Abstract
Furin belongs to the pro‐protein convertases (PCs) family and its aberrant expression has been documented in various types of cancers; however, its role in thyroid cancer remains unclear. We investigated the expression of furin in a large cohort of Middle Eastern papillary thyroid carcinoma (PTC) patient samples and explored its functional role and mechanism in PTC cell lines in vitro and in vivo. Furin overexpression was observed in 44.6% of all PTC cases and was significantly associated with aggressive clinicopathological parameters and poor outcomes. We show that the knockdown of FURIN suppresses tumor growth, proliferation, migration, invasion, spheroid growth, and progression of epithelial‐to‐mesenchymal transition (EMT) in B‐Raf proto‐oncogene, serine/threonine kinase (BRAF) mutant cells, whereas its overexpression in BRAF wild‐type PTC cell lines reversed the effect. FURIN knockdown in the BRAF mutant cell line led to reduced tumor growth and increased apoptosis. Mechanistically, FURIN knockdown led to MEK/ERK pathway suppression in BRAF mutant cells, although inhibition of MEK did not affect furin expression, which suggests that furin acts through the MEK/ERK pathway. Furthermore, our study revealed the synergistic antitumor effect of furin depletion and anti‐MEK inhibitor treatment. Overall, these results indicate that furin is an important prognostic marker in Middle Eastern PTC and that it plays a crucial role in BRAF‐associated MAP/ERK pathway activation and tumorigenesis. Furin inhibition could be a potential therapeutic target for aggressive PTC.
Keywords: apoptosis, BRAF, EMT, Furin, PTC
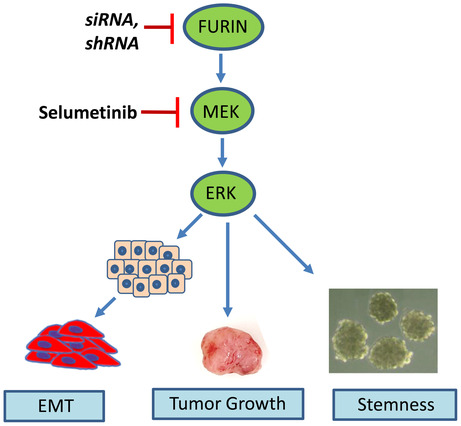
The authors show that furin is overexpressed in papillary thyroid carcinoma (PTC) and is significantly associated with aggressive clinicopathological parameters and poor outcomes. Functional studies show that furin promotes proliferation, tumor growth, migration, invasion, stemness and progression of EMT transition in PTC cell lines via the MEK/ERK pathway. Furin inhibition is a potential therapeutic target for a subset of PTC.
Abbreviations
- ATCC
American‐type culture collection
- DSMZ
Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH
- EMT
epithelial to mesenchymal transition
- ERK1/2
extracellular signal‐regulated protein kinase
- FBS
fetal bovine serum
- IHC
immunohistochemistry
- PARP
Poly (ADP‐ribose) polymerase
- PCR
polymerase chain reaction
- PTC
papillary thyroid cancer
- TMA
tissue microarray
1. Introduction
Papillary thyroid cancer is the most common endocrine malignancy. The incidence of PTC is on the rise worldwide [1, 2, 3]. In Saudi Arabia, PTC is the second commonest cancer affecting women after breast cancer [4]. Despite the indolent nature of PTC, there are considerable percentages (10–15%) of patients who presented with aggressive disease and develop resistance to standard therapeutic approaches [5, 6, 7, 8].
Constitutive activation of the mitogen‐activated protein kinase (MAPK) signaling pathway is a major event in the carcinogenesis of PTC [9]. Mutations in BRAF and KRAS genes are found in ~ 60% of PTC tumors [10]. Several studies have linked PTC's aggressive phenotype and poor clinical outcome to BRAF mutations [11, 12]. This highlights the importance of identifying a new therapeutic target that can interfere with this signaling pathway and inhibit tumor progression.
Furin, pro‐protein convertase subtilisin/kexin family member 3, is a calcium‐dependent protease and it is present in all mammalian cells [13]. Furin with the other six members (PC1/3, PC2, PACE 4, PC4, PC5/6, and PC7/8) constitute the pro‐protein convertases family that is responsible for cleavage and activation of various protein precursors in cells [14, 15]. The maturation of several of these precursors promotes many cancer‐related processes such as proliferation, migration and invasion, and metastasis [16, 17, 18, 19], which strongly indicate that furin could play an important role in tumorigenesis.
Numerous studies have reported the overexpression of furin in several cancers including sarcoma [14], hepatocellular carcinoma [20], breast cancer [21], head and neck [22], ovarian cancer [23], lung cancer [24], skin tumor [25], and colorectal cancer [26]. Interestingly, several studies have suggested that inhibition of furin can suppress the tumorigenic properties and metastatic potential of many cancers [18, 27, 28, 29, 30, 31]. Furthermore, previous studies have indicated that furin inhibition or silencing inhibits the processing of insulin growth factor‐1 receptor (IGF1R), matrix metalloproteinase 2 (MMP‐2) and leads to downregulation of PI3K/AKT and ERK–MAPK pathways [17, 26, 31, 32].
A recent study has shown that furin plays an important role in BRAF‐associated pathway activation in colorectal cancer [26]. This intrigued us to conduct this study to investigate for the first time whether the expression of furin in a large cohort of Middle Eastern PTC and if it plays any role in the activation of the BRAF‐MAPK signaling pathway in thyroid cancer.
2. Materials and methods
2.1. Patient selection
A total of 1479 patient samples diagnosed with primary PTC between 1988 and 2018 at King Faisal Specialist Hospital & Research Centre (Riyadh, Saudi Arabia) and Prince Sultan Military Medical City (Riyadh, Saudi Arabia) were collected from the Department of Pathology. Detailed clinicopathological data, including follow‐up data, were noted from case records and summarized in Table 1. In addition, distant metastatic tissue was available for 15 of these cases. Overall survival was defined as the length of time after primary treatment for cancer ends that the patient survives. The study was conducted according to the guidelines of the Declaration of Helsinki, and approved by the Institutional Review Board (IRB) and Research Ethics Committee (REC) of the King Faisal Specialist Hospital & Research Centre (KFSH&RC). Waiver of consent was obtained for all archived paraffin tissue blocks, including normal tissue blocks used as control, from IRB of the KFSH&RC under Research Advisory Council (RAC) on 07 February 2022 (RAC#2220002).
Table 1.
Patient characteristics for adult PTC (n = 1430).
| Total | ||
|---|---|---|
| No. | % | |
| Total | ||
| Age (in years) | ||
| Median (range) | ||
| < 55 | 1208 | 81.7 |
| ≥ 55 | 271 | 18.3 |
| Sex | ||
| Female | 1124 | 76.0 |
| Male | 355 | 24.0 |
| Histologic subtype | ||
| Classical variant | 976 | 66.0 |
| Follicular variant | 261 | 17.6 |
| Tall cell variant | 133 | 9.0 |
| Other variants | 109 | 7.4 |
| Extrathyroidal extension | ||
| Present | 629 | 42.5 |
| Absent | 850 | 57.5 |
| Tumor focality | ||
| Unifocal | 741 | 50.1 |
| Multifocal | 738 | 49.9 |
| Tumor laterality | ||
| Unilateral | 999 | 67.5 |
| Bilateral | 480 | 32.5 |
| Lymphovascular invasion | ||
| Present | 316 | 21.4 |
| Absent | 1163 | 78.6 |
| pT | ||
| T1 | 575 | 38.9 |
| T2 | 463 | 31.3 |
| T3 | 285 | 19.3 |
| T4 | 110 | 7.4 |
| Unknown | 46 | 3.1 |
| pN | ||
| N0 | 651 | 44.0 |
| N1 | 732 | 49.5 |
| Nx | 96 | 6.5 |
| pM | ||
| M0 | 1402 | 94.8 |
| M1 | 77 | 5.2 |
| TNM stage | ||
| I | 1230 | 83.2 |
| II | 157 | 10.6 |
| III | 20 | 1.4 |
| IV | 50 | 3.4 |
| Unknown | 22 | 1.4 |
| BRAF mutation | ||
| Present | 800 | 54.1 |
| Absent | 640 | 43.3 |
| Unknown | 39 | 2.6 |
| NRAS mutation | ||
| Present | 101 | 6.8 |
| Absent | 1329 | 89.8 |
| Unknown | 49 | 3.3 |
| HRAS mutation | ||
| Present | 33 | 2.2 |
| Absent | 1409 | 95.3 |
| Unknown | 37 | 2.5 |
| KRAS mutation | ||
| Present | 8 | 0.5 |
| Absent | 1426 | 96.5 |
| Unknown | 45 | 3.0 |
2.2. DNA isolation
DNA was isolated from formalin‐fixed, paraffin‐embedded (FFPE) tumor tissues using a Gentra DNA isolation kit (Gentra, Minneapolis, MN, USA), following the manufacturer's recommendations as described previously [33].
2.3. PCR and sanger sequencing
primer 3 software was used to design the primers for entire coding and splicing regions of MAPK genes exons 1 and 2 in HRAS, KRAS, and NRAS and exon 15 in BRAF (primers available upon request). PCR was performed in a total volume of 25 μL with 20 ng of genomic DNA, 2.5 μL 10 × Taq buffer, 2.3 mm dNTPs, 1‐unit Taq polymerase, and 0.2 μm each primer and de‐ionized water. The efficiency and quality of the amplified PCR products were confirmed by loading them on a 2% agarose gel.
The PCR products were subsequently subjected to direct sequencing with BigDye terminator V 3.1 cycle sequencing reagents and analyzed on an ABI 3730XL DNA analyzer (Applied Biosystems, Foster City, CA, USA). Reference sequences were downloaded from NCBI GenBank. Sequencing traces were analyzed with the mutation surveyor v4.04 (Soft Genetics, LLC, State College, PA, USA) as described earlier [33].
2.4. Tissue microarray (TMA) construction and immunohistochemistry
All samples were analyzed in a tissue microarray (TMA) format. Tissue microarray construction was performed from formalin‐fixed, paraffin‐embedded PTC specimens as described previously [34, 35]. Briefly, Tissue cylinders with a diameter of 0.6 mm were punched from representative tumor regions of each donor tissue block and brought into the recipient paraffin block using a modified semiautomatic robotic precision instrument (Beecher Instruments, Woodland, WI, USA). Two cores of papillary thyroid carcinoma were arrayed from each case.
Standard protocol was followed for IHC staining. For antigen retrieval, Dako (Dako Denmark A/S, Glostrup, Denmark) Target Retrieval Solution pH 9.0 (Catalog number S2367) was used, and the slides were placed in a Pascal pressure cooker for 8 min at 120 °C. The slides were incubated with primary antibody against furin (ab183495, Abcam, Cambridge, UK) with a dilution of 1 : 100 (pH 9.0). The Dako Envision Plus System kit was used as the secondary detection system with 3, 3′‐diaminobenzidine as the chromogen. All slides were counterstained with hematoxylin, dehydrated, cleared, and mounted. Negative controls included the omission of the primary antibody. Normal tissues of different organs were also included in the TMA to serve as a control. Only fresh‐cut slides were stained simultaneously to minimize the influence of slide aging and maximize the reproducibility of the experiment.
H score was used to quantify furin protein expression based on intensity and proportion of staining, as described previously by us [36]. The intensity of staining was scored from 0 to 3 (0: absent, 1+: weak, 2+: moderate, 3+: strong), and the proportion of tumor cells staining for that particular intensity was recorded as 5% increments from a range of 0–100. A final H score was assigned using the following formula: H score = [1 × (% cells 1+) + 2 × (% cells 2+) + 3 × (% cells 3+)]. This H score ranges from 0 to 300. Two scores per tumor were analyzed to minimize the number of missing/un‐interpretable spots. However, for the final score, we used the higher of the two scores. X‐tile plots were constructed for the assessment of biomarkers and optimization of cutoff points based on outcome as has been described earlier [37]. Based on X‐tile plots, PTC cases were classified into two subgroups: those with an H score ≤ 75 were defined as low expression of furin, and those with an H score > 75 were defined as over‐expression.
2.5. Cell culture
The PTC cell line, BCPAP (RRID:CVCL_0153), was obtained from DSMZ, and TPC‐1 (RRID:CVCL_6298) was kindly provided by Dr Bryan McIver (Department of Endocrinology, Mayo Clinic, Rochester, Minnesota). The K1 (RRID:CVCL_2537) cell line was purchased from American Type Culture Collection (ATCC). Cell lines were cultured in RPMI 1640 media supplemented with 10% fetal bovine serum (FBS), 100 Units·mL−1 penicillin/streptomycin, and 100 Units·mL−1 Glutamine. These cell lines were tested negative for mycoplasma, and cell line authentication was performed in‐house via short tandem repeat DNA fingerprinting using the AmpFISTR identifier PCR Amplification Kit (Thermo Fisher, Waltham, MA, USA) and the results were in concordance with published data [38, 39]. All experiments were performed using 5% FBS in RPMI 1640 media.
2.6. Reagents and antibodies
Antibodies against Furin (ab183495), N‐cadherin (ab98952), and Twist (ab175430) were purchased from Abcam Inc (Cambridge, MA, USA). Antibodies against pMEK1/2 (9121), MEK1/2 (4694), pERK1/2 (4370), ERK1/2 (4695), PARP (9542), β‐Actin (3700), E‐cadherin (3195), Nanog (4903), CD44 (3570) and CD133 (64326) were purchased from Cell Signaling Technology (Danvers, MA, USA). Zeb1 antibody (NBP1‐05987) was purchased from Novus Biologicals (Minneapolis, MN, USA). Caspase‐3 (sc‐56053) antibody was purchased from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA, USA). Selumetinib (AZD6244) was purchased from Selleck chemicals (Houston, TX, USA) and Mirdametinib (PD0325901) was purchased from Cayman Chemicals (Ann Arbor, MI, USA). The zVAD‐fmk was purchased from Calbiochem (San Diego, CA, USA).
2.7. Clonogenic assay
The single‐cell suspensions of exponentially growing PTC cell lines were seeded into a six‐well plate at a low density of 500 cells per well and allowed to adhere. After attachment, the cell culture medium was refreshed and cells were allowed to grow for 8–10 days. Cell colonies were fixed with formaldehyde (4%) and stained with crystal violet (2% in 10% methanol). The number of colonies in each well was counted and photographed.
2.8. Gene silencing using siRNA
Cells were transfected with two different sequences of FURIN siRNA's SR321213A‐5'r(AUAGUUGAGCCCCAAGUCCUGAAGA)3′ and SR321213B‐5'r(AACAGUAUCUA CACGCUGUCCAUCA)3′ and scrambled negative control siRNA (SR30004) from OriGene (Rockville, MD, USA) using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) for 6 h following which the lipid and siRNA complex was removed and fresh growth medium was added. After 48 h of transfection, cells were used for immunoblotting.
2.9. Plasmid and transfection
FURIN cDNA encoding human FURIN (RC204279) and shRNA's (TR312903A: AAAGTGATTAAACGTGCAGACTATGCAAA and TR312903B: ACGGCATTGTGGTCTCCATTCTGGACGAT) targeting human FURIN were purchased from Origene. The overexpression of FURIN and knockdown of FURIN in PTC cells were performed using Lipofectamine™2000 (Invitrogen) according to the manufacturer's protocol. Briefly, PTC cells were seeded in 6‐well culture plates; when approximately 50% confluent, cells were transfected with 4 μg plasmid for 48 h. Stable overexpression clones resistant to G418 and stable knockdown clones resistant to puromycin were isolated; overexpression and knockdown of furin protein production were confirmed by immunoblotting.
2.10. Cell invasion and migration assays
Cell invasion and migration assays were performed as described previously [38]. Briefly, cells, after treatment were seeded into trans‐well, inserts either uncoated (for migration assay) or coated (for invasion assay) with growth factor‐reduced matrigel for 24 h. After incubation, cells were stained with a Diff‐Quick stain set (Fisher Scientific, Pittsburg, PA, USA), and photographed under a fluorescent microscope.
2.11. Sphere forming assay
Sphere forming assay was performed as described previously [40]. Briefly, PTC cells (500 per well) were plated on Corning 24‐well ultra‐low attachment plates (Corning Inc., Kennebunk, ME, USA) grown in serum‐free DMEM‐F12 (ATCC) supplemented with B27 (Gibco, Grand Island, NY, USA), 20 ng·mL−1 epidermal growth factor (Sigma‐Aldrich, Saint Louis, MO, USA), 0.4% bovine serum albumin (Sigma‐Aldrich, Milwaukee, WI, USA) and 4 μg·mL−1 insulin (Sigma‐Aldrich, Saint Louis, MO, USA). Fresh medium was supplemented every 2 days. The spheroids were counted and photographed on day 14. For secondary spheroid formation, the primary spheroids were dissociated into single cells, and cultured on 24‐well ultra‐low attachment plates using a spheroid culture medium for another 10 days.
2.12. Animals and xenografts study
Six‐week‐old female nude mice (NU/J) were obtained from Jackson Laboratories (Bar Harbor, ME, USA) and maintained in a sterile and pathogen‐free environment (12 h light/12 h darkness) with food and water ad libitum at least 1 week before use. All animal studies were done in accordance with institutional guidelines as described previously [41] and the study was approved by the animal use and care committee (ACUC) of King Faisal Specialist Hospital and Research Center on 07 February 2022 (Approval No: RAC#2220002).
For the xenograft study, PTC cells (5 × 106 cells per mouse) were re‐suspended in serum‐free medium with matrigel basement membrane matrix (BD Biosciences, Bedford, MA, USA) at a 1 : 1 ratio (total volume = 100 μL) and subcutaneously injected into the flanks of NU/J mice (n = 5). After tumors grew to about 100 mm3, mice were treated intraperitoneally with selumetinib (10 mg·kg−1) and vehicle (0.1% DMSO), twice a week for 30 days. The body weight and tumor volume of each mouse were monitored weekly. After 4 weeks of treatment, mice were sacrificed and individual tumors were weighed, then snap‐frozen in liquid nitrogen for storage.
2.13. Statistical analysis
Contingency table analysis and chi‐square tests were used to study the relationship between clinicopathological variables and furin expression. Survival curves were generated using the Kaplan–Meier method, with significance evaluated using the Mantel‐Cox log‐rank test. Cox proportional hazards regression model was used for multivariate analysis. The limit of significance for all analyses was defined as a P‐value of 0.05; two‐sided tests were used in all calculations. The jmp11.0 (SAS Institute, Inc., Cary, NC, USA) software package was used for data analyses.
For all functional studies, data presented are means ± SD of triplicates in an independent experiment, which was repeated at least two times with the same results. Student t‐test (two‐tailed) was performed for statistical significance with a P < 0.05 used as the cut‐off.
3. Results
3.1. Furin expression in normal thyroid, primary tumor, and distant metastatic tumor tissues
Furin was primarily expressed in the cytoplasm (Fig. 1A,B). Furin over‐expression was noted in 44.6% (659/1479) of primary PTCs compared to 5.8% (14/242) of normal thyroid tissues and the difference was statistically significant (P < 0.0001). We also analyzed the expression of furin in 15 distant metastatic tissues and found that furin was overexpressed in 73.3% (11/15) of metastatic tissues, which was significantly higher than that noted in primary PTC (P = 0.0262).
Fig. 1.
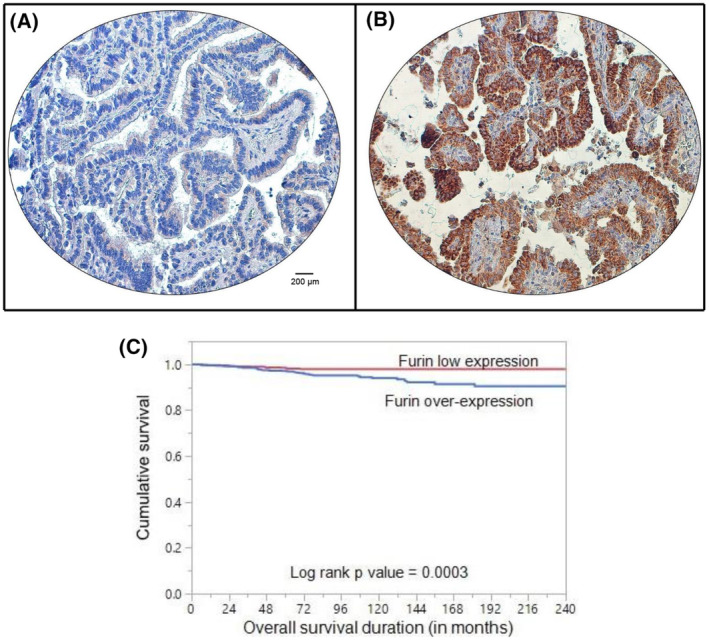
Immunohistochemical analysis of furin expression in papillary thyroid cancer (PTC) TMA. Representative examples of tumors showing (A) low expression and (B) high expression of furin. (20×/0.70 objectives on an Olympus BX 51 microscope (Olympus America Inc, Center Valley, PA, USA) with the inset showing a 40× 0.85 aperture magnified view of the same TMA spot; scale bar in A and B = 200 μm). (C) Kaplan–Meier survival analysis for the prognostic significance of furin expression in PTC. PTC patients with overexpression of furin had reduced disease‐free survival at 5 years compared to tumors showing low expression of Furin (P = 0.0003).
3.2. Association of furin expression with clinicopathological factors and outcome in papillary thyroid carcinoma
Furin overexpression was significantly associated with adverse clinicopathological parameters such as older age (P < 0.0001), tall cell variant (P < 0.0001), extrathyroidal extension (P < 0.0001), T4 tumors (P = 0.0004), advanced stage (P < 0.0001) and proliferation marker Ki‐67 (P = 0.0007). Interestingly, we also found a significant association between furin over‐expression and BRAF mutation (P < 0.0001) in our cohort (Table 2).
Table 2.
Association of clinico‐pathological characteristics with Furin expression in PTC.
| Total | Furin expression | P value | |||||
|---|---|---|---|---|---|---|---|
| High | Low | ||||||
| No. | % | No. | % | No. | % | ||
| No. of patients | 1479 | 659 | 44.6 | 820 | 55.4 | ||
| Age (in years) | |||||||
| < 55 | 1208 | 81.7 | 490 | 74.4 | 718 | 87.6 | < 0.0001 |
| ≥ 55 | 271 | 18.3 | 169 | 25.6 | 102 | 12.4 | |
| Sex | |||||||
| Female | 1124 | 76.0 | 507 | 76.9 | 617 | 75.2 | 0.4488 |
| Male | 355 | 24.0 | 152 | 23.1 | 203 | 24.8 | |
| Histology subtype | |||||||
| Classical variant | 976 | 66.0 | 422 | 64.0 | 554 | 67.6 | < 0.0001 |
| Follicular variant | 261 | 17.6 | 81 | 12.3 | 180 | 21.9 | |
| Tall cell variant | 133 | 9.0 | 101 | 15.3 | 32 | 3.9 | |
| Other variants | 109 | 7.4 | 55 | 8.4 | 54 | 6.6 | |
| Extrathyroidal extension | |||||||
| Present | 629 | 42.5 | 321 | 48.7 | 308 | 37.6 | < 0.0001 |
| Absent | 850 | 57.5 | 338 | 51.3 | 512 | 62.4 | |
| Tumor focality | |||||||
| Unifocal | 741 | 50.1 | 343 | 52.0 | 398 | 48.5 | 0.1794 |
| Multifocal | 738 | 49.9 | 316 | 48.0 | 422 | 51.5 | |
| Tumor laterality | |||||||
| Unilateral | 999 | 67.5 | 456 | 69.2 | 543 | 66.2 | 0.2239 |
| Bilateral | 480 | 32.5 | 203 | 30.8 | 277 | 33.8 | |
| Lymphovascular invasion | |||||||
| Present | 316 | 21.4 | 147 | 22.3 | 169 | 20.6 | 0.4292 |
| Absent | 1163 | 78.6 | 512 | 77.7 | 651 | 79.4 | |
| pT | |||||||
| pT1 | 575 | 40.1 | 246 | 38.5 | 329 | 41.4 | 0.0004 |
| pT2 | 463 | 32.3 | 195 | 30.5 | 268 | 33.8 | |
| pT3 | 285 | 19.9 | 128 | 20.0 | 157 | 19.8 | |
| pT4 | 110 | 7.7 | 70 | 11.0 | 40 | 5.0 | |
| pN | |||||||
| pN0 | 651 | 47.1 | 272 | 44.2 | 379 | 49.4 | 0.0514 |
| pN1 | 732 | 52.9 | 344 | 55.8 | 388 | 50.6 | |
| pM | |||||||
| pM0 | 1402 | 94.8 | 629 | 95.4 | 773 | 94.3 | 0.3078 |
| pM1 | 77 | 5.2 | 30 | 4.6 | 47 | 5.7 | |
| Stage | |||||||
| I | 1230 | 84.4 | 512 | 79.3 | 718 | 88.5 | < 0.0001 |
| II | 157 | 10.8 | 85 | 13.2 | 72 | 8.9 | |
| III | 20 | 1.4 | 16 | 2.5 | 4 | 0.5 | |
| IV | 50 | 3.4 | 33 | 5.1 | 17 | 2.1 | |
| Ki‐67 expression | |||||||
| Present | 251 | 17.4 | 136 | 21.1 | 115 | 14.3 | 0.0007 |
| Absent | 1195 | 82.6 | 508 | 78.9 | 687 | 85.7 | |
| BRAF mutation | |||||||
| Present | 800 | 55.6 | 431 | 67.2 | 369 | 46.2 | < 0.0001 |
| Absent | 640 | 44.4 | 210 | 32.8 | 430 | 53.8 | |
| NRAS mutation | |||||||
| Present | 101 | 7.1 | 37 | 5.8 | 64 | 8.1 | 0.0862 |
| Absent | 1329 | 92.9 | 603 | 94.2 | 726 | 91.9 | |
| HRAS mutation | |||||||
| Present | 33 | 2.3 | 15 | 2.3 | 18 | 2.3 | 0.9132 |
| Absent | 1409 | 97.7 | 627 | 97.7 | 782 | 97.7 | |
| KRAS mutation | |||||||
| Present | 8 | 0.6 | 3 | 0.5 | 5 | 0.6 | 0.6849 |
| Absent | 1426 | 99.4 | 636 | 99.5 | 790 | 99.4 | |
We also found a significant association between furin expression and overall survival (P = 0.0003; Fig. 1C), which remained significant even after adjusting for other clinicopathological parameters on multivariate Cox proportional hazards model (Hazard ratio = 2.43; 95% confidence interval = 1.09–6.01; P‐value = 0.0298; Table 3), thus establishing furin as an independent predictor of overall survival in PTC for this cohort.
Table 3.
Multivariate analysis of overall survival using Cox Proportional Hazard Model.
| Clinico‐pathological variables | Overall survival | ||
|---|---|---|---|
| Hazard ratio | 95% confidence interval | P‐value | |
|
Age Above ≥ 55 years (vs. < 55 years) |
10.41 |
4.19–31.55 |
< 0.0001 |
|
Sex Male (vs. Female) |
1.38 |
0.63–2.86 |
0.4058 |
|
Histology Tall cell variant (vs. other variants) |
2.71 |
1.16–5.93 |
0.0220 |
|
Extrathyroidal extension Present (vs. Absent) |
0.86 |
0.31–2.56 |
0.7815 |
|
pT T3 & T4 (vs. T1 & T2) |
1.68 |
0.57–5.08 |
0.3471 |
|
Lymph node metastasis N1 (vs. N0) |
1.62 |
0.76–3.69 |
0.2183 |
|
Distant metastasis Present (vs. absent) |
3.76 |
1.08–12.30 |
0.0376 |
|
Stage III‐IV (vs. I–II) |
0.92 |
0.26–2.83 |
0.8939 |
|
BRAF mutation Present (vs. Absent) |
0.91 |
0.41–2.19 |
0.8347 |
|
Furin expression High (vs. Low) |
2.43 |
1.09–6.01 |
0.0298 |
3.3. Furin promotes PTC cell growth in vitro
Our clinical data showed furin overexpression in PTC cases and was significantly associated with BRAF mutation. To test this association and function of furin in vitro, we first analyzed the basal expression of furin in three PTC cell lines by immunoblotting. As shown in Fig. 2A, we found high expression of furin along with pMEK1/2 and pERK1/2 in BRAF mutated PTC cell lines (BCPAP and K1) compared to BRAF wild‐type PTC cell line (TPC‐1). Next, we overexpressed furin in the TPC‐1 cell line (Fig. 2B) and evaluated the cell growth by clonogenic assay. As shown in Fig. 2C,D, forced expression of furin significantly increased cell growth. Furthermore, ectopic expression of furin significantly increased the cell proliferation after 48 h of seeding as measured by MTT assay (Fig. 2E) and comparative viable cell counting analysis using Bio‐Rad TC20 automated cell counter (data not shown).
Fig. 2.
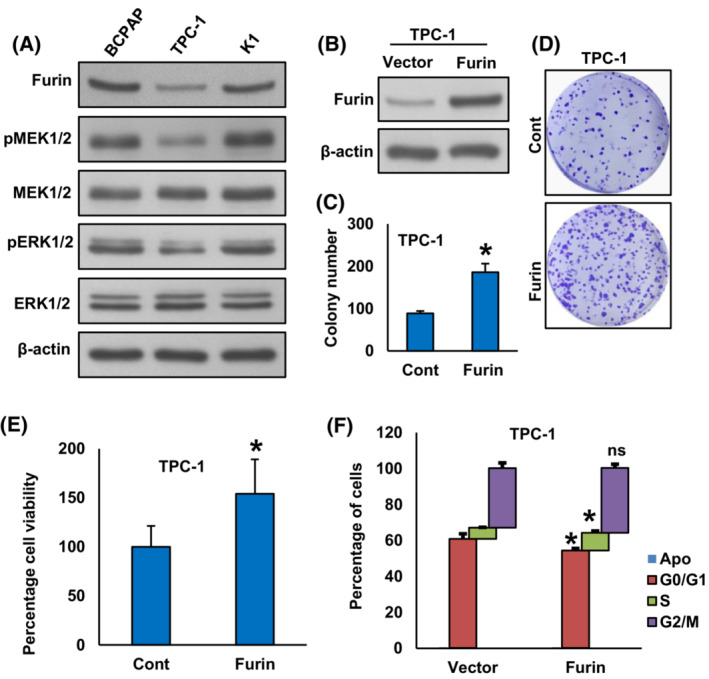
Furin promotes PTC cell growth in vitro. (A) Basal expression of furin in PTC cell lines. Proteins were isolated from three PTC cell lines and immunoblotted with antibodies against Furin, pMEK1/2, MEK1/2, pERK1/2, ERK1/2 and β‐Actin (n = 3). (B) Forced expression of furin in low‐expressing cells. TPC‐1 cells were transfected with either an empty vector or FURIN cDNA and overexpression was confirmed by immunoblotting (n = 3). (C, D) Forced expression of furin increases clonogenicity. TPC‐1 cells were transfected with either an empty vector or FURIN cDNA for 48 h. Selected clones were seeded at a density of 500 cells per well in a 6‐well plate, and grew for an additional 10 days, then stained with crystal violet, and colonies were counted. Data were presented as mean ± SD (n = 3). (E) Furin promotes TPC‐1 cell proliferation. TPC‐1 cells carrying empty vector and FURIN cDNA were grown for 48 h and cell proliferation was measured by MTT assay. Data were presented as mean ± SD (n = 6). (F) TPC‐1 cells were transfected with either an empty vector or FURIN cDNA for 48 h. Cells were stained with propidium iodide (PI) followed by flow cytometry analysis. Data were presented as mean ± SD (n = 3). Statistical analyses were performed using two‐tailed Student's t‐tests. *P < 0.05.
In addition, we investigated whether the overexpression of furin affects the distribution of cells in the different cell cycle phases (Fig. 2F). We observed a significant increase in the percentage of cells in the S phase, the most proliferative phase, from 6.1 ± 0.2% in empty vector‐transfected TPC‐1 cells to 9.9 ± 0.9% in furin overexpressing TPC‐1 cells. At the same time, cells that overexpress furin showed significantly fewer cells in the G0/G1 phase of the cell cycle (Fig. 2F). These results indicate that the upregulation of furin increases the cell proliferation ability of PTC cells.
On the contrary, the knockdown of FURIN in BCPAP and K1 significantly decreased cell growth (Fig. 3A,B) and induced apoptosis (Fig. 3C). We also investigated the expression of apoptotic protein markers after FURIN knockdown on these cells by immunoblotting. As shown in Fig. 3D, the knockdown of FURIN also induced the cleavage of PARP and caspase‐3 in these cells. To confirm whether the decrease in cell growth after furin depletion was due to apoptosis, PTC cells were pre‐treated with a universal inhibitor of apoptosis, zVAD‐fmk (80 μm) for 3 h followed by transfection with FURIN siRNA for 48 h. As shown in Fig. S1A, there was a significant inhibition of apoptosis in zVAD‐fmk pre‐treated cells as compared to FURIN siRNA transfected cells alone. Furthermore, zVAD‐fmk pre‐treatment appreciably improved the cell growth in FURIN knockdown cells (Fig. S1B).
Fig. 3.
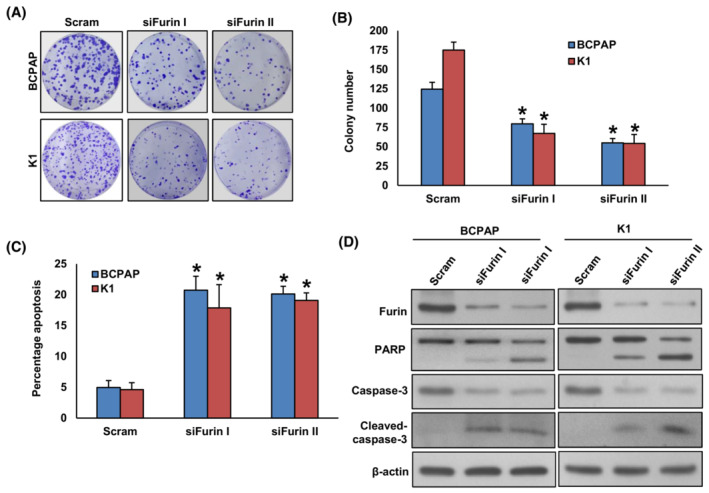
Knockdown of FURIN decrease PTC cell growth and induce apoptosis. (A, B) Silencing of FURIN attenuates clonogenicity. PTC cells were transfected with scrambled siRNA and two different FURIN siRNA sequences (25 nm). After 48 h, cells were seeded at a density of 500 cells per well in a 6‐well plate and grown for an additional 10 days, then stained with crystal violet, and colonies were counted. Data were presented as mean ± SD (n = 3). (C) Depletion of furin induces apoptosis in PTC cell lines. After 48 h of transfection, cells were stained with fluorescein‐conjugated annexin‐V and propidium iodide (PI) and analyzed by flow cytometry. Data were presented as mean ± SD (n = 3). (D) Depletion of furin induces the cleavage of caspase‐3 and PARP. After 48 h of transfection, cells were lysed and proteins were immuno‐blotted with antibodies against Furin, PARP, casapse‐3, cleaved casapse‐3, and β‐Actin as indicated (n = 3). Statistical analyses were performed using two‐tailed Student's t‐tests. *P < 0.05.
To identify whether inhibition of cell growth after FURIN knockdown was associated with cell cycle arrest, we performed cell cycle analysis on PTC cell lines following transfection with two separate FURIN siRNA sequences for 48 h (Fig. S1C). There was a significant increase in the sub‐G1 (Apo) fraction of cells in both FURIN siRNA transfected PTC cell lines compared to scramble siRNA transfected controls suggesting that these cells were dying of apoptosis (Fig. S1C).
3.4. Furin activates MEK/ERK pathway
In an attempt to investigate the mechanism of furin‐induced cell growth, we overexpressed furin in the TPC‐1 cell line and analyzed the expression of MEK/ERK pathway proteins. As shown in Fig. 4A, forced expression of furin markedly increased the phosphorylation of MEK1/2 and ERK1/2 in the TPC‐1 cell line. However, stable knockdown of FURIN noticeably decreased the phosphorylation of MEK1/2 and ERK1/2 in BCPAP and K1 cell lines (Fig. 4B). To test whether MEK inhibition has any effect on furin expression, BRAF mutated cell lines were treated with pharmacological inhibitors for MEK, mirdametinib (10 nM), and selumetinib (20 nM) for 48 h and analyzed the expression of furin, pMEK1/2, and pERK1/2 by immunoblotting. As shown in Fig. 4C, inhibition of MEK by both mirdametinib and selumetinib prominently downregulated the expression of pMEK1/2 and pERK1/2 but did not affect furin expression. We also show that selumetinib treatment downregulated the phosphorylation of MEK1/2 and ERK1/2 in both furin expressing (Fig. 4D) and knockdown (Fig. 4E) cells with no further change in the furin expression. All these data indicate that furin acts through MEK/ERK signaling cascade.
Fig. 4.
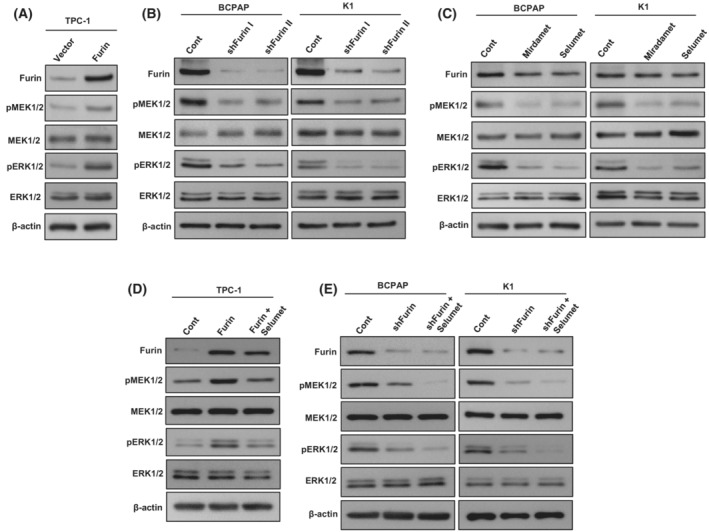
Furin activates MEK/ERK pathway. (A) Forced expression of furin stimulates MEK/ERK pathway. TPC‐1 cells were transfected with either an empty vector or FURIN cDNA for 48 h. Cells were lysed and proteins were immuno‐blotted with antibodies against Furin, pMEK1/2, MEK1/2, pERK1/2, ERK1/2, and β‐Actin as indicated (n = 3). (B) Depletion of furin inhibits MEK/ERK activation. PTC cells were transfected with a control shRNA vector or two different FURIN shRNA constructs. After transfection, cells were lysed and proteins were immuno‐blotted with antibodies against Furin, pMEK1/2, MEK1/2, pERK1/2, ERK1/2, and β‐Actin as indicated (n = 3). (C) The effect of MEK inhibitors on furin expression. PTC cell lines were treated with a pharmacological inhibitor for MEK, mirdametinib (10 nM), and selumetinib (20 nM) for 48 h and analyzed the expression of Furin, pMEK1/2, MEK1/2, pERK1/2, ERK1/2 by immunoblotting (n = 3). (D) Selumetinib downregulates MEK/ERK signaling cascade in furin‐expressing cells. TPC‐1 cells carrying FURIN cDNA were treated with selumetinib (20 nM) for 48 h. Cells were lysed and proteins were immuno‐blotted with antibodies against Furin, pMEK1/2, MEK1/2, pERK1/2, ERK1/2, and β‐Actin as indicated (n = 3). (E) The co‐inhibition of MEK and furin further downregulate MEK/ERK signaling cascade in PTC cells. PTC cells were transfected with FURIN shRNA, and treated with selumetinib (20 nM) for 48 h. Cells were lysed and proteins were immuno‐blotted with antibodies against Furin, pMEK1/2, MEK1/2, pERK1/2, ERK1/2, and β‐Actin as indicated (n = 3).
To test the role of the MEK/ERK signaling pathway in furin‐mediated cell growth, we treated selumetinib in both furin‐expressing and knockdown cells and performed clonogenic assay and annexin V/PI dual staining for apoptosis. As expected, inhibition of MEK by selumetinib significantly decreased cell growth (Fig. 5A,B) and induced apoptosis (Fig. 5C,D) in both furin expressing and knockdown cells. These data demonstrate the important role of the MEK/ERK signaling pathway in furin‐mediated cell growth and apoptosis.
Fig. 5.
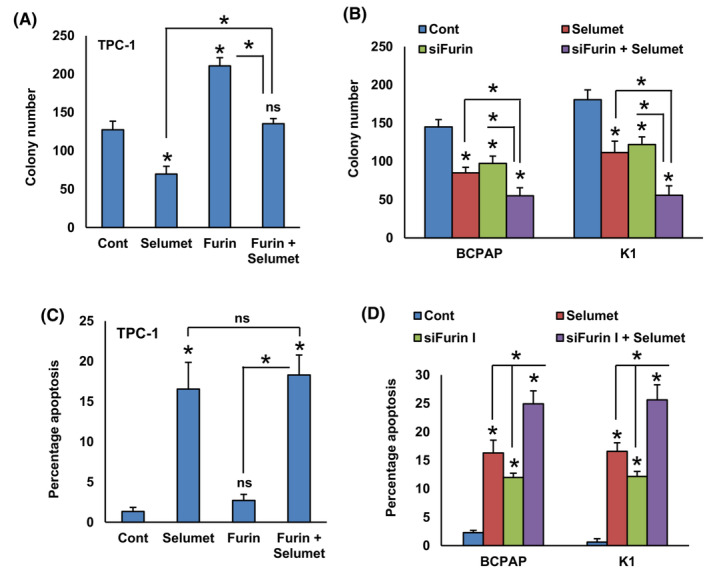
The MEK/ERK signaling pathway plays an important role in furin‐mediated cell growth and apoptosis. (A) TPC‐1 cells carrying empty vector or FURIN cDNA were treated with selumetinib (20 nM) for 48 h and the cells were subjected to clonogenic assay. Data were presented as mean ± SD (n = 3). (B) PTC cells were transfected with FURIN siRNA and treated with selumetinib (20 nM) for 48 h and the cells were subjected to clonogenic assay. Data were presented as mean ± SD (n = 3). (C) TPC‐1 cells carrying empty vector or FURIN cDNA were treated with selumetinib (20 nM). After 48 h, cells were analyzed for apoptosis by annexin‐V/PI. Data were presented as mean ± SD (n = 3). (D) PTC cells were transfected with FURIN siRNA and treated with selumetinib (20 nM). After 48 h, cells were analyzed for apoptosis. Data were presented as mean ± SD (n = 3). Statistical analyses were performed using two‐tailed Student's t‐tests. *P < 0.05.
3.5. Furin promotes cancer cell stemness in PTC cells
Furin has been associated with the stemness of colon cancer stem cells [42]. However, the role of furin in PTC stemness has not yet been investigated. To study the role of furin in spheroid growth in PTC, we overexpressed furin in TPC‐1 cells and were cultured in a spheroid medium. Forced expression of furin significantly increased the growth of spheroids generated from TPC‐1 cells (Fig. 6A,B). Also, furin overexpression elevated the levels of stem cell markers like CD44, CD133, and NANOG as compared to empty vector‐transfected cells (Fig. 6C). To validate the above findings, we stably silenced FURIN in BCPAP and K1 cell lines and grew them in a spheroid medium. As shown in Fig. 6D,E, the depletion of furin significantly reduced the spheroid growth in these cells and decreased the expression of stem cell markers (Fig. 6F). These results reveal the role of furin in maintaining cancer stemness properties in PTC cells. We also show that pharmacological inhibition of MEK by selumetinib significantly decreased spheroid growth (Fig. 7A,B) and stemness (Fig. 7C,D) in both furin expressing and knockdown cells, showing the role of MEK/ERK signaling pathway in furin mediated cancer stemness maintenance.
Fig. 6.
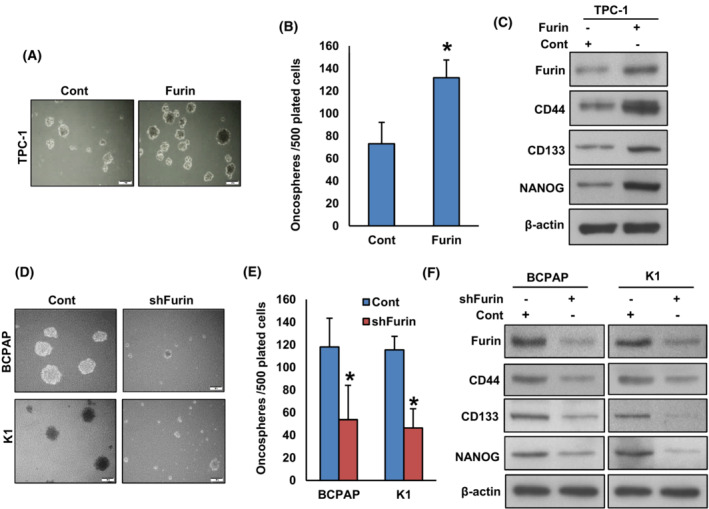
Furin promotes the self‐renewal ability of spheroids generated from PTC cells. (A, B) Forced expression of furin increases spheroid growth. TPC‐1 cells were transfected with either empty vector or FURIN cDNA and cells were subjected to sphere‐forming assay, (scale bar = 1 mm). Spheroids in the entire dish were counted. Data were presented as mean ± SD (n = 3). (C) Forced expression of furin increases the stemness of spheroids as confirmed by immunoblotting using stem cell markers. TPC‐1 cells were transfected with either empty vector or FURIN cDNA and grown in a sphere medium. Proteins were isolated from spheroids and immunoblotted with antibodies against Furin, CD44, CD133, NANOG, and β‐Actin (n = 3). (D, E) Depletion of furin attenuates the self‐renewal ability of spheroids. PTC cells were transfected with FURIN shRNA and cells were subjected to a sphere‐forming assay, (scale bar = 1 mm). Spheroids in the entire well were counted. Data were presented as mean ± SD (n = 3). (F) Depletion of furin reduces the stemness of spheroids. PTC cells were transfected with FURIN shRNA and grown in a sphere medium. Proteins were isolated from spheroids and immunoblotted with antibodies against Furin, CD44, CD133, NANOG, and β‐Actin (n = 3). Statistical analyses were performed using two‐tailed Student's t‐tests. *P < 0.05.
Fig. 7.
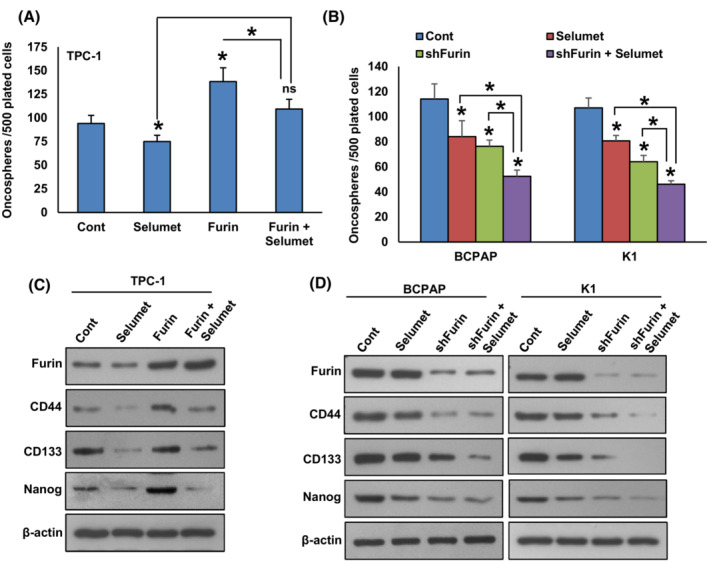
Inhibition of MEK decreases spheroid growth and stemness in both furin expressing and knockdown cells. (A) TPC‐1 cells carrying either empty vector or FURIN cDNA were treated with selumetinib (20 nM) for 48 h and the cells were subjected to sphere‐forming assay. Data were presented as mean ± SD (n = 3). (B) PTC cells carrying either empty vector or FURIN shRNA were treated with selumetinib (20 nM) for 48 h and the cells were subjected to sphere‐forming assay. Data were presented as mean ± SD (n = 3). (C) TPC‐1 cells carrying either empty vector or FURIN cDNA were treated with selumetinib (20 nM) for 48 h and grown in a sphere medium. Proteins were isolated from spheroids and immunoblotted with antibodies against Furin, CD44, CD133, NANOG, and β‐Actin (n = 3). (D) PTC cells carrying either empty vector or FURIN shRNA were treated with selumetinib (20 nM) for 48 h and grown in a sphere medium. Proteins were isolated from spheroids and immunoblotted with antibodies against Furin, CD44, CD133, NANOG, and β‐Actin (n = 3). Statistical analyses were performed using two‐tailed Student's t‐tests. *P < 0.05.
3.6. Furin activates EMT and metastatic potential in PTC cells
It has been reported that furin promotes EMT and its inhibition attenuates metastatic potential in pancreatic cancer cells [19]. The downregulation of furin inhibits migration and invasion in lung cancer [43] and rhabdomyosarcoma [44] cells. Therefore, we wanted to determine whether furin has any effect on inducing EMT and metastatic potential in PTC cell lines. We found that forced expression of furin increased the expression of Furin, N‐cadherin, Zeb1, and Twist with an associated downregulation of E‐cadherin expression in TPC‐1 cells (Fig. 8A). In addition, forced expression of furin also increased invasion (Fig. 8B,C) and migration (Fig. 8D) in TPC‐1 cells. In addition, treatment with selumetinib downregulated the expression of N‐cadherin, Zeb1, and Twist with a concomitant upregulation of E‐cadherin expression in furin‐expressing cells (Fig. 8E). Inhibition of MEK by selumetinib also significantly decreased invasion (Fig. 8F) and migration (Fig. 8G) of these cells.
Fig. 8.
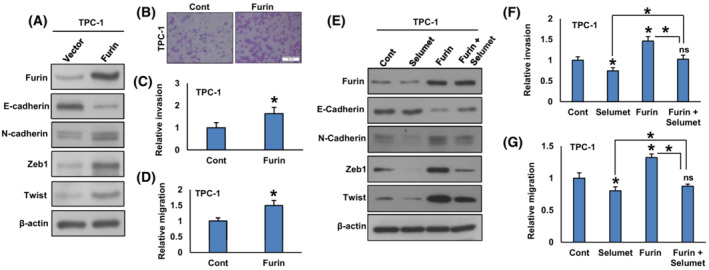
Forced expression of furin promotes EMT and metastatic potential of PTC cells. (A) TPC‐1 cells were transfected with either an empty vector or FURIN cDNA. After cell lysis, proteins were immuno‐blotted with antibodies against Furin, E‐cadherin, N‐cadherin, Zeb1, Twist, and β‐Actin as indicated (n = 3). (B, C) Forced expression of furin increases invasion. TPC‐1 cells were transfected with either an empty vector or FURIN cDNA. Cells were seeded into the upper compartment of invasion chambers. The bottom chambers were filled with RPMI media. After 24 h incubation, invaded cells were fixed, stained, and quantified (scale bar = 1 mm). Data were presented as mean ± SD (n = 3). (D) Forced expression of furin increases migration. After transfection, cells were seeded into the upper compartment of migration chambers. The bottom chambers were filled with RPMI media. After 24 h incubation, migrated cells were fixed, stained, and quantified. Data were presented as mean ± SD (n = 3). (E) Inhibition of MEK reduces EMT in furin‐expressing cells. TPC‐1 cells carrying either empty vector or FURIN cDNA were treated with selumetinib (20 nM) for 48 h. After cell lysis, proteins were immuno‐blotted with antibodies against Furin, E‐cadherin, N‐cadherin, Zeb1, Twist, and β‐Actin as indicated (n = 3). (F, G) Selumetinib decreases the invasion and migration of Furin expressing cells. TPC‐1 cells carrying either empty vector or FURIN cDNA were treated with selumetinib (20 nM) for 48 h and the cells were subjected to invasion and migration assay. Data were presented as mean ± SD (n = 3). Statistical analyses were performed using two‐tailed Student's t‐tests. *P < 0.05.
On the contrary, silencing of furin downregulated the expression of Furin, N‐cadherin, Zeb1, and Twist with a concomitant upregulation of E‐cadherin expression (Fig. 9A) in BCPAP and K1 cell lines. Depletion of furin also decreased invasion (Fig. 9B,C) and migration (Fig. 9D) of these cells. We show that inhibition of MEK along with furin further reduced the expression of N‐cadherin, Zeb1, and Twist with an increased E‐cadherin expression (Fig. 9E). Furthermore, co‐inhibition of MEK and furin significantly decreased invasion (Fig. 9F) and migration (Fig. 9G) of these cells. All these results demonstrate that furin promotes EMT and metastatic potential in PTC cells, where MEK/ERK signaling plays an important role.
Fig. 9.
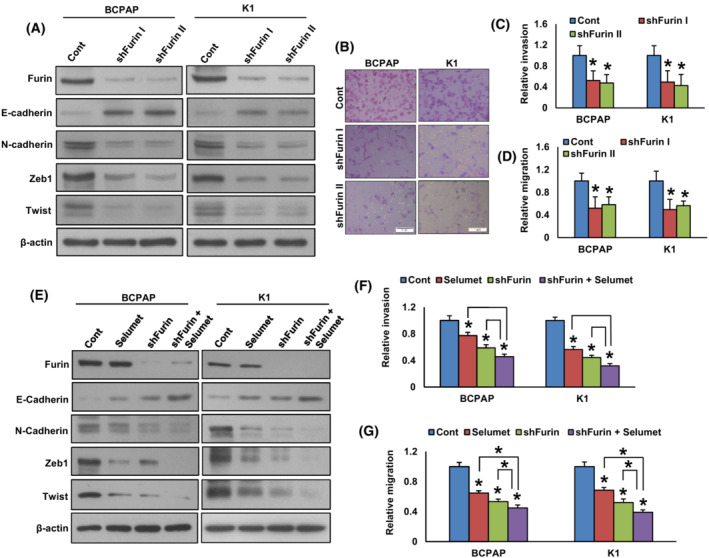
Depletion of furin attenuates EMT and metastatic potential of PTC cells. (A) PTC cells were transfected with a control shRNA vector or two different FURIN shRNA constructs. After transfection, cells were lysed and proteins were immuno‐blotted with antibodies against Furin, E‐cadherin, N‐cadherin, Zeb1, Twist, and β‐Actin as indicated (n = 3). (B, C) Knockdown of FURIN decreases the invasive ability of PTC cells. PTC cells were transfected with FURIN shRNA's and cells were subjected to invasion assay. (scale bar = 1 mm). Data were presented as mean ± SD (n = 3). (D) Knockdown of FURIN decreases the migratory ability of PTC cells. PTC cells were transfected with FURIN shRNA's and cells were subjected to migration assay. Data were presented as mean ± SD (n = 3). (E) The co‐inhibition of MEK and furin further reduce EMT in PTC cells. PTC cells carrying either empty vector or FURIN shRNA were treated with selumetinib (20 nM) for 48 h. After cell lysis, proteins were immuno‐blotted with antibodies against Furin, E‐cadherin, N‐cadherin, Zeb1, Twist, and β‐Actin as indicated (n = 3). (F, G) The co‐inhibition of MEK and furin further decreases the invasion and migration of PTC cells. PTC cells carrying either empty vector or FURIN shRNA were treated with selumetinib (20 nM) for 48 h and the cells were subjected to invasion and migration assay. Data were presented as mean ± SD (n = 3). Statistical analyses were performed using two‐tailed Student's t‐tests. *P < 0.05.
3.7. Furin promotes tumor growth in vivo
We showed that furin promotes PTC growth in vitro. To confirm these findings in vivo, TPC‐1 cells were transfected with empty vector (TPC‐1‐pCMV6‐myc‐DDK) and furin cDNA (TPC‐1‐pCMV6‐myc‐DDK‐furin), subcutaneously inoculated into the right flanks of NU/J mice. After 4 weeks, the animals were sacrificed and their tumor weight was measured and proteins isolated. We found that forced expression of furin significantly increased tumor volume (Fig. 10A,B) and tumor weight (Fig. 10C) in NU/J mice. There was a noticeable increase in the expression of Furin, pMEK1/2, pERK1/2, N‐cadherin, Zeb1 and Twist with an associated downregulation of E‐cadherin expression in furin‐expressing tumors (Fig. 10D). Conversely, depletion of furin in the BCPAP cell line showed delayed tumor growth in NU/J mice as shown by a reduction in tumor volume (Fig. 11A,B) and weight (Fig. 11C). Also, downregulation in the expression of Furin, pMEK1/2, pERK1/2, N‐cadherin, Zeb1, and Twist with upregulation of E‐cadherin expression (Fig. 11D) was observed in furin‐depleted tumors. All these results indicate that furin promotes tumor growth in vivo and is a potential therapeutic target.
Fig. 10.
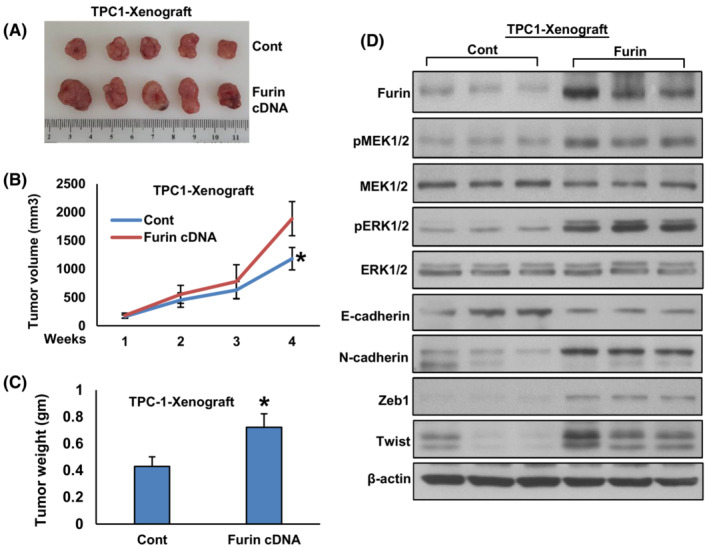
Forced expression of furin promotes tumor growth in vivo. (A) TPC‐1 cells were transfected with empty vector and FURIN cDNA, subcutaneously inoculated into the right flanks of NU/J mice (n = 5). The tumor volume (B) and mouse body weight were monitored weekly. After 4 weeks, mice were sacrificed and individual tumors were weighed (C), then snap‐frozen in liquid nitrogen for storage. Data were presented as mean ± SD (n = 5). (D) Tumors were lysed and proteins were immuno‐blotted with antibodies against Furin, pMEK1/2, MEK1/2, pERK1/2, ERK1/2, E‐cadherin, N‐cadherin, Zeb1, Twist and β‐Actin as indicated (n = 3). Statistical analyses were performed using two‐tailed Student's t‐tests. *P < 0.05.
Fig. 11.
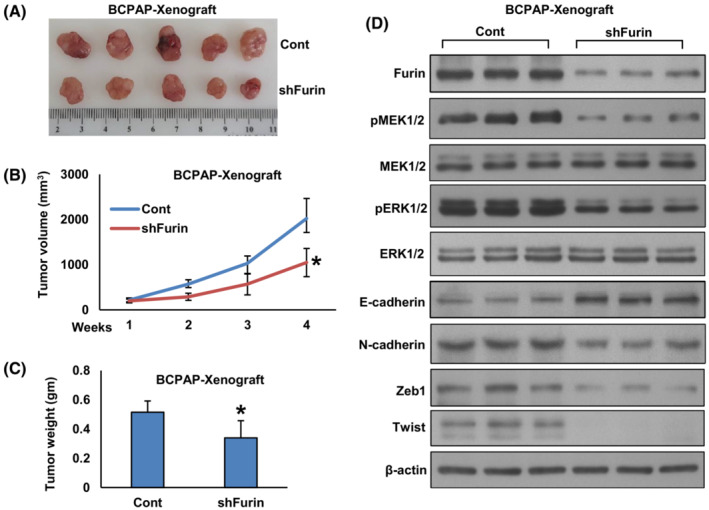
Depletion of furin delayed tumor growth in vivo. (A) BCPAP cells were transfected with empty vector and FURIN shRNA, subcutaneously inoculated into the right flanks of NU/J mice (n = 5). The tumor volume (B) and mouse body weight were monitored weekly. After 4 weeks, mice were sacrificed and individual tumors were weighed (C). Data were presented as mean ± SD (n = 5). (D) Tumors were lysed and proteins were immuno‐blotted with antibodies against Furin, pMEK1/2, MEK1/2, pERK1/2, ERK1/2, E‐cadherin, N‐cadherin, Zeb1, Twist and β‐Actin as indicated (n = 3). Statistical analyses were performed using two‐tailed Student's t‐tests. *P < 0.05.
3.8. Depletion of Furin potentiated the antitumor effect of selumetinib in vivo
We showed that MEK/ERK signaling cascade has a crucial role in furin‐induced PTC cell growth and furin functions upstream to this pathway. Therefore, we wanted to investigate whether the depletion of furin potentiates the antitumor effect of selumetinib (MEK inhibitor) in vivo. For this experiment, NU/J mice were divided into four groups (5 mice per group) and the first two groups of mice were inoculated with BCPAP‐pRS cells (empty vector) and were treated with vehicle (0.1% DMSO, i.p) and selumetinib (10 mg·kg−1, i.p) respectively. The third and fourth groups of mice were inoculated with BCPAP‐pRS‐shfurin cells and treated with vehicle (0.1% DMSO, i.p) and selumetinib (10 mg·kg−1, i.p) respectively. After 4 weeks, the animals were sacrificed and their tumor weight was measured. We found that depletion of furin significantly potentiated the antitumor effect of selumetinib as shown by slower tumor growth (Fig. 12A) and tumor weight (Fig. 12B). However, there was no significant delay in tumor growth in the selumetinib alone treated group.
Fig. 12.
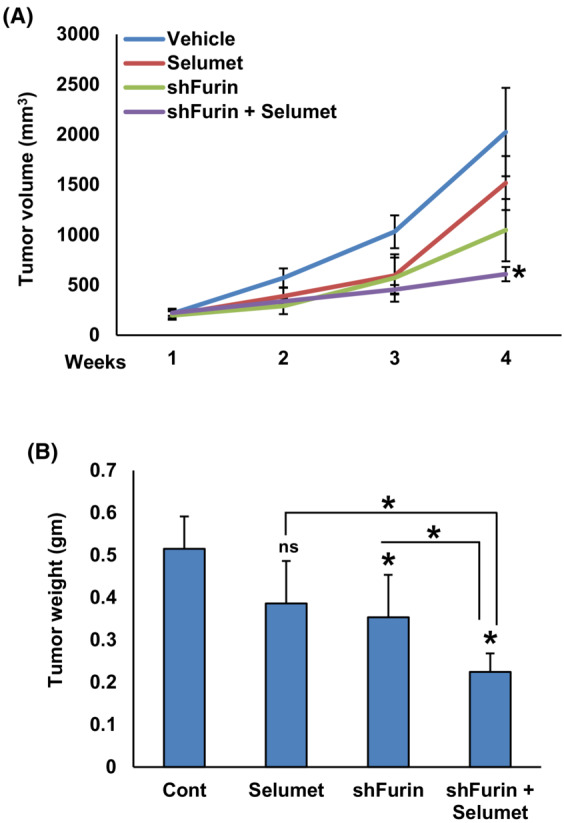
Depletion of furin potentiated the antitumor effect of selumetinib in vivo. NU/J mice were divided into four groups (n = 5) and the first two groups of mice were inoculated with BCPAP‐pRS cells (empty vector) and were treated with vehicle (0.1% DMSO, i.p) and selumetinib (10 mg·kg−1, i.p) respectively. The third and fourth groups of mice were inoculated with BCPAP‐pRS‐shfurin cells and treated with vehicle (0.1% DMSO, i.p) and selumetinib (10 mg·kg−1, i.p) respectively. After tumors grew to about 100 mm3, mice were treated intraperitoneally with selumetinib (10 mg·kg−1) and vehicle (0.1% DMSO), twice a week for 30 days. The tumor volume (A) and mouse body weight were monitored weekly. After 4 weeks, mice were sacrificed and individual tumors were weighed (B). Data were presented as mean ± SD (n = 5). Statistical analyses were performed using two‐tailed Student's t‐tests. *P < 0.05.
4. Discussion
In this study, we revealed that furin is highly expressed in 44.6% of PTC tumor samples (659/1479). The upregulation of furin is found to be significantly associated with several aggressive clinicopathological parameters such as advanced stage, tall cell variant, extrathyroidal extension, and high ATA risk. Furthermore, furin expression is also associated with BRAF mutation. Interestingly multivariant variant analysis shows furin expression is an independent marker for poor overall survival in PTC patients in this cohort.
We demonstrate that targeting furin impairs the malignant phenotype of the PTC cell line by activating the BRAF mutation. Mechanistically, in vitro analysis shows that overexpression of furin increased cell growth, while its knockdown in BCPAP and K1 cell lines significantly inhibited cell growth and induced apoptosis. Furthermore, ectopic expression of furin increased the invasive and migratory potential as well as activated EMT in the PTC cell line, whereas its depletion reversed the effect, which further confirms the role of furin not only in tumor growth but also promoting metastatic potential of PTC cells. These findings are consistent with previous studies that highlight the oncogenic role of furin in cancers from the ovarian [23] and lung [29].
Interestingly, our study also revealed a potential role of furin in the regulation of cancer stem cell‐like cells in PTC. Silencing of furin significantly suppressed the spheroid growth and downregulated stem cell markers such as CD44, CD133, and NANOG in PTC. In an attempt to investigate the mechanism of furin‐induced cell growth, we overexpressed furin in the TPC‐1 cell line and analyzed the expression of MEK/ERK pathway proteins. Interestingly, we find that forced expression of furin markedly increased the phosphorylation of MEK1/2 and ERK1/2 in that cell line, while the opposite effect is seen with stable knockdown of furin.
Our data also demonstrate that furin functions as upstream to MEK/ERK signaling cascade by threatening BRAF mutated cell lines with a pharmacological inhibitor of MEK, mirdametinib, and selumetinib. These inhibitors suppress the activation of MEK1/2 and ERK1/2 with no effect on furin expression. We also show that co‐inhibition of furin and MEK prominently reduced cell proliferation, migration, invasion, EMT, spheroid growth, and stemness in PTC cells, showing the role of MEK/ERK signaling pathway in furin‐mediated cancer stemness maintenance.
The oncogenic role of furin in PTC cell is also suggested by in vivo analysis where forced expression of furin significantly increase tumor growth in NU/J mice, while its depletion showed the opposite effect. Since in vitro knockdown of FURIN also inhibited activation of the MEK/ERK signaling cascade which plays an important role in PTC. Therefore, we studied whether depletion of furin could potentiate the antitumor effect of selumetinib (MEK inhibitor) in vivo, and indeed we were able to see the synergistic antitumor effect of furin depletion and selumetinib in NU/J mice.
The previous report has shown that furin has many additional substrates that might contribute to the observed oncogenic effect of many cancers like pancreatic cancer [19] and colorectal cancer [26]. So, it would not be surprising that our results show the synergistic effect of FURIN knockdown and inhibition of MEK oncogenic pathway in PTC. This synergistic strategy could potentially enhance therapeutic efficacy in treating aggressive or metastatic PTC. Since data from this study and previous studies indicate that furin inhibition has a promising antitumor effect and can simultaneously inhibit multiple oncogenic pathways in several cancers.
Our studies have several limitations, the expression of Furin has been studied retrospectively on a single institute cohort from specific ethnicity. Additional studies are needed to determine the extent of furin overexpression in PTC tumor samples.
5. Conclusion
Our study shows that furin is upregulated in clinical PTC samples and it could be used as a prognostic marker for poor survival. In vitro and in vivo analysis demonstrates that furin inhibition can be used as a therapeutic strategy in PTC.
Author contributions
PKP and AKS: Designed, performed experiments, and wrote the manuscript. DP: Performed experiments and data analysis. SKP: Prepared the TMA and conducted all the immunohistochemistry experiments and scoring of IHC spots. RD, ST, RB, and WH: Performed experiments. FHA: Technical help for animal experiments. SSA and FA: Contributed samples and analyzed clinical data. KSA: Designed, implemented the study, wrote, and critically reviewed the manuscript. This is to confirm that all authors read and approved the final manuscript.
Conflict of interest
The authors declare no conflict of interest.
Peer review
The peer review history for this article is available at https://publons.com/publon/10.1002/1878‐0261.13396.
Supporting information
Fig. S1. Decrease in cell growth after furin depletion was due to apoptosis.
Acknowledgements
This study was not supported by any funding agency therefore the authors declare that there is no funding information to be disclosed for this manuscript.
Pratheesh Kumar Poyil and Abdul K. Siraj contributed equally to this article
Data accessibility
All data generated or analyzed during this study are included in this published article.
References
- 1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. [DOI] [PubMed] [Google Scholar]
- 2. Lim H, Devesa SS, Sosa JA, Check D, Kitahara CM. Trends in thyroid cancer incidence and mortality in the United States, 1974–2013. JAMA. 2017;317:1338–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Wiltshire JJ, Drake TM, Uttley L, Balasubramanian SP. Systematic review of trends in the incidence rates of thyroid cancer. Thyroid. 2016;26:1541–52. [DOI] [PubMed] [Google Scholar]
- 4. Bazarbashi S, Al Eid H, Minguet J. Cancer incidence in Saudi Arabia: 2012 data from the Saudi cancer registry. Asian Pac J Cancer Prev. 2017;18:2437–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Nixon IJ, Ganly I, Patel SG, Palmer FL, Di Lorenzo MM, Grewal RK, et al. The results of selective use of radioactive iodine on survival and on recurrence in the management of papillary thyroid cancer, based on Memorial Sloan‐Kettering Cancer Center risk group stratification. Thyroid. 2013;23:683–94. [DOI] [PubMed] [Google Scholar]
- 6. Ritter A, Mizrachi A, Bachar G, Vainer I, Shimon I, Hirsch D, et al. Detecting recurrence following lobectomy for thyroid cancer: role of thyroglobulin and thyroglobulin antibodies. J Clin Endocrinol Metabol. 2020;105:dgaa152. [DOI] [PubMed] [Google Scholar]
- 7. Xing M, Alzahrani AS, Carson KA, Shong YK, Kim TY, Viola D, et al. Association between BRAF V600E mutation and recurrence of papillary thyroid cancer. J Clin Oncol. 2015;33:42–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Xing M, Liu R, Liu X, Murugan AK, Zhu G, Zeiger MA, et al. BRAF V600E and TERT promoter mutations cooperatively identify the most aggressive papillary thyroid cancer with highest recurrence. J Clin Oncol. 2014;32:2718–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zafon C, Obiols G. The mitogen‐activated protein kinase (MAPK) signaling pathway in papillary thyroid cancer. From the molecular bases to clinical practice. Endocrinol Nutr. 2009;56:176–86. [DOI] [PubMed] [Google Scholar]
- 10. Xing M. BRAF mutation in thyroid cancer. Endocr Relat Cancer. 2005;12:245–62. [DOI] [PubMed] [Google Scholar]
- 11. Abubaker J, Jehan Z, Bavi P, Sultana M, Al‐Harbi S, Ibrahim M, et al. Clinicopathological analysis of papillary thyroid cancer with PIK3CA alterations in a middle eastern population. J Clin Endocrinol Metabol. 2008;93:611–8. [DOI] [PubMed] [Google Scholar]
- 12. Kim TH, Park YJ, Lim JA, Ahn HY, Lee EK, Lee YJ, et al. The association of the BRAFV600E mutation with prognostic factors and poor clinical outcome in papillary thyroid cancer: a meta‐analysis. Cancer. 2012;118:1764–73. [DOI] [PubMed] [Google Scholar]
- 13. Solovyeva N, Gureeva T, Timoshenko O, Moskvitina T, Kugaevskaya E. Furin as proprotein convertase and its role in normal and pathological biological processes. Biomed Khim. 2016;62:609–21. [DOI] [PubMed] [Google Scholar]
- 14. Jaaks P, Bernasconi M. The proprotein convertase furin in tumour progression. Int J Cancer. 2017;141:654–63. [DOI] [PubMed] [Google Scholar]
- 15. Seidah NG, Prat A. The biology and therapeutic targeting of the proprotein convertases. Nat Rev Drug Discov. 2012;11:367–83. [DOI] [PubMed] [Google Scholar]
- 16. Bassi DE, Fu J, Lopez de Cicco R, Klein‐Szanto AJ. Proprotein convertases:“master switches” in the regulation of tumor growth and progression. Mol Carcinog. 2005;44:151–61. [DOI] [PubMed] [Google Scholar]
- 17. He Z, Khatib A‐M, Creemers JW. The proprotein convertase furin in cancer: more than an oncogene. Oncogene. 2022;41:1252–62. [DOI] [PubMed] [Google Scholar]
- 18. Scamuffa N, Siegfried G, Bontemps Y, Ma L, Basak A, Cherel G, et al. Selective inhibition of proprotein convertases represses the metastatic potential of human colorectal tumor cells. J Clin Invest. 2008;118:352–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zhang Y, Zhou M, Wei H, Zhou H, He J, Lu Y, et al. Furin promotes epithelial‐mesenchymal transition in pancreatic cancer cells via hippo‐YAP pathway. Int J Oncol. 2017;50:1352–62. [DOI] [PubMed] [Google Scholar]
- 20. Huang Y‐H, Lin K‐H, Liao C‐H, Lai M‐W, Tseng Y‐H, Yeh C‐T. Furin overexpression suppresses tumor growth and predicts a better postoperative disease‐free survival in hepatocellular carcinoma. PLoS ONE. 2012;7:e40738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cheng M, Watson P, Paterson J, Seidah N, Chrétien M, Shiu RP. Pro‐protein convertase gene expression in human breast cancer. Int J Cancer. 1997;71:966–71. [DOI] [PubMed] [Google Scholar]
- 22. Bassi DE, Mahloogi H, De Cicco RL, Klein‐Szanto A. Increased furin activity enhances the malignant phenotype of human head and neck cancer cells. Am J Pathol. 2003;162:439–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Page RE, Klein‐Szanto AJ, Litwin S, Nicolas E, Al‐Jumaily R, Alexander P, et al. Increased expression of the pro‐protein convertase furin predicts decreased survival in ovarian cancer. Anal Cell Pathol. 2007;29:289–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Mbikay M, Sirois F, Yao J, Seidah N, Chretien M. Comparative analysis of expression of the proprotein convertases furin, PACE4, PC1 and PC2 in human lung tumours. Br J Cancer. 1997;75:1509–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Fu J, Bassi DE, Zhang J, Li T, Nicolas E, Klein‐Szanto AJ. Transgenic overexpression of the proprotein convertase furin enhances skin tumor growth. Neoplasia. 2012;14:271–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. He Z, Thorrez L, Siegfried G, Meulemans S, Evrard S, Tejpar S, et al. The proprotein convertase furin is a pro‐oncogenic driver in KRAS and BRAF driven colorectal cancer. Oncogene. 2020;39:3571–87. [DOI] [PubMed] [Google Scholar]
- 27. Bassi DE, Lopez De Cicco R, Mahloogi H, Zucker S, Thomas G, Klein‐Szanto AJ. Furin inhibition results in absent or decreased invasiveness and tumorigenicity of human cancer cells. Proc Natl Acad Sci USA. 2001;98:10326–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Coppola JM, Bhojani MS, Ross BD, Rehemtulla A. A small‐molecule furin inhibitor inhibits cancer cell motility and invasiveness. Neoplasia. 2008;10:363–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ma Y‐C, Fan W‐J, Rao S‐M, Gao L, Bei Z‐Y, Xu S‐T. Effect of Furin inhibitor on lung adenocarcinoma cell growth and metastasis. Cancer Cell Int. 2014;14:1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Mercapide J, Lopez De Cicco R, Bassi DE, Castresana JS, Thomas G, Klein‐Szanto AJ. Inhibition of furin‐mediated processing results in suppression of astrocytoma cell growth and invasiveness. Clin Cancer Res. 2002;8:1740–6. [PubMed] [Google Scholar]
- 31. Zhou M, Zhang Y, Wei H, He J, Wang D, Chen B, et al. Furin inhibitor D6R suppresses epithelial‐mesenchymal transition in SW1990 and PaTu8988 cells via the hippo‐YAP signaling pathway. Oncol Lett. 2018;15:3192–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. He Z, Khatib A‐M, Creemers JW. Loss of proprotein convertase furin in mammary gland impairs proIGF1R and proIR processing and suppresses tumorigenesis in triple negative breast cancer. Cancers (Basel). 2020;12:2686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Siraj AK, Parvathareddy SK, Siraj N, Al‐Obaisi K, Aldughaither SM, AlManea HM, et al. Loss of ZNF677 expression is a predictive biomarker for lymph node metastasis in middle eastern colorectal cancer. Sci Rep. 2021;11:22346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Siraj AK, Bu R, Arshad M, Iqbal K, Parvathareddy SK, Masoodi T, et al. POLE and POLD1 pathogenic variants in the proofreading domain in papillary thyroid cancer. Endocr Connect. 2020;9:923–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Siraj AK, Parvathareddy SK, Bu R, Iqbal K, Siraj S, Masoodi T, et al. Germline POLE and POLD1 proofreading domain mutations in endometrial carcinoma from middle eastern region. Cancer Cell Int. 2019;19:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Siraj AK, Pratheeshkumar P, Parvathareddy SK, Bu R, Masoodi T, Iqbal K, et al. Prognostic significance of DNMT3A alterations in middle eastern papillary thyroid carcinoma. Eur J Cancer. 2019;117:133–44. [DOI] [PubMed] [Google Scholar]
- 37. Camp RL, Dolled‐Filhart M, Rimm DL. X‐tile: a new bio‐informatics tool for biomarker assessment and outcome‐based cut‐point optimization. Clin Cancer Res. 2004;10:7252–9. [DOI] [PubMed] [Google Scholar]
- 38. Bu R, Siraj AK, Divya SP, Kong Y, Parvathareddy SK, Al‐Rasheed M, et al. Telomerase reverse transcriptase mutations are independent predictor of disease‐free survival in M iddle E astern papillary thyroid cancer. Int J Cancer. 2018;142:2028–39. [DOI] [PubMed] [Google Scholar]
- 39. Zhao M, Sano D, Pickering CR, Jasser SA, Henderson YC, Clayman GL, et al. Assembly and initial characterization of a panel of 85 genomically validated cell lines from diverse head and neck tumor sites. Clin Cancer Res. 2011;17:7248–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Siraj AK, Pratheeshkumar P, Divya SP, Parvathareddy SK, Alobaisi KA, Thangavel S, et al. Krupple‐like factor 5 is a potential therapeutic target and prognostic marker in epithelial ovarian cancer. Front Pharmacol. 2020;11:598880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Siraj AK, Pratheeshkumar P, Parvathareddy SK, Divya SP, Al‐Dayel F, Tulbah A, et al. Overexpression of PARP is an independent prognostic marker for poor survival in middle eastern breast cancer and its inhibition can be enhanced with embelin co‐treatment. Oncotarget. 2018;9:37319–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Descarpentrie J, Araúzo‐Bravo MJ, He Z, François A, González Á, Garcia‐Gallastegi P, et al. Role of Furin in colon cancer stem cells malignant phenotype and expression of LGR5 and NANOG in KRAS and BRAF‐mutated colon tumors. Cancers (Basel). 2022;14:1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Bassi DE, Zhang J, Renner C, Klein‐Szanto AJ. Targeting proprotein convertases in furin‐rich lung cancer cells results in decreased in vitro and in vivo growth. Mol Carcinog. 2017;56:1182–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Jaaks P, D'Alessandro V, Grob N, Büel S, Hajdin K, Schäfer BW, et al. The proprotein convertase furin contributes to rhabdomyosarcoma malignancy by promoting vascularization, migration and invasion. PLoS ONE. 2016;11:e0161396. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Decrease in cell growth after furin depletion was due to apoptosis.
Data Availability Statement
All data generated or analyzed during this study are included in this published article.


