Abstract
Nonionic surfactants are often used as general reagents for cell lysis enabling protein extraction, stabilization, and purification under non-denaturing conditions for downstream analysis in structural biology. However, the presence of surfactants in the sample matrix often has a deleterious effect on electrospray ionization (ESI)-mass spectrometry (MS) analysis of proteins and complexes. Here, we report a nonionic, cleavable surfactant, n-decyl-disulfide-β-D-maltoside (DSSM), for top-down proteomics. DSSM was designed to mimic the properties of one of the most common surfactants used in structural biology, n-dodecyl-β-D-maltoside (DDM) but contains a disulfide bond that allows for facile cleavage and surfactant removal before or during MS analysis. We have shown that DSSM is compatible with direct electrospray ionization (ESI)-MS analysis and reversed-phase liquid chromatography (RPLC)-MS analysis of proteins and protein complexes. We have demonstrated that DSSM can facilitate top-down proteomic characterization of membrane proteins such as a model ion channel protein and a G protein-coupled receptor as well as endogenous proteins from cell lysates for the determination of sequence variations and posttranslational modifications (PTMs). Conceivably, DSSM could serve as a general replacement for DDM in proteomics experiments and structural biology studies.
Graphical Abstract
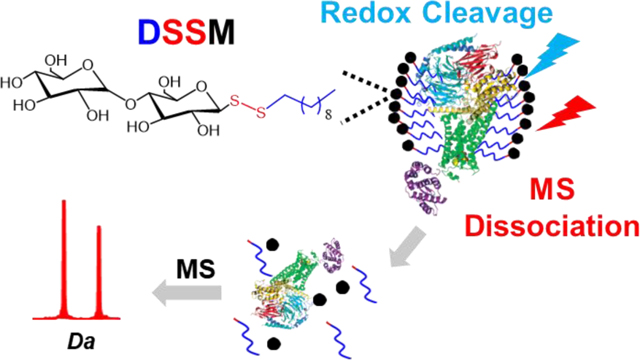
Nonionic surfactants are versatile tools for the solubilization and purification of proteins from cells and are critical reagents used in structural biology.1,2 One of the most popular nonionic surfactants for extracting proteins from their native environment and stabilizing them for downstream biophysical techniques such as crystallography and cryogenic electron microscopy is n-dodecyl-β-D-maltoside (DDM).3
However, the presence of surfactants, even mild ones like DDM, often has a deleterious effect on top-down proteomics for protein sequencing to identify posttranslational modifications (PTMs) and sequence variations.4–6 Surfactant-related signal suppression is generally caused by the higher ionization efficiency and signal-to-noise ratio of the low molecular weight species. Moreover, the presence of surfactant can negatively impact common front-end protein separation techniques such as reversed-phase liquid chromatography (RPLC), which could cause potential problems in reproducibility and robustness.5,6
One approach to overcome the incompatibility of the surfactants for downstream proteomic analysis is to insert a cleavable bond (e.g., acid7–9 or light-labile10,11) that allows for controlled degradation of the molecule into innocuous byproducts before MS analysis. Cleavable surfactants commonly used for proteomics contain denaturing, anionic head groups, such as sulfate, that preclude their use for applications where non-denaturing conditions are desirable.12,13 Thus, there is an urgent need for cleavable surfactants that can aid in traditional biochemical preparation methods under non-denaturing conditions yet are still amenable for downstream proteomic applications. Here, we demonstrate for the first time the use of n-decyl-disulfide-β-D-maltoside (DSSM), a nonionic, cleavable surfactant, for top-down proteomics (Figure 1).
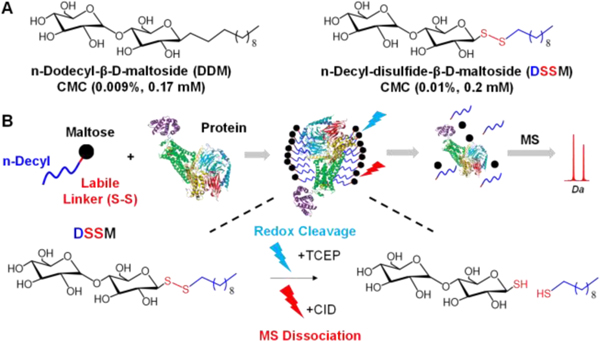
Figure 1.
(A) Comparison of the chemical structures of n-Dodecyl-β-D-maltoside (DDM) and n-Decyl-disulfide-β-D-maltoside (DSSM). (B) Overview of using a DSSM for proteomics
DSSM was originally developed to mimic the properties of DDM while providing a platform for high-throughput detergent exchange for biophysical assays.14 The nonionic maltose head group resembles DDM, but the addition of the disulfide bond between the sugar and the hydrophobic decyl tail imparts cleavable properties. After synthesis and characterization of DSSM (Figure S1-2), we evaluated its compatibility with electrospray-ionization (ESI)- MS analysis for intact proteins.
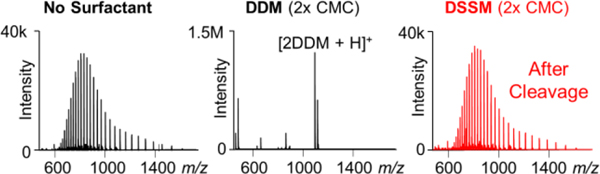
The compatibility of DSSM with direct ESI-MS analysis was evaluated using carbonic anhydrase (29.1 kDa) in denaturing conditions. The surfactant was degraded with 5 equivalence of TCEP (tris(2-carboxyethyl)phosphine) at 4 °C for 2 h (Figure S3). Insoluble degradation products, which commonly pose an issue for acid-cleavable surfactants like RapiGest,8 were not observed after DSSM degradation and centrifugation (Figure S4). We observe no difference in signal between the control sample and that with DSSM after degradation (Figure 2).
Figure 2. Compatibility of DSSM with ESI-MS of intact proteins.
MS spectra of carbonic anhydrase (left), carbonic anhydrase with DDM (2× CMC) (middle), and carbonic anhydrase in DSSM after degradation with TCEP (right).
In contrast, even at the relatively low concentration of 2× CMC (0.02%), DDM is the dominant species suppressing intact-mass analysis of carbonic anhydrase (Figure 2 middle panel). When a large excess of DSSM was used (20× CMC), a species corresponding to the maltose head group was observed as the dominant peak (Figure S5). No deleterious effects were observed from the inclusion of TCEP. Nonetheless, this is a critical consideration for experiments using a redox reaction to degrade the surfactant.
Next, we tested if ion activation (i.e. collision-induced dissociation [CID]) could be used to dissociate the surfactant from the proteins and protein complexes for ESI-MS analysis under non-denaturing conditions13 (Figure S6). MS analysis of carbonic anhydrase (29.1 kDa) ammonium acetate with and without DSSM at a concentration of 2× CMC (critical micelle concentration) yielded spectra with significant signal suppression from DSSM monomers at lower collisional activation (2–10 V) (Figure S6). When higher collisional activation (20–30 V) was applied, quality MS spectra were observed with a similar charge state distribution to the sample in ammonium acetate alone (Figure S6). Similar results were observed for carbonic anhydrase in DDM at 2× CMC (Figure S7).
For the tetramer forming complex, alcohol dehydrogenase (147.5 kDa), quality spectra could be obtained at a low collisional voltage (10 V) with and without DSSM at 2× CMC (Figure S8). Therefore, we further tested DSSM at a higher concentration (5× CMC) using collisional activation energies of 20–30 V to achieve direct ESI-MS analysis (Figure S6). Alternative activation methods, such as surface-induced dissociation (SID),15 ultraviolet photodissociation (UVPD), 16,17 infrared laser activation (IRMPD),18 may be implemented to remove DSSM in the gas phase for direct ESI-MS analysis.13,18,19
To evaluate the surfactant’s compatibility with RPLC-MS, we analyzed a mixture of standard proteins (ribonuclease A, myoglobin, and carbonic anhydrase) with and without DSSM or DDM. DSSM did not influence the separation or the signal intensity of the standard proteins even at 20x CMC (Figure S9). The improved compatibility with RPLC-MS compared to direct ESI-MS analysis results from the fact that the maltose head group after the degradation of DSSM elutes in the void volume before the proteins during LC separation (Figure S10). Similarly, the addition of TCEP did not appear to have a deleterious effect. DDM, on the other hand, led to significant signal suppression in the chromatogram and mass spectra (Figure S9). This demonstrates the promise of DSSM as a general replacement for nonionic surfactants like DDM for RPLC-MS applications.
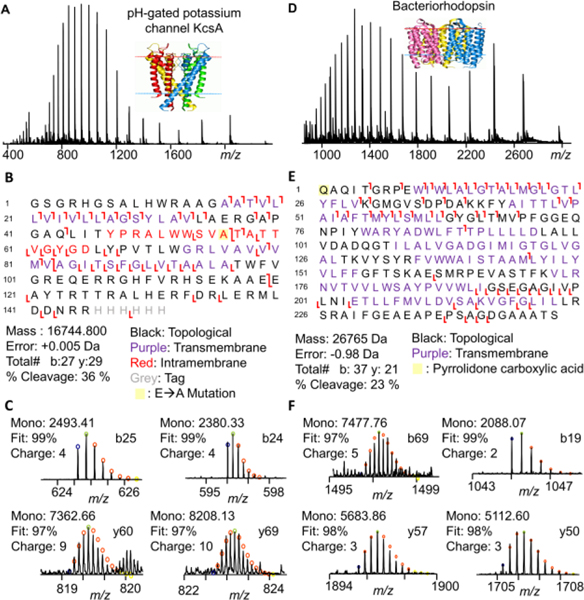
We further assessed DSSM’s compatibility with RPLC-MS to study membrane proteins, an important class of drug targets that are generally difficult to study using top-down proteomics owing to their inherent insolubility outside the plasma membrane and low abundance.1,20,21 We performed DSSM-aided membrane proteomic analysis on a model ion channel protein, a pH-gated potassium channel (KcsA). After removing incompatible buffer components (salts, detergent, etc.) using a chloroform:methanol:water precipitation,22 KcsA was solubilized in the DSSM (2× CMC). The surfactant was degraded with TCEP (in water or 50% isopropanol) and RPLC-MS/MS was performed using CID for fragmentation (Figure 3A-C). Using MASH Explorer23 for peak assignment and validation, we observed good sequence coverage on an LC-MS time scale with 27b ions and 29y ions representing 36% residue cleavage. Many of the bond cleavages were found in the transmembrane domains (TMD), in line with previous studies that characterized the fragmentation trends of intact integral membrane proteins.24,25 Furthermore, we were able to successfully map a mutation (E71A) that prevents channel inactivation (Figure 3).26
Figure 3. Top-down proteomics of DSSM solubilized membrane proteins.
Intact mass spectra, fragmentation map, and representative ions (with theoretical fits) for analysis of KcsA (A-C) and bacteriorhodopsin (D-F). Proteins were solubilized in DSSM and analyzed by LC-MS/MS after surfactant degradation.
Furthermore, we demonstrated that DSSM could enable the top-down analysis of bacteriorhodopsin,27,28 a commercially available GPCR. After bacteriorhodopsin was solubilized in DSSM and degraded using TCEP (in water or 50% isopropanol), RPLC-MS/MS yielded 37b ions and 21y ions corresponding to 23% residue cleavage (Figure 3D-F). A pyrrolidone carboxylic acid modification was localized to the N-terminus of the protein (Figure 3E)
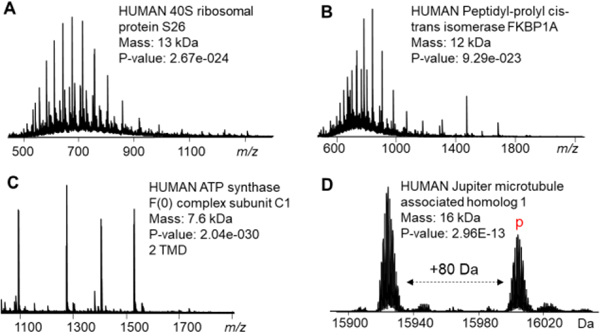
Finally, DSSM was used to extract endogenous protein from mammalian cells using and directly analyzed using RPLC-MS/MS after surfactant degradation. Following TopPIC data analysis,29 we identified a total of 276 proteoforms30 from 206 protein groups over four LC-MS/MS experiments (Figure 4, Table S1). Additionally, PTMs such as phosphorylation, methylation, and trimethylation were successfully localized using CID (Table S1). Overall, we demonstrated DSSM is a valuable surfactant for cell lysis and enables proteoform identification using RPLC-MS/MS analysis.
Figure 4. Top-down proteomics of endogenous proteins extracted from cell lysate using DSSM.
(A-D) Representative proteins were confidently identified from HEK whole cell or crude membrane lysate (A, B, and D from whole cell lysate and C from crude membrane lysate).
In summary, we presented the first demonstration of n-Decyl-disulfide-β-D-maltoside (DSSM), a cleavable DDM mimic, for direct ESI-MS analysis of intact proteins and top-down proteomics. DSSM was generally compatible with ESI-MS as well as RPLC-MS analysis circumventing the characteristic signal suppression typically observed for surfactants. We demonstrated that DSSM enables the top-down proteomic characterization of a model ion channel (KcsA), GPCR (bacteriorhodopsin), and endogenous proteins extracted from cell lysates. DSSM represents an important and versatile surfactant that can facilitate protein sample preparation under non-denaturing conditions for a myriad of proteomic and structural biology applications and acts as a general replacement for DDM.
Supplementary Material
ACKNOWLEDGMENT
This research was supported by National Institutes of Health (NIH) R01 GM117058 (to both S.J. and Y. G.). We would like to acknowledge funding from the HL096971, HL109810, GM125085, and S10 OD018475 (to Y.G.). K.A.B. would like to acknowledge the Vascular Surgery Research Training Program Grant T32 HL110853.
Footnotes
ASSOCIATED CONTENT
Supporting Information
The Supporting Information is available free of charge at. Summary of TopPIC results (Table S1). NMR of DSSM (Figure S1); ESI-MS for DSSM (Figure S2); Time-course of DSSM degradation (Figure S3); Picture of DSSM after degradation and centrifugation (Figure S4); ESI-MS of carbonic anhydrase in DDMS at 2x and 20x CMC (Figure S5); Removal of DSSM by CID for ESI-MS of proteins and protein complexes (Figure S6); ESI-MS carbonic anhydrase with and without DSSM (Figure S7); ESI-MS of alcohol dehydrogenase with and without DSSM (Figure S8); Comparison of DDM and DSSM for RPLC-MS (Figure S9); Base peak chromatogram of standard protein mixture in DSSM (Figure S10).
The University of Wisconsin-Madison has filed a provisional patent application (P220246US01) on the nonionic, cleavable surfactant DSSM. Y.G., S.J., M.K.G., and K.A.B. are named as inventors.
REFERENCES
- 1.Keener JE, Zhang G. & Marty MT Native Mass Spectrometry of Membrane Proteins. Analytical Chemistry 93, 583–597 (2021). 10.1021/acs.analchem.0c04342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lee HJ, Lee HS, Youn T, Byrne B. & Chae PS Impact of novel detergents on membrane protein studies. Chem (2022). 10.1016/j.chempr.2022.02.007 [DOI] [PubMed]
- 3.Stetsenko A. & Guskov A. An Overview of the Top Ten Detergents Used for Membrane Protein Crystallization. Crystals 7 (2017). 10.3390/cryst7070197 [DOI] [Google Scholar]
- 4.Loo RR, Dales N. & Andrews PC Surfactant effects on protein structure examined by electrospray ionization mass spectrometry. Protein Sci 3, 1975–1983 (1994). 10.1002/pro.5560031109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Deschamps JR Detergent Mediated Effects on the High-Performance Liquid Chromatography of Proteins. Journal of Liquid Chromatography 9, 1635–1653 (1986). 10.1080/01483918608076709 [DOI] [Google Scholar]
- 6.Brown KA, Melby JA, Roberts DS & Ge Y. Top-down proteomics: challenges, innovations, and applications in basic and clinical research. Expert Rev Proteomics, 1–15 (2020). 10.1080/14789450.2020.1855982 [DOI] [PMC free article] [PubMed]
- 7.Chang Y-H et al. New Mass-Spectrometry-Compatible Degradable Surfactant for Tissue Proteomics. Journal of Proteome Research 14, 1587–1599 (2015). 10.1021/pr5012679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yu YQ, Gilar M, Lee PJ, Bouvier ES & Gebler JC Enzyme-friendly, mass spectrometry-compatible surfactant for in-solution enzymatic digestion of proteins. Anal Chem 75, 6023–6028 (2003). 10.1021/ac0346196 [DOI] [PubMed] [Google Scholar]
- 9.Saveliev SV et al. Mass Spectrometry Compatible Surfactant for Optimized In-Gel Protein Digestion. Analytical Chemistry 85, 907–914 (2013). 10.1021/ac302423t [DOI] [PubMed] [Google Scholar]
- 10.Brown KA et al. A photocleavable surfactant for top-down proteomics. Nature Method. 16, 417–420 (2019). 10.1038/s41592-019-0391-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brown K. et al. High-throughput Proteomics Enabled by a Photocleavable Surfactant. Angewandte Chemie International Edition 59 (2020). 10.1002/anie.201915374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang X. Less is More: Membrane Protein Digestion Beyond Urea-Trypsin Solution for Next-level Proteomics. Molecular & cellular proteomics : MCP 14, 2441–2453 (2015). 10.1074/mcp.R114.042572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Laganowsky A, Reading E, Hopper JT & Robinson CV Mass spectrometry of intact membrane protein complexes. Nat Protoc 8, 639–651 (2013). 10.1038/nprot.2013.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xue D. et al. A Chemical Strategy for Amphiphile Replacement in Membrane Protein Research. Langmuir 35, 4319–4327 (2019). 10.1021/acs.langmuir.8b04072 [DOI] [PubMed] [Google Scholar]
- 15.Snyder DT, Panczyk EM, Somogyi A, Kaplan DA & Wysocki V. Simple and Minimally Invasive SID Devices for Native Mass Spectrometry. Anal Chem 92, 11195–11203 (2020). 10.1021/acs.analchem.0c01657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Greisch JF et al. Expanding the mass range for UVPD-based native top-down mass spectrometry. Chem Sci 10, 7163–7171 (2019). 10.1039/c9sc01857c [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Crittenden CM et al. Structural Evaluation of Protein/Metal Complexes via Native Electrospray Ultraviolet Photodissociation Mass Spectrometry. Journal of the American Society for Mass Spectrometry 31, 1140–1150 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mikhailov VA et al. Infrared Laser Activation of Soluble and Membrane Protein Assemblies in the Gas Phase. Anal Chem 88, 7060–7067 (2016). 10.1021/acs.analchem.6b00645 [DOI] [PubMed] [Google Scholar]
- 19.Calabrese AN, Watkinson TG, Henderson PJF, Radford SE & Ashcroft AE Amphipols Outperform Dodecylmaltoside Micelles in Stabilizing Membrane Protein Structure in the Gas Phase. Analytical Chemistry 87, 1118–1126 (2015). 10.1021/ac5037022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Helbig AO, Heck AJ & Slijper M. Exploring the membrane proteome--challenges and analytical strategies. J Proteomics 73, 868–878 (2010). 10.1016/j.jprot.2010.01.005 [DOI] [PubMed] [Google Scholar]
- 21.Brown KA et al. Top-Down Proteomics of Endogenous Membrane Proteins Enabled by Cloud Point Enrichment and Multidimensional Liquid Chromatography–Mass Spectrometry. Analytical Chemistry 92, 15726–15735 (2020). 10.1021/acs.analchem.0c02533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Folch J, Ascoli I, Lees M, Meath JA & Le BN Preparation of lipide extracts from brain tissue. J Biol Chem 191, 833–841 (1951). [PubMed] [Google Scholar]
- 23.Wu Z. et al. MASH Explorer: A Universal Software Environment for Top-Down Proteomics. Journal of Proteome Research 19, 3867–3876 (2020). 10.1021/acs.jproteome.0c00469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Skinner OS et al. Fragmentation of integral membrane proteins in the gas phase. Anal Chem 86, 4627–4634 (2014). 10.1021/ac500864w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cohn W, Huguet R, Zabrouskov V. & Whitelegge J. Dissociation Strategies to Maximize Coverage of α-Helical Domains in Top-Down Mass Spectrometry of Integral Membrane Proteins. J Am Soc Mass Spectrom 32, 1380–1387 (2021). 10.1021/jasms.1c00031 [DOI] [PubMed] [Google Scholar]
- 26.Vales E. & Raja M. The “flipped” state in E71A-K+-channel KcsA exclusively alters the channel gating properties by tetraethylammonium and phosphatidylglycerol. J Membr Biol 234, 1–11 (2010). 10.1007/s00232-010-9234-9 [DOI] [PubMed] [Google Scholar]
- 27.Whitelegge JP, Gundersen CB & Faull KF Electrospray-ionization mass spectrometry of intact intrinsic membrane proteins. Protein science : a publication of the Protein Society 7, 1423–1430 (1998). 10.1002/pro.5560070619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ryan CM et al. Post-translational modifications of integral membrane proteins resolved by top-down Fourier transform mass spectrometry with collisionally activated dissociation. Molecular & cellular proteomics : MCP 9, 791–803 (2010). 10.1074/mcp.M900516-MCP200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kou Q, Xun L. & Liu X. TopPIC: a software tool for top-down mass spectrometry-based proteoform identification and characterization. Bioinformatics 32, 3495–3497 (2016). 10.1093/bioinformatics/btw398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smith LM & Kelleher NL Proteoform: a single term describing protein complexity. Nat Methods 10, 186–187 (2013). 10.1038/nmeth.2369 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.