Abstract
Background
People living with HIV may present co-morbidities requiring the initiation and subsequently the discontinuation of medications with inducing properties. The time to reach maximal enzyme induction and to return to baseline enzyme levels has not been thoroughly characterized.
Objective
The aim of this study was to evaluate the onset and disappearance of dolutegravir [uridine diphosphate glucuronosyltransferase (UGT) 1A1 and cytochrome P450 (CYP) 3A4 substrate] and raltegravir (UGT1A1 substrate) induction with strong and moderate inducers using physiologically based pharmacokinetic (PBPK) modeling.
Methods
The predictive performance of the PBPK model to simulate dolutegravir and raltegravir pharmacokinetics and to reproduce the strength of induction was verified using clinical drug–drug interaction studies (steady-state induction) and switch studies (residual induction). The model was considered verified when the predictions were within 2-fold of the observed data. One hundred virtual individuals (50% female) were generated to simulate the unstudied scenarios. The results were used to calculate the fold-change in CYP3A4 and UGT1A1 enzyme levels upon initiation and discontinuation of strong (rifampicin) or moderate (efavirenz or rifabutin) inducers.
Results
The time for reaching maximal induction and subsequent disappearance of CYP3A4 induction was 14 days for rifampicin and efavirenz but 7 days for rifabutin. The distinct timelines for the moderate inducers relate to their different half-lives and plasma concentrations. The induction and de-induction processes were more rapid for UGT1A1.
Conclusions
Our simulations support the common practice of maintaining the adjusted dosage of a drug for another 2 weeks after stopping an inducer. Furthermore, our simulations suggest that an inducer should be administered for at least 14 days before conducting interaction studies to reach maximal induction.
Supplementary Information
The online version contains supplementary material available at 10.1007/s13318-023-00833-9.
Key Points
| Our simulations indicate that at least 14 days are needed to reach maximal induction and to return to CYP3A4 baseline enzymes levels in the presence of an inducer. This implies that an inducer should be administered for at least 14 days before conducting drug–drug interaction studies to reach maximal induction. |
| Our results also support the common practice of maintaining the adjusted dosage of a drug for another 2 weeks upon discontinuation of an inducer. |
| The time to reach maximal induction and resolution of induction is shorter for UGT1A1. |
Introduction
Potent antiretroviral drugs (ARVs) dramatically changed the prognosis of HIV-infection. Treated people living with HIV (PLWH) have a life expectancy that is close, if not identical, to people without HIV infection [1]. Thus, the management of drug–drug interactions (DDIs) with ARVs represents a key clinical activity considering that most PLWH may develop at least one comorbidity during their lifespan. ARVs have considerably improved over the years and have a notably lower propensity to cause DDIs; however, several first-line ARVs are subject to DDIs with inducers of drug metabolizing enzymes [i.e., cytochrome P450 (CYP) and uridine diphosphate glucuronosyltransferase (UGT)] [2].
The induction of drug metabolizing enzymes occurs upon the binding of a drug to a nuclear receptor [i.e., pregnane X receptor (PXR), constitutive androstane receptor (CAR) or aryl hydrocarbon receptor (AhR)]. PXR is involved in the upregulation of CYP3A4 and UGTs expression. Following the heterodimerization with the 9-cis-retinoic acid receptor (RXR), the RXR-PXR complex interacts with response elements of target genes leading to the transcription of mRNA encoding the drug metabolizing enzymes [3, 4]. Thus, induction takes several days to fully develop and to disappear upon initiation and discontinuation of an inducer. This statement is supported by a clinical DDI study showing notably that the moderate inducer efavirenz reduced rilpivirine (CYP3A4 substrate) area under the concentration–time curve (AUCτ) by 46% and 18% at days 1 and 14, respectively, post-discontinuation of efavirenz [5]. However, it is unclear whether the time to reach maximal induction and resolution of induction varies depending on the inducer strength and elimination half-life (t½) or on the degradation rate of the metabolic enzyme. This question is of particular interest as the first-line ARVs dolutegravir (metabolism by UGT1A1 > CYP3A4) or raltegravir (UGT1A1) require a dose adjustment when co-administered with strong inducers as does doravirine (CYP3A4) when given with moderate inducers [6–8]. It is common practice to maintain the adjusted dosage of the ARV for another 2 weeks after the last dose of the inducer, regardless of the properties of the inducer or the enzyme involved in the metabolism of the ARV. However, data to support this practice are missing.
Physiologically based pharmacokinetic (PBPK) modeling has demonstrated its predictive power to simulate clinically relevant yet unstudied DDI scenarios [9, 10]. PBPK modeling combines in vitro data and clinically observed data to simulate pharmacokinetics in virtual individuals. The virtual population used to inform the PBPK model is developed based on observed blood flows, organ weights, and other physiological parameters required to simulate drug disposition. To date, PBPK modeling has been applied to investigate the time needed for CYP3A4 enzyme levels to return to baseline after stopping the strong inducer rifampicin; however, no data are available for moderate inducers or for the recovery of UGT1A1 abundance [11, 12].
Since dolutegravir is metabolized by UGT1A1 and CYP3A4 [13] and raltegravir by UGT1A1 [14], we selected these ARVs to investigate the time required for CYP3A4 and UGT1A1 abundances to return to baseline after stopping rifampicin (t½ 2–3 h [15]) or after discontinuing two moderate inducers with distinct half-lives, namely efavirenz (t½ 40–55 h after multiple doses [16]) and rifabutin (t½ 45 h [17]). We also investigated the time to reach maximal induction of CYP3A4 and UGT1A1 upon initiation of rifampicin, efavirenz and rifabutin.
Methods
We took a 3-step approach. First, we evaluated the performance of the PBPK model to simulate the pharmacokinetics of each individual drug against observed clinical data after intravenous and oral administration. Second, we verified that the model correctly reproduces the strength of induction using available DDI studies (reflecting steady-state induction) and switch studies (reflecting residual induction). Third, we used the fully verified PBPK model to simulate the unstudied scenarios. Each scenario included 100 virtual individuals (50% female) between 20 and 50 years of age. For all the scenarios simulated, we considered the changes in enzyme levels rather than the activity (clearance); however, it should be noted that these parameters are correlated (i.e., an increase in enzyme levels correlates with an increase in clearance).
PBPK Model and Drug Model Development
Our in-house PBPK model was built in Matlab®2020a (MathWorks, Natick, MA, USA) [18]. In order to generate a cohort of virtual individuals, the model was informed with equations previously developed by our group describing the physiological changes in a Caucasian healthy population aged 20–99 years [19]. The equations are presented in Supplementary Table 2. The drug models were developed using published in vitro physicochemical parameters of each drug or, if the data were not available, using in silico predictions. The model parameters for dolutegravir, raltegravir, rifampicin, efavirenz, and rifabutin are summarized in Supplementary Table 1. The model performance to predict the pharmacokinetics was evaluated for intravenous and oral administration, and the model was considered verified when the predictions were within 2-fold of the observed clinical data (Data on file; Bettonte et al.) [20, 21].
Model Verification Against Observed DDI and Switch Studies
The fraction of dolutegravir metabolism by UGT1A1 and CYP3A4 and the fraction of raltegravir metabolized by UGT1A1, as well as the strength of induction were verified against published DDI clinical studies in which the pharmacokinetics of dolutegravir or raltegravir were measured in the absence and in the presence of rifampicin, efavirenz, or rifabutin after steady-state administration [22–26]. In addition, published switch studies were used to verify the performance of the PBPK model to predict the residual induction after stopping efavirenz; therefore, the pharmacokinetics of the CYP3A4 substrate rilpivirine dosed at 25 mg and the UGT1A1/CYP3A4 substrate dolutegravir dosed at 50 mg were simulated on different days following efavirenz discontinuation [5, 27]. The model was considered to be verified when the simulations were within 2-fold, as described in the PBPK guidelines [28, 29]. We also used the more stringent criterion proposed by Guest et al. for the verification of all the DDI scenarios [30]. Finally, the change in CYP3A4 and UGT1A1 enzyme levels during and after stopping the administration of efavirenz were visually checked to extrapolate the time needed to return to baseline enzyme levels. The 1.25-fold induction threshold was considered as the limit below which the ratio of the AUC (i.e., AUC in the presence vs. the absence of a perpetrator), was outside the no-boundary efficacy limit of 0.8.
Model Simulations of Unknown Clinical Scenarios
A cohort of 100 virtual individuals aged 20–50 (50% female) was generated to simulate the unknown scenarios presented in Table 1 and Table 2. The study design of de Wet et al. [27], in which dolutegravir (50 mg once daily) was initiated 12 h after stopping efavirenz, was applied first to verify the residual induction on dolutegravir on days 7, 14, and 28 post-efavirenz dosing. This same dosing schedule was subsequently used to simulate dolutegravir pharmacokinetics following the discontinuation of rifampicin or rifabutin, and the pharmacokinetics of raltegravir during the switch scenario from rifampicin, efavirenz, or rifabutin. The DDI ratio (reflecting the residual induction) was calculated as the ratio of the trough concentration (Ctrough) and AUCτ in the presence versus the absence of the residual inducing effect of rifampicin, efavirenz, or rifabutin. As described in Sect. “Model Verification Against Observed DDI and Switch Studies”, the changes in CYP3A4 and UGT1A1 enzyme levels were visually checked and the time to return to baseline (i.e., below the 1.25-fold induction threshold) was extrapolated.
Table 1.
Effect of residual induction on dolutegravir pharmacokinetics at days 1, 7, 14, and 28 post-rifampicin, efavirenz, and rifabutin dosing
| Absence inducer | After stopping inducer | DDI ratio (residual induction) | Reference | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Observed | Predicted | Ratio P/O | Observed | Predicted | Ratio P/O | Observed | Predicted | Ratio P/O | ||
| Dolutegravir—day 1 after stopping rifampicin | ||||||||||
| Ctrough [ng/mL] | – | 749 (50) | – | – | 62 (109) | – | – | 0.08 | – | – |
| AUC0-τ [ng×h/mL] | – | 35,677 (26) | – | – | 15,549 (38) | – | – | 0.44 | – | |
| Dolutegravir—day 7 after stopping rifampicin | ||||||||||
| Ctrough [ng/mL] | – | 1068 (69) | – | – | 431 (79) | – | – | 0.40 | – | – |
| AUC0-τ [ng×h/mL] | – | 52,295 (45) | – | – | 32,154 (42) | – | – | 0.61 | – | |
| Dolutegravir—day 14 after stopping rifampicin | ||||||||||
| Ctrough [ng/mL] | – | 1070 (70) | – | – | 806 (70) | – | – | 0.75 | – | – |
| AUC0-τ [ng×h/mL] | – | 52,391 (46) | – | – | 44,172 (43) | – | – | 0.84 | – | |
| Dolutegravir—day 28 after stopping rifampicin | ||||||||||
| Ctrough [ng/mL] | – | 1070 (70) | – | – | 1046 (69) | – | – | 0.98 | – | – |
| AUC0-τ [ng×h/mL] | – | 52,391 (46) | – | – | 51,624 (46) | – | – | 0.99 | – | |
| Dolutegravir—day 1 after stopping efavirenz | ||||||||||
| Ctrough [ng/mL] | – | 910 ± 456 | – | – | 435 ± 332 | – | – | 0.48 | – | – |
| AUC0-τ [ng×h/mL] | – | 37,681 ± 9941 | – | – | 27,141 ± 8807 | – | – | 0.72 | – | |
| Dolutegravir—day 7 after stopping efavirenz | ||||||||||
| Ctrough [ng/mL] | – | 1497 ± 1050 | – | 938 ± 955 | 948 ± 698 | 1.01 | – | 0.63 | [27] | |
| AUC0-τ [ng×h/mL] | – | 60,683 ± 28,494 | – | – | 45,429 ± 19,778 | – | – | 0.75 | – | |
| Dolutegravir—day 14 after stopping efavirenz | ||||||||||
| Ctrough [ng/mL] | – | 1505 ± 1070 | – | 861 ± 627 | 1217 ± 833 | 1.41 | – | 0.81 | – | [27] |
| AUC0-τ [ng×h/mL] | – | 60,961 ± 29,091 | – | – | 53,111 ± 23,105 | – | – | 0.87 | – | |
| Dolutegravir—day 28 after stopping efavirenz | ||||||||||
| Ctrough [ng/mL] | – | 1505 ± 1071 | – | 1219 ± 710 | 1428 ± 977 | 1.17 | – | 0.95 | – | [27] |
| AUC0-τ [ng×h/mL] | – | 60,966 ± 29,113 | – | - | 58,909 ± 26,763 | - | – | 0.97 | – | |
| Dolutegravir—day 1 after stopping rifabutin | ||||||||||
| Ctrough [ng/mL] | – | 749 (50) | – | – | 554 (54) | – | – | 0.74 | – | – |
| AUC0-τ [ng×h/mL] | – | 35,677 (26) | – | – | 31,539 (27) | – | – | 0.88 | – | |
| Dolutegravir—day 7 after stopping rifabutin | ||||||||||
| Ctrough [ng/mL] | – | 1068 (69) | – | – | 934 (67) | – | – | 0.87 | – | – |
| AUC0-τ [ng×h/mL] | – | 52,294 (45) | – | – | 48,073 (43) | – | – | 0.92 | – | |
| Dolutegravir—day 14 after stopping rifabutin | ||||||||||
| Ctrough [ng/mL] | – | 1070 (70) | – | – | 1029 (69) | – | – | 0.96 | – | – |
| AUC0-τ [ng×h/mL] | – | 52,390 (46) | – | – | 51,090 (45) | – | – | 0.98 | – | |
| Dolutegravir—day 28 after stopping rifabutin | ||||||||||
| Ctrough [ng/mL] | – | 1070 (70) | – | – | 1067 (70) | – | – | 1.00 | – | – |
| AUC0-τ [ng×h/mL] | – | 52,391 (46) | – | – | 52,288 (46) | – | – | 1.00 | – | |
The results are represented as geometric mean (CV), unless otherwise specified as mean ± SD
AUC0-τ area under the concentration-time curve to tau; Ctrough trough concentration; DDI drug–drug interaction; O observed; P predicted
Table 2.
Predicted effect of residual induction on raltegravir pharmacokinetics at days 1, 7, 14, and 28 post-rifampicin, efavirenz, and rifabutin dosing
| Absence inducer | After stopping inducer | DDI ratio (residual induction) | |
|---|---|---|---|
| Raltegravir—day 1 after stopping rifampicin | |||
| Ctrough [ng/mL] | 107 (69) | 84 (51) | 0.78 |
| AUC0-τ [ng×h/mL] | 7312 (38) | 6192 (30) | 0.85 |
| Raltegravir—day 7 after stopping rifampicin | |||
| Ctrough [ng/mL] | 128 (76) | 128 (76) | 1.00 |
| AUC0-τ [ng×h/mL] | 7740 (41) | 7739 (41) | 1.00 |
| Raltegravir—day 14 after stopping rifampicin | |||
| Ctrough [ng/mL] | 128 (76) | 128 (76) | 1.00 |
| AUC0-τ [ng×h/mL] | 7740 (41) | 7740 (41) | 1.00 |
| Raltegravir—day 28 after stopping rifampicin | |||
| Ctrough [ng/mL] | 128 (76) | 128 (76) | 1.00 |
| AUC0-τ [ng×h/mL] | 7740 (41) | 7740 (41) | 1.00 |
| Raltegravir—day 1 after stopping efavirenz | |||
| Ctrough [ng/mL] | 108 (83) | 82 (49) | 0.76 |
| AUC0-τ [ng×h/mL] | 7357 (40) | 6191 (28) | 0.84 |
| Raltegravir—day 7 after stopping efavirenz | |||
| Ctrough [ng/mL] | 129 (82) | 115 (71) | 0.89 |
| AUC0-τ [ng×h/mL] | 7777 (42) | 7243 (38) | 0.93 |
| Raltegravir—day 14 after stopping efavirenz | |||
| Ctrough [ng/mL] | 129 (82) | 123 (79) | 0.95 |
| AUC0-τ [ng×h/mL] | 7778 (42) | 7547 (41) | 0.97 |
| Raltegravir—day 28 after stopping efavirenz | |||
| Ctrough [ng/mL] | 129 (82) | 127 (83) | 0.99 |
| AUC0-τ [ng×h/mL] | 7778 (42) | 7716 (42) | 0.99 |
| Raltegravir—day 1 after stopping rifabutin | |||
| Ctrough [ng/mL] | 107 (82) | 102 (79) | 0.95 |
| AUC0-τ [ng×h/mL] | 7336 (40) | 7098 (38) | 0.97 |
| Raltegravir—day 7 after stopping rifabutin | |||
| Ctrough [ng/mL] | 128 (82) | 128 (82) | 1.00 |
| AUC0-τ [ng×h/mL] | 7553 (42) | 7744 (42) | 1.00 |
| Raltegravir—day 14 after stopping rifabutin | |||
| Ctrough [ng/mL] | 128 (82) | 128 (82) | 1.00 |
| AUC0-τ [ng×h/mL] | 7554 (42) | 7753 (42) | 1.00 |
| Raltegravir—day 28 after stopping rifabutin | |||
| Ctrough [ng/mL] | 128 (82) | 128 (82) | 1.00 |
| AUC0-τ [ng×h/mL] | 7554 (42) | 7754 (42) | 1.00 |
The results are represented as geometric mean (CV)
AUC0-τ area under the concentration–time curve to tau; Ctrough trough concentration; DDI drug-drug interaction
Sensitivity Analysis
The disappearance of the inducing effect is governed by the t½ of the perpetrator and by the degradation constant (Kdeg) of the enzyme, which correlates with t½ of the enzyme [31]. Since this parameter has not been measured in humans, the Kdeg value for UGT1A1 implemented in our in-house PBPK model was derived from a rat model [32]. Therefore, we conducted a sensitivity analysis to determine how changes in the Kdeg UGT1A1 value impact the residual induction effect of rifampicin on raltegravir (UGT1A1 substrate) AUC, and Ctrough at days 1, 7, 14, and 28 post-rifampicin dosing. For this analysis, we considered the situation of a single male individual aged 35 years old [body mass index (BMI) 25 kg/m2] and we modified by 1% the original UGT1A1 Kdeg value reported in the literature [32]. The impact of Kdeg on the PBPK model uncertainty was calculated using Eq. 1 [33]:
| 1 |
where d0 is the model-predicted dose metric of interest (i.e., AUC and Ctrough) with the original parameter value, d1 is the model-predicted dose metric of interest (i.e., AUC and Ctrough) with a 1% increase in parameter value, p0 is the original parameter value (i.e., Kdeg), and p1 is the parameter value with a 1% increase (i.e., Kdeg).
The model uncertainty was evaluated using the criteria defined by Teeguarden et al. [33, 34]:
Low: sensitivity coefficient is between 0.1 and 0.15. Low model uncertainty is obtained when the available data are measured in the correct species, the coefficient of variation (CV) is less than 0.5, or the parameter was successfully verified using PBPK modeling.
Medium: sensitivity coefficient is between 0.15 and 0.5. Medium model uncertainty can present with data scaled from a different species, but for which the scaling holds across species.
High: sensitivity coefficient is greater than 0.5. High model uncertainty can present when the parameter of interest is not available.
Results
Drug Model Development
The PBPK model was able to predict the pharmacokinetics of all evaluated drugs within 2-fold of the observed clinical data. The simulations of the various drugs as well as the observed clinical data have been previously published by our group (Data on file; Bettonte et al.) [20, 21].
Model Verification Against Observed DDI and Switch Studies
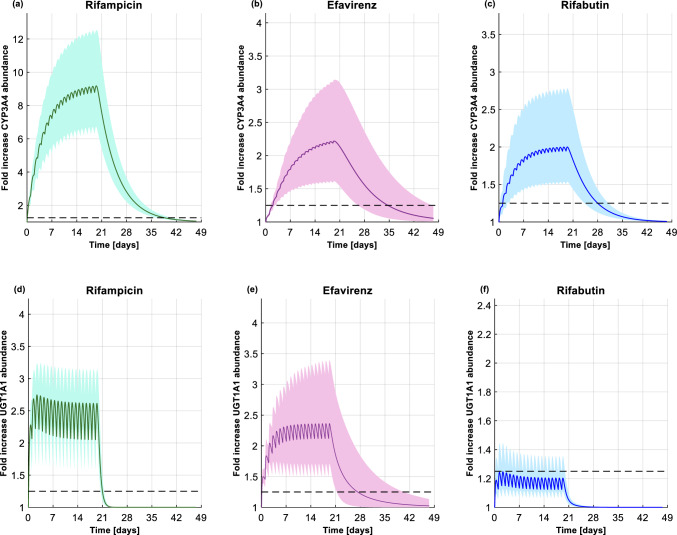
The model correctly predicted the fraction of UGT1A1 and CYP3A4 metabolism for dolutegravir, as the DDI between dolutegravir and rifampicin, efavirenz, or rifabutin were simulated within 2-fold of the observed data, as shown in Supplementary Table 3. Furthermore, the predicted versus observed ratio for AUC, and Ctrough were within the upper and lower limits defined by Guest et al. [30]. However, the ratio for the peak concentration (Cmax) was outside the upper and lower limits because the clinical study paradoxically reported an increase in dolutegravir Cmax [22], even though rifabutin is an inducer (Supplementary Fig. 1). Similarly, for raltegravir, the observed Cmax and AUC used for the criterion proposed by Guest et al. [30] reflected an inhibition effect rather than induction [26] (Supplementary Fig. 1); however, the ratio for Ctrough was within the upper and lower limits proposed by Guest et al. [30] (Supplementary Fig. 1). Finally, as for dolutegravir, all the DDI predictions between raltegravir and rifampicin, efavirenz, or rifabutin were within 2-fold of the clinical observed data (Supplementary Table 3). The PBPK model predicted, also correctly, the residual inducing effect on dolutegravir pharmacokinetics at days 7, 14, and 28 (Table 1) post-efavirenz dosing. The performance of the PBPK model to reproduce residual induction was also verified using clinical data from the efavirenz to rilpivirine switch study [5]. The simulated rilpivirine pharmacokinetic parameters were within 2-fold of the observed clinical data at days 1, 14, 21 post-efavirenz dosing (Supplementary Table 4) and borderline for the upper and lower limits proposed by Guest et al. (Data on file; Bettonte et al) [30] (Supplementary Fig. 2). The simulation results showed that, upon initiation of efavirenz, the mean maximal induction of CYP3A4 was reached after 19 days (Fig. 1b) and the one of UGT1A1 after 7 days (Fig. 1e). Upon discontinuation of efavirenz, 14 days were necessary for the CYP3A4 enzyme level to go below the 1.25-fold threshold (Fig. 1b). However, when considering the population variability, 69% of the individuals were predicted to have reached the 1.25-fold CYP3A4 abundance threshold at this time point. On the other hand, the UGT1A1 enzyme level was predicted to require 7 days to return close to the baseline enzyme level, with 72% of individuals predicted to achieve the 1.25-fold abundance threshold by this time point (Fig. 1e).
Fig. 1.
Fold increase in hepatic CYP3A4 abundance upon initiation and discontinuation of a rifampicin (600 mg once daily), b efavirenz (600 mg once daily), and c rifabutin (300 mg once daily). Fold increase in hepatic UGT1A1 abundance upon initiation and discontinuation of d rifampicin (600 mg once daily), e efavirenz (600 mg once daily), and f rifabutin (300 mg once daily). In all the figures, the solid line, the shaded area, and the dashed line represent the mean, the 90% normal range, and the 1.25-fold cut-off threshold above which the AUC ratio (i.e., in the presence vs. the absence of an inducer) is below 0.8. CYP cytochrome P450; UGT uridine diphosphate glucuronosyltransferase; AUC area under the concentration–time curve
Model Simulations of Unknown Clinical Scenarios
The residual inducing effect of rifampicin was predicted to decrease dolutegravir AUCτ by 56% and 39% at days 1 and 7 post-dosing, respectively. The mitigation of the rifampicin-inducing effect was apparent after 14 days as the decrease in dolutegravir AUCτ was predicted to be minimal (16%), and negligible (1%) after 28 days (Table 1). On the other hand, the residual-inducing effect of rifampicin was more pronounced on dolutegravir Ctrough , as it was predicted to be decreased by 92% and 60% on days 1 and 7, respectively. A 25% reduction in dolutegravir Ctrough was still predicted at day 14 post-dosing, but it was negligible at day 28 (Table 1). The simulation results showed that the residual induction of rifampicin reduced raltegravir AUCτ and Ctrough by 15% and 22%, respectively, on day 1 post-dosing; however, the inducing effect was negligible after 7 days (Table 2). As depicted in Fig. 1a, rifampicin was predicted to require at least 14 days of continuous administration to reach near-maximal induction of CYP3A4. After stopping rifampicin, 18 days were necessary for 55% of the individuals to reach the 1.25-fold abundance threshold. Regarding UGT1A1, the steady-state and the time to go back to the baseline abundance were predicted to be faster than with CYP3A4 (Fig. 1d).
The residual inducing effect of rifabutin was predicted to decrease dolutegravir AUCτ and Ctrough by 12% and 26%, respectively, on day 1 post-dosing. The AUCτ was still reduced by 8% and Ctrough by 13% 7 days after stopping rifabutin. However, the induction was negligible after 14 days (reduction in AUCτ and Ctrough by 2% and 4%, respectively) (Table 1). The simulation results showed that upon discontinuation of rifabutin, the raltegravir AUCτ and Ctrough were reduced by 3% and 5%, respectively, 1 day post-dosing. The residual induction effect of rifabutin is negligible 7 days post-discontinuation (Table 2). Upon initiation of rifabutin, near-maximal CYP3A4 induction was predicted to be reached in 10 days (Fig. 1c) and in 1 day for UGT1A1 (Fig. 1f). However, upon discontinuation of rifabutin, 7 days were necessary for 58% of the individuals to have the CYP3A4 abundance reaching the 1.25-fold threshold (Fig. 1c). UGT1A1 induction by rifabutin was not strong enough to go above this threshold (Fig. 1f).
The residual inducing effect of efavirenz was predicted to decrease the raltegravir AUCτ by 16%, 7%, and 3% at days 1, 7, and 14 post-dosing. The Ctrough was reduced by 24%, 11%, and 5% on days 1, 7, and 14 post-dosing (Table 2). Seven days were necessary to return to UGT1A1 baseline enzyme levels.
Sensitivity Analysis
Modifying the Kdeg of UGT1A1 by 1% resulted in sensitivity coefficients for AUC and Ctrough at days 1, 7, 14, and 28 post-rifampicin dosing which were lower than 0.1, thereby reflecting a low model uncertainty.
Discussion
Contemporary first-line ARVs have a lower potential to cause DDIs; however, these agents can be impacted by other co-medications that a PLWH may be taking, notably those with inducing properties. Thus, the characterization of the onset and disappearance of induction is warranted, not only for the appropriate management of DDIs in clinical practice but also for the correct design of clinical studies.
To date, the onset and disappearance of CYP3A4 enzyme induction has only been characterized for rifampicin. Using PBPK modeling, Kapetas et al. notably showed that the maximal induction of CYP3A4 was achieved faster in the intestine compared to the liver, for which at least a 10-day rifampicin treatment course was needed before reaching maximal induction [11]. Our simulations with rifampicin are in line with the study by Kapetas et al., since hepatic CYP3A4 was predicted to require approximately 14 days to reach near-maximal induction and 18 days to return to baseline abundance after stopping rifampicin. We did not evaluate the induction of intestinal CYP3A4 given that the time to reach maximal induction and de-induction was shown to be quick in the study by Kapetas et al. [11]. The more rapid disappearance of induction in the intestine compared to the liver is explained by the Kdeg of the enzyme which is equal to 0.03 h-1 for intestinal CYP3A4 [9] and 0.0077 h–1 for hepatic CYP3A4 [9]. Our simulations are also supported by clinical data and PBPK modeling studies reporting that induction can take up to 14 days to reach maximal effect [11, 12, 35–43]. In addition, our predictions are consistent with studies evaluating the change in biomarkers (i.e., 6-beta-hydroxy-cortisol excretion and the 6-beta-hydroxy-cortisol/cortisol ratio) at baseline (pre-inducer administration), during the administration of rifampicin, and after stopping the inducer [44, 45]. Our predictions showed that 18 days are necessary to return to baseline CYP3A4 enzyme levels. Our results are consistent with the results published by Reitman et al. [46]. The authors showed that 28 days were enough to completely washout the residual inducing effect of rifampicin on midazolam, since 15 days post-discontinuation of rifampicin, midazolam AUC, and Cmax, were still reduced by 19% and 27%, respectively. However, the authors concluded that the residual inducing effect observed after 15 days may not be clinically significant [46]. Baneyx et al. also demonstrated that the time to return to baseline enzyme levels for CYP3A4 is 15 days [42].
Dolutegravir and raltegravir were chosen as victim drugs in our study to investigate the effect on UGT1A1. Of interest, maximal UGT1A1 induction was achieved faster upon initiation of rifampicin compared to CYP3A4 (Fig. 1a and d). In contrast to CYP3A4, no studies have evaluated the change in UGT1A1 abundance during and after stopping rifampicin. However, a clinical study evaluated the level of bilirubin glucuronidation (UGT1A1 substrate [47]). The clinical study showed that the level of bilirubin conjugation increased at the beginning of the treatment, with rifampicin reaching a plateau after 6 days. On the other hand, upon discontinuation of rifampicin, the level of bilirubin glucuronidation ratios were still significantly higher, indicating that the UGT1A1 activity did not return to baseline level, which could possibly also be explained by lifestyle factors [43]. Another clinical study investigating the DDI between dolutegravir and rifapentine (strong CYP3A4 and UGT1A1 inducer [48]) showed decreases in dolutegravir Ctrough of 23%, 64%, and 56% at days 1, 2 and 6 post-rifapentine administration [48]. Given that UGT1A1 is the major enzyme involved in dolutegravir and raltegravir metabolism, these observed clinical data are supportive of the fast UGT1A1 induction upon initiation of a strong inducer, similar to our model prediction. On the other hand, after discontinuation of rifampicin, our simulations show that the residual inducing effect led to a decrease of 60% in dolutegravir Ctrough. A similar dolutegravir Ctrough reduction has been reported in a clinical study after discontinuation of rifapentine (another strong inducer), in which dolutegravir Ctrough was still reduced by 56% at day 6 post-rifapentine administration [48]. Although our simulations indicate a rapid return to UGT1A1 baseline enzyme level, the longer period for dolutegravir Ctrough recovery could relate to CYP3A4, which contributes to dolutegravir metabolism and which requires more time to recover from induction. Thus, our findings for UGT1A1 pure substrates should be interpreted with caution, since the Kdeg value for UGT1A1 was derived from an animal model and therefore may not be extrapolated to humans.
There is a paucity of data on the timeline of induction for moderate inducers such as efavirenz and rifabutin. Based on our results, rifabutin needs at least 7 days to attain near-maximal CYP3A4 induction, while efavirenz needs at least 14 days. After stopping their administration, rifabutin needs 7 days to return to baseline, while efavirenz needs 14 days. These distinct timelines relate to their different plasma concentrations at the time of stopping their administration. Twenty-four hours after stopping rifabutin, its plasma concentration is lower than its half-maximal inhibitory concentration (IC50) (i.e., 0.3 µm [49]), and therefore the induction effect is minimal. In contrast, 24 h after stopping efavirenz, its concentration is still above its IC50 (i.e., 3.9 µm [50]), and the concentration remains for at least another week around the IC50 value, causing a persisting inducing effect on co-administered drugs. As for rifampicin, the onset of UGT1A1 induction was faster than CYP3A4 upon initiating efavirenz or rifabutin. The DDI between efavirenz and raltegravir has been studied in a clinical trial and, to ensure full induction of UGT1A1, the authors decided to administer efavirenz for 12 days [25]. However, based on Fig. 1e, 7 days would have been sufficient to have near-full induction of UGT1A1. As for CYP3A4, the residual inducing effect on UGT1A1 lasts longer for efavirenz compared to rifabutin, given that the plasma concentration of efavirenz is close to its IC50.
Altogether, available data and the results of our study support the common practice to maintain the adjusted dosage of ARVs for another 2 weeks after the last dose of the inducer. Although our simulations suggest that this time window could be shortened to 1 week after stopping rifabutin, it is preferable to recommend 2 weeks for all inducers, not only to prevent errors but also because the concentrations of inducers, which impact the offset of induction, are not routinely measured. Furthermore, it should be emphasized that the resolution of induction may last longer in some individuals, as demonstrated in clinical trials with CYP3A4 substrates [46, 51], and as indicated by the large variability in enzyme fold-induction of CYP3A4 and UGT1A1 in our virtual cohort representing the healthy Caucasian population (Fig. 1). Our predictions are also supported by the large variability observed in clinical trials [52].
Several limitations should be acknowledged. First, given the limited clinical data for the effect of different inducers on the change in CYP3A4 and UGT1A1 enzyme abundance, we could not fully evaluate our whole-body PBPK model. However, the PBPK model was extensively verified prior to simulating unstudied scenarios, and therefore we are confident that our conclusions are correct. Second, the paucity of biological data for UGT enzymes, particularly data regarding the degradation rate constant in humans, are missing, which is the reason why our in-house PBPK model uses a Kdeg value for UGT1A1 enzyme derived from animal model [32]. Despite this limitation, the sensitivity analysis for this parameter shows a low model uncertainty, but, nevertheless, future studies should determine this parameter in humans.
Conclusions
This study indicates that at least 14 days are necessary to reach the CYP3A4 maximal inducing effect, and to return to baseline enzyme levels upon initiation and discontinuation of an inducer. The induction and de-induction processes are predicted to be faster for UGT1A1. The main driving parameters for onset and resolution of induction include the degradation rate constant of the enzyme, the inducer strength, and its half-life and related concentrations after stopping the administration. Available data and the findings of this study support the common practice to maintain the adjusted dosage of the ARVs for another 2 weeks after stopping an inducer. The findings of this work are particularly relevant for low- and middle-income countries given the HIV/TB syndemic often requiring rifampicin to be initiated and subsequently discontinued in individuals on first-line dolutegravir-containing regimens.
Supplementary Information
Below is the link to the electronic supplementary material.
Declarations
Funding
Open access funding provided by University of Basel. This study was supported by the Swiss National Foundation (Grant number: 188504).
Conflicts of interest
CM has received speaker honoraria from MSD, ViiV and Pfizer unrelated to this work. All other authors have no conflicts of interest relevant to the content of this study.
Ethics approval
Not applicable.
Availability of data and material
The data that support the findings of this study are available upon reasonable request to the corresponding author.
Code availability
May be available upon reasonable request to the corresponding author.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Authors contributions
SB collected the data, ran the simulations, analysed the data, and wrote the first draft of the manuscript. MBe contributed to the data analysis and writing of the manuscript. FS provided modelling input and supervised the data analysis. MBa provided clinical input. CM designed the study, supervised the data analysis, provided clinical input and obtained the funding. All authors contributed to the critical review and approval of the manuscript.
Contributor Information
Sara Bettonte, Email: sara.bettonte@unibas.ch.
Catia Marzolini, Email: catia.marzolini@usb.ch.
References
- 1.Trickey A, Sabin CA, Burkholder G, Crane H, d'Arminio Monforte A, Egger M, et al. Life expectancy after 2015 of adults with HIV on long-term antiretroviral therapy in Europe and North America: a collaborative analysis of cohort studies. Lancet HIV. 2023;10(5):e295–e307. doi: 10.1016/S2352-3018(23)00028-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Back D, Marzolini C. The challenge of HIV treatment in an era of polypharmacy. J Int AIDS Soc. 2020;23(2):e25449. doi: 10.1002/jia2.25449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Urquhart BL, Tirona RG, Kim RB. Nuclear receptors and the regulation of drug-metabolizing enzymes and drug transporters: implications for interindividual variability in response to drugs. J Clin Pharmacol. 2007;47(5):566–578. doi: 10.1177/0091270007299930. [DOI] [PubMed] [Google Scholar]
- 4.Mangelsdorf DJ, Thummel C, Beato M, Herrlich P, Schutz G, Umesono K, et al. The nuclear receptor superfamily: the second decade. Cell. 1995;83(6):835–839. doi: 10.1016/0092-8674(95)90199-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Crauwels H, Vingerhoets J, Ryan R, Witek J, Anderson D. Pharmacokinetic parameters of once-daily rilpivirine following administration of efavirenz in healthy subjects. Antivir Ther. 2012;17(3):439–446. doi: 10.3851/IMP1959. [DOI] [PubMed] [Google Scholar]
- 6.U.S. Food and Drug Administration. Tivicay product label. 2013; Available from: https://www.accessdata.fda.gov/drugsatfda_docs/label/2013/204790lbl.pdf.. Accessed 21 Dec 2022
- 7.U.S. Food and Drug Administration. Isentress product label. 2007; Available from: https://www.accessdata.fda.gov/drugsatfda_docs/label/2011/022145s018lbl.pdf.. Accessed 21 Dec 2022
- 8.U.S. Food and Drug Administration. Pifeltro product label. 2018; Available from: https://www.accessdata.fda.gov/drugsatfda_docs/label/2018/210806s000lbl.pdf.. Accessed 21 Dec 2022
- 9.Rowland Yeo K, Walsky RL, Jamei M, Rostami-Hodjegan A, Tucker GT. Prediction of time-dependent CYP3A4 drug-drug interactions by physiologically based pharmacokinetic modelling: impact of inactivation parameters and enzyme turnover. Eur J Pharm Sci. 2011;43(3):160–173. doi: 10.1016/j.ejps.2011.04.008. [DOI] [PubMed] [Google Scholar]
- 10.Almond LM, Mukadam S, Gardner I, Okialda K, Wong S, Hatley O, et al. Prediction of drug-drug interactions arising from CYP3A induction using a physiologically based dynamic model. Drug Metab Dispos. 2016;44(6):821–832. doi: 10.1124/dmd.115.066845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kapetas AJ, Sorich MJ, Rodrigues AD, Rowland A. Guidance for rifampin and midazolam dosing protocols to study intestinal and hepatic cytochrome P450 (CYP) 3A4 induction and de-induction. AAPS J. 2019;21(5):78. doi: 10.1208/s12248-019-0341-y. [DOI] [PubMed] [Google Scholar]
- 12.Kinvig H, Stader F, Bunglawala FS, Rajoli R, Cottura N, Howarth AE, et al. High-dose rifampicin for the treatment of leprosy in HIV patients taking dolutegravir [abstract n. 450]. Conference on Retroviruses and Opportunistic Infections. Boston, Massachusetts, USA; March 8-11, 2020.
- 13.Song I, Borland J, Chen S, Lou Y, Peppercorn A, Wajima T, et al. Effect of atazanavir and atazanavir/ritonavir on the pharmacokinetics of the next-generation HIV integrase inhibitor, S/GSK1349572. Br J Clin Pharmacol. 2011;72(1):103–108. doi: 10.1111/j.1365-2125.2011.03947.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moss DM, Siccardi M, Back DJ, Owen A. Predicting intestinal absorption of raltegravir using a population-based ADME simulation. J Antimicrob Chemother. 2013;68(7):1627–34. doi: 10.1093/jac/dkt084. [DOI] [PubMed] [Google Scholar]
- 15.U.S. Food and Drug Administration. Rifadin product label. 2010; Available from: https://www.accessdata.fda.gov/drugsatfda_docs/label/2010/050420s073,050627s012lbl.pdf. Accessed 21 Dec 2022
- 16.U.S. Food and Drug Administration. Sustiva product label. 1998; Available from: https://www.accessdata.fda.gov/drugsatfda_docs/label/2011/020972s038lbl.pdf. Accessed 21 Dec 2022
- 17.Pfizer Canada ULC. Product Monograph Mycobutin (rifabutin capsule USP). 2003; Available from: https://www.pfizer.ca/sites/default/files/202102/MYCOBUTIN_PM_E_244143_11Feb2021.pdf. Accessed 21 Dec 2022
- 18.Stader F, Penny MA, Siccardi M, Marzolini C. A comprehensive framework for physiologically based pharmacokinetic modelling in Matlab((R)) CPT Pharmacometrics Syst Pharmacol. 2019;8(7):444–459. doi: 10.1002/psp4.12399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stader F, Siccardi M, Battegay M, Kinvig H, Penny MA, Marzolini C. Repository describing an aging population to inform physiologically based pharmacokinetic models considering anatomical, physiological, and biological age-dependent changes. Clin Pharmacokinet. 2019;58(4):483–501. doi: 10.1007/s40262-018-0709-7. [DOI] [PubMed] [Google Scholar]
- 20.Stader F, Courlet P, Kinvig H, Battegay M, Decosterd LA, Penny MA, et al. Effect of ageing on antiretroviral drug pharmacokinetics using clinical data combined with modelling and simulation. Br J Clin Pharmacol. 2021;87(2):458–470. doi: 10.1111/bcp.14402. [DOI] [PubMed] [Google Scholar]
- 21.Bettonte S, Berton M, Stader F, Battegay M, Marzolini C. Management of drug-drug interactions between long-acting cabotegravir and rilpivirine and comedications with inducing properties: a modelling study. Clin Infect Dis. 2022;76:1225. doi: 10.1093/cid/ciac901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dooley KE, Sayre P, Borland J, Purdy E, Chen S, Song I, et al. Safety, tolerability, and pharmacokinetics of the HIV integrase inhibitor dolutegravir given twice daily with rifampin or once daily with rifabutin: results of a phase 1 study among healthy subjects. J Acquir Immune Defic Syndr. 2013;62(1):21–27. doi: 10.1097/QAI.0b013e318276cda9. [DOI] [PubMed] [Google Scholar]
- 23.Song I, Borland J, Chen S, Guta P, Lou Y, Wilfret D, et al. Effects of enzyme inducers efavirenz and tipranavir/ritonavir on the pharmacokinetics of the HIV integrase inhibitor dolutegravir. Eur J Clin Pharmacol. 2014;70(10):1173–1179. doi: 10.1007/s00228-014-1732-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Taburet AM, Sauvageon H, Grinsztejn B, Assuied A, Veloso V, Pilotto JH, et al. Pharmacokinetics of raltegravir in HIV-infected patients on rifampicin-based antitubercular therapy. Clin Infect Dis. 2015;61(8):1328–1335. doi: 10.1093/cid/civ477. [DOI] [PubMed] [Google Scholar]
- 25.Krishna R, East L, Larson P, Siringhaus T, Herpok L, Bethel-Brown C, et al. Efavirenz does not meaningfully affect the single dose pharmacokinetics of 1200 mg raltegravir. Biopharm Drug Dispos. 2016;37(9):542–549. doi: 10.1002/bdd.2045. [DOI] [PubMed] [Google Scholar]
- 26.Brainard DM, Kassahun K, Wenning LA, Petry AS, Liu C, Lunceford J, et al. Lack of a clinically meaningful pharmacokinetic effect of rifabutin on raltegravir: in vitro/in vivo correlation. J Clin Pharmacol. 2011;51(6):943–950. doi: 10.1177/0091270010375959. [DOI] [PubMed] [Google Scholar]
- 27.de Wet J, DeJesus E, Sloan L, Koteff J, Brennan C, Adkison K, et al. Pharmacokinetics of dolutegravir after switching to abacavir/ dolutegravir/ lamivudine from an efavirenz-based regimen: a PK sub-study from STRIIVING. 17th International Workshop on Clinical Pharmacology of HIV and Hepatitis Therapy. Washington DC; 2016.
- 28.Shebley M, Sandhu P, Emami Riedmaier A, Jamei M, Narayanan R, Patel A, et al. Physiologically based pharmacokinetic model qualification and reporting procedures for regulatory submissions: a consortium perspective. Clin Pharmacol Ther. 2018;104(1):88–110. doi: 10.1002/cpt.1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Abduljalil K, Cain T, Humphries H, Rostami-Hodjegan A. Deciding on success criteria for predictability of pharmacokinetic parameters from in vitro studies: an analysis based on in vivo observations. Drug Metab Dispos. 2014;42(9):1478–1484. doi: 10.1124/dmd.114.058099. [DOI] [PubMed] [Google Scholar]
- 30.Guest EJ, Aarons L, Houston JB, Rostami-Hodjegan A, Galetin A. Critique of the two-fold measure of prediction success for ratios: application for the assessment of drug-drug interactions. Drug Metab Dispos. 2011;39(2):170–173. doi: 10.1124/dmd.110.036103. [DOI] [PubMed] [Google Scholar]
- 31.Hanke N, Frechen S, Moj D, Britz H, Eissing T, Wendl T, et al. PBPK models for CYP3A4 and P-gp DDI prediction: a modeling network of rifampicin, itraconazole, clarithromycin, midazolam, alfentanil, and digoxin. CPT Pharmacometrics Syst Pharmacol. 2018;7(10):647–659. doi: 10.1002/psp4.12343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Suzuki M, Hirata M, Takagi M, Watanabe T, Iguchi T, Koiwai K, et al. Truncated UDP-glucuronosyltransferase (UGT) from a Crigler-Najjar syndrome type II patient colocalizes with intact UGT in the endoplasmic reticulum. J Hum Genet. 2014;59(3):158–162. doi: 10.1038/jhg.2013.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lin Z, Monteiro-Riviere NA, Riviere JE. A physiologically based pharmacokinetic model for polyethylene glycol-coated gold nanoparticles of different sizes in adult mice. Nanotoxicology. 2016;10(2):162–172. doi: 10.3109/17435390.2015.1027314. [DOI] [PubMed] [Google Scholar]
- 34.Teeguarden JG, Deisinger PJ, Poet TS, English JC, Faber WD, Barton HA, et al. Derivation of a human equivalent concentration for n-butanol using a physiologically based pharmacokinetic model for n-butyl acetate and metabolites n-butanol and n-butyric acid. Toxicol Sci. 2005;85(1):429–446. doi: 10.1093/toxsci/kfi103. [DOI] [PubMed] [Google Scholar]
- 35.Lee KH, Shin JG, Chong WS, Kim S, Lee JS, Jang IJ, et al. Time course of the changes in prednisolone pharmacokinetics after co-administration or discontinuation of rifampin. Eur J Clin Pharmacol. 1993;45(3):287–289. doi: 10.1007/BF00315399. [DOI] [PubMed] [Google Scholar]
- 36.Niemi M, Backman JT, Fromm MF, Neuvonen PJ, Kivistö KT. Pharmacokinetic interactions with rifampicin : clinical relevance. Clin Pharmacokinet. 2003;42(9):819–850. doi: 10.2165/00003088-200342090-00003. [DOI] [PubMed] [Google Scholar]
- 37.Chen J, Stringer R, Shah B, Gu J, Zhang Y, Hackling M, et al. Drug-Drug Interaction Studies to Evaluate the Effect of Inhibition of UGT1A1 and CYP3A4 and Induction of CYP3A4 on the Pharmacokinetics of Tropifexor in Healthy Subjects. Clin Pharmacol Drug Dev. 2022;11(11):1253–1263. doi: 10.1002/cpdd.1140. [DOI] [PubMed] [Google Scholar]
- 38.Pithavala YK, Tortorici M, Toh M, Garrett M, Hee B, Kuruganti U, et al. Effect of rifampin on the pharmacokinetics of Axitinib (AG-013736) in Japanese and Caucasian healthy volunteers. Cancer Chemother Pharmacol. 2010;65(3):563–570. doi: 10.1007/s00280-009-1065-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fromm MF, Busse D, Kroemer HK, Eichelbaum M. Differential induction of prehepatic and hepatic metabolism of verapamil by rifampin. Hepatology. 1996;24(4):796–801. doi: 10.1002/hep.510240407. [DOI] [PubMed] [Google Scholar]
- 40.Backman JT, Kivistö KT, Olkkola KT, Neuvonen PJ. The area under the plasma concentration-time curve for oral midazolam is 400-fold larger during treatment with itraconazole than with rifampicin. Eur J Clin Pharmacol. 1998;54(1):53–58. doi: 10.1007/s002280050420. [DOI] [PubMed] [Google Scholar]
- 41.Xu Y, Zhou Y, Hayashi M, Shou M, Skiles GL. Simulation of clinical drug-drug interactions from hepatocyte CYP3A4 induction data and its potential utility in trial designs. Drug Metab Dispos. 2011;39(7):1139–1148. doi: 10.1124/dmd.111.038067. [DOI] [PubMed] [Google Scholar]
- 42.Baneyx G, Parrott N, Meille C, Iliadis A, Lave T. Physiologically based pharmacokinetic modeling of CYP3A4 induction by rifampicin in human: influence of time between substrate and inducer administration. Eur J Pharm Sci. 2014;2(56):1–15. doi: 10.1016/j.ejps.2014.02.002. [DOI] [PubMed] [Google Scholar]
- 43.Chattopadhyay N, Kanacher T, Casjens M, Frechen S, Ligges S, Zimmermann T, et al. CYP3A4-mediated effects of rifampicin on the pharmacokinetics of vilaprisan and its UGT1A1-mediated effects on bilirubin glucuronidation in humans. Br J Clin Pharmacol. 2018;84(12):2857–2866. doi: 10.1111/bcp.13750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ohnhaus EE, Breckenridge AM, Park BK. Urinary excretion of 6 beta-hydroxycortisol and the time course measurement of enzyme induction in man. Eur J Clin Pharmacol. 1989;36(1):39–46. doi: 10.1007/BF00561021. [DOI] [PubMed] [Google Scholar]
- 45.Tran JQ, Kovacs SJ, McIntosh TS, Davis HM, Martin DE. Morning spot and 24-hour urinary 6 beta-hydroxycortisol to cortisol ratios: intraindividual variability and correlation under basal conditions and conditions of CYP 3A4 induction. J Clin Pharmacol. 1999;39(5):487–494. doi: 10.1177/009127009903900508. [DOI] [PubMed] [Google Scholar]
- 46.Reitman ML, Chu X, Cai X, Yabut J, Venkatasubramanian R, Zajic S, et al. Rifampin's acute inhibitory and chronic inductive drug interactions: experimental and model-based approaches to drug-drug interaction trial design. Clin Pharmacol Ther. 2011;89(2):234–242. doi: 10.1038/clpt.2010.271. [DOI] [PubMed] [Google Scholar]
- 47.Miners JO, Mackenzie PI, Knights KM. The prediction of drug-glucuronidation parameters in humans: UDP-glucuronosyltransferase enzyme-selective substrate and inhibitor probes for reaction phenotyping and in vitro-in vivo extrapolation of drug clearance and drug-drug interaction potential. Drug Metab Rev. 2010;42(1):196–208. doi: 10.3109/03602530903210716. [DOI] [PubMed] [Google Scholar]
- 48.Dooley KE, Savic R, Gupte A, Marzinke MA, Zhang N, Edward VA, et al. Once-weekly rifapentine and isoniazid for tuberculosis prevention in patients with HIV taking dolutegravir-based antiretroviral therapy: a phase 1/2 trial. Lancet HIV. 2020;7(6):e401–e409. doi: 10.1016/S2352-3018(20)30032-1. [DOI] [PubMed] [Google Scholar]
- 49.McGinnity DF, Zhang G, Kenny JR, Hamilton GA, Otmani S, Stams KR, et al. Evaluation of multiple in vitro systems for assessment of CYP3A4 induction in drug discovery: human hepatocytes, pregnane X receptor reporter gene, and Fa2N-4 and HepaRG cells. Drug Metab Dispos. 2009;37(6):1259–1268. doi: 10.1124/dmd.109.026526. [DOI] [PubMed] [Google Scholar]
- 50.Molto J, Rajoli R, Back D, Valle M, Miranda C, Owen A, et al. Use of a physiologically based pharmacokinetic model to simulate drug-drug interactions between antineoplastic and antiretroviral drugs. J Antimicrob Chemother. 2017;72(3):805–811. doi: 10.1093/jac/dkw485. [DOI] [PubMed] [Google Scholar]
- 51.Branch RA, Herman RJ. Enzyme induction and beta-adrenergic receptor blocking drugs. Br J Clin Pharmacol. 1984;17(1):77S–84S. doi: 10.1111/j.1365-2125.1984.tb02432.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ramsden D, Fung C, Hariparsad N, Kenny JR, Mohutsky M, Parrott NJ, et al. Perspectives from the innovation and quality consortium induction working group on factors impacting clinical drug-drug interactions resulting from induction: focus on cytochrome 3A substrates. Drug Metab Dispos. 2019;47(10):1206–1221. doi: 10.1124/dmd.119.087270. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.