Abstract
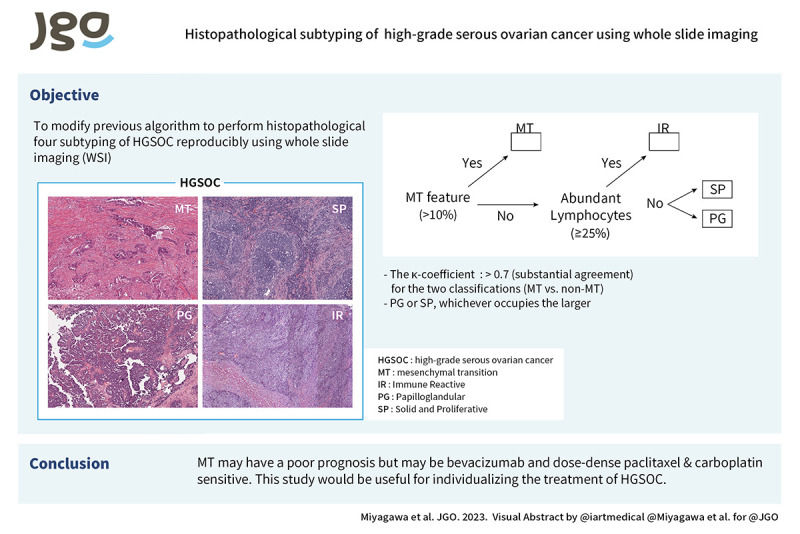
Objective
We have established 4 histopathologic subtyping of high-grade serous ovarian cancer (HGSOC) and reported that the mesenchymal transition (MT) type has a worse prognosis than the other subtypes. In this study, we modified the histopathologic subtyping algorithm to achieve high interobserver agreement in whole slide imaging (WSI) and to characterize the tumor biology of MT type for treatment individualization.
Methods
Four observers performed histopathological subtyping using WSI of HGSOC in The Cancer Genome Atlas data. As a validation set, cases from Kindai and Kyoto Universities were independently evaluated by the 4 observers to determine concordance rates. In addition, genes highly expressed in MT type were examined by gene ontology term analysis. Immunohistochemistry was also performed to validate the pathway analysis.
Results
After algorithm modification, the kappa coefficient, which indicates interobserver agreement, was greater than 0.5 (moderate agreement) for the 4 classifications and greater than 0.7 (substantial agreement) for the 2 classifications (MT vs. non-MT). Gene expression analysis showed that gene ontology terms related to angiogenesis and immune response were enriched in the genes highly expressed in the MT type. CD31 positive microvessel density was higher in the MT type compared to the non-MT type, and tumor groups with high infiltration of CD8/CD103 positive immune cells were observed in the MT type.
Conclusion
We developed an algorithm for reproducible histopathologic subtyping classification of HGSOC using WSI. The results of this study may be useful for treatment individualization of HGSOC, including angiogenesis inhibitors and immunotherapy.
Keywords: Carcinoma, Ovarian Epithelial Cancer, Biomarkers, Histopathology
Synopsis
We developed an algorithm for the histopathological subtyping of high-grade serous ovarian cancer (HGSOC) using whole slide imaging. We found that this classification may be associated with response to bevacizumab and immune-checkpoint inhibitors. This study would be useful for individualizing the treatment of HGSOC.
Graphical Abstract
INTRODUCTION
Ovarian cancer has the poorest prognosis among all gynecologic malignancies with an overall 5-years survival rate of approximately 50% [1]. High-grade serous ovarian cancer (HGSOC) is the most common histopathological type of ovarian cancer [2]. Hallmarks of HGSOC are mainly BRCA1/2 mutations and homologous recombination deficiency (HRD) and maintenance treatment with poly ADP-ribose polymerase (PARP) inhibitor is also used, after first-line chemotherapy [3]. Besides, maintenance therapy with bevacizumab, an anti-vascular endothelial growth factor (VEGF) antibody, is another option. However, several phase III trials showed that bevacizumab only extends the median progression-free survival (PFS) by 3.8 months [4] or 1.5 months [5]. Another option in chemotherapy regimens for ovarian cancer is dose-dense (dd) paclitaxel and carboplatin (TC), which increases the dose of paclitaxel. The Japanese Gynecologic Oncology Group (JGOG) 3016 study showed that ddTC therapy prolonged PFS and overall survival compared with TC therapy [6,7]. However, subsequent clinical trials did not prove the benefit of ddTC [8].
The Cancer Genome Atlas (TCGA) data analysis revealed that HGSOC can be classified into 4 gene expression subtypes [9]: mesenchymal, proliferative, immunoreactive, and differentiated, and the efficacy of bevacizumab was reported to be higher in the first 2 [10]. On the other hand, we previously showed that the mesenchymal type is more sensitive to taxane and ddTC may be useful for patients with this subtype [11]. Thus, gene expression analysis may be useful to select patients who would benefit from bevacizumab or ddTC. However, this is difficult to implement in daily practice because of problems with cost and sample quality assurance. Therefore, we used conventional hematoxylin and eosin (HE) glass slides to histopathologically characterize 4 subtypes of HGSOC, mesenchymal transition (MT), solid and proliferative (SP), immune reactive (IR), and papilloglandular (PG) [12]. Among them, the association between histopathological MT and TCGA Mesenchymal types was strong. We then reviewed the HE slides of HGSOC cases enrolled in the JGOG3016 study and found that ddTC was useful in the MT type [13].
In the research to create a new algorithm for histopathological subtyping, the use of HE slides has limitations in terms of sample transportation, storage, and personal information protection, making it difficult to prove the reproducibility of diagnosis and achieve clinical application. Virtual slides (whole slide imaging, WSI), which are digitalized representative slides of the tumor, can be stored, and copied electronically, and it is easy to evaluate the inter-observer agreement. The WSI data of HGSOC in TCGA are publicly available and can be observed by pathologists around the world. In addition, many recent clinical trials have preserved WSI of representative tumor slides. Therefore, establishing an evaluation method using WSI will allow us to retrospectively investigate the relationship with clinical trial outcomes.
In this study, we demonstrate that histopathological subtyping of HGSOC can be performed using WSI. We also show the relationship between the MT subtype and tumor biology, which may lead to the selection of targeted therapies. These findings will contribute to the implementation of personalized therapy based on the histopathology of HGSOC.
MATERIALS AND METHODS
1. TCGA data analysis
WSI data for ovarian cancer were available for 106 cases in the Genomic Data Commons data portal (https://portal.gdc.cancer.gov/). The annotation of HRD scores and BRCA1/2 alterations (germline and somatic BRCA1/2 mutations and BRCA1 promoter methylations) in TCGA database was based on our previous report [14].
Inter-observer agreement
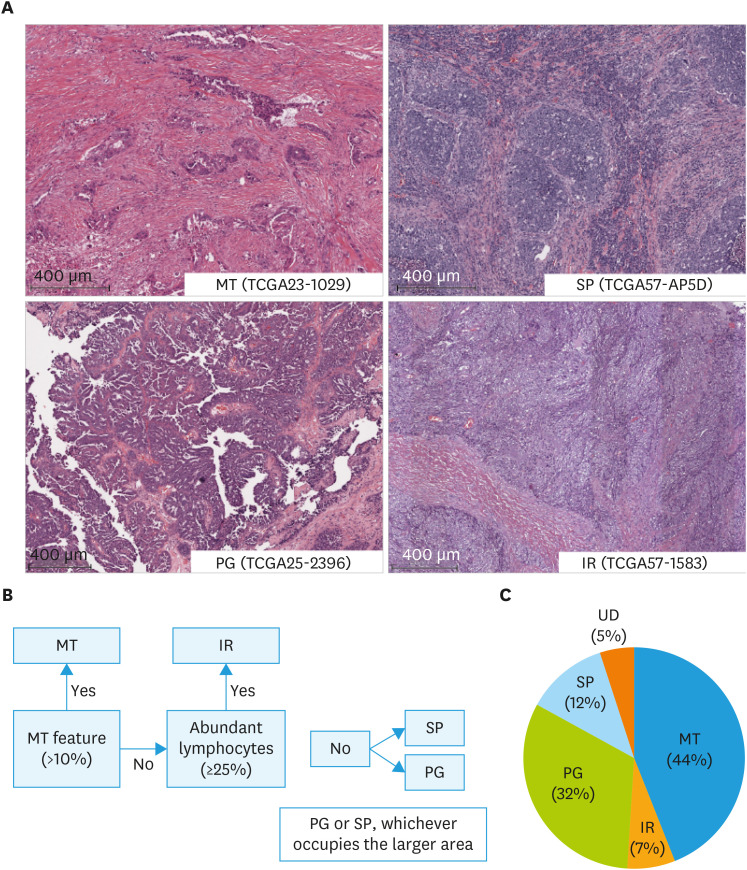
Four observers (M.C., N.H., O.T., and M.R.) specializing in gynecologic pathology and oncology and including one board-certified pathologist (O.T.), evaluated the characteristics of WSI and classified them into MT, IR, PG, and SP histopathological types based on our previously established algorithm [12]. The newly constructed algorithm is detailed in Fig. 1A, B and Data S1. As an overview, first, MT is diagnosed when tumor cell clusters with labyrinthine architecture or cells lacking continuity that infiltrate with stromal reaction are present in more than 10% of the slide. In addition, IR is diagnosed when lymphocytes infiltrating the stroma are present in 25% or more of the slide (Fig. S1). If these are not met, determine the form with the largest occupied area (SP or PG). The discordant cases were discussed, and the diagnostic methods were clarified to increase the agreement rate. Two observers (M.C. and O.T.) assessed Solid, pseudo-Endometrioid, and Transitional cell carcinoma-like morphology (SET) features [15]. Interobserver agreement of the histopathological subtyping was evaluated with Cohen’s (2 observers) or Fleiss’ (4 observers) kappa coefficient using irr package in R (R Foundation, Vienna, Austria). Kappa of 0–0.2 is slight, 0.2–0.6 moderate, 0.6–0.8 substantial, 0.8–1 almost perfect agreement.
Fig. 1. Histopathological subtyping of high-grade serous ovarian cancer using The Cancer Genome Atlas whole slide imaging. (A) Representative images of histopathologic subtypes. MT: glandular ducts and cells infiltrating with stromal reaction occupy ≥10% of the specimen; IR: lymphocytes infiltrating in the stroma occupy ≥25%. (B) Hierarchy of subtyping. (C) Proportion of each subtype.
IR, immune reactive; MT, mesenchymal transition; PG, papillary proliferation; SP, solid proliferation; UD, undetermined.
Gene expression analysis
For gene expression microarray analysis, TCGA HG-U133A data set was obtained from TCGA Data Portal (http://cancergenome.nih.gov). The Affymetrix array data were RMA normalized using the R package affy (http://www.R-project.org). Single-sample Gene Set Enrichment Analysis (ssGSEA) was performed as previously described [12]. For data analyses, the ssGSEA scores were normalized from 0 to 1. Genes highly expressed in the MT type were identified using samroc [16] with false discovery rates (FDR) q<0.05 as a cutoff. The Gene Ontology term (biological process) enriched in the genes was identified using GO net [17], and the network was visualized.
2. The Kindai/Kyoto cohort data analysis
Fifty-nine HGSOC cases that underwent surgery between 2007 and 2018 at Kindai University Hospital and Kyoto University Hospital were included in the study. Formalin-fixed paraffin-embedded samples of untreated tumors were retrieved, and HE slides of representative tumor sections were digitized. The WSI data were observed using the NanoZoomer Digital Pathology System view 2.8.24 (Hamamatsu Photonics K.K., Hamamatsu, Japan).
Inter-observer agreement
Four observers (M.C., N.H., O.T., and M.R.) independently assessed the WSI data, and the agreement rate of histopathological subtyping diagnosis was examined.
Immunohistopathological staining
Formalin-fixed paraffin-embedded sections were cut into sections (3 µm thick) and heated in DAKO Target Retrieval Solution (S1699; Dako, Santa Clara, CA, USA), diluted 1:10 in distilled water pH 6, at 98°C for 50 minutes. Endogenous proteins were blocked with Animal-Free Blocker (SP5030, 1:4; Vector, New York, NY, USA), and samples were incubated for 15 minutes at room temperature (RT).
For immunohistochemical staining of CD31, CD8 and CD103 sections were incubated with CD31 antibody (C31.3, 1:100; Novus Biologicals, Centennial, CO, USA), CD8 antibody (LCL-L-CD8-4B11, 1:250; Leica Biosystems, Nussloch, Germany) and CD103 antibody (ab129202, 1:6,000; Abcam, Cambridge, UK) at 4°C overnight. Sections were washed, and 3% H2O2/PBS was applied for 15 minutes to block peroxidase. DAB (3:100; Vector), Fast Red (415261; Nichirei Biosciences, Tokyo, Japan) and Histgreen (E109; LINARIS Biologische Produkt, Dossenheim, Germany) were added for 3 minutes at RT.
All immunohistochemically stained slides were digitized and quantitatively evaluated using QuPath v0.2.0 (https://qupath.github.io/) (Data S2). Microvessel density (MVD) was determined by counting the number of CD31-positive luminal structures per mm2 at 5 representative tumor sites and calculating the mean value, as described in a previous report [18]. To quantify CD8+ and/or CD103+ staining, the number of positive cells per 1 mm2 was counted for each sample using Qupath, and the mean value of 5 fields of view was calculated. The obtained values were used to perform average linkage clustering using Cluster 3.0, followed by visualization using Java TreeView 1.2.0.
3. Ethics approval
This study was approved by the ethics committees of Kindai University Hospital (the approval number; 31-146). All methods were performed in accordance with the ethical standards of the responsible committee on human experimentation (institutional and national) and with the Helsinki Declaration of 1964 and its later amendments. Informed consent was obtained in the form of an opt-out option on the website of the institution in 2020.
4. Statistical analysis
Statistical analysis was performed using GraphPad Prism ver. 8.2.0 (GraphPad Software, San Diego, CA, USA). Comparison among 3 or more groups was conducted with ordinary one-way analysis of variance and comparison between 2 groups was performed with t-test. p<0.05 indicated a significant difference.
RESULTS
1. Histopathological subtyping of HGSOC using WSI
Histopathological subtyping of TCGA samples
Among the 106 TCGA ovarian cancer cases, 4 cases without TP53 mutation [19] could not be diagnosed as HGSOC by pathomorphology and were excluded. We examined the WSI of the other 102 cases. First, based on the original definitions (Fig. 1) [12] and diagnosed independently by the 4 observers without training, the Fleiss’ kappa coefficient was low with 4 subtypes, 0.348 (fair agreement), and binary classifications of MT vs. non-MT, 0.350 (fair agreement) (Table S1).
To improve the reproducibility of the diagnosis, the algorithm was clarified (Data S1). The summary is as follows: for MT, morphological characteristics of labyrinthine infiltrating cells with marked stromal reaction (i.e., micropapillary pattern, irregular glands, small nests, single cells) were specified. In addition, it was difficult to observe intraepithelial tumor infiltrating lymphocytes (TILs) in WSI. Therefore, based on the evaluation method of the TILs Working Group in Breast Cancer [20,21], only stromal TILs were counted, not intraepithelial TILs or TILs in necrotic tissue, with a cut-off of 25%. Regarding PG, we included not only the classical papillary form, but also low-grade endometrioid carcinoma like and serous borderline tumor like forms.
Next, the WSI data were independently re-evaluated by the 4 observers. The Fleiss’ kappa coefficient increased to 0.549 (moderate agreement) for the 4 subtypes and 0.705 (substantial agreement) for the binary classifications (Table 1). Under the new algorithm, perfect agreement (4 out of 4 observers) for 4 and binary classifications was 48 cases (47%) and 56 cases (55%), majority agreement (3 out of 4 observers) was 77 cases (75%) and 86 cases (84%). Final histopathologic subtyping classification was agreed upon through observer discussion for 97 of the 102 cases and was used for subsequent analysis. MT, IR, PG, and SP accounted for 44%, 7%, 32%, and 12%, respectively (Fig. 1C). The remaining 5 cases could not be classified (Undetermined; UD). Two were cases with histomorphology that could not be classified even with the new classification algorithm; the presence of such cases is acknowledged at the bottom of the subtyping algorithm (Data S1). Three cases were not suitable for histological analysis because 2 had mostly stromal tissue with little or no neoplastic epithelium in the WSI available on the TCGA website and one had only frozen sections (Fig. S2).
Table 1. Inter-observer concordance rates for TCGA samples.
| Observers | Four classifications; Fleiss’ kappa 0.549 | Two classifications (MT vs. non-MT); Fleiss’ kappa 0.705 | ||||
|---|---|---|---|---|---|---|
| N.H. | O.T. | M.R. | N.H. | O.T. | M.R. | |
| M.C. | 70% (0.56) | 87% (0.81) | 62% (0.48) | 82% (0.65) | 92% (0.84) | 78% (0.58) |
| N.H. | 67% (0.51) | 63% (0.51) | 83% (0.68) | 80% (0.63) | ||
| O.T. | 60% (0.46) | 77% (0.57) | ||||
The numbers in parentheses represent Cohen’s kappa coefficient.
TCGA, The Cancer Genome Atlas.
Inter-observer agreement of the Kindai/Kyoto cohort
For assessment of inter-observer agreement and for later analysis of immunohistochemistry, 4 observers independently performed histopathological subtyping of WSI in the Kindai/Kyoto cohort (n=59). The Fleiss’ kappa coefficient was 0.558 (moderate agreement) for the 4 classifications and improved to 0.703 (substantial agreement) for the binary classifications (MT and non-MT) (Table 2). Perfect agreement (4 out of 4 observers) for 4 and binary classifications was 23 cases (39%) and 39 cases (69%), majority agreement (3 out of 4 observers) was 46 cases (78%) and 54 cases (92%).
Table 2. Inter-observer concordance rates for the Kindai/Kyoto cohortThe numbers in parentheses represent Cohen’s kappa coefficient.
| Observers | Four classifications; Fleiss’ kappa 0.588 | Two classifications (MT vs. non-MT); Fleiss’ kappa 0.703 | ||||
|---|---|---|---|---|---|---|
| N.H. | O.T. | M.R. | N.H. | O.T. | M.R. | |
| M.C. | 83% (0.76) | 88% (0.83) | 62% (0.49) | 93% (0.86) | 94% (0.90) | 81% (0.59) |
| N.H. | 67% (0.66) | 58% (0.42) | 92% (0.83) | 78% (0.53) | ||
| O.T. | 54% (0.39) | 75% (0.50) | ||||
MT, mesenchymal transition.
2. Molecular characteristics of HGSOC subtype
Relationship between HRD status and histopathological features
We found no association between the 4 subtypes and HRD scores or BRCA alterations (germline/somatic BRCA1/2 mutations and BRCA1 promoter methylations) (Fig. S3). Then SET features, previously reported to be associated with BRCA mutations [15], were evaluated in the 102 TCGA cases by 2 observers using WSI The concordance rate for SET cases (SET features ≥50%) was 79% (Cohen’s kappa coefficient: 0.44) (Table S2). We compared these matched cases with our HGSOC histopathological subtyping, and the SP type was mostly SET cases (p<0.001, Fig. S3) because the histological images of “solid tumor” overlapped with those defined by SET. There was no association between SET features and BRCA alterations or HRD scores (Fig. S3).
Characterization of MT type by gene expression profiling
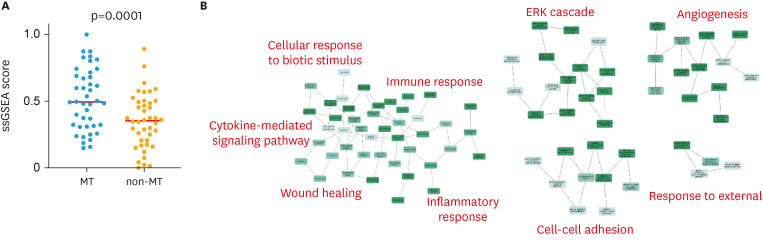
The MT type, which shows the poorest prognosis among the 4 subtypes [12], was investigated for its biological properties, with the aim to explore personalized therapy for MT. Of the 94 cases for which histopathological subtyping was performed, 84 cases had gene expression data available. We examined the ssGSEA scores of HALLMARK_EPITHELIAL_MESENCHYMAL_TRANSITION, genes defining epithelial-mesenchymal transition [22], on these data. The results showed that the MT type had a higher score than the other subtypes (p=0.001) (Fig. 2).
Fig. 2. Characteristics of MT type in gene expression profiles. (A) ssGSEA scores of an epithelial-mesenchymal transition-related gene set (HALLMARK_EPITHELIAL_MESENCHYMAL_TRANSITION). Y axis: normalized value of ssGSEA score. (B) Network diagram of gene ontology terms enriched in genes highly expressed in the MT type in The Cancer Genome Atlas dataset. Representative gene ontology terms are shown in red.
MT, mesenchymal transition; ssGSEA, single-sample Gene Set Enrichment Analysis.
Additionally, we examined differentially expressed genes between MT and non-MT types. A total of 286 genes (380 probes) were highly expressed (FDR q < 0.05) in the MT type (Table S3). The gene ontology terms enriched in these genes (Table S4) were related to immune response, wound healing, ERK cascade, angiogenesis, and cell adhesion (Fig. 2).
Analysis of the angiogenesis pathway
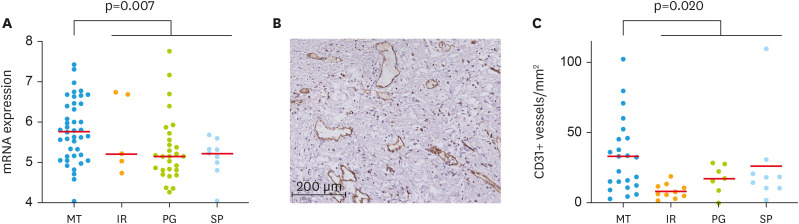
Next, we searched for molecular targets in MT type. As bevacizumab is a clinically used anti-angiogenic drug for ovarian cancer, we focused on the angiogenesis pathway. The PECAM1 gene (208983_s_at), which encodes CD31, a marker of vascular endothelium, was significantly upregulated in the MT type (p=0.007, Fig. 3). We next assessed CD31-positive MVD in 46 patients in the Kindai/Kyoto cohort (Tables S5 and S6) in whom at least 3 out of 4 diagnoses were concordant. We found that MVD was significantly higher in the MT group than in the non-MT group (IR+PG+SP) (p=0.020, Fig. 3).
Fig. 3. Association of histopathologic subtypes of high-grade serous ovarian cancer with MVD. (A) Expression of PECAM1 gene (208983_s_at) in The Cancer Genome Atlas dataset. (B) Representative image of CD31 immunohistochemistry. (C) Comparison of CD31+ MVD among histopathological subtypes.
IR, immune reactive; MT, mesenchymal transition; MVD, microvascular density; PG, papilloglandular; SP, solid and proliferative.
Analysis of tumor immunity
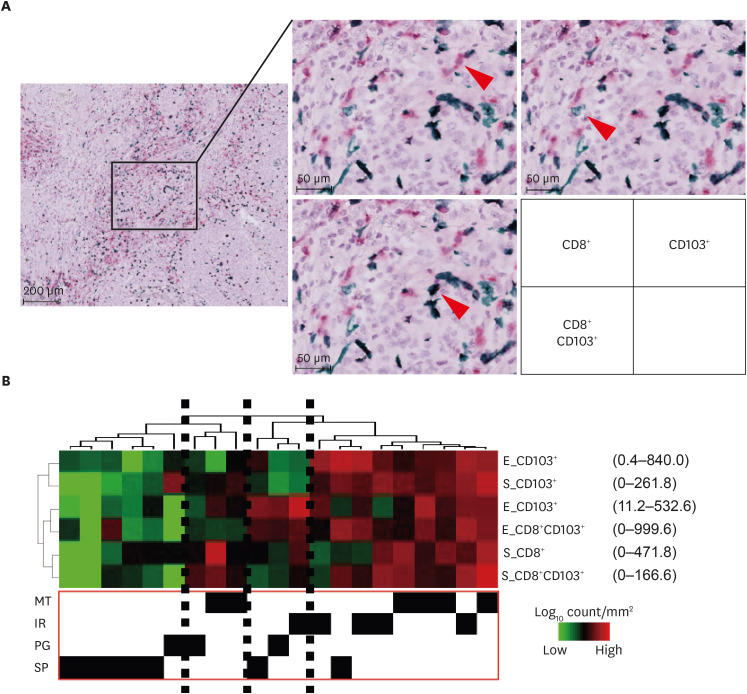
We focused on tissue resident lymphocytes [23] and performed double immunohistochemistry for CD8 and CD103 (Fig. 4). We evaluated the number of CD8 positive, CD103 positive, and CD8 and CD103 double positive cells in the epithelium or stroma in 21 recent cases of the Kindai/Kyoto cohort and performed hierarchical clustering. Tumor clusters with high infiltration of immune cells were observed, which contained almost exclusively MT or IR types (Fig. 4).
Fig. 4. Association of histopathological subtypes of high-grade serous ovarian cancer and tumor-infiltrating lymphocytes. (A) Representative image of immunohistochemical staining. Arrowhead shows lymphocytes. CD8+ T cells; fast red, CD103+ cells; hist green, double positive cells; black. (B) Hierarchical clustering with tumor-infiltrating lymphocytes.
E, epithelial component, IR, immune reactive; MT, mesenchymal transition; MVD, microvascular density; PG, papilloglandular; S, stromal component; SP, solid and proliferative.
DISCUSSION
When we developed the histopathologic subtyping of HGSOC in our previous study, we selected 5 typical cases for each of the 4 subtypes and used a total of 20 glass slides as a training set. We then tested for inter-observer agreement in another 28 cases with 6 observers [12]. The main objective of the current study was to modify the histopathological subtyping of HGSOC into a definitive classification using WSI. Making pathological diagnosis by WSI has significant advantages in terms of consultation, information sharing, and application to AI. However, unlike glass slide, WSI is difficult to observe cells in detail, and therefore, certain modifications are required to use WSI for pathological diagnosis [24,25]. In this study, 102 cases in TCGA were used for training by 4 observers. The Kindai/Kyoto cohort (59 cases) was then independently evaluated by the 4 observers (Table 2). Thus, a total of 161 cases with WSI were used in this study, and our results proved the reproducibility of this pathological subtyping diagnosis by validating inter-observer agreement rate in a much larger number of cases compared with that in our previous study. Recently, 7 gynecologic pathologists independently reviewed HE specimens of 75 cases of endocervical adenocarcinoma and compared the International Endocervical Adenocarcinoma Criteria and Classification with the World Health Organization classification [26]. They reported that the former was superior in terms of inter-observer agreement, with Kappa coefficients of 0.46 vs. 0.3 and perfect agreement (7/7 matched) in 42 cases (56%) vs. 7 cases (10%), which results are similar to the present study. Therefore, the Kappa coefficients and concordance rates in the present study are considered acceptable as a new histopathological diagnosis.
HGSOC is the most frequent cancer type with HRD because of BRCA1/2 mutations, BRCA1 promoter methylation, and alterations in other DNA homologous recombination pathway genes [27]. HRD accounts for approximately half of all HGSOC and leads to platinum and PARP inhibitor sensitivity [3]. Therefore, the ability to determine HRD in pathological slides would be clinically useful. SET features have been defined as morphological features observed in HGSOC with BRCA gene mutations. In a previous study, SET features were examined by a single observer, who found SET features in 22/31 cases with BRCA alterations and 2/12 cases without BRCA alterations in the training set and 9/9 cases with BRCA1 alterations and 9/14 cases without BRCA1 alterations in the validation set [15]. In a subsequent report of cases evaluated by 2 observers, SET was found in 13/26 cases with germline BRCA1/2 mutations and 9/32 cases without germline BRCA1/2 mutations, with no statistically significant difference in their frequencies [28]. When we examined the WSI data of TCGA, we found no association between SET features and BRCA alterations (Fig. S3). In previous papers, all HE slides of the tumor were examined, and our observation of WSI of tumor representative images may have been insufficient to evaluate SET features. However, previous reports have had several problems, including small sample size, insufficient consideration of inter-observer variability, and different frequencies of tumors with SET features between training and test sets and between papers. Hence, we have doubts about the reproducibility of the association between SET and BRCA alterations. At present, it seems difficult to predict BRCA alterations and HRD, at least by observation of WSI.
In this study we focused our analysis specifically on treatment individualization of the MT type. The WSI classification of only a representative part of the tumor may differ from the classification of the whole tumor because of intratumor heterogeneity. However, we previously demonstrated that diagnosis of the MT type is reproducible in the same individual even when the tumor specimen collection sites are different [12]. In this study, we found that the ssGSEA score of HALLMARK_EPITHELIAL_MESENCHYMAL_TRANSITION, which characterizes the mesenchymal gene expression subtype [22], was significantly higher in the MT type (Fig. 2), suggesting that the tumor site used for HE specimen preparation has the same characteristics as the site where gene expression analysis was performed.
A group of genes whose expression was downregulated by anti-VEGF antibody (VEGF-dependent genes), including PECAM1, which encodes CD31, correlated with sensitivity to the anti-VEGF antibody bevacizumab in colorectal cancer [29]. In HGSOC, overexpression of VEGF-dependent genes was found in the mesenchymal type, which correlated with poor prognosis [30]. In our study, we found that genes related to angiogenesis were upregulated in the MT type, and CD31-positive MVD was high (Fig. 3). In the data analysis of the GOG218 study, CD31-positive MVD high was a predictive marker for bevacizumab sensitivity [31], suggesting that bevacizumab may be more effective in the MT type. Furthermore, the mesenchymal type determined by gene expression [32] and the MT type evaluated by histopathology [12] are associated with residual tumor at surgery, and bevacizumab is more effective in patients with more residual tumor at surgery [5]. WSI data from the GOG218 study [4] and the ICON7 study [5], which randomized patients with and without bevacizumab, should be reviewed in future studies to determine whether the effect of bevacizumab is enhanced in the MT type.
It has been reported that the “mesenchymal type” in the TCGA gene expression subtype classification includes cases with a high immune response [33]. In this study, we found an enrichment of gene ontology terms related to immune response in genes that are highly expressed in the histopathological MT type (Fig. 2). Recently, CD103 has been used as a marker for tissue resident T cells and natural killer cells [34], and infiltration of CD103-positive resident memory-like CD8+ T cells is expected to be a biomarker for predicting the effect of immune checkpoint inhibitors (ICIs) [35]. In this study, we found CD8+, CD103+, and CD8+CD103+ immune cell infiltration in MT and IR, and almost no infiltration in SP and PG (Fig. 3). In a recent phase 3 study for ovarian cancer, the efficacy of ICIs was not proven [36,37], but it may be useful to examine whether ICIs show efficacy in MT and IR.
In the JGOG3016 trial of advanced ovarian cancer, ddTC with an increased dose of paclitaxel prolonged PFS compared with TC, but there was no significant difference in the larger phase 3 trial ICON8 [8]. However, in ICON8, the median PFS was prolonged by approximately 3 months in the ddTC group compared with that in the TC group [8]. Because ddTC was more beneficial than TC in MT patients in our JGOG3016A1 study [13], if the ICON8 cohort is divided into MT and non-MT types by WSI, ddTC may prove to be useful in MT type.
The identification of histopathological subtyping by WSI will lead to the establishment of a classification method by AI or machine learning. Convolutional neural networks have been able to classify the entire TCGA slide image into 19 cancer types and even subtypes [38]. The deep learning system also diagnosed Gleason grading of prostate needle biopsy specimens more accurately than a general pathologist [39]. The histotype diagnosis of ovarian cancer was also predicted with high accuracy by machine learning [40]. The results of this study provide basic information for the development of a machine learning program for histopathological subtyping of HGSOC. Although the interobserver agreement based on human observation was not perfect, the establishment of AI diagnostics will allow for more reproducible histopathological subtyping.
In conclusion, we have modified our previous algorithm to perform histopathological subtyping of HGSOC reproducibly using WSI. In this study, we used publicly available TCGA data as a training set so that pathologists around the world can learn this diagnostic method and apply it to clinical practice immediately. We found that the MT type had increased angiogenesis and high tumor immune activity. If we can evaluate the WSI enrolled in bevacizumab, ICI or ddTC clinical trials and classify the cases into MT type and non-MT type, and prove the difference in therapeutic effects, the personalized treatment of HGSOC will be advanced. The application of the results of this study to machine learning is expected to further improve the objectivity of diagnosis and advance its application to general practice.
Footnotes
Funding: This work was supported by the Grant-in-Aid for Scientific Research (KAKENHI) from the Japan Society for the Promotion of Science [Grant-in-Aid for Scientific Research (C): 22K09630, N.H. and (B): 18H02947, M.N.].
Conflict of Interest: No potential conflict of interest relevant to this article was reported.
- Conceptualization: M.C., M.R., M.M., M.N., N.H.
- Data curation: M.C., O.T., M.R., T.S., N.H.
- Formal analysis: M.C., M.R., T.H., M.K., M.N.
- Funding acquisition: M.N., N.H.
- Methodology: M.C., O.T., M.R., M.N., N.H.
- Software: T.H., M.N.
- Supervision: M.M., M.N.
- Writing - original draft: M.C., M.N.
- Writing - review & editing: M.C., O.T., M.R., T.S., T.H., M.K., M.M., M.N., N.H.
SUPPLEMENTARY MATERIALS
New algorithm of histopathological subtyping of high-grade serous ovarian cancer
QuPath analysis of CD31 immunohistochemistry
Initial inter-observer concordance rates for The Cancer Genome Atlas samples
The Cancer Genome Atlas cases
Highly expressed genes in the mesenchymal transition type
Enriched gene ontology terms in the highly expressed genes in the mesenchymal transition type
Clinical characteristics of the Kindai/Kyoto cohort
Kindai/Kyoto cohort
Evaluation of the percentage of lymphocytes infiltrating the tumor stroma.
Undetermined cases in TCGA samples. (TCGA 23-1118) (TCGA 23-1121) Mostly stromal tissue with no epithelial structure. (TCGA VG-A8LO) This was frozen sections and unsuitable for detailed histological analysis. (TCGA 23-1110) Complex epithelial arrangement with relatively little intervening stroma, not easily identified as cribriform, papillary, or solid. (TCGA 23-1120) Similar to many other types of carcinomas, high grade serous carcinomas can have a more ragged architecture in the tumor invasive front and/or around intratumoral fibrosis. In PG/SP pattern-predominant cases, this can result in PG/SP pattern gradually transitioning to mesenchymal transition pattern in the invasive front or around fibrosis.
Histopathological subtype and HRD status. (A) Association of BRCA alterations and HRD scores with the histopathological subtypes. There was no association between them. (B) Association between histopathological subtypes and SET cases (SET feature ≥50%). (C) Association between SET and BRCA alterations. (D) Association between SET and HRD scores.
References
- 1.Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2021. CA Cancer J Clin. 2021;71:7–33. doi: 10.3322/caac.21654. [DOI] [PubMed] [Google Scholar]
- 2.Seidman JD, Horkayne-Szakaly I, Haiba M, Boice CR, Kurman RJ, Ronnett BM. The histologic type and stage distribution of ovarian carcinomas of surface epithelial origin. Int J Gynecol Pathol. 2004;23:41–44. doi: 10.1097/01.pgp.0000101080.35393.16. [DOI] [PubMed] [Google Scholar]
- 3.Nakai H, Matsumura N. Individualization in the first-line treatment of advanced ovarian cancer based on the mechanism of action of molecularly targeted drugs. Int J Clin Oncol. 2022;27:1001–1012. doi: 10.1007/s10147-022-02163-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burger RA, Brady MF, Bookman MA, Fleming GF, Monk BJ, Huang H, et al. Incorporation of bevacizumab in the primary treatment of ovarian cancer. N Engl J Med. 2011;365:2473–2483. doi: 10.1056/NEJMoa1104390. [DOI] [PubMed] [Google Scholar]
- 5.Perren TJ, Swart AM, Pfisterer J, Ledermann JA, Pujade-Lauraine E, Kristensen G, et al. A phase 3 trial of bevacizumab in ovarian cancer. N Engl J Med. 2011;365:2484–2496. doi: 10.1056/NEJMoa1103799. [DOI] [PubMed] [Google Scholar]
- 6.Katsumata N, Yasuda M, Takahashi F, Isonishi S, Jobo T, Aoki D, et al. Dose-dense paclitaxel once a week in combination with carboplatin every 3 weeks for advanced ovarian cancer: a phase 3, open-label, randomised controlled trial. Lancet. 2009;374:1331–1338. doi: 10.1016/S0140-6736(09)61157-0. [DOI] [PubMed] [Google Scholar]
- 7.Katsumata N, Yasuda M, Isonishi S, Takahashi F, Michimae H, Kimura E, et al. Long-term results of dose-dense paclitaxel and carboplatin versus conventional paclitaxel and carboplatin for treatment of advanced epithelial ovarian, fallopian tube, or primary peritoneal cancer (JGOG 3016): a randomised, controlled, open-label trial. Lancet Oncol. 2013;14:1020–1026. doi: 10.1016/S1470-2045(13)70363-2. [DOI] [PubMed] [Google Scholar]
- 8.Clamp AR, James EC, McNeish IA, Dean A, Kim JW, O’Donnell DM, et al. Weekly dose-dense chemotherapy in first-line epithelial ovarian, fallopian tube, or primary peritoneal carcinoma treatment (ICON8): primary progression free survival analysis results from a GCIG phase 3 randomised controlled trial. Lancet. 2019;394:2084–2095. doi: 10.1016/S0140-6736(19)32259-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cancer Genome Atlas Research Network. Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474:609–615. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kommoss S, Winterhoff B, Oberg AL, Konecny GE, Wang C, Riska SM, et al. Bevacizumab may differentially improve ovarian cancer outcome in patients with proliferative and mesenchymal molecular subtypes. Clin Cancer Res. 2017;23:3794–3801. doi: 10.1158/1078-0432.CCR-16-2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Murakami R, Matsumura N, Brown JB, Wang Z, Yamaguchi K, Abiko K, et al. Prediction of taxane and platinum sensitivity in ovarian cancer based on gene expression profiles. Gynecol Oncol. 2016;141:49–56. doi: 10.1016/j.ygyno.2016.02.027. [DOI] [PubMed] [Google Scholar]
- 12.Murakami R, Matsumura N, Mandai M, Yoshihara K, Tanabe H, Nakai H, et al. Establishment of a novel histopathological classification of high-grade serous ovarian carcinoma correlated with prognostically distinct gene expression subtypes. Am J Pathol. 2016;186:1103–1113. doi: 10.1016/j.ajpath.2015.12.029. [DOI] [PubMed] [Google Scholar]
- 13.Murakami R, Matsumura N, Michimae H, Tanabe H, Yunokawa M, Iwase H, et al. The mesenchymal transition subtype more responsive to dose dense taxane chemotherapy combined with carboplatin than to conventional taxane and carboplatin chemotherapy in high grade serous ovarian carcinoma: a survey of Japanese Gynecologic Oncology Group study (JGOG3016A1) Gynecol Oncol. 2019;153:312–319. doi: 10.1016/j.ygyno.2019.02.010. [DOI] [PubMed] [Google Scholar]
- 14.Takaya H, Nakai H, Takamatsu S, Mandai M, Matsumura N. Homologous recombination deficiency status-based classification of high-grade serous ovarian carcinoma. Sci Rep. 2020;10:2757. doi: 10.1038/s41598-020-59671-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Soslow RA, Han G, Park KJ, Garg K, Olvera N, Spriggs DR, et al. Morphologic patterns associated with BRCA1 and BRCA2 genotype in ovarian carcinoma. Mod Pathol. 2012;25:625–636. doi: 10.1038/modpathol.2011.183. [DOI] [PubMed] [Google Scholar]
- 16.Broberg P. Statistical methods for ranking differentially expressed genes. Genome Biol. 2003;4:R41. doi: 10.1186/gb-2003-4-6-r41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pomaznoy M, Ha B, Peters B. GOnet: a tool for interactive Gene Ontology analysis. BMC Bioinformatics. 2018;19:470. doi: 10.1186/s12859-018-2533-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marien KM, Croons V, Waumans Y, Sluydts E, De Schepper S, Andries L, et al. Development and validation of a histological method to measure microvessel density in whole-slide images of cancer tissue. PLoS One. 2016;11:e0161496. doi: 10.1371/journal.pone.0161496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vang R, Levine DA, Soslow RA, Zaloudek C, Shih IM, Kurman RJ. Molecular alterations of tp53 are a defining feature of ovarian high-grade serous carcinoma: a rereview of cases lacking TP53 mutations in The Cancer Genome Atlas ovarian study. Int J Gynecol Pathol. 2016;35:48–55. doi: 10.1097/PGP.0000000000000207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Salgado R, Denkert C, Demaria S, Sirtaine N, Klauschen F, Pruneri G, et al. The evaluation of tumor-infiltrating lymphocytes (TILs) in breast cancer: recommendations by an International TILs Working Group 2014. Ann Oncol. 2015;26:259–271. doi: 10.1093/annonc/mdu450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kos Z, Roblin E, Kim RS, Michiels S, Gallas BD, Chen W, et al. Pitfalls in assessing stromal tumor infiltrating lymphocytes (sTILs) in breast cancer. NPJ Breast Cancer. 2020;6:17. doi: 10.1038/s41523-020-0156-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liberzon A, Birger C, Thorvaldsdóttir H, Ghandi M, Mesirov JP, Tamayo P. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 2015;1:417–425. doi: 10.1016/j.cels.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Anadon CM, Yu X, Hänggi K, Biswas S, Chaurio RA, Martin A, et al. Ovarian cancer immunogenicity is governed by a narrow subset of progenitor tissue-resident memory T cells. Cancer Cell. 2022;40:545–557.e13. doi: 10.1016/j.ccell.2022.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pantanowitz L, Sinard JH, Henricks WH, Fatheree LA, Carter AB, Contis L, et al. Validating whole slide imaging for diagnostic purposes in pathology: guideline from the College of American Pathologists Pathology and Laboratory Quality Center. Arch Pathol Lab Med. 2013;137:1710–1722. doi: 10.5858/arpa.2013-0093-CP. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Evans AJ, Brown RW, Bui MM, Chlipala EA, Lacchetti C, Milner DA, et al. Validating whole slide imaging systems for diagnostic purposes in pathology. Arch Pathol Lab Med. 2022;146:440–450. doi: 10.5858/arpa.2020-0723-CP. [DOI] [PubMed] [Google Scholar]
- 26.Hodgson A, Park KJ, Djordjevic B, Howitt BE, Nucci MR, Oliva E, et al. International endocervical adenocarcinoma criteria and classification: validation and interobserver reproducibility. Am J Surg Pathol. 2019;43:75–83. doi: 10.1097/PAS.0000000000001095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Takamatsu S, Brown JB, Yamaguchi K, Hamanishi J, Yamanoi K, Takaya H, et al. Utility of homologous recombination deficiency biomarkers across cancer types. JCO Precis Oncol. 2022;6:e2200085. doi: 10.1200/PO.22.00085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Howitt BE, Hanamornroongruang S, Lin DI, Conner JE, Schulte S, Horowitz N, et al. Evidence for a dualistic model of high-grade serous carcinoma: BRCA mutation status, histology, and tubal intraepithelial carcinoma. Am J Surg Pathol. 2015;39:287–293. doi: 10.1097/PAS.0000000000000369. [DOI] [PubMed] [Google Scholar]
- 29.Brauer MJ, Zhuang G, Schmidt M, Yao J, Wu X, Kaminker JS, et al. Identification and analysis of in vivo VEGF downstream markers link VEGF pathway activity with efficacy of anti-VEGF therapies. Clin Cancer Res. 2013;19:3681–3692. doi: 10.1158/1078-0432.CCR-12-3635. [DOI] [PubMed] [Google Scholar]
- 30.Yin X, Wang X, Shen B, Jing Y, Li Q, Cai MC, et al. A VEGF-dependent gene signature enriched in mesenchymal ovarian cancer predicts patient prognosis. Sci Rep. 2016;6:31079. doi: 10.1038/srep31079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bais C, Mueller B, Brady MF, Mannel RS, Burger RA, Wei W, et al. Tumor microvessel density as a potential predictive marker for bevacizumab benefit: GOG-0218 biomarker analyses. J Natl Cancer Inst. 2017;109:djx066. doi: 10.1093/jnci/djx066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Riester M, Wei W, Waldron L, Culhane AC, Trippa L, Oliva E, et al. Risk prediction for late-stage ovarian cancer by meta-analysis of 1525 patient samples. J Natl Cancer Inst. 2014;106:dju048. doi: 10.1093/jnci/dju048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Verhaak RG, Tamayo P, Yang JY, Hubbard D, Zhang H, Creighton CJ, et al. Prognostically relevant gene signatures of high-grade serous ovarian carcinoma. J Clin Invest. 2013;123:517–525. doi: 10.1172/JCI65833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dornieden T, Sattler A, Pascual-Reguant A, Ruhm AH, Thiel LG, Bergmann YS, et al. Signatures and specificity of tissue-resident lymphocytes identified in human renal peritumor and tumor tissue. J Am Soc Nephrol. 2021;32:2223–2241. doi: 10.1681/ASN.2020101528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dhodapkar MV, Dhodapkar KM. Tissue-resident memory-like T cells in tumor immunity: clinical implications. Semin Immunol. 2020;49:101415. doi: 10.1016/j.smim.2020.101415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Monk BJ, Colombo N, Oza AM, Fujiwara K, Birrer MJ, Randall L, et al. Chemotherapy with or without avelumab followed by avelumab maintenance versus chemotherapy alone in patients with previously untreated epithelial ovarian cancer (JAVELIN Ovarian 100): an open-label, randomised, phase 3 trial. Lancet Oncol. 2021;22:1275–1289. doi: 10.1016/S1470-2045(21)00342-9. [DOI] [PubMed] [Google Scholar]
- 37.Moore KN, Bookman M, Sehouli J, Miller A, Anderson C, Scambia G, et al. Atezolizumab, bevacizumab, and chemotherapy for newly diagnosed stage III or IV ovarian cancer: placebo-controlled randomized phase III trial (IMagyn050/GOG 3015/ENGOT-OV39) J Clin Oncol. 2021;39:1842–1855. doi: 10.1200/JCO.21.00306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Noorbakhsh J, Farahmand S, Foroughi Pour A, Namburi S, Caruana D, Rimm D, et al. Deep learning-based cross-classifications reveal conserved spatial behaviors within tumor histological images. Nat Commun. 2020;11:6367. doi: 10.1038/s41467-020-20030-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nagpal K, Foote D, Tan F, Liu Y, Chen PC, Steiner DF, et al. Development and validation of a deep learning algorithm for Gleason grading of prostate cancer from biopsy specimens. JAMA Oncol. 2020;6:1372–1380. doi: 10.1001/jamaoncol.2020.2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Farahani H, Boschman J, Farnell D, Darbandsari A, Zhang A, Ahmadvand P, et al. Deep learning-based histotype diagnosis of ovarian carcinoma whole-slide pathology images. Mod Pathol. 2022;35:1983–1990. doi: 10.1038/s41379-022-01146-z. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
New algorithm of histopathological subtyping of high-grade serous ovarian cancer
QuPath analysis of CD31 immunohistochemistry
Initial inter-observer concordance rates for The Cancer Genome Atlas samples
The Cancer Genome Atlas cases
Highly expressed genes in the mesenchymal transition type
Enriched gene ontology terms in the highly expressed genes in the mesenchymal transition type
Clinical characteristics of the Kindai/Kyoto cohort
Kindai/Kyoto cohort
Evaluation of the percentage of lymphocytes infiltrating the tumor stroma.
Undetermined cases in TCGA samples. (TCGA 23-1118) (TCGA 23-1121) Mostly stromal tissue with no epithelial structure. (TCGA VG-A8LO) This was frozen sections and unsuitable for detailed histological analysis. (TCGA 23-1110) Complex epithelial arrangement with relatively little intervening stroma, not easily identified as cribriform, papillary, or solid. (TCGA 23-1120) Similar to many other types of carcinomas, high grade serous carcinomas can have a more ragged architecture in the tumor invasive front and/or around intratumoral fibrosis. In PG/SP pattern-predominant cases, this can result in PG/SP pattern gradually transitioning to mesenchymal transition pattern in the invasive front or around fibrosis.
Histopathological subtype and HRD status. (A) Association of BRCA alterations and HRD scores with the histopathological subtypes. There was no association between them. (B) Association between histopathological subtypes and SET cases (SET feature ≥50%). (C) Association between SET and BRCA alterations. (D) Association between SET and HRD scores.