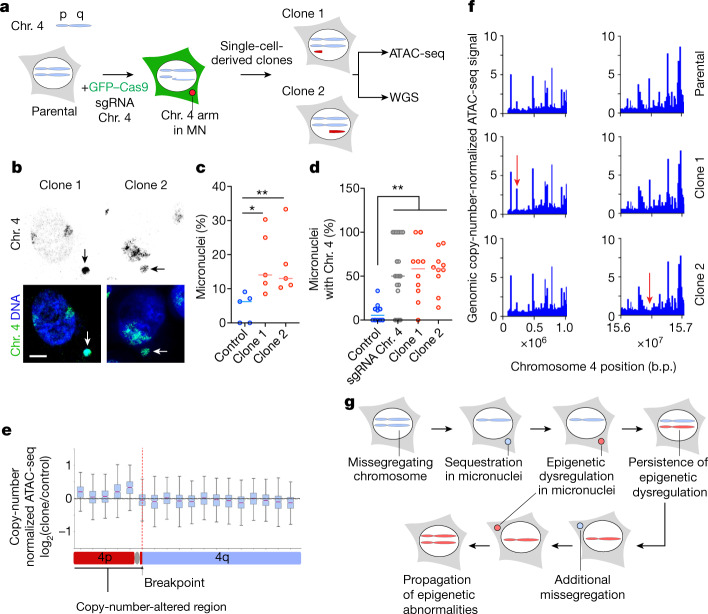
Fig. 5. Chromosomal transit in micronuclei promotes heritable epigenetic abnormalities.
a, Experimental schematic depicting missegregation of Chr. 4 in RPE-1 TP53-knockout cells induced using a chromosome 4 telomeric CRISPR guide; sgRNA, single guide RNA. b, Representative chromosome paint images from 2 biological replicates in RPE-1 TP53-knockout single-cell clones from a chromosome-4 missegregation system showing chromosome 4 (green) and DNA (blue). Scale bar, 5 µm. c, Percentage of micronuclei in single-cell-derived clones derived after transfection of TP53-knockout RPE-1 cells with either chromosome-4-targeting sgRNA or non-targeting control sgRNA; *P < 0.05, **P < 0.01, two-sided Mann–Whitney U-test; bars represent medians; n = 5 fields of view under 63× magnification. d, Percentage of micronuclei that contain chromosome 4 in single-cell-derived clones derived after transfection of TP53-knockout RPE-1 cells with chromosome 4 targeting sgRNA or non-targeting control or the mixed cell population shortly after transfection with the chromosome 4 targeting sgRNA; **P < 0.01, two-sided Mann–Whitney U-test; bars represent median; n = 10 fields of view under 63× magnification. e, Box plot (10-Megabase bin) showing copy-number normalized change of ATAC-seq counts on chromosome 4 between clone 1 and the control clone treated with the non-targeting guide sgRNA. The line represents the median and error bars represent minimum and maximum values; n = 3; solid red line represents the median; the bounds of the box are the interquartile range (Q1 to Q3); error bars are defined by 1.5*interquartile range beyond Q1 and Q3. f, Genome viewer plot showing copy-number-normalized ATAC-seq counts in the region of chromosome 4 that is copy-number altered in clone 1 (left) or clone 2 (right). Red arrows denote differentially accessible peaks compared with the control clone. g, Model showing how continuous chromosomal missegregation followed by micronucleation can introduce heritable epigenetic dysregulation. Because chromosomally unstable cells tend to continuously undergo chromosomal missegregation, this could in turn propagate epigenetic abnormalities through the same mechanism, leading to epigenetic heterogeneity.