Abstract
Genetically encoded biosensor systems operating in living cells are versatile, cheap, and transferable tools for the detection and quantification of a broad range of small molecules. This review presents state-of-the-art biosensor designs and assemblies, featuring transcription factor-, riboswitch-, and enzyme-coupled devices, highly engineered fluorescent probes, and emerging two-component systems. Importantly, (bioinformatic-assisted) strategies to resolve contextual issues, which cause biosensors to miss performance criteria in vivo, are highlighted. The optimized biosensing circuits can be used to monitor chemicals of low molecular mass (<200 g mol–1) and physicochemical properties that challenge conventional chromatographical methods with high sensitivity. Examples herein include but are not limited to formaldehyde, formate, and pyruvate as immediate products from (synthetic) pathways for the fixation of carbon dioxide (CO2), industrially important derivatives like small- and medium-chain fatty acids and biofuels, as well as environmental toxins such as heavy metals or reactive oxygen and nitrogen species. Lastly, this review showcases biosensors capable of assessing the biosynthesis of platform chemicals from renewable resources, the enzymatic degradation of plastic waste, or the bioadsorption of highly toxic chemicals from the environment. These applications offer new biosensor-based manufacturing, recycling, and remediation strategies to tackle current and future environmental and socioeconomic challenges including the wastage of fossil fuels, the emission of greenhouse gases like CO2, and the pollution imposed on ecosystems and human health.
Introduction
Today’s environmental and socioeconomic challenges are multifaceted. Main contributors are the pollution of water, air, and soil, as well as the production of waste, paired with insufficient disposal and recycling strategies. Equally problematic are the depletion of natural resources and a strong dependence on fossil fuels. The latter lead to the ever-rising emission of greenhouse gases such as the single-carbon (C1) compounds carbon dioxide (CO2) or methane (CH4).1,2 To overcome these challenges, robust and efficient tools for the detection and quantification of contaminants as well as unprecedented strategies for the utilization of C1 molecules are highly desired. C1 compounds are considered important carbon sources in the future to both sustainably produce value-added chemicals and generate energy. Additionally, improved recycling and remediation schemes are vital to achieving closed-loop economies and reducing, ideally preventing, the pollution imposed on ecosystems and human health.1,3,4
Complementary to recent reviews highlighting advancements in natural and artificial carbon fixation pathways,1,5,6 this review will focus on genetically encoded biosensor systems operating in living cells, suitable to detect immediate fixation products (e.g., formate and pyruvate) and their industrially relevant derivatives. Physicochemical biosensor designs employed in vitro were recently reviewed by others and are not included herein.7,8 Furthermore, this condensed review will feature devices for the biosensing of (inorganic) contaminants that directly relate to anthropogenic activities, including, but not limited to, heavy metal (HM) and fluoride (F–) ions, as well as chalcogen-containing compounds such as hydrogen sulfide (H2S) and hydrogen peroxide (H2O2). Many of these chemical entities, organic and inorganic, have low molecular masses (<200 g mol–1) and can be difficult to analyze due to their physicochemical properties. Hence, analysis regularly requires laborious sample preparation and specialized instruments for detection and quantification.2
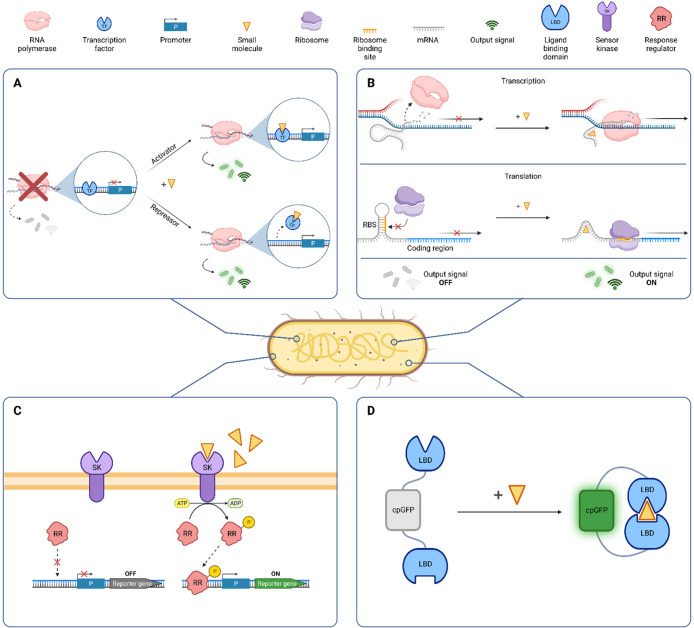
The implementation of genetically encoded biosensor systems offers solutions to these obstacles. Generally, biosensors consist of two functionally linked components: a sensing and a transduction module.9 Transcription factors (TFs; Figure 1A) and riboswitches (RSWs; Figure 1B) can be used as sensory parts to detect the presence of a chemical entity (i.e., the input signal). Minimal TFs consist of a DNA-binding domain (DBD) and a ligand-binding domain (LBD), while RSWs comprise oligonucleotides with a length of 30–80 nucleobases, so-called RNA aptamers that specifically bind a target small molecule. Ligand binding regularly triggers a conformational change in TFs and RSWs, regulating the expression of reporter or pathway genes, which act as transducers. Transducers decode the input into a readable output signal (e.g., fluorescence, luminescence) encoded by different reporter genes (Table 1).9−11 Complementary, two-component systems (TCS) have been introduced only recently as a class of biosensors. TCS consist of a membrane-bound sensor kinase that binds a small molecule in the environment. This input signal results in the activation of the intracellular kinase domain, phosphorylating a response regulator. The activated regulator can bind to its cognate promoter sequence, ultimately, driving reporter gene expression (Figure 1C).12 Lastly, fluorescent probes (Figure 1D) or certain enzymes such as luciferases (Table 1) directly generate readable outputs in the presence of target small molecules and will also be featured in this review.
Figure 1.
Types of genetically encoded biosensor systems. (A) (Allosteric) transcription factors (TFs) can act as activators or repressors. Whereas in the absence of the target small molecule no readable output signal is generated (OFF state), ligand binding to the TF facilitates promoter recognition or promoter clearance, respectively, resulting in the reporter gene transcription (ON state). (B) Riboswitches (RSWs) act on the co- or posttranscriptional level. Regulatory mechanisms involve the formation of hairpin terminators in the absence of the ligand, leading to truncated transcripts, or the sequestration of the RBS, impeding translation (OFF state). Binding of the small molecule, triggers a conformational change in the mRNA (mRNA), enabling transcription and translation, respectively (ON state). (C) Two component systems (TCS) consist of a membrane-bound sensor kinase. Ligand binding outside of the cell activates the kinase domain, subsequently phosphorylating a response regulator at the expense of adenosine triphosphate (ATP). The activated regulator protein recognizes its cognate promoter, enabling reporter gene expression (ON state). (D) Fluorescent probes such as variants of circularly permuted green fluorescent protein (cpGFP) act as sensors on the post-translational level. In cpGFPs, the termini are fused to sensing domains and fluorescent intensity is modulated in the presence of target small molecules, generating the output signal. Additionally, enzyme-based biosensor systems can directly generate output signals by converting target substrates (not shown; see main text). Figure created with BioRender.com.
Table 1. Featured Biosensor Systems.
| sensor | input (ligand) | output (reporter) | host | operational range | ref |
|---|---|---|---|---|---|
| AlkR (TF) | n-alkanes (≥C12) | GFP (fluorescence) | A. baylyi | 1 | (37) |
| AlkS (wildtype TF) | n-alkanes (C5–C10), primary SMCAHs (≥C5) | GFP (fluorescence) | E. coli | 0.5–10 mM2 | (34) |
| AlkS (mutant TF) | n-alkanes (C5–C9), various SMCAHs (C3–C5) | GFP (fluorescence) | E. coli | 1–100 mM3 | (34 and 35) |
| ArsR (TF) | As3+4 | sfGFP (fluorescence) | in vitro5 | 0.125 μM6 | (87) |
| AtoSC (TCS) | acetoacetate (C4) | GFP (fluorescence) | E. coli | 0.001–10 mM | (28) |
| BmoR (TF) | various SMCAHs (C3–C6) | GFP (fluorescence) | E. coli | 0.1–40 mM3 | (33) |
| CadC (TF)7 | Pb2+, Cd2+ | Luc (bioluminescence)8 | E. coli | 0.01–10 μM | (53) |
| CadC (TF)7 | Cd2+9 | sfGFP (fluorescence) | in vitro5 | 0.5 μM6 | (87) |
| CadR (TF)10 | Cd2+10 | GFP (fluorescence) | E. coli | 3 nM6,10 | (54) |
| CadR (TF)11 | Cd2+, Hg2+ | multiple read-outs11 | E. coli | 0.625–2.5 μM12 | (56) |
| ChnR (TF) | various lactams | mCherry (fluorescence) | E. coli | 1–100 mM | (62) |
| Co/Ni apt (RWS) | Co2+, Ni2+ | mCherry (fluorescence) | E. coli | 13 | (58) |
| DmpR (TF) | o-cresol (C7) | mCherry (fluorescence) | E. coli | 0.1–5 μM | (43) |
| FerC (TF) | feruloyl-CoA14 | sfGFP (fluorescence) | E. coli | 0.001–1 mM | (73) |
| FrmR (TF) | formaldehyde (C1) | multiple read-outs15 | E. coli | 1–25 μM | (21) |
| F– apt (RWS)16 | F– | sfGFP (fluorescence) | P. putida | 2.5–10 mM17 | (49) |
| Haa1-BM3R1 (TF)18 | organic acids (C1–C6) | mCherry (fluorescence) | S. cerevisiae | 10–60 mM18 | (19) |
| hsFRET (probe)19 | H2S | cpsGFP-EBFP2 (FRET; fluorescence) | in vitro/in vivo20 | 10–100 μM19 | (45) |
| HyPer7 (TF)21 | H2O2 | cpGFP (fluorescence) | in vitro/S. cerevisiae | 20–100 μM | (39) |
| Hypocrates (TF)22 | (pseudo)hypohalous acids | cpYFP (fluorescence) | in vitro/in vivo20 | 0.1–0.33 μM6 | (41) |
| Leu3p (TF) | 2-IPM (C7)23 | GFP (fluorescence) | S. cerevisiae | 10–80 μM | (32) |
| LuxAB (enzyme) | various aldehydes | LuxAB (bioluminescence) | E. coli or A. baylyi | 0.1 mM6,24 | (36, 37, and 75) |
| MerR (TF) | Hg2+ | sfGFP (fluorescence) | in vitro5 | 0.011 μM6 | (87) |
| MGapt (RWS) | malachite green | GFP (fluorescence) | in vitro5 | 0.0375–3.0 μM25 | (88) |
| OxyR (TF) | hydrogen peroxide (H2O2) | multiple read-outs26 | E. coli | 5–100 μM17 | (43) |
| PbrR (TF)27 | Pb2+ | GFP (fluorescence) | E. coli | 0.001–100 μM28 | (52) |
| PbrR (TF)27 | Pb2+ | Luc (bioluminescence)8 | E. coli | 1–100 μM | (53) |
| PcaV (TF) | protocatechuic acid (C7) | sfGFP (fluorescence) | E. coli | 0.1–1000 μM29 | (73) |
| pnGFP-Ultra (probe) | peroxynitrite (ONO2–) | cpsGFP (fluorescence) | in vitro/in vivo20 | 122 nM6 | (40) |
| PREmR34 (RWS) | protons (pH sensor) | mCherry (fluorescence) | E. coli | pH = 5.0–8.0 | (72) |
| PyronicSF (TF)30 | pyruvate (C3) | cpGFP (fluorescence) | in vitro/in vivo20 | 30 | (22) |
| Tsa2-GFP (enzyme)31 | H2O2 | roGFP (fluorescence) | in vitro/S. cerevisiae | 1–100 μM31 | (39 and 44) |
| War1 (TF) | various SMCFAs (C3–C7) | GFP (fluorescence) | S. cerevisiae | 0.5–25 mM32 | (29 and 30) |
GFP output at 40 mM n-alkane concentration.
Linear range for 1-pentanol.
Linear range for 1-butanol.
Response also to As5+, Cd2+, Hg2+, and Pb2+ ions.
TF and sfGFP transcribed/translated by cell-free expression (CFE) systems.
Lower limit of detection (LOD).
Staphylococcus aureus (S. aureus) CadC.
Luc, firefly luciferase.
CFE also induced by Pb2+ and Hg2+.
Halomonas caseinilytica CadR; biosensor sensitivity increased by ZitB transporter, minimizing Zn2+ interference.
Pseudomonas putida CadR; reporters: fluorescent proteins (GFP or mCherry) and LacZ (β-galactosidase).
For Cd2+; induction by Hg2+ >2.5 μM.
Detection range: Co2+ <1 mM; Ni2+ <2 mM.
Feruloyl-CoA produced from ferulic acid by FerA.
GFP (fluorescence) or LuxAB (bioluminescence).
Apt, aptamer.
Linear range; for maximal nonlinear response, see reference.
BM3R1 DBD fused to the TF Haa1; operational range for acetic acid (C2).
CpsGFP-EBFP2 fusion probe; linear change in FRET ratio in vitro with hsFRET (1 μM) and tested H2S concentrations.
For mammalian cells and/or animal model, see reference.
OxyR-GFP fusion.
NemR-YFP fusion.
2-IPM detected as byproduct or precursor, indicating isobutanol (C4) or isopentanol (C5) production, respectively.
Based linear calibration with purified MGapt-GFP transcript.88
mCherry or GFP.
Cupriavidus metallidurans PbrR.
LOD of most sensitive biosensor variant (S23); sensitivity increased by LuxR as signal amplifier.
Sensitivity enhanced by protocatechuic acid-specific transporter PcaK.
PdhR-GFP fusion; changes in pyruvate concentrations as low as 10 μM detected (e.g., mean mitochondrial pyruvate concentrations = 25 μM).
Thiol peroxidase Tsa2 fused to H2O2-sensitive roGFP;44 detection range according to Pak and co-workers.39
Based on isovaleric acid (C5); concentrations >25 mM not tested.
In the last two decades, the utilization of biosensors has advanced beyond their initial use as analytical tools for the high-throughput (HT) detection and quantification of natural metabolites and xenobiotics. Today, biosensor systems complement well-established but low-throughput chromatographical methods and have facilitated the directed evolution of proteins, the engineering of (natural) metabolic pathways, and their dynamic control. This resulted in the construction of microbial cell factories for the detection of various small molecules and the efficient manufacturing of value-added platform chemicals, of which selected examples, focused on biosensing, will be featured in the following.10,13,14
Biosensors for Carbon Fixation Products and Derivatives
Currently, the fixation of CO2 through the Calvin-Benson-Bassham cycle mainly by autotrophs (e.g., plants, algae, cyanobacteria) and a few recently discovered natural carbon-fixation pathways in bacteria and archaea cannot balance the excessive anthropogenic CO2 emission.5 Very recently, Gleizer et al. constructed and evolved Escherichia coli (E. coli) to produce all its biomass carbon from CO2via the Calvin-Benson-Bassham cycle, enabling autotrophic growth of an otherwise heterotrophic bacterium.15 Complementary, significant efforts have been made not only to transplant and improve natural carbon fixation but also to design artificial assimilation routes.1,5 While many natural and de novo CO2 fixation pathways depend on the same energy carriers such as ATP and (phosphorylated) nicotinamide cofactors, fixation products and pathway intermediates can be fairly distinct.5,6 Besides the challenging implementation of synthetic pathways in microorganisms, biosensors might not have become available to sense associated small molecules.1,5,10 This is also true for the CETCH (crotonyl-CoA/ethylmalonyl-CoA/hydroxybutyryl-CoA) cycle, assembled by Schwander and co-workers, for the fixation of two molecules CO2in vitro.16 Furthermore, many immediate CO2 fixation products are quickly metabolized and do not accumulate in living (micro)organisms, calling for qualitative and quantitative measurement tools that are highly sensitive to evaluate engineering efforts.
Biosensors have been developed and employed to match these requirements but have yet to be implemented to accelerate the optimization of (artificial) CO2 assimilation pathways and to complement (indirect) performance assessments such as growth assays.10,17 Hence, biosensor systems that have been developed for other purposes but are suitable to detect carbon fixation products and related molecules will be highlighted in the following.5,6
One industrially important CO2 fixation product is formate (C1; 46.03 g mol–1), for example. Since it can be produced (enzymatically) from multiple (renewable) sources and further converted into platform chemicals like formaldehyde and pyruvate, formate is of interest to different industries.5,18 Mormino et al. constructed a biosensor, discussed for biosensing formate, by fusing the Saccharomyces cerevisiae (S. cerevisiae) TF Haa1 and the DBD of BM3R1 from Bacillus megaterium. Whereas promotors containing Haa1 binding sites did not drive reporter gene expression, the production of mCherry, a red fluorescent protein (RFP), could be tuned by synthetic promoters containing different numbers of BM3R1 binding sites in the presence of various C1–C6 acids, including formate, acetate (C2; 60.05 g mol–1), propionate (C3; 74.08 g mol–1), and lactate (C3; 90.08 g mol–1).19
Similarly, formaldehyde (C1; 30.03 g mol–1) is a versatile chemical building block. Lu et al. implemented an artificial fixation route for formaldehyde operating in E. coli.20 Their synthetic acetyl-coenzyme A (SACA) cycle condenses two molecules of formaldehyde into glycolaldehyde. The latter is further converted into acetyl-coenzyme A (CoA), which is a central metabolite for the biosynthesis of numerous products as discussed below. Due to its high reactivity, formaldehyde is considered an environmental toxin and must be carefully monitored.2,21 Accordingly, Woolston et al. developed a formaldehyde biosensor based on the FrmR repressor protein and the cognate Pfrm promoter sequence of E. coli. The native TF binding site was rationally engineered and allowed detection as low as 1 μM, utilizing the luciferase LuxAB from Photorhabdus luminescens or GFP as readouts. Ultimately, the biosensor system was used to characterize methanol dehydrogenase variants in vivo.21
Arce-Molina et al. developed a highly responsive sensor for pyruvate (C3; 88.06 g mol–1), another carbon fixation product.22 Pyruvate is a central molecule in carbon metabolism and a precursor for acetyl-CoA, which fuels numerous vital metabolic pathways.5,20 The biosensor PyronicSF comprises the complete sequence of the bacterial TF PdhR, which is linked to a circularly permuted version of green fluorescent protein (cpGFP).22 CpGFPs or other fluorescent proteins contain engineered termini that are fused to sensing domains. The conformational rearrangement of the latter modulates the fluorescent intensity.23 Exposure of the PdhR-GFP sensor to pyruvate causes an increase in fluorescence when excited by blue light. Importantly, their biosensor remained insensitive to acetate, lactate, and other structurally related C2–C6 acids.
As introduced above, acetyl-CoA is ubiquitous in living organisms and a key intermediate in the biosynthesis of (branched) short- and medium-chain fatty acids (SMCFAs), the corresponding alcohols (SMCAHs), and n-alkanes, among other compounds. Particularly, SMCFAs and SMCAHs are considered valuable carbon and energy sources and have applications as drop-in biofuels.24,25 Consequently, the biosensor-based engineering of microbial cells to sustainably produce these molecules is of great interest.
Rutter et al. utilized a TCS from E. coli to detect acetoacetate (C4; 102.09 g mol–1), which can be directly used as a carbon source and plays a crucial role in the synthesis of SMCFAs and poly-(R)-3-hydroxybutyrate, for example.26,27 The whole-cell biosensor consists of the AtoS histidine kinase, which autophosphorylates in the presence of acetoacetate. Subsequently, the phosphate group is transferred to the AtoC response regulator, which triggers the expression of gfp from the native promoter Pato. On the basis of a model-driven sensitivity analysis, Rutter et al. generated a range of input/output responses by tuning the concentration of AtoS/AtoC (genomic versus plasmid-based expression) and Pato (low- versus high-copy number vector), aiming at optimizing the genetic context.28 Since TCS are highly susceptible to contextual effects, their engineering and contextualization will play a crucial role in broader future applications as briefly discussed later.
To directly monitor the production of SMCFAs, Baumann et al. developed a whole-cell biosensor that is based on a multicopy yeast plasmid, encoding gfp as the reporter gene under transcriptional control of the PDR12 promoter (PPDR12) and the constitutively bound TF War1. Upon exposure to SMCFAs (C3–C7), War1 undergoes phosphorylation and conformational changes, initiating GFP expression in S. cerevisiae.29 Similarly, Miyake et al. utilized PPDR12/War1 and GFP to detect the branched SMCFAs isobutyric acid (C4; 88.11 g mol–1), 2-methylbutanoic acid and 3-methylbutanoic acid (both C5; 102.13 g mol–1), as well as unsaturated methacrylic acid (C4; 86.06 g mol–1), and interestingly, benzoic acid (C7; 122.12 g mol–1).30 The authors also altered biosensor performance (e.g., sensitivity and operational range) by varying the expression levels of the weak organic acid transporter PDR12 and suggested the implementation of War1 mutants, displaying different binding affinities of the LBD and the DBD for branched SMCFAs and cognate operator sequences, respectively.31
For the detection of the corresponding (branched-chain) SMCAHs, Zhang et al. developed a TF-based biosensor system in S. cerevisiae.32 The TF Leu3p binds 2-isopropyl malate (2-IPM, C7; 176.17 g mol–1) and controls the expression of GFP. Two distinct biosensor configurations enable the HT screening for the enhanced production of isobutanol (C4; 74.12 g mol–1) or isopentanol (C5; 88.15 g mol–1) by monitoring 2-IPM as a byproduct or as a precursor, respectively.
In contrast, Yu et al. directly monitored the production of isobutanol in E. coli through the alcohol-regulated TF BmoR and its cognate promoter Pbmo, driving the expression of GFP. In the natural host Thauera butanivorans, BmoR regulates an alkane monooxygenase involved in the metabolization of n-alkanes. The TF exhibits high sensitivity to linear and branched-chain SMCAHs (C4–C6 and C3–C5, respectively).33 A complementary approach was utilized by Bahls et al., who engineered the LBD of the TF AlkS by error-prone polymerase chain reaction (epPCR), importantly, without influencing the DBD.34 Wildtype AlkS accepts short- and medium-chain n-alkanes and the corresponding SMCAHs, with 1-pentanol (C5; 88.15 g mol–1) being the shortest alcohol detectable. Mutant AlkS also accepted isopropanol (C3; 60.10 g mol–1), butanol and 2-butanol, as well as isopentanol. E. coli cells expressing AlkS mutants and GFP were enriched by fluorescence-activated cell sorting (FACS) after exposure to the new ligands.34 In another study, epPCR had been used to enhance the binding profile of AlkS to n-alkanes (C5–C9).35
In the context of the detection and manufacturing of biofuels in living cells, the previously introduced monooxygenase LuxAB from Photorhabdus luminescens has been implemented versatilely. Bayer et al. coupled different heterologous oxidoreductases and LuxAB to monitor the production of aldehydes, including hexanal (C6; 100.16 g mol–1) and benzaldehyde (C7; 106.12 g mol–1), from carboxylic acid or primary alcohol substrates in an engineered E. coli K-12 MG1655 strain, exhibiting reduced aldehyde reduction activity (E. coli RARE).36 The structural relation of the investigated substrates to SMCFAs and SMCAHs might qualify this enzyme-coupled biosensor system not only for the HT detection of the corresponding aldehydes but the optimized biobased production of biofuels. Lastly, Lehtinen et al. combined the acetogen Acetobacterium woodii, to produce acetate from CO2 and H2, and an engineered Acinetobacter baylyi ADP1 (A. baylyi) strain. Albeit low-yielding, the second strain converted the produced acetate into n-alkanes. This was monitored by a twin-layer biosensor system: the formation of intermediate aldehydes was followed by LuxAB. A cyanobacterial aldehyde-deformylating oxygenase yielded, besides formate, target n-alkanes, which were sensed by the TF AlkR and the cognate PalkM promoter, regulating the expression of GFP.37
So far, examples of the implementation of genetically encoded biosensors focused on the assessment of strategies for the conversion of C1 compounds into industrially important precursors and diversified products including biofuels (C2–C7). As pointed out earlier, many of these organic compounds do not accumulate in high quantities naturally.10 The C1 molecules methanol, formaldehyde, and formate, as well as n-alkanes exhibit cellular toxicity even at low concentrations and their assimilation as well as their detoxification requires (micro)organisms with unique enzymatic activities.1,4,18,21,36 Similarly, inorganic molecules leaking into the environment, even accumulating as seen with HMs, due to anthropogenic actions can be toxic.2,4,38 Hence, the following section will feature biosensor systems for the detection of selected inorganic compounds.
Biosensing of Inorganic Molecules and Contaminants
Physiologically, many inorganic compounds such as metals, reactive oxygen and nitrogen species (ROS and RNS, respectively), or hypohalous acids are involved in important cellular processes.39−41 Consequently, not only the detection of inorganic pollutants is crucial; robust and highly sensitive biosensor systems enable the monitoring of biological functions and signaling events at single-cell resolution.
Hydrogen peroxide (H2O2; 34.02 g mol–1) is a ROS and a key intermediate of the aerobic metabolism. Different sensitive biosensors for the detection of H2O2, referred to as HyPer variants, have been developed and are based on circularly permuted yellow fluorescent protein (cpYFP) or cpGFP integrated into the regulatory domain of the E. coli TF OxyR, an H2O2-sensing regulator.39 A cysteine residue (Cys199) in OxyR is sensitive to oxidation by H2O2, but not other oxidants, resulting in a disulfide bond formation and a subsequent conformation change.42 Initially, 11 OxyR regulatory domains from different bacterial species were tested for the integration of cpYFP with varying insertion positions to optimize folding of the fluorescence domain. The most sensitive construct, using OxyR from Neisseria meningitidis, was further enhanced by protein engineering, using a combination of rational and random mutations. This optimized biosensor allowed in vitro detection of H2O2 concentrations in the lower micromolar range, while demonstrating high pH stability at physiological relevant conditions, an advantage to previous HyPer variants.39
Kardashliev et al. employed OxyR coupled to fluorescent reporters – GFP or RFP – to monitor H2O2 as a byproduct of the oxidation of glycerol to glyceraldehyde (C3; 90.08 g mol–1) and of toluene to o-cresol (C7; 108.14 g mol–1) by different recombinant oxidoreductases in E. coli. Additionally, o-cresol formation was followed by a second genetically encoded sensor, the phenol-sensitive transcriptional activator DmpR, which drives the expression of an orthogonal fluorescent reporter gene.43
A third biosensor for H2O2 utilizes the thiol peroxidase Tsa2 from S. cerevisiae genetically linked to a redox-sensitive GFP (roGFP). In this redox relay system, Tsa2 catalyzes the transfer of oxidizing equivalents from H2O2 to roGFP. Under endogenous conditions, the biosensor is about 50% oxidized, which allows measuring the increase and decrease of H2O2 in living cells with both high sensitivity and selectivity.44
Like ROS, RNS are associated with signal transduction and stress responses in mammalian cells. Hence, Chen et al. advanced an earlier biosensor design to develop a highly sensitive fluorescence probe for peroxynitrite (ONO2–; 62.01 g mol–1), using a circularly permuted superfolder GFP (cpsGFP) with the noncanonical amino acid p-boronophenylalanine incorporated into the chromophore.40 Several rounds of directed evolution yielded a fluorescence biosensor with a 110-fold turn-on response and high selectivity for peroxynitrite over other ROS/RNS. Peroxynitrite concentrations as low as 122 nM could be detected in vitro and in vivo.
(Pseudo)hypohalous acids, including hypochlorous acid (HOCl; 52.46 g mol–1) and hypobromous acid (HOBr; 96.91 g mol–1), are part of the immune response system that provides an effective defense mechanism against bacteria, fungi, and larger parasites. Kostyuk et al. developed a fluorescence biosensor consisting of cpYFP integrated into the transcriptional repressor NemR from E. coli, which senses hypochlorite anions.41 To optimize the fluorescence response, 16 different constructs with varying linker regions were designed and tested. The best construct, named Hypocrates, showed a 1.6-fold turn-on response and allowed the detection of HOCl, HOBr, N-chlorotaurine, and hypothiocyanous acid with a lower detection limit of 100–330 nM (for NaOCl and NaOBr). Hypocrates enabled measuring (pseudo)hypohalous acid derivatives in different mammalian cell lines and in a zebrafish model.
Recently, Youssef et al. developed a fluorescent biosensor for H2S (34.1 g mol–1), a toxic gas and an important biological signaling molecule. Detection is based on the incorporation of the noncanonical amino acid p-azidophenylalanine in an engineered GFP-blue fluorescent protein (BFP) fusion.45 First, cpsGFP was fused to the BFP EBFP2 by a short, flexible peptide linker and resulted in a Förster resonance energy transfer (FRET) pair. This chimeric fusion was then modified by genetic code expansion to incorporate p-azidophenylalanine into the chromophore of cpsGFP (Tyr154). The construct was improved through several rounds of directed evolution for increased FRET. This yielded the optimized mutant hsFRET, in which the azido group in p-azidophenylalanine is reduced to an amino group by H2S, resulting in enhanced FRET from EBFP2; hsFRET allowed the selective detection of H2S as low as 10 μM in vitro. Importantly, experiments in mammalian cells demonstrated that other redox-active molecules caused no signal response by hsFRET. Noteworthy, other fluorescence proteins incorporating noncanonical amino acids have been reported as well.46,47
Most of the TF-based biosensors detecting the inorganic molecules described above operated in eukaryotic organisms and subcellular compartments (e.g., mitochondria). Whereas their transplantation into microbial host cells remains to be demonstrated, the detection of HM ions, for example, was realized in different E. coli strains.
Many (divalent) ions fulfill essential cellular functions. While free intracellular levels of magnesium ions can reach 5 mM, the same concentration of divalent ions like nickel (Ni2+) or cobalt (Co2+) can inhibit growth; the F– anion is toxic at elevated concentrations.48,49 HMs such as lead (Pb), cadmium (Cd), and mercury (Hg) exert toxic effects on living organisms at very low concentrations and are recalcitrant to degradation. Some of these HMs can be absorbed by plants and, thus, can potentially accumulate in the food chain.38 Pb and Hg have toxic effects on the nervous, digestive and immune systems, lungs, and kidneys. Similarly, exposure to low levels of Cd over time, particularly in tobacco smoke, may cause kidney disease. Cd is also considered a cancer-causing agent.50,51 Consequently, the leakage of HMs from industrial and agricultural activities into the environment is of concern, and biosensors for their sensitive, fast, and low-cost monitoring are desired.
Jia et al. improved a microbial whole-cell biosensor for Pb (207.2 g mol–1).52 In Cupriavidus metallidurans CH34, the TF PbrR regulates a gene operon conferring resistance to enhanced levels of Pb2+ ions from the divergent promoter Ppbr. Synthetic PbrR-based gene circuits were constructed featuring different genetic architectures tuning the expression of gfp as the reporter gene. Furthermore, a positive-feedback amplifier employing a variant of the quorum-sensing regulator LuxR was introduced to improve sensitivity to Pb2+ ions and increase the fluorescent output signal. In genetic configurations featuring positive feedback loops, the divergent promoter Ppbr also controlled the expression of luxR, which binds to its cognate promoter PluxI. The latter drives the expression of the gfp reporter gene and an additional copy of luxR amplifier, which increased the output signal up to 1.9-times compared to biosensors without positive feedback.52 Biosensors operating in E. coli DH5α were able to detect Pb2+ at a concentration as low as 0.01 μM; the standard drinking water quality requirement is <0.05 μM.51
Similarly, Nourmohammadi et al. compared two Pb biosensors with luciferase reporter gene expression either controlled by PbrR/Ppbr from Cupriavidus metallidurans CH34 or CadC/Pcad from Staphylococcus aureus (S. aureus) in E. coli DH5α.53 Whereas the PbrR-based construct detected Pb concentrations of 1–100 μM without interference by other metal ions, including Cd2+, Ni2+, and zinc (Zn2+), the CadC-based biosensor could detect Pb2+ concentrations between 10 nM and 10 μM.
He et al. genetically tuned the metal transport system of E. coli to enrich intracellular Cd2+ ions, while reducing the concentration of other interfering metal species in vivo. The TFs CadR from Pseudomonas aeruginosa and Pseudomonas putida (P. putida) were coupled to GFP as the readout. Although these biosensors were able to detect Cd (112.41 g mol–1) at relatively low concentrations of 1 μM, an unspecific response to Hg2+, Pb2+, and Zn2+ ions, for example, was observed. After an extensive sequence similarity search, 14 more Cd-sensing TFs were selected and tested. The resulting Cd biosensor featured CadR from Halomonas caseinilytica JCM 14802 and the E. coli metal transporter ZitB. The system was highly specific for Cd2+ and showed a detection limit of 3 nM.54 ZitB preferentially ejects Zn2+ ions, thereby, minimizing interference.55
The natural cad operon of P. putida is controlled by the repressor CadR, binding to its cognate divergent promoter (Pcad). In the presence of Cd, the dissociation of CadR increases the transcription of its own gene and a Cd efflux pump. Guo et al. redesigned the operon, generating a novel cell-based bioadsorption device for Cd2+ ions as well as different multiple-signal biosensor modules for their detection in vivo.56 Bioadsorption was facilitated by the Cd-binding domain (CdBD) from CadR fused to the C-terminus of the surface display protein Lpp-OmpA. The chimeric protein Lpp-OmpA-CdBD can bind Cd on the cellular surface. Although it remained unclear whether enriched Cd on the surface of whole-cell biosensors contributed to elevated intracellular concentrations of Cd, and consequently, an increased GFP output, the surface adsorption of Cd2+ ions was up to 8.8-fold higher than with E. coli TOP10 cells lacking CdBD display. Biosensor constructs employed the reporters mCherry, GFP, and LacZ (encoding a β-galactosidase) in different combinations under the control of CadR/Pcad to generate multiple readouts. The lower limit of detection for Cd2+ ions was 0.1 μM; strong output signals were also observed in the presence of Hg2+ ions.
Regarding the detection of the biologically essential metal ions Ni2+ (58.69 g mol–1) and Co2+ (58.93 g mol–1), Wang et al. employed a previously described Co/Ni-specific RSW by Furukawa and co-workers.57 Their sensor responds to increased intracellular ion levels, regulating the expression of mCherry fluorescent protein in E. coli DH5α. Subsequently, the RSW was used to assess the influence of gene deletions, ΔrcnA, ΔrcnB, ΔrcnR, ΔnikA, and ΔnikR, related to Co/Ni homeostasis and detoxification in E. coli K12 strains. Lower limits of reporter gene induction were 50 μM and ≥1 mM for Co2+ and Ni2+, respectively.58
Lastly, Calero et al. constructed synthetic RSWs responsive to F– (18.99 g mol–1). For biosensing, different (artificial) promoters controlled the production of superfolder GFP (sfGFP) in P. putida.49 Discernable sfGFP outputs above background levels could be detected at 2.5 mM sodium fluoride (NaF) and increased 200-fold at 15 mM; deletion of the crcB gene (encoding a F– exporter) inhibited growth at NaF concentrations ≥0.5 mM. Subsequently, Calero et al. coupled the translation of an orthogonal T7 RNA polymerase (RNAP) to the RWS. The T7 RNAP facilitates the T7 promoter (PT7)-controlled expression of various fluorinases and a purine nucleotide phosphorylase to synthesize fluorosugars and fluoronucleotides in vivo.
Identification of Novel Biosensor Systems and Contextualization Strategies In Vivo
Biosensing and signal transduction in response to chemicals in the environment are common features in all living (micro)organisms and essential to regulate cellular functions including growth and survival.10,12 Key players are the naturally evolved sensory devices including TFs, RSWs, and TCS (Figure 1A–C). In particular TF- and RSW-based biosensor systems have been utilized, engineered, and contextualized to efficiently detect (non-natural) compounds as highlighted above. To facilitate the detection of small molecules for which no natural or engineered sensory part has become available yet, tools for the identification of novel biosensors as well as the adaptation of biosensor performance in the cellular context are vital.
In recent years, the number of characterized RSWs as sensing tools has not increased as rapidly compared to TFs although RSWs are predicted to be broadly distributed gene regulatory elements in bacteria, fungi, and plants but are conspicuously absent in animals.10,59 This is also deducible from the overrepresentation of TF-based biosensor systems in this focused review (Table 1). Similarly, TCS are a vast group of proteins, predicted to be the biggest group of signal transduction pathways in biology. Due to their only recent introduction as biosensor devices, conserved regulatory motifs and variability in TCS across (microbial) genomes, determining signaling mechanisms, their integration in gene regulatory networks, and ligand-binding scopes are still uncharacterized. Additionally, first engineering approaches indicate that TCS are highly susceptible to contextual effects, impeding their broader application.12,60,61
So far, TFs have been successfully identified by combined transcriptome and proteome analyses after exposure of (microbial) cells to the desired small molecule as well as the computer-assisted mining of databases. Furthermore, (prokaryotic) TFs can be responsive to different but structurally related compounds (e.g., War1 for SMCFAs,29,30 BmoR,33 and AlkS34,35 for SMCAHs, or CadC for Cd2+, Pb2+, and other HM ions53). This ligand binding promiscuity facilitated the random mutagenesis by epPCR and the rational engineering of various LBDs and DBDs by well-established protein engineering techniques.10,17,31,32,34,35 A complementary approach for the identification of new biosensors was employed by Zhang et al., who scouted chemicals with molecular shapes similar to the target small molecule. Bioinformatic-assisted gene cluster analysis identified the TF ChnR and the cognate promoter Pb as a biosensor for the non-natural γ-butyrolactam (2-pyrrolidone; 85.11 g mol–1), δ-valerolactam (2-piperidone; 99.13 g mol–1), and ε-caprolactam (azepan-2-one; 113.16 g mol–1). Detection of these platform chemicals was based on mCherry with a linear range of induction at 1–100 mM.62
Curated databases for prokaryotic and eukaryotic TFs such as PRODORIC (https://www.prodoric.de)63 and JASPAR (https://jaspar.genereg.net),64 respectively, include ligand binding profiles or the type of regulation (e.g., transcriptional activation and repression) together with cognate gene regulatory elements, which can assist initial biosensor designs. Similarly, databases and prediction tools such as Rfam (http://rfam.org)65 and the Riboswitch Scanner (http://service.iiserkol.ac.in/~riboscan/),66 respectively, are valuable sources of prokaryotic RSWs, providing information about their RNA aptamer sequences, conserved secondary structures, and known ligands. Despite the simple architecture of RSWs, the limited understanding of ligand-induced structural changes and the strong genetic contextual dependency impair their rational design.10,49,67 Hence, engineering examples exist but are scarce.
The systematic evolution of ligands by exponential enrichment (SELEX) has been successfully employed, during which a library of oligonucleotides specifically binding the target ligand can be enriched in vitro.68 During classical SELEX, only RNA aptamers with high-affinity binding were selected, while accompanying essential conformational changes had been neglected. Recently, this was addressed by various groups through RNA Capture-SELEX protocols, for example.69−71 This strategy involves the insertion of a small defined motif within the randomized region of RNA aptamer libraries. This “docking sequence” is hybridized through base pairing with a complementary oligonucleotide containing a linker molecule and biotin, which is anchored on streptavidin-coated magnetic beads. Only aptamers undergoing a structural rearrangement upon addition of the target ligand will be eluted from the beads.
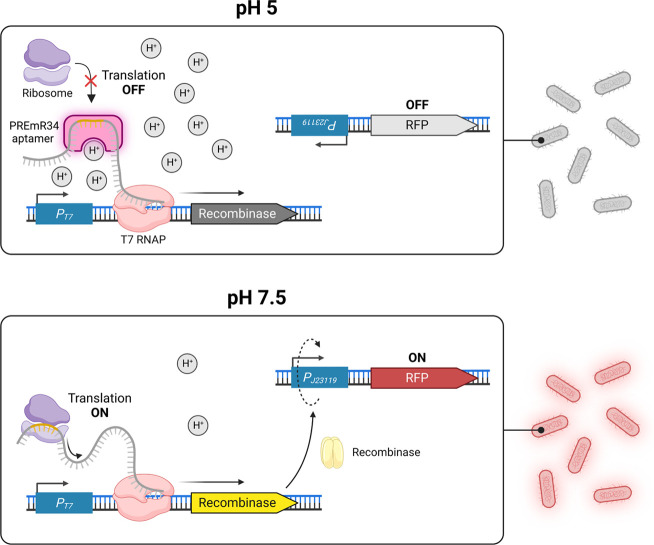
One prominent engineering example by Pham et al. features a set of pH-responsive RSWs, enabling the production of RFP in response to changing environmental pH conditions (Figure 2).72 At an extracellular pH of 6.8, the wild-type RWS adopts a structure with a (weak) ribosome binding site (RBS), inaccessible for the ribosome, yielding translationally inactive transcripts (OFF state). Under an extracellular pH of 8.0, conformational changes lead to the formation of translationally active transcripts with an accessible RBS (ON state; Figure 2, bottom). For optimized biosensor performance, Pham et al. had to employ multiple strategies. First, the weak natural RBS of the RWS was replaced by a strong artificial one. Alleviating the leakiness of the pH-responsive sensor devices was equally important. Whereas tuning the PT7 strength was less effective, the implementation of a T7 RNAP with mutations in the active site (F644A and Q649S) to modulate transcription speed was crucial. Orthogonal expression of the T7 RNAP mutant was controlled by the inducer l-arabinose and carefully adjusted. Lastly, the engineered pH-sensitive RWS (PREmR34) downstream of PT7 regulated the expression of a recombinase. At a low extracellular pH of 5.0, the recombinase is repressed, maintaining E. coli cells in their OFF state (no fluorescence; Figure 2, top). At elevated pH, PREmR34 allows recombinase expression. This triggers the switching of the physical direction constitutive promoter J23119 (PJ23119), embedded within the recombinase recognition sites (attB and attP). An insulator sequence upstream of the RFP-coding region was inserted to minimize potential context effects of the long 5′-untranslated region (UTR) introduced by the attB and attP flanking sites. Transcription of PJ23119 and subsequent translation led to high red fluorescence (ON state; Figure 2, bottom). In the final genetic circuit, PREmR34 increased the OFF/ON fluorescence output up to 31-fold with a broadened dynamic range (pH 5.0–8.0). To illustrate the practicality of the engineered RWS system, pH sensing was linked to an artificial error-prone genome replication machinery, representing a novel RSW-based directed evolution (RiDE) protocol. Pham et al. employed RiDE, assisted by automated sampling, HT flow cytometry, and FACS, to isolate E. coli mutants tolerant to increased amounts of industrially important organic acids (C2–C5). This is a requisite for E. coli strains to be employed as production hosts.72
Figure 2.
Contextualization of the pH-responsive RWS-based biosensor PREmR34. Transcripts are produced by a (mutant) T7 RNAP in E. coli. At low external pH, the strong RBS encoded within the aptamer (region highlighted in pink) is inaccessible for the ribosome (OFF state; top). At elevated pH, translation of a recombinase occurred, switching the orientation of the PJ23119 promoter. Subsequent transcription and translation yield RFP as the readable output (ON state, bottom). Multilevel optimization approaches greatly increased the dynamic range of the target RWS.72 Figure created with BioRender.com.
The development of PREmR34 (Figure 2) and selected examples featuring TFs above highlight the importance of contextuality for biosensors to operate optimally in vivo. Biosensor performance is not only affected by the attributes of the sensory part including the ligand specificity of TFs and RSWs. Sensitivity and operational range is defined as the concentration of the target small molecule (i.e., input signal) required for the biosensor to provide a significant change in the output signal (e.g., fluorescence) above host background.10,11,13
Obviously, adjusting biosensor expression is as important as tuning reporter gene levels and can be achieved by engineering the regulatory elements in the 5′ and 3′ UTR, including (natural and synthetic) promoters,19,21,49 transcriptional terminators, as well as the composition of RBS and adjacent nucleotides. This genetic contextualization is crucial to optimize the functionality of the biosensor.9−11,72,73 Additionally, the stoichiometry of sensory modules (TFs and RSWs) and binding targets (e.g., small molecules for TFs and RSWs or DNA binding sites for TFs) greatly influences sensitivity and the operational window.10,19
The necessity of iterative rounds permutating different combinations of genetic parts is laborious, time-consuming, and results are often nonintuitive.10,11,73 For example, the PbrR-based biosensor variant S23 showed the highest sensitivity for Pb2+ ions when production of the PbrR TF and the GFP reporter was controlled by the same (unidirectional) promoter.52
Berepiki et al. addressed this issue and used a design of experiments (DoE) methodology to systematically map combinations of genetic elements to greatly improve the performance of TF-based biosensor systems for protocatechuic acid (C7; 154.12 g mol–1) and ferulic acid (C10; 194.18 g mol–1), two catabolic breakdown products of lignin biomass. Noteworthy, Berepiki et al. could engineer the strong digital behavior of the investigated biosensor systems, characterized by high signal-to-noise ratios and a sigmoid dose–response curve, into a more analogue dose response. While the first allows one to confidently assign small molecule concentrations above a required threshold as desired in the initial screening of genetic libraries or the environmental monitoring of pollutants, analogue biosensor behavior (i.e., shallow or linear dose–response curve) is more appropriate to distinguish different variants or samples with similar activities, for example, as a result of protein engineering campaigns. Hence, the latter allows one to accurately distinguish subtle changes in analyte concentrations.73 In summary, the presented DoE approach promises to be a generalizable methodology to optimize other genetically encoded biosensor systems.
Furthermore, the cellular environment of host cells (i.e., host context) can strongly influence biosensor performance. Woolston et al. reduced the lower detection limit of formaldehyde in E. coli S1030 ΔfrmA from 10 μM to 5 μM compared to the wildtype strain, in which FrmA is important for the detoxification of formaldehyde. However, variability in the GFP output signal was observed in the ΔfrmA background.21 Furthermore, Bayer et al. showed that the use of E. coli RARE, exhibiting increased intracellular aldehyde persistence due to the knockout of endogenous alcohol dehydrogenases and aldo-keto reductases,74 improved the fold-increase in bioluminescence above background emitted by the luciferase LuxAB in the presence of various aldehydes.36,75 While the detection of terephthalic acid (TPA)-derived aldehydes such as 4-formylbenzoic acid and terephthalaldehyde (C8; 150.13 g mol–1 and 134.13 g mol–1, respectively) was feasible in different E. coli strains, the semiquantitative correlation between bioluminescence output and TPA concentration only succeeded in E. coli RARE.75 This enzyme-coupled biosensor system was one of the first to assess the activity of different (engineered) enzymes for the degradation of poly(ethylene terephthalate) and highlights the importance of host context for biosensor performance.
The knockout of genes otherwise responsible for the metabolization of target compounds contributed to increased intracellular concentrations, ultimately, increasing sensitivity. Similarly, Miyake et al. enhanced the intracellular concentration of SMCFAs by adjusting expression levels of PDR12, a transporter for organic acids. This strategy increased the sensitivity and the operational window of their SMCFA-responsive biosensor (PPDR12/War1 coupled to GFP).30 Complementary, export proteins can be used to reduce intracellular concentrations of potentially interfering chemical entities. One example from above featured the reduction of intracellular Zn2+ ions by the overexpression of the ZitB metal transporter to minimize interference with the CadR-based detection of Cd2+ ions in E. coli.54 Related knockout and knock-in strategies have also enabled the engineering of the central carbon metabolism in biotechnological hosts like E. coli and S. cerevisiae. Since the resulting strains can accumulate different carbon metabolites, including CO2 fixation products such as pyruvate (see above), they might provide suitable hosts to put related biosensor systems to the test.10,76,77
Lastly, the nature of reporter genes dictates their applicability and can strongly influence biosensor performance, particularly in the context of living cells. Commonly used reporter proteins include fluorescent proteins, (bacterial) luciferases, and metabolic enzymes such as LacZ (Table 1), and their unique advantages and disadvantages have been discussed earlier.78−81 Although dependent on the excitation by an external light source and potential interference with background fluorescence, one major advantage of autofluorescent proteins like GFP is that the generated amount of fluorescence is independent of exogenous substrates and usually stable over the monitoring time. Depending on the biosensor design, the fluorescent output can be correlated to the concentration of the target analyte, allowing quantification. Hence, variants of fluorescent proteins have become indispensable reporters in biosensing and HT applications including FACS.10,34,72
Bacterial luciferases are encoded by the luxAB genes and dependent on oxygen and reduced flavin mononucleotide cofactors.81,82 They emit bioluminescence in the presence of decanal (C10; 156.20 g mol–1) and other aldehyde substrates, either added directly or produced enzymatically from precursors.36,37,75 This allows the (near) real-time monitoring of aldehydes in living cells. Furthermore, bioluminescence can be autonomously produced if the complete luxCDABE operon is expressed. Coupled with a biosensor, such fully autonomous bioluminescent reporters can be employed remotely since aldehyde addition or in situ production are not required for expression.78,81 In general, luciferase enzymes, also including the well-known firefly luciferase, are good photoemitters in terms of quantum yield, and most cells are not luminescent, yielding highly sensitive detection tools with high signal-to-background ratios.36,81,82 However, the transient nature of bioluminescence signals might complicate quantification but examples exist.53,75,78
Summary and Future Directions
The presented examples for TF- and RSW-based biosensors as well as enzyme-coupled systems and emerging TCS illustrate that biological parts are capable of “sensing the smallest”, compounds with low to average molecular masses and physicochemical properties that require time-consuming sample preparation and specialized instrumentation for their detection and quantification, rendering the analysis of these chemicals inflexible and costly. Contrarily, genetically encoded biosensors operating in whole cells are versatile, transferable, and cheap tools for the qualitative and (semi)quantitative analysis of small molecules.10,12 Advancements in the omics fields (e.g., transcriptomics, proteomics, and metabolomics) and bioinformatics have contributed to the discovery of TFs and RSWs, as well as their cognate natural genetic regulatory sequences. Improved DNA synthesis and recombineering technologies, together with HT screening strategies such as FACS, have enabled the permutation of natural and synthetic genetic parts to improve the performance of biosensor systems in vivo.9−11,13 The advent of synthetic biology has enabled the design, construction, and engineering of both sensory and transduction modules. Examples in this condensed review featured TFs with altered ligand specificities10,31,32,34,35 and chimeric TFs. The latter featured the combination of LBDs and DBDs from different regulatory proteins19,56,83 or covalent fusions of TFs to functional proteins including surface proteins56 and reporters.22 The engineering of reporters has yielded (circularly permuted) fluorescent proteins with customized properties, including varied excitation/emission wavelengths, for example, or split-systems being highly sensitive for target ligands.10,12,23,41 Recently, the incorporation of noncanonical amino acids further extended the repertoire of customized reporter proteins.40,45−47 The implementation of RSWs as biosensors is still underrepresented but protocols like Capture-SELEX69−71 and RiDE72 have been employed successfully to engineer RSWs to meet the required performance criteria.
In this regard, (genetic) contextualization remains a major challenge in biosensor development and (industrial) application. DoE methodologies, for example, already offer a valuable solution to greatly reduce the number of iterative rounds of permutating different genetic parts to customize biosensor performance.10,73 Furthermore, combinations of TFs and RSWs have been realized, yielding hybrid regulators in which the TF can compensate for the low dynamic range of the RSW and amplify the output signal.84 Current and future research will certainly target a more systematic and in-depth characterization of TCS, including the elucidation of ligand binding profiles, for applications in non-natural contexts and species including important biotechnological hosts.12,60,61 So far, the swapping of sensor kinase domains has been performed to engineer the detection scope toward the desired inputs.12,85,86 Nonetheless, success has been mere due to a limited understanding of the regulation and interactions of sensory domains and downstream response regulators. Although the latter contribute greatly to the variability and modularity in TCS families, they share high conservation in structure and regulation, suggesting that DBDs can be swapped to utilize alternative but well-characterized output promoters, driving the expression of reporter genes of choice.12,84
A major drawback of using living cells as sensing chasses is the inherent delay between input sensing and output generation, which is dictated by cell growth including protein synthesis and viability, for example.10,13,87 Some applications like assessing the quality of drinking water might require very short analysis times. For such purposes, cell-free expression (CFE) systems, harnessing the transcription/translation machinery of living cells to synthesize proteins in vitro, can be used.10 Beabout et al. developed three CFE biosensors to detect the HMs arsenic (As), Cd, and Hg.87 The TF/promoter pairs ArsR/P70a (E. coli), CadC/Pcad (S. aureus), and MerR/PBBa_J23104 (Shigella flexneri) were coupled to sfGFP under the control of the cognate promoters Pars, Pcad (TF and reporter in operon configuration), and PmerT, respectively. Noteworthy, successful transcription was determined by MGapt, a malachite green-sensitive RWS introduced previously to monitor RNA dynamics (Table 1).88 Engineering of these CFE biosensors led to the detection of 0.125 μM As3+, 0.5 μM Cd2+, and 0.011 μM Hg2+ ions in less than 30 min. Despite the low selectivity of the ArsR- and CadC-based biosensors, characterized by the detection of other HM ions like Pb2+ or Hg2+, output signals were obtained rapidly and meeting the recommendations by the World Health Organization for drinking water quality.51,87
In summary, the established biosensor systems are versatile analytical tools for the real-time monitoring of various metabolites and contaminants that are related to anthropogenic activities. Continuous advancements in bioinformatics and synthetic biology not only promise the discovery of novel sensor devices including their cognate gene regulatory parts but also customization strategies will certainly yield biosensor systems for the detection and quantification of non-natural small molecules including value-added platform chemicals and new-to-nature metabolites from synthetic CO2 fixation pathways, for example. Current biosensor applications showcased their use beyond mere sensing devices and included feedback control, the coordinated expression of multiple pathway genes, and the synchronization of cell populations, yielding microbial cell factories for the production of highly desired platform chemicals from nonfossil resource stocks.10,13,14,37 Furthermore, biosensors have been suggested to assess the degradation efficiency of plastic waste75 or to sequester highly toxic HM ions from the environment and adsorb them on the surface of bacterial cells.56 This points toward the integration of biosensors with new recycling and remediation strategies, tackling current and future environmental and socioeconomic challenges.
Glossary
Abbreviations
- 2-IPM
2-isopropyl malate
- ATP
adenosine triphosphate
- A. baylyi
Acinetobacter baylyi ADP1
- BFP
blue fluorescent protein
- CdBD
cadmium-binding domain
- CETCH
crotonyl-CoA/ethylmalonyl-CoA/hydroxybutyryl-CoA
- CFE
cell-free expression
- CoA
coenzyme A
- cpGFP
circularly permuted green fluorescent protein
- cpsGFP
circularly permuted superfolder GFP
- cpYFP
circularly permuted yellow fluorescent protein
- DBD
DNA-binding domain
- DNA
deoxyribonucleic acid
- DoE
design of experiments
- epPCR
error-prone polymerase chain reaction
- E. coli
Escherichia coli
- FACS
fluorescence-activated cell sorting
- FRET
Förster resonance energy transfer
- GFP
green fluorescent protein
- HM
heavy metal
- HT
high-throughput
- LBD
ligand-binding domain
- LOD
lower limit of detection
- P
promoter
- RARE
reduced aldehyde reduction activity
- RBS
ribosome binding site
- RiDE
riboswitch-based directed evolution
- RFP
red fluorescent protein
- (m)RNA
(messenger) ribonucleic acid
- RNAP
RNA polymerase
- RNS
reactive nitrogen species
- roGFP
redox-sensitive GFP
- ROS
reactive oxygen species
- RSW
riboswitch
- SACA
synthetic acetyl-CoA
- SELEX
systematic evolution of ligands by exponential enrichment
- sfGFP
superfolder GFP
- SMCAH
short-/medium-chain alcohol
- SMCFA
short-/medium-chain fatty acid
- S. cerevisiae
Saccharomyces cerevisiae
- TCS
two-component systems
- TF
transcription factor
- TPA
terephthalic acid
- UTR
untranslated region
Biographies
Thomas Bayer graduated in the Molecular Biology program from the University of Vienna and obtained his PhD in (Bio)-organic Chemistry at the TU Wien (Austria). Thomas Bayer received the Erwin Schrödinger fellowship by The Austrian Science Fund (FWF), which included scientific stays at the University of Greifswald, the Albert Einstein College of Medicine in New York City (USA), and the TU Graz (Austria) in 2019–2021. In 2022, he rejoined the Bornscheuer group, focusing on the functional annotation of proteins and the development of biosensor systems as a means to accelerate the engineering of enzymes and metabolic pathways for the production of value-added chemicals.
Luise Hänel is a biochemist with expertise in biocatalysis and pharmaceutical biology. She earned her diploma in Biochemistry from the University of Greifswald, where she conducted research on protein engineering. Currently, she is pursuing her Ph.D. at the Institute of Biochemistry in the Protein Biochemistry group, where she is focusing on the development of biocatalysts for industrial applications.
Jana Husarcikova studied Biotechnology at Slovak Technical University in Bratislava. In 2020, she received her Ph.D. from TU Braunschweig for her research on ligninolytic enzymes, followed by a postdoctoral stay in the Bornscheuer group at the University of Greifswald, focusing on marine polysaccharide degradation. Currently, she is working as a Research Scientist at BRAIN Biotech.
Andreas Kunzendorf studied Technical Biology at the University of Stuttgart. He received his Ph.D. from the University of Groningen (The Netherlands) in 2021 with a focus on enzyme engineering of non-natural carboligases for the synthesis of fine chemicals and pharmaceutical intermediates. As a postdoctoral fellow at the University of Greifswald in 2022, he worked on the optimization and application of methyltransferases for the flavor and fragrance industries.
Uwe T. Bornscheuer studied Chemistry and received his Ph.D. in 1993 at Hannover University, followed by a postdoc at the Nagoya University (Japan). In 1998, he completed his Habilitation about the use of lipases and esterases in organic synthesis at Stuttgart University. He has been Professor at the Institute of Biochemistry at Greifswald University since 1999. Besides other awards, he received the “Enzyme Engineering Award” in 2022. His current research interest focuses on the discovery and engineering of enzymes from various classes for applications in organic synthesis, lipid modification, degradation of plastics, and complex marine polysaccharides.
Author Contributions
The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
Open Access is funded by the Austrian Science Fund (FWF).
The authors declare no competing financial interest.
References
- Bae J.; Jin S.; Kang S.; Cho B.-K.; Oh M.-K. Recent Progress in the Engineering of C1-Utilizing Microbes. Curr. Opin. Biotechnol. 2022, 78, 102836. 10.1016/j.copbio.2022.102836. [DOI] [PubMed] [Google Scholar]
- Moreno-Bondi M. C.; Le X. C.; Field J. A.; Richardson S. D.; Li X.-F.; Diamond M. L.; Li X.; Goring P. D. From Detection to Remediation: Analytical Science at the Forefront of Environmental Research. ACS Omega 2022, 7, 38105–38108. 10.1021/acsomega.2c06631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kara S.; Hauschild M.; Sutherland J.; McAloone T. Closed-Loop Systems to Circular Economy: A Pathway to Environmental Sustainability?. CIRP Ann. 2022, 71, 505–528. 10.1016/j.cirp.2022.05.008. [DOI] [Google Scholar]
- Yaashikaa P. R.; Devi M. K.; Kumar P. S. Engineering Microbes for Enhancing the Degradation of Environmental Pollutants: A Detailed Review on Synthetic Biology. Environ. Res. 2022, 214, 113868. 10.1016/j.envres.2022.113868. [DOI] [PubMed] [Google Scholar]
- Santos Correa S.; Schultz J.; Lauersen K. J.; Soares Rosado A. Natural Carbon Fixation and Advances in Synthetic Engineering for Redesigning and Creating New Fixation Pathways. J. Adv. Res. 2023, 47, 75. 10.1016/j.jare.2022.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bierbaumer S.; Nattermann M.; Schulz L.; Zschoche R.; Erb T. J.; Winkler C. K.; Tinzl M.; Glueck S. M.. Enzymatic Conversion of CO2: From Natural to Artificial Utilization. Chem. Rev. 2023, in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piroozmand F.; Mohammadipanah F.; Faridbod F. Emerging Biosensors in Detection of Natural Products. Synth. Syst. Biotechnol. 2020, 5, 293–303. 10.1016/j.synbio.2020.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naresh V.; Lee N. A Review on Biosensors and Recent Development of Nanostructured Materials-Enabled Biosensors. Sensors 2021, 21, 1109. 10.3390/s21041109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia P.-F.; Ling H.; Foo J. L.; Chang M. W. Synthetic Genetic Circuits for Programmable Biological Functionalities. Biotechnol. Adv. 2019, 37, 107393. 10.1016/j.biotechadv.2019.04.015. [DOI] [PubMed] [Google Scholar]
- Yi D.; Bayer T.; Badenhorst C. P. S.; Wu S.; Doerr M.; Höhne M.; Bornscheuer U. T. Recent Trends in Biocatalysis. Chem. Soc. Rev. 2021, 50, 8003–8049. 10.1039/D0CS01575J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hossain G. S.; Saini M.; Miyake R.; Ling H.; Chang M. W. Genetic Biosensor Design for Natural Product Biosynthesis in Microorganisms. Trends Biotechnol. 2020, 38 (7), 797–810. 10.1016/j.tibtech.2020.03.013. [DOI] [PubMed] [Google Scholar]
- Hicks M.; Bachmann T. T.; Wang B. Synthetic Biology Enables Programmable Cell-Based Biosensors. ChemPhysChem 2020, 21, 132–144. 10.1002/cphc.201900739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma B. K.; Mannan A. A.; Zhang F.; Oyarzún D. A. Trade-Offs in Biosensor Optimization for Dynamic Pathway Engineering. ACS Synth. Biol. 2022, 11, 228–240. 10.1021/acssynbio.1c00391. [DOI] [PubMed] [Google Scholar]
- Markel U.; Essani K. D.; Besirlioglu V.; Schiffels J.; Streit W. R.; Schwaneberg U. Advances in Ultrahigh-Throughput Screening for Directed Enzyme Evolution. Chem. Soc. Rev. 2020, 49, 233–262. 10.1039/C8CS00981C. [DOI] [PubMed] [Google Scholar]
- Gleizer S.; Ben-Nissan R.; Bar-On Y. M.; Antonovsky N.; Noor E.; Zohar Y.; Jona G.; Krieger E.; Shamshoum M.; Bar-Even A.; Milo R. Conversion of Escherichia coli to Generate All Biomass Carbon from CO2. Cell 2019, 179, 1255. 10.1016/j.cell.2019.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwander T.; Schada von Borzyskowski L.; Burgener S.; Cortina N. S.; Erb T. J. A Synthetic Pathway for the Fixation of Carbon Dioxide In Vitro. Science 2016, 354, 900–904. 10.1126/science.aah5237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayer T.; Balke K.; Hamnevik E.; Bornscheuer U. T.. Protein Engineering. In The Autotrophic Biorefinery: Raw Materials from Biotechnology; Kourist R., Schmidt S., Eds.; De Gruyter: Berlin, 2021; pp 47–84. [Google Scholar]
- Bar-Even A. Formate Assimilation: The Metabolic Architecture of Natural and Synthetic Pathways. Biochemistry 2016, 55, 3851–3863. 10.1021/acs.biochem.6b00495. [DOI] [PubMed] [Google Scholar]
- Mormino M.; Siewers V.; Nygård Y. Development of an Haa1-Based Biosensor for Acetic Acid Sensing in Saccharomyces cerevisiae. FEMS Yeast Res. 2021, 21, foab049. 10.1093/femsyr/foab049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X.; Liu Y.; Yang Y.; Wang S.; Wang Q.; Wang X.; Yan Z.; Cheng J.; Liu C.; Yang X.; Luo H.; Yang S.; Gou J.; Ye L.; Lu L.; Zhang Z.; Guo Y.; Nie Y.; Lin J.; Li S.; Tian C.; Cai T.; Zhuo B.; Ma H.; Wang W.; Ma Y.; Liu Y.; Li Y.; Jiang H. Constructing a Synthetic Pathway for Acetyl-Coenzyme A from One-Carbon through Enzyme Design. Nat. Commun. 2019, 10, 1378. 10.1038/s41467-019-09095-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolston B. M.; Roth T.; Kohale I.; Liu D. R.; Stephanopoulos G. Development of a Formaldehyde Biosensor with Application to Synthetic Methylotrophy. Biotechnol. Bioeng. 2018, 115, 206–215. 10.1002/bit.26455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arce-Molina R.; Cortés-Molina F.; Sandoval P. Y.; Galaz A.; Alegría K.; Schirmeier S.; Barros L. F.; San Martín A. A Highly Responsive Pyruvate Sensor Reveals Pathway-Regulatory Role of the Mitochondrial Pyruvate Carrier MPC. eLife 2020, 9, e53917. 10.7554/eLife.53917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostyuk A. I.; Demidovich A. D.; Kotova D. A.; Belousov V. V.; Bilan D. S. Circularly Permuted Fluorescent Protein-Based Indicators: History, Principles, and Classification. Int. J. Mol. Sci. 2019, 20, 4200. 10.3390/ijms20174200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peralta-Yahya P. P.; Zhang F.; del Cardayre S. B.; Keasling J. D. Microbial Engineering for the Production of Advanced Biofuels. Nature 2012, 488, 320–328. 10.1038/nature11478. [DOI] [PubMed] [Google Scholar]
- Marella E. R.; Holkenbrink C.; Siewers V.; Borodina I. Engineering Microbial Fatty Acid Metabolism for Biofuels and Biochemicals. Energy Biotechnol. Environ. Biotechnol. 2018, 50, 39–46. 10.1016/j.copbio.2017.10.002. [DOI] [PubMed] [Google Scholar]
- Theodorou E. C.; Theodorou M. C.; Kyriakidis D. A. Regulation of Poly-(R)-(3-hydroxybutyrate-co-3-hydroxyvalerate) Biosynthesis by the AtoSCDAEB Regulon in PhaCAB+Escherichia coli. Appl. Microbiol. Biotechnol. 2013, 97, 5259–5274. 10.1007/s00253-013-4843-8. [DOI] [PubMed] [Google Scholar]
- Pilalis E.; Chatziioannou A. A.; Grigoroudis A. I.; Panagiotidis C. A.; Kolisis F. N.; Kyriakidis D. A. Escherichia coli Genome-Wide Promoter Analysis: Identification of Additional AtoC Binding Target Elements. BMC Genomics 2011, 12, 238. 10.1186/1471-2164-12-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutter J. W.; Dekker L.; Fedorec A. J. H.; Gonzales D. T.; Wen K. Y.; Tanner L. E. S.; Donovan E.; Ozdemir T.; Thomas G. M.; Barnes C. P. Engineered Acetoacetate-Inducible Whole-Cell Biosensors Based on the AtoSC Two-Component System. Biotechnol. Bioeng. 2021, 118, 4278–4289. 10.1002/bit.27897. [DOI] [PubMed] [Google Scholar]
- Baumann L.; Rajkumar A. S.; Morrissey J. P.; Boles E.; Oreb M. A Yeast-Based Biosensor for Screening of Short- and Medium-Chain Fatty Acid Production. ACS Synth. Biol. 2018, 7, 2640–2646. 10.1021/acssynbio.8b00309. [DOI] [PubMed] [Google Scholar]
- Miyake R.; Ling H.; Foo J. L.; Fugono N.; Chang M. W. Transporter-Driven Engineering of a Genetic Biosensor for the Detection and Production of Short-Branched Chain Fatty Acids in Saccharomyces cerevisiae. Front. Bioeng. Biotechnol. 2022, 10, 838732. 10.3389/fbioe.2022.838732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregori C.; Schüller C.; Frohner I. E.; Ammerer G.; Kuchler K. Weak Organic Acids Trigger Conformational Changes of the Yeast Transcription Factor War1 In Vivo to Elicit Stress Adaptation. J. Biol. Chem. 2008, 283, 25752–25764. 10.1074/jbc.M803095200. [DOI] [PubMed] [Google Scholar]
- Zhang Y.; Cortez J. D.; Hammer S. K.; Carrasco-López C.; García Echauri S. Á.; Wiggins J. B.; Wang W.; Avalos J. L. Biosensor for Branched-Chain Amino Acid Metabolism in Yeast and Applications in Isobutanol and Isopentanol Production. Nat. Commun. 2022, 13, 270. 10.1038/s41467-021-27852-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H.; Wang N.; Huo W.; Zhang Y.; Zhang W.; Yang Y.; Chen Z.; Huo Y.-X. Establishment of BmoR-Based Biosensor to Screen Isobutanol Overproducer. Microb. Cell Factories 2019, 18, 30. 10.1186/s12934-019-1084-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahls M. O.; Platz L.; Morgado G.; Schmidt G. W.; Panke S. Directed Evolution of Biofuel-Responsive Biosensors for Automated Optimization of Branched-Chain Alcohol Biosynthesis. Metab. Eng. 2022, 69, 98–111. 10.1016/j.ymben.2021.10.014. [DOI] [PubMed] [Google Scholar]
- Reed B.; Blazeck J.; Alper H. Evolution of an Alkane-Inducible Biosensor for Increased Responsiveness to Short-Chain Alkanes. J. Biotechnol. 2012, 158, 75–79. 10.1016/j.jbiotec.2012.01.028. [DOI] [PubMed] [Google Scholar]
- Bayer T.; Becker A.; Terholsen H.; Kim I. J.; Menyes I.; Buchwald S.; Balke K.; Santala S.; Almo S. C.; Bornscheuer U. T. LuxAB-Based Microbial Cell Factories for the Sensing, Manufacturing, and Transformation of Industrial Aldehydes. Catalysts 2021, 11 (8), 953. 10.3390/catal11080953. [DOI] [Google Scholar]
- Lehtinen T.; Virtanen H.; Santala S.; Santala V. Production of Alkanes from CO2 by Engineered Bacteria. Biotechnol. Biofuels 2018, 11, 228. 10.1186/s13068-018-1229-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sodhi K. K.; Mishra L. C.; Singh C. K.; Kumar M. Perspective on the Heavy Metal Pollution and Recent Remediation Strategies. Curr. Res. Microb. Sci. 2022, 3, 100166. 10.1016/j.crmicr.2022.100166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pak V. V.; Ezerina D.; Lyublinskaya O. G.; Pedre B.; Tyurin-Kuzmin P. A.; Mishina N. M.; Thauvin M.; Young D.; Wahni K.; Martínez Gache S. A.; Demidovich A. D.; Ermakova Y. G.; Maslova Y. D.; Shokhina A. G.; Eroglu E.; Bilan D. S.; Bogeski I.; Michel T.; Vriz S.; Messens J.; Belousov V. V. Ultrasensitive Genetically Encoded Indicator for Hydrogen Peroxide Identifies Roles for the Oxidant in Cell Migration and Mitochondrial Function. Cell Metab. 2020, 31, 642–653. 10.1016/j.cmet.2020.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z.; Zhang S.; Li X.; Ai H. A High-Performance Genetically Encoded Fluorescent Biosensor for Imaging Physiological Peroxynitrite. Cell Chem. Biol. 2021, 28, 1542–1553. 10.1016/j.chembiol.2021.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostyuk A. I.; Tossounian M.-A.; Panova A. S.; Thauvin M.; Raevskii R. I.; Ezerina D.; Wahni K.; Van Molle I.; Sergeeva A. D.; Vertommen D.; Gorokhovatsky A. Yu.; Baranov M. S.; Vriz S.; Messens J.; Bilan D. S.; Belousov V. V. Hypocrates Is a Genetically Encoded Fluorescent Biosensor for (Pseudo)Hypohalous Acids and Their Derivatives. Nat. Commun. 2022, 13, 171. 10.1038/s41467-021-27796-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilan D. S.; Belousov V. V. HyPer Family Probes: State of the Art. Antioxid. Redox Signal. 2016, 24, 731–751. 10.1089/ars.2015.6586. [DOI] [PubMed] [Google Scholar]
- Kardashliev T.; Weingartner A.; Romero E.; Schwaneberg U.; Fraaije M.; Panke S.; Held M. Whole-Cell Screening of Oxidative Enzymes Using Genetically Encoded Sensors. Chem. Sci. 2021, 12, 14766–14772. 10.1039/D1SC02578C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan B.; Van Laer K.; Owusu T. N. E.; Ezerina D.; Pastor-Flores D.; Amponsah P. S.; Tursch A.; Dick T. P. Real-Time Monitoring of Basal H2O2 Levels with Peroxiredoxin-Based Probes. Nat. Chem. Biol. 2016, 12, 437–443. 10.1038/nchembio.2067. [DOI] [PubMed] [Google Scholar]
- Youssef S.; Zhang S.; Ai H. A Genetically Encoded, Ratiometric Fluorescent Biosensor for Hydrogen Sulfide. ACS Sens. 2019, 4, 1626–1632. 10.1021/acssensors.9b00400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S.; Chen Z.; Ren W.; Ai H. Reaction-Based Genetically Encoded Fluorescent Hydrogen Sulfide Sensors. J. Am. Chem. Soc. 2012, 134, 9589–9592. 10.1021/ja303261d. [DOI] [PubMed] [Google Scholar]
- Chen Z.; Ai H. A Highly Responsive and Selective Fluorescent Probe for Imaging Physiological Hydrogen Sulfide. Biochemistry 2014, 53, 5966–5974. 10.1021/bi500830d. [DOI] [PubMed] [Google Scholar]
- Wedekind J. E.; Dutta D.; Belashov I. A.; Jenkins J. L. Metalloriboswitches: RNA-Based Inorganic Ion Sensors That Regulate Genes. J. Biol. Chem. 2017, 292, 9441–9450. 10.1074/jbc.R117.787713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calero P.; Volke D. C.; Lowe P. T.; Gotfredsen C. H.; O’Hagan D.; Nikel P. I. A Fluoride-Responsive Genetic Circuit Enables In Vivo Biofluorination in Engineered Pseudomonas putida. Nat. Commun. 2020, 11, 5045. 10.1038/s41467-020-18813-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godt J.; Scheidig F.; Grosse-Siestrup C.; Esche V.; Brandenburg P.; Reich A.; Groneberg D. A. The Toxicity of Cadmium and Resulting Hazards for Human Health. J. Occup. Med. Toxicol. 2006, 1 (22), 1–6. 10.1186/1745-6673-1-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guidelines for Drinking-Water Quality: First Addendum to the Fourth Edition; World Health Organization (WHO), 2017. [PubMed] [Google Scholar]
- Jia X.; Zhao T.; Liu Y.; Bu R.; Wu K. Gene Circuit Engineering to Improve the Performance of a Whole-Cell Lead Biosensor. FEMS Microbiol. Lett. 2018, 365, fny157. 10.1093/femsle/fny157. [DOI] [PubMed] [Google Scholar]
- Nourmohammadi E.; Hosseinkhani S.; Nedaeinia R.; Khoshdel-Sarkarizi H.; Nedaeinia M.; Ranjbar M.; Ebrahimi N.; Farjami Z.; Nourmohammadi M.; Mahmoudi A.; Goli M.; Ferns G. A.; Sadeghizadeh M. Construction of a Sensitive and Specific Lead Biosensor Using a Genetically Engineered Bacterial System with a Luciferase Gene Reporter Controlled by PbrR and CadA Promoters. Biomed. Eng. OnLine 2020, 19, 79. 10.1186/s12938-020-00816-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He M.-Y.; Lin Y.-J.; Kao Y.-L.; Kuo P.; Grauffel C.; Lim C.; Cheng Y.-S.; Chou H.-H. D. Sensitive and Specific Cadmium Biosensor Developed by Reconfiguring Metal Transport and Leveraging Natural Gene Repositories. ACS Sens. 2021, 6, 995–1002. 10.1021/acssensors.0c02204. [DOI] [PubMed] [Google Scholar]
- Grass G.; Fan B.; Rosen B. P.; Franke S.; Nies D. H.; Rensing C. ZitB (YbgR), a Member of the Cation Diffusion Facilitator Family, Is an Additional Zinc Transporter in Escherichia coli. J. Bacteriol. 2001, 183, 4664–4667. 10.1128/JB.183.15.4664-4667.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y.; Hui C.; Zhang N.; Liu L.; Li H.; Zheng H. Development of Cadmium Multiple-Signal Biosensing and Bioadsorption Systems Based on Artificial Cad Operons. Front. Bioeng. Biotechnol. 2021, 9, 585617. 10.3389/fbioe.2021.585617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furukawa K.; Ramesh A.; Zhou Z.; Weinberg Z.; Vallery T.; Winkler W. C.; Breaker R. R. Bacterial Riboswitches Cooperatively Bind Ni2+ or Co2+ Ions and Control Expression of Heavy Metal Transporters. Mol. Cell 2015, 57, 1088–1098. 10.1016/j.molcel.2015.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X.; Wei W.; Zhao J. Using a Riboswitch Sensor to Detect Co2+/Ni2+ Transport in E. coli. Front. Chem. 2021, 9, 631909. 10.3389/fchem.2021.631909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallberg Z. F.; Su Y.; Kitto R. Z.; Hammond M. C. Engineering and In Vivo Applications of Riboswitches. Annu. Rev. Biochem. 2017, 86, 515–539. 10.1146/annurev-biochem-060815-014628. [DOI] [PubMed] [Google Scholar]
- Xie Y.; Li J.; Ding Y.; Shao X.; Sun Y.; Xie F.; Liu S.; Tang S.; Deng X. An Atlas of Bacterial Two-Component Systems Reveals Function and Plasticity in Signal Transduction. Cell Rep. 2022, 41, 111502. 10.1016/j.celrep.2022.111502. [DOI] [PubMed] [Google Scholar]
- Jacob-Dubuisson F.; Mechaly A.; Betton J.-M.; Antoine R. Structural Insights into the Signalling Mechanisms of Two-Component Systems. Nat. Rev. Microbiol. 2018, 16, 585–593. 10.1038/s41579-018-0055-7. [DOI] [PubMed] [Google Scholar]
- Zhang J.; Barajas J. F.; Burdu M.; Ruegg T. L.; Dias B.; Keasling J. D. Development of a Transcription Factor-Based Lactam Biosensor. ACS Synth. Biol. 2017, 6, 439–445. 10.1021/acssynbio.6b00136. [DOI] [PubMed] [Google Scholar]
- Dudek C.-A.; Jahn D. PRODORIC: State-of-the-Art Database of Prokaryotic Gene Regulation. Nucleic Acids Res. 2022, 50, D295–D302. 10.1093/nar/gkab1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandelin A.; Alkema W.; Engström P.; Wasserman W. W.; Lenhard B. JASPAR: An Open-access Database for Eukaryotic Transcription Factor Binding Profiles. Nucleic Acids Res. 2004, 32, D91–D94. 10.1093/nar/gkh012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalvari I.; Nawrocki E. P.; Argasinska J.; Quinones-Olvera N.; Finn R. D.; Bateman A.; Petrov A. I. Non-Coding RNA Analysis Using the Rfam Database. Curr. Protoc. Bioinforma. 2018, 62, e51. 10.1002/cpbi.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee S.; Sengupta S. Riboswitch Scanner: An Efficient PHMM-Based Web-Server to Detect Riboswitches in Genomic Sequences. Bioinformatics 2016, 32, 776–778. 10.1093/bioinformatics/btv640. [DOI] [PubMed] [Google Scholar]
- Strobel B.; Spöring M.; Klein H.; Blazevic D.; Rust W.; Sayols S.; Hartig J. S.; Kreuz S. High-Throughput Identification of Synthetic Riboswitches by Barcode-Free Amplicon-Sequencing in Human Cells. Nat. Commun. 2020, 11, 714. 10.1038/s41467-020-14491-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuerk C.; Gold L. Systematic Evolution of Ligands by Exponential Enrichment: RNA Ligands to Bacteriophage T4 DNA Polymerase. Science 1990, 249, 505–510. 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- Nutiu R.; Li Y. In Vitro Selection of Structure-Switching Signaling Aptamers. Angew. Chem., Int. Ed. 2005, 44, 1061–1065. 10.1002/anie.200461848. [DOI] [PubMed] [Google Scholar]
- Boussebayle A.; Torka D.; Ollivaud S.; Braun J.; Bofill-Bosch C.; Dombrowski M.; Groher F.; Hamacher K.; Suess B. Next-Level Riboswitch Development: Implementation of Capture-SELEX Facilitates Identification of a New Synthetic Riboswitch. Nucleic Acids Res. 2019, 47, 4883–4895. 10.1093/nar/gkz216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramat J.; Suess B.. Efficient Method to Identify Synthetic Riboswitches Using RNA-Based Capture-SELEX Combined with In Vivo Screening. In Riboregulator Design and Analysis; Chappell J., Takahashi M. K., Eds.; Springer US: New York, NY, 2022; pp 157–177. [DOI] [PubMed] [Google Scholar]
- Pham H. L.; Wong A.; Chua N.; Teo W. S.; Yew W. S.; Chang M. W. Engineering a Riboswitch-Based Genetic Platform for the Self-Directed Evolution of Acid-Tolerant Phenotypes. Nat. Commun. 2017, 8, 411. 10.1038/s41467-017-00511-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berepiki A.; Kent R.; Machado L. F. M.; Dixon N. Development of High-Performance Whole-Cell Biosensors Aided by Statistical Modeling. ACS Synth. Biol. 2020, 9, 576–589. 10.1021/acssynbio.9b00448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunjapur A. M.; Tarasova Y.; Prather K. L. J. Synthesis and Accumulation of Aromatic Aldehydes in an Engineered Strain of Escherichia coli. J. Am. Chem. Soc. 2014, 136, 11644–11654. 10.1021/ja506664a. [DOI] [PubMed] [Google Scholar]
- Bayer T.; Pfaff L.; Branson Y.; Becker A.; Wu S.; Bornscheuer U. T.; Wei R. Biosensor and Chemo-Enzymatic One-Pot Cascade Applications to Detect and Transform PET-Derived Terephthalic Acid in Living Cells. iScience 2022, 25, 104326. 10.1016/j.isci.2022.104326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z.; Liang X.; Li M.; Ma M.; Zheng Q.; Li D.; An T.; Wang G. Advances in the Optimization of Central Carbon Metabolism in Metabolic Engineering. Microb. Cell Factories 2023, 22, 76. 10.1186/s12934-023-02090-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Falco B.; Giannino F.; Carteni F.; Mazzoleni S.; Kim D.-H. Metabolic Flux Analysis: A Comprehensive Review on Sample Preparation, Analytical Techniques, Data Analysis, Computational Modelling, and Main Application Areas. RSC Adv. 2022, 12, 25528–25548. 10.1039/D2RA03326G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Close D. M.; Ripp S.; Sayler G. S. Reporter Proteins in Whole-Cell Optical Bioreporter Detection Systems, Biosensor Integrations, and Biosensing Applications. Sensors 2009, 9, 9147–9174. 10.3390/s91109147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hossain G. S.; Saini M.; Miyake R.; Ling H.; Chang M. W. Genetic Biosensor Design for Natural Product Biosynthesis in Microorganisms. Spec. Issue Metab. Eng. 2020, 38, 797–810. 10.1016/j.tibtech.2020.03.013. [DOI] [PubMed] [Google Scholar]
- Gui Q.; Lawson T.; Shan S.; Yan L.; Liu Y. The Application of Whole Cell-Based Biosensors for Use in Environmental Analysis and in Medical Diagnostics. Sensors 2017, 17, 1623. 10.3390/s17071623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleiss A.; Sarkisyan K. S. A Brief Review of Bioluminescent Systems. Curr. Genet. 2019, 65 (4), 877–882. 10.1007/s00294-019-00951-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. Perspectives on Bioluminescence Mechanisms. Photochem. Photobiol. 2017, 93, 389–404. 10.1111/php.12650. [DOI] [PubMed] [Google Scholar]
- Schmidl S. R.; Ekness F.; Sofjan K.; Daeffler K. N.-M.; Brink K. R.; Landry B. P.; Gerhardt K. P.; Dyulgyarov N.; Sheth R. U.; Tabor J. J. Rewiring Bacterial Two-Component Systems by Modular DNA-Binding Domain Swapping. Nat. Chem. Biol. 2019, 15, 690–698. 10.1038/s41589-019-0286-6. [DOI] [PubMed] [Google Scholar]
- Wang Y.-H.; McKeague M.; Hsu T. M.; Smolke C. D. Design and Construction of Generalizable RNA-Protein Hybrid Controllers by Level-Matched Genetic Signal Amplification. Cell Syst. 2016, 3, 549–562. 10.1016/j.cels.2016.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B.; Barahona M.; Buck M.; Schumacher J. Rewiring Cell Signalling through Chimaeric Regulatory Protein Engineering. Biochem. Soc. Trans. 2013, 41, 1195–1200. 10.1042/BST20130138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao N.; Cubillos-Ruiz A.; Cameron D. E.; Collins J. J. Probiotic Strains Detect and Suppress Cholera in Mice. Sci. Transl. Med. 2018, 10, eaao2586. 10.1126/scitranslmed.aao2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beabout K.; Bernhards C. B.; Thakur M.; Turner K. B.; Cole S. D.; Walper S. A.; Chávez J. L.; Lux M. W. Optimization of Heavy Metal Sensors Based on Transcription Factors and Cell-Free Expression Systems. ACS Synth. Biol. 2021, 10, 3040–3054. 10.1021/acssynbio.1c00331. [DOI] [PubMed] [Google Scholar]
- Siegal-Gaskins D.; Tuza Z. A.; Kim J.; Noireaux V.; Murray R. M. Gene Circuit Performance Characterization and Resource Usage in a Cell-Free “Breadboard. ACS Synth. Biol. 2014, 3, 416–425. 10.1021/sb400203p. [DOI] [PubMed] [Google Scholar]