Abstract
Hepatocellular carcinoma (HCC) exhibits a remarkable degree of heterogeneity, not only at an inter-patient level but also between and within tumors in the same patient. The advent of next-generation sequencing (NGS)-based technologies has allowed the creation of high-resolution atlases of HCC. This review outlines recent findings from genomic, epigenomic, transcriptomic, and proteomic sequencing that have yielded valuable insights into the spatial and temporal heterogeneity of HCC. The high heterogeneity of HCC has both clinical and therapeutic implications. The challenges in prospectively validating molecular classifications for HCC either for prognostication or for prediction of therapeutic response are partly due to the immense heterogeneity in HCC. Moreover, the heterogeneity of HCC tumors combined with the lack of commonly mutated, druggable targets severely limits treatment options for HCC. Recently, immune checkpoint inhibitors and combination therapies have shown promise for advanced HCC, while T cell therapies and vaccines are currently being investigated. Yet, immunotherapies show benefit only in a limited subset of patients, making it imperative to decipher tumor heterogeneity in HCC in order to enable optimal patient selection. This review summarizes the cutting-edge research on heterogeneity in HCC and explores the implications of heterogeneity on stratifying patients and developing biomarkers and therapies for HCC.
1. Introduction
Liver cancer causes over 800,000 deaths annually and is the third leading cause of cancer-related mortality in the world (Llovet et al., 2021; Sung et al., 2021). Furthermore, the incidence of liver cancer is on the rise in the Americas and Europe (Petrick et al., 2020). In the US, for instance, HCC incidence rose by 4% from 1978 to 2012 (Petrick et al., 2020). Hepatocellular carcinoma (HCC), the most common form of liver cancer, represents an average of 75–85% of all liver cancer cases (Thun, Linet, Cerhan, Haiman, & Schottenfeld, 2017). Patients with HCC can have variable clinical outcomes depending on the stage of tumor diagnosis and severity of the underlying liver disease. In general, patients with advanced HCC have a dismal prognosis with 5-year survival ranging from 15% to 20% (Siegel, Miller, & Jemal, 2020).
A key factor that complicates the diagnosis and treatment of HCC is tumor heterogeneity. Relative to other tumors, HCC exhibits a remarkable degree of heterogeneity (Khatib, Pomyen, Dang, & Wang, 2020). This heterogeneity makes HCC extremely resistant to most conventional chemotherapy. In 2007, the tyrosine kinase inhibitor, sorafenib, was approved for use in advanced HCC. Today, more treatment options exist for patients with advanced HCC in addition to sorafenib including regorafenib, lenvatinib, cabozantinib, and ramucirumab. However, all these options still provide limited clinical benefits. In 2020, the combination of atezolizumab (an antibody against PDL-1) and bevacizumab (an antibody against VEGF) was approved for patients with unresectable HCC (Finn, Qin, et al., 2020). Yet, this combination immunotherapy beat the median progression-free survival of sorafenib by only 2.5 months (Finn, Qin, et al., 2020; Sukowati, El-Khobar, & Tiribelli, 2021). Thus, there is a vast unmet need to develop novel therapies for HCC. We believe that a deeper understanding of tumor heterogeneity is necessary to develop successful therapeutic solutions and improve clinical outcomes for patients with HCC.
The complexity of HCC arises from several sources including diverse etiologies, varying geographical trends, and also molecular heterogeneity. Several different etiological risk factors are strongly associated with HCC development like chronic hepatitis B, chronic hepatitis C, alcohol-related or nonalcoholic fatty liver disease, aflatoxin exposure, hemochromatosis, and other causes of cirrhosis (Ghouri, Mian, & Rowe, 2017; Fujiwara, Friedman, Goossens, & Hoshida, 2018). The specific etiologies of HCC also drive the geographical trends in HCC incidence and outcomes in different parts of the world. For instance, HCC incidence is high in East and Southeast Asia due to the high prevalence of the hepatitis B virus (HBV) (Jiang, Han, Zhao, & Zhang, 2021; Yang et al., 2019). Even with falling HBV rates accompanied by a decline in HCC in these regions, liver cancer incidence remains very high (Dasgupta et al., 2020). On the other hand, metabolic risk factors for HCC like obesity and diabetes rates are increasing in many parts of the world including the US. In 2016, an alarming 1.9 billion adults worldwide were classified as overweight or obese, and this is a major contributor to the rising incidence of liver cancer (Sun & Karin, 2012; World Health Organization, 2018). Thus, disparate etiological and geographical factors contribute to the heterogeneity seen in HCC incidence.
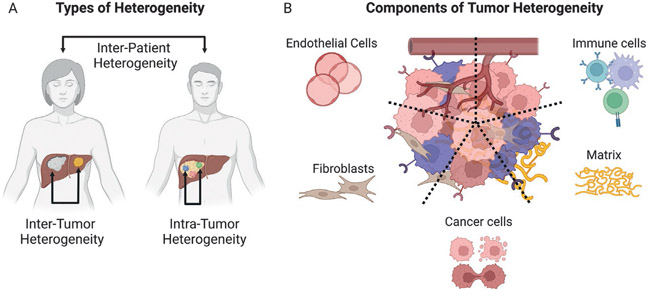
Molecular heterogeneity is a critical area of research in HCC and will be the focus of the rest of this chapter. HCC tumor progression frequently occurs in the context of inflammation and fibrosis in a cirrhotic liver (Llovet et al., 2021; Villanueva, 2019). This environment is permissive to synchronous and metachronous tumor-initiating events and acts as a catalyst for molecular heterogeneity (Dhanasekaran, Bandoh, & Roberts, 2016). Molecular heterogeneity occurs on different levels. 1. on the interpatient level, there are molecular variations between tumors in different patients, and 2. on the intrapatient level, there are differences between tumors in the same patient. Moreover, intratumorally, there are variations in different regions within one tumor in a given patient. Further, a tumor is not just a mass of genetically mutated cancer cells; it is infiltrated by a variety of immune cells, structurally supported by the extracellular matrix, and nourished with oxygen and nutrients by the vasculature. These components, therefore, are also important considerations in the discussion of HCC molecular heterogeneity (Fig. 1).
Fig. 1.
Types and components of tumor heterogeneity in hepatocellular carcinoma (HCC). (A) The different types of heterogeneity in HCC are shown. Inter-patient—molecular differences between tumors of different patients, inter-tumor—variations between different tumors in the same patient, and intra-tumor heterogeneity—variations across different regions of an individual tumor in the same patient. (B) The different components of heterogeneity in HCC are shown. Cancer-cell intrinsic variations, differences in tumor immune cell infiltrates, and changes in endothelial cells, fibroblasts, and the matrix.
2. Approaches to studying heterogeneity in HCC
Prior to the advancement of next-generation sequencing technologies, experimental schemes were largely hypothesis-driven and mostly only allowed investigators to confirm or refute a hypothesis. However, this approach limited our exploration of tumor heterogeneity because it necessitated the targets of interest to be known a priori. The advent of next-generation sequencing-based technologies for multi-omic and single-cell analysis has greatly accelerated a discovery-based understanding of tumor heterogeneity. Genome, epigenome, transcriptome, and proteome sequencing of serial and multi-region samples has allowed us to create high-resolution atlases of tumors and yielded several valuable insights into the spatial and temporal heterogeneity of HCC.
An important approach to studying heterogeneity in HCC is single-cell sequencing. In this approach, bulk tumors are dissociated into single cells, and DNA or RNA is extracted and sequenced from each cell. Additionally, the study of single-cell epigenetics is made possible by technologies like DNA methylome sequencing and transposase-accessible chromatin with sequencing (ATAC-seq). Today, the profiling of tens of thousands of cells is both technically feasible and relatively cost-effective. Thus, there is an explosion of research performing single-cell analysis in different cancers. Single-cell technology is powerful because of the resolution it offers. Single-cell RNA sequencing, for example, allows the investigation of the transcriptome of individual cancer, immune and stromal cells, and the tracking of subclones and their evolution. Several of the studies discussed in Section 3 leveraged single-cell sequencing for these purposes.
A drawback of single-cell sequencing is that it cannot capture the spatial distribution of different cell types. Given that paracrine signaling operates over short distances of less than 200μm, knowing regional distributions of sequenced cells is critical in disentangling crosstalk between cells (Longo, Guo, Ji, & Khavari, 2021). Spatial transcriptomics offers a way to gain transcriptional information while maintaining tissue architecture. Two major techniques are used for spatial transcriptomics: high-plex RNA imaging and spatial barcoding. While high-plex RNA imaging has single-cell resolution and a greater depth of sequencing, spatial barcoding has a lower depth per transcript. On the other hand, spatial barcoding can be used on a larger field of view and has a greater transcriptome coverage compared to high-plex RNA imaging (Longo et al., 2021). The study by Brady et al exemplifies the strength of spatial transcriptomics in deciphering tumor heterogeneity. The researchers sampled different primary and metastatic tumor lesions in patients with prostate cancer. They found significant intrapatient heterogeneity when comparing tumor lesions within each patient, including in the gene expression of the therapeutically relevant, NCAM1 for which antibodies and CAR-T cells exist. This showed that metastatic lesions in prostate cancer are not monoclonal and have distinct mutation profiles. Within a given tumor lesion, too, spatially distinct regions did not always have similar gene expression profiles. One metastasis, for instance, had two separate phenotypes within it that would have been misattributed by a bulk sequencing approach (Brady et al., 2021).
Liquid biopsies are a growing area of interest in the study of tumor heterogeneity. The term ‘liquid biopsy’ refers to analyzing tumors through circulating tumor cells (CTCs), which are cells that break off from tumors and enter circulation, or circulating tumor DNA (ctDNA), which is a type of fragmented DNA in blood circulation that is derived from tumors. Liquid biopsies offer many benefits over traditional needle biopsies which have long been used to detect genomic alterations in HCC. Needle biopsies are an invasive approach, requiring sampling from the tumor, while liquid biopsies are comparatively noninvasive and only require a blood draw. The lower risk profile of liquid biopsies means it is feasible to repeat them over time to track temporal changes and use them in surveillance for recurrence. Further, because needle biopsies sample from one tumor lesion at a singular point in time, they may fail to reflect tumor burden and fully capture genomic changes across the tumor (Swanton, 2012; Zhang et al., 2021). Liquid biopsies, on the other hand, measure CTCs or ctDNA that enter the blood-stream from all major tumor regions. Thus, liquid biopsies capture genetic changes from a majority of the tumor rather than from a specific region. Cai et al investigated the utility of ctDNA as a biomarker in HCC and found the ctDNA adequately reflected tumor subclonal mutations, thus capturing tumor heterogeneity (Cai et al., 2017). The researchers studied mutations in 574 cancer genes from tissue and plasma samples of three patients. Overall, over 200 subclonal mutations were discovered in the three patients. Intrapatient heterogeneity was also found: only 50–80% of subclonal mutations were overlapping between the primary tumor and portal vein tumor thrombus in each patient. Notwithstanding, the researchers reported that over 98% of detected subclonal mutations were also detected in ctDNA, indicating the ability of ctDNA to capture tumor heterogeneity (Cai et al., 2017).
Sun et al. investigated the spatial and temporal features of CTCs in the blood (Sun et al., 2018). The study used blood drawn from the peripheral vein, peripheral artery, hepatic veins, infrahepatic inferior vena cava, and portal vein of patients prior to tumor resection and during follow-up. A majority of CTCs at sites close to the tumor were epithelial while a majority of CTCs in systemic blood circulation had activated epithelial-mesenchymal transition (EMT). The total number of CTCs in the hepatic vein blood correlated with EMT activation in the primary tumor. Follow-up analysis showed that an increased number of CTCs in the peripheral vein or peripheral artery indicated an increased risk of intrahepatic recurrence (Sun et al., 2018). Overall, this study demonstrated the importance of the location of liquid biopsy and showcased the potential of CTCs in guiding HCC clinical progress. However, despite the promise of CTC, the lack of specific biomarkers to detect CTCs remains a challenge in HCC. Many studies have used the epithelial cell adhesion molecule (EpCAM) as a marker, however, this alone may not be ideal to detect CTCs that undergo EMT and lose their epithelial phenotype (Maravelia et al., 2021; Plaks, Koopman, & Werb, 2013). Combinations of biomarkers like EpCAM with hepatocyte-specific asialoglycoprotein receptor (ASGPR) and Twist1 have also been explored in other studies (Li et al., 2014; Sun et al., 2018). Another obstacle to the utility of CTCs is that CTCs levels can be particularly low in patients with small or early tumors, making this technique difficult to use in detection or early-disease monitoring (Chen, Zhong, Tan, Wang, & Feng, 2020; Mocan et al., 2020).
Generation of patient-derived primary cell lines, organoids, and xenografts from resected or biopsied HCC tumors offers another way to study HCC tumor heterogeneity. Gao et al performed multi-region sampling on 55 regions from 10 patients and maintained the isolated cells in culture. Whole-exome sequencing, copy-number analysis, and high-throughput screening were performed. The researchers discovered a high degree of intratumoral heterogeneity, with around 40% of mutations being heterogeneous. They also discovered that cells from some subregions had genetic aberrations in FGF19, DDR2, PDGFRA, and TOP1, and showed sensitivity to the application of a therapeutic treatment. However, subregions from the same tumor without these mutations did not show sensitivity. This experiment shows an in vitro approach to study HCC tumor heterogeneity and drug responses (Gao et al., 2017).
Organoids are models where cancer cells are cultured as three-dimensional structures (Tuveson & Clevers, 2019). Zhao et al generated seven hepatobiliary tumor organoids and studied them using single-cell RNA sequencing (Zhao et al., 2021). The researchers noted that there was inter-organoid transcriptional and therapeutic heterogeneity. One of the organoids, HC272, for example, showed enrichment of HIF-1, MAPK, and PI3K-Akt signaling pathways compared to the other organoids. Consequently, this organoid also displayed resistance to the eleven tyrosine kinase inhibitors (TKIs) tested in the study, while other organoids showed sensitivity to one or more TKIs (Zhao et al., 2021).
Patient-derived xenografts have also been investigated to identify biomarkers, study tumor progression, and conduct personalized drug screens. Blumer et al validated that PDXs generated from biopsies of Grade III or IV HCC preserved tumor markers, gene signatures, and copy-number alterations from the original tumor over 6 generations of retransplantation (Blumer et al., 2019). Hu et al. studied the effects of three drugs in sixteen PDX models. They discovered inter-individual heterogeneity as well as intra-individual differences in sensitivity to the agents tested. One of their findings was that impaired sorafenib response was correlated with low MAP3K1 expression in patients. The investigators further studied this finding in their PDX model and cell lines and found that MAP3K1 overexpression resulted in a more robust response to sorafenib treatment while MAP3K1 inhibition increased cell proliferation during sorafenib treatment (Hu et al., 2020). Taken together, these results indicate the ability of organoids and PDXs to recapitulate tumor biology and study drug responses (see also chapter “Molecular therapeutic targets for cholangiocarcinoma: Present challenges and future possibilities” by Anderson et al.) (Fig. 2).
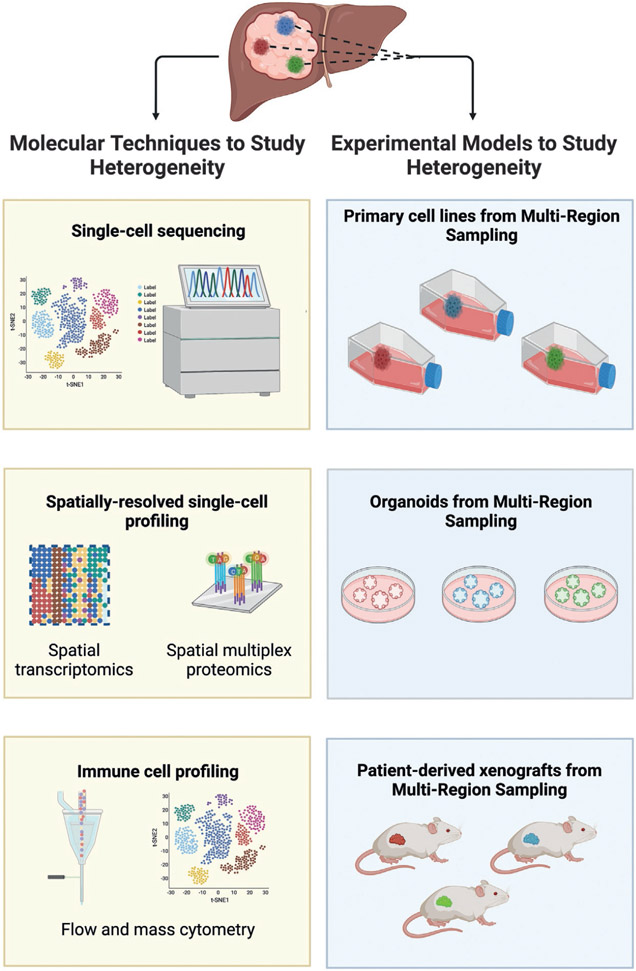
Fig. 2.
Molecular techniques (left) and experimental models (right) to study tumor heterogeneity in hepatocellular carcinoma (HCC) are shown. On the left, extracts from the tumor or liquid biopsy can be used to perform single-cell sequencing, spatial transcriptomics and multiplex proteomics, and flow and mass cytometry. On the right, multi-region samples from tumors can be used to establish primary cell lines, organoids, and patient-derived xenografts in immunocompromised mice.
3. Molecular heterogeneity in HCC
Tumors are composed of more than just clusters of cancer cells: they are complex quasi-organs that are infiltrated by immune cells, structurally supported by the extracellular matrix, and supplied with oxygen and nutrients from the vasculature. All of these components contribute to molecular heterogeneity in HCC tumors. Here, we discuss three contributors to molecular heterogeneity: cancer cells, immune cells, and stromal cells.
3.1. Tumor heterogeneity
Next-generation sequencing (NGS) has revolutionized our understanding of cancer genetics. DNA sequencing of bulk HCC tumors has revealed that they have a median of 50–70 protein-altering mutations and an average of 2–6 driver mutations across all tumor stages (Schulze et al., 2015; The Cancer Genome Atlas Research Network, 2017; Nault et al., 2020; Ahn et al., 2013). The most prevalent driver gene mutations are in the TERT promoter, TP53, CTNNB1, AXIN1, ARID1A, and ARID2 which cause changes in the activation of several pathways including telomere maintenance, P53 cell regulation, Wnt/β-catenin, Akt/mTOR, MAP kinase, and oxidative stress (Ahn et al., 2014; The Cancer Genome Atlas Research Network, 2017; Fujimoto et al., 2012; Guichard et al., 2012; Nault et al., 2013; Schulze et al., 2015; Totoki et al., 2014). Some driver gene mutations tend to co-occur, suggesting a synergistic effect in tumorigenesis and tumor progression such as CTNNB1 and TERT promoter mutations with ARID2 and NFE2L2 mutations while others like AXIN1 and TP53 mutations are mutually exclusive suggesting redundancy or disadvantageous effects when co-occurring (Schulze et al., 2015). These findings highlight the variety of mutations and mutation combinations that are present in different tumors that contribute to inter-patient heterogeneity in HCC.
On an intrapatient level too, there can be incredible heterogeneity between tumors in the same patient (Xue et al., 2016). Exome and whole-genome sequencing of 43 lesions from 10 patients with HCC revealed that, in general, intrahepatic metastases and tumor thrombi tended to have different mutations and copy number variations compared to the primary lesion. However, the percentage of shared mutations between lesions in a given patient varied widely from 8% to 97%. This broad range of intrapatient heterogeneity suggests that a single lesion may not always sufficiently capture the genomic features of HCC in every patient (Xue et al., 2016).
In about half of patients with recurrent HCC, the primary and recurrent tumors have dissimilar genetic features, according to a study by Ding et al. (2019). By sequencing 824 HCC-relevant genes in multi-region samples from 41 matched primary and recurrent tumor pairs, the investigators found that recurrent HCC fell into two broad categories: multicentric and progressive. Multicentric HCC comprised 48% of recurrent tumors and had an independent lineage from the primary tumor. Progressive HCC, on the other hand, comprised the remaining 52% of recurrent tumors, shared several mutations with the primary tumor, and were derived from the same clonal lineage. The recurrence time between these two categories also appeared distinct—though not statistically significant—with the multicentric type generally recurring after 2 years and progressive type recurring earlier. Thus, the similarity between the primary and recurrent HCC tumors is another contributor to interpatient and intrapatient heterogeneity (Ding et al., 2019).
3.2. Cancer cell heterogeneity
Cancer cells are the primary contributor to tumor heterogeneity because of their high degree of genome instability. Several studies have explored cancer cell heterogeneity present in HCC tumors using single-cell sequencing and/or multi-region sampling.
Guo et al used single-cell DNA sequencing to study copy number alterations (CNAs), which occur due to genome instability and contribute to cancer cell heterogeneity (Guo et al., 2022). The percentage of amplifications and deletions in the genome of cells from a given patient varied widely with standard deviations between 1% and 30%. There were also differences between patients with the average percentage of genomic alterations ranging from 15% to 60% and the percentage of non-diploid cells ranging from 34% to 94%. These findings indicated a variable degree of intrapatient and interpatient heterogeneity in cancer cell genomes (Guo et al., 2022).
Single-cell sequencing studies using multi-region sampling have further revealed the extraordinary diversity of cancer cells present in HCC tumors. Ling et al performed whole-genome and whole-exome sequencing on 286 regions from a 35mm, histopathological grade III HCC tumor (Ling et al., 2015). Their results revealed a high degree of genetic diversity in the tumor with over 100 million estimated coding region mutations, including 6 driver gene mutations and 209 single-nucleotide variations (SNVs). A portion of the cancer cells with no driver gene mutations was discovered that defined a whopping 20 distinct cell clones in the whole-exome sequenced regions. Thus, the study reported extreme heterogeneity in the HCC tumor, much higher than would be predicted by Darwinian evolution (Ling et al., 2015).
Zhai et al also used single-cell sequencing of multi-regional samples to investigate the evolution and spatial organization of intratumoral heterogeneity in HCC (Zhai et al., 2017). A series of sections were sampled along the diameter of nine early HCC tumors and either whole-genome or whole-exome sequenced. The authors found that observed variability within each tumor increased substantially with an increasing number of sampled regions, suggesting that single samples of HCC tumors likely fail to fully capture tumor heterogeneity. Phylogenetic trees for each tumor were constructed using variations in somatic mutations across sections and showed that sections from one end of the tumor on the central axis were consistently more similar than those from the other end. This led to the suggestion that HCC ancestral clones may be found in the tumor center, with new clades arising as the tumor expands outwards (Zhai et al., 2017).
Wu et al. performed high-resolution spatial transcriptomics to study 3 distinct regions–nontumor, leading-edge, and tumor–in seven HCC patients (Wu et al., 2021). One aspect of the study investigated cancer stem cell (CSC) niches and found that the distribution of CSCs varies from the leading-edge to the tumor and constituted diverse cells and activated pathways. The PROM1+ and CD47+ CSC niche, for example, was found to increase in abundance from the leading-edge to the tumor and the portal vein thrombus and was implicated in immunosuppression and vascular metastasis. The preservation of spatial information in the study allowed the investigation of CSC niche composition and distribution within tumors (Wu et al., 2021). Overall, the aforementioned studies reveal a rich diversity of cancer cells in HCC tumors both on the interpatient and intrapatient levels.
3.3. Immune cell heterogeneity
HCC tumors vary widely in their immune subpopulation frequency and distribution, the activation and functional states of immune cells, and the overall degree of immune infiltration. With the recent approval of combination immunotherapies for HCC, atezolizumab/bevacizumab and nivolumab/ipilimumab, there is a renewed interest in immune cells in the tumor microenvironment.
Zhang et al. analyzed immune heterogeneity in HCC using mass cytometry (Zhang, Lou, et al., 2019). They identified 40 distinct clusters of immune cells within the tumor microenvironment and noted differences between immune environments in their eight patients and between lesions within the same tumor in each patient. Certain immune cell populations were positively correlated such as PD-L1+ macrophages with PD-1+ CD4 + T cells, and dendritic cells with CD11b + CD4 + T cells. The immune environments were clustered into three categories with distinct cell frequencies, gene expression of immune suppressive genes and metabolomes. Their study of the immune landscape of tumors revealed notable interpatient and intrapatient immune heterogeneity (Zhang, Lou, et al., 2019).
The immune environment is dynamically regulated through the stages of tumor progression. Song et al. used single-cell RNA sequencing of immune cells in hepatitis B/C-related HCC tumors and found 29 unique immune cell subsets of myeloid, lymphoid, and NK cells that shifted between functional states and interacted with other subsets (Song et al., 2020). Effector CD8+ T cells showcased distinct transcriptomes and cytotoxicity that varied from early to advanced HCC. A subset of M2 macrophages with high expression of CCL18 and CREM was elevated in patients with advanced HCC (Song et al., 2020). The findings of this paper provide insight into the changes in the HCC immune environment with tumor progression.
Several studies have studied specific immune cell populations and their functional states in HCC tumors. Losic et al. investigated T cell clonality in samples from spatially different regions of HCC tumors (Losic et al., 2020). Using a combination of DNA, RNA, and T cell receptor (TCR) sequencing, they found significant differences in the number of unique T cell expansions, and density of immune cells in different regions of tumors. They also found that some regions within a given tumor had tertiary lymphoid structures while other regions did not, demonstrating another instance of intratumoral immune heterogeneity. They determined that tumor neoantigens primarily recruit tumor-infiltrating lymphocytes (B and T cells). Immune checkpoint genes such as CTLA4 and CD274 also tended to be upregulated in tumor cells in regions with more tumor-infiltrating lymphocytes, suggesting regional differences in immunosuppressive effect on T cell activation (Losic et al., 2020).
T cell heterogeneity was further explored by Hung et al who investigated the impact of tumor methionine metabolism on T cell exhaustion in HCC. They used ATAC-seq to uncover the chromatin accessibility of T cells during activation and treatment with two products of methionine metabolism. In their results, they show that the products of tumor methionine metabolism induce significant changes in chromatin in T cells that inhibit effector T cell function and potentially contribute to exhaustion (Hung et al., 2021).
Immune cell distribution and properties were explored by Zhang et al. by RNA sequencing of immune cells at five regions: tumor, adjacent liver, hepatic lymph node, blood, and ascites (Zhang, He, et al., 2019). The study found a population of LAMP3+ dendritic cells had the potential to migrate from the tumor to the lymph node and expressed immune-modulating ligands that could regulate various lymphocytes. Macrophages with distinct transcriptional states were discovered, and the expression of SLC40A1 and GPNMB in tumor-associated macrophages (TAMs) was found to play an inflammatory role in the tumor. The origins of myeloid and lymphoid cells in the ascites were traced to the tumor and blood, respectively. These findings reveal the diversity of immune cell functions in the regions in and around the tumor (Zhang, He, et al., 2019).
The heterogeneity of the HCC immune microenvironment has implications on patient prognosis. Kurebayashi et al. studied the immune microenvironment in 919 regions from 158 HCC tumors and classified them into three subtypes: immune-high, immune-mid and immune-low. The immune-high subtype was characterized by increased B cell and T cell infiltration. Importantly, they found that high-grade tumors of the immune high subtype had a significantly better prognosis (Kurebayashi et al., 2018). In a similar vein, Calderaro et al. investigated intratumoral tertiary lymphoid structures (TLSs) in HCC. TLSs are sites for immune response initiation and maintenance present in nonlymphoid tissue. The presence of TLSs within HCC tumors was found to be associated with a decreased risk of early HCC recurrence in two cohorts consisting of 273 and 225 tumors respectively (Calderaro et al., 2019). The above studies showcase the incredible heterogeneity in immune cell frequency, distribution, and activation in HCC.
3.4. Stromal heterogeneity
Stromal cells, including cancer-associated fibroblasts (CAFs) and endothelial cells, make up important components of the tumor, though their contribution to HCC tumor heterogeneity remains an active area of investigation.
CAFs in the HCC microenvironment display heterogeneity in tissue distribution, cellular origin, and function. CAFs are often present in the HCC fibrous septum, fibrous capsule, and hepatic blood sinusoids (Yin et al., 2019). CAFs originate from a variety of cells including hepatic stellate cells, mesenchymal stem cells, and cancer cells. Secretory products from cancer cells can cause these various cell types to take on a CAF role. A growing body of evidence suggests that CAFs play a pro-tumor role in HCC. Liu et al showed that CAFs secrete chemokines including CCL2, CCL5, CCL7, and CXCL16 which promote cancer cell migration, invasion, and epithelial-mesenchymal transformation (EMT) in vitro, and promote HCC metastasis in vivo through the activation of the hedgehog (Hh) and transforming growth factor-β (TGFβ) pathways (Liu et al., 2016). Some CAFs induce the chemotaxis of immune cells such as neutrophils, dendritic cells, and monocytes to the tumor and promote immunosuppressive phenotypes through IL6-STAT3 signaling (Cheng et al., 2018). Thus, CAFs display heterogeneity in the tumor through their presence in a variety of tumor regions, having multiple cellular origins, and activating different pro-tumor and immunomodulatory signaling pathways.
The role of fibroblasts in HCC tumorigenesis from non-alcoholic steatohepatitis (NASH)-related fibrosis has been explored (Asakawa et al., 2019). RNA sequencing was used to study the gene expression profiles of fibroblasts in a NASH mouse model of fibrosis. Compared to a chemical-induced fibrosis model, fibroblasts in the NASH model showed a robust upregulation in cancer-related pathways including fibroblast growth factor 9 (FGF9) expression. Follow-up experiments validated the role of FGF9 in promoting tumorigenesis to drive HCC in the NASH fibrotic liver (Asakawa et al., 2019). This study suggests that there may be heterogeneity in the transcriptional state and roles of fibroblasts in HCC of different etiologies, such as in NASH-driven HCC.
Overall, however, the heterogeneity of CAF phenotypes and functions in HCC remains to be thoroughly investigated. Studies from other cancers indicate that the functional states and spatial distribution of CAFs are important contributors to tumor heterogeneity (see also chapter “Cancer-associated fibroblasts in intrahepatic cholangiocarcinoma progression and therapeutic resistance” by Affò et al.). Sebastian et al explored the molecular heterogeneity of CAFs in triple-negative breast cancer in a mouse model using single-cell RNA sequencing and found six distinct subpopulations such as (i) myofibroblastic CAFs enriched in contractile proteins, (ii) inflammatory CAFs with increased expression of inflammatory cytokines, and (iii) CAFs expressing major histocompatibility complex (MHC) class II, which are generally found on antigen-presenting cells (Sebastian et al., 2020). The spatial distribution of CAFs in pancreatic cancer was explored by a study that found that the myofibroblast CAF population is frequently in regions close to the pancreatic tumor, while inflammatory CAFs are found in regions distant from the tumor, though TGF-β or JAK/STAT signaling can facilitate their interconversion (Biffi et al., 2019; Öhlund et al., 2017). The differing transcriptional states and spatial distribution of different CAF subclasses suggest that they may have differing contributions to tumorigenesis and tumor progression. Comparing HCC CAF phenotypes and distribution in different patients and between lesions in a given patient will further elucidate their contribution to interpatient and intrapatient heterogeneity.
Endothelial cells (ECs) which typically line blood vessels are another source of heterogeneity in HCC. Sharma et al performed single-cell RNA sequencing on HCC tumors and discovered eleven unique clusters of ECs including a cluster of CD9+ ECs found primarily in the adjacent liver, a cluster of phospholipid phosphatase 3-positive (PLPP3+) ECs present in the peripheral tumor, and a cluster of plasmalemma vesicle-associated protein-positive (PLVAP+) ECs present in the core tumor (Sharma et al., 2020). This study sheds light on the rich diversity of spatially distributed endothelial cells present within HCC tumors. The roles of these EC subpopulations in HCC pathophysiology remain to be explored. However, some EC subpopulations are likely pro-tumorigenic: ECs are known to engage in crosstalk with cancer cells that contribute to epithelial to mesenchymal transition, cancer stem cell phenotype, and metastasis (Alsina-Sanchis, Mülfarth, & Fischer, 2021; Maishi et al., 2016; McCoy et al., 2019).
The abundance of tumor vasculature, which is composed of ECs, shows interpatient differences and impacts HCC prognosis. Several studies have reported the biomarker potential of vessels that encapsulated-tumor clusters (VETCs)–detected through immunostaining for vascular ECs (Fang et al., 2015, 2019; Renne et al., 2020). Renne et al report the presence of VETCs in only a subset of patients, about 19%, where their presence was associated with several factors including tumor sizes greater than 5cm, a poorly differentiated phenotype, microvascular invasion, and early recurrence (Renne et al., 2020). Overall, this study exemplifies the interpatient heterogeneity in HCC vasculature and the role of vascular endothelial cells in HCC pathophysiology and prognostication.
4. Clinical implications of genetic heterogeneity in HCC
4.1. Challenges in stratification of HCC
The understanding and classification of tumors into subtypes based on molecular or mutational signatures has improved patient outcomes for several cancers. The advances in single-cell sequencing technologies carry the promise of deepening our understanding of the characteristics of, and interactions between, cancer, immune, and stromal cells. NGS technologies offer high-plex information at a significantly higher sensitivity than traditional methods like immunohistochemistry, and this could, in turn, facilitate the identification of molecular signatures for distinct classes of HCC.
Several studies have used transcriptome analysis to categorize HCC tumors into subtypes based on hierarchical clustering or correlations with clinical variables (Boyault et al., 2007; Chiang et al., 2008; Hoshida et al., 2009, 2010; Lachenmayer et al., 2012). The analysis of multiple, genome-wide studies reveals that HCC tumors can be categorized into two major classes: proliferative and non-proliferative (Boyault et al., 2007; The Cancer Genome Atlas Research Network, 2017; Chiang et al., 2008; Hoshida et al., 2009; Lee et al., 2004). The proliferative class consists of tumors of a more aggressive and poorly-differentiated phenotype. These tumors showcase high chromosomal instability, global DNA hypomethylation, TP53 inactivation, and CCND1 and FGF19 amplification. Pro-survival pathways are frequently activated including cell cycle, MET, RAS-MAPK, and mTOR (Boyault et al., 2007; Calvisi et al., 2007; Chiang et al., 2008; Hoshida et al., 2010; Lee et al., 2004). The proliferative class can be further subdivided into the Wnt-TGFβ subclass, characterized by an exhausted immune response, and the progenitor subclass, consisting of hepatic progenitor marker overexpression and ERK hyperphosphorylation (Boyault et al., 2007; Calderaro et al., 2017; Hoshida et al., 2009; Sia et al., 2017).
The non-proliferative class consists of tumors with Wnt/β-catenin pathway activation, higher genome stability, and a less aggressive, more differentiated phenotype (The Cancer Genome Atlas Research Network, 2017; Hoshida et al., 2009, 2010; Lee et al., 2004). However, even within tumors of this class, there is a significant degree of heterogeneity and tumors can be subdivided into those with CTNNB1 mutations and “G4” subclasses. The CTNNB1 subclass show activation of the Wnt/β-catenin pathway, TERT promoter mutations, and CDKN2A and CDH1 promoter hypermethylation (The Cancer Genome Atlas Research Network, 2017; Chiang et al., 2008; Xu et al., 2018). Tumors of this subclass tend to be “cold”, with reduced T cell infiltration and an increased presence of regulatory T cells (Calderaro et al., 2017; Sia et al., 2017). The “G4” subclass shows activation of the IL6/JAK-STAT pathway and tends to be “hot”, with an active immune response (Boyault et al., 2007; Calderaro et al., 2017; Chiang et al., 2008; Sia et al., 2017). Tumors of the non-proliferative class have alternatively been grouped depending on their metabolic functions into periportal and perivenous subclasses. The periportal subclass expresses the HNF4A gene signature and is associated with the least likelihood of early recurrence and the highest survival rate (Chan, Tsui, Ho, & Ng, 2021; Desert et al., 2017).
Another approach has been to use cell deconvolution methods on transcriptomic data from HCC tumors. This approach has allowed the identification of the proportion of different immune cells within HCC and yielded two immune classes of HCC, immune-high and immune-low (Sia et al., 2017). About 25% of HCC tumors fall in the immune-high class and show macrophage, T cell, cytotoxic cell, and tertiary lymphoid structure enrichment and have elevated PD1 signaling. The immune-high subclass can be further divided into those that show an active or exhausted T cell response, with the exhausted response subclass showing TGFβ1-mediated gene regulation and immunosuppression.
HCC is yet to have a molecular classification system to prospectively stratify patients and guide clinical care. A central challenge to validating and implementing a classification system is that the current guidelines for HCC diagnosis and treatment do not require tissue biopsies. This limits the systematic collection of tissue samples and the correlation of molecular features with clinical variables.
4.2. Challenges in developing biomarkers for HCC
Biomarkers can serve as useful tools to guide clinical care for patients with HCC. One area of interest has been the discovery of biomarkers that predict response to therapies, particularly to tyrosine kinase inhibitors which constitute a major portion of the available therapies for patients with advanced HCC. Sorafenib is a multikinase inhibitor that is used as a first-line treatment in patients with advanced HCC. Fang et al. report that primary tumors with CD34+ vessels that encapsulate tumor clusters (VETC) pattern show a better response to sorafenib treatment upon HCC recurrence than tumors without VETC pattern (Fang et al., 2019). Other factors including FGF3/FGF4 amplification, VEGFA amplification, VEGFR overexpression, and Mapk14-Atf2 signaling elevation have also been associated with better sorafenib response (Arao et al., 2013; Horwitz et al., 2014; Peng et al., 2014; Rudalska et al., 2014). Harding et al performed NGS on 81 tumors from patients with advanced HCC who received sorafenib treatment. They found that the activation of the P13K-mTOR pathway in patients receiving sorafenib treatment was associated with poorer outcomes including shorter median progression-free survival and shorter median overall survival (Harding et al., 2019). Similarly, another study found that a 9-marker myeloid signature predicted sorafenib efficacy in recurrent HCC (Wu et al., 2020). Despite these findings, there is no validated biomarker that is currently used in patient care to predict sorafenib response. Similarly, a retrospective analysis revealed that 5 proteins and 9 miRNAs present in plasma could predict response to regorafenib, a tyrosine kinase inhibitor that is used as second-line treatment (Teufel et al., 2019). However, this finding also needs validation in a prospective cohort. Phase III of the REACH-2 study validated the first predictive biomarker for an HCC drug that is usable in clinical practice. It was found that high serum alpha-fetoprotein is associated with sensitivity to ramucirumab (Zhu, Kang, et al., 2019).
Immune checkpoint inhibitors have shown major benefits in the treatment of several cancers. Given that HCC arises in the context of inflammation and immunosuppression, immunotherapies are an exciting therapeutic prospect. Despite these facts, a sizable portion of HCC patients does not respond to ICIs. For example, in a phase II trial for nivolumab, a PD-1 inhibitor, the objective response rate was only 20%. Further, tumoral PD-L1 expression did not correlate with the effectiveness of PD-1 inhibition (El-Khoueiry et al., 2017). This suggests a need to identify biomarkers that can predict benefit from PD-1 inhibition, or other ICIs. One study found that a higher proportion of CD38+ cells in the tumor correlated with better response to PD-1 inhibition, while another study showed that the upregulation of indoleamine 2,3-dioxygenase (IDO) was associated with poorer response to PD-1 inhibition (Brown et al., 2018; Garnelo et al., 2017). It is also possible that the immune classification of HCC, discussed in Section 4.1, could predict response to PD-1 therapy. The “G4” subclass, which has high immune infiltration and enriched PD1 signaling, is likely to be more responsive to ICI, especially PD-1 inhibition. However, the association between this HCC subclass and ICI response is yet to be confirmed. On the other hand, the CTNNB1 subclass, which shows activation of the Wnt/β-catenin pathway and has reduced immune infiltration, is likely to be resistant to ICI. Indeed, a study found that in patients receiving PD-1 inhibition, Wnt/β-catenin pathway mutations were associated with poorer outcomes including shorter median progression-free survival and shorter median overall survival (Harding et al., 2019). For the combination therapy of PD-1/PD-L1 inhibition, many studies across different cancers have validated the biomarker potential of tumor mutational burden, a measure of the total number of somatic mutations per megabase. These studies point to the association between a high tumor mutational burden and greater benefit from PD-1/PD-L1 inhibition (Zhu, Zhang, et al., 2019; Forschner et al., 2019; Lee, Samstein, Valero, Chan, & Morris, 2020; Chen et al., 2019). Several factors including genetic heterogeneity, activated pathways, tumor microenvironment, systemic immunity status and metastases can impact the efficacy of immunotherapy (Siu et al., 2018; Sukowati et al., 2021). However, there is still much progress to be made in identifying predictive biomarkers for response to immune checkpoint inhibitor therapies for patients with HCC.
Tumor biopsy remains the most reliable source for identifying biomarkers. However, given the difficulty in obtaining single and longitudinal tissue biopsies as in HCC, a liquid biopsy is a lucrative, non-invasive intervention that allows temporal and some degree of spatial monitoring of tumor heterogeneity. A study by Kim et al. points to the ability of ctDNA to reflect tumor heterogeneity and predict prognosis in HCC. The researchers first investigated common SNVs in ctDNA using a digital droplet PCR panel of 2924 SNVs in 69 genes. There was considerable inter-patient heterogeneity in detected SNVs but the researchers determined that the top four most frequent SNVs in ctDNA from HCC patients were MLH1, STK11, PTEN, and CTNNB1. The biomarker potential of these four candidates was then validated in a separate cohort of 62 patients. The researchers found that the presence of MLH1 SNV and elevated ctDNA levels together predicted poor overall survival (Kim et al., 2020). Liquid biopsies hold incredible potential to aid the management and treatment of many cancers. However, this method has a long way to go before it can reach the bedside, including the need to improve sensitivity and specificity (Rebouissou & Nault, 2020).
5. Therapeutic implications of genetic heterogeneity in HCC
Tumor heterogeneity has important implications for predicting response to therapy. In general, tumors with a high degree of heterogeneity do not respond well to therapies because the selection pressure can lead to the expansion of resistant sub-clones or the emergence of new drug-resistant clones. HCC tumors are highly heterogeneous, as described in detail in Section 3. The study of the genomic landscape of HCC has revealed that HCC tumors often present with loss-of-function mutations in tumor-suppressor genes like P53, AXIN1, ARID1A, and TSC1/2 (The Cancer Genome Atlas Research Network, 2017; Guichard et al., 2012; Ho et al., 2017). While gain-of-function mutations result in activated pathways and present clear potential targets for therapy, loss-of-function mutations are more difficult to target therapeutically. Though a subset of patients has aberrant activation of pathways from TERT promoter and CTNNB1 mutations, these targets are largely considered undruggable (Chan et al., 2021). Thus, the high heterogeneity of HCC tumors combined with the lack of commonly mutated, druggable targets severely limits treatment options in HCC. For patients with early or intermediate stage HCC, local radio- or chemo-embolization, surgical resection, and liver transplantation are curative treatment options. However, for patients with advanced HCC that is inoperable, first-line therapy includes protein kinase inhibitors like sorafenib and lenvatinib which have limited survival benefits, high toxicity, and a high likelihood of resistance (Kudo et al., 2018; Llovet et al., 2008). There is a need to not only identify more therapeutic targets but also to select appropriate therapies for each patient’s specific tumor biology.
5.1. Personalized medicine
Affected pathways and druggable targets for individual patients have the potential to be discovered using a personalized medicine approach where the genomic and transcriptomic alterations from biopsy- or liquid biopsy-obtained cancer cells are investigated. Such findings can be used to select appropriate anti-tumor therapies. While individual patients may have druggable mutations, the use of bulk sequencing or single location biopsy approaches may make it difficult to disentangle whether the mutation is shared by cancer cells in different tumor regions. Thus, it is likely that truncal or driver mutations need to be targeted to account for otherwise heterogeneous mutations between subclones (Lohr et al., 2014; McGranahan et al., 2015).
While a personalized medicine approach could help select therapies for individual patients, several approaches have been suggested for treatment regimens to address tumor heterogeneity. McQuerry et al suggest that a strategy could be to sequentially cycle different treatments for tumors consisting of heterogeneous subclones (McQuerry, Chang, Bowtell, Cohen, & Bild, 2017). In patients with multiple tumor lesions, intrapatient intertumor heterogeneity can present as a mixed response to therapy with some tumors shrinking and others growing (McQuerry et al., 2017). In such cases, there may be a benefit in obtaining multiple and/or serial biopsies from a patient in order to select appropriate treatments. However, HCC, relative to other cancers, is unique in that there is currently no clinical indication to perform a tumor biopsy to confirm an HCC diagnosis (Dhanasekaran, 2021). This somewhat hinders the ability to obtain tissue samples to study tumor progression and evolution in the context of treatment response. An alternate approach could be to use liquid biopsy instead, though the low concentration of ctDNA and CTCs and the high sensitivity required for their detection may be limiting factors (Maravelia et al., 2021; Mocan et al., 2020).
5.2. Novel targets for therapies and combination therapies
The liver is a highly vascularized organ that is exposed to many antigens due to its physiological functions (Thomson & Knolle, 2010). This feature requires the liver to be immunologically tolerant, a characteristic that may impede anti-tumor immunity. In HCC following viral hepatitis infection, there is immune exhaustion as seen by an exhausted T cell phenotype that may further impede an anti-tumor immune response (Harding, El Dika, & Abou-Alfa, 2016). This unique immune environment makes immunotherapies, and specifically immune checkpoint inhibitors (ICIs) an exciting prospect for the treatment of HCC. Nivolumab and pembrolizumab are PD-1 receptor inhibitors that were both shown to have benefits for HCC patients as second-line treatment following sorafenib failure or toxicity (El-Khoueiry et al., 2017; Finn, Ryoo, et al., 2020; Zhu et al., 2018). However, as with most ICIs, nivolumab and pembrolizumab only show benefits for a limited subset of patients. These therapies had a 15-20% rate of objective remissions with only a 1-5% rate of complete response (Sangro et al., 2017; Zhu et al., 2018). Further, a study found that tumoral PD-L1 expression did not correlate with the effectiveness of PD-1 inhibition by nivolumab (El-Khoueiry et al., 2017). Thus, there is a pressing need to develop molecular classifications so that patients can be matched to therapies that they are more likely to benefit from.
Surgical resection, radio- or chemo-embolization, and liver transplantation are curative treatment options that remain the standard of care for patients with early or intermediate-stage HCC. An ongoing pilot study is investigating the use of perioperative immunotherapy as part of curative treatment for patients with resectable HCC (Kaseb et al., 2019). The clinical outcomes of perioperative immunotherapy remain to be seen, however, mass cytometry analysis of pre- and post-treatment lesions reveals an increase in two subsets of CD8 + T cells. One subset showed robust anti-tumor immunity with a twofold increase in CD45RO and a threefold increase in granzyme B which are markers of T cell activation. The other subset possessed immune-modulatory and suppressive phenotypes with the elevation of Foxp3 and PD-L1 expression (Kaseb et al., 2019). The immune profiling of patients in this study could prove to be a powerful tool for stratifying patient responses and aid prospective patient selection for perioperative immunotherapy in the future.
Single-agent therapies have not been able to bypass the median overall survival of less than one year (Abou-Alfa & Venook, 2013). As a result, combination therapies have been evaluated in different combinations. High intra-tumoral heterogeneity in patients with HCC may also be an indication for combination therapy over single-agent therapies (Barcena-Varela & Lujambio, 2021). Some of the currently investigated combination therapies include VEGF inhibitors with ICIs, TKIs with ICIs, and ICIs with other ICIs. The rationale for the combination of VEGF inhibitors with ICIs is as follows. It has been shown that anti-VEGF therapies inhibit angiogenesis, inducing hypoxia in the tumor, which in turn upregulates the immune checkpoint protein PD-L1 (Kimura et al., 2018). Further, VEGF inhibition may improve tumor-specific T cell activity (Noman et al., 2014). The combination of VEGF inhibitors and PD-1/PD-L1 inhibitors has already been shown to be beneficial in lung and genitourinary cancers, and likewise as first-line treatment in HCC as well through the combination of atezolizumab, a PD-L1 inhibitor, and bevacizumab, an anti-VEGF agent (Finn, Qin, et al., 2020; Reck et al., 2019; Rini et al., 2019). Further, combinations of immunotherapies such as anti-PD1/PD-L1 and CTLA4 are also being shown to be clinically beneficial in HCC (Sangro, Sarobe, Hervás-Stubbs, & Melero, 2021).
Chimeric antigen receptor (CAR) engineered T cell therapy may be an upcoming treatment option for HCC. Preclinical studies have shown the adoptive glypican-3 (GPC3) CAR T therapy in patient-derived xenografts (PDX) models slowed down tumor growth in tumors with high expression of GPC3 (Jiang et al., 2016). Other targets such as alpha-fetoprotein (AFP), human TERT, and melanoma antigen gene (MAGE3) are also being investigated. Early phase clinical trials for patients with HCC are currently identifying different CAR targets (NCT02905188, NCT03884751, NCT03980288, NCT03993743). Other immune-based therapies such as adoptive T cell transfer, vaccination and virotherapy, and the combination of locoregional therapies with ICIs are also being studied (Sangro et al., 2021).
While many treatment options and combinations emerge, there is still a need to fully understand how to sequence treatments. Further, many treatments only show responses in a subset of patients, likely due to inter-patient and intra-tumor heterogeneity. Thus, it is important to develop biomarkers to predict response to the array of treatments available, so that the most appropriate therapy can be selected. Achieving this necessitates the molecular profiling of patients in clinical trials so that clinical endpoints can be determined for distinct subgroups and reanalyzed in the future as more evidence emerges from basic and clinical research studies (Fig. 3).
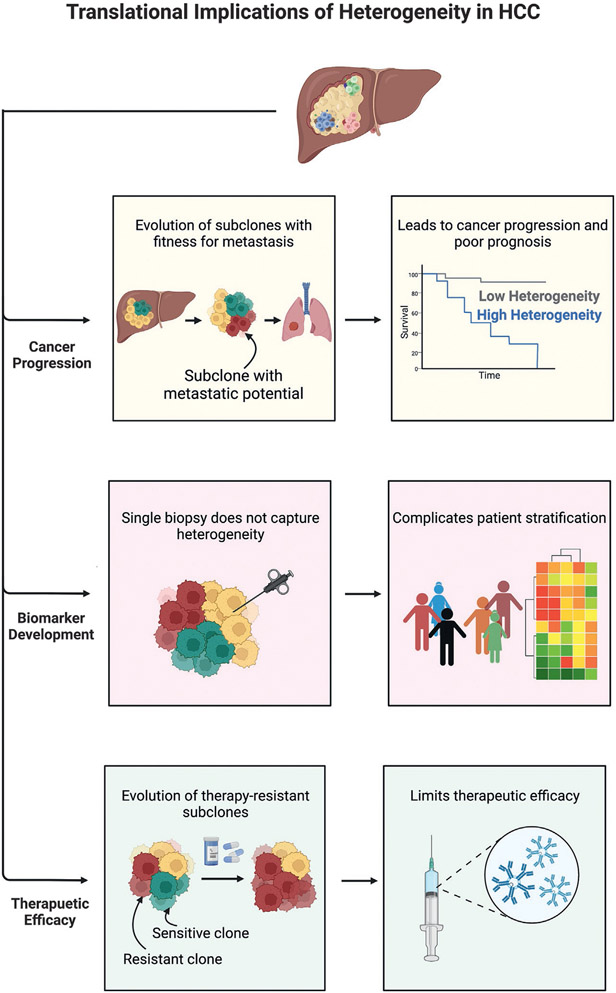
Fig. 3.
Translational implications of heterogeneity in hepatocellular carcinoma (HCC). HCC heterogeneity promotes cancer progression—multiple subclones are present and the evolution of subclones with metastatic potential can lead to metastasis. Consequently, patients with high heterogeneity have a greater degree of cancer progression and poorer prognoses. Heterogeneity impedes biomarker development—a single tissue biopsy often doesn’t capture tumor heterogeneity, and this limited insight into the tumor complicates patient stratification and biomarker development. Heterogeneity limits therapeutic efficacy—therapies can apply selective pressure on the tumor, leading to the evolution of resistant subclones.
6. Current perspectives and future directions
HCC exhibits a remarkable degree of heterogeneity not just in etiology and geographic prevalence, but also in the molecular signatures within and between tumors. The advent of next-generation sequencing-based technologies for multi-omic and single-cell analysis has greatly accelerated the understanding of tumor heterogeneity. Genome, epigenome, transcriptome, and proteome sequencing of serial and multi-region samples has allowed us to create high-resolution atlases of tumors and yielded several valuable insights into the spatial and temporal heterogeneity of HCC. However, an obstacle in the implementation of NGS sequencing for HCC tumors is that there is currently no clinical indication to biopsy an HCC tumor for diagnosis or treatment. This limits the systematic access to HCC tumor samples, and researchers who wish to study tumors must continue collecting tumor samples through approved research protocols with patient consent. Liquid biopsies are a developing area of interest and are a non-invasive approach to studying and monitoring tumor cells in circulation. However, issues related to sensitivity, specificity, and validity in larger cohorts need to be addressed before liquid biopsy can be adopted in clinical practice.
NGS sequencing has paved the way for the development of several classification systems for HCC. Tumors have been classified based on driver mutations, pathway activation, and immune infiltration. However, a classification system is yet to be approved for use in clinical care, whether to predict prognosis or decide a treatment course. Additional validation of classification systems is awaited for these tools to be translated.
Expanding therapeutic options for advanced HCC and developing guidelines to select appropriate therapies for each patient’s tumor biology remains an active area of research. Tumor heterogeneity is postulated to be a major reason for the failure of multiple therapies in HCC clinical trials. While there is no direct evidence to support this claim in HCC, experimental data have linked heterogeneity to drug resistance in multiple cancers (Dagogo-Jack & Shaw, 2018; Turner & Reis-Filho, 2012). Thus, there is a pressing need to explicitly study and account for tumor heterogeneity both in research and in clinical trials. For instance, research protocols that biopsy tumors before and after treatment could help identify tumor evolution and therapy resistance. Additionally, stratifying patients in clinical trials by genetic signatures or other biomarkers could help identify therapies that may provide benefit to patients with specific tumor biology, even if the average benefit across all patients is minimal.
Thus, single cell sequencing and spatial biology techniques have allowed the creation of high-resolution tumor landscapes that are ushering in a new era of personalized medicine. We believe that deciphering the molecular heterogeneity of HCC will ultimately lead to the discovery of novel biomarkers and therapeutic strategies, and thus improve the outcomes of patients with HCC.
Acknowledgements
All figures were created with BioRender.com.
Abbreviations
- CAF
cancer-associated fibroblast
- CTC
circulating tumor cell
- ctDNA
circulating tumor DNA
- EMT
epithelial-mesenchymal transformation
- HCC
hepatocellular carcinoma
- ICI
immune checkpoint inhibitor
- NGS
next-generation sequencing
- PDX
patient-derived xenograft
- SNV
single-nucleotide variation
- TGFβ
transforming growth factor-β
- TKI
tyrosine kinase inhibitor
- TLS
tertiary lymphoid structure
Footnotes
Conflict of interest statement
A.S. and R.D. have no financial or personal disclosures relevant to the contents of this manuscript.
References
- Abou-Alfa GK, & Venook AP (2013). The antiangiogenic ceiling in hepatocellular carcinoma: does it exist and has it been reached? The Lancet Oncology, 14(7), e283–e288. [DOI] [PubMed] [Google Scholar]
- Ahn S-M, Jang SJ, Shim JH, Kim D, Hong S-M, Sung CO, et al. (2014). Genomic portrait of resectable hepatocellular carcinomas: implications of RB1 and FGF19 aberrations for patient stratification. Hepatology, 60(6), 1972–1982. [DOI] [PubMed] [Google Scholar]
- Alsina-Sanchis E, Mülfarth R, & Fischer A (2021). Control of Tumor Progression by Angiocrine Factors. Cancers, 13(11). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arao T, Ueshima K, Matsumoto K, Nagai T, Kimura H, Hagiwara S, et al. (2013). FGF3/FGF4 amplification and multiple lung metastases in responders to sorafenib in hepatocellular carcinoma. Hepatology, 57(4), 1407–1415. [DOI] [PubMed] [Google Scholar]
- Asakawa M, Itoh M, Suganami T, Sakai T, Kanai S, Shirakawa I, et al. (2019). Upregulation of cancer-associated gene expression in activated fibroblasts in a mouse model of non-alcoholic steatohepatitis. Scientific Reports, 9(1), 19601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barcena-Varela M, & Lujambio A (2021). The Endless Sources of Hepatocellular Carcinoma Heterogeneity. Cancers, 13(11). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biffi G, Oni TE, Spielman B, Hao Y, Elyada E, Park Y, et al. (2019). IL1-Induced JAK/STAT Signaling Is Antagonized by TGFβ to Shape CAF Heterogeneity in Pancreatic Ductal Adenocarcinoma. Cancer Discovery, 9(2), 282–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumer T, Fofana I, Matter MS, Wang X, Montazeri H, Calabrese D, et al. (2019). Hepatocellular Carcinoma Xenografts Established From Needle Biopsies Preserve the Characteristics of the Originating Tumors. Hepatology Communications, 3(7), 971–986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyault S, Rickman DS, de Reyniès A, Balabaud C, Rebouissou S, Jeannot E, et al. (2007). Transcriptome classification of HCC is related to gene alterations and to new therapeutic targets. Hepatology, 45(1), 42–52. [DOI] [PubMed] [Google Scholar]
- Brady L, Kriner M, Coleman I, Morrissey C, Roudier M, True LD, et al. (2021). Inter- and intra-tumor heterogeneity of metastatic prostate cancer determined by digital spatial gene expression profiling. Nature Communications, 12(1), 1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown ZJ,Yu SJ, Heinrich B, Ma C, Fu Q, Sandhu M, et al. (2018). Indoleamine 2,3-dioxygenase provides adaptive resistance to immune checkpoint inhibitors in hepatocellular carcinoma. Cancer Immunology, Immunotherapy: CII, 67(8), 1305–1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Z-X, Chen G, Zeng Y-Y, Dong X-Q, Lin M-J, Huang X-H, et al. (2017). Circulating tumor DNA profiling reveals clonal evolution and real-time disease progression in advanced hepatocellular carcinoma. International Journal of Cancer. Journal International Du Cancer, 141(5), 977–985. [DOI] [PubMed] [Google Scholar]
- Calderaro J, Couchy G, Imbeaud S, Amaddeo G, Letouzé E, Blanc J-F, et al. (2017). Histological subtypes of hepatocellular carcinoma are related to gene mutations and molecular tumour classification. Journal of Hepatology, 67(4), 727–738. [DOI] [PubMed] [Google Scholar]
- Calderaro J, Petitprez F, Becht E, Laurent A, Hirsch TZ, Rousseau B, et al. (2019). Intra-tumoral tertiary lymphoid structures are associated with a low risk of early recurrence of hepatocellular carcinoma. Journal of Hepatology, 70(1), 58–65. [DOI] [PubMed] [Google Scholar]
- Calvisi DF, Ladu S, Gorden A, Farina M, Lee J-S, Conner EA, et al. (2007). Mechanistic and prognostic significance of aberrant methylation in the molecular pathogenesis of human hepatocellular carcinoma. The Journal of Clinical Investigation, 117(9), 2713–2722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan L-K, Tsui Y-M, Ho DW-H, & Ng IO-L (2021). Cellular heterogeneity and plasticity in liver cancer. Seminars in Cancer Biology. [DOI] [PubMed] [Google Scholar]
- Chen H, Chong W, Teng C, Yao Y, Wang X, & Li X (2019). The immune response-related mutational signatures and driver genes in non-small-cell lung cancer. Cancer Science, 110(8), 2348–2356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F, Zhong Z, Tan H-Y, Wang N, & Feng Y (2020). The significance of circulating tumor cells in patients with hepatocellular carcinoma: Real-time monitoring and moving targets for cancer therapy. Cancers, 12(7). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y, Li H, Deng Y, Tai Y, Zeng K, Zhang Y, et al. (2018). Cancer-associated fibroblasts induce PDL1+ neutrophils through the IL6-STAT3 pathway that foster immune suppression in hepatocellular carcinoma. Cell Death & Disease, 9(4), 422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang DY, Villanueva A, Hoshida Y, Peix J, Newell P, Minguez B, et al. (2008). Focal gains of VEGFA and molecular classification of hepatocellular carcinoma. Cancer Research, 68(16), 6779–6788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dagogo-Jack I, & Shaw AT (2018). Tumour heterogeneity and resistance to cancer therapies. Nature Reviews. Clinical Oncology, 15(2), 81–94. [DOI] [PubMed] [Google Scholar]
- Dasgupta P, Henshaw C, Youlden D, Clark P, Aitken J, & Baade P (2020). Global trends in incidence rates of primary adult liver cancers: A systematic review and meta-analysis. Frontiers in Oncology, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Désert R, Rohart F, Canal F, Sicard M, Desille M, Renaud S, et al. (2017). Human hepatocellular carcinomas with a periportal phenotype have the lowest potential for early recurrence after curative resection. Hepatology, 66(5), 1502–1518. [DOI] [PubMed] [Google Scholar]
- Dhanasekaran R. (2021). Deciphering tumor heterogeneity in hepatocellular carcinoma (HCC)-multi-omic and singulomic approaches. Seminars in Liver Disease, 41(1), 9–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhanasekaran R, Bandoh S, & Roberts LR (2016). Molecular pathogenesis of hepatocellular carcinoma and impact of therapeutic advances. F1000Research, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding X, He M, Chan AWH, Song QX, Sze SC, Chen H, et al. (2019). Genomic and epigenomic features of primary and recurrent hepatocellular carcinomas. Gastroenterology, 157(6). 1630–1645.e6. [DOI] [PubMed] [Google Scholar]
- El-Khoueiry AB, Sangro B, Yau T, Crocenzi TS, Kudo M, Hsu C, et al. (2017). Nivolumab in patients with advanced hepatocellular carcinoma (CheckMate 040): an open-label, non-comparative, phase 1/2 dose escalation and expansion trial. The Lancet, 389(10088), 2492–2502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang J-H, Xu L, Shang L-R, Pan C-Z, Ding J, Tang Y-Q, et al. (2019). Vessels that encapsulate tumor clusters (VETC) pattern is a predictor of sorafenib benefit in patients with hepatocellular carcinoma. Hepatology, 70(3), 824–839. [DOI] [PubMed] [Google Scholar]
- Fang J-H, Zhou H-C, Zhang C, Shang L-R, Zhang L, Xu J, et al. (2015). A novel vascular pattern promotes metastasis of hepatocellular carcinoma in an epithelial-mesenchymal transition-independent manner. Hepatology, 62(2), 452–465. [DOI] [PubMed] [Google Scholar]
- Finn RS, Qin S, Ikeda M, Galle PR, Ducreux M, Kim T-Y, et al. (2020). Atezolizumab plus Bevacizumab in Unresectable Hepatocellular Carcinoma. The New England Journal of Medicine, 382(20), 1894–1905. [DOI] [PubMed] [Google Scholar]
- Finn RS, Ryoo B-Y, Merle P, Kudo M, Bouattour M, Lim HY, et al. (2020). Pembrolizumab as second-line therapy in patients with advanced hepatocellular carcinoma in KEYNOTE-240: A randomized, double-blind, phase III trial. Journal of Clinical Oncology: Official Journal of the American Society of Clinical Oncology, 38(3), 193–202. [DOI] [PubMed] [Google Scholar]
- Forschner A, Battke F, Hadaschik D, Schulze M, Weißgraeber S, Han C-T, et al. (2019). Tumor mutation burden and circulating tumor DNA in combined CTLA-4 and PD-1 antibody therapy in metastatic melanoma - results of a prospective biomarker study. Journal for Immunotherapy of Cancer, 7(1), 180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimoto A, Totoki Y,Abe T, Boroevich KA, Hosoda F, Nguyen HH, et al. (2012). Whole-genome sequencing of liver cancers identifies etiological influences on mutation patterns and recurrent mutations in chromatin regulators. Nature Genetics, 44(7), 760–764. [DOI] [PubMed] [Google Scholar]
- Fujiwara N, Friedman S, Goossens N, & Hoshida Y (2018). Risk factors and prevention of hepatocellular carcinoma in the era of precision medicine. Journal of Hepatology, 68(3), 526–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Q, Wang Z-C, Duan M, Lin Y-H, Zhou X-Y, Worthley DL, et al. (2017). Cell culture system for analysis of genetic heterogeneity within hepatocellular carcinomas and response to pharmacologic agents. Gastroenterology, 152(1). 232–242.e4. [DOI] [PubMed] [Google Scholar]
- Garnelo M, Tan A, Her Z, Yeong J, Lim CJ, Chen J, et al. (2017). Interaction between tumour-infiltrating B cells and T cells controls the progression of hepatocellular carcinoma. Gut, 66(2), 342–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghouri YA, Mian I, & Rowe JH (2017). Review of hepatocellular carcinoma: Epidemiology, etiology, and carcinogenesis. Journal of Carcinogenesis, 16(1), 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guichard C, Amaddeo G, Imbeaud S, Ladeiro Y, Pelletier L, Maad IB, et al. (2012). Integrated analysis of somatic mutations and focal copy-number changes identifies key genes and pathways in hepatocellular carcinoma. Nature Genetics, 44(6), 694–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L, Yi X, Chen L, Zhang T, Guo H, Chen Z, et al. (2022). Single-cell DNA sequencing reveals punctuated and gradual clonal evolution in hepatocellular carcinoma. Gastroenterology, 162(1), 238–252. [DOI] [PubMed] [Google Scholar]
- Harding JJ, El Dika I, & Abou-Alfa GK (2016). Immunotherapy in hepatocellular carcinoma: Primed to make a difference? Cancer, 122(3), 367–377. [DOI] [PubMed] [Google Scholar]
- Harding JJ, Nandakumar S, Armenia J, Khalil DN, Albano M, Ly M, et al. (2019). Prospective genotyping of hepatocellular carcinoma:Clinical implications of next-generation sequencing for matching patients to targeted and immune therapies. Clinical Cancer Research: An Official Journal of the American Association for Cancer Research, 25(7), 2116–2126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho DWH, Chan LK, Chiu YT, Xu IMJ, Poon RTP, Cheung TT, et al. (2017). TSC1/2 mutations define a molecular subset of HCC with aggressive behaviour and treatment implication. Gut, 66(8), 1496–1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horwitz E, Stein I, Andreozzi M, Nemeth J, Shoham A, Pappo O, et al. (2014). Human and mouse VEGFA-amplified hepatocellular carcinomas are highly sensitive to sorafenib treatment. Cancer Discovery, 4(6), 730–743. [DOI] [PubMed] [Google Scholar]
- Hoshida Y, Nijman SMB, Kobayashi M, Chan JA, Brunet J-P, Chiang DY, et al. (2009). Integrative transcriptome analysis reveals common molecular subclasses of human hepatocellular carcinoma. Cancer Research, 69(18), 7385–7392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshida Y, Toffanin S, Lachenmayer A, Villanueva A, Minguez B, & Llovet JM (2010). Molecular classification and novel targets in hepatocellular carcinoma: recent advancements. Seminars in Liver Disease, 30(1), 35–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu B,Li H, Guo W, Sun Y-F, Zhang X, Tang W-G, et al. (2020). Establishment of a hepatocellular carcinoma patient-derived xenograft platform and its application in biomarker identification. International Journal of Cancer. Journal International Du Cancer, 146(6), 1606–1617. [DOI] [PubMed] [Google Scholar]
- Hung MH, Lee JS, Ma C, Diggs LP, Heinrich S, Chang CW, et al. (2021). Tumor methionine metabolism drives T-cell exhaustion in hepatocellular carcinoma. Nature Communications, 12(1), 1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y, Han Q, Zhao H, & Zhang J (2021). The mechanisms of HBV-induced hepatocellular carcinoma. Journal of Hepatocellular Carcinoma, 8, 435–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Z,Jiang X, Chen S, Lai Y, Wei X, Li B, et al. (2016). Anti-GPC3-CAR T cells suppress the growth of tumor cells in patient-derived xenografts of hepatocellular carcinoma. Frontiers in Immunology, 7, 690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaseb AO, Vence L, Blando J, Yadav SS, Ikoma N, Pestana RC, et al. (2019). Immunologic correlates of pathologic complete response to preoperative immunotherapy in hepatocellular carcinoma. Cancer Immunology Research, 7(9), 1390–1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khatib S, Pomyen Y, Dang H, & Wang XW (2020). Understanding the cause and consequence of tumor heterogeneity. Trends in Cancer Research, 6(4), 267–271. [DOI] [PubMed] [Google Scholar]
- Kim SS, Eun JW, Choi J-H, Woo HG, Cho HJ, Ahn HR, et al. (2020). MLH1 single-nucleotide variant in circulating tumor DNA predicts overall survival of patients with hepatocellular carcinoma. Scientific Reports, 10(1), 17862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura T, Kato Y, Ozawa Y, Kodama K, Ito J, Ichikawa K, et al. (2018). Immunomodulatory activity of lenvatinib contributes to antitumor activity in the Hepa1-6 hepatocellular carcinoma model. Cancer Science, 109(12), 3993–4002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudo M, Finn RS, Qin S, Han K-H, Ikeda K, Piscaglia F, et al. (2018). Lenvatinib versus sorafenib in first-line treatment of patients with unresectable hepatocellular carcinoma: A randomised phase 3 non-inferiority trial. The Lancet, 391(10126), 1163–1173. [DOI] [PubMed] [Google Scholar]
- Kurebayashi Y, Ojima H, Tsujikawa H, Kubota N, Maehara J, Abe Y, et al. (2018). Landscape of immune microenvironment in hepatocellular carcinoma and its additional impact on histological and molecular classification. Hepatology, 68(3), 1025–1041. [DOI] [PubMed] [Google Scholar]
- Lachenmayer A, Alsinet C, Savic R, Cabellos L, Toffanin S, Hoshida Y, et al. (2012). Wnt-pathway activation in two molecular classes of hepatocellular carcinoma and experimental modulation by sorafenib. Clinical Cancer Research: An Official Journal of the American Association for Cancer Research, 18(18), 4997–5007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J-S, Chu I-S, Heo J, Calvisi DF, Sun Z, Roskams T, et al. (2004). Classification and prediction of survival in hepatocellular carcinoma by gene expression profiling. Hepatology, 40(3), 667–676. [DOI] [PubMed] [Google Scholar]
- Lee M, Samstein RM, Valero C, Chan TA, & Morris LGT (2020). Tumor mutational burden as a predictive biomarker for checkpoint inhibitor immunotherapy. Human Vaccines & Immunotherapeutics, 16(1), 112–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Chen L, Zhang X, Zhang Y, Liu H, Sun B, et al. (2014). Detection of circulating tumor cells in hepatocellular carcinoma using antibodies against asialoglycoprotein receptor, carbamoyl phosphate synthetase 1 and pan-cytokeratin. PLoS One, 9(4), e96185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling S, Hu Z, Yang Z, Yang F, Li Y, Lin P, et al. (2015). Extremely high genetic diversity in a single tumor points to prevalence of non-Darwinian cell evolution. Proceedings of the National Academy of Sciences of the United States of America, 112(47), E6496–E6505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Chen S, Wang W, Ning B-F, Chen F, Shen W, et al. (2016). Cancer-associated fibroblasts promote hepatocellular carcinoma metastasis through chemokine-activated hedgehog and TGF-β pathways. Cancer Letters, 379(1), 49–59. [DOI] [PubMed] [Google Scholar]
- Llovet JM, Kelley RK, Villanueva A, Singal AG, Pikarsky E, Roayaie S, et al. (2021). Hepatocellular carcinoma. Nature Reviews. Disease Primers, 7(1), 6. [DOI] [PubMed] [Google Scholar]
- Llovet JM, Ricci S, Mazzaferro V, Hilgard P, Gane E, Blanc J-F, et al. (2008). Sorafenib in advanced hepatocellular carcinoma. The New England Journal of Medicine, 359(4), 378–390. [DOI] [PubMed] [Google Scholar]
- Lohr JG, Stojanov P, Carter SL, Cruz-Gordillo P, Lawrence MS, Auclair D, et al. (2014). Widespread genetic heterogeneity in multiple myeloma: implications for targeted therapy. Cancer Cell, 25(1), 91–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longo SK, Guo MG, Ji AL, & Khavari PA (2021). Integrating single-cell and spatial transcriptomics to elucidate intercellular tissue dynamics. Nature Reviews. Genetics, 22(10), 627–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losic B, Craig AJ, Villacorta-Martin C, Martins-Filho SN, Akers N, Chen X, et al. (2020). Intratumoral heterogeneity and clonal evolution in liver cancer. Nature Communications, 11(1), 291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maishi N, Ohba Y, Akiyama K, Ohga N, Hamada J-I, Nagao-Kitamoto H, et al. (2016). Tumour endothelial cells in high metastatic tumours promote metastasis via epigenetic dysregulation of biglycan. Scientific Reports, 6, 28039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maravelia P, Silva DN, Rovesti G, Chrobok M, Stål P, Lu Y-C, et al. (2021). Liquid biopsy in hepatocellular carcinoma: Opportunities and challenges for immunotherapy. Cancers, 13(17). [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy MG, Nyanyo D, Hung CK, Goerger JP, R Zipfel W, Williams RM, et al. (2019). Endothelial cells promote 3D invasion of GBM by IL-8-dependent induction of cancer stem cell properties. Scientific Reports, 9(1), 9069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGranahan N, Favero F, de Bruin EC, Birkbak NJ, Szallasi Z, & Swanton C (2015). Clonal status of actionable driver events and the timing of mutational processes in cancer evolution. Science Translational Medicine, 7(283), 283ra54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McQuerry JA, Chang JT, Bowtell DDL, Cohen A, & Bild AH (2017). Mechanisms and clinical implications of tumor heterogeneity and convergence on recurrent phenotypes. Journal of Molecular Medicine, 95(11), 1167–1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mocan T, Simão AL, Castro RE, Rodrigues CMP, Słomka A, Wang B, et al. (2020). Liquid biopsies in hepatocellular carcinoma: Are we winning? Journal of Clinical Medicine Research, 9(5). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nault JC, Mallet M, Pilati C, Calderaro J, Bioulac-Sage P, Laurent C, et al. (2013). High frequency of telomerase reverse-transcriptase promoter somatic mutations in hepatocellular carcinoma and preneoplastic lesions. Nature Communications, 4, 2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nault J-C, Martin Y, Caruso S, Hirsch TZ, Bayard Q, Calderaro J, et al. (2020). Clinical Impact of Genomic Diversity From Early to Advanced Hepatocellular Carcinoma. Hepatology, 71(1), 164–182. [DOI] [PubMed] [Google Scholar]
- Noman MZ, Desantis G, Janji B, Hasmim M, Karray S, Dessen P, et al. (2014). PD-L1 is a novel direct target of HIF-1α, and its blockade under hypoxia enhanced MDSC-mediated T cell activation. The Journal of Experimental Medicine, 211(5), 781–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Öhlund D, Handly-Santana A, Biffi G, Elyada E, Almeida AS, Ponz-Sarvise M, et al. (2017). Distinct populations of inflammatory fibroblasts and myofibroblasts in pancreatic cancer. The Journal of Experimental Medicine, 214(3), 579–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng S, Wang Y, Peng H, Chen D, Shen S, Peng B, et al. (2014). Autocrine vascular endothelial growth factor signaling promotes cell proliferation and modulates sorafenib treatment efficacy in hepatocellular carcinoma. Hepatology, 60(4), 1264–1277. [DOI] [PubMed] [Google Scholar]
- Petrick JL, Florio AA, Znaor A, Ruggieri D, Laversanne M, Alvarez CS, et al. (2020). International trends in hepatocellular carcinoma incidence, 1978–2012. International Journal of Cancer. Journal International Du Cancer, 147(2), 317–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plaks V, Koopman CD, & Werb Z (2013). Cancer. Circulating tumor cells. Science, 341(6151), 1186–1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebouissou S, & Nault J-C (2020). Advances in molecular classification and precision oncology in hepatocellular carcinoma. Journal of Hepatology, 72(2), 215–229. [DOI] [PubMed] [Google Scholar]
- Reck M, Mok TSK, Nishio M, Jotte RM, Cappuzzo F, Orlandi F, et al. (2019). Atezolizumab plus bevacizumab and chemotherapy in non-small-cell lung cancer (IMpower150): key subgroup analyses of patients with EGFR mutations or baseline liver metastases in a randomised, open-label phase 3 trial. The Lancet. Respiratory Medicine, 7(5), 387–401. [DOI] [PubMed] [Google Scholar]
- Renne SL, Woo HY, Allegra S, Rudini N, Yano H, Donadon M, et al. (2020). Vessels encapsulating tumor clusters (VETC) is a powerful predictor of aggressive hepatocellular carcinoma. Hepatology, 71(1), 183–195. [DOI] [PubMed] [Google Scholar]
- Rini BI, Powles T, Atkins MB, Escudier B, McDermott DF, Suarez C, et al. (2019). Atezolizumab plus bevacizumab versus sunitinib in patients with previously untreated metastatic renal cell carcinoma (IMmotion151): a multicentre, open-label, phase 3, randomised controlled trial. The Lancet, 393(10189), 2404–2415. [DOI] [PubMed] [Google Scholar]
- Rudalska R, Dauch D, Longerich T, McJunkin K, Wuestefeld T, Kang T-W, et al. (2014). In vivo RNAi screening identifies a mechanism of sorafenib resistance in liver cancer. Nature Medicine, 20(10), 1138–1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sangro B, Melero I, Yau T, Hsu C, Kudo M, Kim T-Y, et al. (2017). Nivolumab in sorafenib-naive and-experienced patients with advanced hepatocellular carcinoma (HCC): Survival, hepatic safety, and biomarker assessments in CheckMate 040. [DOI] [PubMed] [Google Scholar]
- Sangro B, Sarobe P, Hervás-Stubbs S, & Melero I (2021). Advances in immunotherapy for hepatocellular carcinoma. Nature Reviews. Gastroenterology & Hepatology, 18(8), 525–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze K, Imbeaud S, Letouzé E, Alexandrov LB, Calderaro J, Rebouissou S, et al. (2015). Exome sequencing of hepatocellular carcinomas identifies new mutational signatures and potential therapeutic targets. Nature Genetics, 47(5), 505–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebastian A, Hum NR, Martin KA, Gilmore SF, Peran I, Byers SW, et al. (2020). Single-cell transcriptomic analysis of tumor-derived fibroblasts and normal tissue-resident fibroblasts reveals fibroblast heterogeneity in breast cancer. Cancers, 12(5). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma A, Seow JJW, Dutertre C-A, Pai R, Blériot C, Mishra A, et al. (2020). Onco-fetal reprogramming of endothelial cells drives immunosuppressive macrophages in hepatocellular carcinoma. Cell, 183(2). 377–394.e21. [DOI] [PubMed] [Google Scholar]
- Sia D, Jiao Y, Martinez-Quetglas I, Kuchuk O, Villacorta-Martin C, Castro de Moura M, et al. (2017). Identification of an immune-specific class of hepatocellular carcinoma, based on molecular features. Gastroenterology, 153(3), 812–826. [DOI] [PubMed] [Google Scholar]
- Siegel RL, Miller KD, & Jemal A (2020). Cancer statistics, 2020. CA: a Cancer Journal for Clinicians, 70(1), 7–30. [DOI] [PubMed] [Google Scholar]
- Siu EH-L, Chan AW-H, Chong CC-N, Chan SL, Lo K-W, & Cheung ST (2018). Treatment of advanced hepatocellular carcinoma: immunotherapy from checkpoint blockade to potential of cellular treatment. Translational Gastroenterology and Hepatology, 3, 89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song G, Shi Y, Zhang M, Goswami S, Afridi S, Meng L, et al. (2020). Global immune characterization of HBV/HCV-related hepatocellular carcinoma identifies macrophage and T-cell subsets associated with disease progression. Cell Discovery, 6(1), 90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sukowati CHC, El-Khobar KE, & Tiribelli C (2021). Immunotherapy against programmed death-1/programmed death ligand 1 in hepatocellular carcinoma: Importance of molecular variations, cellular heterogeneity, and cancer stem cells. World Journal of Stem Cells, 13(7), 795–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y-F, Guo W, Xu Y, Shi Y-H, Gong Z-J, Ji Y, et al. (2018). Circulating tumor cells from different vascular sites exhibit spatial heterogeneity in epithelial and mesenchymal composition and distinct clinical significance in hepatocellular carcinoma. Clinical Cancer Research: An Official Journal of the American Association for Cancer Research, 24(3), 547–559. [DOI] [PubMed] [Google Scholar]
- Sun B, & Karin M (2012). Obesity, inflammation, and liver cancer. Journal of Hepatology, 56(3), 704–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. (2021). Global Cancer Statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA: a Cancer Journal for Clinicians, 71(3), 209–249. [DOI] [PubMed] [Google Scholar]
- Swanton C. (2012). Intratumor heterogeneity: Evolution through space and time. Cancer Research, 72(19), 4875–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teufel M, Seidel H, Köchert K, Meinhardt G, Finn RS, Llovet JM, et al. (2019). Biomarkers associated with response to regorafenib in patients with hepatocellular carcinoma. Gastroenterology, 156(6), 1731–1741. [DOI] [PubMed] [Google Scholar]
- The Cancer Genome Atlas Research Network. (2017). Comprehensive and Integrative Genomic Characterization of Hepatocellular Carcinoma. Cell, 169(7), 1327–1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson AW, & Knolle PA (2010). Antigen-presenting cell function in the tolerogenic liver environment. Nature Reviews. Immunology, 10(11), 753–766. [DOI] [PubMed] [Google Scholar]
- Thun M, Linet MS, Cerhan JR, Haiman CA, & Schottenfeld D (2017). Cancer Epidemiology and Prevention. Oxford University Press. [Google Scholar]
- Totoki Y, Tatsuno K, Covington KR, Ueda H, Creighton CJ, Kato M, et al. (2014). Trans-ancestry mutational landscape of hepatocellular carcinoma genomes. Nature Genetics, 46(12), 1267–1273. [DOI] [PubMed] [Google Scholar]
- Turner NC, & Reis-Filho JS (2012). Genetic heterogeneity and cancer drug resistance. The Lancet Oncology, 13(4), e178–e185. [DOI] [PubMed] [Google Scholar]
- Tuveson D, & Clevers H (2019). Cancer modeling meets human organoid technology. Science, 364(6444), 952–955. [DOI] [PubMed] [Google Scholar]
- Villanueva A. (2019). Hepatocellular Carcinoma. The New England Journal of Medicine, 380, 1450–1462. [DOI] [PubMed] [Google Scholar]
- World Health Organization. (2018). Noncommunicable diseases country profiles 2018. https://apps.who.int/iris/bitstream/handle/10665/274512/9789241514620-eng.pdf.
- Wu R, Guo W, Qiu X, Wang S, Sui C, Lian Q, et al. (2021). Comprehensive analysis of spatial architecture in primary liver cancer. Science Advances, 7(51). eabg3750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C, Lin J, Weng Y, Zeng D-N, Xu J, Luo S, et al. (2020). Myeloid signature reveals immune contexture and predicts the prognosis of hepatocellular carcinoma. The Journal of Clinical Investigation, 130(9), 4679–4693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Tao Y, Shan L, Chen R, Jiang H, Qian Z, et al. (2018). The role of MicroRNAs in hepatocellular carcinoma. Journal of Cancer, 9(19), 3557–3569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue R, Li R, Guo H, Guo L, Su Z, Ni X, et al. (2016). Variable intra-tumor genomic heterogeneity of multiple lesions in patients with hepatocellular carcinoma. Gastroenterology, 150(4), 998–1008. [DOI] [PubMed] [Google Scholar]
- Yang JD, Hainaut P, Gores GJ, Amadou A, Plymoth A, & Roberts LR (2019). A global view of hepatocellular carcinoma: Trends, risk, prevention and management. Nature Reviews Gastroenterology & Hepatology, 16, 589–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Z, Dong C, Jiang K, Xu Z, Li R, Guo K, et al. (2019). Heterogeneity of cancer-associated fibroblasts and roles in the progression, prognosis, and therapy of hepatocellular carcinoma. Journal of Hematology & Oncology, 12(1), 101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai W, Lim TK-H, Zhang T, Phang S-T, Tiang Z, Guan P, et al. (2017). The spatial organization of intra-tumour heterogeneity and evolutionary trajectories of metastases in hepatocellular carcinoma. Nature Communications, 8, 4565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, He Y, Luo N, Patel SJ, Han Y, Goa R, et al. (2019). Landscape and dynamics of single immune cells in hepatocellular carcinoma. Cell, 179(4), 829–845. [DOI] [PubMed] [Google Scholar]
- Zhang Q, Lou Y, Yang J, Wang J, Feng J, Zhao Y, et al. (2019). Integrated multiomic analysis reveals comprehensive tumour heterogeneity and novel immunophenotypic classification in hepatocellular carcinomas. Gut, 68(11), 2019–2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu Z, Ji K, Li X, Wang C, Ren Z, et al. (2021). Clinical application value of circulating cell-free DNA in hepatocellular carcinoma. Frontiers in Molecular Biosciences, 8, 736330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Li Z-X, Zhu Y-J, Fu J, Zhao X-F, Zhang Y-N, et al. (2021). Single-cell transcriptome analysis uncovers intratumoral heterogeneity and underlying mechanisms for drug resistance in hepatobiliary tumor organoids. Advancement of Science, 8(11), e2003897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu AX, Finn RS, Edeline J, Cattan S, Ogasawara S, Palmer D, et al. (2018). Pembrolizumab in patients with advanced hepatocellular carcinoma previously treated with sorafenib (KEYNOTE-224): a non-randomised, open-label phase 2 trial. The Lancet Oncology, 19(7), 940–952. [DOI] [PubMed] [Google Scholar]
- Zhu AX, Kang Y-K, Yen C-J, Finn RS, Galle PR, Llovet JM, et al. (2019). Ramucirumab after sorafenib in patients with advanced hepatocellular carcinoma and increased α-fetoprotein concentrations (REACH-2): a randomised, double-blind, placebo-controlled, phase 3 trial. The Lancet Oncology, 20(2), 282–296. [DOI] [PubMed] [Google Scholar]
- Zhu J, Zhang T, Li J, Lin J, Liang W, Huang W, et al. (2019). Association between tumor mutation burden (TMB) and outcomes of cancer patients treated with PD-1/PD-L1 inhibitions: A meta-analysis. Frontiers in Pharmacology, 10, 673. [DOI] [PMC free article] [PubMed] [Google Scholar]