Abstract
Objective
The National Institutes of Health (NIH) HEAL Initiative is making data findable, accessible, interoperable, and reusable (FAIR) to maximize the value of the unprecedented federal investment in pain and opioid-use disorder research. This involves standardizing the use of common data elements (CDE) for clinical research.
Methods
This work describes the process of the selection, processing, harmonization, and design constraints of CDE across a pain and opioid use disorder clinical trials network.
Results
The network alignment allowed for incorporation of newer data standards across the clinical trials. Specific advances included geographic coding (RUCA), deidentified patient identifiers (GUID), shareable clinical survey libraries (REDCap), and concept mapping to standardized concepts (UMLS).
Conclusions
While complex, harmonization across a network of chronic pain and opioid use disorder clinical trials with separate interventions can be optimized through use of CDEs and data standardization processes. This standardization process will support the robust secondary data analyses. Scaling this process could standardize CDE results across interventions or disease state which could help inform insurance companies or government organizations about coverage determinations. The development of the HEAL CDE program supports connecting isolated studies and solutions to each other, but the practical aspects may be challenging for some studies to implement. Leveraging tools and technology to simplify process and create ready to use resources may support wider adoption of consistent data standards.
Background
The National Institutes of Health (NIH) Helping to End Addiction Long-term® Initiative, or NIH HEAL Initiative®, is a crosscutting funding portfolio that supports more than 40 programs across the NIH with an investment of 2.5 billion dollars to date.1 HEAL is a trans-agency effort to speed scientific solutions to stem the national opioid public health crisis. Its goals are to increase the understanding and treatment of pain and improve the treatment and prevention of opioid use disorder and overdose. While the initiative has funded 1000 projects that are unique solutions working on different approaches to preventing and treating chronic pain and addiction, the NIH HEAL Initiative recognized the need to connect these projects to increase the impact and synergy of the programmatic interventions. The NIH HEAL Initiative recognized that making data findable, accessible, interoperable, and reusable (FAIR) would maximize the value of the unprecedented federal investment in pain and opioid-use disorder research.2 The overarching approach to accomplishing this ambitious goal reflects the inclusion of FAIR data principles and specifically through the use of common data elements (CDEs) for pain clinical trials and clinical studies (Figure 1).3
Figure 1.
NIH HEAL required chronic pain CDE.
Use of the FAIR principles can help ensure that data is disseminated to the appropriate communities and can facilitate the emergence of new findings from previously generated data. Therefore, as part of the NIH HEAL Initiative’s Public Access and Data Sharing Plan,4 clinical pain research funded by HEAL must collect a core group of patient-reported outcomes (PROs) for nine important pain domains.3,5 Core CDEs are defined as a minimal and defined set of pain domains and questionnaires that all HEAL pain clinical studies or trials are required to collect.3 Each pain domain has a prescribed questionnaire and accompanying common data elements. The HEAL CDE program also has supplemental CDEs which include a comprehensive set of domains and PROs used in individual studies but made available for others to facilitate uniform data collection for studies with shared domains outside of the core selected by HEAL investigators for use in their studies.6–15
In addition to being a new process, it has yet to be described how CDEs are implemented in a network with diverse studies. One of the newer NIH HEAL clinical trials networks, the Integrative Management of chronic Pain and OUD for Whole Recovery (IMPOWR) Network, has the novel charge of studying comorbid chronic pain and opioid use disorder across numerous sites and studies under the IMPOWR Dissemination Education And Coordination Center (IDEA-CC).16 Originally funded in September 2021, the network had four primary sites and nine large clinical studies along with the IDEA-CC and currently includes 11 studies across six sites. The goal of the IDEA-CC and the network was to connect these diverse approaches to assess the impact of combined treatment for chronic pain and opioid use disorder using unified variable selections (common data elements).
The IDEA-CC focus on harmonizing CDEs in the network moved beyond the minimum standards of the NIH HEAL Initiative’s required CDEs.16 The studies in the network center on the two-way impact of chronic pain and opioid use disorder on the whole patient and include individual factors such as mental health and social support and systemic domains such as stigma, health equity, and economic influences on health care. Furthermore, to increase the impact of the studies, CDEs were required to assess cost effectiveness, implementation barriers, and partner perspectives (people with lived experiences and public/private partners). Individually, the patient centered research of the network would improve care for people, but the IDEA-CC’s combination of these data at the variable level using FAIR principles, is anticipated to deliver greater impact and synergy of these solutions.
The network, as early adopters of the harmonized HEAL CDEs, has incorporated required CDEs, adapted existing surveys, and developed new CDE variables to reflect the projects' scientific and partner-driven goals.16 While there have been networks separately focused on pain and opioid use disorder, IDEA-CC, IMPOWR projects and CDE is one of the first combined projects, providing a unique perspective for the larger HEAL Data Ecosystem.4 Ultimately, the goal of these harmonized data is to provide meta-data and secondary data analyses to provide more dimensional and comprehensive understanding of prevention and treatment solutions. This work provides an overview of the implementation process, challenges, and design practices to support successful completion of the CDE harmonization process.
Methods
CDE selection process
While the model for many NIH and NIH HEAL Initiative clinical trials networks involves the design and implementation of a single study across numerous sites and patient populations, the design of this network inverts that approach. By allowing diverse sites and studies to implement variable level CDEs across 13 different domains, the results would be unified dataset across different populations and interventions (Figure 2). Informed by the IDEA-CC IMPOWR Network Kickoff Meeting (10/2021) and the proposed study approaches, the IMPOWR CDE working group (ICDEWG) harmonized along the proposed domains of pain, substance use, mood, sleep, and functioning that are required by HEAL (Figure 1).3,17 A best practice recommendation would be to start the licensing process for any potential CDE as early as possible as it can take 1 to 12 months.
Figure 2.
NIH HEAL IMPOWR required common data elements.
In addition to these, the network partners (researchers and people with lived experience) brought a focus on the need to assess trauma,17 social determinants of health,18 and stigma19 as cross-cutting themes for the network. The network also had an implementation focus,20 supported by the Implementation Science working group, developing a collection of patient-facing and provider-facing surveys. The Health Economics working group developed both core and supplemental studies to address cost effectiveness21 that were adapted from the NIH HEAL Justice Community Opioid Innovation Network (JCOIN) network’s CDEs.22–24 To further support integration and future applicability of studies, we developed surveys that reflected key pain diagnoses.25 The ICDEWG also adopted COVID-19 focused questions for both chronic pain and opioid use disorder as a reflection of current events during the study.26
Additionally, the ICDEWG met from November 2021 to February 2022 to establish the network's first combined pain and OUD CDE. Once the ICDEWG determined the required and supplemental CDE for the network projects, the IDEA-CC worked with the NIH HEAL CDE team to evaluate and process of the network specific CDEs. This team was focused on the alignment of the selected variables/surveys with existing case report forms (CRFs) in the NIH HEAL Initiative database. Of note, while the existing HEAL Initiative CRFs are readily accessible in the NIH CDE database stored in BOX,2,27 some CDEs require licensing for viewing and use which can be coordinated with the HEAL Initiative CDE team.2,4,5
HEAL initiative data ecosystem CDE processing
While many of the CDEs used by the studies may already have existing CRFs in the HEAL Initiative database, there may be customizations to existing surveys that would represent a change from existing variable assignment. For example, in the standard HEAL Initiative CDE demographics gender identity is represented through the options: male, female, unknown, and other. But the network, in conjunction with network partners, wanted to expand that to include a wider range of gender identity for participants to select. In the existing CRF, each answer has a unique variable identification to allow for granular analyses of responses. These variable names had to be generated by the IDEA-CC for each of the new possible choices. This process needed to be repeated for each unique question or survey that was added to the network REDCap library. For the network, this resulted in a library of approximately 800 variables. The IDEA-CC submitted the new variables to the HEAL Initiative CDE team for review and incorporation as a new CRF. When new validated surveys are being added to the HEAL Initiative Box, the HEAL Initiative CDE team also requests references for validation of the survey to include in the CRF development. The HEAL Initiative CDE team also does a review to make sure that the validated measure has not been altered, and that references, scoring instructions, and Spanish CRFs are available as applicable. Although the network did not use any copyrighted supplemental measures, many investigators within the HEAL Initiative are using copyrighted supplemental questionnaires. Since it can take many months to receive a copyrighted license from a copyright license holder (1–12 months), HEAL Initiative researchers are encouraged to order their copyrighted licenses as soon as they are funded and/or at the beginning of their planning year.
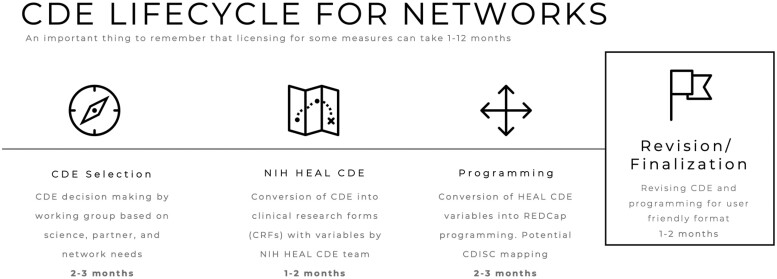
The standardized labeling of individual variables allows those variables to be compared across studies even when the whole variable set is not implemented in the study. The future benefit of labeling of each variable is that it will build the framework for cross-cutting analyses of variables as these data are combined in new ways to hopefully provide more impact for the existing work and secondary analysis. The challenge of this approach is that it introduces a rate limiting step between selection of the CDEs for a network/project and the programming of the variables into whatever format is being used for the study (Figure 3). This step can take from 1 to 3 months, depending on the workflow of the HEAL Initiative CDE group and timing of other studies having CDE processed.
Figure 3.
Lifecycle and timeline for common data element (CDE) network harmonization.
GUID and RUCA
Some of the newer factors incorporated into the network implementation of the CDE process by the IDEA-CC are the NIH unique patient identifier (GUID) and geographic (RUCA) coding.28–30 Given the complexity of the network’s numerous studies and working goal of having a unified data storage and platform for the network data, we adopted the evolving data standards that would help support participant privacy with data sharing (Figure 4).
Figure 4.
Definitions of Global Unique Identifier (GUID) and Rural Urban Coding (RUCA).
Incorporating a unique standardized patient identifier, GUID, solves several data sharing issues and allows for data storage (eg, NIMH NDA) in repositories that require a standardized identifier. The network’s front-end planning for the use of GUID allowed for the language to be incorporated both in patient consent and data sharing agreements, minimizing later revisions and back-end work. The personal health information collected to generate the GUID is kept locally at the study and will not be stored as part of the cloud computing platform. However, the use of GUID identifier should allow for the combination of de-identified participant data across projects in a larger data ecosystem. The use of the RUCA code allowed for the inclusion of understanding about the rural and urban distribution of patients while protecting individual participant location privacy.30 This innovation was welcomed by our research teams and partners to meet the scientific goals of the project balanced with patient privacy concerns. In addition, a HEAL Initiative Data Ecosystem team developed a tool to generate the RUCA codes for participants that is available from the HEAL Initiative CDE group.
HEAL initiative, CDE limitations, and design considerations
Selecting, processing, and programming CDE to meet scientific, administrative, and participant design constraints is a challenging and lengthy process. Our team’s approach was to address as many design challenges as possible in the front end for both efficiency and simplification. As a shared CDE library across a network that evolves as additional grants are funded, minor changes later could have significant impact. While it was never going to be perfect, our goal was to get as many details addressed upfront rather than after completion of the data acquisition or back-end programming, which would complicate the data storage process. Various studies are being implemented across the HEAL Initiative ecosystem to meet the HEAL Initiative CDE requirements for the use of standardized variable names. We decided to incorporate these names into the programming of REDCap to eliminate the need for postprocessing or back-end programming. In implementing this approach, we discovered challenges. The first issue is that by using these variable names, our studies were unable to use the PROMIS measures' psychometrically validated computer adaptive testing (CAT) versions.31 Our investigators were concerned about the additional patient burden associated with the longer version of the studies but were not able to bypass the requirement for variable level fidelity. Ultimately, our design focus was on meeting the scientific needs of the network while including the HEAL Initiative variable names in as user-friendly surveys as possible. Following the programming of the network CDE library into REDCap, there was two-month period of evaluation of content, question wording, and REDCap format editing while the study teams evaluated and gave feedback on the library. The IDEA-CC and HEAL Initiative CDE collaboration allowed for bidirectional updates as our team edited CRFs submitted to the HEAL Initiative CDE team or found problems in existing CRFs. Approximately 26% of the library underwent shifts during this period that needed to be updated by the HEAL Initiative CDE team.
REDCap: Strengths and limitations
A major source of debate across our investigative teams was the use of REDCap versus Qualtrics at the time of this discussion, Qualtrics has a user-friendly and more responsive user interface and would have been the obvious choice if these were the primary issues. We chose REDCap for several key reasons.32 Metadata is the information about the data collected and therefore essential for reusing and interpreting data.2,4 The HEAL Initiative Data ecosystem is currently implementing study level metadata and is using the CDE program as the first step of implementing variable level metadata.4 The REDCap platform includes and collects variable level metadata, which was the deciding factor. Qualtrics is unable to do so, ruling it out as a data collection method. Another advantage of using REDCap for our research team was the ability to deploy our own REDCap application programming interface (API) tools. API tools are a set of tools, or computer protocols, that allow two systems to communicate (eg, Wake Forest REDCap to “talk” to the REDCap at a different institution. Wake Forest developed tools that can directly pull the data collected in the study site’s REDCap into our data system for processing and eventual capture in a cloud computing data ecosystem. This eliminates the need for manual data entry and/or manual data transfer that can provide challenges for study teams. The second tool enabled by using REDCap is a CDE checking mechanism that can detect and report deviation from a study protocol (eg, changes in question language or variable components). This adds a layer of automated continuity verification and logging of changes or alterations for a study codebook.
CDISC and mapping to larger data frameworks
An additional component we are building with the HEAL Data Ecosystem is variable level CDE connections to larger data frameworks. As our team connects clinical data from studies, the ability to connect to existing data from claims, public health, and other sources will allow us to maximize the impact of our work. The network data will have numerous pathways to connect with other data sources. This network is using the standardized variable names which would allow connection with other studies and research networks (eg, JCOIN) using these data standards. The HEAL CDE team started to map some of the CDEs to Clinical Data Interchange Standards Consortium (CDISC) codes.33,34 The IDEA-CC brings together different interventions to address combined chronic pain and opioid use disorder, which overlaps with numerous other studies with a primary pain or primary opioid use disorder focus. Additionally, our network has a focus on cost effectiveness and is partnered with the Centers for Medicare and Medicaid Services (CMS) and commercial insurance groups. To translate our findings to have maximum impact on implementation, we wanted our data to be flexible and harmonizable with other sources. To accomplish this goal, we mapped the entire network library of CDEs to the Unified Medical Language System (UMLS) concept unique identifier (CUI) codes.35 This mapping conceptually links our data to other coding systems used in electronic health record data, claims, and insurance data, and builds the framework to connect to larger common data models.36,37 This will allow us to connect our findings to both other clinical studies as well as connect these data with other claims data sources to amplify impact.
CDE and cloud computing
One of the unique roles of the IDEA-CC is building a data dashboard for the storage of network related data. Our original plan included the use of the Liferay platform38 but transitioned to the Gen3 platform to harmonize with other NIH HEAL Initiative programs. The University of Chicago data team’s work through the HEAL Data Ecosystem has built a cloud computing data framework on the Gen3 platform that allows for visualization of the data generated by JCOIN. This team has extended this Gen3 cloud computing work for the HEAL Data Ecosystem, and currently the platform is being used to start collecting study level meta-data for HEAL projects.4 Our implementation of this platform is centered on providing study investigators with access to a dashboard view of their CDE’s, as well as the ability to combine data for larger evaluations of study themes such as stigma, health equity, or economic impact across projects. The use of CDE with standardized HEAL Initiative variable names will allow for simpler connections between these data and the ability to enhance secondary data analyses. The IDEA-CC team worked with the HEAL Initiative CDE team to develop and adjust these variable names. While this will necessitate additional work by the network in terms of data use agreements and patient consents, it has resulted in a one-of-a-kind repository and data set.
Discussion
As scientists and policy makers look for solutions and systems to improve our understanding and treatment of pain as well as the prevention and treatment of addiction, we need cohesive approaches to care built on the best possible science. While the NIH is looking at this problem from numerous perspectives, connecting these studies may allow for faster development of solutions. The development of the HEAL common data element (CDE) program is the next iterative step in connecting isolated studies and solutions to each other. The most established roadmap for this process is the NIH National Cancer Institute’s Metadata Services for Cancer, which includes CDEs,39 standards registry and repository (caDSR), standard collection CDEs, and template forms.40 The current state of standardized pain and addiction data does not have the legacy development of widely adopted terminologies. However, through the HEAL CDE program, the pain and addiction clinical domains can develop FAIR data and infrastructure that eases adoption of these CDEs.
The experience of the IDEA-CC reflects an early adoption of the HEAL CDE process and will provide a template for the adoption of the HEAL CDEs by sharing the REDCap programming for the HEAL CDE used in the network. While REDCap can be a less user-friendly interface than other commercially available platforms such as Qualtrics, there are programming approaches that significantly ease REDCap use for the study participant. For example, the network CDEs included the Michigan Body Map (MBM).41 This implementation involved significant programming and score calculation by the IDEA-CC, but on the participant side, this allows for a simplified process where patients only need to click on their affected body parts.42 On the analytic side, our REDCap will allow automated generation of the Michigan Body Map Score, Widespread pain region scores, and the Widespread pain index (WPI) score.43 This modification does not impact the study participant, but creates connections between data generated by these measures and studies using the Michigan Body Map, chronic overlapping pain condition regions, or WPI. We plan to share the programming done by the IDEA-CC to allow ease of use and improve data harmonization across studies.
Prior to the HEAL standardization, the wider pain and addiction research community has not adopted uniform standards for data collection. As the opioid and pain crises continue to worsen, new approaches that leverage the collective findings of the HEAL Initiative portfolio will allow for better understanding of how to prevent and treat pain and addiction more effectively. While the burdens of data standardization are newer to our research community, the IDEA-CC investigators recommend the front-end adoption of FAIR principles and HEAL Initiative CDE variable names to simplify the post processing work on projects and allow for faster dissemination of results.
Future directions
While the core domains, questionnaires, and CDEs have been finalized, work continues to expand the scope of the available supplemental questionnaires and CDEs, and to make them all accessible to the pain management community. NIH staff, working with the HEAL Initiative Data Coordinating Center at the University of Utah’s Trial Innovation Center, are preparing the HEAL Initiative CDEs to be included in a broader CDE repository managed by the National Library of Medicine (NLM).44 Simultaneously, HEAL is planning for a website that will be available to the public to search for the questionnaires and CDEs that have been used by HEAL-funded clinical pain research studies.
As HEAL programs launch and expand, more studies are entering the HEAL Data Ecosystem, and many of these studies are measuring outcomes that others have not yet investigated. While HEAL is currently collaborating with all study teams to ensure that their supplemental questionnaires are incorporated into the HEAL CDEs, the HEAL CDE program is also developing an automated ticketing system to track the questionnaires that the new studies intend to use and process new supplemental measures. A more systematic process would make it easier to understand which questionnaires HEAL investigators are using throughout the HEAL clinical pain research program; submit new questionnaires, scoring instructions, and references; and track measures, their copyright status, and any other elements that HEAL investigators must submit or request. The HEAL CDE program plans to share at least the Core CDE REDCap data dictionaries so that current and future investigators working with the same questionnaires can program their data collection in a way that will make it ready for a seamless submission to the HEAL Data Ecosystem.
Policy and public health implications
The NIH HEAL Initiative decided it was important to establish comprehensive core outcome sets for chronic and acute pain that all HEAL-funded pain researchers could use within their studies. The value of standardizing CDEs has numerous secondary applications and implications for policy and public health. Specifically, this type of standardization could help promote secondary data analyses across studies and networks, which could allow the pain research community to access quality and meaningful data across pain conditions, in diverse populations, and multiple interventions. Similarly, the secondary data analyses could facilitate cross-study comparisons, improve interpretability of findings from patient-reported outcome measures, and will allow for the comparability of results across trials and quantify the impact of interventions. Being able to standardize CDE results across interventions or disease states could help inform insurance companies or government organizations about coverage determinations. The use of core CDEs may also encourage clinical pain studies outside of HEAL, or clinicians to adopt the core CDEs in research and clinical practice. By taking the additional time and team effort, coding upfront makes it easier to deposit data into the HEAL Data Ecosystem, which may make it easier to translate research results into clinical practice and shorten the research to clinical practice timeline. This will likely reduce the delay in applying research innovations to clinical practice, which will have practical implications for people living with pain. In summary, the goal of the HEAL CDE program is to enhance and connect the work of existing and new studies to accelerate the development of cohesive solutions for the prevention and treatment of pain and addiction.
Supplementary Material
Acknowledgements
Authors within the ICDEWG consist of: Adams, Meredith C.B.; Arnsten, Julia; Bao, Yuhua; Barry, Declan; Becker, William C.; Fiellin, David; Fox, Aaron (COI: Royalties from Wolters Kluwer); Ghiroli, Megan; Hanmer, Janel; Horn, Brady; Hurlocker, Margo; Jalal, Hawre; Joseph, Verlin; Merlin, Jessica (COI: Research Grant from Cambia Health Foundation); Murray-Krezan, Christina; Pearson, Matthew; Rogal, Shari; Starrels, Joanna (COI: Research funding from FDA, royalties from Wolters); Bachrach, Rachel; Witkiewitz, Katie; Vasquez, Angel. Site affiliation is listed in the supplementary table.
Conflicts of interest: There are no conflicts of interest related to this work to report.
Contributor Information
Meredith C B Adams, Departments of Anesthesiology, Biomedical Informatics, and Public Health Sciences, Wake Forest University School of Medicine, Medical Center Boulevard, Winston-Salem, NC 27157, United States.
Robert W Hurley, Departments of Anesthesiology, Translational Neuroscience, and Public Health Sciences, Wake Forest University School of Medicine, Winston-Salem, NC 27157, United States.
Andrew Siddons, National Institute of Neurological Disorders and Stroke, Bethesda, MD, United States.
Umit Topaloglu, Department of Cancer Biology, Wake Forest University School of Medicine, Winston-Salem, NC 27157, United States.
Laura D Wandner, National Institute of Neurological Disorders and Stroke, Bethesda, MD, United States.
ICDEWG:
Meredith C B Adams, Julia Arnsten, Yuhua Bao, Declan Barry, William C Becker, David Fiellin, Aaron Fox, Megan Ghiroli, Janel Hanmer, Brady Horn, Margo Hurlocker, Hawre Jalal, Verlin Joseph, Jessica Merlin, Christina Murray-Krezan, Matthew Pearson, Shari Rogal, Joanna Starrels, Rachel Bachrach, Katie Witkiewitz, and Angel Vasquez
Funding
Research reported in this publication was supported by the NIH HEAL Initiative through the National Institute of Biomedical Imaging and Bioengineering of the National Institutes of Health under grant number K08EB022631 (MCBA) and the National Institute of Drug Abuse under grant number R24DA055306 (MCBA, RWH), R24DA055306-01S1 (MCBA). Additionally, the ICDEWG was supported by the NIH HEAL Initiative under awards numbered RM1DA055301, RM1DA055310, RM1DA055311 and RM1DA055437.
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. The contents of this work do not represent the views of the Department of Veterans Affairs, Department of Defense, or the U.S. Government.
References
- 1. NIH HEAL Initiative. Accessed January 28, 2023. https://heal.nih.gov/about.
- 2. HEAL Data FAIR Principles. Accessed January 28, 2023. https://www.healdatafair.org/.
- 3. Wandner LD, Domenichiello AF, Beierlein J, et al. NIH's helping to end addiction long-term(SM) initiative (NIH HEAL Initiative) clinical pain management common data element program. J Pain. 2022;23(3):370–378. [DOI] [PubMed] [Google Scholar]
- 4. HEAL Data Ecosystem. Accessed January 28, 2023. https://heal.nih.gov/data/heal-data-ecosystem.
- 5. HEAL Common Data Elements. Accessed January 28, 2023. https://heal.nih.gov/data/common-data-elements.
- 6. Cook KF, Dunn W, Griffith JW, et al. Pain assessment using the NIH toolbox. Neurology. 2013;80(11 suppl 3):S49–S53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Amtmann D, Cook KF, Jensen MP, et al. Development of a PROMIS item bank to measure pain interference. Pain. 2010;150(1):173–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Pogatzki-Zahn EM, Liedgens H, Hummelshoj L, et al. ; Panel IM-PPc. Developing consensus on core outcome domains for assessing effectiveness in perioperative pain management: results of the PROMPT/IMI-PainCare Delphi Meeting. Pain. 2021;162(11):2717–2736. [DOI] [PubMed] [Google Scholar]
- 9. Harvey AG, Stinson K, Whitaker KL, Moskovitz D, Virk H.. The subjective meaning of sleep quality: a comparison of individuals with and without insomnia. Sleep. 2008;31(3):383–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sullivan MJL, Bishop SR, Pivik J.. The pain catastrophizing scale: development and validation. Psychol Assess. 1995;7(4):524–532. [Google Scholar]
- 11. Plummer F, Manea L, Trepel D, McMillan D.. Screening for anxiety disorders with the GAD-7 and GAD-2: a systematic review and diagnostic meta-analysis. Gen Hosp Psychiatry. 2016;39:24–31. [DOI] [PubMed] [Google Scholar]
- 12. Levis B, Benedetti A, Thombs BD; DEPRESsion Screening Data (DEPRESSD) Collaboration. Accuracy of Patient Health Questionnaire-9 (PHQ-9) for screening to detect major depression: individual participant data meta-analysis. BMJ. 2019;365:l1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Perrot S, Lantéri-Minet M.. Patients’ Global Impression of Change in the management of peripheral neuropathic pain: clinical relevance and correlations in daily practice. Eur J Pain. 2019;23(6):1117–1128. [DOI] [PubMed] [Google Scholar]
- 14. Gryczynski J, McNeely J, Wu L-T, et al. Validation of the TAPS-1: a four-item screening tool to identify unhealthy substance use in primary care. J Gen Intern Med. 2017;32(9):990–996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hølen JC, Hjermstad MJ, Loge JH, et al. Pain assessment tools: is the content appropriate for use in palliative care? J Pain Symptom Manage. 2006;32(6):567–580. [DOI] [PubMed] [Google Scholar]
- 16. NIH HEAL IMPOWR Network. Accessed January 28, 2023. https://heal.nih.gov/research/clinical-research/integrative-management-chronic-pain.
- 17. Salahuddin D, Conti T.. Trauma and behavioral health care for patients with chronic pain. Prim Care. 2022;49(3):415–423. [DOI] [PubMed] [Google Scholar]
- 18. Karran EL, Grant AR, Moseley GL.. Low back pain and the social determinants of health: a systematic review and narrative synthesis. Pain. 2020;161(11):2476–2493. [DOI] [PubMed] [Google Scholar]
- 19. Perugino F, De Angelis V, Pompili M, Martelletti P.. Stigma and chronic pain. Pain Ther. 2022;11(4):1085–1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Volkow ND, Blanco C.. The changing opioid crisis: development, challenges and opportunities. Mol Psychiatry. 2021;26(1):218–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Fairley M, Humphreys K, Joyce VR, et al. Cost-effectiveness of treatments for opioid use disorder. JAMA Psychiatry. 2021;78(7):767–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ducharme LJ, Wiley TRA, Mulford CF, Su ZI, Zur JB.. Engaging the justice system to address the opioid crisis: the Justice Community Opioid Innovation Network (JCOIN). J Subst Abuse Treat. 2021;128:108307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Murphy SM, Laiteerapong N, Pho MT, et al. Health economic analyses of the justice community opioid innovation network (JCOIN). J Subst Abuse Treat. 2021;128:108262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Knight D, Becan J, Olson D, et al. Justice community opioid innovation network (JCOIN): the TCU research hub. J Subst Abuse Treat. 2021;128:108290- [DOI] [PubMed] [Google Scholar]
- 25. Treede RD, Rief W, Barke A, et al. Chronic pain as a symptom or a disease: the IASP Classification of Chronic Pain for the International Classification of Diseases (ICD-11). Pain. 2019;160(1):19-27. [DOI] [PubMed] [Google Scholar]
- 26. Kemp HI, Corner E, Colvin LA.. Chronic pain after COVID-19: implications for rehabilitation. Br J Anaesth. 2020;125(4):436–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Common Data Elements (CDEs): Getting More Common All the Time. Accessed January 28, 2023. https://www.youtube.com/watch?v=l2oRKT07RFo.
- 28. Johnson SB, Whitney G, McAuliffe M, et al. Using global unique identifiers to link autism collections. J Am Med Inform Assoc. 2010;17(6):689–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. NIH NIMH NDA GUID and GUID TOOL. Accessed January 28, 2023. https://nda.nih.gov/s/guid/nda-guid.html.
- 30. Hart LG, Larson EH, Lishner DM.. Rural definitions for health policy and research. Am J Public Health. 2005;95(7):1149–1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Broderick JE, DeWitt EM, Rothrock N, Crane PK, Forrest CB.. Advances in patient-reported outcomes: the NIH PROMIS((R)) measures. EGEMS (Wash DC)). 2013;1(1):1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Harris PA, Taylor R, Thielke R, Payne J, Gonzalez N, Conde JG.. Research electronic data capture (REDCap)–a metadata-driven methodology and workflow process for providing translational research informatics support. J Biomed Inform. 2009;42(2):377–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hume S, Aerts J, Sarnikar S, Huser V.. Current applications and future directions for the CDISC Operational Data Model standard: a methodological review. J Biomed Inform. 2016;60:352–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hume S, Sarnikar S, Becnel L, Bennett D.. Visualizing and validating metadata traceability within the CDISC standards. AMIA Jt Summits Transl Sci Proc. 2017;2017:158–165. [PMC free article] [PubMed] [Google Scholar]
- 35. Varghese J, Dugas M.. Frequency analysis of medical concepts in clinical trials and their coverage in MeSH and SNOMED-CT. Methods Inf Med. 2015;54(1):83–92. [DOI] [PubMed] [Google Scholar]
- 36. Fung KW, Bodenreider O.. Utilizing the UMLS for semantic mapping between terminologies. AMIA Annu Symp Proc. 2005;2005:266–270. [PMC free article] [PubMed] [Google Scholar]
- 37. Despotou G, Korkontzelos I, Arvanitis TN.. Bottom-up natural language processing based evaluation of the fitness of UMLS as a semantic source for a computer interpretable guidelines ontology. Stud Health Technol Inform. 2022;290:1060–1061. [DOI] [PubMed] [Google Scholar]
- 38. Ganzinger M, Gietzelt M, Karmen C, Firnkorn D, Knaup P.. An IT architecture for systems medicine. Stud Health Technol Inform. 2015;210:185–189. [PubMed] [Google Scholar]
- 39. Komatsoulis GA, Warzel DB, Hartel FW, et al. caCORE version 3: implementation of a model driven, service-oriented architecture for semantic interoperability. J Biomed Inform. 2008;41(1):106–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Jiang G, Solbrig HR, Chute CG.. Quality evaluation of value sets from cancer study common data elements using the UMLS semantic groups. J Am Med Inform Assoc. 2012;19(e1):e129-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Brummett CM, Bakshi RR, Goesling J, et al. Preliminary validation of the Michigan Body Map. Pain. 2016;157(6):1205–1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Adams MCB, Brummett CM, Wandner LD, Topaloglu U, Hurley RW. Michigan body map: connecting the NIH HEAL IMPOWR network to the HEAL ecosystem. Pain Med. Jul 5 2023;24(7):907–909. doi:10.1093/pm/pnad028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Galvez-Sanchez CM, de la Coba P, Duschek S, Del Paso R, Reliability GA.. Factor structure and predictive validity of the widespread pain index and symptom severity scales of the 2010 american college of rheumatology criteria of fibromyalgia. J Clin Med. 2020;9(8). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. NIH CDE Repository. Accessed January 28, 2023. https://cde.nlm.nih.gov/home.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.