Abstract
Background: The role of microsomal glutathione S-transferase 1 (MGST1) underlying gastric cancer (GC) is unclear. The purpose of this research was to study the expression level and biological functions of MGST1 in GC cells. Methods: Expression of MGST1 was detected by RT-qPCR, Western blot (WB), and immunohistochemical staining. MGST1 was knockdown and overexpression by short hairpin RNA lentivirus in GC cells. Cell proliferation was evaluated by the CCK-8 assay and EDU assay. The cell cycle was detected by flow cytometry. The TOP-Flash reporter assay was used to examine the activity of T-cell factor/lymphoid enhancer factor transcription based on β-catenin. WB was performed to assess the protein levels involved in the cell signaling pathway and ferroptosis. The MAD assay and C11 BODIPY 581/591 lipid peroxidation probe assay were performed to determine the reactive oxygen species lipid level in GC cells. Results: MGST1 expression was upregulated in GC and it was correlated with poor overall survival of GC patients. MGST1 knockdown significantly inhibited GC cell proliferation and cell cycle by regulating the AKT/GSK-3β/β-catenin axis. In addition, we found that MGST1 inhibits ferroptosis in GC cells. Conclusion: These findings suggested that MGST1 played a confirmed role in promoting GC development and serving as a possible independent prognostic factor for GC.
1. Introduction
It was reported that gastric cancer (GC) has induced high carcinoma-associated mortality worldwide.17 The development of GC is driven by external risk factors5 and internal genetics,30 and Helicobacter pylori infection,33 age, and improper diets43 are included in the main risk factors. The main treatment approaches are endoscopic resection9 and chemotherapy.18 Therefore, future treatment will focus on genetic therapy. Microsomal glutathione S-transferase 1 (MGST1) is a member of the MAPEG (membrane-associated proteins in eicosanoid and glutathione metabolism) family, and its proteins are integral membrane proteins localized to the membrane of organelles.27 The main functions of MGST1 proteins are the reduction of lipid peroxidation and protection of intracellular membranes from oxidative stress.4
According to previous studies, MGST1 is widely expressed. For example, it is expressed significantly in pancreatic cancer,23 glioma,49 neuroblastoma,20 and colorectal cancer.1 However, it has not been well studied in GC. Through genome-wide analysis, high-expression MGST1 has been demonstrated to contribute to higher metastatic potential.3,8,57 MGST1 promotes proliferation and apoptosis of adenocarcinoma cells via inactivating the AKT/GSK-3β pathway.54 Substantial evidence shows that overexpression of MGST1 is associated with poor prognosis in many tumors,28 and overexpression of this enzyme has been reported to be a tumor marker in early tumors.21,25 In addition, high expression of MGST1 was correlated to resistance against anticarcinogens in several tumors.2,36,39 Therefore, it has been suggested that MGST1 plays pivotal roles in drug resistance40 and targeted therapies.37
Wnt/β-catenin signaling is one of the most important signaling pathways. The destruction complex is composed of the membrane proteins,β-catenin protein, and a degradation complex.41 T-cell factor/lymphoid enhancer factor (TCF/LEF) plays a crucial role in activating the intracellular parts of this signaling pathway.22 This pathway is also known as a β-catenin-dependent pathway because of the central function of β-catenin in activating the downstream signaling cascade,44 and this pathway is switched on by phosphorylation of GSK3 kinases.51 Abundant findings have suggested that Wnt/β-catenin signaling was correlated with tumorigenesis of many cancer types.29,34 For example, in cutaneous melanoma, mutational β-catenin leads to incorrect transcription and consequently initiates the expression of downstream factors.12 In consequence, we should focus on WNT-driven cancers and translate theories into effective therapies.
Ferroptosis is a new form of cell death characterized by the accumulation of iron-dependent lipid reactive oxygen species (ROS).10 Simultaneously, ferroptosis can be activated under oxidative turbulence stimuli, which is mainly triggered by glutathione peroxidase 4 (GPX4).24,42 Emerging evidence has indicated that ferroptosis is related to physiological processes or various diseases including diverse cancer types.53 For example, inhibition of dipeptidyl-peptidase-4 (DDP4) activity by TP53 results in colorectal cancer.47 Another study reported that Erastin-induced ferroptosis in GC cells resulted from abnormal GPX4, ROS, and GSH levels.14 Collectively, ferroptosis is a type of programmed necrosis, and targeting the cell death process is a hot topic in anti-tumor therapies.
In this study, we revealed the possible role of MGST1 in the development of GC. Our results found a relationship between the expression of MGST1 and the overall survival (OS) of patients with gastric carcinoma. Furthermore, MGST1-silenced GC cells proliferated and invaded slower than normal cancer cells, and MGST1-overexpressed GC cells had contrary outcomes. We studied the connection between MGST1, AKT, and Wnt/β-catenin pathway using both MGST1 knockdown and overexpressing GC cell lines. Additionally, the relationship between ferroptosis and MGST1 in GC was well investigated in this study.
2. Materials and Methods
2.1. Clinical Specimens and Cell Lines
The clinical tumor specimens were collected from 107 patients with GC hospitalized in the First Affiliated Hospital of Anhui Medical University between October 2012 and December 2013, and a follow-up was conducted in December 2020. All specimens included in this study were recognized by pathologists, and no patient had received targeted molecular therapy, chemotherapy, or radiotherapy before surgery. The tissue microarray was composed of 107 GC tissues and 19 randomly selected corresponding adjacent normal tissues. GC tissues and paired normal tissues, which were adjacent to the tumors, were all stored at −80 °C until use.
Five GC cell lines (AGS, BGC-823, MGC-803, HGC-27, and SGC-7901) and a normal human stomach cell line (GES-1) were all provided by GENEPharma (Shanghai, China) and cultured in a thermostatic Incubator at 37 °C with 5% carbon dioxide.
2.2. Immunohistochemistry
Immunohistochemical (IHC) staining was used to analyze the expressions of MGST1 (1:800, ZEN-BIOSCIENCE, China). The IHC clinical scoring process was based on both the staining intensity and the proportion of staining. The staining intensity had four tiers ranging from 0 to 3 (0 points, negative; 1 point, light yellow; 2 points, claybank; and 3 points, tan). The proportion of staining was assigned 0 (0%), 1 (1–25%), 2 (26–50%), 3 (51–75%), or 4 (76–100%). Score calculation is the product of the score of staining intensity and the score of the proportion of staining. The final score was assigned as ≤4 points (0, 1, 2, 3, and 4) and was negative or of low expression, and a final score of ≥5 points (6, 8, 9, and 12) was of high expression.
2.3. RNA Extraction, cDNA Synthesis, and RT-qPCR
Total RNAs from clinical specimens or cells were extracted by the TRIzol reagent, and cDNA was synthesized according to total RNA as the template. RT-qPCR was performed using an SYBR Green mixture kit (Thermo, USA) by cDNA, and GAPDH was used as a normalization to analyze relative gene expression by Ct data. The primer pairs are as follows: GAPDH forward 5′-GCA TCC TGG GCT ACACT-3′ and reverse 5′-CAC CAC CCT GTT GCTGT-3′; MGST1 forward 5′-TGA CCT CAC CCA GGT AAT GGAT-3′ and reverse 5′-CTG CGT ACA CGT TCT ACT CTGT3’.
2.4. Western Blotting Assay
Proteins from clinical specimens or cells were separated on 12% SDS-PAGE gels and transferred to polyvinylidene fluoride membranes. Afterward, they were blocked for 1 h with 5% skim milk at 24 °C and incubated with primary antibodies overnight at 4 °C. Then, the membranes were incubated with the HRP-conjugated secondary antibodies. ECL reagents were used for signal detection and quantified by ImageJ. Primary and secondary antibodies were obtained from Abcam (MGST1, E-cadherin, N-cadherin, Slug, Snail, Vimentin, β-catenin, GPX4, GAPDH and Anti-Rabbit IgG antibody) and Cell Signaling Technology (AKT, pAKT, GSK-3β, pGSK-3β). The pAKT antibody detects endogenous levels of Akt when phosphorylated at Ser473. The pGSK-3β antibody was produced against a chemically synthesized phosphopeptide derived from a region of human GSK-3β that contains tyrosine 279/216.
2.5. Cell Viability Assay
The CCK-8 assay and the EdU assay were used to estimate cell viability. The CCK-8 assay was implemented using 10 μL of CCK-8 reagent with a density of 105 cells per well in 96-well plates and measured by a universal microplate reader at 450 nm after cultivating. The EdU assay was executed using an Edu cell proliferation detection kit (Beyotime) following the manuals provided with the product.
2.6. Cells and Virus Transfection
We purchased MGST1-silencing and MGST1-overexpressing vectors (including blank vectors, control vectors, and target vectors) from GENEPharma (Shanghai, China), and those vectors were transfected to AGS and SGC-7901 in line with the protocol supplied by the manufacturer. In the inhibitor experiments, 4 μM XAV-939 (MedChemExpress, cat. no. HY-15147) and 1 μM MK-2206 (MedChemExpress, cat. no. HY-108232) inhibitors were added after adenovirus transduction for 48 h.
2.7. Transwell Migration and Invasion Assay
In the migration assay, 3.5 × 104 cells per well were cultured in 500 μL of culture medium without fetal bovine serum (FBS) in 6-well plates and moved to top chambers, and the bottom chamber was placed in 20% FBS (AGS) or 40% FBS (SGC-7901). After incubation for 24 h, the migrated cells were fixed with 4% paraformaldehyde and stained with 0.1% crystal violet for visualization. Cells were counted in random fields was counted. In the invasion assay, the Matrigel (BD Biosciences, Germany) was prepared on the surface of the upper chamber, and the following steps were similar to those of the migration assay.
2.8. Flow Cytometry
Flow cytometry analysis can be used in cell cycle analysis by measuring the DNA content of cells. For cell cycle analysis, cells were fixed in 75% ethanol at −20 °C until use. Cell cycles were examined by flow cytometry (BD Biosciences, USA) according to the directions. A negative control was used to confirm the correct gate. Cells in “Q2 + Q3” were regarded as apoptotic GC cells.
2.9. TOP-Flash Reporter Assay (Luciferase Reporter Assay)
The TOP-Flash reporter assay was used to examine the activity of TCF/LEF transcription based on β-catenin. The M50 Super 8x TOP-Flash vector (YOUBIO, China) provides a promoter driving expression of the firefly luciferase gene, and 7 TCF/LEF binding sites (AGATCAAAGGgggta) were cloned in this vector. For this assay, the Super8XFOPflash reporter vector, MGST1 short hairpin RNA (shRNA) and negative control vectors were co-transfected with GC cells by a superTOP-Flash transfection reagent. After 48 h of co-culture, cells were harvested and detected luciferase activity by a Firefly Luciferase Reporter Gene Assay Kit (Beyotime, China) as the manufacturers described.
2.10. Determination of Malonaldehyde Expression Assay and C11 BODIPY 581/591 Lipid Peroxidation Probe Assay
The malonaldehyde (MDA) assay and lipid peroxidation probe assay are both used to reflect the lipid peroxidation level of cells. The MDA level was measured by Lipid Peroxidation MDA Assay Kit (Beyotime, China) manually and the absorbance was measured at 450 nm.
For the lipid peroxidation probe assay, 1.5 × 105 cells were incubated with 2 μL of 1.5 mM BODIPY-C11 solution for 24 h, and cells were disposed of following the manual. The lipid ROS level was measured by a flow cytometry machine.
2.11. Statistical Analysis
All data were analyzed using IBM SPSS Statistics 23.0 and GraphPad Prism 8.0.1 system software. In this study, the final data were expressed as mean ± SD (*p < 0.05, **p < 0.01, ***p < 0.001), and when the P-values of data were less than 0.05, it was considered statistically significant.
3. Results
3.1. Upregulated MGST1 Level Was Observed in GC and Predicted a Poor Prognosis
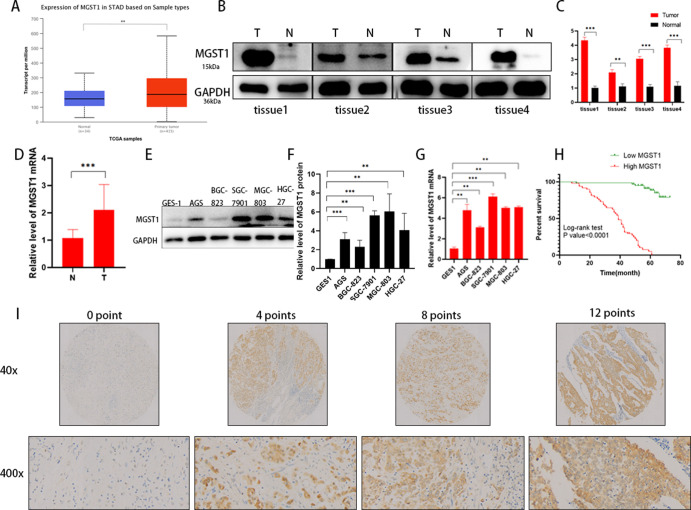
To explore the role of MGST1 in prognosis, we analyzed the MGST1 level in GC using the UALCAN database, which indicated that MGST1 was upregulated in GC tissue (Figure 1A). Moreover, using Western blot (WB) and PCR, we found that levels of MGST1 in GC tissue were higher than in adjacent normal tissue at both protein level in 4 GC tissues (Figure 1B,C) and the mRNA level in 48 fresh frozen GC tissues (Figure 1D). To further investigate the expression of MGST1 in GC, we detected MGST1 mRNA and protein level in gastric epithelial cell lines (GES-1) and five GC cell lines (AGS, BGC-823, SGC-7901, MGC-803, and HGC-27), and MGST1 expression was prominently increased in all GC cell lines at both the protein level (Figure 1E,F) and the mRNA level (Figure 1G). AGS and HGC-27 GC cell lines were chosen to prefer further research because of the high MGST1-protein expression and better invasion ability in the follow-up experiments. Tissue microarray analysis was used to determine MGST1 expression in GC. We analyzed 107 GC patients with detailed clinicopathological information. Kaplan–Meier analysis based on the tissue microarray analysis suggested that a high MGST1 expression presented a shorter OS time than those with a low MGST1 level (p < 0.001; Figure 1H). The part vision of the tissue microarray is shown in Figure 1I. Subsequently, the associations between MGST1 expression and clinical pathological features are listed in Table 1. Univariate analysis showed that lymph node metastasis, tumor node metastasis (TNM) stage, and MGST1 expression were prominently associated with the OS of GC patients. Taken together, these results verify that an upregulated MGST1 level is observed in GC and predicted poor prognosis.
Figure 1.
Expression of MGST1 is elevated in GC cells. (A) MGST1 mRNA levels were significantly increased in GC by TCGA database analysis. (B) MGST1 protein levels were increased in tumor tissues compared to normal tissues. (C) Relative expressions of B in bar graphs. (D) MGST1 mRNA levels were increased in tumor tissues compared to normal tissues. (E) MGST1 protein levels were increased in GC cell lines compared to GES-1. (F) Relative expressions of (E) in bar graphs. (G) MGST1 mRNA levels were increased in GC cell lines compared to GES-1. (H) OS curve of GC patients with high and low MGST1 expressions (log-rank test: p < 0.0001). (I) Representative images of MGST1 expression in tissue arrays.
Table 1. Clinicopathological Parameters and Cox Regression Analysis of the GC Patients (n = 107).
| univariate
analysis |
multivariate
analysis |
||||
|---|---|---|---|---|---|
| clinicopathological variables | total | HR (95% CI) | p-value | HR (95% CI) | p-value |
| gender | 1.039 (0.582–1.853) | 0.897 | |||
| male | 71 | ||||
| female | 36 | ||||
| age (y) | 1.192 (0.684–2.077) | 0.536 | |||
| <61 | 43 | ||||
| ≥61 | 64 | ||||
| tumor size (cm) | 0.693 (0.404–1.188) | 0.182 | |||
| <6 | 48 | ||||
| ≥6 | 59 | ||||
| differentiation | 1.670 (0.928–3.005) | 0.087 | |||
| high/moderate | 41 | ||||
| low/undifferentiated | 66 | ||||
| depth of invasion | 1.694 (0.869–3.300) | 0.122 | |||
| T1/T2 | 29 | ||||
| T3/T4 | 78 | ||||
| lymph node metastasis | 3.820 (1.513–9.644) | 0.005 | 1.476 (0.428–5.085) | 0.573 | |
| yes | 84 | ||||
| no | 23 | ||||
| TNM | 2.189 (1.125–4.260) | 0.021 | 0.553 (0.241–1.271) | 0.163 | |
| I/II | 34 | ||||
| III/IV | 73 | ||||
| location | 1.125 (0.626–2.023) | 0.693 | |||
| proximal | 36 | ||||
| distal | 71 | ||||
| MGST1 expression | 31.502 (12.671–78.320) | <0.001 | 0.029 (0.011–0.078) | <0.001 | |
| negative | 59 | ||||
| positive | 48 | ||||
3.2. MGST1-Silenced GC Cells Regulated Proliferation Negatively
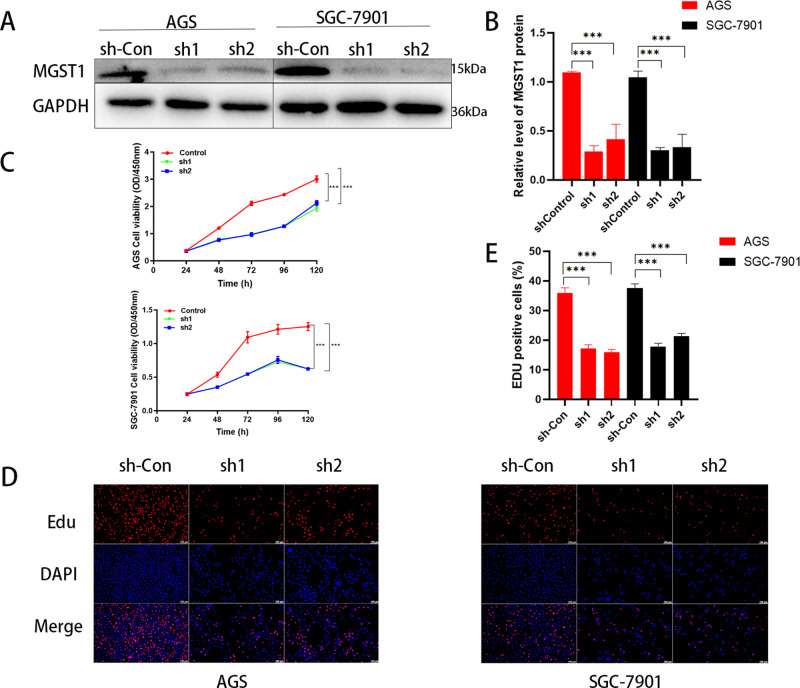
To evaluate the biology of MGST1 in GC cells, we transfected the AGS and SGC-7901 with MGS1-specific shRNA interference lentiviral vectors or a control shRNA vector. The knockdown efficiency in MGST1 shRNA in cells was measured by WB, and the result indicated that the protein level of MGST1 was markedly decreased in both AGS and SGC-7901 cells (Figure 2A,B). In light of the effect of MGST1 on cell proliferation, we performed CCK-8 and EDU assays. As is shown in CCK-8 curves (Figure 2C), the growth rate of shMGST1 groups were prominently lower than that of sh-control groups. Subsequently, the EDU results revealed that the number of cells of the shMGST1 groups was significantly less than that of the sh-control groups (Figure 2D,E).
Figure 2.
MGST1-silenced GC regulated proliferation negatively. (A) WB was used to verify the efficiency of MGST1 knockdown in both AGS and SGC-7901 cells. (B) Relative expressions of (A) in bar graphs. (C) Cell activity of GC cells was prominently decreased in MGST1 knockdown groups evaluated by CCK-8 assays. (D,E). EdU assays for GC cells after MGST1 silencing showed that MGST1 knockdown notably reduced cell growth in both AGS and SCC-7901 cells.
3.3. Silencing MGST1 Affected Migration, Invasion, and Cell Cycle of GC Cells
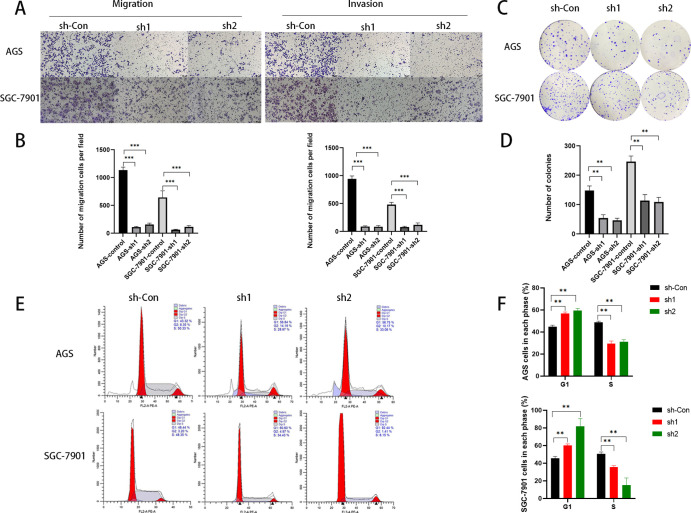
To further explore how MGST1 affects the progress of GC, we next performed transwell migration and invasion assays. In both transwell migration and invasion assays, the number of cells in sh-MGST1 groups was significantly less than the number in sh-control groups in both AGS and SGC-7901 cells (Figure 3A,B). These results indicated that depletion of MGST1 potently inhibited cell migration and invasion. The colony formation assay was operated to investigate the ability of MGST1 to migrate and invade further. In the colony formation assay, the number of sh-MGST1 cells was significantly less than sh-control cells (Figure 3C,D), and the results revealed that low expression of MGST1 suppresses the migration and invasion of GC cells. Moreover, in the cell cycle assay, MGST1 knockdown induced cell cycle arrest in the G1/S phase (Figure 3E,F).
Figure 3.
Silencing MGST1 affected migration, invasion, and cell cycle of GC cells. (A,B) Cell migration and invasion capacities were inhibited in MGST1 knockdown GC cells (both AGS and SGC-7901 cells). (C,D) In colony formation assays, the inhibition of cell clone formation was shown after MGST1 knockdown in both AGS and SGC-7901 cells. (E,F) The proportion of S phase cells was decreased and the proportion of G1 phase cells was increased after MGST1 knockdown in AGS and SGC-7901 cells.
3.4. MGST1 Regulated the Wnt/β-Catenin Pathway in GC Cells
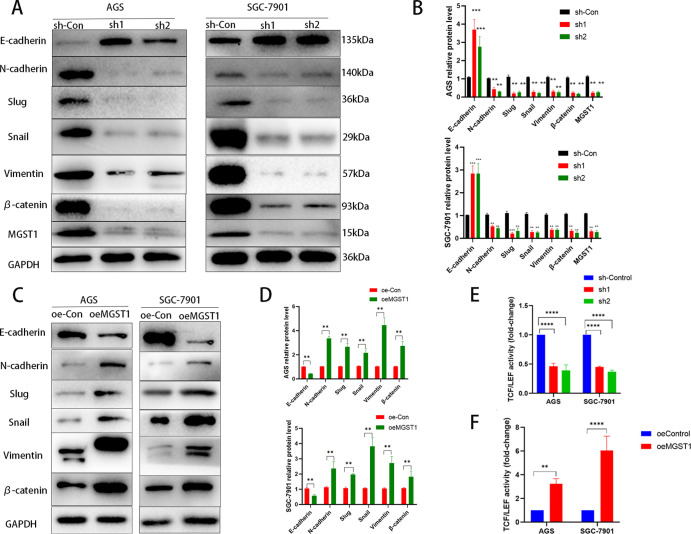
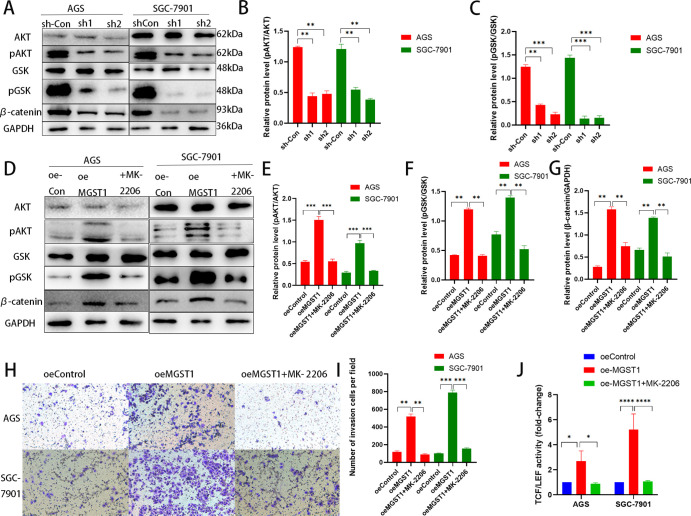
To explore whether factors in the Wnt/β-catenin pathway were regulated by MGST1 of GC cells, we performed a WB to determine protein levels in this pathway in sh-MGST1 GC cells. We found that knockdown of MGST1 significantly reduced the protein expression levels of N-cadherin, Slug, Snail, Vimentin, and β-catenin and increased the protein level of E-cadherin in the AGS and SGC-7901 cells (Figure 4A,B). Contrary to the results above, overexpression of MGST1 enhanced the expression of N-cadherin, Slug, Snail, Vimentin, and β-catenin and decreased the protein level of E-cadherin in the AGS and SGC-7901 cells (Figure 4C,D). TOP-Flash is a reporter plasmid used to detect β-catenin-mediated TCF/LEF transcriptional activity in the Wnt signaling pathway. The TOP-Flash reporter assay was used to further study the relationship between MGST1 and the Wnt/β-catenin pathway. Silencing of MGST1 reduced the activity of TCF/LEF transcription (Figure 4E) and over-expressing of MGST1 enhanced the activity of TCF/LEF transcription (Figure 4F). In this assay, the TCF/LEF transcription level of the control group was normalized to one. This suggested that MGST1 played an important role in activating the Wnt/β-catenin signaling pathway.
Figure 4.
MGST1 regulated the Wnt/β-catenin pathway in GC cells. (A) WB assays showed that knockdown of MGST1 significantly reduced the protein expression levels of N-cadherin, Slug, Snail, Vimentin, and β-catenin and increased the protein level of E-cadherin in the AGS and SGC-7901 cells. (B) Relative expressions of (A) in bar graphs (**: p < 0.01 compared with the sh-Con group; ***: p < 0.001 compared with the sh-Con group). (C) WB assays showed that overexpression of MGST1 enhanced the expression of N-cadherin, Slug, Snail, Vimentin, and β-catenin and decreased the protein level of E-cadherin in the AGS and SGC-7901 cells. (D) Relative expressions of C in bar graphs. (E) In the TOP-Flash reporter assay, TCF/LEF transcription was decreased in MGST1 knockdown GC cells. (F) Overexpression of MGST1 enhanced the activity of TCF/LEF transcription.
3.5. MGST1 Contributed to β-Catenin Activation through AKT
MK-2206 is an Allosteric AKT Inhibitor, which has the ability to suppress AKT phosphorylation.15 To deeper investigate MGST1 regulated Wnt/β-catenin pathway by regulating AKT/GSK-3β axis, knockdown of MGST1 reduced the phosphorylation of AKT and GSK-3β (Figure 5A,C). While forced expression of MGST1 enhanced the active β-catenin and phosphorylation of both AKT and GSK-3, treating GC cells with MK-2206 markedly reversed the enhancing effects on the Wnt/β-catenin pathway of MGST1 overexpression (Figure 5D–G). The transwell invasion assay of MK-2206 treatment showed that the increasing influence on proliferation and invasion of MGST1 overexpression in GC cells were both reversed (Figure 5H,I). In addition, the effect of MGST1 overexpression on the activity of TCF/LEF transcription was reversed the same as above (Figure 5J). In this assay, the TCF/LEF transcription level of the control group was normalized to one. Taken together, MGST1 contributed to β-catenin activation through AKT.
Figure 5.
MGST1 contributed to β-catenin activation through AKT. (A–C) In WB assay, knockdown of MGST1 reduced the phosphorylation of AKT and GSK-3β. In other words, pAKT/AKT and pGSK-3β/GSK-3β were reduced in MGST1-silencing GC cells. (D–G) Forced expression of MGST1 enhanced the pAKT/AKT, pGSK-3β/GSK-3β, and active β-catenin, and GC cells treated with MK-2206 markedly reversed these enhancing effects. (H,I) The transwell invasion assay of MK-2206 treating showed that the increasing influence on proliferation and invasion of MGST1 overexpression in GC cells were both reversed. (J) The increased activity of TCF/LEF transcription in MGST1-overexpressing GC cells was reversed after MK-2206 treatment (oe-Con: cells in the control group; oe-MGST1: MGST1-overexpressing cells; +MK-2206: MGST1-overexpressing cells treated with MK-2206).
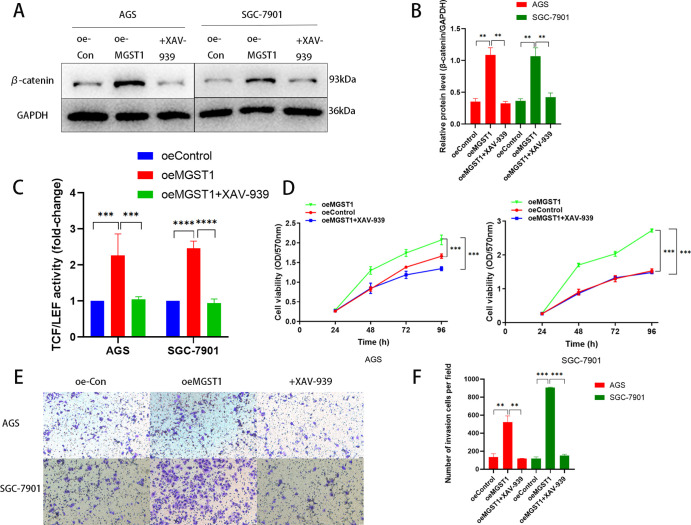
3.6. Inhibition of Wnt/β-Catenin Reserved the Promoting Effect of MGST1 in GC Cells
XAV-939 is a potent β-catenin inhibitor in the Wnt/β-catenin pathway, which has a remarkable role in inhibiting the activation of Wnt/β-catenin in MGST1-overexpressing cells (Figure 6A,B). As expected, inhibition of Wnt/β-catenin drastically reversed the MGST1-overexpressed enhancing effect on the activity of TCF/LEF transcription in the TOP-Flash assay (Figure 6C). In this assay, the TCF/LEF transcription level of the control group was normalized to one. Furthermore, in the CCK-8 curve, XAV-939 played a part in inhibiting proliferation (Figure 6D). The transwell invasion assay revealed that the function of invasion in MGST1-overexpressing cells was reversed by XAV-939 (Figure 6E,F). Collectively, these results indicated that MGST1 has a crucial role in pro-tumor by modulating Wnt/β-catenin through AKT in GC cells.
Figure 6.
Inhibiting Wnt/β-catenin reversed the promoting effect of MGST1 in GC cells. (A,B) In the WB assay, the level of β-catenin rose in MGST1-overexpressing GC cells, and this promoting effect was reversed after treating XAV-939 in AGS and SGC-7901 cells. (C) The enhanced effect of TCF/LEF transcription activity was inhibited by XAV-939 in AGS and SGC-7901 cells. (D) The CCK-8 assay showed that cell proliferation rose in MGST1-overexpressing GC cells, and this promoting effect was reversed after treating XAV-939 in AGS and SGC-7901 cells. (E,F) In the transwell invasion assay, the promoting effect of invasion in MGST1-overexpressing cells was reversed by XAV-939 in AGS and SGC-7901 cells (oe-Con: cells in the control group; oe-MGST1: MGST1-overexpressing cells; +XAV-939: MGST1-overexpressing cells treated with XAV-939).
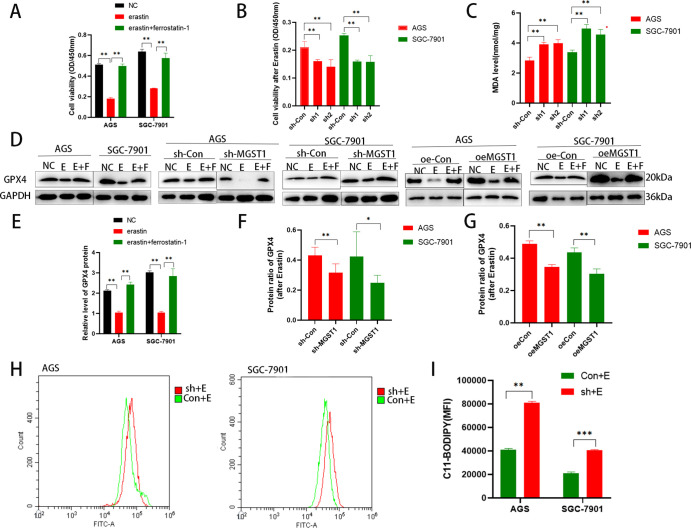
3.7. MGST1 Is a Negative Factor of Ferroptosis in GC Cells
To determine the association between MGST1 and ferroptosis, we analyzed the Erastin activity in AGS and SGC-7901 cells. We treated GC cells with Erastin (10 μM, 24 h), which is a ferroptosis inducer through triggering iron-dependent cell death11 and Ferrostatin-1 (10 μM, 24 h), which plays a potent role in suppressing Erastin-induced ferroptosis.38 As shown in the CCK-8 assay, Erastin-induced cell death was inhibited by the ferroptosis inhibitors Ferrostatin-1 in AGS and SGC-7901 cells (Figure 7A). Next, we studied in detail the mechanism of MGST1 in ferroptosis. After treating GC cells with Erastin, the cell viability of shMGST1 cells was significantly lower than that of the cells in the corresponding control (sh-Con) group (Figure 7B). To further study the role of MGST1 in ferroptosis, we analyzed lipid peroxidation, which is a key factor of ferroptosis. After treating GC cells with Erastin, the knockdown of MGST1 increased the MDA level of GC cells compared to the sh-Con groups (Figure 7C), which indicated that the lipid peroxidation increased. GPX4 is a ferroptosis marker, which protects cells from lipid peroxidation of the membrane. After Erastin treatment of GC cells, the GPX4 protein levels also declined in the MGST1 knockdown and MGST1-overexpressed GC cells. WB further confirmed that the decline of GPX4 protein in the MGST1 silencing group was more significant than that of the corresponding control group. However, the decline of the GPX4 protein in the MGST1 overexpression group was smaller than in the corresponding control group (Figure 7D–G). These above results showed that knockdown MGST1 promoted ferroptosis and overexpression of MGST1 inhibited ferroptosis, which suggested that MGST1 may have an inhibitory effect on ferroptosis in GC cells. The C11 BODIPY 581/591 lipid peroxidation probe assay further verified the increase in lipid ROS in MGST1-knockdown GC cells (Figure 7H,I). Taken together, these results showed that compared with the NC group, sh-MGST1 cells had increased and oeMGST1 cells had decreased sensitivity to the ferroptosis activator (Erastin). Therefore, MGST1 is a negative factor in ferroptosis in GC cells.
Figure 7.
MGST1 is a negative factor of ferroptosis in GC cells. (A) The CCK-8 assay showed that cell proliferation rose after Erastin treatment and was reversed by Ferrostatin-1 in GC cells. (B) After treating GC cells with Erastin, the cell viability of shMGST1 cells was significantly lower than that of the cells in the corresponding control (sh-Con) group. (C) After treating GC cells with Erastin, the knockdown of MGST1 increased the MDA level of GC cells compared to sh-Con groups. (D) In the WB assay, the level of GPX4 rose after Erastin treatment, and the protein level could be reversed after Ferrostatin-1 treatment in GC cells. (E) Quantification of GPX4 protein levels of normal untreated GC cells in histograms. (F) Quantification of GPX4 protein levels of MGST1 knockdown GC cells. The protein ratio of GPX4 referred to the ratio of GPX4 protein levels after Erastin treatment divided by the GPX4 protein levels in the NC group. (G) Quantification of GPX4 protein levels of MGST1-overexpressing GC cells. (H,I) The C11 BODIPY 581/591 lipid peroxidation probe assay showed an increase in lipid ROS in MGST1-knockdown GC cells (NC: Negative control group; E: erastin group; E + F: both erastin and ferrostatin-1 group; sh-Con: control group in downregulation; sh-MGST1: MGST1-knockdown group; oe-Con: control group in overexpression; oeMGST1: MGST1-overexpressing group).
4. Discussion
MGST1 has a confirmed role in protecting membranes from membrane-embedded lipid peroxidation and cells from oxidative stress by biotransformation of lipophilic reactive electrophiles.19 Furthermore, there are a mass of cellular nucleophiles that are vulnerable to being affected by electrophiles, leading to immunogenic responses and drug resistance.31 Previous data show that the MGST1 is upregulated in several human cancer types, such as retinal tumor, glioma, and esophageal adenocarcinoma.6,16,26 In addition, MGST1 determinately has an association with ferroptosis according to recent research.55 Until now, whether the MGST1 level is altered in GC has not been thoroughly studied.
To discover the expression of MGST1 in GC cells, we investigated it via the TCGA database and found that high MGST1 expression was found in GC tissues compared with non-tumor tissues. RT-qPCR and WB were done to support evidence that MGST1 mRNA and protein levels were upregulated in both GC tissues and GC cell lines compared to normal stomach tissues and gastric mucosa epithelial cells. Therefore, this study proved that MGST1 is upregulated in GC. Subsequently, we analyzed tissue microarray cores in 107 GC patients. High MGST1 level was associated with declined overall survival of patients, suggesting a prognostic value of MGST1. In short, MGST1 was expressed highly in GC and associated with poor prognosis.
Next, we investigated the function of MGST1 in GC cells through CCK-8 assays, EdU assays, Transwell migration and invasion assays, and cell cycle assays. The results showed that the downregulation of MGST1 prominently inhibited cell proliferation, invasion, migration, and cell cycle in AGS and SGC-7901 cells, and these were reversed in upregulating MGST1 GC cells. Previous studies showed that MGST1 downregulation inhibits the proliferation invasion and migration of lung adenocarcinoma cells56 and glioma.50 However, the effect of MGST1 on the cell cycle has been not studied. In our study, we found that MGST1 knockdown induced cell cycle arrest in the G1/S phase. Therefore, a decrease in MGST1 expression may influence preparing of DNA synthesis in the G1 phase and DNA synthesis in the S phase. To sum up, MGST1 is equipped to affect the cell functions of GC cells.
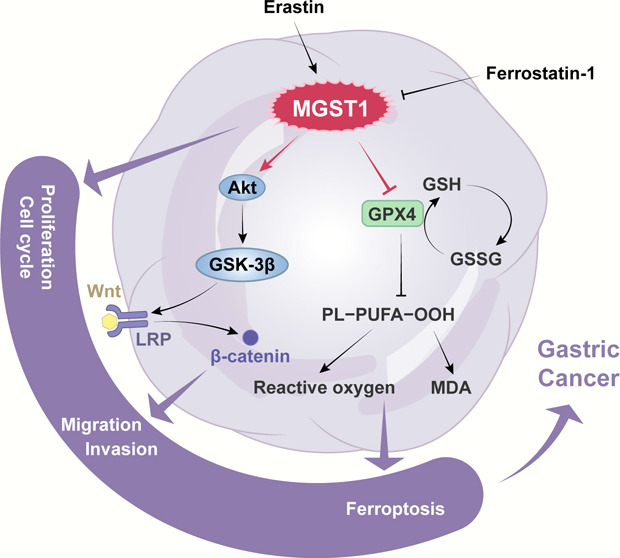
Moreover, emerging data showed that MGST1 had a confirmed effect on escape to anti-growth signal pathways,7 and MGST1 was a novel regulator of the Wnt/β-catenin pathway.4 We examined the proteins in the AKT/GSK-3β signaling pathway of MGST1 downregulated and upregulated AGS and SGC-7901 cells, and the β-catenin protein expressions were reduced in MGST1 knockdown cells and increased in MGST1 upregulated cells. That result indicated that MGST1 had a regulatory effect on the Wnt/β-catenin pathway in GC cells. Further, AKT/GSK-3β has a well-determined association with MGST1 in lung cancer.55 In our research, inhibiting AKT by MK-2206 abolished the regulatory effect of MGST1 on the Wnt/β-catenin pathway, and inactivating β-catenin by XAV-939 got a similar result. Activated β-catenin enters into the cell nucleus and combines with the TCF/LEF family of transcription factors to increase the expression of target genes.45 According to existing research, when the Wnt signaling pathway is activated, β-catenin cannot be phosphorylated and is transported into the nucleus.13 β-Catenin and TCF/LEF form a transcription complex in the nucleus, which activates the Wnt signaling pathway and the downstream genes in the Wnt signaling pathway such as C-myC and cyclin D1.48 Therefore, determining the activity of TCF/LEF shows the activation of the Wnt/β-catenin signaling pathway. Our results indicated that silencing of MGST1 declined the activity of β-catenin-TCF/LEF-mediated transcription and upregulating of MGST1 reversed this result. In summary, our study proves that MGST1 has a tumor-promoting function in GC cells by regulating the Wnt/β-catenin pathway via the effects on AKT. In other words, MGST1 modulated cellular functions by the AKT/GSK-3β/β-catenin axis in GC cells.
A previous study has shown that MGST1 had an important role in inhibiting ferroptosis in pancreatic cancer cells.35 Ferroptosis is a type of programmed cell death, which regulates cell death depending on iron and ROS accumulation.32 GPX4 transforms lipid hydroperoxides to lipid alcohols,46 and low activity of GPX4 triggers a high level of lipid ROS in the cells.52 Our current study explored the relationship between MGST1 and ferroptosis in the GC cells. In our study, the cell viability in Erastin-induced GC cells, proteins in ferroptosis, MDA level, and lipid ROS was reduced in GC cells under the influence of MGST1. In addition, Erastin was used in our study to induce ferroptosis and Ferrostatin-1 was used to reverse ferroptosis induced by Erastin, and the results showed that ferroptosis was more prominently in the sh-MGST1 GC cell and Erastin induced lower level of ferroptosis in MGST1-overexpressing GC cells compared to GC cells in the control group. These results indicated that MGST1 is a negative regulator of ferroptosis through limiting lipid peroxidation during ferroptosis in GC cells. In our current study, therefore, targeting the MGST1 may enhance ferroptosis-based treatment, such as Erastin and sorafenib.
5. Conclusions
In conclusion, MGST1 expression is associated with poor prognosis, enhancing the Wnt/β-catenin pathway via regulating AKT, and inhibiting ferroptosis in GC cells. Our results suggested that MGST1 may be a potential therapeutic target in GC.
Acknowledgments
This work was supported by Wang Mingliang, Li Jing, Wu Youliang, and Wang Huizhen.
Author Contributions
Yaxian Li. completed the conceptualization, methodology, and software. X.X. carried out the data curation and writing—original draft preparation. X.W. performed the visualization and investigation. C.Z. executed the supervision. A.H. implemented the software and validation. Yongxiang Li conducted the writing—reviewing and editing.
This research was funded by the National Natural Science Foundation, grant number 81874063. This work was also supported by the grants from Scientific Research Project Plan for Anhui Universities (2022AH051192).
Informed Consent Statement: Informed consent was obtained from all subjects involved in the study. All 107 patients who were investigated in this study or their family members signed the Statement of Written Informed Consent Form according to the Research Ethics Review Committee (ERC).
Ethics Approval Statement: The study approval of the first affiliated hospital of Anhui medical university the committee on medical ethics (approval code: Quick-PJ 2021-13-23). According to the Ministry of Health Ethics review on biomedical research involving human subjects, the declaration of Helsinki and International ethical guidelines for biomedical research involving the committee has agreed to report the review results, carry out the research, and publish the research results.
The authors declare no competing financial interest.
References
- Akil F.; Akil H. A.; Lutfie A. M.; Wibowo W. S.; Miskad U.; Yusuf I. The Role of Xenobotic Metabolism Mgst1 Gene Polymorphism in Colorectal Cancer Patients. Acta Med. Indones. 2012, 44, 284–289. [PubMed] [Google Scholar]
- Bai J.; Sata N.; Nagai H. Gene Expression Analysis for Predicting Gemcitabine Sensitivity in Pancreatic Cancer Patients. HPB 2007, 9, 150–155. 10.1080/13651820601175918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertucci F.; Pages C.; Finetti P.; Rochaix P.; Lamant L.; Devilard E.; Nguyen C.; Houlgatte R.; Birnbaum D.; Xerri L.; Brousset P. Gene Expression Profiling of Human Melanoma Cell Lines with Distinct Metastatic Potential Identifies New Progression Markers. Anticancer Res. 2007, 27, 3441–3449. [PubMed] [Google Scholar]
- Brautigam L.; Zhang J.; Dreij K.; Spahiu L.; Holmgren A.; Abe H.; Tew K. D.; Townsend D. M.; Kelner M. J.; Morgenstern R.; Johansson K. Mgst1, a Gsh Transferase/Peroxidase Essential for Development and Hematopoietic Stem Cell Differentiation. Redox Biol. 2018, 17, 171–179. 10.1016/j.redox.2018.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray F.; Ferlay J.; Soerjomataram I.; Siegel R. L.; Torre L. A.; Jemal A. Global Cancer Statistics 2018: Globocan Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. Ca - Cancer J. Clin. 2018, 68, 394–424. 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- Buas M. F.; He Q.; Johnson L. G.; Onstad L.; Levine D. M.; Thrift A. P.; Gharahkhani P.; Palles C.; Lagergren J.; Fitzgerald R. C.; Ye W.; Caldas C.; Bird N. C.; Shaheen N. J.; Bernstein L.; Gammon M. D.; Wu A. H.; Hardie L. J.; Pharoah P. D.; Liu G.; Iyer P.; Corley D. A.; Risch H. A.; Chow W. H.; Prenen H.; Chegwidden L.; Love S.; Attwood S.; Moayyedi P.; MacDonald D.; Harrison R.; Watson P.; Barr H.; DeCaestecker J.; Tomlinson I.; Jankowski J.; Whiteman D. C.; MacGregor S.; Vaughan T. L.; Madeleine M. M. Germline Variation in Inflammation-Related Pathways and Risk of Barrett’s Oesophagus and Oesophageal Adenocarcinoma. Guttmacher Policy Rev. 2017, 66, 1739–1747. 10.1136/gutjnl-2016-311622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calcaterra V.; Chiricosta L.; Mazzon E.; Gugnandolo A.; Alberti D.; Maestri L.; Meroni M.; Vestri E.; Verduci E.; Dilillo D.; Zuccotti G.; Pelizzo G. Determining Oncogenic Patterns and Cancer Predisposition through the Transcriptomic Profile in Mitchell-Riley Syndrome with Heterotopic Gastric Mucosa and Duodenal Atresia: A Case Report. Orphanet J. Rare Dis. 2021, 16, 455. 10.1186/s13023-021-02093-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaib H.; Cockrell E. K.; Rubin M. A.; Macoska J. A. Profiling and Verification of Gene Expression Patterns in Normal and Malignant Human Prostate Tissues by Cdna Microarray Analysis. Neoplasia 2001, 3, 43–52. 10.1038/sj.neo.7900126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis J. L.; Ripley R. T. Postgastrectomy Syndromes and Nutritional Considerations Following Gastric Surgery. Surg. Clin. North Am. 2017, 97, 277–293. 10.1016/j.suc.2016.11.005. [DOI] [PubMed] [Google Scholar]
- Dixon S. J.; Lemberg K. M.; Lamprecht M. R.; Skouta R.; Zaitsev E. M.; Gleason C. E.; Patel D. N.; Bauer A. J.; Cantley A. M.; Yang W. S.; Morrison B. R.; Stockwell B. R. Ferroptosis: An Iron-Dependent Form of Nonapoptotic Cell Death. Cell 2012, 149, 1060–1072. 10.1016/j.cell.2012.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon S. J.; Lemberg K. M.; Lamprecht M. R.; Skouta R.; Zaitsev E. M.; Gleason C. E.; Patel D. N.; Bauer A. J.; Cantley A. M.; Yang W. S.; Morrison B. R.; Stockwell B. R. Ferroptosis: An Iron-Dependent Form of Nonapoptotic Cell Death. Cell 2012, 149, 1060–1072. 10.1016/j.cell.2012.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan G.; Ye D.; Zhu S.; Xi J.; Guo X.; Qiao J.; Wu Y.; Jia W.; Wang G.; Fan G.; Kang J. Rtl1 Promotes Melanoma Proliferation by Regulating Wnt/Beta-Catenin Signalling. Oncotarget 2017, 8, 106026–106037. 10.18632/oncotarget.22523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flentke G. R.; Garic A.; Hernandez M.; Smith S. M. Camkii Represses Transcriptionally Active Beta-Catenin to Mediate Acute Ethanol Neurodegeneration and Can Phosphorylate Beta-Catenin. J. Neurochem. 2014, 128, 523–535. 10.1111/jnc.12464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao S.; Yu J.; He W.; Huang Q.; Zhao Y.; Liang B.; Zhang S.; Wen Z.; Dong S.; Rao J.; Liao W.; Shi M. Cysteine Dioxygenase 1 Mediates Erastin-Induced Ferroptosis in Human Gastric Cancer Cells. Neoplasia 2017, 19, 1022–1032. 10.1016/j.neo.2017.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirai H.; Sootome H.; Nakatsuru Y.; Miyama K.; Taguchi S.; Tsujioka K.; Ueno Y.; Hatch H.; Majumder P. K.; Pan B. S.; Kotani H. MK-2206, an Allosteric Akt Inhibitor, Enhances Antitumor Efficacy by Standard Chemotherapeutic Agents Or Molecular Targeted Drugs in Vitro and in Vivo. Mol. Cancer Therapeut. 2010, 9, 1956–1967. 10.1158/1535-7163.mct-09-1012. [DOI] [PubMed] [Google Scholar]
- Johansson K.; Ito M.; Schophuizen C. M.; Mathew Thengumtharayil S.; Heuser V. D.; Zhang J.; Shimoji M.; Vahter M.; Ang W. H.; Dyson P. J.; Shibata A.; Shuto S.; Ito Y.; Abe H.; Morgenstern R. Characterization of New Potential Anticancer Drugs Designed to Overcome Glutathione Transferase Mediated Resistance. Mol. Pharm. 2011, 8, 1698–1708. 10.1021/mp2000692. [DOI] [PubMed] [Google Scholar]
- Kakeji Y.; Yoshida K.; Kodera Y.; Kochi M.; Sano T.; Ichikawa W.; Lee S. W.; Shibahara K.; Shikano T.; Kataoka M.; Ishiguro A.; Ojima H.; Sakai Y.; Musha N.; Takase T.; Kimura T.; Takeuchi M.; Fujii M. Three-Year Outcomes of a Randomized Phase Iii Trial Comparing Adjuvant Chemotherapy with S-1 Plus Docetaxel Versus S-1 Alone in Stage Iii Gastric Cancer: Jaccro Gc-07. Gastric Cancer 2022, 25, 188. 10.1007/s10120-021-01224-2. [DOI] [PubMed] [Google Scholar]
- Kang Y. K.; Yook J. H.; Park Y. K.; Lee J. S.; Kim Y. W.; Kim J. Y.; Ryu M. H.; Rha S. Y.; Chung I. J.; Kim I. H.; Oh S. C.; Park Y. S.; Son T.; Jung M. R.; Heo M. H.; Kim H. K.; Park C.; Yoo C. H.; Choi J. H.; Zang D. Y.; Jang Y. J.; Sul J. Y.; Kim J. G.; Kim B. S.; Beom S. H.; Cho S. H.; Ryu S. W.; Kook M. C.; Ryoo B. Y.; Kim H. K.; Yoo M. W.; Lee N. S.; Lee S. H.; Kim G.; Lee Y.; Lee J. H.; Noh S. H. Prodigy: A Phase Iii Study of Neoadjuvant Docetaxel, Oxaliplatin, and S-1 Plus Surgery and Adjuvant S-1 Versus Surgery and Adjuvant S-1 for Resectable Advanced Gastric Cancer. J. Clin. Oncol. 2021, 39, 2903–2913. 10.1200/jco.20.02914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelner M. J.; Bagnell R. D.; Montoya M. A.; Estes L. A.; Forsberg L.; Morgenstern R. Structural Organization of the Microsomal Glutathione S-Transferase Gene (Mgst1) On Chromosome 12P13.1-13.2. Identification of the Correct Promoter Region and Demonstration of Transcriptional Regulation in Response to Oxidative Stress. J. Biol. Chem. 2000, 275, 13000–13006. 10.1074/jbc.275.17.13000. [DOI] [PubMed] [Google Scholar]
- Kelner M. J.; Diccianni M. B.; Yu A. L.; Rutherford M. R.; Estes L. A.; Morgenstern R. Absence of Mgst1 Mrna and Protein Expression in Human Neuroblastoma Cell Lines and Primary Tissue. Free Radic. Biol. Med. 2014, 69, 167–171. 10.1016/j.freeradbiomed.2014.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan M. I.; Debski K. J.; Dabrowski M.; Czarnecka A. M.; Szczylik C. Gene Set Enrichment Analysis and Ingenuity Pathway Analysis of Metastatic Clear Cell Renal Cell Carcinoma Cell Line. Am. J. Physiol. Ren. Physiol. 2016, 311, F424–F436. 10.1152/ajprenal.00138.2016. [DOI] [PubMed] [Google Scholar]
- Kretzschmar K.; Clevers H. Wnt/Beta-Catenin Signaling in Adult Mammalian Epithelial Stem Cells. Dev. Biol. 2017, 428, 273–282. 10.1016/j.ydbio.2017.05.015. [DOI] [PubMed] [Google Scholar]
- Kuang F.; Liu J.; Xie Y.; Tang D.; Kang R. Mgst1 is a Redox-Sensitive Repressor of Ferroptosis in Pancreatic Cancer Cells. Cell Chem. Biol. 2021, 28, 765–775.e5. 10.1016/j.chembiol.2021.01.006. [DOI] [PubMed] [Google Scholar]
- Li C.; Zhang G.; Zhao L.; Ma Z.; Chen H. Metabolic Reprogramming in Cancer Cells: Glycolysis, Glutaminolysis, and Bcl-2 Proteins as Novel Therapeutic Targets for Cancer. World J. Surg. Oncol. 2016, 14, 15. 10.1186/s12957-016-0769-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linnerth N. M.; Sirbovan K.; Moorehead R. A. Use of a Transgenic Mouse Model to Identify Markers of Human Lung Tumors. Int. J. Cancer 2005, 114, 977–982. 10.1002/ijc.20814. [DOI] [PubMed] [Google Scholar]
- Maeda A.; Crabb J. W.; Palczewski K. Microsomal Glutathione S-Transferase 1 in the Retinal Pigment Epithelium: Protection Against Oxidative Stress and a Potential Role in Aging. Biochemistry 2005, 44, 480–489. 10.1021/bi048016f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgenstern R.; Zhang J.; Johansson K. Microsomal Glutathione Transferase 1: Mechanism and Functional Roles. Drug Metab. Rev. 2011, 43, 300–306. 10.3109/03602532.2011.558511. [DOI] [PubMed] [Google Scholar]
- Morgenstern R.; Zhang J.; Johansson K. Microsomal Glutathione Transferase 1: Mechanism and Functional Roles. Drug Metab. Rev. 2011, 43, 300–306. 10.3109/03602532.2011.558511. [DOI] [PubMed] [Google Scholar]
- Nusse R.; Clevers H. Wnt/Beta-Catenin Signaling, Disease, and Emerging Therapeutic Modalities. Cell 2017, 169, 985–999. 10.1016/j.cell.2017.05.016. [DOI] [PubMed] [Google Scholar]
- Oliveira C.; Pinheiro H.; Figueiredo J.; Seruca R.; Carneiro F. Familial Gastric Cancer: Genetic Susceptibility, Pathology, and Implications for Management. Lancet Oncol. 2015, 16, e60–e70. 10.1016/s1470-2045(14)71016-2. [DOI] [PubMed] [Google Scholar]
- Pumford N. R.; Halmes N. C. Protein Targets of Xenobiotic Reactive Intermediates. Annu. Rev. Pharmacol. Toxicol. 1997, 37, 91–117. 10.1146/annurev.pharmtox.37.1.91. [DOI] [PubMed] [Google Scholar]
- Qiao Y.; Wang B.; Yang J. J.; Fan Y. F.; Guo Q.; Dou Z. J.; Huang Y. Q.; Feng T. T.; Wang S. J.; An D. D.; Gao X. L. Bone Metabolic Markers in Patients with Obstructive Sleep Apnea Syndrome. Chin. Med. J. 2018, 131, 1898–1903. 10.4103/0366-6999.238149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren S.; Cai P.; Liu Y.; Wang T.; Zhang Y.; Li Q.; Gu Y.; Wei L.; Yan C.; Jin G. Prevalence of Helicobacter Pylori Infection in China: A Systematic Review and Meta-Analysis. J. Gastroenterol. Hepatol. 2022, 37, 464. 10.1111/jgh.15751. [DOI] [PubMed] [Google Scholar]
- Reya T.; Clevers H. Wnt Signalling in Stem Cells and Cancer. NATURE 2005, 434, 843–850. 10.1038/nature03319. [DOI] [PubMed] [Google Scholar]
- Sacks D.; Baxter B.; Campbell B.; Carpenter J. S.; Cognard C.; Dippel D.; Eesa M.; Fischer U.; Hausegger K.; Hirsch J. A.; Hussain M. S.; Jansen O.; Jayaraman M. V.; Khalessi A. A.; Kluck B. W.; Lavine S.; Meyers P. M.; Ramee S.; Rufenacht D. A.; Schirmer C. M.; Vorwerk D. Multisociety Consensus Quality Improvement Revised Consensus Statement for Endovascular Therapy of Acute Ischemic Stroke. Int. J. Stroke 2018, 13, 612–632. 10.1177/1747493018778713. [DOI] [PubMed] [Google Scholar]
- Scotlandi K.; Remondini D.; Castellani G.; Manara M. C.; Nardi F.; Cantiani L.; Francesconi M.; Mercuri M.; Caccuri A. M.; Serra M.; Knuutila S.; Picci P. Overcoming Resistance to Conventional Drugs in Ewing Sarcoma and Identification of Molecular Predictors of Outcome. J. Clin. Oncol. 2009, 27, 2209–2216. 10.1200/jco.2008.19.2542. [DOI] [PubMed] [Google Scholar]
- Scotlandi K.; Remondini D.; Castellani G.; Manara M. C.; Nardi F.; Cantiani L.; Francesconi M.; Mercuri M.; Caccuri A. M.; Serra M.; Knuutila S.; Picci P. Overcoming Resistance to Conventional Drugs in Ewing Sarcoma and Identification of Molecular Predictors of Outcome. J. Clin. Oncol. 2009, 27, 2209–2216. 10.1200/jco.2008.19.2542. [DOI] [PubMed] [Google Scholar]
- Skouta R.; Dixon S. J.; Wang J.; Dunn D. E.; Orman M.; Shimada K.; Rosenberg P. A.; Lo D. C.; Weinberg J. M.; Linkermann A.; Stockwell B. R. Ferrostatins Inhibit Oxidative Lipid Damage and Cell Death in Diverse Disease Models. J. Am. Chem. Soc. 2014, 136, 4551–4556. 10.1021/ja411006a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith P. J.; Furon E.; Wiltshire M.; Campbell L.; Feeney G. P.; Snyder R. D.; Errington R. J. Abcg2-Associated Resistance to Hoechst 33342 and Topotecan in a Murine Cell Model with Constitutive Expression of Side Population Characteristics. Cytometry, Part A 2009, 75, 924–933. 10.1002/cyto.a.20800. [DOI] [PubMed] [Google Scholar]
- Sobczak A.; Zajac M.; Malinka W.; Redzicka A. Mechanism of Solvolysis of N-[2-(4-O-Fluorophenylpiperazin-1-Yl)Ethyl]-2,5-Dimethyl-1-Phenylpyrrole-3,4-Dica Rboximide (Pdi). Acta Pol. Pharm. 2010, 67, 225–232. [PubMed] [Google Scholar]
- Taciak B.; Pruszynska I.; Kiraga L.; Bialasek M.; Krol M.. Wnt Signaling Pathway in Development and Cancer. J. Physiol. Pharmacol. 2018, 69(), 10.26402/jpp.2018.2.07. [DOI] [PubMed] [Google Scholar]
- Torresano L.; Nuevo-Tapioles C.; Santacatterina F.; Cuezva J. M. Metabolic Reprogramming and Disease Progression in Cancer Patients. Biochim. Biophys. Acta, Mol. Basis Dis. 2020, 1866, 165721. 10.1016/j.bbadis.2020.165721. [DOI] [PubMed] [Google Scholar]
- van der Post R. S.; Oliveira C.; Guilford P.; Carneiro F. Hereditary Gastric Cancer: What’s New? Update 2013-2018. Fam. Cancer 2019, 18, 363–367. 10.1007/s10689-019-00127-7. [DOI] [PubMed] [Google Scholar]
- van Kappel E. C.; Maurice M. M. Molecular Regulation and Pharmacological Targeting of the Beta-Catenin Destruction Complex. Br. J. Pharmacol. 2017, 174, 4575–4588. 10.1111/bph.13922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veeman M. T.; Axelrod J. D.; Moon R. T. A Second Canon. Functions and Mechanisms of Beta-Catenin-Independent Wnt Signaling. Dev. Cell 2003, 5, 367–377. 10.1016/s1534-5807(03)00266-1. [DOI] [PubMed] [Google Scholar]
- Xie Y.; Hou W.; Song X.; Yu Y.; Huang J.; Sun X.; Kang R.; Tang D. Ferroptosis: Process and Function. Cell Death Differ. 2016, 23, 369–379. 10.1038/cdd.2015.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Y.; Zhu S.; Song X.; Sun X.; Fan Y.; Liu J.; Zhong M.; Yuan H.; Zhang L.; Billiar T. R.; Lotze M. T.; Zeh H. J.; Kang R.; Kroemer G.; Tang D. The Tumor Suppressor P53 Limits Ferroptosis by Blocking Dpp4 Activity. Cell Rep. 2017, 20, 1692–1704. 10.1016/j.celrep.2017.07.055. [DOI] [PubMed] [Google Scholar]
- Yamaguchi K.; Zhu C.; Ohsugi T.; Yamaguchi Y.; Ikenoue T.; Furukawa Y. Bidirectional Reporter Assay Using Hal Promoter and Topflash Improves Specificity in High-Throughput Screening of Wnt Inhibitors. Biotechnol. Bioeng. 2017, 114, 2868–2882. 10.1002/bit.26394. [DOI] [PubMed] [Google Scholar]
- Yang B.; Xia S.; Ye X.; Jing W.; Wu B. Mir-379-5P Targets Microsomal Glutathione Transferase 1 (Mgst1) to Regulate Human Glioma in Cell Proliferation, Migration and Invasion and Epithelial-Mesenchymal Transition (Emt). Biochem. Biophys. Res. Commun. 2021, 568, 8–14. 10.1016/j.bbrc.2021.05.099. [DOI] [PubMed] [Google Scholar]
- Yang B.; Xia S.; Ye X.; Jing W.; Wu B. Mir-379-5P Targets Microsomal Glutathione Transferase 1 (Mgst1) to Regulate Human Glioma in Cell Proliferation, Migration and Invasion and Epithelial-Mesenchymal Transition (Emt). Biochem. Biophys. Res. Commun. 2021, 568, 8–14. 10.1016/j.bbrc.2021.05.099. [DOI] [PubMed] [Google Scholar]
- Yang J.; Han F.; Liu W.; Chen H.; Hao X.; Jiang X.; Yin L.; Huang Y.; Cao J.; Zhang H.; Liu J. Alx4, an Epigenetically Down Regulated Tumor Suppressor, Inhibits Breast Cancer Progression by Interfering Wnt/Beta-Catenin Pathway. J. Exp. Clin. Cancer Res. 2017, 36, 170. 10.1186/s13046-017-0643-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W. S.; SriRamaratnam R.; Welsch M. E.; Shimada K.; Skouta R.; Viswanathan V. S.; Cheah J. H.; Clemons P. A.; Shamji A. F.; Clish C. B.; Brown L. M.; Girotti A. W.; Cornish V. W.; Schreiber S. L.; Stockwell B. R. Regulation of Ferroptotic Cancer Cell Death by Gpx4. Cell 2014, 156, 317–331. 10.1016/j.cell.2013.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying S.-Y.; Chang D. C.; Lin S. L.. The Microrna. Methods in Molecular Biology; Humana Press, 2018, 1733. [DOI] [PubMed] [Google Scholar]
- Zeng B.; Ge C.; Li R.; Zhang Z.; Fu Q.; Li Z.; Lin Z.; Liu L.; Xue Y.; Xu Y.; He J.; Guo H.; Li C.; Huang W.; Song X.; Huang Y. Knockdown of Microsomal Glutathione S-Transferase 1 Inhibits Lung Adenocarcinoma Cell Proliferation and Induces Apoptosis. Biomed. Pharmacother. 2020, 121, 109562. 10.1016/j.biopha.2019.109562. [DOI] [PubMed] [Google Scholar]
- Zeng B.; Ge C.; Li R.; Zhang Z.; Fu Q.; Li Z.; Lin Z.; Liu L.; Xue Y.; Xu Y.; He J.; Guo H.; Li C.; Huang W.; Song X.; Huang Y. Knockdown of Microsomal Glutathione S-Transferase 1 Inhibits Lung Adenocarcinoma Cell Proliferation and Induces Apoptosis. Biomed. Pharmacother. 2020, 121, 109562. 10.1016/j.biopha.2019.109562. [DOI] [PubMed] [Google Scholar]
- Zeng B.; Ge C.; Li R.; Zhang Z.; Fu Q.; Li Z.; Lin Z.; Liu L.; Xue Y.; Xu Y.; He J.; Guo H.; Li C.; Huang W.; Song X.; Huang Y. Knockdown of Microsomal Glutathione S-Transferase 1 Inhibits Lung Adenocarcinoma Cell Proliferation and Induces Apoptosis. Biomed. Pharmacother. 2020, 121, 109562. 10.1016/j.biopha.2019.109562. [DOI] [PubMed] [Google Scholar]
- Zhang H.; Liao L. H.; Liu S. M.; Lau K. W.; Lai A. K.; Zhang J. H.; Wang Q.; Chen X. Q.; Wei W.; Liu H.; Cai J. H.; Lung M. L.; Tai S. S.; Wu M. Microsomal Glutathione S-Transferase Gene Polymorphisms and Colorectal Cancer Risk in a Han Chinese Population. Int. J. Colorectal Dis. 2007, 22, 1185–1194. 10.1007/s00384-007-0308-9. [DOI] [PubMed] [Google Scholar]