Abstract
Genomic alterations (GA) in NF2 tumor-suppressor gene have been associated with aggressive behavior in kidney tumors. We used comprehensive genomic profiling (CGP) to evaluate the frequencies of NF2 GA in histologic subtypes of kidney tumors and co-occurring GA in other genes and biomarkers. Advanced kidney tumors included 1875 clear cell (ccRCC), 405 papillary (pRCC), 108 chromophobe (chRCC), 171 sarcomatoid (sRCC), 61 collecting duct (cdRCC), 49 medullary (mRCC), 134 unclassified (uRCC), 906 urothelial carcinoma of renal pelvis (UC), and 147 Wilms tumors underwent hybrid-capture based CGP to evaluate all classes of GA. 192 (4.9%) of kidney tumors featured NF2 GA which were predominantly structural variant mutations (89%), followed by copy number alterations (9%). Gender and age were similar between NF2-mutant (NF2mut) and NF2-wild type (NF2wt) cohorts with male preponderance. NF2 GA frequency was highest in cdRCC (30%), sRCC (21%), uRCC (15%), and pRCC (12%) while lowest in ccRCC (3%), UC (3%) Wilms tumor (1%), and chRCC (0%). NF2 mutational status was associated with loss of Ch 22 (P < .001). NF2mut RCC harbored co-occurring GA including CDKN2A, CDKN2B, SETD2, and BAP1. VHL, PBRM1, PTEN, and FGFR3 GA were significantly more frequent in NF2wt than in NF2mut tumors. MTOR pathway GAs were uncommon in NF2mut tumors. No NF2 mutated RCC featured MSI-high or high TMB. sRCC was associated with high PD-L1 expression. PD-L1 SP142 tumoral (P = .04) and immune cells (P = .013) were more frequent in NF2mut as compared to NF2wt group. Among histologic subtypes of RCC, cdRCC, sRCC, pRCC, and uRCC are enriched in NF2 GA. Co-occurrent GA in CDKN2A/B, SETD2, and BAP1 may represent potential therapeutic targets. Higher level of PD-L1 expression in NF2mut cohort suggests that these tumors might be sensitive to immune checkpoint inhibitor therapies.
Keywords: NF2, genomic alteration, kidney tumors, renal cell carcinoma, comprehensive genomic profiling, PD-L1
In this study, comprehensive genomic profiling (CGP) was used to evaluate the frequency of NF2 genomic alterations in histologic subtypes of kidney tumors and co-occurring genomic alterations in other genes and biomarkers.
Implications for Practice.
In this study, 192 (4.9%) kidney tumors that featured NF2 genomic alterations (GA) were found. Among histologic subtypes of renal cell carcinoma (RCC), aggressive variants, such as collecting duct RCC, sarcomatoid RCC, papillary RCC, and unclassified RCC were found to be enriched in NF2 GA (30%, 21%, 12%, and 15%, respectively). In these RCC subtypes, NF2 genomic alteration appears to serve as predominant driver mutation, with corresponding suppression of additional driver mutations in the MTOR pathway and other targetable kinases. Co-occurrent GA in CDKN2A/B, SETD2, and BAP1 may represent potential therapeutic targets. The higher level of PD-L1 expression seen in NF2-mutated kidney tumors suggests they may be sensitive to immune checkpoint inhibitor therapies.
Introduction
Kidney tumors are heterogeneous and categorized by distinct histopathological features and genomic alterations.1 Renal cell carcinoma (RCC) is a the most common kidney malignancy and is classified into clear cell RCC (ccRCC, 75%), and more rare histologic variants collectively grouped as non-clearcell RCC (nccRCC, 25%). nccRCC include papillary RCC (pRCC, 15%), chromophobe RCC (chRCC, 5%), unclassified RCC (uRCC, 5%), and other rare subtypes such as medullary (mRCC, <1%) and collecting duct (cdRCC, <1%).1. Sarcomatoid differentiation (sRCC) is a morphologic change that can be seen in all subtypes and typically portends a poor prognosis.2
Molecular profiles have long been known to correlate with histologic kidney cancer subtypes.3 There is a well-established genotype-phenotype association between VHL alterations and ccRCC, cMET protooncogene activation in low grade pRCC, fumarate hydratase (FH) inactivating mutations and hereditary leiomyomatosis and RCC (HLRCC) syndrome-associated renal cancer, succinate dehydrogenase (SDH)-inactivating mutations and SDH-deficient RCC, amongst others.4 In the most recent 2022 WHO classification of renal tumors, molecularly driven subtypes have been introduced including SMARCB1-deficient medullary RCC, TFEB-rearranged RCC, ALK-rearranged RCC, and ELOC-mutated RCC.5
Recently, a RCC with NF2 genomic alteration (GA) gained attention not only due to morphologic features6,7 but also due to its association with treatment responses in nccRCC.6 In a recent phase II clinical trial of advanced nccRCC, 5 of 6 patients with NF2 mutations achieved an objective response to multi-targeted tyrosine kinase inhibitor cabozantinib plus human programmed death receptor-1 (PD-1) blocker nivolumab.8
NF2 gene on chromosome 22q encodes the tumor suppressor protein moesin-ezrin-radixin-like protein (merlin), also known as schwannomin important for the function of various mitogenic signaling pathways, including receptor tyrosine kinases (RTKs), Rac/p-21 activated kinase (RAK), mammalian target of rapamycin (mTOR), and the Hippo pathway.9,10 Neurofibromatosis type 2 syndrome is caused by heterozygous germline NF2 loss or inactivation and results in the development of vestibular schwannomas, meningiomas, ependymomas, and ocular disturbances.11NF2 GA have been postulated to defect the NF2 protein at the interface between the plasma membrane and the cytoskeleton, leading to dysfunction in adhesion, which is essential to cellular development and regeneration.12,13
Currently, there is expanding interest in defining predictive and prognostic role of PD-L1 expression for immune checkpoint inhibitors therapies.14-16 Across 15 tumor types including RCC, Davis et al. demonstrated PD-L1 was predictive for sensitivity to immune checkpoint blockade in 28.9% of cases, not predictive in 53.3%, and not tested in the remaining 17.8% of cases.16 A retrospective study of 306 ccRCC patients revealed PD-L1 (B7-H1) expression to be significantly associated with poorer cancer survival rates (41.9%) when compared to those whose tumors did not express PD-L1 (82.9%).17 PD-1-specific therapies including nivolumab18,19 and pembrolizumab,20 along with the PD-L1 antibody avelumab,21 have received FDA approval in metastatic RCC.
In this study, we performed comprehensive genomic profiling (CGP) of a large cohort of 3919 clinically advanced kidney tumors. Our findings demonstrate that NF2 GA are frequent in nccRCCs, and especially enriched in cdRCC. NF2-mutant (NF2mut) tumors often harbor CDKN2A/B, SETD2, and BAP1 GA, which are potentially amenable to targeted therapies. Higher frequencies of PD-L1 expression in NF2mut group suggest that these patients may benefit from immune checkpoint inhibitors.
Materials and Methods
Patient Selection
Approval for this study was obtained from the Western Institutional Review Board (Protocol No. 20152817). We reviewed the Foundation Medicine, Inc. (Cambridge, MA) database to retrieve all kidney tumors tested between 2015 and 2021. All cases submitted to Foundation Medicine were reviewed by pathologist with genitourinary expertise. All cases were clinically advanced, and the vast majority were stage IV. These cases were analyzed by CGP and PD-L1 immunohistochemistry (IHC) during routine clinical care. Demographic data were extracted from pathology reports.
This study received approval by the Institutional Review Board at Foundation Medicine, Inc. (Cambridge, MA).
A second validation cohort from The Cancer Genome Atlas (TCGA) included 1486 patients across the following publicly available datasets. Cases of ccRCC were retrieved from (1) The Cancer Genome Atlas, Firehose Legacy; (2) Nat Genet 2014; (3) Beijing Genome Institute, Nat Genet 2012; (4) Dana-Farber Cancer Institute, Science 2019; (5) University of Tokyo, Nat Genet 2013, chRCC; (6) TCGA, Firehose Legacy, pRCC; (7) TCGA, Firehose Legacy, renal non-clear cell carcinoma; (8) Genentech, Nat Genet 2014, uRCC; (9) MSK, Nature 2016, Pediatric Rhabdoid Tumor; (10) TARGET, 2018, Rhabdoid Cancer; (11) BCGSC, Cancer Cell 2016, Pediatric Wilms Tumor; and (12) TARGET, 2018. Care was taken during cohort creation to not select overlapping patients and samples were excluded if they were unprofiled for NF2. The bookmark query for this study from CBioPortal is listed here: https://www.cbioportal.org/study?id=62070c1f0934121b56de2448.
Comprehensive Genomic Profiling
CGP was performed using the FDA-approved FoundationOne CDx assay (Foundation Medicine, Cambridge, MA) in a Clinical Laboratory Improvement Amendments (CLIA)-certified and College of American Pathologists (CAP)-accredited laboratory using previously described methods.22 Prior to nucleic acid extraction, hematoxylin and eosin-stained slides were reviewed to confirm the presence of tumor. DNA extracted from formalin-fixed paraffin-embedded tissues underwent hybrid-capture based next generation sequencing using the FoundationOne platform which interrogates all coding exons of 324 cancer-related genes and introns from 31 genes commonly rearranged in cancer. Data were analyzed for all types of genomic alterations, including base substitutions, insertions/deletions, copy number alterations, and gene rearrangements. In addition, variant-level loss of heterozygosity (LOH), tumor mutational burden (TMB), and microsatellite instability (MSI) were determined. TMB was evaluated on up to 1.1 Mb of sequenced DNA, and MSI was assessed from DNA sequencing across 95 loci as previously described.23,24 TMB ≥ 20 mutations/Mb was considered High (TMB-High), >10 mutations/Mb considered intermediate (TMB-Int), and 0-9 mutations/Mb to be TMB low (TMB-low).
Immunohistochemistry
PD-L1 testing was performed according to individual standard of care and clinical requirements. IHC for PD-L1 was performed according to the manufactures instructions and guidelines using the DAKO PD-L1 22C3 PharmDx assay (Agilent Technologies, Santa Clara, CA) or Ventana PD-L1 SP142 companion diagnostics (CDx) assay (Roche, Tucson, AZ) in a CLIA-certified and CAP-accredited reference laboratory (Foundation Medicine, Morrisville, NC).
For DAKO 22C3 PD-L1 assay was evaluated using the tumor proportion score (TPS) of any intensity, and the combined positive score (CPS). PD-L1 expressing tumor cells were categorized as negative (<1%), low positive (1%-49%), or high positive (≥50%). CPS was calculated as the number of PD-L1 stained cells including tumor cells and immune cells, divided by the total number of tumor cells multiplied by 100. For Ventana SP142, evaluation was based on either the proportion of tumor area occupied by PD-L1 expressing tumor-infiltrating immune cells (IC) of any intensity or the percentage of PD-L1 expressing tumor cells (TPS) of any intensity. PDL-1 IC were scored as negative (IC < 1%), low positive (IC ≥ 1%), and high positive (IC ≥ 10%).
Merlin immunohistochemistry was performed using Ventana Discovery XT autostainer (Roche Diagnostics, Indianapolis, IN). Tissue sections were deparaffinized and pretreated in CC1 solution (EDTA, pH8). The primary anti-Merlin antibody (clone D3S3W, rabbit monoclonal, Cell Signaling Technology, Danvers, MA) was used at 1:100 dilution.
Statistical Analysis
To examine the landscape of genomic biomarkers in our patient cohort, we extracted the top 50 genes with GA and compared these between the NF2mut and NF2-wild type (NF2wt) tumor subsets. Descriptive statistics such as frequencies and percentages were calculated for the GA in each cohort. Statistical analysis was performed using ANOVA, χ2 contingency test, or Fisher’s exact test as appropriate. Analysis was performed using SPSS 1.0.0.1508. A critical P value of <.05 was used to indicate statistical significance. We also performed Bonferroni correction for multiple testing by dividing the critical P value by the number of cooccurring gene comparisons (30), allowing for a modified P value of .00167.
Results
Clinicopathologic and Molecular Characteristics
The study cohort of 3919 patients included 1875 ccRCC, 405 pRCC, 108 chRCC, 171 sRCC, 61cdRCC, 49 mRCC, 134 uRCC, 906 urothelial carcinoma (UC), and 147 Wilms tumors (Table 1). The median age of the cohort was 62 years (Supplemental Table S1). No significant difference in age of the patients was observed between ccRCC and nccRCC histologic types except patients with mRCC and Wilms tumor. The patients with mRCC and Wilms tumor were younger (median 27 and 6 years, respectively) as compared to other RCC subtypes. In all histologic subtypes except Wilms tumor male patients outnumbered the female ones. The age of patients with NF2mut tumors was 60 years (15->89; Table 1). There was a male predominance in the NF2mut ccRCC, pRCC, cdRCC, and sRCC, while in mRCC and WT the gender was equal, and in UC female patients slightly predominated.
Table 1.
Clinical, molecular, and immune biomarkers in NF2-mutated kidney tumors.
| Tumor | ccRCC | pRCC | sRCC | cdRCC | mRCC | uRCC | UC | Wilms |
|---|---|---|---|---|---|---|---|---|
| Number of cases with NF2 GA | 55/1820 (3%) | 43/362 (12%) | 30/141 (21%) | 14/47 (30%) | 2/47 (4%) | 25/172 (15%) | 30/885 (3%) | 2/145 (1%) |
| Gender | F35%/M65% | F26%/M74% | F23%/M77% | F36%/M64% | F50%/M50% | F12%/M88% | 57%F/43%M | F50%/M50% |
| Median age years (range) | 63 (31-83) | 61 (23-78) | 58 (26-78) | 58 (32-72) | 40 (15-65) | 58 (35->89) | 67 (36->89) | 26 (20-30) |
| GA/tumor | 4.5 | 3.1 | 5.1 | 2.7 | 2.0 | 3.6 | 11.4 | 3 |
| NF2 zygosity status | ||||||||
| SV homozygous | 29 | 33 | 15 | 10 | 0 | 19 | 10 | 1 |
| SV heterozygous | 7 | 1 | 1 | 0 | 1 | 1 | 4 | 1 |
| SV unknown | 10 | 6 | 8 | 2 | 1 | 4 | 7 | 0 |
| CN | 8 | 2 | 6 | 1 | 0 | 0 | 0 | 0 |
| RE | 1 | 1 | 0 | 1 | 0 | 1 | 0 | 0 |
| NF2 GA Type | ||||||||
| Substitutions | 18 | 22 | 13 | 4 | 1 | 12 | 12 | 2 |
| Ins/del | 28 | 18 | 11 | 8 | 1 | 12 | 9 | 0 |
| Loss | 8 | 2 | 6 | 1 | 0 | 0 | 0 | 0 |
| RE | 1 | 1 | 0 | 1 | 0 | 1 | 0 | 0 |
| Most common co-alterations |
VHL substitution (44%) CDKN2A loss (35%) CDKN2B loss (29%) SETD2 indel (27%) VHL indel (24%) MTAP loss (11%) |
CDKN2A loss (19%) FH substitution (14%) CDKN2B loss (12%) SETD2 (12%) BAP1 substitution (9%) KMT2D indel (7%) |
CDKN2A loss (57%) CDKN2B loss (53%) VHL indel (30%) TP53 (27%) BAP1 indel (13%) MTAP loss (13%) |
CDKN2A loss (14%) SETD2 substitution (14%) CDKN2B loss (7%) BAP1 indel (7%) MTAP loss (7%) PBMR1 indel (7%) |
FBXW7 substitution (100%) FAT1 indel (50%) |
CDKN2A loss (32%) CDKN2B loss (24%) SETD2 indel (20%) TERT substitution (12%) MTAP loss (12%) PBRM1 indel (12%) |
TERT substitution (37%) TP53 substitution (33%) CDKN2A loss (20%) CDKN2B loss (20%) HRAS substitution (17%) MTAP loss (13%) |
KMT2C substitution (50%) FBXW7 substitution (50%) ASXL1 indel (50%) FUBP1 indel (50%) |
| MSI-high total | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 |
| TMB low | 47 | 37 | 25 | 13 | 2 | 21 | 13 | 1 |
| TMB int | 8 | 6 | 5 | 1 | 0 | 4 | 2 | 1 |
| TMB high | 0 | 0 | 0 | 0 | 0 | 0 | 6 | 0 |
Abbreviations: ccRCC, clear cell RCC; cdRCC, collecting duct; chRCC, chromophobe RCC; CNA, copy number alteration; GA, genomic alteration; MSI, microsatellite instable; mRCC, medullary; pRCC, papillary RCC; RE, rearrangement; sRCC, sarcomatoid; SV, structural variation; TMB, tumor mutational burden; UC, urothelial carcinoma; uRCC, unclassified; Wilms, Wilms tumor.
One hundred ninety-two of kidney tumors featured NF2 GA (4.9%), while 3727 (95.1%) did not. NF2 GA frequency was highest in cdRCC (30%) and sRCC (21%) and lowest in ccRCC (3%) and UC (3%) (Table1). Of note, in cdRCC most common GA were involving NF2 gene. No NF2 GA were identified in chRCC and therefore this cohort was excluded from further analysis. The most common type of GA in the NF2 gene was structural variation mutations (89%), followed by copy number alterations (homozygous deletions and amplifications) (9%) and gene rearrangements (2%). Loss of chromosome 22q harboring the NF2 gene was found in the majority (79%) of NF2mut tumors. Overall, 69% of NF2mut specimens were under LOH, either with one mutant allele remaining or with multiple copies of mutant allele (homozygous mutations), 9% were heterozygous mutations, and zygosity was unknown in 22%. All NF2 GA were predicted to be inactivating based on disruption of the FERM domain (amino acids 22-311), which includes in-frame deletions that disrupt the Paxillin-binding region (aa 50-70) of the FERM domain2 as well as the C-terminal region (amino acids 506-547).
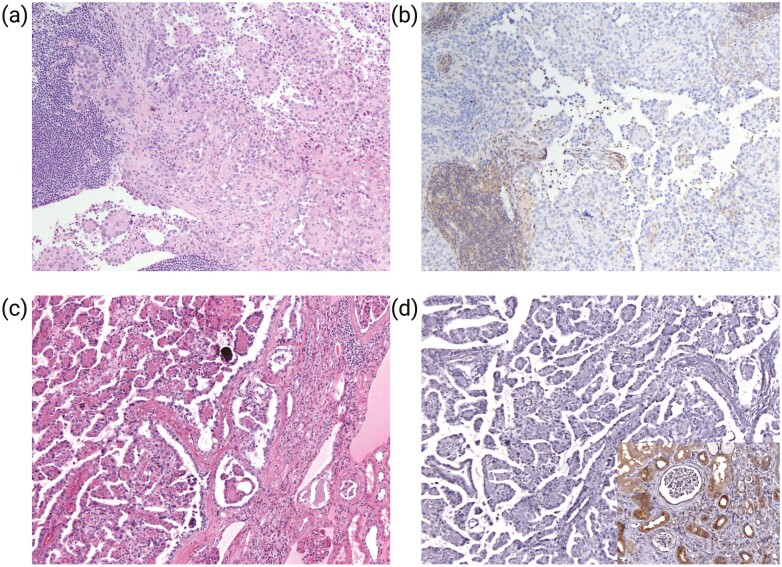
To confirm that NF2 GA result in protein loss, we evaluated merlin expression by immunohistochemistry in 2 cases in which tissue was available. Both tumors demonstrated complete loss of merlin expression (Fig. 1). In contrast, inflammatory cells and non-neoplastic kidney tissue adjacent to tumors showed retained merlin expression in renal tubules and Bowman capsule (Fig. 1d, inset).
Figure 1.
Immunohistochemical analysis of merlin expression in NF2-mutated RCC. Representative histologic sections of tumors (H&E) show solid and papillary architecture with small cells clustering around hyaline material and forming micropapillae surrounded by larger cells and scattered calcifications (a, c). Negative staining of tumor cells for merlin (b, d). The inflammatory cell (b, lower left) renal tubules and Bowman capsule (d, inset) show retained merlin immunoreactivity.
Analysis of co-mutated genes revealed that VHL was the most common co-altered gene in NF2mut ccRCC. Deletion of cyclin-dependent kinase inhibitor CDKN2A was the most common co-alteration in pRCC (19%), sRCC (57%), cdRCC (14%), and uRCC (32%). CDKN2B GA co-occurred with CDKN2A in slightly lower frequencies. FBXW7 was the most altered gene in mRCC (100%). TERT was the most commonly co-altered GA in UC (37%). The most commonly co-mutated genes across kidney tumor subtypes can be found in Supplemental Fig. S1.
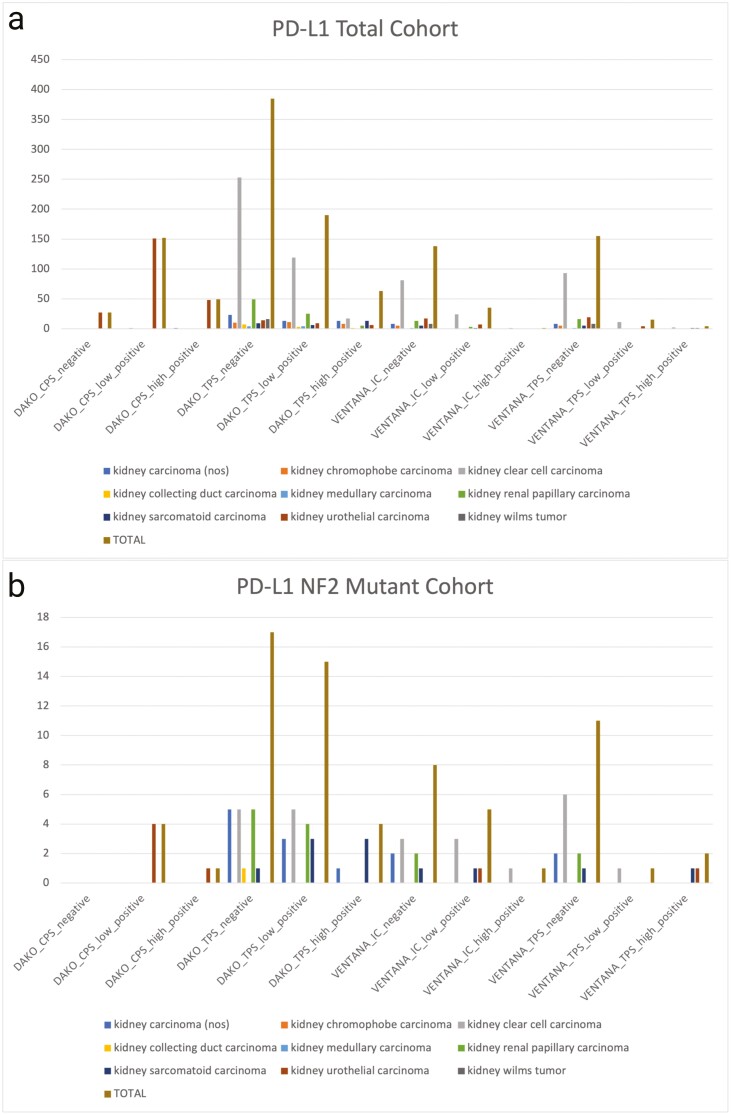
Two NF2mut kidney tumors featured MSI high and 6 featured TMB high status, although exclusively in UC. Variations in PD-L1 positivity were found across different kidney tumor types (Fig. 2). sRCC was found to be associated with strong positivity for PD-L1 expression in tumor cells (TPS) with both DAKO SP22C3 (43%) and Ventana SP142 (50%). One uRCC case was found to have strong SP22C3 TPS staining (11%), while one case of ccRCC (14%) and one case of UC (20%) were found to have a high IC (SP142), and one case of UC (20%) was found to have a high CPS (SP22C3).
Figure 2.
PDL1 scoring with DAKO PD-L1 22C3 and Ventana PD-L1 SP142 in the total cohort (a) and NF2-mutated cohort (b). TPS, tumor proportion score; CPS, combined positive score; IC, immune cells.
The total number of GA per tumor in the NF2mut cohort including NF2 GA and other co-occurring GA was 4.74, for a total of 911 GA. Seven hundred twenty-five GA were co-occurring including 331 base substitutions (46%), 182 insertion/deletions (25%), 141 homozygous deletions (19%), 41 amplifications (6%), and 30 gene rearrangements (4%).
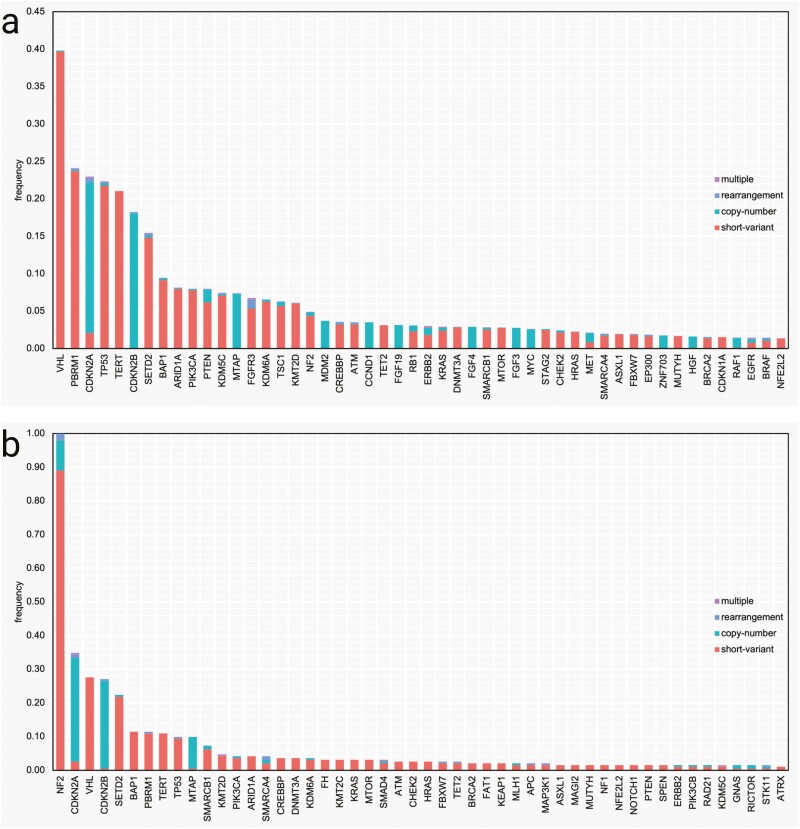
Genomic landscape showing co-mutation plots of the top 50 genes with GA in total cohort and NF2mut subset can be seen in Fig. 3. The 4 most common GA in the total disease cohort were: VHL, PBRM1, CDKN2A, and TP53; which contrasted with the NF2mut disease cohort: NF2, CDKN2A, VHL, and CDKN28.
Figure 3.
Frequency of pathogenic gene mutations in the total cohort (a) and NF2-mutated kidney tumors (b).
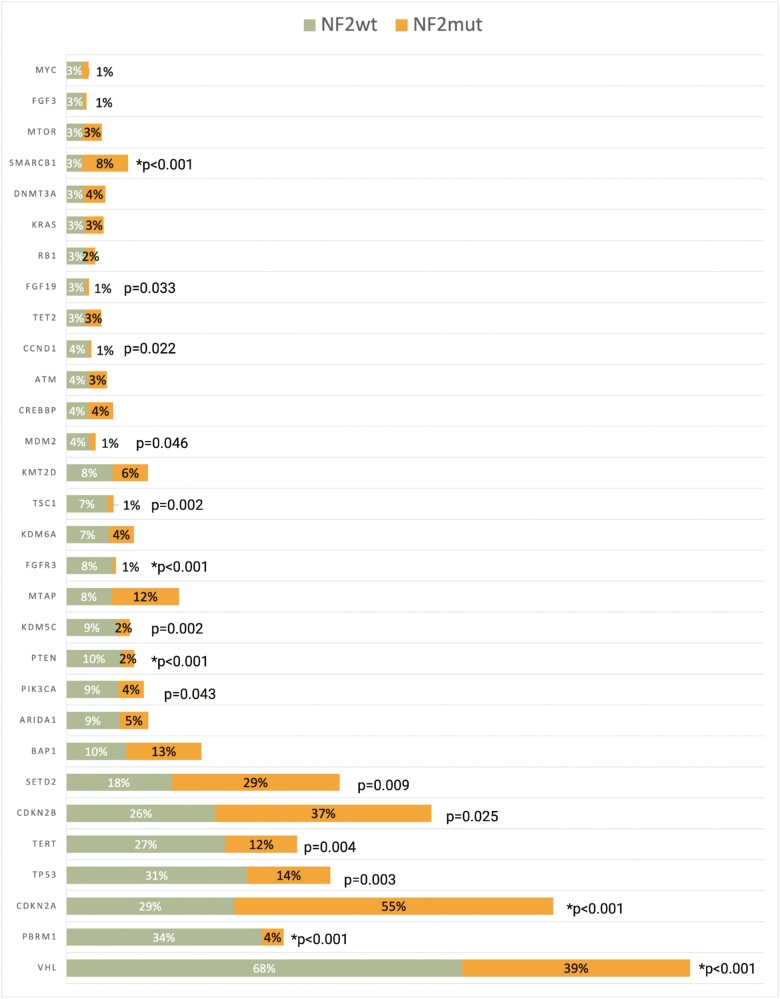
Of the top 50 genes in the total cohort, 30 were genes with shared GA in the NF2wt and NF2mut cohorts. The frequencies of 16 of the 30 shared genes were significantly different between the NF2wt and NF2mut subsets (Fig. 4). The following GA were found to be enriched in NF2mut tumors: CDKN2A (P < .001), CDKN2B (P = .025), SETD2 (P = .009), and SMARCB1 (P < .001). In contrast, NF2wt tumors featured GA in VHL (P < .001), PBRM1 (P < .001), TP53 (P = .003), TERT (P = .004), PIK3CA (P = .043), PTEN (P < .001), KDM5C (P = .002), FGFR3 (P < .001), TSC1 (P = .002), MDM2 (P = .046), CCND1 (P = .022), and FGF19 (P = .033). The remaining GA were not found to be associated with NF2 mutational status. Only rare co-occurring mutations were identified in mTOR pathway in NF2mut tumors, including PIK3CA (4%), MTOR (3%), TSC1 (1%), and PTEN (2%). Following Bonferroni correction and a modified P value of .00167, only CDKN2A (P < .001), SMARCB1 (P < .001), VHL (P < .001), PBRM1 (P < .001), PTEN (P < .001), and FGFR3 (P < .001) were found to be significant.
Figure 4.
Frequency of top 30 co-occurring genomic alterations (GA) between the NF2wt and NF2mut tumors. *Significant based on a modified P value of .00167 following Bonferroni correction.
The results for comparisons between NF2wt and NF2mut cohorts are presented in Table 2. TMB was not associated with NF2 mutational status (P = .619). Chromosome 22 was more likely to be lost in the NF2mut cohort when compared to NF2wt tumors (P < .001). There was no association between MSI and NF2 mutational status (P = .651).
Table 2.
Comparisons between NF2wt and NF2mut cohorts.
| Patient characteristics | NF2 wild type (n = 3727) | NF2 mutant (n = 192) | P value |
|---|---|---|---|
| Age, years | — | ||
| Median | 62 | 60 | |
| Gender | |||
| Male | 2504 | 133 | .548b |
| Female | 1223 | 59 | |
| TMB (mutations/Mb) | .619b | ||
| TMB-high | 89 | 6 | |
| TMB-int | 603 | 27 | |
| TMB-low | 3035 | 159 | |
| Ch 22 status | <.001 b | ||
| Lost | 528 | 89 | |
| Retained | 1770 | 33 | |
| MSI | .651b | ||
| MSI-H | 27 | 2 | |
| MSS | 3276 | 168 | |
| PD-L1 DAKO 22C3 | |||
| TPS positive | 59 | 4 | .228a |
| TPS low positive | 175 | 15 | |
| TPS negative | 368 | 17 | |
| CPS positive | 48 | 1 | 1.000b |
| CPS low positive | 148 | 4 | |
| CPS negative | 27 | 0 | |
| PD-L1 Ventana SP142 | |||
| TPS positive | 2 | 2 | .040 b |
| TPS low positive | 14 | 1 | |
| TPS negative | 144 | 11 | |
| IC positive | 0 | 1 | .013 b |
| IC low positive | 30 | 5 | |
| IC negative | 130 | 8 |
a χ2 contingency test.
bFisher’s exact test.
Bolded values are significant based on a P value of <.05.
Abbreviations: CPS, combined positive score; IC, immune cells; MSI, microsatellite instable; MSS, microsatellite stable; TMB, tumor mutational burden; TPS, tumor proportion score.
Analysis of PD-L1 expression revealed no difference in PD-L1 22C3 assay TPS (P = .228) and CPS (P = 1.00) between NF2wt and NF2mut groups. However, PD-L1 SP142 assay TPS (P = .040) and IC (P = .013) were found more frequent in NF2mut kidney tumors.
Findings from the Combined TCGA Cohort
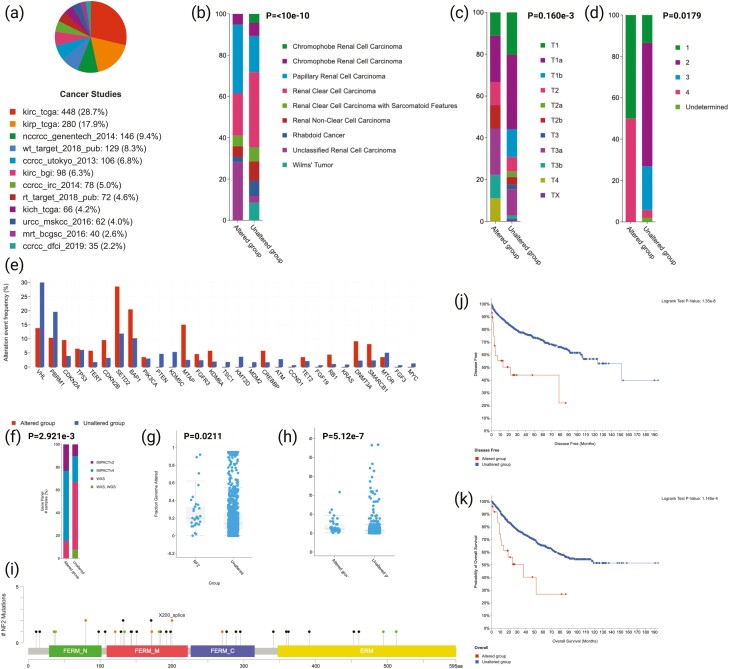
In the TCGA validation cohort, 35/1486 (2.4%) kidney tumors harbored NF2 GA and 37 types of different mutations were seen. Briefly, 32 were driver mutations: 24 truncating and 8 splice, while 5 were variant of undetermined significance (VUS), all of which were missense. The breakdown of patients according to corresponding study percentages can be seen in Fig. 5a. Regarding tumor subtype, NF2 GA were found to be more common in uRCC and pRCC (P < 10e-10) as compared to other histologic types (Fig. 5b). High pathologic stage (P = .160e-3) and high WHO/ISUP histologic grade (P = .179) were characteristic of tumors with NF2 GA (Fig. 4c, 4d). The top 30 genes found in the Foundation Medicine cohort were analyzed in relation to NF2 GA (Fig. 4e). The fraction of GA was higher in the NF2mut cohort (median = 0.2) compared to NF2wt tumors (median = 0.14; P = .0211); a finding also seen with TMB (P = 5.12e-7; Fig. 4g, 4h). Mutation diagram circles are colored with respect to the corresponding mutation types can be seen in Fig. 4i. Regarding survival, NF2mut kidney tumors featured lower disease-free (P = 1.35e-8) and overall survival (P = 1.145e-4) when compared to the NF2wt group (Fig. 4j, 4k). VHL was validated as being more commonly mutated in the NF2wt cohort (P = .0393), while NF2mut tumors harbored SETD2 (P < .001) and BAP1 (P = .0344) GA, similarly to Foundation Medicine cohort. Following Bonferroni correction and a modified P value of .00167, only SETD2 was found to be significant. Findings from the validation of the top 30 co-occurring genes can be found in Supplemental Table S2.
Figure 5.
Findings from the TCGA cohort. (a) Breakdown of patients according to corresponding originating cancer study. (b) Histologic subtypes according to NF2 mutational status. (c) AJCC pathologic stage according to NF2 mutational status. (d) Histologic grade according to NF2 mutational status. (e) The top 30 co-altered genes found in the Foundation Medicine cohort were analyzed and demonstrated in relation to NF2 mutational status. (f) NF2 mutational status in relation to gene panels. (g) The fraction of genomic alteration according to NF2 mutational status (h) TMB according to NF2 mutational status. (i) Mutation diagram circles are colored with respect to the corresponding mutation types. In case of different mutation types at a single position, color of the circle is determined with respect to the most frequent mutation type. Mutation types and corresponding color codes are as follows: green, missense mutations; black, truncating mutations; red, inframe mutations; orange, splice mutations; purple, fusion mutations; pink, other mutations (for colour figure refer to online version). (j) Kaplan-Meier for disease-free-survival between NF2mut and WT groups. (k) Kaplan-Meier for overall survival between NF2mut and NF2wt groups.
Discussion
In this study, we characterized the genomic landscape of NF2-mutated kidney tumors in a large cohort of 3919 cases. Germline NF2 loss or inactivation is associated with neurofibromatosis type 2 syndrome, which results in the development of bilateral vestibular schwannomas, meningiomas, and ependymomas.10 Loss of merlin encoded by NF2 gene is found also in 40%-60% of sporadic meningiomas.25 In addition to tumors of the nervous system, NF2 GA alterations and merlin inactivation also occur in a large proportion of malignant mesothelioma (MM) patients. NF2 GA were found as the most frequent GA in asbestos nonexposed patients with a third of the patients carrying NF2 mutations.26NF2 GA are less frequent in ovarian serous carcinoma, glioblastoma multiforme, breast, colorectal, skin, hepatic, medullary thyroid, prostate cancer, and melanoma.10
Inactivating NF2 GA have been described in spectrum of kidney tumors including aggressive variants such as cdRCC (29%),27 pRCC (12%),28,29 sRCC (19.2%),30 and uRCC (18%),31 as well as in more indolent mucinous and spindle cell carcinoma of the kidney.32 The frequencies of GA in histologic subtypes of renal tumors in our cohort are similar to previous studies.
Differences in NF2 GA frequencies between the Foundation Medicine (4.9%) and TCGA (2.4%) cohorts could be secondary to selection bias since most tumors being tested in the Foundation Medicine were advanced stage IV kidney tumors in contrast to the limited number of patients with confirmed stage IV disease in TCGA cohort. Based on a modified P value of .00167, only SETD2 was found to be significant. Many genes trended towards being significant (CDKN2A, CDKN2B, and SMARCB1) but the cohort was small (35 patients).
The relatively high prevalence of NF2 GA in a subset of nccRCC, lack of other driver genes, and low NF2 GA frequency (3%) in ccRCC suggest its driving role in the tumorigenesis. Our finding of low incidence of NF2 mutations in ccRCC is congruent with previous cohort of 220 metastatic ccRCC in RECORD3 study (4%).33 Co-occurrence of NF2 GA with VHL mutations in ccRCC cohort suggests that NF2 GA may be a secondary event in ccRCC, similarly to co-occurrence of TSC1 and TSC2 GA in VHL driven ccRCC.34 In the study of sarcomatoid ccRCC Malouf et al. presented one tumor with deleterious NF2 mutation in its sarcomatoid component only, suggesting that NF2 GA may represent a late event in ccRCC with sarcomatoid differentiation.35
The significant proportion of NF2mut renal tumors in our series have co-occurring inactivating GA in other tumor suppressor genes. These include cell cycle regulator genes CDKN2A/2B, chromatin remodeler genes BAP1, SETD2, and SWI/SNF-related chromatin remodeler SMARCB1. In the study of uRCC by Chen et al., NF2 GA also co-occurred with SETD2 and BAP1, and the occurrence of SETD2 mutations was significantly higher in uRCC tumors with NF2 loss than in remaining uRCC tumors (44% vs 9%.).31 In recent study of 14 NF2-mutated RCC cases, co-occurrence of NF2 and chromatin modulator PBRM1 GA was found in 5 (42%) cases.6 However, no NF2wt group was included in this study for comparison, and the number of cases was relatively small in contrast to our series.6 Although PBRM1 GA were found in 3.6% of NF2mut tumors in our study, the prevalence of this alteration was significantly lower as opposed to NF2wt group (25.1%).
NF2 tumor suppressor gene inactivation along with mutations in CDKN2A/B, and chromatin modulators BAP1, SETD2, and SMARCB1 has been described as driving GA in high-grade/progressive meningioma, and MM similarly to NF2mut RCC.36-38 All these are highly aggressive tumors refractory to conventional therapies. Our analysis of TCGA data supports aggressive behavior of NF2mut renal tumors. Mutations in CDKN2A/B were found to be the most associated co-alteration in aggressive NF2mut meningiomas, seen in 24% of cases.37SMARCB1 mutations were also found in NF2mut intraventricular meningioma.39 Recently, a mouse model of MM was generated based upon disruption of the NF2, BAP1, and CDKN2A/B tumor suppressor loci in various combinations as also frequently observed in human MM.40 Inactivation of all 3 loci in the mesothelial lining of the thoracic cavity led to a highly aggressive MM that recapitulates the histologic features and gene expression profile observed in human MM.
As all major GA in NF2mut RCC are tumor suppressor genes, targeted therapies that exploit abnormal tumor suppressor genes have proven far more difficult as opposed to inhibition of oncoproteins. It is important to mention that the loss or inactivation of NF2 may have the ability to predict sensitivity to focal adhesion kinase inhibitors, this is based on strong preclinical data from malignant pleural mesothelioma.41 Preclinical mouse models of NF2mut meningiomas have shown overexpression of the mTOR signaling complex 1 pathway, which can be suppressed by mTOR inhibitors.42 Limited preclinical and clinical evidence in vestibular schwannoma suggest possible sensitivity of NF2-deficient tumors to the pan-ERBB inhibitor lapatinib.43 Similarly, based on limited clinical and preclinical evidence, NF2 inactivation may predict sensitivity to MEK inhibitors, such as approved agents trametinib and cobimetinib.44 Data from a Chinese breast cancer cohort suggest that NF2 loss-of-function mutations may increase sensitivity to Hippo-targeting strategies.45 Targeting the Hippo pathway including downstream effectors YAP/TAZ can be a valid approach in renal tumors as well.35 In a preclinical model of NF2mut pRCC, inhibition of the YAP1 partner YES1 by dasatinib or sarcatinib led to repression of Hippo transcriptional targets and provided potent antitumor activity.46CDKN2A/B, BAP1, and SETD2 may also represent potential therapeutic targets, as demonstrated in preclinical studies of other tumor types.25 These potential therapeutic strategies warrant further investigation in clinical trials.
Immunotherapy is another potential target for investigation in NF2mut RCC, as we demonstrated higher level of PD-L1 expression in NF2mut cohort. Expression of PD-L1 on tumor and immune cells appears to impact efficacy of PD-L1 inhibitor pembrolizumab. In the KEYNOTE-427 trial from advanced non-clear cell RCC the response rate was 35.3% with a CPS ≥1 as opposed to 12.1% in patients with CPS less than 1.47 Combinations of immune checkpoint inhibitors with TKIs such as cabozantinib and axitinib have higher anti-tumor activity and are currently approved for treatment of metastatic clear cell RCC.48,49 Selecting tumors with higher PD-L1 expression such as those with NF2 GA might expand the benefit of these combinations to non-clear cell RCC. In a phase II trial of cabozantinib that targets MET, AXL, and VEGFR2 plus nivolumab, a human PD-1 blocking antibody, NF2 GA were found in 19% of unclassified/papillary, and translocation-associated RCC (6). Of note, objective tumor responses were seen in 5/6 patients with tumors harboring NF2 mutations (6). Although conclusions are limited by small sample size, they suggest that NF2 GA may predict treatment responses in non-clear cell RCC. Paintal et al. reported 2 cases of NF2mut RCC with dramatic response to immune checkpoint inhibitors (ipilimumab/nivolumab).6 Our findings of more frequent PD-L1 tumor and immune cell expression in NF2mut tumors support that these patients may benefit form immune checkpoint inhibitors.
There are several limitations of this study. First, the study suffers from selection bias, as it includes only samples sent to molecular analysis, and therefore, the results may not be representative of general population. Similar studies, including consecutive unselected cases of kidney tumors, are needed to further characterize RCCs harboring NF2 GA. Second, although the FoundationOne panel of the 324 genes is quite comprehensive, it is possible that there may be other important genes that were simply not included in the testing panel, thus limiting our findings. Third, epigenetic mechanisms of NF2 inactivation were not addressed in this study. Comprehensive studies of promoter methylation and epigenetic inactivation of NF2 gene are needed. Fourth, histology of the NF2mut tumors was not evaluated. Argani et al. described a series of histologically distinct NF2mut that they termed biphasic hyalinizing psammomatous RCC.7 Paintal et al. described common morphologic features of NF2mut RCC in a series of 14 cases.6 While the individual morphologic features seen in these cases are non-specific in isolation, the presence of the typical morphologic constellation (eosinophilic cytology, high nuclear grade, tubulopapillary architecture, sclerotic stroma, microscopic coagulative necrosis, and psammomatous calcifications) can allow for their prospective identification and triage for confirmatory molecular studies. The utility of ancillary techniques such as immunohistochemical detection of NF2 protein expression is limited.7,29NF2 gene deletion can be detected by fluorescent in situ hybridization in MM50; however, this has not been validated in renal tumors. Currently, comprehensive genomic profiling proved to be a reliable platform for detection of NF2 GA in RCC.
In conclusion, the present study is the largest to characterize genomic findings in NF2-mutated kidney tumors. Although these aggressive tumors are driven by tumor-suppressor genes, they harbor potentially targetable genomic alterations. Higher frequencies of PD-L1 expression in NF2mut tumors suggest that these patients may benefit from immune checkpoint inhibitors. Further studies and clinical trials implementing these therapies are warranted to confirm the clinical relevance and benefit.
Supplementary Material
Acknowledgment
Figures were created at Biorender.com.
Contributor Information
Sean M Hacking, Department of Pathology and Laboratory Medicine, Rhode Island Hospital, Warren Alpert Medical School of Brown University, Providence, RI, USA.
Dean Pavlick, Foundation Medicine, Inc., Cambridge, MA, USA.
Yihong Wang, Department of Pathology and Laboratory Medicine, Rhode Island Hospital, Warren Alpert Medical School of Brown University, Providence, RI, USA.
Benedito A Carneiro, Lifespan Cancer Institute, Legorreta Cancer Center at Brown University, Providence, RI, USA.
Matthew Mullally, Department of Pathology and Laboratory Medicine, Rhode Island Hospital, Warren Alpert Medical School of Brown University, Providence, RI, USA.
Shaolei Lu, Department of Pathology and Laboratory Medicine, Rhode Island Hospital, Warren Alpert Medical School of Brown University, Providence, RI, USA.
Mariana Canepa, Department of Pathology and Laboratory Medicine, Rhode Island Hospital, Warren Alpert Medical School of Brown University, Providence, RI, USA.
Gennady Bratslavsky, Department of Urology, State University of New York (SUNY), Upstate Medical University, Syracuse, NY, USA.
Joseph Jacob, Department of Urology, State University of New York (SUNY), Upstate Medical University, Syracuse, NY, USA.
Andrea Necchi, Department of GU Medical Oncology, San Raffaele University, Milan, Italy.
Philippe E Spiess, Department of Genitourinary Oncology, Moffitt Cancer Center, Tampa, FL, USA.
Li Wang, Department of Pathology and Laboratory Medicine, Rhode Island Hospital, Warren Alpert Medical School of Brown University, Providence, RI, USA.
Evgeny Yakirevich, Department of Pathology and Laboratory Medicine, Rhode Island Hospital, Warren Alpert Medical School of Brown University, Providence, RI, USA.
Jeffrey Ross, Foundation Medicine, Inc., Cambridge, MA, USA.
Funding
No specific funding was provided to produce this manuscript.
Conflict of Interest
Andrea Necchi receives honoraria from Roche, MSD, AstraZeneca, Janssen, Foundation Medicine, BMS, and Astellas, has consulting/advisory roles with MSD, Roche, Bayer, AstraZeneca, Clovis Oncology, Janssen, Incyte, Seattle Genetics/Astellas, Bristol-Myers Squibb, Rainier Therapeutics, Bicycle Therapeutics, GlaxoSmithKline, Basilea Pharmaceutica, and Catalym, receives research funding from MSD, AstraZeneca, Ipsen, and Gilead, receives travel accommodations and expenses from Roche, MSD, AstraZeneca, Janssen, Rainer Therapeutics, and Pfizer, and has the following employment and stock ownership spouse disclosures: Bayer. Jeffrey Ross is an employee of Foundation Medicine, an equity owner in Roche Holdings, and a consultant and equity owner for Tango Therapeutics and Celsius Therapeutics. The other authors indicated no financial relationships.
Author Contributions
Conception/design: S.M.H., D.P., E.Y., J.R. Provision of study material or patients: S.M.H., D.P., E.Y., J.R. Collection and/or assembly of data: S.H., D.P., E.Y., J.R. Data analysis and interpretation: All authors. Manuscript writing: S.M.H., M.M., E.Y., J.R. Final approval of manuscript: All authors.
Data Availability
All data generated and analyzed in this study can be provided by the corresponding author upon reasonable request.
References
- 1. Moch H HP, Ulbright TM, Reuter VE.. WHO Classification of Tumours of the Urinary System and Male Genital Organs. 4th ed. Vol 8; 2016. https://books.google.com.mx/books/about/WHO_Classification_of_Tumours_of_the_Uri.html?id=qxQyjgEACAAJ&redir_esc=y [DOI] [PubMed] [Google Scholar]
- 2. Blum KA, Gupta S, Tickoo SK, et al. Sarcomatoid renal cell carcinoma: biology, natural history and management. Nat Rev Urol. 2020;17(12):659-678. 10.1038/s41585-020-00382-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. D’Avella C, Abbosh P, Pal SK, Geynisman DM.. Mutations in renal cell carcinoma. Urol Oncol. 2020;38(10):763-773. 10.1016/j.urolonc.2018.10.027 [DOI] [PubMed] [Google Scholar]
- 4. Pavlovich CP, Schmidt Laura S., Schmidt LS.. Searching for the hereditary causes of renal-cell carcinoma. Nat Rev Cancer. 2004;4(5):381-393. 10.1038/nrc1364 [DOI] [PubMed] [Google Scholar]
- 5. Lobo J, Ohashi R, Amin MB, et al. WHO 2022 landscape of papillary and chromophobe renal cell carcinoma. Histopathology. 2022;81(4):426-438. [DOI] [PubMed] [Google Scholar]
- 6. Paintal A, Tjota MY, Wang P, et al. NF2-mutated renal carcinomas have common morphologic features which overlap with biphasic hyalinizing psammomatous renal cell carcinoma: a comprehensive study of 14 cases. Am J Surg Pathol. 2022;46(5):617-627. [DOI] [PubMed] [Google Scholar]
- 7. Argani P, Reuter VE, Eble JN, et al. Biphasic hyalinizing psammomatous renal cell carcinoma (BHP RCC): a distinctive neoplasm associated with somatic NF2 mutations. Am J Surg Pathol. 2020;44(7):901-916. 10.1097/PAS.0000000000001467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lee CH, Voss MH, Carlo MI, et al. Phase II trial of cabozantinib plus nivolumab in patients with non-clear-cell renal cell carcinoma and genomic correlates. J Clin Oncol. 2022;40(21):2333-2341. 10.1200/JCO.21.01944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Beltrami S, Kim R, Gordon J.. Neurofibromatosis type 2 protein, NF2: an uncoventional cell cycle regulator. Anticancer Res. 2013;33(1):1-11. [PMC free article] [PubMed] [Google Scholar]
- 10. Petrilli AM, Fernández-Valle C.. Role of Merlin/NF2 inactivation in tumor biology. Oncogene. 2016;35(5):537-548. 10.1038/onc.2015.125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Evans DGR. Neurofibromatosis type 2 (NF2): a clinical and molecular review. Orphanet J Rare Dis. 2009;4:16-16. 10.1186/1750-1172-4-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Maitra S, Kulikauskas RM, Gavilan H, Fehon RG.. The tumor suppressors Merlin and expanded function cooperatively to modulate receptor endocytosis and signaling. Curr Biol. 2006;16(7):702-709. 10.1016/j.cub.2006.02.063 [DOI] [PubMed] [Google Scholar]
- 13. Ramesh V. Merlin and the ERM proteins in Schwann cells, neurons and growth cones. Nat Rev Neurosci. 2004;5(6):462-470. 10.1038/nrn1407 [DOI] [PubMed] [Google Scholar]
- 14. Weinstock M, McDermott D.. Targeting PD-1/PD-L1 in the treatment of metastatic renal cell carcinoma. Ther Adv Urol. 2015;7(6):365-377. 10.1177/1756287215597647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Aggen DH, Drake CG, Rini BI.. Targeting PD-1 or PD-L1 in metastatic kidney cancer: combination therapy in the first-line setting. Clin Cancer Res. 2020;26(9):2087-2095. 10.1158/1078-0432.CCR-19-3323 [DOI] [PubMed] [Google Scholar]
- 16. Davis AA, Patel VG.. The role of PD-L1 expression as a predictive biomarker: an analysis of all US Food and Drug Administration (FDA) approvals of immune checkpoint inhibitors. J ImmunoTher Cancer. 2019;7(1):278-278. 10.1186/s40425-019-0768-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Thompson RH, Kuntz SM, Leibovich BC, et al. Tumor B7-H1 is associated with poor prognosis in renal cell carcinoma patients with long-term follow-up. Cancer Res. 2006;66(7):3381-3385. 10.1158/0008-5472.CAN-05-4303 [DOI] [PubMed] [Google Scholar]
- 18. Motzer RJ, Escudier B, McDermott DF, et al. Nivolumab versus everolimus in advanced renal-cell carcinoma. N Engl J Med. 2015;373(19):1803-1813. 10.1056/nejmoa1510665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Motzer RJ, Tannir NM, McDermott DF, et al. Nivolumab plus ipilimumab versus sunitinib in advanced renal-cell carcinoma. N Engl J Med. 2018;378(14):1277-1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Rini BI, Plimack ER, Stus V, et al. Pembrolizumab plus axitinib versus sunitinib for advanced renal-cell carcinoma. N Engl J Med. 2019;380(12):1116-1127. 10.1056/nejmoa1816714 [DOI] [PubMed] [Google Scholar]
- 21. Motzer RJ, Penkov K, Haanen J, et al. Avelumab plus axitinib versus sunitinib for advanced renal-cell carcinoma. N Engl J Med. 2019;380(12):1103-1115. 10.1056/nejmoa1816047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Frampton GM, Fichtenholtz A, Otto GA, et al. Development and validation of a clinical cancer genomic profiling test based on massively parallel DNA sequencing. Nat Biotechnol. 2013;31(11):1023-1031. 10.1038/nbt.2696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chalmers ZR, Connelly CF, Fabrizio D, et al. Analysis of 100,000 human cancer genomes reveals the landscape of tumor mutational burden. Genome Med. 2017;9(1):34-34. 10.1186/s13073-017-0424-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Trabucco SE, Gowen K, Maund SL, et al. A novel next-generation sequencing approach to detecting microsatellite instability and pan-tumor characterization of 1000 microsatellite instability-high cases in 67,000 patient samples. J Mol Diagn. 2019;21(6):1053-1066. 10.1016/j.jmoldx.2019.06.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Venur VA, Santagata S, Galanis E, Brastianos PK.. New molecular targets in meningiomas: the present and the future. Curr Opin Neurol. 2018;31(6):740-746. 10.1097/wco.0000000000000615 [DOI] [PubMed] [Google Scholar]
- 26. Quetel L, Meiller C, Assié JB, et al. Genetic alterations of malignant pleural mesothelioma: association with tumor heterogeneity and overall survival. Mol Oncol. 2020;14(6):1207-1223. 10.1002/1878-0261.12651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Pal SK, Choueiri TK, Wang K, et al. Characterization of clinical cases of collecting duct carcinoma of the kidney assessed by comprehensive genomic profiling. Eur Urol. 2016;70(3):516-521. 10.1016/j.eururo.2015.06.019 [DOI] [PubMed] [Google Scholar]
- 28. Pal SK, Ali SM, Yakirevich E, et al. Characterization of clinical cases of advanced papillary renal cell carcinoma via comprehensive genomic profiling. Eur Urol. 2018;73(1):71-78. 10.1016/j.eururo.2017.05.033 [DOI] [PubMed] [Google Scholar]
- 29. Yakirevich E, Perrino C, Necchi A, et al. NF2 mutation-driven renal cell carcinomas (RCC): A comprehensive genomic profiling (CGP) study. J Clin Oncol. 2020;38(6_suppl):726-726. 10.1200/jco.2020.38.6_suppl.726 [DOI] [Google Scholar]
- 30. Malouf GG, Ali SM, Wang K, et al. Genomic characterization of renal cell carcinoma with sarcomatoid dedifferentiation pinpoints recurrent genomic alterations. Eur Urol. 2016;70(2):348-357. 10.1016/j.eururo.2016.01.051 [DOI] [PubMed] [Google Scholar]
- 31. Chen Y-B, Xu J, Skanderup AJ, et al. Molecular analysis of aggressive renal cell carcinoma with unclassified histology reveals distinct subsets. Nat Commun. 2016;7:13131-13131. 10.1038/ncomms13131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mehra R, Vats P, Cieslik M, et al. Biallelic alteration and dysregulation of the hippo pathway in mucinous tubular and spindle cell carcinoma of the kidney. Cancer Discov. 2016;6(11):1258-1266. 10.1158/2159-8290.CD-16-0267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hsieh JJ, Chen D, Wang PI, et al. Genomic biomarkers of a randomized trial comparing first-line everolimus and sunitinib in patients with metastatic renal cell carcinoma. Eur Urol. 2017;71(3):405-414. 10.1016/j.eururo.2016.10.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Fisher R, Horswell S, Rowan A, et al. Development of synchronous VHL syndrome tumors reveals contingencies and constraints to tumor evolution. Genome Biol. 2014;15(8):433-433. 10.1186/s13059-014-0433-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Malouf GG, Flippot R, Dong Y, et al. Molecular characterization of sarcomatoid clear cell renal cell carcinoma unveils new candidate oncogenic drivers. Sci Rep. 2020;10(1):701-701. 10.1038/s41598-020-57534-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kukuyan AM, Sementino E, Kadariya Y, et al. Inactivation of Bap1 cooperates with losses of Nf2 and Cdkn2a to drive the development of pleural malignant mesothelioma in conditional mouse models. Cancer Res. 2019;79(16):4113-4123. 10.1158/0008-5472.CAN-18-4093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Williams EA, Santagata S, Wakimoto H, et al. Distinct genomic subclasses of high-grade/progressive meningiomas: NF2-associated, NF2-exclusive, and NF2-agnostic. Acta Neuropathol Commun. 2020;8(1):171-171. 10.1186/s40478-020-01040-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ugurluer G, Chang K, Mayeda M, et al. A comprehensive genome-based mutational analysis by next generation sequencing technology in patients with malignant pleural and peritoneal mesothelioma. Int J Rad Oncol Biol Phys. 2015;93(3):S185-S186. [Google Scholar]
- 39. Ammendola S, Simbolo M, Ciaparrone C, et al. Intraventricular meningiomas: clinical-pathological and genetic features of a monocentric series. Curr Oncol. 2022;29(1):178-185. 10.3390/curroncol29010017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Badhai J, Pandey GK, Song JY, et al. Combined deletion of Bap1, Nf2, and Cdkn2ab causes rapid onset of malignant mesothelioma in mice. J Exp Med. 2020;217(6):e20191257-e20191257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Shapiro IM, Kolev VN, Vidal CM, et al. Merlin deficiency predicts FAK inhibitor sensitivity: a synthetic lethal relationship. Sci Transl Med. 2014;6(237):237ra268-237ra268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Pachow D, Andrae N, Kliese N, et al. mTORC1 inhibitors suppress meningioma growth in mouse models. Clin Cancer Res. 2013;19(5):1180-1189. 10.1158/1078-0432.CCR-12-1904 [DOI] [PubMed] [Google Scholar]
- 43. Bush ML, Burns SS, Oblinger J, et al. Treatment of vestibular schwannoma cells with ErbB inhibitors. Otol Neurotol. 2012;33(2):244-257. 10.1097/MAO.0b013e31823e287f [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Fuse MA, Dinh CT, Vitte J, et al. Preclinical assessment of MEK1/2 inhibitors for neurofibromatosis type 2-associated schwannomas reveals differences in efficacy and drug resistance development. Neuro Oncol. 2019;21(4):486-497. 10.1093/neuonc/noz002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lang GT, Jiang YZ, Shi JX, et al. Characterization of the genomic landscape and actionable mutations in Chinese breast cancers by clinical sequencing. Nat Commun. 2020;11(1):5679-5679. 10.1038/s41467-020-19342-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Sourbier C, Liao PJ, Ricketts CJ, et al. Targeting loss of the Hippo signaling pathway in NF2-deficient papillary kidney cancers. Oncotarget. 2018;9(12):10723-10733. 10.18632/oncotarget.24112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. McDermott DF, Lee JL, Bjarnason GA, et al. Open-label, single-arm phase II study of pembrolizumab monotherapy as first-line therapy in patients with advanced clear cell renal cell carcinoma. J Clin Oncol. 2021;39(9):1020-1028. 10.1200/JCO.20.02363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Powles T, Plimack ER, Soulières D, et al. Pembrolizumab plus axitinib versus sunitinib monotherapy as first-line treatment of advanced renal cell carcinoma (KEYNOTE-426): extended follow-up from a randomised, open-label, phase 3 trial. Lancet Oncol. 2020;21(12):1563-1573. 10.1016/S1470-2045(20)30436-8 [DOI] [PubMed] [Google Scholar]
- 49. Choueiri TK, Powles T, Burotto M, et al. ; CheckMate 9ER Investigators. Nivolumab plus cabozantinib versus sunitinib for advanced renal-cell carcinoma. N Engl J Med. 2021;384(9):829-841. 10.1056/NEJMoa2026982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Kinoshita Y, Hamasaki M, Yoshimura M, et al. Hemizygous loss of NF2 detected by fluorescence in situ hybridization is useful for the diagnosis of malignant pleural mesothelioma. Mod Pathol. 2020;33(2):235-244. 10.1038/s41379-019-0309-6 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated and analyzed in this study can be provided by the corresponding author upon reasonable request.