A hallmark of aggressive meningiomas is their genomic instability [2, 3]. In fact, the World Health Organization (WHO) recently added homozygous loss of cyclin-dependent kinase inhibitor 2A and/or B (CDKN2A, CDKN2B)—two tumor suppressor genes adjacent to one another on chromosome 9p21—to their grade 3 (aggressive) meningioma classification [6]. Although not required for grade 3 designation, homozygous loss of either gene is associated with high mitotic count and shorter recurrence-free survival. It is unclear, however, how practical this criterion is for prognosis. First, CDKN2A/B deletions seem to be rare [9]. Second, detecting CDKN2A/B loss can be difficult, as immunohistochemistry for the protein product may not be reliable [5]. Third, many of these deletions occur in tumors already classifiable as grade 3, most of which are recurrences, meaning that they have declared themselves to be aggressive by their behavior even before pathological analysis. To clarify the prognostic value of CDKN2A/B loss, we therefore used integrated molecular classification, which predicts outcome much more reliably than WHO grade [1, 4, 7, 8].
We examined 776 tumors from four institutions in total [1, 4, 7], grading them using the 2016 WHO guidelines. We then used methylation (Illumina Infinium MethylationEPIC (850 k) BeadChip) and RNA profiling to classify the tumors as Meningioma Group (MenG) A (benign, merlin-intact), B (benign, merlin-deficient), or C (aggressive) [1, 4, 8]; we determined CDKN2A/B status from the methylation data [4]. Two investigators examined CNV traces highlighting the CDKN2A/B site [10] to identify either focal CDKN2A/B deletions or chromosome 9p loss (Fig. 1a); the senior author adjudicated any inter-rater discrepancies. We used R [RRID:SCR_021094] to analyze recurrence-free survival (RFS) and statistics.
Fig. 1.
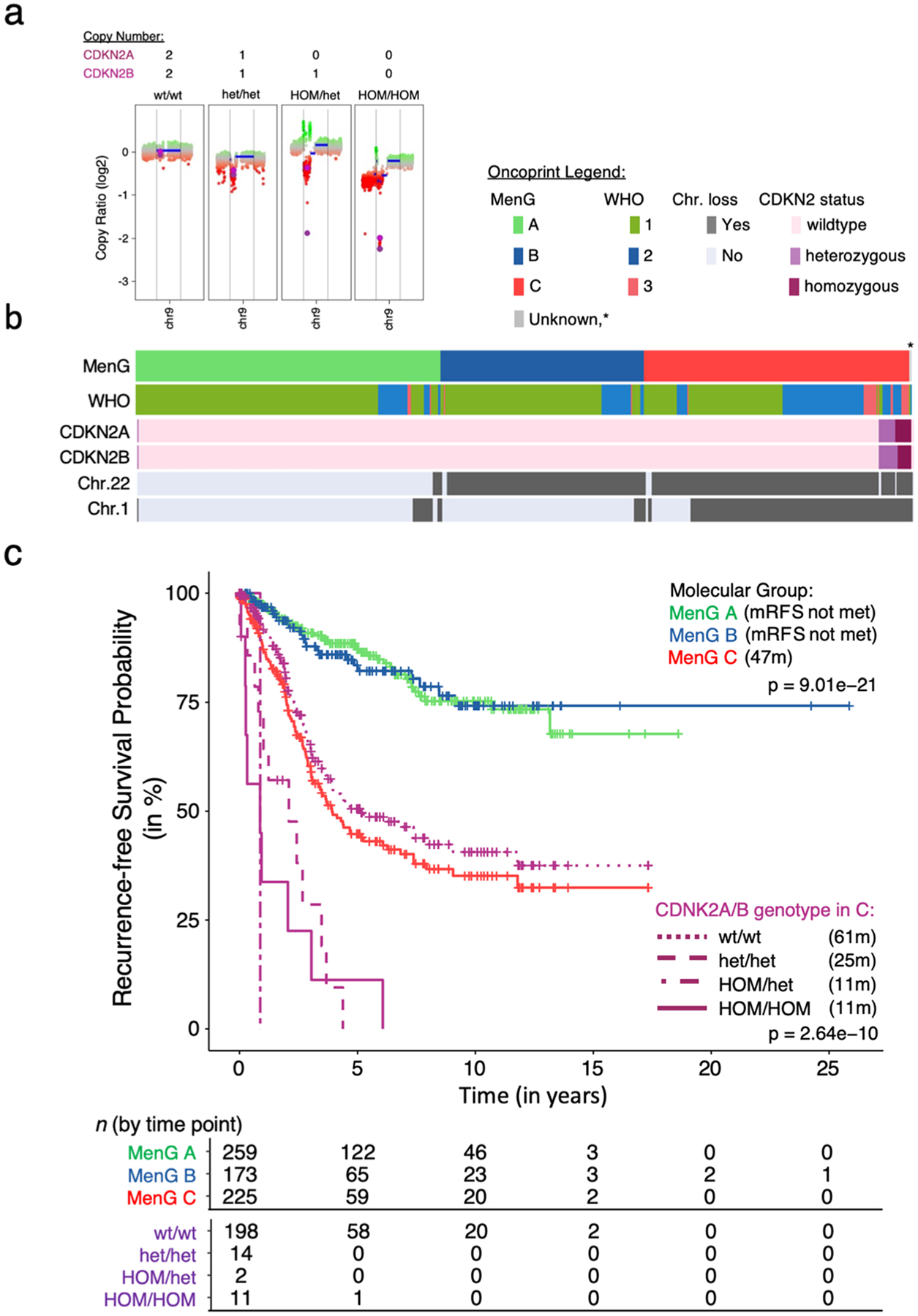
CDKN2A/B heterozygous loss shortens RFS. a CDKN2A/B scoring based on copy number counts of CNV traces. b Oncoprint of the 659 tumors with clinical data, clustered by molecular Groups. The asterisk marks the nonrecurrent tumor that had CDK2A/B loss. c Median RFS by molecular group and CDKN2A/B status within Group C, omitting two unclassifiable tumors and the nonrecurrent group A tumor. mRFS in months of each group in parenthesis. Kruskal–Wallis ANOVA by group as well as genotype
Thirty-eight tumors (4.9%) showed partial or complete loss of CDKN2A and/or CDKN2B. Considering only the 659 tumors for which we had clinical data [1, 4], 28 (4.2%) showed partial or complete loss of CDKN2A, CDKN2B, or both. Of these 28, 11 tumors showed homozygous loss of both genes, 15 showed heterozygous loss of both, and two tumors had homozygous loss of CDKN2A with heterozygous CDKN2B loss (Supplementary Table 1 online resource). Although tumors with any loss of CDKN2A/B were more proliferative, less likely to be primary tumors, and more likely to recur, they appeared in all three WHO grades (Fig. 1b, Supplementary Fig. 1, Supplementary Table 2). Molecular classification, however, showed that CDKN2A/B losses were almost exclusive to Group C, excepting one primary skull base tumor with heterozygous CDKN2A/B loss classified as MenG A. This tumor came from a 62-year-old woman who remained recurrence-free at last follow-up (29 months) (Fig. 1b, Supplementary Table S2).
Even in this aggressive class of C tumors, CDKN2A/B loss shortened mRFS: the mRFS was 11 months, and no patient with any type of CDKN2A/B loss survived more than 73 months without recurrence (Fig. 1c, Supplementary Table 1). Even heterozygous loss reduced mRFS to 25 months.
Without examining other causes of gene dysfunction, such as single nucleotide variants [5], we may have missed some instances of reduced CDKN2A and/or B protein activity. Nonetheless, deletions involving CDKN2A/B were rare even in this sizeable cohort, consistent with previous reports [9], and homozygosity was even more rare. We therefore propose that the most pragmatic course would be to first identify Group C tumors—50% of which will recur by 47 months (Fig. 1c)—and then analyze CDKN2A/B status to refine the prognosis.
Supplementary Material
Acknowledgements
This work was supported by the NINDS (K08NS102474 to AJP and R25NS070694 to JCB), the Roderick D. MacDonald Fund (AJP), the Hamill Foundation (AJP), the Northwestern Medicine Malnati Brain Tumor Institute of the Lurie Cancer Center (STM), P50 CA097257 (WCC, DRR), R01 CA262311 (DRR), F32 CA213944 (STM), F30 CA246808 and T32 GM007618 (AC), a UCSF Catalyst Program Award (WCC, DRR), and the UCSF Wolfe Meningioma Program Project (DRR). This work was also supported by a Conquer Cancer Herman H. Freckman, MD, Endowed Young Investigator Award from the American Society of Clinical Oncology (WCC). Any opinions, findings, and conclusions expressed in this material are those of the author(s) and do not necessarily reflect those of Conquer Cancer or the American Society of Clinical Oncology.
Footnotes
Supplementary Information The online version contains supplementary material available at https://doi.org/10.1007/s00401-023-02543-7.
Data availability
Data used in this study is available in the Gene Expression Omnibus database, https://ncbi.nlm.nih.gov/geo, under the following accession numbers: from GSE84465, GSE136661, GSE183656, GSE101638, GSE101638, GSE139652, GSE180061 and from the authors by request.
References
- 1.Bayley JC, Hadley CC, Harmanci AO, Harmanci AS, Klisch TJ, Patel AJ (2022) Multiple approaches converge on three biological subtypes of meningioma and extract new insights from published studies. Sci Adv 8:eabm6247. 10.1126/sciadv.abm6247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bi WL, Abedalthagafi M, Horowitz P, Agarwalla PK, Mei Y, Aizer AA et al. (2016) Genomic landscape of intracranial meningiomas. J Neurosurg 125:525–535. 10.3171/2015.6.JNS15591 [DOI] [PubMed] [Google Scholar]
- 3.Bi WL, Greenwald NF, Abedalthagafi M, Wala J, Gibson WJ, Agarwalla PK et al. (2017) Genomic landscape of high-grade meningiomas. NPJ Genomic Med 2:15. 10.1038/s41525-017-0014-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Choudhury A, Magill ST, Eaton CD, Prager BC, Chen WC, Cady MA et al. (2022) Meningioma DNA methylation groups identify biological drivers and therapeutic vulnerabilities. Nat Genet 54:649–659. 10.1038/s41588-022-01061-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guyot A, Duchesne M, Robert S, Lia A-S, Derouault P, Scaon E et al. (2019) Analysis of CDKN2A gene alterations in recurrent and non-recurrent meningioma. J Neuro-Oncol 145:449–459. 10.1007/s11060-019-03333-6 [DOI] [PubMed] [Google Scholar]
- 6.Louis DN, Perry A, Wesseling P, Brat DJ, Cree IA, Figarella-Branger D et al. (2021) The 2021 who classification of tumors of the central nervous system: a summary. Neuro Oncol. 10.1093/neuonc/noab106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nassiri F, Liu J, Patil V, Mamatjan Y, Wang JZ, Hugh-White R et al. (2021) A clinically applicable integrative molecular classification of meningiomas. Nature 597:119–125. 10.1038/s41586-021-03850-3 [DOI] [PubMed] [Google Scholar]
- 8.Patel AJ, Wan Y-W, Al-Ouran R, Revelli J-P, Cardenas MF, Oneissi M et al. (2019) Molecular profiling predicts meningioma recurrence and reveals loss of DREAM complex repression in aggressive tumors. Proc Natl Acad Sci USA 116:21715–21726. 10.1073/pnas.1912858116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sievers P, Hielscher T, Schrimpf D, Stichel D, Reuss DE, Berghoff AS et al. (2020) CDKN2A/B homozygous deletion is associated with early recurrence in meningiomas. Acta Neuropathol 140:409–413. 10.1007/s00401-020-02188-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhou W, Triche TJ, Laird PW, Shen H (2018) SeSAMe: reducing artifactual detection of DNA methylation by Infinium BeadChips in genomic deletions. Nucleic Acids Res 46:e123. 10.1093/nar/gky691 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data used in this study is available in the Gene Expression Omnibus database, https://ncbi.nlm.nih.gov/geo, under the following accession numbers: from GSE84465, GSE136661, GSE183656, GSE101638, GSE101638, GSE139652, GSE180061 and from the authors by request.


