Abstract
Progressive pulmonary fibrosis results from a dysfunctional tissue repair response and is characterized by fibroblast proliferation, activation, and invasion and extracellular matrix accumulation. Lung fibroblast heterogeneity is well recognized. With single-cell RNA sequencing, fibroblast subtypes have been reported by recent studies. However, the roles of fibroblast subtypes in effector functions in lung fibrosis are not well understood. In this study, we incorporated the recently published single-cell RNA-sequencing datasets on murine lung samples of fibrosis models and human lung samples of fibrotic diseases and analyzed fibroblast gene signatures. We identified and confirmed the novel fibroblast subtypes we reported recently across all samples of both mouse models and human lung fibrotic diseases, including idiopathic pulmonary fibrosis, systemic sclerosis–associated interstitial lung disease, and coronavirus disease (COVID-19). Furthermore, we identified specific cell surface proteins for each fibroblast subtype through differential gene expression analysis, which enabled us to isolate primary cells representing distinct fibroblast subtypes by flow cytometry sorting. We compared matrix production, including fibronectin, collagen, and hyaluronan, after profibrotic factor stimulation and assessed the invasive capacity of each fibroblast subtype. Our results suggest that in addition to myofibroblasts, lipofibroblasts and Ebf1+ (Ebf transcription factor 1+) fibroblasts are two important fibroblast subtypes that contribute to matrix deposition and also have enhanced invasive, proliferative, and contraction phenotypes. The histological locations of fibroblast subtypes are identified in healthy and fibrotic lungs by these cell surface proteins. This study provides new insights to inform approaches to targeting lung fibroblast subtypes to promote the development of therapeutics for lung fibrosis.
Keywords: pulmonary fibrosis, fibroblast subtype, single-cell RNA-sequencing, cell surface marker, extracellular matrix
Clinical Relevance
We defined lung fibroblast subtypes and identified cell surface markers for directly isolating primary cells of fibroblast subtypes from human idiopathic pulmonary fibrosis lungs. Multiple lung fibroblast subtypes secreted matrix proteins and had different capacities for invasion, proliferation, and contraction. In this study, we document the direct contributions of fibroblast subtypes to lung matrix gene expression and production and identify new insights into approaches to target fibroblast subpopulations.
Pulmonary fibrosis occurs in a variety of clinical settings, constitutes a major cause of morbidity and mortality, and represents an enormous unmet medical need (1). The development of lung fibrosis is an essential response of the human lung to pathogens and noninfectious injuries (2), leading to downstream fibrotic tissue remodeling and extracellular matrix deposition, which in turn perpetuates fibrosis formation (3). As a criterion for a large group of interstitial lung disease and usual interstitial pneumonia in human lungs, pulmonary fibrosis is usually categorized into lung diseases including idiopathic pulmonary fibrosis (IPF), chronic hypersensitivity pneumonitis, systemic sclerosis–associated interstitial lung disease (SSc-ILD), rheumatoid arthritis, lupus erythematosus, and other rheumatic diseases (4). Nintedanib and pirfenidone have been approved by the U.S. Food and Drug Administration and the European Medicines Agency for patients with pulmonary fibrosis, including IPF and SSc-ILD, but many questions remain regarding the lack of long-term lung functional improvement, effects on mortality, and significant tolerability issues associated with these therapies (2).
The major effector cells in fibrotic diseases are fibroblasts, which have long been suggested to be heterogeneous in mammalian lungs (5). Myofibroblasts, one of the major components of fibroblasts, have been believed to be the major sources of matrix production in lung fibrosis (6), although their contractile properties were initially identified to be critical for normal wound healing (7). Increased expression of α-SMA (α-smooth muscle actin) has been a major indication of fibroblast activation. However, recent studies suggest that α-SMA is an inconsistent marker of fibroblast activation (8). In addition to myofibroblasts, we and others have identified multiple fibroblast subtypes in lungs, including lipofibroblasts, EBF1+ fibroblasts, smooth muscle cells, pericytes, and mesothelial cells (9–13), although the genes for the definitions might differ.
Except for myofibroblasts, other fibroblast subtypes might not express high levels of α-SMA under homeostatic conditions. However, their contributions to matrix gene expression in fibrotic conditions have been reported (14–16). For example, the Adrp+ lipogenic fibroblasts (lipofibroblasts) were found to be located in dense fibrotic areas in bleomycin-treated mouse lungs and showed an upregulated expression of matrix genes, such as Acta2 and Col1a1, after injury (17). WT1 (Wilms’ tumor 1), a mesothelial cell–specific gene, was necessary for the maintenance of the human lung mesothelial cell phenotypes. Loss of WT1 upregulated the expression of extracellular matrix markers α-SMA and fibronectin and increased the migration and contractility of the primary mesothelial cells isolated from human lungs (18). In the microenvironment of the IPF lungs, microvascular pericytes from human lungs adopted the expression of matrix genes, such as α-SMA. Pericytes from IPF lungs responding to TGF-β1 adopted functional characteristics, including the ability to produce a collagen- and fibronectin-rich extracellular matrix (19). Mouse lung pericytes derived from Foxd1 progenitors expanded after bleomycin lung injury and activated expression of Col1a1 and α-SMA in the fibrotic foci of mouse lungs (15). These studies shed light on the potential contribution of these fibroblast subpopulations to matrix secretion in fibrotic conditions (10), but the direct or comprehensive evidence of these hypotheses has yet to be fully established.
To further delineate the biological function of fibroblast subtypes in lung fibrosis, we incorporated several single-cell RNA-sequencing (scRNA-seq) datasets on lung fibrosis from both mouse models and human diseases published recently. We defined lung fibroblast subtypes with a common feature of collagen gene expression and assessed differentially expressed genes in each subtype. Most importantly, we identified cell surface markers for directly isolating primary cells of fibroblast subtypes from human IPF lungs. With further in vitro study, we showed that multiple lung fibroblast subtypes secreted matrix proteins. Interestingly, we found that fibroblast subtypes had different capacities for invasion, proliferation, and contraction. In this study, we document the direct contributions of fibroblast subtypes to lung matrix gene expression and production and identify new insights into approaches to target fibroblast subpopulations.
Methods
See the data supplement for detailed methods.
Study Approval
All the experiments on human lungs were approved by the Institutional Review Board (IRB) of Cedars-Sinai Medical Center and were in accordance with the guidelines outlined by the IRB (IRB number: Pro00032727). Informed consent was obtained from each subject.
scRNA-seq Analysis and Data Accessibility
scRNA-seq data of freshly isolated lung single cells from mice, human patients, and healthy donors were obtained from publicly available datasets, and the gene expression omnibus (GEO) accession numbers are presented in Table 1 (20–30). Details for data analysis and cell surface marker identification are described in the data supplement.
Table 1.
Accessibility of the Single-Cell RNA-Sequencing Data on Murine and Human Lung Tissues Used in This Study
| Species | Treatment/Model/Disease | GEO ID | Numbers of Tissues (n) |
Collection Timepoint | Dissociation Reagent | Instrument Model | References | |
|---|---|---|---|---|---|---|---|---|
| Fibrosis | Normal | |||||||
| Murine | Bleomycin/adult | GSE111664 | 3 | 7 | Day 14 | Liberase | MiSeq | 20 |
| GSE129605 | 4 | 4 | Day 11 | Collagenase A | NextSeq 500 | 21 | ||
| GSE131800 | 1 | 1 | Day 21 | Elastase | NovaSeq 6000 | 22 | ||
| GSE104154 | 1 | 1 | Day 21 | Elastase | NextSeq 500 | 9 | ||
| GSE132771 | 4 | 4 | Day 14 | Collagenase A | HiSeq 4000 | 23 | ||
| Bleomycin/aged | GSE157379 | 3 | 3 | Day 14 | Collagenase IV | NextSeq 500 | 10 | |
| Silica/adult | GSE184854 | 12 (Days 7 and 21) | 5 (Day 3) | Days 3, 7, and 21 | Liberase | NovaSeq 6000 | 24 | |
| Human | IPF | GSE136831 | 32 | 28 | NA | Elastase/liberase | HiSeq 4000 | 12 |
| GSE157376 | 5 | 5 | NA | Elastase | NextSeq 500 | 10 | ||
| GSE128169 | 0 | 5 | NA | Collagenase A | NextSeq 500 | 25 | ||
| GSE135893 | 12 | 10 | NA | Collagenase I | NovaSeq 6000 | 11 | ||
| GSE128033 | 8 | 10 | NA | Collagenase A | NextSeq 500 | 26 | ||
| GSE122960 | 4 | 8 | NA | Collagenase D | HiSeq 4000 | 27 | ||
| GSE132771 | 6 | 6 | NA | Collagenase A | NovaSeq 6000 | 23 | ||
| SSc-ILD | GSE159354 | 2 | 5 | NA | Collagenase I | HiSeq 4000 | 28 | |
| GSE132771 | 6 | 4 | NA | Collagenase A | NovaSeq 6000 | 23 | ||
| COVID-19 | GSE171524 | 19 | 7 | NA | NA | NovaSeq 6000 | 29 | |
| COVID-19 late-stage | GSE158127 | 3 | 3 | NA | Collagenase D | HiSeq 4000 | 30 | |
Definition of abbreviations: COVID-19 = coronavirus disease; GEO = gene expression omnibus; IPF = idiopathic pulmonary fibrosis; NA = not associated; SSc-ILD = systemic sclerosis–associated interstitial lung disease.
Bleomycin/adult indicates bleomycin-induced lung fibrosis in adult mice; bleomycin/aged indicates bleomycin-induced lung fibrosis in aged mice; silica/adult indicates silica-induced mouse lung fibrosis.
Human Lung Fibroblast Culture
Human lung fibroblasts were isolated from surgical lung biopsies or lung transplant explants obtained from patients with IPF. Detailed isolation protocols were previously described (31, 32). Detailed methods can be found in the data supplement.
Immunofluorescence Staining
Immunofluorescence staining was performed and processed on fixed human lung tissues embedded in optimal cutting temperature compound (OCT) or paraffin. Detailed methods can be found in the data supplement.
ELISA
ELISA was performed according to the manufacturer’s instructions. For the prefibrotic treatment of the fibroblasts, TGF-β1 or fibrosis cocktail (FC) (33) was used. Detailed methods can be found in the data supplement.
Fibroblast Invasion
Fibroblast invasion was performed as previously described (31, 32). Detailed methods can be found in the data supplement.
Cell Contraction Assay
Fibroblast-induced polymerized collagen matrix contraction was performed with a cell contraction assay kit. Detailed methods can be found in the data supplement.
Statistical Analysis
The statistical difference between two groups in the bioinformatics analysis of the scRNA-seq data was calculated using the Wilcoxon signed-rank test, and the lowest P value calculated in Seurat was P < 2.2 × 10−16. For all other data, the statistical differences between groups were calculated using GraphPad. The exact values are shown by mean ± SEM with P values. Unpaired Student’s two-tailed t test was used for comparing differences between two groups. Two-way ANOVA followed by Tukey’s multiple comparison test was used for multiple comparisons.
Results
Collagen Contribution by Fibroblast Subtypes in Fibrotic Murine Lungs
We reported several collagen gene–expressing lung fibroblast subtypes previously (10). To analyze fibroblast subtypes comprehensively in lung fibrosis, we incorporated recently published scRNA-seq datasets on fibrotic and normal murine lungs of different pulmonary fibrosis murine models (Table 1). In the bleomycin-induced lung fibrosis model, five datasets composed of 13 fibrotic and 17 normal samples from adult mice were integrated (9, 10, 20–23), and 3 fibrotic and 3 normal samples from aged mice were integrated (10). Fibroblast components were subset from the corresponding integrated datasets (Figures 1A and 1F), and the purities were confirmed (see Figures E1A and E1C in the data supplement). Using the fibroblast subtype features (average expression of the gene sets) we identified previously (10) (Figures E1B and E1D), we defined the fibroblast cell subtypes in adult and aged lungs with the bleomycin lung injury model (Figures 1B and 1G). Quantification of each fibroblast subtype suggested a dramatic decrease in lipofibroblast percentage in both adult and aged fibrotic lungs and an increased intermediate fibroblast subpopulation in aged fibrotic lung (Figures 1C and 1H). We then checked the expression of common matrix genes, including Col1a1, Col1a2, Col3a1, and Fn1 (Figures E2A and E2C), as well as other matrix component genes (Figures E2B and E2D), and found elevated expression of these genes in total fibroblasts and fibroblast subtypes from fibrotic lungs compared with the cells from control lungs. To simplify the data presentation, we then defined the average expression of common matrix genes, Col1a1, Col1a2, Col3a1, and Fn1, as the collagen score. Both the adult and aged fibrotic fibroblasts demonstrated dramatically elevated collagen scores (Figures 1D and 1I) compared with the matched control fibroblasts. As expected, all the fibroblast subtypes contributed to the increased collagen expression in the total fibroblasts (Figures 1E, 1J, and E2), although the contribution ratios varied. Consistent with our previous findings, these observations demonstrated that all fibroblast subtypes have elevated matrix gene expression in bleomycin-induced lung fibrosis in adult and aged mice.
Figure 1.
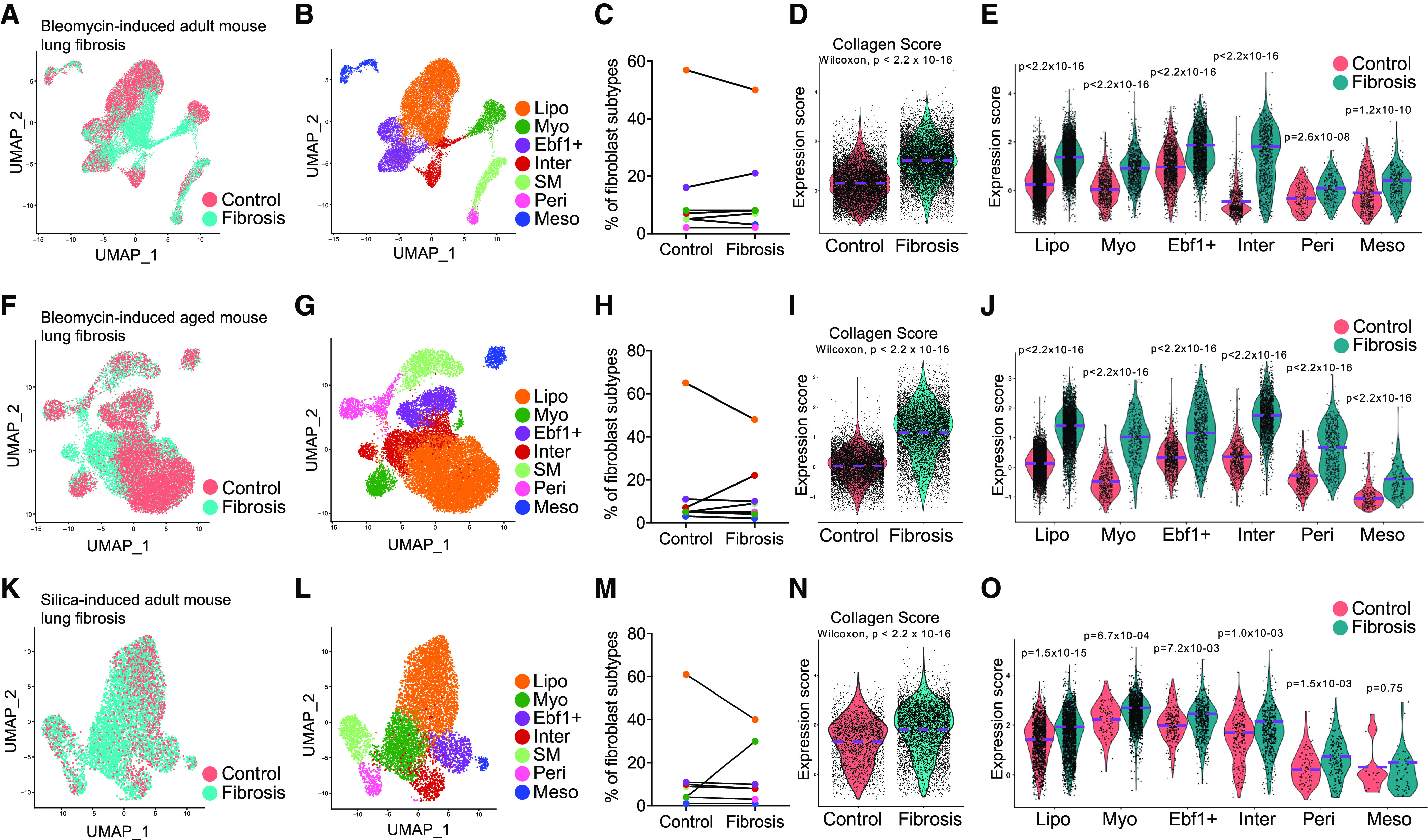
Matrix contribution by lung fibroblast subpopulations in mouse fibrotic lungs. (A–J) Single-cell transcriptomics analysis of the adult mice (A–E) or aged mice (F–J) with bleomycin-induced lung fibrosis. (K–O) Single-cell transcriptomics analysis of mice with silica-induced lung fibrosis. (A, F, and K) Distribution of the mesenchymal cells from normal and fibrotic lungs of adult mice (A) and aged mice (F) in bleomycin- and silica-induced (K) lung fibrosis models. (B, G, and L) Identification of the fibroblast subtypes in lungs from adult (B) or aged (G) mice in bleomycin- and silica-induced (L) lung fibrosis models. (C, H, and M) Percentage of fibroblast subtypes in lungs from adult (C) or aged (H) mice in bleomycin- and silica-induced (M) lung fibrosis models. (D, I, and N) Violin plot comparisons of the collagen score (average expression of the matrix genes: Col1a1, Col1a2, Col3a1, and Fn1) in lung fibroblasts of adult (D) and aged (I) mice in bleomycin- and silica-induced (N) lung fibrosis models. (E, J, and O) Violin plot comparisons of the collagen score in fibroblast subtypes of adult (E) and aged (J) mice in bleomycin- and silica-induced (O) lung fibrosis models. Wilcoxon P < 0.05 is statistically significant by Wilcoxon rank sum test (D, E, I, J, N, and O). Ebf1+ = Ebf1+ fibroblast; Inter = intermediate fibroblast; Lipo = lipofibroblast; Meso = mesothelial cell; Myo = myofibroblast; Peri = pericyte; SM = smooth muscle cell; UMAP = uniform manifold approximation and projection.
To further validate this finding, we accessed another scRNA-seq dataset of a murine lung fibrosis model induced by silica (Table 1) (24), which is commonly used to model fibrotic lung diseases caused by free crystalline silica particles, such as silicosis (34). The cells of mouse lungs on Days 3, 7, and 21 after silica treatment were integrated, and fibroblasts were subset and clustered (Figures E3A–E3C). As the cells from Day 3 after silica treatment mostly reflect acute inflammatory response and epithelial cell injury (35), we identified the fibroblast component mainly from Days 7 and 21 as fibrotic cells and fibroblasts from Day 3 as nonfibrotic or control cells (Figure E3B). The fibroblasts were subset (Figure 1K) and the purity was confirmed (Figure E3D), and fibroblast subtypes were identified by the expression of subtype features and individual genes (Figures 1L and E3E). Similarly, the ratio of lipofibroblasts decreased and that of myofibroblasts increased in the fibrotic lungs (Figure 1M). The collagen expression of the fibroblasts from late silica-treated fibrotic lungs was significantly elevated compared with the fibroblasts from early injured control lungs (Figures 1N and E3F), although the increase in ratio was not as high as in the lung fibrosis model induced by bleomycin. Increased collagen expression in all the fibroblast subtypes from fibrotic lungs was also observed (Figures 1O, E3F, and E3G), suggesting that extracellular matrix deposition in this model was contributed by all the fibroblast subtypes.
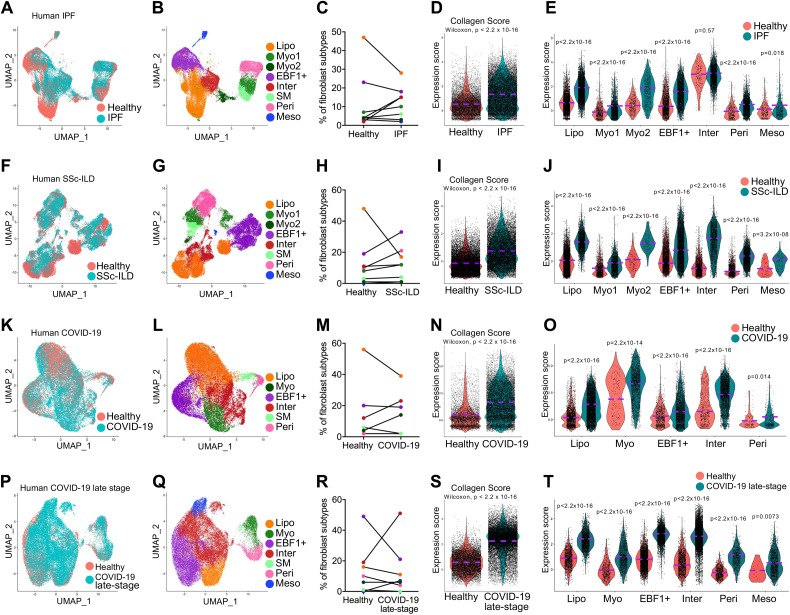
Extracellular Matrix Deposition by Fibroblast Subtypes in Fibrotic Human Lungs
As elevated collagen expression was observed in all fibroblast subtypes in murine lungs, we next attempted to determine if this is also present in human fibrotic lung diseases. We first incorporated recently published datasets on human IPF and healthy lung samples (10–12, 23, 25–27) (Table 1) and distinguished the purified fibroblast compartments using the same criterion as used with mouse fibroblasts (Figures 2A and E4A). We then identified fibroblast subtypes from IPF and healthy control lungs (Figures 2B and E4B). Again, the lipofibroblast percentage decreased while myofibroblast and intermediate fibroblast percentages increased in the IPF lungs (Figure 2C). The fibroblasts from IPF lungs showed significantly upregulated collagen gene expression (Figures 2D, E4C, and E4D), and all IPF fibroblast subtypes demonstrated increased collagen gene expression (Figures 2E, E4C, and E4D) compared with that of healthy fibroblasts. These results were consistent with what we observed with mouse lung fibrosis and indicated that the collagen deposition in IPF lungs was contributed by all the fibroblast subtypes.
Figure 2.
Matrix contribution by lung fibroblast subpopulations in human fibrotic lung diseases. Single-cell transcriptomics analysis of the lung fibroblasts from patients with (A–E) idiopathic pulmonary fibrosis (IPF), (F–J) systemic sclerosis–associated interstitial lung disease (SSc-ILD), (K–O) coronavirus disease (COVID-19), and (P–T) late-stage COVID-19 with confirmed pulmonary fibrosis and matched healthy donors. (A, F, K, and P) Distribution of the mesenchymal cells from human lungs of patients with IPF (A), SSc-ILD (F), COVID-19 (K), and late-stage COVID-19 with confirmed pulmonary fibrosis (P) and matched healthy donors. (B, G, L, and Q) Identification of the fibroblast subtype in human lungs of patients with IPF (B), SSc-ILD (G), COVID-19 (L), and late-stage COVID-19 with confirmed pulmonary fibrosis (Q) and matched healthy donors. (C, H, M, and R) Percentage of fibroblast subtypes in human lungs of patients with IPF (C), SSc-ILD (H), COVID-19 (M), and late-stage COVID-19 with confirmed pulmonary fibrosis (R) and matched healthy donors. (D, I, N, and S) Violin plot comparisons of the collagen score (average expression of the matrix genes: COL1A1, COL1A2, COL3A1, and FN1) in lung fibroblasts from patients with IPF (D), SSc-ILD (I), COVID-19 (N), and late-stage COVID-19 with confirmed pulmonary fibrosis (S) and matched healthy donors. (E, J, O, and T) Violin plot comparisons of the collagen score in fibroblast subtypes from patients with IPF (E), SSc-ILD (J), COVID-19 (O), and late-stage COVID-19 with confirmed pulmonary fibrosis (T) and matched healthy donors. Wilcoxon P < 0.05 is statistically significant by Wilcoxon rank sum test (D, E, I, J, N, O, S, and T).
SSc is a multidisciplinary disease with a wide range of systemic complications. The complications in the pulmonary system include ILD and pulmonary arterial hypertension, which are the main causes of morbidity and mortality in the course of SSc (36). Here we accessed two scRNA-seq datasets on human lung tissues of patients with SSc-ILD (Table 1) (23, 28). The pure fibroblasts were subset and clustered (Figures 2F and E5A–E5D), and major fibroblast subtypes were identified (Figures 2G and E5E). Similarly, the lipofibroblast ratio decreased, whereas myofibroblast and EBF1+ fibroblast ratios were dramatically elevated (Figure 2H). Significantly upregulated collagen gene expression was evidenced in total fibroblasts (Figures 2I and E5F) and in each fibroblast subtype (Figures 2J, E5F, and E5G) from the SSc-ILD lungs compared with that of fibroblasts from control lungs, suggesting all fibroblast subtypes contributed to lung fibrosis in the disease of SSc-ILD.
With the increasing impact of coronavirus disease (COVID-19) worldwide, progressive pulmonary fibrosis has been found in patients with severe COVID-19, and COVID-19–associated lung fibrosis shares features with IPF (30, 37). In this study, we retrieved a dataset on lung samples from individuals who died from COVID-19 and control individuals (29), as well as a dataset on lung samples of patients with late-stage COVID-19 with confirmed progressive lung fibrosis and control lung samples (Table 1) (30). The pure fibroblasts were clustered (Figures 2K, 2P, E6A–E6D, and E7A–E7D), and major fibroblast subtypes were identified in these two datasets (Figures 2L, 2Q, E6E, and E7E). The ratio changes in the fibroblast subtypes were similar to those of other fibrotic lungs (Figures 2M and 2R). The fibroblasts from the lungs of patients with COVID-19 showed significantly upregulated collagen gene expression (Figures 2N and E6F), contributed by all the fibroblast subtypes (Figures 2O, E6F, and E6G). Furthermore, fibroblasts from lungs of patients with late-stage COVID-19 with confirmed lung fibrosis showed much higher expression of collagen genes (Figures 2S and E7F). Notably, all the fibroblast subtypes showed significant contribution to collagen gene expression (Figures 2T, E7F, and E7G).
In summary, we have identified novel fibroblast subtypes in lungs of human fibrotic diseases, including IPF, SSc-ILD, and COVID-19. All the fibroblast subtypes in fibrotic lungs showed elevated matrix gene expression.
Identification of Cell Surface Markers for Fibroblast Subtypes
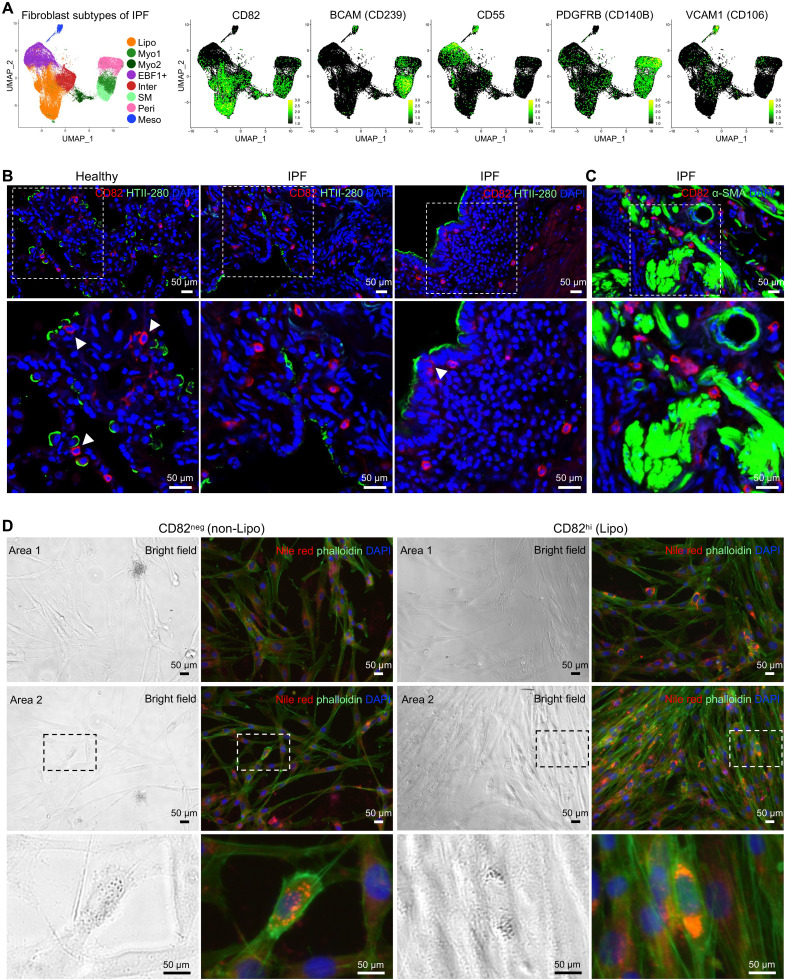
Next, we attempted to determine cell surface markers for each fibroblast subtype. We analyzed differentially expressed genes in IPF lung fibroblast subtypes and focused on those genes encoding cell surface proteins. We first identified specific cell surface protein genes in each subtype, including FGFR4, ITGA8, SLC40A1, and CD82 for lipofibroblasts; BCAM for myofibroblasts; CD55 and CD248 for EBF1+ fibroblasts; PDGFRB for pericytes; and VCAM1, CXADR, TM4SF1, and CLDN1 for mesothelial cells (Figures 3A and E8A).
Figure 3.
Identification of cell surface marker genes of each fibroblast subpopulation in human lungs. (A) Cell surface marker genes were identified in the integrated single-cell RNA-sequencing data of mesenchymal cells from IPF and healthy human lungs: CD82 for lipofibroblasts, BCAM (CD239) for myofibroblasts, CD55 for EBF1+ fibroblasts, PDGFRB (CD140B) for pericytes, and VCAM1 (CD106) for mesothelial cells. (B and C) Costaining of CD82 (lipofibroblast marker) and HTII-280 (human alveolar type II cell marker) (B) and α-SMA (α-smooth muscle actin) (myofibroblast marker) (C) on healthy and IPF lung sections. (D) Bright-field and fluorescence images of the CD82neg (nonlipofibroblasts) and CD82hi (lipofibroblasts) fibroblasts after visualization of the intracellular lipid droplets by Nile red staining. Higher magnifications of the boxed regions are shown (B–D). Scale bars, 50 μm (B and D).
We then validated the cell surface protein expression in primary lung fibroblasts from patients with IPF by commercial antibodies using flow cytometry. Of these, BioLegend monoclonal antibody, clone ASL-24, against CD82 worked well in the identification of lipofibroblasts in primary IPF lungs (Figures 3A and E8B).
Pulmonary lipofibroblasts are recognizable by their characteristic lipid droplets, which endow them with the role as an accessory cell to the type II epithelial cells in the synthesis of surfactant proteins during alveolar development and regeneration (38). To localize lipofibroblasts anatomically in human lungs, we did immunostaining of CD82 with human alveolar type II (AT2) marker HTII-280 and activated myofibroblast marker α-SMA. Most of the CD82+ fibroblasts were juxtaposed to AT2s in healthy human lungs (Figure 3B), suggesting a basic function of lipofibroblasts: transferring neutral lipids to AT2s. In IPF lungs, these CD82+ fibroblasts were sporadically distributed in the lung (Figure 3B). Although CD82+ fibroblasts can be seen in fibrotic foci, CD82 immunostaining was not colocalizated with α-SMA+ (Figure 3C). To confirm the practicality of the cell surface markers we identified above in vitro, we stained the sorted CD82 negative (CD82neg) and CD82 high (CD82hi) fibroblasts with Nile red for lipid droplets and phalloidin for cytoskeleton. Although scattered lipid droplets could be found in very few CD82neg fibroblasts, potentially owing to the active proliferation and lipid metabolism of these cells to generate energy or produce the structural components of cell membranes (Figure 3D, boxed regions of left panels), most of the CD82hi fibroblasts showed lipid droplet pools (Figure 3D, boxed regions of right panels), suggesting these cells contained abundant lipid storage.
Of the antibodies tested to identify other fibroblast subtypes, the following antibodies worked in primary IPF lung fibroblasts: Phycoerythrin (PE) anti-human CD239 (BCAM) antibody (Miltenyi Biotec, 130-126-539) for myofibroblasts, PE anti-human CD55 antibody (BD Biosciences, 561901) for EBF1+ fibroblasts, PE anti-human CD140b (PDGFRB) antibody (BioLegend, 323605) for pericytes, and PE anti-human CD106 (VCAM1) antibody (BioLegend, 305806) for mesothelial cells (Figures 3A and E8B). Immunostaining suggested that many of the CD239+ fibroblasts (myofibroblasts) were colocalized with SMA+ fibroblasts, especially in the IPF lung sections (Figure E9A), and CD55+ fibroblasts (EBF1+ fibroblasts, previously identified as pericyte progenitor cells [10]) were mostly localized surrounding the vascular-like structures in IPF lung sections (Figure E9B). Notably, in normal human lung sections, CD55 was also detected in the alveolar epithelial layers, suggesting that CD55 was also potentially expressed in human AT1 cells (Figure E9B). CD140B+ cells were localized adjacent to CD31+ endothelial cells (Figure E9C), and CD106+ cells were localized in the pleural mesothelial layers in both healthy and IPF lungs (Figure E9D).
To test the practicality of these markers in mouse datasets, we checked their transcription in the mouse datasets and found most of the human cell surface marker genes also showed specific transcription in the matched fibroblast subtypes in mouse lungs (Figures E10A and E10B). Specifically, Cd82/Fgfr4(Cd334)/Igta8 were specific to lipofibroblasts, Cd55/Cd248 were specific to Ebf1+ fibroblasts, and Cxadr/Tm4 sf1 were specific to mesothelial cells, whereas Gpc3/Slc40a1/Bcam(Cd239)/Pdgfra(Cd140)/Vcam1(Cd106)/Cldn1 showed either low or unspecific transcriptions in the fibroblast subtypes and should not be considered as cell surface markers in the mouse datasets (Figures E10A and E10B).
Matrix Secretion Capacities of Fibroblast Subtypes
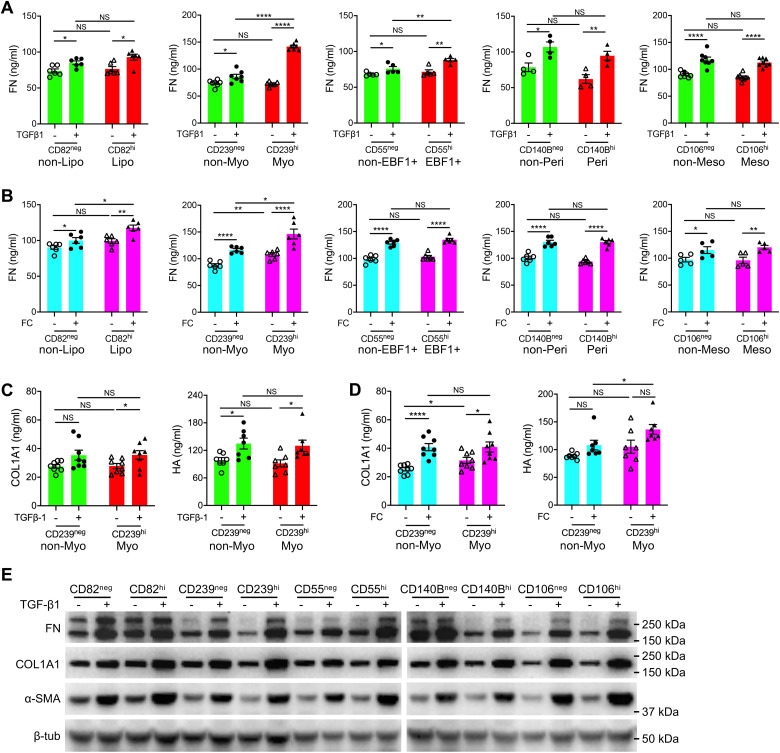
To further validate the matrix contribution by the fibroblast subtypes in IPF lungs, the primary IPF lung fibroblasts with negative or high expression of the above cell surface markers were sorted (Figure E8B) and treated with profibrotic factors TGF-β1 or FC (33).
Secreted matrix proteins, including FN (Fibronectin), COL1A1 (collagen), and HA (hyaluronan), in the culture medium were examined by ELISA assays (Figures 4A–4D). As the assay for FN showed fewer variations (Figures 4A–4D), we used the ELISA for FN to represent the matrix secretion for most of the analyses. As expected, both TGF-β1 (Figures 4A and 4C) and FC (Figures 4B and 4D) treatment significantly increased the secretion of FN (Figures 4A and 4B), collagen, and hyaluronan (Figures 4C and 4D) in all fibroblasts. Notably, CD239hi fibroblasts (myofibroblasts) showed much higher FN and HA secretion after TGF-β1 or FC treatment than the negative control fibroblasts (nonmyofibroblasts). CD55hi fibroblasts (EBF1+ fibroblasts) secreted higher levels of FN protein than negative fibroblasts after TGF-β1 treatment, whereas CD82hi fibroblasts (lipofibroblasts) secreted higher levels of FN protein than negative fibroblasts after FC treatment. The other two subtypes, pericytes (CD140hi) and mesothelial cells (CD106hi), showed comparable secretion of FN with negative control fibroblasts after TGF-β1 or FC treatment (Figures 4A and 4B).
Figure 4.
Matrix secretion by each fibroblast subpopulation after profibrotic treatment. (A and B) Quantification of fibronectin secretion by each human fibroblast subtype stimulated by TGF-β1 (A) and fibrosis cocktail (FC) (B). n = 6 per group for CD82neg and CD82hi (A and B); n = 7 (A) or 6 (B) per group for CD239neg and CD239hi; n = 5 (A) or 6 (B) per group for CD55neg and CD55hi; n = 4 (A) or 6 (B) per group for CD140Bneg and CD140Bhi; n = 8 (A) or 5 (B) per group for CD106neg and CD106hi. (C and D) Quantification of COL1A1 and hyaluronan (HA) secretion by sorted myofibroblasts (CD239hi) and control fibroblasts (CD239neg) stimulated by TGF-β1 (C) and FC (D). n = 8 per group for COL1A1 secretion and n = 7 per group for HA secretion (C and D). (E) Western blot for total protein levels of FN, COL1A1, and α-SMA in each human fibroblast subtype after TGF-β1 stimulation. Quantitative data are represented as mean ± SEM. *P < 0.05, **P < 0.01, and ****P < 0.0001 by one-way ANOVA. hi = high; neg = negative; NS = not significant.
Western blotting was then applied to examine the total matrix protein levels in these fibroblast subtypes with or without profibrotic treatments. Most of the fibroblast subtypes showed comparable expression levels of these proteins before TGF-β1 treatment, whereas after TGF-β1 stimulation, all the fibroblast subtypes showed elevated protein levels of FN1, COL1A1, and α-SMA. Notably, CD82hi (lipofibroblast), CD239hi (myofibroblast), and CD55hi (EBF1+ fibroblast) fibroblasts showed higher expression levels of these proteins than the corresponding cell surface marker–low fibroblasts, suggesting these fibroblast subtypes contributed more collagen in the fibrotic conditions (Figures 4E and E11A–E11C). These data confirmed the observations in the scRNA-seq analysis that all the fibroblast subtypes contributed extracellular matrix production in fibrotic lungs, but the contribution ratios differed, with highest matrix expression in myofibroblasts.
Functional Characterizations of Fibroblast Subtypes
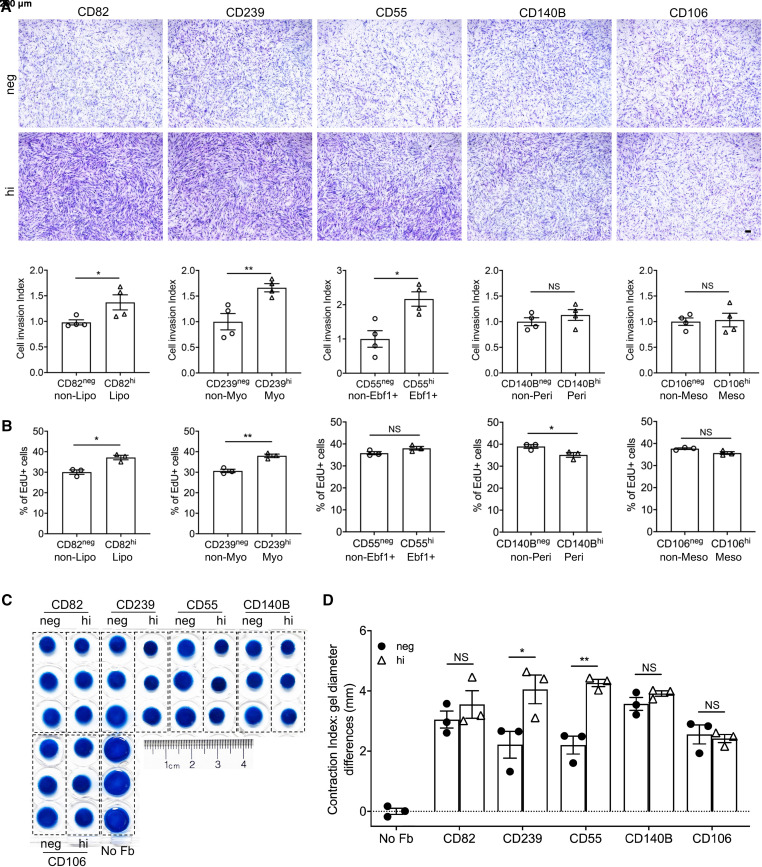
We and others reported that the invasive phenotype of lung fibroblasts promotes severe fibrosis (39). To determine the invasive capacities of fibroblast subtypes, we performed invasion assays on fibroblast subtypes sorted by cell surface markers we identified. Lipofibroblasts, myofibroblasts, and EBF+ fibroblasts enriched by high expression of cell surface CD82, CD239, and CD55, respectively, showed elevated invasion indices compared with the corresponding negative fibroblasts, whereas pericytes (CD140Bhi) and mesothelial cells (CD106hi) showed similar invasion capacities to those of the respective control fibroblasts (Figure 5A). Fibroblast invasion is associated with the degradation of the extracellular matrix (ECM) by matrix-degrading enzymes, such as MMP genes (40). The MMP genes showed elevated expression in IPF fibroblasts compared with normal fibroblasts and in lipofibroblast, myofibroblast, EBF1+ fibroblast, and intermediate fibroblast subtypes compared with other fibroblast subtypes (Figures E11D–E11F), which was consistent with the observation that these subtypes showed higher invasion capacities. These data suggested that the invasive capacity of fibroblast subtypes is correlated to the matrix-degrading enzyme gene expression of the cells.
Figure 5.
Invasion, proliferation, and contraction of each fibroblast subpopulation. (A) Representative images and quantification of invasion of each fibroblast subpopulation. n = 4 per group. Scale bars, 200 μm. (B) Cell proliferation rates of the cell surface marker–negative and -positive fibroblast subtypes by EdU assay. n = 3 per group. (C) Collagen contraction induced by each fibroblast subpopulation and (D) quantifications of the contraction diameters. n = 3 per group. Quantitative data are represented as mean ± SEM. *P < 0.05, **P < 0.01 by Student’s t test (A and B) or one-way ANOVA (D). No Fb = no fibroblast control.
We next attempted to investigate the proliferation of the fibroblast subtypes. Although most of the cell-surface negative fibroblasts had comparable proliferation rates, the cell-surface positive fibroblasts showed different proliferation rates (Figure E12A). Notably, CD82- and CD239-high fibroblasts showed higher proliferation rates relative to respective cell-surface negative fibroblast subtypes (Figure 5B).
Fibroblasts presented a contractility property, which is an important factor in regulating tissue injury repair and fibrosis (41). Here we applied the cell contraction assay to evaluate the contractility properties of each fibroblast subtype. A significant contractility was only found in myofibroblasts (CD239hi) and EBF1+ fibroblasts (CD55hi) compared with the matched control fibroblasts (Figures 5C, 5D, and E12B).
These data established the well-described functional characterizations of each fibroblast subtype in healthy and IPF lungs. Expanded studies on these fibroblast features might provide novel insights in investigating the involvements of fibroblast subtypes in lung fibrosis initiation and progression.
Discussion
ILDs, a set of progressive lung diseases with high morbidity and mortality, are a heterogeneous group of diffuse parenchymal lung disorders of known or unknown causes with varying degrees of inflammation or fibrosis (42). IPF is the most common fibrotic ILD and is characterized by an imaging and pathological pattern of pulmonary fibrosis of unidentifiable causes (2). Lung fibroblasts, the key fibrotic effector cells responsible for the synthesis of extracellular matrix proteins of lung fibrosis (43), have been better characterized in recent years, thanks to the development and application of single-cell omics technologies, such as scRNA-seq. The heterogeneity of fibroblasts has been well described, and the involvement of lung fibroblast subpopulations and their lineages, together with those of other lung compartments, in lung fibrosis have been well studied at single-cell levels (9–12, 27). Up to now, the studies of fibroblast contributions to lung fibrosis have been mostly based on two aspects: the genetic deficiency or activation-induced fibroblast–myofibroblast transition, and the fibroblast-directed cell–cell communications proposed by digital analyses. Myofibroblasts have been believed to be the major contributor of matrix deposition in fibrosis. However, the functions of fibroblast subtypes have not been as well determined as other lung cellular components, such as epithelial and immune cells (44). We previously reported the contribution of collagen genes by all the major fibroblast components at mRNA levels, not only myofibroblasts, in murine and human fibrotic lungs (10). To address this issue more comprehensively and to examine effector functions, the current study incorporated more data to include more samples from different datasets and different murine lung fibrosis models and human fibrotic lung diseases. As expected, all the fibroblast subtypes in both murine and human lungs showed contributions of matrix gene expression in fibrotic lungs. This observation was further confirmed at protein levels by in vitro profibrotic fibroblast models.
Myofibroblasts, one of the major fibroblast subtypes we identified, have been believed to be the key effector cells in fibrotic tissues and, as such, are responsible for the synthesis of extracellular matrix proteins (45). Elevated collagen expression was found in the myofibroblast subtypes of murine and human fibrotic lungs at single-cell mRNA levels. Moreover, the sorting-enriched human myofibroblasts showed much higher fibronectin protein secretion than nonmyofibroblast components when treated with antifibrosis factors TGF-β1 or FC. These data further supported the fact that myofibroblasts are one of the major contributors of matrix protein expression in fibrotic lungs.
In addition to myofibroblasts, reports on the contribution of other lung fibroblast components are more limited. Lipofibroblasts are another major subset of lung fibroblasts (10) and contributions to alveolar development largely rely on the transportation of neutral lipids to AT2 cells to support surfactant and phospholipid synthesis (38). Initially, lipofibroblasts, residing next to AT2 cells, were considered to be supportive stem niche cells of AT2 cells during development and regeneration (46). However, a recent study also suggested lipofibroblasts as one of the sources of activated myofibroblasts in mouse lung after bleomycin-induced injury, although a portion of myofibroblasts reverted to quiescent state characteristics of the preexisting lipofibroblasts during fibrosis resolution (17). In our current study, lipofibroblasts were one of the major contributors of collagen gene expression in fibrosis conditions at both mRNA and protein levels both in vivo and in vitro, although we could not deny the existence of lipofibroblast–myofibroblast transition when lipofibroblasts were treated with profibrotic mediators in vitro.
EBF1+ fibroblasts were suggested to be the potential progenitor cells of lung pericytes in early lung development (10), although lineage-tracing experiments are ultimately needed to clarify this. Both EBF1+ fibroblasts and pericytes also contributed to the expression of collagen genes directly in fibrotic conditions in both in vitro and in vivo models. Supporting evidence in the literature included murine Foxd1 lineage+ and human α-SMA+ lung pericytes showed elevated collagen gene expression in fibrotic conditions (15, 19). This observation has also been believed to be an important source of matrix deposition in the studies of fibrosis of other organs, especially in the kidney (47).
Intermediate fibroblasts were another fibroblast subtype contributing a significant amount of collagen in the fibrotic conditions in both human and mouse lungs. However, it is hard to identify any markers that were consistent at different development stages (10) or among different datasets. This subtype is close to the adventitial fibroblasts in normal human lung tissues (13). Recently, a fibroblast subtype marked by the gene Cthrc1/CTHRC1 has been reported to be a collagen-producing fibroblast subpopulation in mouse and human lungs (23). In our analysis, this gene showed specific transcription in the intermediate fibroblast clusters in the bleomycin-induced adult and aged fibrotic lungs and human IPF lungs. However, in the other datasets, including silica-induced mouse fibrotic lungs, human SSc-ILD lungs, and COVID-19–caused human fibrotic lungs, the expression of this gene was either low or not specific in intermediate fibroblast clusters. We assumed that this subset of fibroblasts might show some further heterogeneity, but this assumption needs further investigation in a future study.
In addition to the above fibroblast sublineages, mesothelial cells, although they account for a small percentage of the lung fibroblasts in the fibrotic conditions (10), have been reported to be of importance in the development of tissue fibrosis after injury, in not only lung but also other mesothelial cell–abundant tissues such as pleura and peritoneum (16). Mesothelial–myofibroblast transition has been suggested, but because of the small numbers of mesothelial cells in lung fibroblast compartments, their contribution to the myofibroblast burden and to lung fibrosis is limited (18). We found this to be true in our in vitro profibrotic models on human lung fibroblasts.
Cell–cell interactions between fibroblasts and other cell types may importantly contribute to the homeostasis and pathogenesis of fibrotic diseases in the lung. During lung development and maturation, alveolar myofibroblasts are essential for secondary septation, a process critical for the development of the gas-exchange region of the lung (48). Alveolar epithelial cell injury and death provoke the proliferation and activation of lung myofibroblasts, leading to accumulation of these pathogenetic myofibroblasts and the extracellular matrix that they synthesize. In turn, these activated myofibroblasts induce further alveolar epithelial cell injury and death, thereby creating a vicious cycle of profibrotic epithelial cell–fibroblast interactions (49). Pulmonary lipofibroblasts participate in the synthesis of extracellular matrix structural proteins during alveolar development and transfer neutral lipids to AT2 cells to support surfactant and phospholipid synthesis in normal lung homeostasis (38); however, their participation in lung fibrosis has been rarely investigated. Our previous study has suggested EBF1+ fibroblasts as the potential progenitor cells of lung pericytes in the developing lungs (10). Pericytes play crucial key roles in lung morphogenesis and vascular homeostasis by regulating the epithelial and endothelial morphogenesis, and in fibrosis models, early ablation of pericytes reduced acute lung inflammation (50). The mesothelial cells, although at low proportion in the lungs, also play important roles in the fibrotic process through interaction with inflammatory cells, profibrotic mediators, and both the coagulation and fibrinolytic pathways (16). These studies have enumerated a great deal of data evidencing the involvement of the lung fibroblast subtypes in the fibrotic process not only by direct matrix contribution but also by regulating cell–cell communications and interactions.
In summary, this functional investigation extends our previous transcriptomics study (10), solidifying the concept that all fibroblast subpopulations, not only myofibroblasts, contribute to matrix production and to lung fibrosis. Although the fibroblasts are believed to be the key effectors in lung fibrosis, only myofibroblasts and the transition to the myofibroblast phenotype have drawn significant attention in research and in drug development. In addition, we found that, in contrast to matrix production, the emergence of the invasive fibroblast phenotype was more relegated to a few fibroblast subpopulations. Collectively, these data can inform future therapeutic approaches to targeting progressive pulmonary fibrosis.
Acknowledgments
Acknowledgment
The authors thank the Genomic Core, Biobank and Pathology Research Core, and Flow Cytometry Core of the Cedars-Sinai Medical Center for technical assistance.
Footnotes
Supported by National Institutes of Health grants R35-HL150829, R01-AI052201 (P.W.N.), and P01-HL108793 (P.W.N. and D.J.).
Author Contributions: D.J. and P.W.N. conceived this study. X.L. and J.L. performed single-cell RNA-sequencing data analyses. X.L., K.D., X.Z., G.H., H.L., and A.R. designed and performed the experiments. X.L., K.D., J.L., P.W.N., and D.J. prepared the figures and wrote the manuscript. All authors reviewed and approved the final version of the manuscript.
This article has a data supplement, which is accessible from this issue’s table of contents at www.atsjournals.org.
Originally Published in Press as DOI: 10.1165/rcmb.2022-0292OC on March 16, 2023
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1. Noble PW, Barkauskas CE, Jiang D. Pulmonary fibrosis: patterns and perpetrators. J Clin Invest . 2012;122:2756–2762. doi: 10.1172/JCI60323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Wijsenbeek M, Cottin V. Spectrum of fibrotic lung diseases. N Engl J Med . 2020;383:958–968. doi: 10.1056/NEJMra2005230. [DOI] [PubMed] [Google Scholar]
- 3. Distler JHW, Györfi AH, Ramanujam M, Whitfield ML, Königshoff M, Lafyatis R. Shared and distinct mechanisms of fibrosis. Nat Rev Rheumatol . 2019;15:705–730. doi: 10.1038/s41584-019-0322-7. [DOI] [PubMed] [Google Scholar]
- 4. Wu B, Tang L, Kapoor M. Fibroblasts and their responses to chronic injury in pulmonary fibrosis. Semin Arthritis Rheum . 2021;51:310–317. doi: 10.1016/j.semarthrit.2020.12.003. [DOI] [PubMed] [Google Scholar]
- 5. Jordana M, Schulman J, McSharry C, Irving LB, Newhouse MT, Jordana G, et al. Heterogeneous proliferative characteristics of human adult lung fibroblast lines and clonally derived fibroblasts from control and fibrotic tissue. Am Rev Respir Dis . 1988;137:579–584. doi: 10.1164/ajrccm/137.3.579. [DOI] [PubMed] [Google Scholar]
- 6. Phan SH. The myofibroblast in pulmonary fibrosis. Chest . 2002;122:286S–289S. doi: 10.1378/chest.122.6_suppl.286s. [DOI] [PubMed] [Google Scholar]
- 7. Blankesteijn WM, Essers-Janssen YP, Verluyten MJ, Daemen MJ, Smits JF. A homologue of Drosophila tissue polarity gene frizzled is expressed in migrating myofibroblasts in the infarcted rat heart. Nat Med . 1997;3:541–544. doi: 10.1038/nm0597-541. [DOI] [PubMed] [Google Scholar]
- 8. Sun KH, Chang Y, Reed NI, Sheppard D. α-Smooth muscle actin is an inconsistent marker of fibroblasts responsible for force-dependent TGFβ activation or collagen production across multiple models of organ fibrosis. Am J Physiol Lung Cell Mol Physiol . 2016;310:L824–L836. doi: 10.1152/ajplung.00350.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Xie T, Wang Y, Deng N, Huang G, Taghavifar F, Geng Y, et al. Single-cell deconvolution of fibroblast heterogeneity in mouse pulmonary fibrosis. Cell Rep . 2018;22:3625–3640. doi: 10.1016/j.celrep.2018.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Liu X, Rowan SC, Liang J, Yao C, Huang G, Deng N, et al. Categorization of lung mesenchymal cells in development and fibrosis. iScience . 2021;24:102551. doi: 10.1016/j.isci.2021.102551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Habermann AC, Gutierrez AJ, Bui LT, Yahn SL, Winters NI, Calvi CL, et al. Single-cell RNA sequencing reveals profibrotic roles of distinct epithelial and mesenchymal lineages in pulmonary fibrosis. Sci Adv . 2020;6:eaba1972. doi: 10.1126/sciadv.aba1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Adams TS, Schupp JC, Poli S, Ayaub EA, Neumark N, Ahangari F, et al. Single-cell RNA-seq reveals ectopic and aberrant lung-resident cell populations in idiopathic pulmonary fibrosis. Sci Adv . 2020;6:eaba1983. doi: 10.1126/sciadv.aba1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Travaglini KJ, Nabhan AN, Penland L, Sinha R, Gillich A, Sit RV, et al. A molecular cell atlas of the human lung from single-cell RNA sequencing. Nature . 2020;587:619–625. doi: 10.1038/s41586-020-2922-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lv YQ, Dhlamini Q, Chen C, Li X, Bellusci S, Zhang JS. Fgf10 and lipofibroblasts in lung homeostasis and disease: insights gained from the adipocytes. Front Cell Dev Biol . 2021;9:645400. doi: 10.3389/fcell.2021.645400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hung C, Linn G, Chow YH, Kobayashi A, Mittelsteadt K, Altemeier WA, et al. Role of lung pericytes and resident fibroblasts in the pathogenesis of pulmonary fibrosis. Am J Respir Crit Care Med . 2013;188:820–830. doi: 10.1164/rccm.201212-2297OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. von Gise A, Stevens SM, Honor LB, Oh JH, Gao C, Zhou B, et al. Contribution of fetal, but not adult, pulmonary mesothelium to mesenchymal lineages in lung homeostasis and fibrosis. Am J Respir Cell Mol Biol . 2016;54:222–230. doi: 10.1165/rcmb.2014-0461OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. El Agha E, Moiseenko A, Kheirollahi V, De Langhe S, Crnkovic S, Kwapiszewska G, et al. Two-way conversion between lipogenic and myogenic fibroblastic phenotypes marks the progression and resolution of lung fibrosis. Cell Stem Cell . 2017;20:261–273.e3. doi: 10.1016/j.stem.2016.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Karki S, Surolia R, Hock TD, Guroji P, Zolak JS, Duggal R, et al. Wilms’ tumor 1 (Wt1) regulates pleural mesothelial cell plasticity and transition into myofibroblasts in idiopathic pulmonary fibrosis. FASEB J . 2014;28:1122–1131. doi: 10.1096/fj.13-236828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sava P, Ramanathan A, Dobronyi A, Peng X, Sun H, Ledesma-Mendoza A, et al. Human pericytes adopt myofibroblast properties in the microenvironment of the IPF lung. JCI Insight . 2017;2:e96352. doi: 10.1172/jci.insight.96352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Aran D, Looney AP, Liu L, Wu E, Fong V, Hsu A, et al. Reference-based analysis of lung single-cell sequencing reveals a transitional profibrotic macrophage. Nat Immunol . 2019;20:163–172. doi: 10.1038/s41590-018-0276-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Peyser R, MacDonnell S, Gao Y, Cheng L, Kim Y, Kaplan T, et al. Defining the activated fibroblast population in lung fibrosis using single-cell sequencing. Am J Respir Cell Mol Biol . 2019;61:74–85. doi: 10.1165/rcmb.2018-0313OC. [DOI] [PubMed] [Google Scholar]
- 22. Parimon T, Yao C, Habiel DM, Ge L, Bora SA, Brauer R, et al. Syndecan-1 promotes lung fibrosis by regulating epithelial reprogramming through extracellular vesicles. JCI Insight . 2019;5:e129359. doi: 10.1172/jci.insight.129359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Tsukui T, Sun KH, Wetter JB, Wilson-Kanamori JR, Hazelwood LA, Henderson NC, et al. Collagen-producing lung cell atlas identifies multiple subsets with distinct localization and relevance to fibrosis. Nat Commun . 2020;11:1920. doi: 10.1038/s41467-020-15647-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ogawa T, Shichino S, Ueha S, Bando K, Matsushima K. Profibrotic properties of C1q+ interstitial macrophages in silica-induced pulmonary fibrosis in mice. Biochem Biophys Res Commun . 2022;599:113–119. doi: 10.1016/j.bbrc.2022.02.037. [DOI] [PubMed] [Google Scholar]
- 25. Valenzi E, Bulik M, Tabib T, Morse C, Sembrat J, Trejo Bittar H, et al. Single-cell analysis reveals fibroblast heterogeneity and myofibroblasts in systemic sclerosis-associated interstitial lung disease. Ann Rheum Dis . 2019;78:1379–1387. doi: 10.1136/annrheumdis-2018-214865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Morse C, Tabib T, Sembrat J, Buschur KL, Bittar HT, Valenzi E, et al. Proliferating SPP1/MERTK-expressing macrophages in idiopathic pulmonary fibrosis. Eur Respir J . 2019;54:1802441. doi: 10.1183/13993003.02441-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Reyfman PA, Walter JM, Joshi N, Anekalla KR, McQuattie-Pimentel AC, Chiu S, et al. Single-cell transcriptomic analysis of human lung provides insights into the pathobiology of pulmonary fibrosis. Am J Respir Crit Care Med . 2019;199:1517–1536. doi: 10.1164/rccm.201712-2410OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sun T, Huang Z, Liang WC, Yin J, Lin WY, Wu J, et al. TGFβ2 and TGFβ3 isoforms drive fibrotic disease pathogenesis. Sci Transl Med . 2021;13:eabe0407. doi: 10.1126/scitranslmed.abe0407. [DOI] [PubMed] [Google Scholar]
- 29. Melms JC, Biermann J, Huang H, Wang Y, Nair A, Tagore S, et al. A molecular single-cell lung atlas of lethal COVID-19. Nature . 2021;595:114–119. doi: 10.1038/s41586-021-03569-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bharat A, Querrey M, Markov NS, Kim S, Kurihara C, Garza-Castillon R, et al. Lung transplantation for patients with severe COVID-19. Sci Transl Med . 2020;12:eabe4282. doi: 10.1126/scitranslmed.abe4282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Geng Y, Liu X, Liang J, Habiel DM, Kulur V, Coelho AL, et al. PD-L1 on invasive fibroblasts drives fibrosis in a humanized model of idiopathic pulmonary fibrosis. JCI Insight . 2019;4:e125326. doi: 10.1172/jci.insight.125326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Liu X, Geng Y, Liang J, Coelho AL, Yao C, Deng N, et al. HER2 drives lung fibrosis by activating a metastatic cancer signature in invasive lung fibroblasts. J Exp Med . 2022;219:e20220126. doi: 10.1084/jem.20220126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Gerckens M, Schorpp K, Pelizza F, Wögrath M, Reichau K, Ma H, et al. Phenotypic drug screening in a human fibrosis model identified a novel class of antifibrotic therapeutics. Sci Adv . 2021;7:eabb3673. doi: 10.1126/sciadv.abb3673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Leung CC, Yu IT, Chen W. Silicosis. Lancet . 2012;379:2008–2018. doi: 10.1016/S0140-6736(12)60235-9. [DOI] [PubMed] [Google Scholar]
- 35. Arras M, Huaux F, Vink A, Delos M, Coutelier JP, Many MC, et al. Interleukin-9 reduces lung fibrosis and type 2 immune polarization induced by silica particles in a murine model. Am J Respir Cell Mol Biol . 2001;24:368–375. doi: 10.1165/ajrcmb.24.4.4249. [DOI] [PubMed] [Google Scholar]
- 36. Steen VD, Medsger TA. Changes in causes of death in systemic sclerosis, 1972-2002. Ann Rheum Dis . 2007;66:940–944. doi: 10.1136/ard.2006.066068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. George PM, Wells AU, Jenkins RG. Pulmonary fibrosis and COVID-19: the potential role for antifibrotic therapy. Lancet Respir Med . 2020;8:807–815. doi: 10.1016/S2213-2600(20)30225-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. McGowan SE, Torday JS. The pulmonary lipofibroblast (lipid interstitial cell) and its contributions to alveolar development. Annu Rev Physiol . 1997;59:43–62. doi: 10.1146/annurev.physiol.59.1.43. [DOI] [PubMed] [Google Scholar]
- 39. Li Y, Jiang D, Liang J, Meltzer EB, Gray A, Miura R, et al. Severe lung fibrosis requires an invasive fibroblast phenotype regulated by hyaluronan and CD44. J Exp Med . 2011;208:1459–1471. doi: 10.1084/jem.20102510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zhou Y, Horowitz JC, Naba A, Ambalavanan N, Atabai K, Balestrini J, et al. Extracellular matrix in lung development, homeostasis and disease. Matrix Biol . 2018;73:77–104. doi: 10.1016/j.matbio.2018.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Vallée A, Lecarpentier Y. TGF-β in fibrosis by acting as a conductor for contractile properties of myofibroblasts. Cell Biosci . 2019;9:98. doi: 10.1186/s13578-019-0362-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Renzoni EA, Poletti V, Mackintosh JA. Disease pathology in fibrotic interstitial lung disease: is it all about usual interstitial pneumonia? Lancet . 2021;398:1437–1449. doi: 10.1016/S0140-6736(21)01961-9. [DOI] [PubMed] [Google Scholar]
- 43. Rockey DC, Bell PD, Hill JA. Fibrosis—a common pathway to organ injury and failure. N Engl J Med . 2015;372:1138–1149. doi: 10.1056/NEJMra1300575. [DOI] [PubMed] [Google Scholar]
- 44. Gieseck RL, III, Wilson MS, Wynn TA. Type 2 immunity in tissue repair and fibrosis. Nat Rev Immunol . 2018;18:62–76. doi: 10.1038/nri.2017.90. [DOI] [PubMed] [Google Scholar]
- 45. Hinz B, Phan SH, Thannickal VJ, Galli A, Bochaton-Piallat ML, Gabbiani G. The myofibroblast: one function, multiple origins. Am J Pathol . 2007;170:1807–1816. doi: 10.2353/ajpath.2007.070112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. El Agha E, Herold S, Al Alam D, Quantius J, MacKenzie B, Carraro G, et al. Fgf10-positive cells represent a progenitor cell population during lung development and postnatally. Development . 2014;141:296–306. doi: 10.1242/dev.099747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Kramann R, Schneider RK, DiRocco DP, Machado F, Fleig S, Bondzie PA, et al. Perivascular Gli1+ progenitors are key contributors to injury-induced organ fibrosis. Cell Stem Cell . 2015;16:51–66. doi: 10.1016/j.stem.2014.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Li R, Li X, Hagood J, Zhu MS, Sun X. Myofibroblast contraction is essential for generating and regenerating the gas-exchange surface. J Clin Invest . 2020;130:2859–2871. doi: 10.1172/JCI132189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Sakai N, Tager AM. Fibrosis of two: epithelial cell-fibroblast interactions in pulmonary fibrosis. Biochim Biophys Acta . 2013;1832:911–921. doi: 10.1016/j.bbadis.2013.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Hung CF, Wilson CL, Chow YH, Liles WC, Gharib SA, Altemeier WA, et al. Effect of lung pericyte-like cell ablation on the bleomycin model of injury and repair. Am J Physiol Lung Cell Mol Physiol . 2022;322:L607–L616. doi: 10.1152/ajplung.00392.2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
scRNA-seq data of freshly isolated lung single cells from mice, human patients, and healthy donors were obtained from publicly available datasets, and the gene expression omnibus (GEO) accession numbers are presented in Table 1 (20–30). Details for data analysis and cell surface marker identification are described in the data supplement.
Table 1.
Accessibility of the Single-Cell RNA-Sequencing Data on Murine and Human Lung Tissues Used in This Study
| Species | Treatment/Model/Disease | GEO ID | Numbers of Tissues (n) |
Collection Timepoint | Dissociation Reagent | Instrument Model | References | |
|---|---|---|---|---|---|---|---|---|
| Fibrosis | Normal | |||||||
| Murine | Bleomycin/adult | GSE111664 | 3 | 7 | Day 14 | Liberase | MiSeq | 20 |
| GSE129605 | 4 | 4 | Day 11 | Collagenase A | NextSeq 500 | 21 | ||
| GSE131800 | 1 | 1 | Day 21 | Elastase | NovaSeq 6000 | 22 | ||
| GSE104154 | 1 | 1 | Day 21 | Elastase | NextSeq 500 | 9 | ||
| GSE132771 | 4 | 4 | Day 14 | Collagenase A | HiSeq 4000 | 23 | ||
| Bleomycin/aged | GSE157379 | 3 | 3 | Day 14 | Collagenase IV | NextSeq 500 | 10 | |
| Silica/adult | GSE184854 | 12 (Days 7 and 21) | 5 (Day 3) | Days 3, 7, and 21 | Liberase | NovaSeq 6000 | 24 | |
| Human | IPF | GSE136831 | 32 | 28 | NA | Elastase/liberase | HiSeq 4000 | 12 |
| GSE157376 | 5 | 5 | NA | Elastase | NextSeq 500 | 10 | ||
| GSE128169 | 0 | 5 | NA | Collagenase A | NextSeq 500 | 25 | ||
| GSE135893 | 12 | 10 | NA | Collagenase I | NovaSeq 6000 | 11 | ||
| GSE128033 | 8 | 10 | NA | Collagenase A | NextSeq 500 | 26 | ||
| GSE122960 | 4 | 8 | NA | Collagenase D | HiSeq 4000 | 27 | ||
| GSE132771 | 6 | 6 | NA | Collagenase A | NovaSeq 6000 | 23 | ||
| SSc-ILD | GSE159354 | 2 | 5 | NA | Collagenase I | HiSeq 4000 | 28 | |
| GSE132771 | 6 | 4 | NA | Collagenase A | NovaSeq 6000 | 23 | ||
| COVID-19 | GSE171524 | 19 | 7 | NA | NA | NovaSeq 6000 | 29 | |
| COVID-19 late-stage | GSE158127 | 3 | 3 | NA | Collagenase D | HiSeq 4000 | 30 | |
Definition of abbreviations: COVID-19 = coronavirus disease; GEO = gene expression omnibus; IPF = idiopathic pulmonary fibrosis; NA = not associated; SSc-ILD = systemic sclerosis–associated interstitial lung disease.
Bleomycin/adult indicates bleomycin-induced lung fibrosis in adult mice; bleomycin/aged indicates bleomycin-induced lung fibrosis in aged mice; silica/adult indicates silica-induced mouse lung fibrosis.