bhat KG, Khot P, Patil S, Pattar G, Majukar S, J. Oral Maxillofac. Pathol 2019, 23, 231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Åberg CH, Kelk P, Johansson A, Virulence 2015, 6, 188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Bittinger K, Zhao C, Li Y, Ford E, Friedman ES, Ni J, Kulkarni CV, Cai J, Tian Y, Liu Q, Patterson AD, Sarkar D, Chan SHJ, Maranas C, Saha-Shah A, Lund P, Garcia BA, Mattei LM, Gerber JS, Elovitz MA, Kelly A, DeRusso P, Kim D, Hofstaedter CE, Goulian M, Li H, Bushman FD, Zemel BS, Wu GD, Nat Microbiol 2020, 5, 838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Quinn RA, Melnik AV, Vrbanac A, Fu T, Patras KA, Christy MP, Bodai Z, Belda-Ferre P, Tripathi A, Chung LK, Downes M, Welch RD, Quinn M, Humphrey G, Panitchpakdi M, Weldon KC, Aksenov A, da Silva R, Avila-Pacheco J, Clish C, Bae S, Mallick H, Franzosa EA, Lloyd-Price J, Bussell R, Thron T, Nelson AT, Wang M, Leszczynski E, Vargas F, Gauglitz JM, Meehan MJ, Gentry E, Arthur TD, Komor AC, Poulsen O, Boland BS, Chang JT, Sandborn WJ, Lim M, Garg N, Lumeng JC, Xavier RJ, Kazmierczak BI, Jain R, Egan M, Rhee KE, Ferguson D, Raffatellu M, Vlamakis H, Haddad GG, Siegel D, Huttenhower C, Mazmanian SK, Evans RM, Nizet V, Knight R, Dorrestein PC, Nature 2020, 579, 123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Jarmusch AK, Wang M, Aceves CM, Advani RS, Aguire S, Aksenov AA, Aleti G, Aron AT, Bauermeister A, Bolleddu S, Others, bioRxiv 2019. [Google Scholar]
- [79].Wang M, Jarmusch AK, Vargas F, Aksenov AA, Gauglitz JM, Weldon K, Petras D, da Silva R, Quinn R, Melnik AV, van der Hooft JJJ, Caraballo-Rodrguez AM, Nothias LF, Aceves CM, Panitchpakdi M, Brown E, Di Ottavio F, Sikora N, Elijah EO, Labarta-Bajo L, Gentry EC, Shalapour S, Kyle KE, Puckett SP, Watrous JD, Carpenter CS, Bouslimani A, Ernst M, Swafford AD, Zúñiga EI, Balunas MJ, Klassen JL, Loomba R, Knight R, Bandeira N, Dorrestein PC, Nat. Biotechnol 2020, 38, 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Thompson LR, The Earth Microbiome Project Consortium, Sanders JG, McDonald D, Amir A, Ladau J, Locey KJ, Prill RJ, Tripathi A, Gibbons SM, Ackermann G, Navas-Molina JA, Janssen S, Kopylova E, Vázquez-Baeza Y, González A, Morton JT, Mirarab S, Xu ZZ, Jiang L, Haroon MF, Kanbar J, Zhu Q, Song SJ, Kosciolek T, Bokulich NA, Lefler J, Brislawn CJ, Humphrey G, Owens SM, Hampton-Marcell J, Berg-Lyons D, McKenzie V, Fierer N, Fuhrman JA, Clauset A, Stevens RL, Shade A, Pollard KS, Goodwin KD, Jansson JK, Gilbert JA, Knight R, Nature 2017, 551, 457.29088705 [Google Scholar]
- [81].Walters W, Hyde ER, Berg-Lyons D, Ackermann G, Humphrey G, Parada A, Gilbert JA, Jansson JK, Gregory Caporaso J, Fuhrman JA, Apprill A, Knight R, mSystems 2016, 1, DOI 10.1128/msystems.00009-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Amir A, McDonald D, Navas-Molina JA, Kopylova E, Morton JT, Zech Xu Z, Kightley EP, Thompson LR, Hyde ER, Gonzalez A, Knight R, mSystems 2017, 2, DOI 10.1128/mSystems.00191-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, Huber T, Dalevi D, Hu P, Andersen GL, Appl. Environ. Microbiol 2006, 72, 5069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Bolyen E, Rideout JR, Dillon MR, Bokulich NA, Abnet CC, Al-Ghalith GA, Alexander H, Alm EJ, Arumugam M, Asnicar F, Bai Y, Bisanz JE, Bittinger K, Brejnrod A, Brislawn CJ, Brown CT, Callahan BJ, Caraballo-Rodríguez AM, Chase J, Cope EK, Da Silva R, Diener C, Dorrestein PC, Douglas GM, Durall DM, Duvallet C, Edwardson CF, Ernst M, Estaki M, Fouquier J, Gauglitz JM, Gibbons SM, Gibson DL, Gonzalez A, Gorlick K, Guo J, Hillmann B, Holmes S, Holste H, Huttenhower C, Huttley GA, Janssen S, Jarmusch AK, Jiang L, Kaehler BD, Kang KB, Keefe CR, Keim P, Kelley ST, Knights D, Koester I, Kosciolek T, Kreps J, Langille MGI, Lee J, Ley R, Liu Y-X, Loftfield E, Lozupone C, Maher M, Marotz C, Martin BD, McDonald D, McIver LJ, Melnik AV, Metcalf JL, Morgan SC, Morton JT, Naimey AT, Navas-Molina JA, Nothias LF, Orchanian SB, Pearson T, Peoples SL, Petras D, Preuss ML, Pruesse E, Rasmussen LB, Rivers A, Robeson MS 2nd, Rosenthal P, Segata N, Shaffer M, Shiffer A, Sinha R, Song SJ, Spear JR, Swafford AD, Thompson LR, Torres PJ, Trinh P, Tripathi A, Turnbaugh PJ, Ul-Hasan S, van der Hooft JJJ, Vargas F, Vázquez-Baeza Y, Vogtmann E, von Hippel M, Walters W, Wan Y, Wang M, Warren J, Weber KC, Williamson CHD, Willis AD, Xu ZZ, Zaneveld JR, Zhang Y, Zhu Q, Knight R, Caporaso JG, Nat. Biotechnol 2019, 37, 852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Mirarab S, Nguyen N, Warnow T, Pac. Symp. Biocomput 2012, 247. [DOI] [PubMed] [Google Scholar]
- [86].Pluskal T, Castillo S, Villar-Briones A, Oresic M, BMC Bioinformatics 2010, 11, 395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Nothias L-F, Petras D, Schmid R, Dührkop K, Rainer J, Sarvepalli A, Protsyuk I, Ernst M, Tsugawa H, Fleischauer M, Aicheler F, Aksenov AA, Alka O, Allard P-M, Barsch A, Cachet X, Caraballo-Rodriguez AM, Da Silva RR, Dang T, Garg N, Gauglitz JM, Gurevich A, Isaac G, Jarmusch AK, Kameník Z, Kang KB, Kessler N, Koester I, Korf A, Le Gouellec A, Ludwig M, Martin H C, McCall L-I, McSayles J, Meyer SW, Mohimani H, Morsy M, Moyne O, Neumann S, Neuweger H, Nguyen NH, Nothias-Esposito M, Paolini J, Phelan VV, Pluskal T, Quinn RA, Rogers S, Shrestha B, Tripathi A, van der Hooft JJJ, Vargas F, Weldon KC, Witting M, Yang H, Zhang Z, Zubeil F, Kohlbacher O, Böcker S, Alexandrov T, Bandeira N, Wang M, Dorrestein PC, Nat. Methods 2020, 17, 905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].MSI Board Members, Sansone S-A, Fan T, Goodacre R, Griffin JL, Hardy NW, Kaddurah-Daouk R, Kristal BS, Lindon J, Mendes P, Morrison N, Nikolau B, Robertson D, Sumner LW, Taylor C, van der Werf M, van Ommen B, Fiehn O, Nat. Biotechnol 2007, 25, 846. [DOI] [PubMed] [Google Scholar]
- [89].Shannon CE, The Bell System Technical Journal 1948, 27, 379. [Google Scholar]
- [90].Aitchison J, J. R. Stat. Soc. Series B Stat. Methodol 1982, 44, 139. [Google Scholar]
- [91].Aitchison J, Biometrika 1983, 70, 57. [Google Scholar]
- [92].Fedarko MW, Martino C, Morton JT, González A, Rahman G, Marotz CA, Minich JJ, Allen EE, Knight R, NAR Genom Bioinform 2020, 2, lqaa023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Knights D, Kuczynski J, Charlson ES, Zaneveld J, Mozer MC, Collman RG, Bushman FD, Knight R, Kelley ST, Nat. Methods 2011, 8, 761. [DOI] [PMC free article] [PubMed] [Google Scholar]


