ABSTRACT
Horizontal basal cells (HBCs) residing within severely damaged olfactory epithelium (OE) mediate OE regeneration by differentiating into odorant-detecting olfactory sensory neurons (OSNs) and other tissue supporting non-neuronal cell types. Depending on both tissue type and integrity, the Notch signaling pathway can either positively or negatively regulate resident stem cell activity. Although Notch1 specifies HBC dormancy in the uninjured OE, little is known about how HBCs are influenced by the Notch pathway following OE injury. Here, we show that HBCs depend on a functional inversion of the Notch pathway to appropriately mediate OE regeneration. At 24 h post-injury, HBCs enhance Notch1-mediated signaling. Moreover, at 3 days post-injury when the regenerating OE is composed of multiple cell layers, HBCs enrich both Notch1 and the Notch ligand, Dll1. Notably, HBC-specific Notch1 knockout increases HBC quiescence and impairs HBC differentiation into neuronal progenitors and OSNs. Interestingly, complete HBC knockout of Dll1 only decreases differentiation of HBC-derived OSNs. These data underscore the context-dependent nature of Notch signaling. Furthermore, they reveal that HBCs regulate their own neurogenic potential after OE injury.
Keywords: Horizontal basal cell, Stem cell, Olfactory epithelium, Regeneration
Summary: Functional inversion of the Notch signaling pathway underlies the ability of horizontal basal cells to regulate their own differentiation into olfactory sensory neurons during injury-induced regeneration of the olfactory epithelium.
INTRODUCTION
The olfactory epithelium (OE) harbors a population of reserve resident stem cells called horizontal basal cells (HBCs) that contribute to OE regeneration after severe tissue injury (Leung et al., 2007). The ability of HBCs to activate and differentiate into neurosensory and non-neuronal cell types begins with downregulation of p63, a transcription factor that specifies HBC dormancy (Packard et al., 2011b; Schnittke et al., 2015). This cell autonomous mechanism, along with others (Gadye et al., 2017; Fletcher et al., 2011; Herrick et al., 2017), complements our increasing appreciation that the local OE environment also plays a critical role in regulating HBC fate following tissue injury. For example, inflammatory cytokines within the OE post-injury have been identified as a mediator of HBC proliferation and contribute to tissue repair (Chen et al., 2017). Moreover, HBC-specific ablation of cilia mitigates tissue repair (Joiner et al., 2015), suggesting that HBCs must integrate extrinsic cues to support OE regeneration.
An aspect of the post-injury environment that has yet to be fully considered is the cell-cell interactions that occur within the regenerating OE itself shortly after injury. The Notch signaling pathway is a well-conserved motif that regenerative tissues utilize to specify stem cell fate. Within the intestine, Notch signaling prompts intestinal stem cell progenitors to proliferate and specifies enterocyte differentiation (Pellegrinet et al., 2011; Fre et al., 2005; Beumer and Clevers, 2021). The functional effect of Notch signaling, however, demonstrates tissue specificity, as the pathway can also prevent proliferation and differentiation of subventricular zone neural stem cells and skeletal muscle satellite cells (Kawaguchi et al., 2013; Bjornson et al., 2012).
Interestingly, the influence that Notch signaling has on resident stem cells can also depend on tissue integrity. While pathway activity continues to positively regulate crypt base columnar cell proliferation and differentiation following intestinal injury, Notch signaling functionally inverts after skeletal muscle injury. Rather than continuing to suppress satellite cell activity, Notch signaling becomes critical for the ability of satellite cells to contribute to skeletal muscle regeneration (Conboy and Rando, 2002; Luo et al., 2005).
In the uninjured OE, Notch1-mediated signaling specifies HBC dormancy (Herrick et al., 2017). Following OE injury, however, little is known about how the Notch pathway specifies HBC fate during tissue regeneration. To address this, we interrogated the OE at various time points after injury. Our spatiotemporal analyses revealed that during acute regeneration, when the OE is only a few cells thick but already composed of multiple cell populations, the Notch receptor Notch1 and the Notch ligand Dll1 concentrate to HBCs. Subsequent in vivo HBC-specific genetic recombination experiments demonstrated that both Notch1 and Dll1 specify HBC differentiation into olfactory sensory neurons (OSNs). Interestingly, only the former promotes HBC proliferation and neuronal progenitor formation during early OE regeneration, thus suggesting that Notch1 integrates multiple specification cues. In sum, our data demonstrate that disrupting HBC enrichment of Notch1 or Dll1 alters the fate of these resident stem cells, thereby revealing HBCs to be a self-governing stem cell population that regulates its own neurogenic potential during OE regeneration.
RESULTS
HBCs enhance Notch pathway activation during acute regeneration at 24 h post-injury
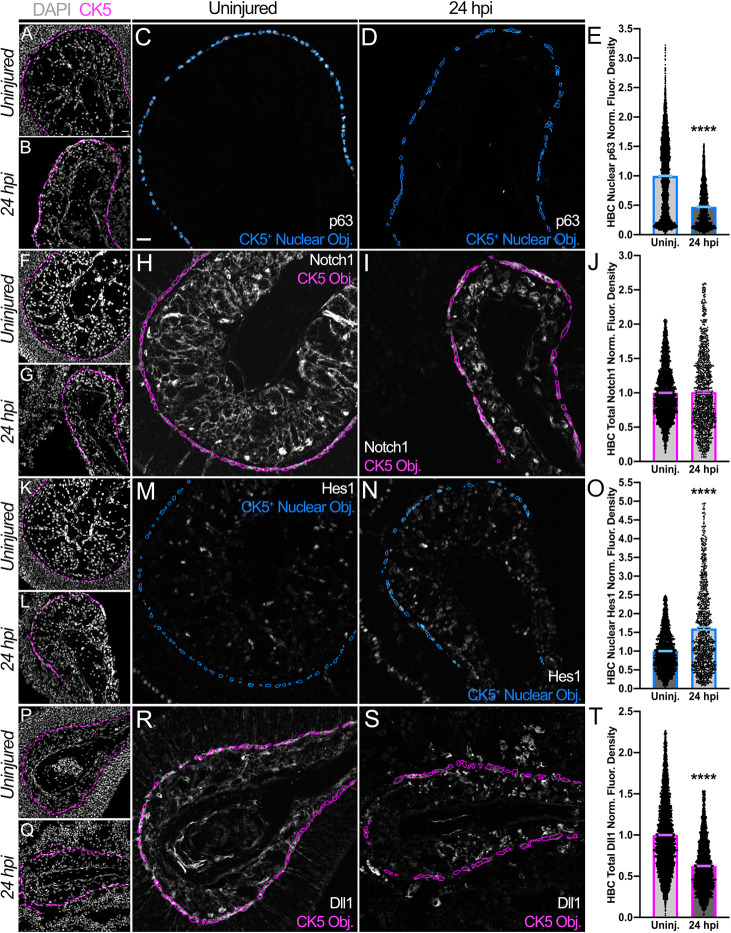
Following intra-peritoneal injection of wild-type (WT) mice with methimazole (MTZ), a compound known to induce OE injury and HBC-mediated regeneration (Leung et al., 2007; Genter et al., 1995), we analyzed the dynamics of Notch pathway proteins expressed by cytokeratin 5+ (CK5+) HBCs during acute regeneration. Analyses were first performed at 24 h post-injury (24 hpi) when HBCs are activated – a process marked by p63 downregulation (Fig. 1A-E) (Packard et al., 2011b; Schnittke et al., 2015; Fletcher et al., 2011). Given that Notch1 promotes HBC dormancy prior to OE injury (Herrick et al., 2017), HBCs at 24 hpi surprisingly express Notch1 at levels comparable to HBCs within uninjured OE (Fig. 1F-J). More surprisingly, HBCs enhance Notch pathway activation at 24 hpi, as evidenced by a 60.6% increase in nuclear Hes1 (Fig. 1K-O), a transcription factor whose expression is predicated on Notch signaling (Ohtsuka et al., 1999; Fryer et al., 2004). HBC upregulation of Notch pathway activity at 24 hpi is also demonstrated by a 63.1% increase in nuclear Notch1 intracellular domain (NICD) (Fig. S1), a region of Notch1 that undergoes proteolytic cleavage and nuclear translocation following receptor activation (Schroeter et al., 1998). Given this increased pathway activity, we characterized the expression of the two predominant Notch ligands found in the OE: jagged 1 (Jag1) and Dll1 (Herrick et al., 2017). Jag1 is largely expressed by sustentacular (Sus) cells, glial-like cells (Breipohl et al., 1974; Dahl et al., 1982; Suzuki et al., 1996; Rafols and Getchell, 1983) whose somatic apices establish the OE-nasal cavity interface and extend processes basally to contact the basal lamina adjacent to HBCs (Holbrook et al., 1995). Sus cells, however, are destroyed following MTZ-induced tissue injury (Gadye et al., 2017; Chen et al., 2017), thus relegating Jag1 to insignificance within the acutely regenerating OE (Fig. S2). As such, we expected Dll1 expressed primarily by HBCs (Herrick et al., 2017) (Fig. 1R) to faciliate the observed Notch signaling enhancement. Surprisingly, HBCs at 24 hpi downregulate Dll1 expression by nearly 40% (Fig. 1P-T). These data, therefore, intimate that Notch pathway activation is indeed critical to HBC-mediated OE regeneration. Additionally, they suggest that HBCs downregulate Dll1 to potentially promote Notch activity.
Fig. 1.
HBCs at 24 hpi exhibit dynamic Notch pathway protein expression that occurs concomitant with their activation. (A-D) Relative to their counterparts within uninjured (uninj.) OE (A,C), HBCs at 24 hpi (B,D) are activated, given the decrease in nuclear p63. Each CK5+ Nuclear Object (Obj.) is derived from a DAPI+ nucleus within a CK5+ HBC. (E) Quantification of HBC nuclear p63 normalized (norm.) fluorescence (fluor.) density, each point represents a CK5+ Nuclear Object (as represented by blue outlines in C and D) (n=3 mice, 3829 uninjured HBCs and 1419 24 hpi HBCs). (F-I) Total HBC-expressed Notch1 (within magenta outlines, H and I) is unchanged at 24 hpi. (J) Quantification of HBC total Notch1 normalized fluorescence density, each point represents a CK5 Object (as represented by magenta outlines in H and I) (n=3 mice, 2833 uninjured HBCs and 1126 24 hpi HBCs). (K-N) Relative to their counterparts within uninjured OE (K,M), HBCs at 24 hpi (L,N) demonstrate an increase in nuclear Hes1. Each CK5+ Nuclear Object is derived from a DAPI+ nucleus within a CK5+ HBC. (O) Quantification of HBC nuclear Hes1 normalized fluorescence density, each point represents a CK5+ Nuclear Object (as represented by blue outlines in M and N) (n=3 mice, 2201 uninjured HBCs and 983 24 hpi HBCs). (P-S) Total HBC-expressed Dll1 (within magenta outlines, R and S) is significantly decreased at 24 hpi. (T) Quantification of HBC total Dll1 normalized fluorescence density, each point represents a CK5 Object (as represented by magenta outlines in R and S) (n=3 mice, 4739 uninjured HBCs and 2688 24 hpi HBCs). Uninjured was used as baseline, data are mean±s.e.m., Mann–Whitney test, ****P<0.0001 (E,J,O,T). Scale bars: 20 μm.
Notch signaling pathway proteins enrich to HBCs as multiple cell layers emerge during OE regeneration
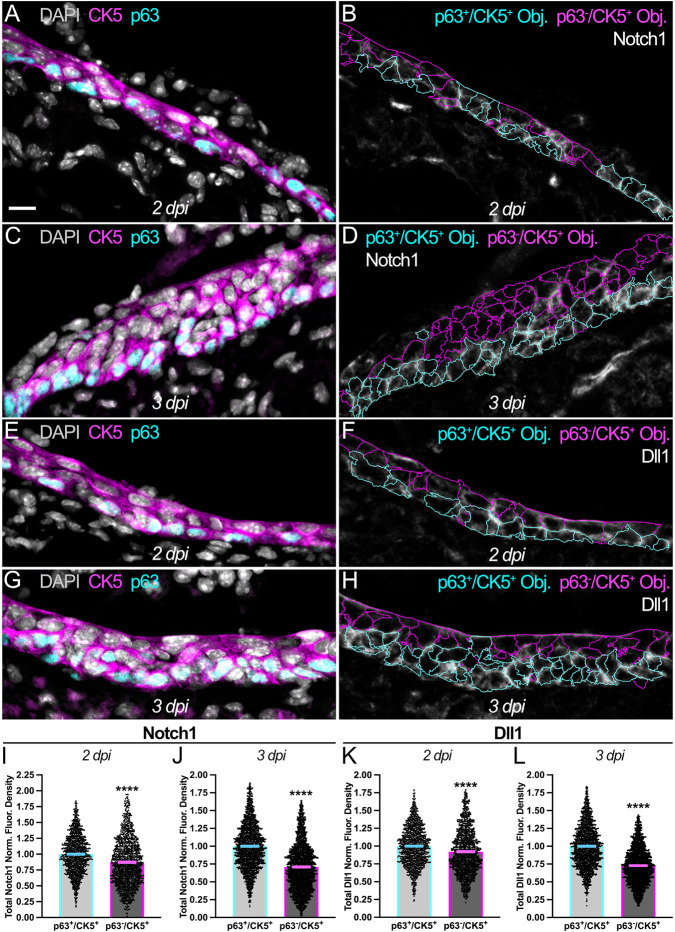
By 2 days post-injury (2 dpi), the regenerating OE consists of multiple cell layers comprising molecularly distinct CK5+ populations. p63+/CK5+ HBCs, which are primarily found atop the basal lamina (Holbrook et al., 2011; Gadye et al., 2017), have enriched expression of Notch1 (13.1% increase; Fig. 2A,B,I) and Dll1 (8.5% increase; Fig. 2E,F,K) relative to the p63−/CK5+ cells that generally overlie them. As OE regeneration proceeds, p63+/CK5+ HBCs at 3 dpi increase their concentration of Notch1 (39.3% increase; Fig. 2C,D,J) and Dll1 (37.4% increase; Fig. 2G,H,L) relative to p63−/CK5+ cells. Taken together, the increasing enhancement of Notch1 and Dll1 to HBCs within the reforming tissue further highlights the potential role that these Notch pathway proteins may play in specifying HBC fate during OE regeneration.
Fig. 2.
HBCs enrich Notch1 and Dll1 during acute OE regeneration. (A-D) Relative to p63−/CK5+ cells (magenta outlines in B and D), p63+/CK5+ HBCs (cyan outlines in B and D) become increasingly enriched for Notch1 [2 dpi (A,B), 13.1% increase; 3 dpi (C,D), 39.3% increase]. (E-H) Relative to p63−/CK5+ cells (magenta outlines in F and H), p63+/CK5+ HBCs (cyan outlines in F and H) become increasingly enriched for Dll1 [2 dpi (E,F), 8.5% increase; 3 dpi (G,H), 37.4% increase]. (I,J) Quantification of total Notch1 normalized fluorescence density within p63+/CK5+ HBCs and p63−/CK5+ cells, each point represents an analyzed Object (as represented by cyan and magenta outlines, respectively, in B and D) [2 dpi (B,I): n=3 mice, 1481 p63+/CK5+ HBCs and 1224 p63−/CK5+ cells; 3 dpi (D,J): n=3 mice, 2389 p63+/CK5+ HBCs and 2485 p63−/CK5+ cells]. (K,L) Quantification of total Dll1 normalized fluorescence density within p63+/CK5+ HBCs and p63−/CK5+ cells, each point represents an analyzed Object (as represented by cyan and magenta outlines, respectively, in F and H) [2 dpi (F,K): n=3 mice, 1470 p63+/CK5+ HBCs and 1226 p63−/CK5+ cells; 3 dpi (H,L): n=3 mice, 2143 p63+/CK5+ HBCs and 2333 p63−/CK5+ cells]. p63+/CK5+ HBCs used as baseline, data are mean±s.e.m., Mann–Whitney test, ****P<0.0001 (I-L). Scale bar: 10 μm.
HBC-specific conditional knockout of Notch1 diminishes Notch signaling pathway activity at 24 hpi and functionally disrupts HBCs
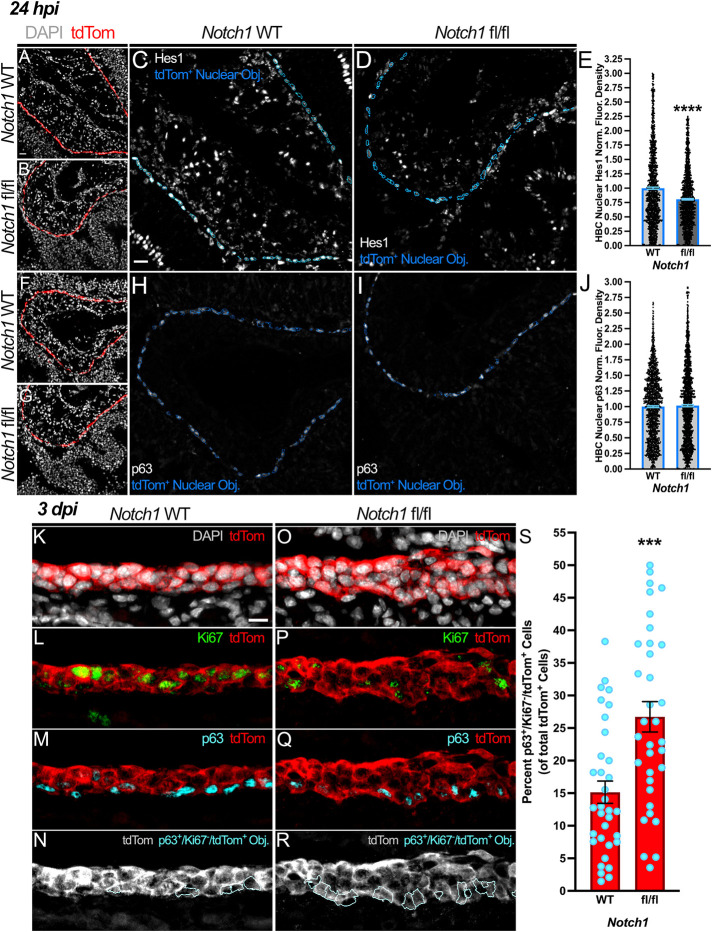
Given that HBCs maintain Notch1 receptor expression (Fig. 1J) and enhance Notch pathway activity at 24 hpi (Fig. 1O; Fig. S1E), we investigated whether Notch1 transduces cues that promote HBCs to contribute to OE regeneration. To do so, we used tri-genic mice in which a tamoxifen-inducible Cre recombinase driven by the HBC-specific CK5 promotor simultaneously facilitates Notch1 conditional knockout (cKO) (Fig. S3) and labels HBC-derived progeny with a tdTomato (tdTom) fluorescent reporter [i.e. CK5CreERT2; Notch1fl/fl; Rosa26R(tdTomato), hereafter referred to as Notch1 fl/fl]. Relative to their counterparts within OE of CK5CreERT2; Notch1WT/WT; Rosa26R(tdTomato) mice (hereafter referred to as Notch1 WT), HBCs within Notch1 fl/fl OE at 24 hpi attenuated Notch signaling pathway activity, as indicated by ∼34% and ∼20% decreases in nuclear NICD (Fig. S4) and Hes1 (Fig. 3A-E), respectively. Interestingly, this reduction was not accompanied by a significant change in p63 expression (Fig. 3F-J). In total, these data demonstrate that HBCs indeed engage in Notch1-mediated signaling at 24 hpi, thus further potentiating the possibility that Notch1 specifies HBC contribution to OE repair.
Fig. 3.
Attenuated Notch1-mediated signaling during OE regeneration at 24 hpi correlates with increased HBC quiescence at 3 dpi. (A-D) At 24 hpi, HBCs within Notch1 fl/fl OE (B,D) relative to those within Notch1 WT OE (A,C) demonstrate diminished Notch signaling activity, as evidenced by decreased nuclear Hes1 (within blue outlines, C and D). (E) Quantification of HBC nuclear Hes1 normalized fluorescence density, each point represents a tdTom+ Nuclear Object (as represented by blue outlines in C and D) (n=4 mice, 1596 Notch1 WT HBCs and 1562 Notch1 fl/fl HBCs). (F-I) Relative to control (F,H), HBCs within Notch1 fl/fl OE (G,I) demonstrate no significant change in nuclear p63 expression (within blue outlines, H and I) at 24 hpi. (J) Quantification of HBC nuclear p63 normalized fluorescence density, each point represents a tdTom+ Nuclear Object (as represented by blue outlines in H and I) (n=4 mice, 1670 Notch1 WT HBCs and 1510 Notch1 fl/fl HBCs). (K-R) Relative to control (K-N), the proportion of quiescent HBCs (cyan outlines, N and R) amongst all HBC-derived cells within the regenerating OE at 3 dpi decreases following HBC-specific Notch1 cKO (O-R). (S) Each circle represents an analyzed region, as represented in N and R (n=3 mice, 34 Notch1 WT regions and 34 Notch1 fl/fl regions). Notch1 WT is used as baseline (E,J), data are mean±s.e.m., Mann–Whitney test, ***P<0.001, ****P<0.0001 (E,J,S). Scale bars: 20 μm (A,C); 10 μm (K).
Therefore, to determine the functional impact of Notch1 cKO on HBC-mediated OE regeneration, we first assessed whether HBC fate is affected at 3 dpi when multiple cell layers become clearly evident. Notably, there are significantly more quiescent, p63+/Ki67−/tdTom+ HBCs within the regenerating Notch1 fl/fl OE at 3 dpi (26.7%) than in Notch1 WT controls (15.1%) (Fig. 3K-S). The finding that Notch1 cKO disrupts HBC biology during acute regeneration was corroborated by negative marker selection of HBCs with Sec8. Within uninjured OE, Sec8 localizes to the stem cell compartment where it exclusively labels globose basal cells (GBCs), the active multipotent OE resident stem cell population responsible for homeostatic maintenance (Caggiano et al., 1994; Chen et al., 2014), and excludes HBCs (Joiner et al., 2015). As HBCs within uninjured OE are also overwhelmingly Ki67− (Fletcher et al., 2011), the increase in quiescent HBCs resulting from Notch1 cKO (Fig. 3K-S) was mirrored by a significant increase in p63+/Sec8−/tdTom+ HBCs (23.7% in Notch1 fl/fl OE versus 11.2% in Notch1 WT OE) (Fig. S5A-I).
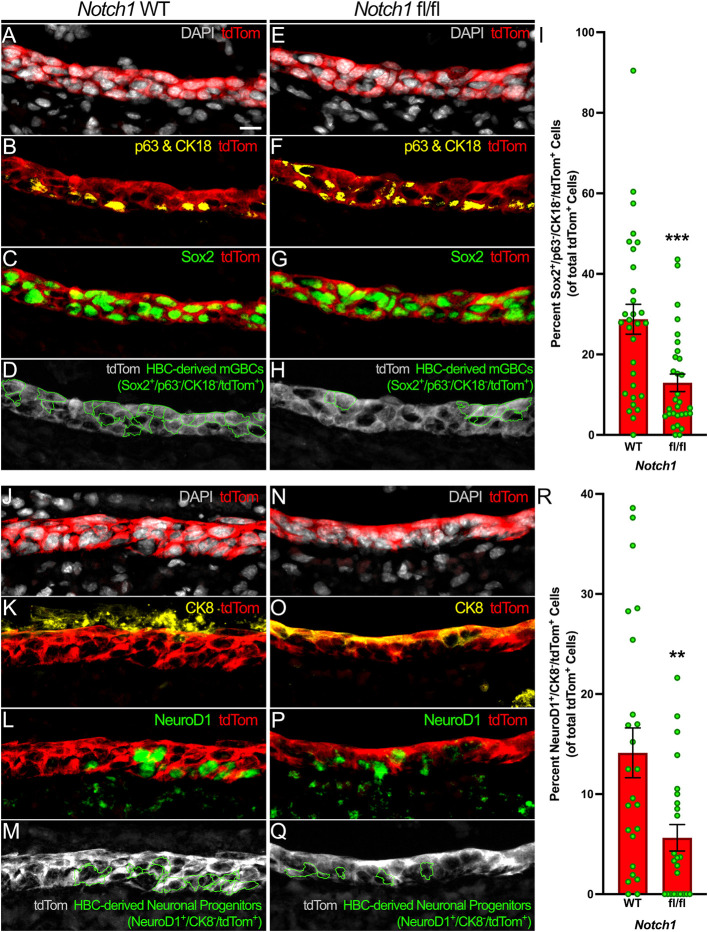
As transcriptome-based modeling suggests that HBCs can differentiate directly into GBCs (Gadye et al., 2017), we interrogated whether HBC bias towards quiescence following HBC-specific Notch1 cKO also disrupts GBC formation. As Sec8 has currently only been established as a GBC marker within uninjured OE, we used a constellation of established molecular markers to identify HBC-derived multipotent GBCs (mGBCs) centered around the transcription factor Sox2 (Guo et al., 2010; Zunitch et al., 2023). Because HBCs and Sus cells are also Sox2+ (Guo et al., 2010) and present within the acutely regenerating OE (Gadye et al., 2017; Guo et al., 2010), we excluded HBCs and Sus cells by their expression of p63 and CK18, respectively. After eliminating p63+/Sox2+/tdTom+ HBCs and CK18+/Sox2+/tdTom+ Sus cells from consideration, quantification of Sox2+/p63−/CK18−/tdTom+ mGBCs at 3 dpi demonstrated that Notch1 cKO significantly diminishes the degree of mGBCs arising from HBCs (13.0% in Notch1 fl/fl OE versus 28.8% in Notch1 WT OE) (Fig. 4A-I). In total, these data indicate that Notch1 limits HBC quiescence and specifies HBCs towards mGBC differentiation during acute injury-induced regeneration.
Fig. 4.
Notch1 cKO decreases HBC differentiation towards mGBCs and neuronal progenitors. (A-H) Relative to control (A-D), the proportion of HBCs that differentiate into Sox2+/p63−/CK18−/tdTom+ mGBCs (green outlines, D and H) within the regenerating OE at 3 dpi decreases following HBC-specific Notch1 cKO (E-H). (I) Each circle represents an analyzed region, as represented in D and H (n=3 mice, 30 Notch1 WT regions and 30 Notch1 fl/fl regions). (J-Q) Relative to control (J-M), the proportion of HBCs that differentiate into NeuroD1+/CK8−/tdTom+ neuronal progenitors (green outlines, M and Q) within the regenerating OE at 3 dpi decreases following HBC-specific Notch1 cKO (N-Q). (R) Each circle represents an analyzed region, as represented in M and Q (n=3 mice, 24 Notch1 WT regions and 24 Notch1 fl/fl regions). Data are mean±s.e.m., Mann–Whitney test, **P<0.01, ***P<0.001 (I,R). Scale bar: 10 μm.
Notch1 specifies HBC neuronal progenitor differentiation during acute OE regeneration
During acute regeneration, Notch1 cKO significantly compromises both OE stem cell populations by increasing quiescent HBC (Fig. 3K-S) and decreasing mGBC (Fig. 4A-I) tissue compositions. Therefore, we investigated what effect Notch1 cKO may have on the differentiation of two major cell types within the OE, namely OSNs and Sus cells, the latter of which can also be identified by their expression of CK8. OSNs are derived from determined, NeuroD1+ neuronal progenitors (Packard et al., 2011a), which themselves arise from mGBCs (Gadye et al., 2017; Zunitch et al., 2023). Notably, decreased differentiation of HBC-derived mGBCs (Fig. 4A-I) correlated with a significant decrease in NeuroD1+/CK8−/ tdTom+ HBC-derived neuronal progenitors (5.6% in Notch1 fl/fl OE versus 14.1% in Notch1 WT OE) (Fig. 4J-R). On the other hand, Sus cells can arise directly from HBCs (Gadye et al., 2017); however, increased HBC quiescence as a consequence of Notch1 cKO (Fig. 3K-S) did not yield a significant change in NeuroD1−/CK8+/ tdTom+ HBC-derived Sus cells at 3 dpi (54.7% in Notch1 fl/fl OE versus 51.1% in Notch1 WT OE) (Fig. S5J-R).
Notch1-mediated specification of HBC neuronal progenitor differentiation correlates with differentiation of HBC-derived OSNs in regenerated OE
To investigate whether the cell type deficits that result from Notch1 cKO persist beyond the acute phase of OE regeneration, we assessed linear cellular density of HBC-derived Sus cells and OSNs at 28 dpi when uninjured OE morphology has regenerated. At this time point, both immature and mature OSNs express PGP9.5 (Holbrook et al., 2011), whereas Sus cell bodies form a distinct and morphologically identifiable layer at the apical aspect of the OE (Graziadei and Graziadei, 1979) and are molecularly defined by CK8 and IL33 (Gadye et al., 2017; Maurya et al., 2015). Within Notch1 fl/fl OE, HBC-derived CK8+/apical IL33+/tdTom+ Sus cell linear density remained unchanged relative to control (19.5 HBC-derived Sus cells per 100 μm Notch1 fl/fl OE versus 19.3 HBC-derived Sus cells per 100 μm Notch1 WT OE) (Fig. 5F-J). Notably, however, the linear density of HBC-derived PGP9.5+/tdTom+ OSNs within mutant OE decreased by more than half (14.6 HBC-derived OSNs per 100 μm Notch1 fl/fl OE versus 31.3 HBC-derived OSNs per 100 μm Notch1 WT OE) (Fig. 5A-E). To determine whether this overall OSN deficit resulted from decreased mature and/or immature OSN (mOSN and iOSN, respectively) differentiation, we used immunofluorescent labeling of olfactory marker protein (OMP) to identify the former population (Holbrook et al., 2011; Gadye et al., 2017). To demarcate iOSNs, we leveraged their expression of β-III tubulin (Tuj1) (Coleman et al., 2019; Durante et al., 2020). However, as an appreciable percentage of Tuj1+ OSNs also express OMP (Gavid et al., 2023), we defined definitive iOSNs as Tuj1+/OMP−. Interestingly, Notch1 fl/fl OE demonstrated a decrease in HBC-derived Tuj1+/OMP−/tdTom+ iOSNs relative to Notch1 WT OE (5.5 HBC-derived iOSNs per 100 μm Notch1 fl/fl OE versus 10.2 HBC-derived iOSNs per 100 μm Notch1 WT OE) (Fig. S6F-J). This reduction was also seen among HBC-derived OMP+/tdTom+ mOSNs (7.2 HBC-derived mOSNs per 100 μm Notch1 fl/fl OE versus 12.2 HBC-derived mOSNs per 100 μm Notch1 WT OE) (Fig. S6A-E). Collectively, these data demonstrate that Notch1 specifies HBC neuronal differentiation during injury-induced OE regeneration.
Fig. 5.
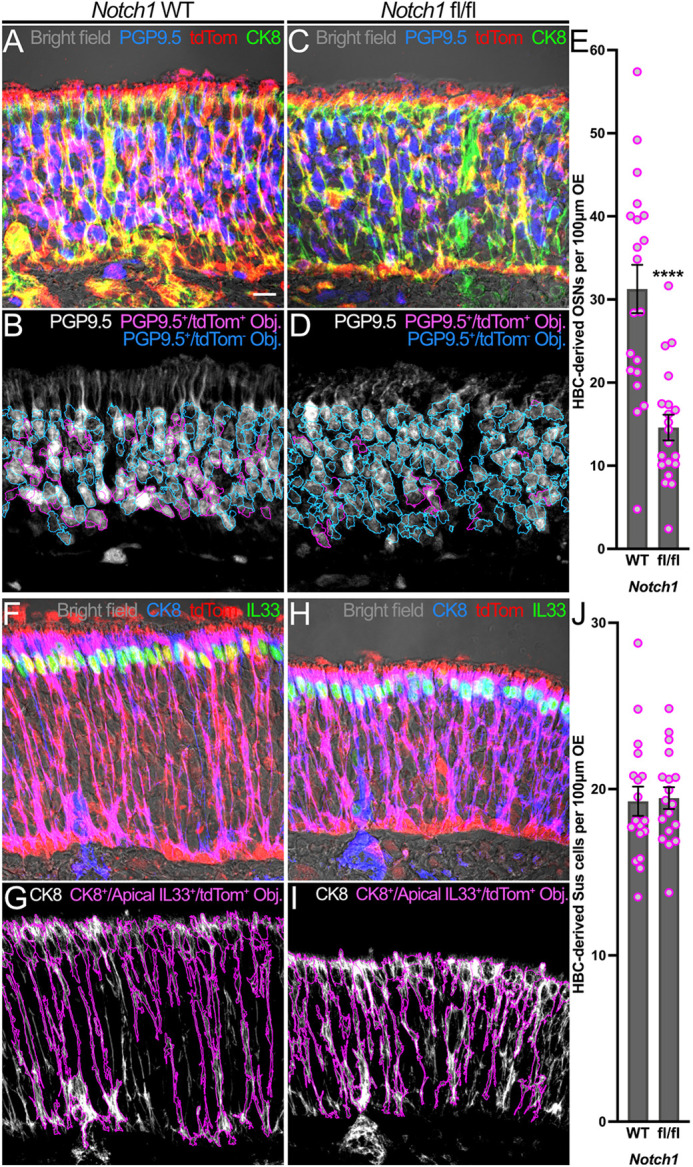
HBC-mediated neurogenesis following OE injury is attenuated by Notch1 cKO. (A-D) Relative to control (A,B), fewer HBC-derived PGP9.5+/tdTom+ OSNs (magenta in A and C; magenta outlines in B and D) constitute the morphologically regenerated OE at 28 dpi following HBC-specific Notch1 cKO (C,D). (E) Each circle represents an analyzed region, as represented in B and D (n=3 mice, 20 Notch1 WT regions and 20 Notch1 fl/fl regions). (F-I) Relative to control (F,G), OE composition of HBC-derived CK8+/apically localized IL33+/tdTom+ Sus cells (magenta in F and H; magenta outlines in G and I) at 28 dpi demonstrates no significant change following HBC-specific Notch1 cKO (H,I). (J) Each circle represents an analyzed region, as represented in G and I (n=3 mice, 18 Notch1 WT regions and 18 Notch1 fl/fl regions). Data are mean±s.e.m., unpaired t-test, ****P<0.0001 (E). Scale bar: 10 μm.
Dll1 specification of HBC OSN differentiation occurs independently of progenitor differentiation
The aforementioned data demonstrate that Notch1 transduces signals specifying HBC neuronal progenitor and OSN differentiation. However, the source of these cues remains unknown. Jag1 and Dll1 are the predominant Notch ligands within uninjured OE; however, expression of the former during early tissue regeneration becomes negligible (Fig. S2). Dll1, on the other hand, exhibits a degree of dynamism (Fig. 1P-T) and becomes increasingly enriched in HBCs during the acute stages of tissue regeneration (Fig. 2E-H,K,L) when HBCs also demonstrate functional sensitivity to perturbations in Notch1-mediated signal transduction (Figs. 3-5). As such, we interrogated the influence that Dll1 has on HBC biology during OE regeneration. To achieve adequate HBC-specific Dll1 recombination, we bred mice expressing tamoxifen-inducible CK5-dependent Cre recombinase and a conditional tdTom fluorescent reporter that identifies HBC-derived progeny to obtain animals that additionally harbored one floxed Dll1 allele and one constitutively null Dll1 allele due to lacZ knock in [i.e. CK5CreERT2; Dll1fl/lacZ; Rosa26R(tdTomato), hereafter referred to as Dll1 fl/lacZ] (Fig. S7). Dll1 fl/lacZ OE were compared to OE from CK5CreERT2; Dll1WT/lacZ; Rosa26R(tdTomato) mice (hereafter referred to as Dll1 WT/lacZ). Surprisingly, Dll1 fl/lacZ OE did not significantly affect HBCs at 3 dpi in terms of their quiescence (20.1% p63+/Ki67−/tdTom+ HBCs in Dll1 fl/lacZ OE versus 25.2% p63+/Ki67−/tdTom+ HBCs in Dll1 WT/lacZ OE) (Fig. S8A-E) nor differentiation into Sox2+/p63−/CK18−/tdTom+ mGBCs (22.9% in Dll1 fl/lacZ OE versus 19.3% in Dll1 WT/lacZ OE) (Fig. S8F-J). Dll1 fl/lacZ OE at 3 dpi similarly did not perturb HBC differentiation into NeuroD1+/CK8−/tdTom+ neuronal progenitors (3.8% in Dll1 fl/lacZ OE versus 3.7% in Dll1 WT/lacZ OE) (Fig. S9A-E) nor NeuroD1−/CK8+/tdTom+ Sus cells (56.9% in Dll1 fl/lacZ OE versus 59.5% in Dll1 WT/lacZ OE) (Fig. S9F-J). Interestingly, despite no appreciable deficit in Dll1 fl/lacZ mice during acute tissue regeneration, nearly 40% fewer HBC-derived PGP9.5+/tdTom+ OSNs populated the mutant OE at 28 dpi (13.4 HBC-derived OSNs per 100 μm Dll1 fl/lacZ OE versus 21.4 HBC-derived OSNs per 100 μm Dll1 WT/lacZ OE) (Fig. 6A-E). This overall OSN differentiation deficit was recapitulated by both OSN subpopulations, as mutant OE demonstrated decreases in HBC-derived Tuj1+/OMP−/tdTom+ iOSNs (2.7 HBC-derived iOSNs per 100 μm Dll1 fl/lacZ OE versus 5.9 HBC-derived iOSNs per 100 μm Dll1 WT/lacZ OE) (Fig. S10F-J) and HBC-derived OMP+/tdTom+ mOSNs (7.8 HBC-derived mOSNs per 100 μm Dll1 fl/lacZ OE versus 13.2 HBC-derived mOSNs per 100 μm Dll1 WT/lacZ OE) (Fig. S10A-E). These decreases in differentiation did not extend to HBC-derived CK8+/apical IL33+/tdTom+ Sus cells (16.1 HBC-derived Sus cells per 100 μm Dll1 fl/lacZ OE versus 17.8 HBC-derived Sus cells per 100 μm Dll1 WT/lacZ OE) (Fig. 6F-J). Taken together, these data demonstrate that Dll1 specifies HBC neurogenesis independently of upstream progenitor formation.
Fig. 6.
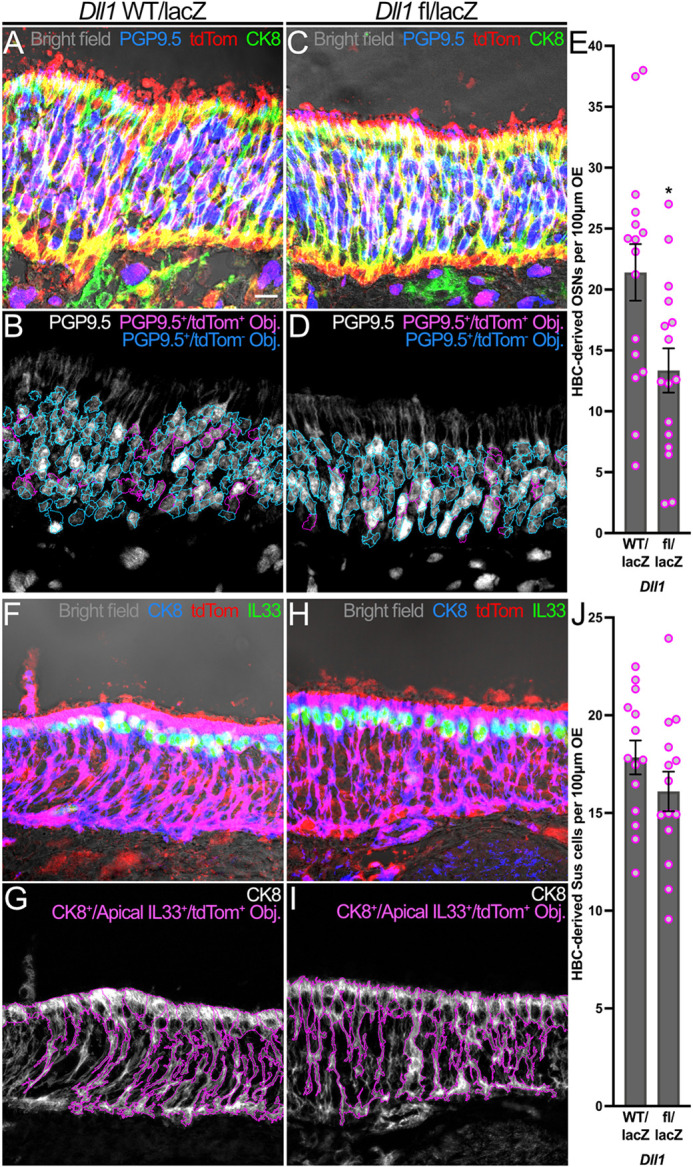
HBC-mediated neurogenesis following OE injury is attenuated by complete Dll1 KO. (A-D) Relative to control (A,B), HBCs give rise to fewer PGP9.5+/tdTom+ OSNs (magenta in A and C; magenta outlines in B and D) at 28 dpi following complete Dll1 KO. (E) Each circle represents an analyzed region, as represented in B and D (n=3 mice, 16 Dll1 WT/lacZ regions and 16 Dll1 fl/lacZ regions). (F-I) Relative to control (F,G), HBC differentiation into CK8+/apically localized IL33+/tdTom+ Sus cells (magenta in F and H; magenta outlines in G and I) at 28 dpi demonstrates no significant change following complete Dll1 KO. (J) Each circle represents an analyzed region, as represented in G and I (n=3 mice, 14 Dll1 WT/lacZ regions and 14 Dll1 fl/lacZ regions). Data are mean±s.e.m., unpaired t-test, *P<0.05 (E). Scale bar: 10 μm.
DISCUSSION
Our data demonstrate that OE integrity influences how the Notch pathway differentially specifies HBC fate. Within the uninjured OE, Notch1 promotes HBC dormancy (Herrick et al., 2017). Here, however, data gathered after OE injury demonstrate that Notch1 functionally inverts to promote HBC-mediated tissue regeneration. In contrast to previous descriptions of Notch1 expression within the acutely regenerating OE (Herrick et al., 2018), this study quantified Notch1 spatial localization to demonstrate its enrichment in HBCs (Fig. 2B,D,I,J). Furthermore, Notch1 cKO to perturb the expression of this receptor specifically within the HBC population demonstrated for the first time that Notch1 limits HBC quiescence following OE injury (Fig. 3K-S and Fig. S5A-I) and specifies HBC differentiation into neuronal progenitors (Fig. 4) and OSNs (Fig. 5A-E) during injury-induced OE regeneration. These disparate effects exhibited by Notch1 before and after OE injury highlight the need to consider tissue status when studying the role of molecular players. A similar context dependency is evident in skeletal muscle, wherein Notch signaling promotes satellite cell quiescence during homeostasis (Bjornson et al., 2012), but functionally inverts after muscle injury to drive satellite cell proliferation and differentiation (Conboy and Rando, 2002; Luo et al., 2005). This study also examined the Notch ligand Dll1 and similarly quantified its enrichment to HBCs (Fig. 2F,H,K,L) and demonstrated its role in specifying HBC neurogenesis (Fig. 6A-E) during injury-induced OE regeneration. Interestingly, it remains to be seen whether Dll1 also specifies disparate HBC fate depending on OE integrity.
The positive regulatory role that Notch1 plays in HBC-mediated OE regeneration contrasts with spinal cord regeneration during which Notch1 inhibits spinal progenitor proliferation and motor neuron differentiation (Dias et al., 2012), thus underscoring the need to analyze this well-conserved stem cell regulatory pathway in a niche-specific manner. In addition to emphasizing the importance of tissue type and integrity, OE regeneration highlights the need to consider temporality. As such, it would be worthwhile to determine at what point during OE regeneration does Notch1 once again specify HBC dormancy, and elucidate the mechanisms that underlie the functional reversion. Indeed, a time-sensitive transition point is apparent during the OE injury inflammatory response and its influence on HBC biology. After abrupt OE injury, acute inflammation promotes HBC-mediated tissue regeneration (Chen et al., 2017); however, chronic inflammation that characterizes chronic rhinosinusitis hinders HBC differentiation (Chen et al., 2019).
HBC integration of environmental cues such as inflammatory cytokines has been further implicated by demonstration that cilia play a significant role in promoting HBC-mediated OE regeneration (Joiner et al., 2015). Subsequent studies have identified additional extrinsic factors as positive regulators of HBC activity following tissue injury. This includes the sphingolipid metabolite S1P, which similarly limits HBC quiescence and promotes iOSN and mOSN differentiation (Wu et al., 2022). Another, the chitinase-like protein Ym2, is expressed by Sus cells in the uninjured OE and positively regulates OE regeneration after injury (Wang et al., 2021). As a secreted protein (Ward et al., 2001), Ym2 raises the prospect that Sus cells act as a reservoir of factors crucial to OE regeneration. Although Jag1 is significantly diminished during acute OE regeneration (Fig. S2) and, unlike Ym2, is a transmembrane protein (Lindsell et al., 1995; Grochowski et al., 2016), tissue injury may free Jag1 stored in destroyed Sus cells. Therefore, in addition to potentially establishing the biological activity of non-membrane integrated Jag1 during OE regeneration, Sus cell-specific Jag1 cKO may add Jag1 to the growing list of environmentally derived cues that promote tissue repair, thereby complementing this current work demonstrating how the HBC population expresses and utilizes Notch1 and Dll1 to regulate its own neurogenic potential during injury-induced OE regeneration.
Reinforcing the crucial role of this receptor during HBC-mediated OE regeneration, complete knockout (KO) of a canonical Notch1 ligand, Dll1, yielded a similar OSN deficit within regenerated OE (Fig. 6A-E). By enriching for both of these Notch proteins that regulate their own neuronal differentiation, HBCs, interestingly, recapitulate the self-regulatory nature demonstrated by Drosophila ectodermal cells (Heitzler and Simpson, 1991). Despite Notch1 and Dll1 concentrating within the stem cell population during acute regeneration (Fig. 2), HBCs do not solely differentiate into OSNs. Therefore, it remains to be seen whether the cellular diversity arising from HBCs after OE injury similarly depends on variable expression of basic helix-loop-helix transcription factors, such as Hes1, that drives differential cell fates within the Drosophila ectoderm (Kunisch et al., 1994).
Surprisingly, Notch1 cKO and complete Dll1 KO did not have the same effect on HBC differentiation into Sox2+/p63−/CK18−/tdTom+ mGBCs nor NeuroD1+/CK8−/ tdTom+ neuronal progenitors (Fig. 4, Figs S8 and S9). This discrepancy may reflect functions that Dll1 performs independently of engaging Notch receptors. In Drosophila, the Dll1 homolog Delta has been implicated in cell autonomously promoting the expression of daughterless (Mok et al., 2005), a transcription factor that positively regulates neuronal differentiation (Caudy et al., 1988; Vaessin et al., 1994). As such, daughterless mutants demonstrate severe reduction in all peripheral neurons during fruit fly embryogenesis (Caudy et al., 1988). Interestingly, daughterless mutants continue to form early neuronal precursors but exhibit defective formation of late neuronal precursors (Vaessin et al., 1994). Within the regenerating OE, Dll1 may function similarly, thereby raising the possibility that HBCs require unique, step-wise input in order to assume a determined neuronal fate.
Despite downregulating Dll1 (Fig. 1P-T), HBCs at 24 hpi enhance Notch signaling (Fig. 1K-O; Fig. S1). As pre-existing transcriptomic data indicate that other canonical Notch ligands are not upregulated at 24 hpi (Gadye et al., 2017), Notch1 may transduce early specification cues via non-canonical means. This includes non-canonical ligands (D'Souza et al., 2010), such as the microfibrillar protein MAGP1 (Miyamoto et al., 2006), which has been shown to promote skin wound healing (Weinbaum et al., 2008). Alternatively, Notch1 may also influence HBCs soon after OE injury in a ligand-independent manner (Palmer et al., 2014). Interestingly, receptor auto-activation can be attenuated by Dll1 (Palmer et al., 2014), thereby suggesting that HBCs may downregulate Dll1 at 24 hpi to safeguard their differentiation into neuronal progenitors.
In total, these data demonstrate that tissue status affects how the Notch pathway influences HBCs. They also reveal that HBCs regulate their own neuronal differentiation during OE regeneration. Furthermore, this work provides a basis for future studies to investigate how HBC heterogeneity shortly after OE injury regulates tissue regeneration. This, in turn, will continue to enhance our ability to leverage these resident stem cells in therapies designed to treat olfactory deficits.
MATERIALS AND METHODS
Mice
All mice (Mus musculus) were maintained on ad libitum chow and water in a heat- and humidity-controlled AALAC-accredited vivarium. All protocols for the use of vertebrate animal were approved by the Committee for the Humane Use of Animals at Tufts University School of Medicine. WT mice were F2 generation mice from an initial cross of C57BL/6J (Jax Stock 000664) and 129S1/SvlmJ (Jax Stock 002448) mice performed in-house. CK5CreERT2 mice were generously provided by Dr Pierre Chambon (Indra et al., 1999). Rosa26R(tdTomato) (Jax Stock 007909), B6.129X1-Notch1tm2Rko/GridJ (Jax Stock 007181) and B6.129-Dll1tm1Gos/J (Jax Stock#002957) mice were obtained from Jackson Labs. B6.129-Dll1tm1Mjo mice were generously transferred from the Lucy Liaw Lab, Maine Medical Center.
Tri-genic CK5CreERT2; Notch1tm2Rko/ tm2Rko /GridJ; Rosa26R(tdTomato) mice (Notch1 fl/fl mice) enabling HBC-specific inducible Notch1 KO and lineage tracing were generated in-house. As tri-genic CK5CreERT2; B6.129-Dll1tm1Mjo/ tm1Mjo; Rosa26R(tdTomato) mice (Dll1 fl/fl mice) did not efficiently recombine Dll1 on induction, these animals were bred in-house with CK5CreERT2; B6.129-Dll1tm1Gos/J; Rosa26R(tdTomato) mice (Dll1 WT/lacZ). Resulting tri-genic progeny that were CK5CreERT2; B6.129-Dll1tm1Mjo/tm1Gos/J; Rosa26R(tdTomato) mice (Dll1 fl/lacZ mice) enabled adequate HBC-specific inducible Dll1 KO and lineage tracing. All control animals for gene manipulation experiments possessed CK5CreERT2 and Rosa26R(tdTomato) alleles. Control animals for Notch1 recombination maintained both Notch1 WT alleles (Notch1 WT). Control animals for Dll1 recombination maintained one WT and possessed one constitutively inactive Dll1 allele (Dll1 WT/lacZ mice). All experiments used sex- and age-matched mice between 8 and 12 weeks old when initiated.
Tamoxifen induction of Cre-mediated recombination
Tamoxifen (Sigma-Aldrich, T5648) was dissolved in corn oil (Spectrum Chemical, CO136) at a stock concentration of 30 mg/ml. This tamoxifen stock was used to perform intra-peritoneal injections of Notch1 WT and Notch1 fl/fl mice at 300 mg/kg for 2 consecutive days before subsequent experimentation. Given sub-optimal recombination with tamoxifen, Dll1 WT/lacZ and fl/lacZ mice were injected with 4-hydroxytamoxifen (Hello Bio, HB6040), which was initially dissolved in 100% ethanol (ThermoFisher Scientific, BP2818) at a stock concentration of 40 mg/ml. This stock was then emulsified in an oil mixture of four parts sunflower oil (Sigma-Aldrich, S5007) and one part castor oil (Sigma-Aldrich, 259853) before intra-peritoneal injection. Each mouse received 2.5 mg 4-hydroxytamoxifen per day for 5 consecutive days.
Methimazole-induced OE injury
Methimazole (Sigma-Aldrich, Cat. M8506) was dissolved in PBS (Gibco, Cat. 10010-023) at a stock concentration of 5 mg/ml. WT mice were injected intra-peritoneally at 10 μl MTZ stock/g mouse to induce OE injury. Uninjured WT control mice were injected with 10 μl PBS vehicle/g mouse. Because of over-injury at 24 hpi as suggested by the absence of tdTom+ HBCs, presumably due to increased metabolic load of the 300 mg/kg tamoxifen required to induce Notch1 recombination, Notch1 WT and Notch1 fl/fl mice were injected intra-peritoneally at 5 μl MTZ stock/g mouse 13 days after the initial tamoxifen injection. Given similar issues with over injuring the OE when injected with 10 μl MTZ stock/g mouse, Dll1 WT/lacZ and Dll1 fl/lacZ mice were intra-peritoneally injected with 7.5 μl MTZ stock/g mouse 21 days after the last 4-hydroxytamoxifen injection.
Tissue processing
Mice were euthanized with an intra-muscular injection of a ketamine (37.5 mg/kg), xylazine (7.5 mg/kg) and acepromazine (1.25 mg/kg) triple cocktail at indicated time points. Mice were subsequently trans-cardially perfused, first with 20 ml PBS (Gibco, 10010-023) and followed by 40 ml 1% PLP fixative (1% paraformaldehyde, 0.1 M monobasic and dibasic phosphates, 90 mM lysine and 0.1 M meta-sodium periodate). After dissection of surrounding cranial structures, the isolated OE was post-fixed for 1.5 h under vacuum in 1% PLP, briefly rinsed three times in PBS, and incubated overnight at 4°C in decalcification buffer [0.35 M EDTA (Sigma-Aldrich, ED2SS) and 0.35 M NaOH in distilled water]. For subsequent cryoprotection, the OE was incubated in 30% sucrose overnight at 4°C, embedded in OCT and stored at −80°C until cryosectioning. A Leica cryostat was used to cut 10 μm coronal sections onto positively charged glass slides.
Immunofluorescence
Please see Table S1 for a list of antibodies used and their associated labeling conditions. Normal donkey block (NDB) [10% normal donkey serum, 5% non-fat powdered milk, 4% bovine serum albumin (BSA) and 0.1% Triton X-100 in 1× PBS] was used as diluent for all primary and secondary antibodies. Species-specific fluorophore-conjugated donkey secondary antibodies (Jackson ImmunoResearch) were diluted at 1:150. If an antigen detection required tyramide signal amplification (TSA), then species-specific biotin-conjugated donkey secondary antibodies and streptavidin-conjugated horseradish peroxidase (SA-HRP) tertiary antibody (Jackson ImmunoResearch) were diluted 1:100 in NDB and 1:400 in TNB [0.5% blocking reagent (Akoya Biosciences, FP1020), 0.15 M NaCl and 0.1 M Tris-HCl (pH 7.5)], respectively. Day 1 and day 2 washes were performed in 1× PBS and 3× PBS, respectively, with subsequent antibody incubation carried out for 3×5 min. After drying on a slide warmer for 45-60 min at ∼37°C, slides were washed for 10-15 min to remove OCT. If antigen retrieval was necessary, slides covered with antigen retrieval buffer (0.1 M sodium citrate, pH 6) were placed in a pre-heated commercial steamer for 10 min and then washed for 10 min. If an antigen required TSA, sections were then permeabilized in 3% H2O2 diluted in methanol for 5 min. If TSA was not required, sections were permeabilized in 90% methanol for 5 min. After permeabilization, slides were washed for 3×5 min and sections were blocked in NDB for 1 h at room temperature. If mouse antibodies were to be used, sections were next incubated for 1 h at room temperature in mouse-on-mouse IgG blocking reagent (Invitrogen, R37621) as per the manufacturer's protocol. Overnight primary antibody incubation at 4°C was followed by incubation with secondary antibodies at room temperature in the dark for 1 h. For antigens requiring TSA, appropriate sections were blocked in TNB for 30 min before incubation with SA-HRP at room temperature for 1 h in the dark. A solution composed of 98% TSA buffer [0.1 M boric acid (pH 8.5) and 0.003% H2O2], 1% 10% w/v BSA in 1× PBS and 1% FITC-tyramide was used to achieve TSA. After nuclear counterstaining with DAPI, slides were coverslipped in NPG (0.5% N-propyl gallate in 90% glycerol) mounting media, sealed with nail polish and stored in the dark at 4°C until imaging.
Imaging and imaging analyses
All images were taken on a Zeiss LSM800 confocal microscope. During each imaging session, identical excitation and emission capture settings were used to quantify protein expression via fluorescence intensity of control and experimental conditions that were processed in parallel. Z-planes were stepped at 2 μm intervals to sample through a cell approximately five times. For each captured channel, ImageJ was used to sum project the uncompressed, 16-bit tiff files and subtract background via its built-in Rolling Ball function. Quantification of total fluorescence intensity was performed using CellProfiler. For CellProfiler to process and demarcate cellular objects or landmarks, images of channels that did not require quantification were converted to 16-bit. To calculate relative fluorescence between control and experimental conditions for all performed experiments, the mean fluorescence density of the control condition was set as the denominator to derive the normalized fluorescence density of each quantified object. Basal lamina length of each region imaged at 28 dpi was determined morphologically in the tdTom channel using the freehand line tool in ImageJ. Although pseudo-colored to the same color for illustrative clarity of HBC-derived Tuj1+/OMP−/tdTom+ iOSNs at 28 dpi, OMP and CK8 signals for these experiments were captured in separate channels.
Statistical analyses and data presentation
GraphPad Prism Version 8 and Version 9 were used to generate graphs and perform statistical analyses as described in the figure legends. ImageJ, Adobe Photoshop CS6 and Affinity Designer were used to globally adjust the brightness and contrast of representative images. Adobe Photoshop CS6 and Affinity Designer were used to construct all figures.
Supplementary Material
Acknowledgements
We thank Po Kwok-Tse for her exceptional technical assistance.
Footnotes
Author contributions
Conceptualization: J.D.L., J.E.S.; Methodology: J.D.L., M.J.Z.; Formal analysis: J.D.L., M.J.Z.; Investigation: J.D.L., B.H.B., M.J.Z.; Resources: J.D.L., J.E.S.; Writing - original draft: J.D.L.; Writing - review & editing: J.D.L., B.H.B., J.E.S.; Visualization: J.D.L.; Funding acquisition: J.D.L., J.E.S.
Funding
Financial support was provided by the National Institutes of Health (F30 DC018450 to J.D.L. and R01 DC017869 to J.E.S.). Deposited in PMC for release after 12 months.
Data availability
All relevant data can be found within the article and its supplementary information.
References
- Beumer, J. and Clevers, H. (2021). Cell fate specification and differentiation in the adult mammalian intestine. Nat. Rev. Mol. Cell Biol. 22, 39-53. 10.1038/s41580-020-0278-0 [DOI] [PubMed] [Google Scholar]
- Bjornson, C. R., Cheung, T. H., Liu, L., Tripathi, P. V., Steeper, K. M. and Rando, T. A. (2012). Notch signaling is necessary to maintain quiescence in adult muscle stem cells. Stem Cells 30, 232-242. 10.1002/stem.773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breipohl, W., Laugwitz, H. J. and Bornfeld, N. (1974). Topological relations between the dendrites of olfactory sensory cells and sustentacular cells in different vertebrates. An ultrastructural study. J. Anat. 117, 89-94. [PMC free article] [PubMed] [Google Scholar]
- Caggiano, M., Kauer, J. S. and Hunter, D. D. (1994). Globose basal cells are neuronal progenitors in the olfactory epithelium: a lineage analysis using a replication-incompetent retrovirus. Neuron 13, 339-352. 10.1016/0896-6273(94)90351-4 [DOI] [PubMed] [Google Scholar]
- Caudy, M., Vassin, H., Brand, M., Tuma, R., Jan, L. Y. and Jan, Y. N. (1988). daughterless, a Drosophila gene essential for both neurogenesis and sex determination, has sequence similarities to myc and the achaete-scute complex. Cell 55, 1061-1067. 10.1016/0092-8674(88)90250-4 [DOI] [PubMed] [Google Scholar]
- Chen, M., Tian, S., Yang, X., Lane, A. P., Reed, R. R. and Liu, H. (2014). Wnt-responsive Lgr5(+) globose basal cells function as multipotent olfactory epithelium progenitor cells. J. Neurosci. 34, 8268-8276. 10.1523/JNEUROSCI.0240-14.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, M., Reed, R. R. and Lane, A. P. (2017). Acute inflammation regulates neuroregeneration through the NF-kappaB pathway in olfactory epithelium. Proc. Natl. Acad. Sci. USA 114, 8089-8094. 10.1073/pnas.1620664114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, M., Reed, R. R. and Lane, A. P. (2019). Chronic inflammation directs an olfactory stem cell functional switch from neuroregeneration to immune defense. Cell Stem Cell 25, 501-513 e5. 10.1016/j.stem.2019.08.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman, J. H., Lin, B., Louie, J. D., Peterson, J., Lane, R. P. and Schwob, J. E. (2019). Spatial determination of neuronal diversification in the olfactory epithelium. J. Neurosci. 39, 814-832. 10.1523/JNEUROSCI.3594-17.2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conboy, I. M. and Rando, T. A. (2002). The regulation of Notch signaling controls satellite cell activation and cell fate determination in postnatal myogenesis. Dev. Cell 3, 397-409. 10.1016/S1534-5807(02)00254-X [DOI] [PubMed] [Google Scholar]
- Dahl, A. R., Hadley, W. M., Hahn, F. F., Benson, J. M. and Mcclellan, R. O. (1982). Cytochrome P-450-dependent monooxygenases in olfactory epithelium of dogs: possible role in tumorigenicity. Science 216, 57-59. 10.1126/science.7063870 [DOI] [PubMed] [Google Scholar]
- Dias, T. B., Yang, Y. J., Ogai, K., Becker, T. and Becker, C. G. (2012). Notch signaling controls generation of motor neurons in the lesioned spinal cord of adult zebrafish. J. Neurosci. 32, 3245-3252. 10.1523/JNEUROSCI.6398-11.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Souza, B., Meloty-Kapella, L. and Weinmaster, G. (2010). Canonical and non-canonical Notch ligands. Curr. Top. Dev. Biol. 92, 73-129. 10.1016/S0070-2153(10)92003-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durante, M. A., Kurtenbach, S., Sargi, Z. B., Harbour, J. W., Choi, R., Kurtenbach, S., Goss, G. M., Matsunami, H. and Goldstein, B. J. (2020). Single-cell analysis of olfactory neurogenesis and differentiation in adult humans. Nat. Neurosci. 23, 323-326. 10.1038/s41593-020-0587-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher, R. B., Prasol, M. S., Estrada, J., Baudhuin, A., Vranizan, K., Choi, Y. G. and Ngai, J. (2011). p63 regulates olfactory stem cell self-renewal and differentiation. Neuron 72, 748-759. 10.1016/j.neuron.2011.09.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fre, S., Huyghe, M., Mourikis, P., Robine, S., Louvard, D. and Artavanis-Tsakonas, S. (2005). Notch signals control the fate of immature progenitor cells in the intestine. Nature 435, 964-968. 10.1038/nature03589 [DOI] [PubMed] [Google Scholar]
- Fryer, C. J., White, J. B. and Jones, K. A. (2004). Mastermind recruits CycC:CDK8 to phosphorylate the Notch ICD and coordinate activation with turnover. Mol. Cell 16, 509-520. 10.1016/j.molcel.2004.10.014 [DOI] [PubMed] [Google Scholar]
- Gadye, L., Das, D., Sanchez, M. A., Street, K., Baudhuin, A., Wagner, A., Cole, M. B., Choi, Y. G., Yosef, N., Purdom, E.et al. (2017). injury activates transient olfactory stem cell states with diverse lineage capacities. Cell Stem Cell 21, 775-790.e9. 10.1016/j.stem.2017.10.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavid, M., Coulomb, L., Thomas, J., Aouimeur, I., Verhoeven, P., Mentek, M., Dumollard, J. M., Forest, F., Prades, J. M., Thuret, G.et al. (2023). Technique of flat-mount immunostaining for mapping the olfactory epithelium and counting the olfactory sensory neurons. PLoS One 18, e0280497. 10.1371/journal.pone.0280497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genter, M. B., Deamer, N. J., Blake, B. L., Wesley, D. S. and Levi, P. E. (1995). Olfactory toxicity of methimazole: dose-response and structure-activity studies and characterization of flavin-containing monooxygenase activity in the Long-Evans rat olfactory mucosa. Toxicol. Pathol. 23, 477-486. 10.1177/019262339502300404 [DOI] [PubMed] [Google Scholar]
- Graziadei, P. P. and Graziadei, G. A. (1979). Neurogenesis and neuron regeneration in the olfactory system of mammals. I. Morphological aspects of differentiation and structural organization of the olfactory sensory neurons. J. Neurocytol. 8, 1-18. 10.1007/BF01206454 [DOI] [PubMed] [Google Scholar]
- Grochowski, C. M., Loomes, K. M. and Spinner, N. B. (2016). Jagged1 (JAG1): structure, expression, and disease associations. Gene 576, 381-384. 10.1016/j.gene.2015.10.065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, Z., Packard, A., Krolewski, R. C., Harris, M. T., Manglapus, G. L. and Schwob, J. E. (2010). Expression of pax6 and sox2 in adult olfactory epithelium. J. Comp. Neurol. 518, 4395-4418. 10.1002/cne.22463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heitzler, P. and Simpson, P. (1991). The choice of cell fate in the epidermis of Drosophila. Cell 64, 1083-1092. 10.1016/0092-8674(91)90263-X [DOI] [PubMed] [Google Scholar]
- Herrick, D. B., Lin, B., Peterson, J., Schnittke, N. and Schwob, J. E. (2017). Notch1 maintains dormancy of olfactory horizontal basal cells, a reserve neural stem cell. Proc. Natl. Acad. Sci. USA 114, E5589-E5598. 10.1073/pnas.1701333114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrick, D. B., Guo, Z., Jang, W., Schnittke, N. and Schwob, J. E. (2018). Canonical Notch signaling directs the fate of differentiating neurocompetent progenitors in the mammalian olfactory epithelium. J. Neurosci. 38, 5022-5037. 10.1523/JNEUROSCI.0484-17.2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holbrook, E. H., Szumowski, K. E. and Schwob, J. E. (1995). An immunochemical, ultrastructural, and developmental characterization of the horizontal basal cells of rat olfactory epithelium. J. Comp. Neurol. 363, 129-146. 10.1002/cne.903630111 [DOI] [PubMed] [Google Scholar]
- Holbrook, E. H., Wu, E., Curry, W. T., Lin, D. T. and Schwob, J. E. (2011). Immunohistochemical characterization of human olfactory tissue. Laryngoscope 121, 1687-1701. 10.1002/lary.21856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Indra, A. K., Warot, X., Brocard, J., Bornert, J. M., Xiao, J. H., Chambon, P. and Metzger, D. (1999). Temporally-controlled site-specific mutagenesis in the basal layer of the epidermis: comparison of the recombinase activity of the tamoxifen-inducible Cre-ER(T) and Cre-ER(T2) recombinases. Nucleic Acids Res. 27, 4324-4327. 10.1093/nar/27.22.4324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joiner, A. M., Green, W. W., Mcintyre, J. C., Allen, B. L., Schwob, J. E. and Martens, J. R. (2015). Primary cilia on horizontal basal cells regulate regeneration of the olfactory epithelium. J. Neurosci. 35, 13761-13772. 10.1523/JNEUROSCI.1708-15.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaguchi, D., Furutachi, S., Kawai, H., Hozumi, K. and Gotoh, Y. (2013). Dll1 maintains quiescence of adult neural stem cells and segregates asymmetrically during mitosis. Nat. Commun. 4, 1880. 10.1038/ncomms2895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunisch, M., Haenlin, M. and Campos-Ortega, J. A. (1994). Lateral inhibition mediated by the Drosophila neurogenic gene delta is enhanced by proneural proteins. Proc. Natl. Acad. Sci. USA 91, 10139-10143. 10.1073/pnas.91.21.10139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung, C. T., Coulombe, P. A. and Reed, R. R. (2007). Contribution of olfactory neural stem cells to tissue maintenance and regeneration. Nat. Neurosci. 10, 720-726. 10.1038/nn1882 [DOI] [PubMed] [Google Scholar]
- Lindsell, C. E., Shawber, C. J., Boulter, J. and Weinmaster, G. (1995). Jagged: a mammalian ligand that activates Notch1. Cell 80, 909-917. 10.1016/0092-8674(95)90294-5 [DOI] [PubMed] [Google Scholar]
- Luo, D., Renault, V. M. and Rando, T. A. (2005). The regulation of Notch signaling in muscle stem cell activation and postnatal myogenesis. Semin. Cell Dev. Biol. 16, 612-622. 10.1016/j.semcdb.2005.07.002 [DOI] [PubMed] [Google Scholar]
- Maurya, D. K., Henriques, T., Marini, M., Pedemonte, N., Galietta, L. J., Rock, J. R., Harfe, B. D. and Menini, A. (2015). Development of the olfactory epithelium and nasal glands in TMEM16A-/- and TMEM16A+/+ Mice. PLoS One 10, e0129171. 10.1371/journal.pone.0129171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto, A., Lau, R., Hein, P. W., Shipley, J. M. and Weinmaster, G. (2006). Microfibrillar proteins MAGP-1 and MAGP-2 induce Notch1 extracellular domain dissociation and receptor activation. J. Biol. Chem. 281, 10089-10097. 10.1074/jbc.M600298200 [DOI] [PubMed] [Google Scholar]
- Mok, L. P., Qin, T., Bardot, B., Lecomte, M., Homayouni, A., Ahimou, F. and Wesley, C. (2005). Delta activity independent of its activity as a ligand of Notch. BMC Dev. Biol. 5, 6. 10.1186/1471-213X-5-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtsuka, T., Ishibashi, M., Gradwohl, G., Nakanishi, S., Guillemot, F. and Kageyama, R. (1999). Hes1 and Hes5 as notch effectors in mammalian neuronal differentiation. EMBO J. 18, 2196-2207. 10.1093/emboj/18.8.2196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Packard, A., Giel-Moloney, M., Leiter, A. and Schwob, J. E. (2011a). Progenitor cell capacity of NeuroD1-expressing globose basal cells in the mouse olfactory epithelium. J. Comp. Neurol. 519, 3580-3596. 10.1002/cne.22726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Packard, A., Schnittke, N., Romano, R. A., Sinha, S. and Schwob, J. E. (2011b). DeltaNp63 regulates stem cell dynamics in the mammalian olfactory epithelium. J. Neurosci. 31, 8748-8759. 10.1523/JNEUROSCI.0681-11.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer, W. H., Jia, D. and Deng, W. M. (2014). Cis-interactions between Notch and its ligands block ligand-independent Notch activity. Elife 3, e04415. 10.7554/eLife.04415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellegrinet, L., Rodilla, V., Liu, Z., Chen, S., Koch, U., Espinosa, L., Kaestner, K. H., Kopan, R., Lewis, J. and Radtke, F. (2011). Dll1- and dll4-mediated notch signaling are required for homeostasis of intestinal stem cells. Gastroenterology 140, 1230-1240.e1-7. 10.1053/j.gastro.2011.01.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rafols, J. A. and Getchell, T. V. (1983). Morphological relations between the receptor neurons, sustentacular cells and Schwann cells in the olfactory mucosa of the salamander. Anat. Rec. 206, 87-101. 10.1002/ar.1092060111 [DOI] [PubMed] [Google Scholar]
- Schnittke, N., Herrick, D. B., Lin, B., Peterson, J., Coleman, J. H., Packard, A. I., Jang, W. and Schwob, J. E. (2015). Transcription factor p63 controls the reserve status but not the stemness of horizontal basal cells in the olfactory epithelium. Proc. Natl. Acad. Sci. USA 112, E5068-E5077. 10.1073/pnas.1512272112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeter, E. H., Kisslinger, J. A. and Kopan, R. (1998). Notch-1 signalling requires ligand-induced proteolytic release of intracellular domain. Nature 393, 382-386. 10.1038/30756 [DOI] [PubMed] [Google Scholar]
- Suzuki, Y., Takeda, M. and Farbman, A. I. (1996). Supporting cells as phagocytes in the olfactory epithelium after bulbectomy. J. Comp. Neurol. 376, 509-517. [DOI] [PubMed] [Google Scholar]
- Vaessin, H., Brand, M., Jan, L. Y. and Jan, Y. N. (1994). daughterless is essential for neuronal precursor differentiation but not for initiation of neuronal precursor formation in Drosophila embryo. Development 120, 935-945. 10.1242/dev.120.4.935 [DOI] [PubMed] [Google Scholar]
- Wang, L., Ren, W., Li, X., Zhang, X., Tian, H., Bhattarai, J. P., Challis, R. C., Lee, A. C., Zhao, S., Yu, H.et al. (2021). Chitinase-like protein Ym2 (Chil4) regulates regeneration of the olfactory epithelium via interaction with inflammation. J. Neurosci. 41, 5620-5637. 10.1523/JNEUROSCI.1601-20.2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward, J. M., Yoon, M., Anver, M. R., Haines, D. C., Kudo, G., Gonzalez, F. J. and Kimura, S. (2001). Hyalinosis and Ym1/Ym2 gene expression in the stomach and respiratory tract of 129S4/SvJae and wild-type and CYP1A2-null B6, 129 mice. Am. J. Pathol. 158, 323-332. 10.1016/S0002-9440(10)63972-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinbaum, J. S., Broekelmann, T. J., Pierce, R. A., Werneck, C. C., Segade, F., Craft, C. S., Knutsen, R. H. and Mecham, R. P. (2008). Deficiency in microfibril-associated glycoprotein-1 leads to complex phenotypes in multiple organ systems. J. Biol. Chem. 283, 25533-25543. 10.1074/jbc.M709962200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, Q., Xu, X., Miao, X., Bao, X., Li, X., Xiang, L., Wang, W., Du, S., Lu, Y., Wang, X.et al. (2022). YAP signaling in horizontal basal cells promotes the regeneration of olfactory epithelium after injury. Stem Cell Rep. 17, 664-677. 10.1016/j.stemcr.2022.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zunitch, M. J., Fisch, A. S., Lin, B., Barrios-Camacho, C. M., Faquin, W. C., Tachie-Baffour, Y., Louie, J. D., Jang, W., Curry, W. T., Gray, S. T.et al. (2023). Molecular evidence for olfactory neuroblastoma as a tumor of malignant globose basal cells. Mod. Pathol. 36, 100122. 10.1016/j.modpat.2023.100122 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.