Abstract
Comprehensive genomic profiling (CGP) tests have been nationally reimbursed in Japan since June 2019 under strict restrictions, and over 46,000 patients have taken the test. Core Hospitals and Designated Hospitals host molecular tumor boards, which is more time‐consuming than simply participating in them. We sent a questionnaire to government‐designated Cancer Genomic Medicine Hospitals, including all 12 Core Hospitals, all 33 Designated Hospitals, and 117 of 188 Cooperative Hospitals. The questionnaire asked how much time physicians and nonphysicians spent on administrative work for cancer genomic medicine. For every CGP test, 7.6 h of administrative work was needed. Physicians spent 2.7 h/patient, while nonphysicians spent 4.9 h/patient. Time spent preparing for molecular tumor boards, called Expert Panels, was the longest, followed by time spent participating in Expert Panels. Assuming an hourly wage of ¥24,000/h for physicians and ¥2800/h for nonphysicians, mean labor cost was ¥78,071/patient. On a monthly basis, more time was spent on administrative work at Core Hospitals compared with Designated Hospitals and Cooperative Hospitals (385 vs. 166 vs. 51 h/month, respectively, p < 0.001). Consequently, labor cost per month was higher at Core Hospitals than at Designated Hospitals and Cooperative Hospitals (¥3,951,854 vs. ¥1,687,167 vs. ¥487,279/month, respectively, p < 0.001). Completing a CGP test for a cancer patient in Japan is associated with significant labor at each hospital, especially at Core Hospitals. Streamlining the exchange of information and simplifying Expert Panels will likely alleviate this burden.
Keywords: administrative personnel, cancer genomic medicine, comprehensive genomic profiling, cost analysis, workforce
We sent a questionnaire to government‐designated Cancer Genomic Medicine Hospitals in Japan. For every comprehensive genomic profiling (CGP) test, 7.6 h of administrative work was needed, with an estimated labor cost of ¥78,071/test. Streamlining the administrative work for CGP tests and subsidies are necessary for expansion of CGP testing.
Abbreviations
- AI
artificial intelligence
- C‐CAT
Center for Cancer Genomics and Advanced Therapeutics
- CGP
comprehensive genomic profiling
- CI
confidence interval
- ctDNA
circulating tumor DNA
- MHLW
Ministry of Health Labour and Welfare
- MTB
molecular tumor board
- PGPV
presumed germline pathogenic variant
1. INTRODUCTION
Comprehensive CGP tests have been implemented worldwide with hopes of improving prognosis of the patients and promoting development of novel therapy. Currently, five CGP tests are approved in Japan: OncoGuide NCC Oncopanel (Sysmex Corporation), FoundationOne CDx (Foundation Medicine, Inc.), FoundationOne Liquid CDx (Foundation Medicine, Inc.), Guardant360 CDx (Guardant Health Inc), and GenMine TOP (Konica Minolta, Inc.). 1 , 2 , 3 , 4 Currently, CGP tests have a fixed rate of 560,000 yen (¥560,000 = $4308 at ¥130/dollar and 4000 euros at ¥140/euro) in Japan, and 70%–90% is reimbursed. 5 For reimbursement under the national health insurance, the following conditions must be met: (1) the patient has a solid tumor in which standard therapy is expected to finish or is unavailable; (2) the patient is well enough to receive treatment if it is recommended by a molecular tumor board (MTB), specifically called Expert Panels; (3) the CGP tests must be done at Designated Core Hospitals for Cancer Genomic Medicine (Core Hospitals; responsible for training and research in cancer genomic medicine as well as holding Expert Panels), Designated Hospitals for Cancer Genomic Medicine (Designated Hospitals; responsible for holding Expert Panels), or Cooperative Hospitals for Cancer Genomic Medicine (Cooperative Hospitals), all of which are assigned by the Ministry of Health, Labour and Welfare (MHLW), 6 (4) a pre‐specified informed consent procedure is followed, (5) every test result must be discussed at MTBs, which can only be hosted by Core or Designated Hospitals, and (6) clinical and genomic data from every patient must be submitted to C‐CAT. 7 , 8 In an Expert Panel, specialists in clinical oncology, clinical genetics, genetic counseling, pathology, cancer genomic medicine, and bioinformatics are all required to participate and discuss each test result. As of October 2022, more than 46,000 patients have undergone CGP testing using the national health insurance system.
A CGP test is a multistep process: (1) informed consent, (2) determining whether pre‐existing surgery or biopsy samples are sufficient for analysis, (3) preparing and sending samples to the test company, (4) input of data to the C‐CAT electronic data capture system, (5) input of clinical data to a testing company portal, (6) uploading to and downloading files from the C‐CAT file exchange portal, (7) preparing for Expert Panels for multiple institutions to participate, (8) participating in Expert Panels, and (9) referral of patients to clinical trials and/or genetic counseling, if applicable. In addition, only 12 Core Hospitals and 33 Designated Hospitals are approved, and therefore, Expert Panels for tens of thousands of cases each year are only held at the 45 institutions. Consequently, the adoption of CGP tests in Japan has resulted in an increased burden for health‐care workers at approved Cancer Genome Medicine Hospitals, including physicians, nurses, pharmacists, genetic counselors, laboratory technicians, and health information managers, as well as administrative staff. In March 2022, MHLW announced that Expert Panels could be simplified if test results were returned only with gene mutations of high evidence level; however, they constitute <10% of all test results.
Overall, the universal health‐care in Japan has significantly contributed to the standardization and expansion of the CGP tests, however, they might not be sustainable under current constraints. In order to increase access to CGP tests and improve survival of patients with advanced cancer, action is needed to decrease the burden of hospitals. However, the scope of this cost in terms of hours of labor is unknown. In addition, which steps are more time‐consuming than others and whether measures can be taken to ease the burden are also unknown. Here, we sent a questionnaire to designated Core, Designated, and Cooperative Hospitals asking for the amount of time spent for each administrative step of CGP testing according to profession.
2. MATERIALS AND METHODS
2.1. Institutions
Between January 11 and January 18, 2022, questionnaires were sent by e‐mail to all 12 Core Hospitals and all 33 Designated Hospitals. We also asked each Core and Designated Hospitals to send questionnaires to all Cooperative Hospitals affiliated with their institution. Because we did not get a response from some Designated and Cooperative Hospitals, we sent a reminder between June 7 and June 11, 2022.
2.2. Questionnaire
In the questionnaire, we asked questions on organizational structure, data management, and communication between approved Cancer Genomic Medicine Hospitals, preparing and running an Expert Panel, and employment and labor costs (Figure 1). The questionnaire was in Japanese, and the English translation is shown in Table S1. We excluded time spent on informed consent or explanation of test results (including disclosure of germline findings and/or clinical trial information, if any), because they are part of routine clinical practice. This study was approved by the Research Ethics Committee of the Faculty of Medicine of The University of Tokyo (#2020378NI) and all institutional review boards at participating hospitals.
FIGURE 1.

Steps necessary to carry out a comprehensive genomic profiling test. The steps do not include informed consent or explanation of test results, including disclosure of germline findings and/or clinical trial information.
2.3. Labor cost
Japanese Health Insurance Federation for Surgery calculated the hourly wage of physicians as ¥24,000/h (¥24,000 = $185 at ¥130/dollar and €171 at ¥140/euro) and nonphysicians as ¥2400/h (¥2400 = $18 at ¥130/dollar and €17 at ¥140/euro).
2.4. Statistics
Categorical data were compared by the χ‑‐test, and continuous data were compared by ANOVA. p values of <0.05 were considered statistically significant. If a statistically significant difference was detected, multiple comparisons were carried out by the Tukey test. All analyses were undertaken using Sigma Plot version 12.3 (Systat Software).
3. RESULTS
3.1. Questionnaire responses
Between January 14 and February 22, 2022, responses were received from all 12 Core Hospitals, 25 Designated Hospitals, and 117 Cooperative Hospitals. After sending out reminders, we received responses from two additional Designated Hospitals. We did not receive responses from six Designated Hospitals or 67 Cooperative Hospitals. The number of hospitals that participated according to hospital type and regions of Japan are in Table 1. Responses came from more than 50% of the hospitals in every region.
TABLE 1.
Number of questionnaire responses regarding administrative work related to comprehensive genomic profiling in Japan, according to hospital type and region
| Hokkaido | Tohoku | Kanto | Chubu | Kinki | Chugoku/Shikoku | Kyushu | Total | |
|---|---|---|---|---|---|---|---|---|
| Core | 1 | 1 | 4 | 2 | 2 | 1 | 1 | 12 |
| Designated | 1 | 1 | 7 | 5 | 6 | 3 | 4 | 27 |
| Cooperative | 5 | 4 | 23 | 27 | 22 | 20 | 16 | 117 |
| Total | 7 | 6 | 34 | 34 | 30 | 24 | 21 | 156 |
| All | 9 | 10 | 63 | 51 | 41 | 36 | 23 | 233 |
| % | 78 | 60 | 54 | 67 | 73 | 67 | 91 | 67 |
3.2. Organizational structure
Eleven of 12 (92%) Core Hospitals had a department dedicated to cancer genomic medicine, while 24 of 27 (89%) Designated Hospitals and 68 of 117 (58%) Cooperative Hospitals had one (Table 2, p = 0.002). The numbers of dedicated physicians and nonphysicians working for the department are shown in Figure 2A and Figure 2B, respectively; Core Hospitals had significantly more physicians and nonphysicians compared with Designated Hospitals, which in turn had significantly more physicians and nonphysicians compared with Cooperative Hospitals. The number of persons employed for each job type are shown in Table S2. Ten of 12 (83%) Core Hospitals had a department dedicated to data management for cancer genomic medicine, while 21 of 27 (78%) Designated Hospitals and 70 of 113 (62%) Cooperative Hospitals had one (p = 0.13).
TABLE 2.
Number of Core Hospitals, Designated Hospitals, or Cooperative Hospitals in Japan with a dedicated department for cancer genomic medicine and data management
| Core | Designated | Cooperative | |
|---|---|---|---|
| (a) Department dedicated to cancer genomic medicine | |||
| Yes | 11 | 24 | 68 |
| No | 1 | 3 | 47 |
| (b) Department dedicated to CGP test data management | |||
| Yes | 10 | 21 | 70 |
| No | 2 | 6 | 43 |
Note: (a) All: p = 0.002. Designated vs. Cooperative: p = 0.007. (b) All: p = 0.128.
Abbreviation: CGP, cancer genomic profiling.
FIGURE 2.

Percentages of Cancer Genomic Hospitals in Japan with no dedicated personnel, one dedicated personnel member, or two or more dedicated personnel working on cancer genomic medicine. (A) Percentages of dedicated physicians according to hospital type (Core, Designated, or Cooperative Hospitals). (B) Percentages of dedicated nonphysicians according to hospital type. **p < 0.01, ***p < 0.001
3.3. Labor cost of data management
In Japan, for CGP tests to proceed, clinical data must be filled on both the C‐CAT electronic data capture system and the testing company portal. In addition, reports from the testing company and C‐CAT must be exchanged on the C‐CAT portal so staff from Core, Designated, and Cooperative Hospitals can access them. For input of clinical data to the C‐CAT electronic data capture system, 1.6 h/patient (95% CI, 1.3–1.8 h/patient) was spent, with physicians spending 0.6 h/patient (95% CI, 0.5–0.7 h/patient) and nonphysicians spending 1.0 h/patient (95% CI, 0.8–1.2 h/patient). No difference was seen between hospital type (Core Hospitals, 1.3 h/patient [95% CI, 0.5–2.2 h/patient], Designated Hospitals, 1.4 h/patient [95% CI, 1.0–1.9 h/patient], and Cooperative Hospitals 1.6 p‐h [95% CI, 1.4–1.9 h/patient]) (Figure 3A).
FIGURE 3.

Numbers of hours/patient spent by physicians and nonphysicians on each step that is necessary to perform a comprehensive genomic profiling test in Japan, according to hospital type (Core Hospitals, Designated Hospitals, or Cooperative Hospitals). Hours spent on (A) inputting clinical data for the Center for Cancer Genomics and Advanced Therapeutics (C‐CAT) database, (B) inputting clinical data for the testing company portal, (C) exchanging files on the C‐CAT portal, (D) communication between hospitals, (E) preparing for Expert Panels, and (F) spent during Expert Panels. ***p < 0.001
For input of clinical data to the testing company portal, 0.6 h/patient (95% CI, 0.5–0.6 h/patient) was spent, with physicians spending 0.2 h/patient (95% CI, 0.1–0.2 h/patient) and nonphysicians spending 0.4 h/patient (95% CI, 0.4–0.5 h/patient). No difference was seen between hospital type (Core Hospitals, 0.4 h/patient (95% CI, 0.2–0.5 h/patient), Designated Hospitals, 0.6 h/patient (95% CI, 0.4–0.8 h/patient), and Cooperative Hospitals 0.6 h/patient (95% CI, 0.5–0.7 h/patient)) (Figure 3B).
The time spent on the C‐CAT portal was 1.0 h/patient (95% CI, 0.9–1.2 h/patient) per CGP test, with physicians spending 0.3 h/patient (95% CI, 0.2–0.3 h/patient) and nonphysicians spending 0.7 h/patient (95% CI, 0.6–0.9 h/patient). No difference was seen between hospital type (Core Hospitals, 0.6 h/patient [95% CI, 0.3–0.9 h/patient], Designated Hospitals, 1.0 h/patient [95% CI, 0.6–1.4 h/patient], and Cooperative Hospitals, 1.1 h/patient [95% CI, 0.9–1.3 h/patient]) (Figure 3C).
3.4. Labor cost preparing and running an Expert Panel
Because Expert Panels can only be hosted by Core or Designated Hospitals, Core/Designated Hospitals and Cooperative Hospitals must communicate and exchange patient information, test results, and specifics of the meeting. The time spent communicating between Cancer Genomic Medicine Hospitals was 1.2 h/patient (95% CI, 0.8–1.6 h/patient) per CGP test, with physicians spending 0.4 h/patient (95% CI, 0.1–0.7 h/patient) and nonphysicians spending 0.8 h/patient (95% CI, 0.5–1.0 h/patient). No difference was seen between hospital type (Core Hospitals, 0.9 h/patient (95% CI, 0.3–1.5 h/patient), Designated Hospitals, 0.9 h/patient (95% CI, 0.5–1.2 h/patient), and Cooperative Hospitals 1.3 h/patient (95% CI, 0.7–1.9 h/patient)) (Figure 3D).
Core Hospitals and Designated Hospitals spend time preparing for Expert Panels and then host them, while Cooperative Hospitals participate. The time spent preparing for Expert Panels was 1.7 h/patient (95% CI, 1.3–2.1 h/patient), with physicians spending 0.7 h/patient (95% CI, 0.5–0.9 h/patient) and nonphysicians spending 0.9 h/patient (95% CI, 0.7–1.2 h/patient). Core Hospitals spent 2.0 h/patient (95% CI, 0.8–3.2 h/patient), Designated Hospitals spent 2.9 h/patient (95% CI, 1.6–4.2 h/patient), and Cooperative Hospitals spent 1.3 h/patient (95% CI, 0.9–1.8 h/patient) (p < 0.001 for differences between hospitals) (Figure 3E). Post hoc analysis revealed that Designated Hospitals spent significantly more time compared with Cooperative Hospitals (p < 0.001), whereas no difference was seen between Core and Designated Hospitals or Core and Cooperative Hospitals.
The time spent running Expert Panels was 1.6 h/patient (95% CI, 1.2–2.0 h/patient), with physicians spending 0.5 h/patient (95% CI, 0.4–0.7 h/patient) and nonphysicians spending 1.1 h/patient (95% CI, 0.8–1.3 h/patient). No difference was seen between hospital type Core Hospitals, 1.2 h/patient (95% CI, 0.7–1.7 h/patient), Designated Hospitals, 2.1 h/patient (95% CI, 1.3–2.9 h/patient), and Cooperative Hospitals, 1.5 h/patient (95% CI, 1.1–1.9 h/patient) (Figure 3F).
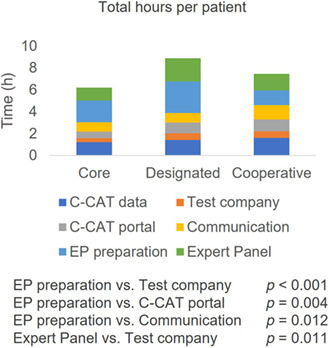
3.5. Total hours per patient
For each patient, an average of 7.6 h/patient (95% CI, 6.7–8.6 h/patient) of work was needed to complete a CGP test. Physicians spent an average of 2.7 h/patient (95% CI, 2.2–3.2 h/patient), while nonphysicians spent 4.9 h/patient (95% CI, 4.2–5.6 h/patient). No difference was seen between hospital type (Core Hospitals, 6.2 h/patient [95% CI, 4.3–8.0 h/patient], Designated Hospitals, 8.9 h/patient [95% CI, 6.4–11.4 h/patient], and Cooperative Hospitals, 7.4 h/patient [95% CI, 6.3–8.5 h/patient]) (Figure 4A). Time spent preparing for Expert Panels was statistically longer than time spent on the testing company portal (p < 0.001), time spent on the C‐CAT portal (p = 0.004), and time hospitals spent communicating with each other (p = 0.012) (Figure 4B). Time spent on Expert Panels was statistically longer than time spent on the testing company portal (p = 0.011). No difference was seen between the other steps.
FIGURE 4.

Total number of hours/patient spent on a comprehensive genomic profiling (CGP) test in Japan, according to hospital type (Core, Designated, or Cooperative Hospitals). (A) Total number of hours/patient spent by physicians and nonphysicians. (B) Total number of hours/patient spent on a CGP test according to each step.
3.6. Hours of work per month
At Core Hospitals, an average of 66.5 (95% CI, 40.7–92.4) CGP test results were discussed per month, while 19.1 (95% CI 14.6–23.6) and 8.2 (5.0–11.5) test results per month were discussed at Designated and Cooperative Hospitals, respectively (Figure 5A) (p < 0.001 between Core and Designated Hospitals and Core and Cooperative Hospitals, p = 0.024 between Designated and Cooperative Hospitals). Multiplying the number of hours spent per patient by the number of CGP tests per month gives the number of hours of work per month. In total, an average of 97 h were spent per month at Cancer Genomic Medicine Hospitals (95% CI, 70–123 h/month). More time was spent per month at Core Hospitals (385 h/month, 95% CI 338–433 h/month) compared with Designated Hospitals (166 h/month, 95% CI 132–199 h/month, p < 0.001) and Cooperative Hospitals (51 h/month, 95% CI 31–71 h/month, p < 0.001) (Figure 5B). The number of hours/month spent at Designated Hospitals was statistically longer than that at Cooperative Hospitals (p < 0.001).
FIGURE 5.

Amount of work required for comprehensive genomic profiling (CGP) tests done at Core Hospitals, Designated Hospitals, or Cooperative Hospitals in Japan. (A) Number of tests done per month. (B) Number of hours/month spent by physicians and nonphysicians on CGP tests. *p < 0.05, ***p < 0.001
3.7. Economic cost
Assuming an hourly wage of ¥24,000/h for physicians and ¥2800/h for nonphysicians, mean labor cost was ¥78,071/patient (95% CI, ¥65,203–¥90,938/patient). Physicians accounted for ¥64,298/patient (95% CI, ¥51,937–¥76,660/patient), and nonphysicians accounted for ¥13,772/patient (95% CI, ¥11,865–¥15,679/patient) (p < 0.001). No difference in labor cost per patient was seen between Core Hospitals (¥63,830/patient, 95% CI ¥56,832–¥70,827/patient), Designated Hospitals (¥88,704/patient, 95% CI ¥75,636–¥101,772/patient), and Cooperative Hospitals (¥77,077/patient, 95% CI ¥61,202–¥92,953/patient) was seen (Figure 6A).
FIGURE 6.

Cost of labor for comprehensive genomic profiling tests at Core, Designated, or Cooperative Hospitals according to physicians and nonphysicians. (A) Cost of labor per test. (B) Total cost of labor per month. ***p < 0.001
Multiplying the labor cost per patient by the number of CGP tests per month gives the labor cost per month. In total, an average of ¥961,457/month was spent at Cancer Genomic Medicine Hospitals (95% CI, ¥689,304–¥1,233,611/month). Physicians accounted for ¥782,255/month (95% CI, ¥552,420–¥1,012,090/month), and nonphysicians accounted for ¥179,203/month (95% CI, ¥127,739–¥230,666/month). Labor cost per month was higher at Core Hospitals (¥3,951,854/month, 95% CI ¥3,493,253–¥4,410,455/month) compared with Designated Hospitals (¥1,687,167/month, 95% CI ¥1,328,560–¥2,045,774/month; p < 0.001) and Cooperative Hospitals (¥487,279/month, 95% CI ¥284,395–¥690,163/month; p < 0.001) (Figure 6B). Labor cost per month at Designated Hospitals was statistically higher than that at Cooperative Hospitals (p < 0.001).
4. DISCUSSION
Many steps are necessary to undertake a CGP test and undergo an MTB, which are complex and time‐consuming. 9 , 10 , 11 , 12 , 13 , 14 Many institutions that carry out CGP testing have MTBs available, 13 and most have reported on MTBs done at single institutions. 15 , 16 , 17 , 18 , 19 , 20 The MTBs encompassing multiple institutions are reported less frequently, 21 and reports on multiple MTBs are rare. 22 , 23 , 24 We have for the first time, to our knowledge, quantified the time spent on each administrative step necessary to undergo CGP testing in Japan, excluding the clinical steps. We have also analyzed the differences in time spent at Core, Designated, and Cooperative Hospitals, as well as between physicians and nonphysicians.
On average, 7.6 h/patient of work was needed to complete a CGP test. Nonphysicians spent more time (4.9 h/patient) than physicians (2.7 h/patient). No difference was seen between hospital type. Among the six steps, most time was spent preparing for Expert Panels (1.7 h/patient) and participating in Expert Panels (1.6 h/patient). Therefore, simplifying Expert Panels should directly lead to a decrease in time spent, especially for physicians.
One possible reason of the long time spent preparing for and participating in Expert Panels could be interpretation of germline findings. Because pathogenicity of somatic mutations is considered independent of race, annotations are made using global cancer knowledge databases and are usually straightforward. However, annotation of germline variants depends not only on global databases but also on race‐specific genome variation data and past and family history, which requires expertise and effort. Furthermore, tumor‐only panels can complicate discussion because somatic and germline origin cannot be distinguished, and therefore, PGPVs are an aggregate of somatic and true germline pathogenic variants. Indeed, confirmatory tests from tumor‐only panels showed that 21 of 36 PGPVs were somatic. 25 This means the discussion of variant origin for some PGPVs can be omitted if they are tested by a tumor–normal paired CGP. 26 , 27 Artificial intelligence‐assisted annotation for each genetic variant can help finalize Expert Panel reports by incorporating biologic, clinical, and therapeutic information through public knowledge databases. However, Expert Panels made changes in 33% of all AI‐assisted reports and the most common type of change was germline disclosure, which accounted for 29%. 28 Although a common criteria for evaluation of germline findings is broadly applied in each hospital in Japan, 29 AI‐assisted annotation alone might not be sufficient for simplification of the Expert Panels.
The number of CGP tests done per month was higher at Core Hospitals (66.5 tests/month) than Designated Hospitals (19.1 tests/month) and Cooperative Hospitals (8.2 tests/month). Consequently, Core Hospitals (385 h/month) spent more hours per month on CGP tests compared with Designated Hospitals (166 h/month) and Cooperative Hospitals (51 h/month). Again, simplifying Expert Panels should directly lead to less time spent, especially at Core Hospitals where the highest number of test results are done and discussed. In addition, because only Core and Designated Hospitals can hold Expert Panels, allowing high‐volume Cooperative Hospitals to hold Expert Panels should lessen the burden for both Core/Designated Hospitals and Cooperative Hospitals.
Mean labor cost was calculated as ¥78,071/patient, or $601/patient at ¥130/dollar or €558/patient at ¥140/euro. Because physicians are assumed to have higher wages than nonphysicians, the economic cost on physicians was higher than that of nonphysicians, even though labor in terms of hours of work was longer in nonphysicians. Because more tests are done at Core Hospitals, cost per month was highest at Core Hospitals at nearly ¥4 million/month ($30,769/month at ¥130/dollar or €28,571/patient at ¥140/euro), followed by Designated Hospitals, and finally Cooperative Hospitals.
Of the nationally fixed rate of ¥560,000 for all CGP tests in Japan, 70%–90% is reimbursed by the national health‐care system, while co‐pay for patients is 10%–30%. 5 It is important to note that most of the ¥560,000 paid to hospitals are then used to pay the testing companies, and the remainder is considered fees for clinical practice such as informed consent and explanation of CGP test results, including disclosure of germline findings, if applicable. Because cancer genomic medicine requires significant administrative work outside of usual clinical practice, the actual labor cost is higher than a typical laboratory test.
It is probably not feasible to increase the nationally fixed rate of ¥560,000; therefore, our result indicates that measures to decrease the burden to hospitals, such as streamlining the exchange of information between hospitals, testing companies, and C‐CAT, and simplifying Expert Panels are necessary. Natural language processing of textual data in electronic health records can dramatically facilitate data input. 30 , 31 In addition, the Japanese Society of Medical Oncology has issued a manual to run an Expert Panel efficiently and effectively. 32 One proposed method is that a limited number of Expert Panel members can precheck the test results and determine the ones that are not necessary to undergo a full Expert Panel. Another is increasing the use of AI to assist the generation of Expert Panel reports.
There are several limitations to this study. We did not ask for the difference in time and labor required for tissue DNA and ctDNA CGP tests in our questionnaire. Evaluation and preparation of tissue specimen by pathologists and technicians require additional time and cost compared with ctDNA, which we did not include in our analysis. In addition, estimation of somatic or germline origin of variants is often easier with ctDNA because variant allele frequency is often lower. However, ctDNA requires more time to distinguish somatic variants of cancer cells from clonal hematopoiesis. In addition, there are two limitations inherent in a questionnaire survey: some cancer genomic medicine hospitals did not respond, and the accuracy of responses might have been variable.
Decreasing the burden of hospitals and health‐care professionals is necessary to improve survival and maintain quality of life of advanced cancer patients. In Japan, 7.7% of the patients have been reported to undergo genomically matched treatment. 24 This will likely improve if CGP tests can be given before standard treatment is expected to finish, because: (1) more patients will be physically strong enough to receive subsequent treatment, and (2) many clinical trials limit the number of previous treatments given as an inclusion/exclusion criterion. In order to allow more patients to receive CGP tests in Japan, the burden on hospitals and health‐care professionals must be alleviated. These efforts to decrease the hospital burden will hopefully result in a more sustainable cancer genomic medicine and an increased capacity to evaluate the genomic status of cancer cells in advanced cancer patients.
FUNDING INFORMATION
This study was supported by a grant from the Ministry of Health, Labour and Welfare (20EA1006).
CONFLICT OF INTEREST STATEMENT
H.K. and K.O. received research funds from Konica Minolta, Inc. K.O. received research funds and lecture fees from Chugai Pharmaceutical Co. M.M. received lecture fees from Chugai Pharmaceutical Co. and MSD, and research funds from Chugai Pharmaceutical Co. and Sysmex Corporation. Y.O. received research funds and lecture fees from Chugai Pharmaceutical Co. A.H. received lecture fees from Chugai Pharmaceutical Co. K.O., K.T., and K.M. are Associate Editors of Cancer Science. The other authors have no conflict of interest.
ETHICS STATEMENTS
Approval of the research protocol by an Institutional Review Board: This study was approved by the Research Ethics Committee of the Faculty of Medicine of The University of Tokyo (#2020378NI) and all institutional review boards at participating hospitals.
Informed consent: N/A.
Registry and registration no. of the study/trial: This study is registered with the Japan Registry of Clinical Trials, registration no. jRCT1030210547.
Animal studies: N/A.
Supporting information
Table S1
Table S2
ACKNOWLEDGMENTS
We would like to thank all the hospitals that participated in this study. We would like to thank the members of informed consent working group, secondary findings working group, repository working group, expert panel working group, and drug discovery working group, and Dr. Takayuki Yoshino for their suggestions.
Kage H, Oda K, Muto M, et al. Human resources for administrative work to carry out a comprehensive genomic profiling test in Japan. Cancer Sci. 2023;114:3041‐3049. doi: 10.1111/cas.15833
REFERENCES
- 1. Naito Y, Aburatani H, Amano T, et al. Clinical practice guidance for next‐generation sequencing in cancer diagnosis and treatment (edition 2.1). Int J Clin Oncol. 2021;26:233‐283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Imai M, Nakamura Y, Sunami K, et al. Expert panel consensus recommendations on the use of circulating tumor DNA assays for patients with advanced solid tumors. Cancer Sci. 2022;113:3646‐3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Sunami K, Ichikawa H, Kubo T, et al. Feasibility and utility of a panel testing for 114 cancer‐associated genes in a clinical setting: a hospital‐based study. Cancer Sci. 2019;110:1480‐1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kage H, Shinozaki‐Ushiku A, Ishigaki K, et al. Clinical utility of Todai OncoPanel in the setting of approved comprehensive cancer genomic profiling tests in Japan. Cancer Sci. 2023;114:1710‐1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Yoshii Y, Okazaki S, Takeda M. Current status of next‐generation sequencing‐based cancer genome profiling tests in Japan and prospects for liquid biopsy. Life. 2021;11:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Mukai Y, Ueno H. Establishment and implementation of cancer genomic medicine in Japan. Cancer Sci. 2021;112:970‐977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kohno T, Kato M, Kohsaka S, et al. C‐CAT: the National Datacenter for cancer genomic medicine in Japan. Cancer Discov. 2022;12:2509‐2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Mano H. Cancer genomic medicine in Japan. Proc Jpn Acad Ser B Phys Biol Sci. 2020;96:316‐321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Tamborero D, Dienstmann R, Rachid MH, et al. Support systems to guide clinical decision‐making in precision oncology: the cancer core Europe molecular tumor board portal. Nat Med. 2020;26:992‐994. [DOI] [PubMed] [Google Scholar]
- 10. Rao S, Pitel B, Wagner AH, et al. Collaborative, multidisciplinary evaluation of cancer variants through virtual molecular tumor boards informs local clinical practices. JCO Clin Cancer Inform. 2020;4:602‐613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Luchini C, Lawlor RT, Milella M, Scarpa A. Molecular tumor boards in clinical practice. Trends Cancer. 2020;6:738‐744. [DOI] [PubMed] [Google Scholar]
- 12. Schmid S, Jochum W, Padberg B, et al. How to read a next‐generation sequencing report‐what oncologists need to know. ESMO Open. 2022;7:100570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. van der Velden DL, van Herpen CML, van Laarhoven HWM, et al. Molecular tumor boards: current practice and future needs. Ann Oncol. 2017;28:3070‐3075. [DOI] [PubMed] [Google Scholar]
- 14. Russo A, Incorvaia L, Capoluongo E, et al. The challenge of the molecular tumor board empowerment in clinical oncology practice: a position paper on behalf of the AIOM‐ SIAPEC/IAP‐SIBioC‐SIC‐SIF‐SIGU‐SIRM Italian scientific societies. Crit Rev Oncol Hematol. 2022;169:103567. [DOI] [PubMed] [Google Scholar]
- 15. Schwaederle M, Parker BA, Schwab RB, et al. Molecular tumor board: the University of California‐San Diego Moores Cancer Center experience. Oncologist. 2014;19:631‐636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Harada S, Arend R, Dai Q, et al. Implementation and utilization of the molecular tumor board to guide precision medicine. Oncotarget. 2017;8:57845‐57854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Basse C, Morel C, Alt M, et al. Relevance of a molecular tumour board (MTB) for patients' enrolment in clinical trials: experience of the Institut Curie. ESMO Open. 2018;3:e000339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Moore DA, Kushnir M, Mak G, et al. Prospective analysis of 895 patients on a UK Genomics Review Board. ESMO Open. 2019;4:e000469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Reitsma M, Fox J, Borre PV, et al. Effect of a collaboration between a health plan, oncology practice, and comprehensive genomic profiling company from the payer perspective. J Manag Care Spec Pharm. 2019;25:601‐611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. VanderWalde A, Grothey A, Vaena D, et al. Establishment of a molecular tumor board (MTB) and uptake of recommendations in a community setting. J Pers Med. 2020;10:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bryce AH, Egan JB, Borad MJ, et al. Experience with precision genomics and tumor board, indicates frequent target identification, but barriers to delivery. Oncotarget. 2017;8:27145‐27154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sunami K, Naito Y, Aimono E, et al. The initial assessment of expert panel performance in core hospitals for cancer genomic medicine in Japan. Int J Clin Oncol. 2021;26:443‐449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Naito Y, Sunami K, Kage H, et al. Concordance between recommendations from multidisciplinary molecular tumor boards and central consensus for cancer treatment in Japan. JAMA Netw Open. 2022;5:e2245081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sunami K, Naito Y, Komine K, et al. Chronological improvement in precision oncology implementation in Japan. Cancer Sci. 2022;113:3995‐4000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Minamoto A, Yamada T, Shimada S, et al. Current status and issues related to secondary findings in the first public insurance covered tumor genomic profiling in Japan: multi‐site questionnaire survey. J Hum Genet. 2022;67:557‐563. [DOI] [PubMed] [Google Scholar]
- 26. Klek S, Heald B, Milinovich A, et al. Genetic counseling and germline testing in the era of tumor sequencing: a cohort study. JNCI Cancer Spectr. 2020;4:pkaa018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Fishler KP, Breese EH, Walters‐Sen L, McGowan ML. Experiences of a multidisciplinary genomic tumor board interpreting risk for underlying germline variants in tumor‐only sequencing results. JCO Precis Oncol. 2019;3:1‐8. [DOI] [PubMed] [Google Scholar]
- 28. Kage H, Aoki T, Shinozaki‐Ushiku A, et al. Performance of an artificial intelligence‐based annotation algorithm for reporting cancer genomic profiling tests. J Clin Oncol. 2022;40:suppl.1551. [Google Scholar]
- 29. AMED Program for Promoting Platform of Genomics based Drug Discovery . Proposal concerning the information transmission process in genomic medicine. 2019. Accessed February 6, 2023. https://www.amed.go.jp/content/000064662.pdf
- 30. Kawazoe Y, Shibata D, Shinohara E, Aramaki E, Ohe K. A clinical specific BERT developed using a huge Japanese clinical text corpus. PLoS One. 2021;16:e0259763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wang L, Fu S, Wen A, et al. Assessment of electronic health record for cancer research and patient care through a scoping review of cancer natural language processing. JCO Clin Cancer Inform. 2022;6:e2200006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Japanese Society of Clinical Oncology . A manual to run an expert panel efficiently and effectively in cancer genomic medicine Japanese. 2022. Accessed January 31, 2023. https://www.jsmo.or.jp/about/doc/guideline20220704.pdf
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1
Table S2


