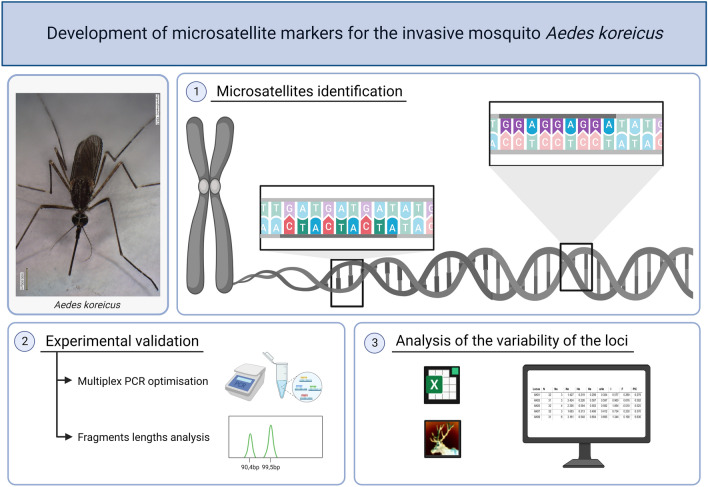
Abstract
Background
Aedes koreicus is a mosquito species native to East Asia which has recently invaded several countries in Europe. In Italy, this mosquito was first detected in the North-East in 2011 and is now widely distributed in the entire northern part of the country. The development of specific genetic markers, such as microsatellites, is necessary to uncover the dispersal routes of this mosquito from its native areas and, eventually, to plan future control interventions.
Methods
Available raw sequences of genomic DNA of Ae. koreicus were screened in silico using BLASTn to identify possible microsatellite-containing sequences. Specific primer pairs were then designed, and their efficiency was determined through polymerase chain reaction (PCR) on 32 individuals of Ae. koreicus collected in Italy. PCR conditions were optimised in three multiplex reactions. Genotyping of individual mosquitoes was performed on both single and multiplex PCR reactions. Finally, analysis of intra-population variation was performed to assess the level of polymorphism of the markers.
Results
Mosquito genotyping provided consistent results in both single and multiplex reactions. Out of the 31 microsatellite markers identified in the Ae. koreicus genome raw sequences, 11 were polymorphic in the examined mosquito samples.
Conclusions
The results show that the 11 microsatellite markers developed here hold potential for investigating the genetic structure of Ae. koreicus populations. These markers could thus represent a novel and useful tool to infer the routes of invasion of this mosquito species into Europe and other non-native areas.
Graphical Abstract
Supplementary Information
The online version contains supplementary material available at 10.1186/s13071-023-05823-z.
Keywords: Invasive mosquitoes, Aedes mosquitoes, Simple sequence repeats (SSRs), Population genetics, Monitoring
Background
Invasive Aedes mosquitoes represent a global concern for public health due to their role as vectors of several pathogens and their ability to colonise new territories [1, 2]. Their dispersal in non-native regions is mainly associated with climate change, and the increasing movement of people and goods (e.g. used tires, ornamental plants, used machineries) [3, 4] that facilitates the importation of alien species into new territories. In the globalisation era, the trade of goods develops into intricate patterns, and the tracking back of invasion routes becomes quite difficult.
In this context, population genetics is a powerful tool: determining the “genetic makeup” of a species can help, for example, in locating the putative origin(s) of multiple introductions, revealing species expansion phenomena, and providing clues on the potential vectorial capacity of different populations of invasive species [5–10].
Aedes koreicus, the Korean bush mosquito, is a species native to temperate areas of Asia, particularly Korea, China, and Far East Russia [11]. It was described for the first time outside its endemic territory in Europe, in 2008 in Belgium [12]. Since then, this alien mosquito species has been found in several territories in Southern, Central, and Eastern European countries [13, 14], with the last finding in the Czech Republic in 2022 [15]. The invasiveness success of this Korean mosquito is evident in Italy, where the mosquito populations did not remain confined to relatively small areas, as has instead been observed in other European countries [16]. Indeed, after the first description in Belluno district in 2011 [17], Ae. koreicus mosquitoes were collected in all the Italian North-East regions (as recently reviewed in [18, 19]), in the municipality of Genoa (2015) [20], in five districts of the Lombardy region, namely Como (2013), Sondrio (2015), Bergamo (2020), Brescia and Lecco (both in 2021) [14, 21, 22], and in three districts of the Piedmont region, Alessandria, Asti, and Torino (all in 2021) [14, 23].
Despite the large number of reports of Ae. koreicus in Europe, and in particular in Italy, possible entry routes into the continent have not been reconstructed so far, nor it is clear how the mosquito spread after its first introduction(s). Filling this knowledge gap is important, considering that the Korean bush mosquito has the potential to further spread in currently uncolonised European countries [14]. Indeed, the study of the genetic structure of populations of Ae. koreicus and the understanding of the genetic relatedness between individuals from different locations would permit us to hypothesise how this mosquito spread in Northern Italy, and even in other European countries [24].
To achieve this goal, genetic markers are needed, and microsatellites were selected as the markers of choice. Indeed, simple sequence repeats (SSRs) are characterised by high variability level and relative ease of use at low cost [25]; moreover, microsatellites have been successfully used for revealing the genetic structure of other Aedes native and invasive mosquito species, such as Ae. albopictus, Ae. japonicus, Ae. taeniorhynchus, Ae. polynesiensis, Ae. sticticus, Ae. fluviatilis, and Ae. aegypti [8, 10, 26–30]. Particularly, SSRs developed for Ae. japonicus were indicated as polymorphic also in Ae. koreicus [31]. For this reason, we preliminarily assayed these microsatellites using DNA samples from Ae. koreicus, but the amplification resulted ineffective (data not shown). Since no other nuclear genetic markers have been developed so far for the study of the genetic structure of Ae. koreicus, SSR markers for this mosquito species have been developed in the present study.
Methods
Putative SSR-containing sequences were identified in the DNA of Ae. koreicus thanks to the screening of raw sequencing data available in the Sequence Read Archive (SRA; Runs: ERR7598205–ERR7598207 [32, 33]) with the nucleotide Basic Local Alignment Search Tool (BLASTn), while primers for their amplification were designed using the web tool Primer3Plus [34, 35]. The amplification efficiency of each primer pair was tested with polymerase chain reaction (PCR). First, single PCRs were performed using DNA samples purified from individuals of Ae. koreicus. Primer pairs that proved to be effective were then labelled at the 5′ end with three different fluorophores (Table 1), and different combinations of multiplex PCRs were tested.
Table 1.
Microsatellite sequences identified in the nuclear genome of Aedes koreicus, and organisation of the validated markers as multiplex PCR
| Locus name | Reference sequence (Run number.read number) | (Repeat motif)na | Primer sequence (5′-3′) | PCR fluorophoresb | Size range (bp) | |
|---|---|---|---|---|---|---|
| Multiplex A | AK01 | ERR7598207.14364316 | (CAA)7 |
F: GCCGCTACAAACAGCTCAGT R: TCGTAAATTCGACGGGGTAG |
TAMRA | 121–127 |
| AK05 | ERR7598207.14346661 | (CAA)6 |
F: AACCGACTACGATGCACACA R: TAAACACAGAGCCAGCCACA |
HEX | 104–116 | |
| AK07 | ERR7598207.13874909 | (GGC)6 |
F: ATCGAGCACACCGTCGTT R: ATCCATTTTTCTTTCTTTCGCTGT |
6-FAM | 144–150 | |
| AK13 | ERR7598207.13190639 | (GTT)6 |
F:CCATTAAAAGGTTTTACTCAATGTTGT R: AAGAGACGGCTGTGGTCCT |
TAMRA | 141–144 | |
| Multiplex B | AK03 | ERR7598207.14368852 | (GAA)6 |
F: CGACTCCCTGAGCATAGTGT R: CATGCTATGATGAGCACATTGA |
6-FAM | 133–139 |
| AK09 | ERR7598207.14366337 | (ACG)11 |
F: TACCCGAATCCAGCAAACAT R: GCCAGAAGGACATCGTCAC |
TAMRA | 90–102 | |
| AK10 | ERR7598207.14369161 | (ATC)6 |
F: AACTTGCCGCAATAGATGGT R: CCAAGACCTATTCTGGAAGCA |
HEX | 117–126 | |
| AK12 | ERR7598207.14357357 | (ACA)6 |
F: CCAAAACGTATATCATCCGAAG R: GGAGAATCATCTGTGATAGTTTTGTG |
6-FAM | 91–97 | |
| Multiplex C | AK15 | ERR7598207.14247464 | (TAA)7 |
F: CCCAGTCCCAGCTTGAATAA R: AGGCATCAAGCTGCATCCTA |
HEX | 103–106 |
| AK23 | ERR7598207.14317350 | (CCTT)5 |
F: GGGGTTAACCAACCGAACC R: CGTTTGCCTTATACTGACAAATCT |
6-FAM | 134–145 | |
| AK28 | ERR7598207.14270387 | (GGTT)8 |
F: GCAGCAACTTCCTTCCGTAG R: AACTGGCTGACCAGCGTAAC |
6-FAM | 80–104 | |
| Discarded | AK02 | ERR7598207.1435939 | (GCA)10 |
F: GGTGCGTCAGCAGCAGTAGT R: GGGATGAGAGACAGAGGGAAA |
||
| AK04 | ERR7598207.14356098 | (TGA)8 |
F: GATTAGATAATCCGGCATGGA R: TTAAGATCAACCAGTGGCATAAGA |
|||
| AK06 | ERR7598207.14366465 | (CGT)6 |
F: CTCCGCCACCATTTTAGTGT R: TCCAGTTTTTCAAGAGCCATA |
|||
| AK08 | ERR7598207.12531158 | (GGA)7 |
F: GCAGCCGGTAGTGGTGAC R: GCAGAATTGCTTGCTGCAC |
|||
| AK11 | ERR7598207.14345973 | (AAT)6 |
F: TACCCGAATCCAGCAAACAT R: GCCAGAAGGACATCGTCAC |
|||
| AK14 | ERR7598207.14353635 | (GAT)8 |
F: CGGCTTTGATGACAGGAAAC R: TGTTTACCAGACCGCACTGT |
|||
| AK16 | ERR7598207.14084369 | (GAA)6 |
F: ACAATCCCTGGCAATGTCC R: CAGCCTGCACAAACAACAC |
|||
| AK17 | ERR7598207.14057111 | (TCT)6 |
F: AGCAATGATGGGTGACTATTGTT R: AGCAATGATGGGTGACTATTGTT |
|||
| AK18 | ERR7598207.14365408 | (GTC)7 |
F: CGATGTGGCCTTTTGTCG R: CTTCCAAAATTTCATAAAACACTGC |
|||
| AK19 | ERR7598207.14367708 | (CA)7 |
F: CAACTGAAGAGTGAATTCCAAAA R: AGGGCGATACGGTCAAAAT |
|||
| AK20 | ERR7598207.8923957 | (CG)10 |
F: GGTTTTCCCCGAGTTCGT R: GGGTGGGTGGGTTAATTTTC |
|||
| AK21 | ERR7598207.13745169 | (AATT)5 |
F: GCTTTTTATCTGGTGAAATGCT R: AGCAGCAACACCATCATGC |
|||
| AK22 | ERR7598207.14102450 | (GGAA)5 |
F: CGACTCGGTACGAGTTCACA R: CAACCGAACCAACCAATAGT |
|||
| AK24 | ERR7598207.14368656 | (CT)17 |
F: GATTTCGAAACATGGTGAAAG R: GATGTAGCCATGATTGCAAGTAG |
|||
| AK25 | ERR7598207.14357143 | (GA)12 |
F: AACTGTTCGCAATTGGCTTT R: ATTCATAGCACTCGGCGAAA |
|||
| AK26 | ERR7598207.14364132 | (TG)8 |
F: CGCTCCGATTTTCGTATTCA R: CGTCGGGCTCAGACTATTTG |
|||
| AK27 | ERR7598207.14359268 | (TA)7 |
F: GTCGGTGATTGTCACCATGT R: CATCCAGAGTGCATCAATCG |
|||
| AK29 | ERR7598206.10239627 | (GGCC)4 |
F: CTACCCTTGCTTGGAGGTTG R: GTCGAGACGTGTGAGAGTGC |
|||
| AK30 | ERR7598207.11336441 | (GC)5 |
F: TGAGTAACTGCGAGCTTGTCTC R: GAGATTGATTGTAAATACACACACACA |
|||
| AK31 | ERR7598207.14342765 | (TGA)6 |
F: TCGCTGGAATGGTATAAGGAA R: TTGCCTTGCTACATTAGATGGT |
Indicated for each locus: the reference microsatellite-containing sequence (available in the Sequence Read Archive, SRA); the repeat motifs; forward (F) and reverse (R) primer sequences. In addition, for the 11 functional microsatellite markers, details about fluorophores used to mark primers, and the size ranges of the detected alleles (base pairs [bp]) are specified
an = number of in tandem repeats of the microsatellite motive in the reference sequence
bFluorophores were all added at 5′-forward primers, except for AK15, which has been marked on a 5′-reverse primer
The obtained microsatellite loci were assayed on a panel of 32 mosquito individuals collected in four different locations, representative of different areas of Northern Italy: the districts of Asti, Como, and Vicenza (collected in autumn 2021) and Bergamo (collected in summer 2022). Mosquitoes were identified as Ae. koreicus using the methodology described by Arnoldi et al. [14]. In particular, the DNA of eight mosquito samples from each location was extracted with the DNeasy Blood & Tissue kit (Qiagen, Hilden, Germany) and used in PCR. The PCR protocol that we developed for the single PCR is as follows. Final concentrations of PCR reagents in a 25 µl volume: 1 × Colorless GoTaq® Reaction Buffer (Promega, Milan, Italy), 0.6 U GoTaq® DNA polymerase (Promega, Milan, Italy), 0.2 mM dNTPs (Promega, Milan, Italy), 0.5 μM of each primer, and ~10 ng of DNA as template. Concerning multiplex PCRs, the final concentrations of the reagents is the same, except for primer pairs. Particularly, in multiplex A and B (both with four primer pairs), primer concentration was 0.125 μM; on the contrary, in multiplex C (three primer pairs), primer concentration was 0.167 μM (see Table 1). The thermal protocol for the amplification is identical for single and multiplex PCR reactions. The latter started with an initial denaturation at 94 °C for 2 min, followed by 40 cycles of denaturation at 94 °C for 30 s, primer annealing at 57 °C for 30 s and elongation at 72 °C for 30 s; a final extension of 5 min at 72 °C concluded the reaction.
The amplicons were visualised on 1.5% agarose gel, and only loci that showed clear single bands for all the amplified samples were considered for further testing (Additional File 1: Figure S1). Finally, as the last step of experimental validation, the precise length of the amplified fragments was determined, thus genotyping the examined mosquito individuals. Briefly, PCR products were directly sent to the laboratory of Eurofins Genomics—Europe Applied Genomics GmbH (Ebersberg, Germany) for fragment length analysis (FLA service). Amplicon lengths were determined by capillary electrophoresis with an ABI 3130 XL sequencing machine, using Promega ILS600 as standard. The SSR fragment data were analysed by the company using GeneMapper v.5 software (Applied Biosystem, Waltham, MA, USA). Before statistical analysis, all results were manually checked using Peak Scanner v.2 software (Applied Biosystem, Waltham, MA, USA), and allelic length rounded to the integer.
To obtain preliminary data about SSR polymorphism, the genotypes of all 32 individuals were analysed considering all mosquitoes as part of a unique population. Parameters of intra-population genetic diversity, such as number of alleles (Na), number of effective alleles (Ne), observed and expected heterozygosity (Ho and He, respectively), unbiased expected heterozygosity (uHe), Shannon diversity index (I), and inbreeding coefficient (F) were computed using GenAlEx 6.5 software [36]; in addition, Polymorphic Information Content (PIC) was computed using Cervus v. 3.0.7 software [37, 38].
Results and discussion
Our initial in silico screening identified 31 putative SSR-containing sequences, named AK01–AK31 (see Table 1). All the loci showed perfect repetitions of di-, tri-, and tetranucleotides; specifically, the majority of the loci are trimeric, while AK19, AK20, AK24, AK25, AK26, AK27, and AK30 are characterised by dimeric motifs, and AK21, AK22, AK23, AK28, and AK29 are tetrameric.
Primers designed for SSR amplification were assayed in eight mosquito samples using PCR, and for 16 primers pairs the amplification was successful. After PCR validation, FLA of these 16 loci was performed on the 32 mosquito samples (Table 1). Most of the SSR electropherograms showed clearly distinguishable peaks, and the length of the fragments was in line with the expected range. However, two SSRs were discarded after this analysis: AK02 and AK24. Peaks of the AK02 locus were altered by the presence of stutters (both in n − 1 and n + 1 positions), and length was shorter than the expected size (observed: ~73 base pairs [bp]; expected: 101 bp). The size range of the AK24 locus was broad, and the presence of spurious peaks made correct peak calling impossible. Although the AK11, AK19, and AK21 loci showed no technical problems, they were not further considered because they were not polymorphic in the 32 samples analysed.
Primers for the amplification of the remaining 11 loci were combined in three multiplex PCRs (Table 1). Results from the analysis of the multiplexes were congruent with those of the electropherograms of the single loci (see Additional File 2: Figure S2 for some representative electropherograms of both single and multiplex reactions). This suggests that the combination of primer pairs does not affect the amplification efficiency of multiplexes, and this was true even among samples from different locations. Particularly, all individuals were successfully genotyped for most of the 11 loci. Missing values have been observed just in two samples: one mosquito from Asti at locus AK03, and another individual from Como in AK09.
The genotyping results that we obtained for the 32 individual mosquitoes were then used to evaluate the level of SSR variation in this test population sample (Table 2).
Table 2.
Data of microsatellite variation in Aedes koreicus individuals
| Locus | N | Na | Ne | Ho | He | uHe | I | F | PIC |
|---|---|---|---|---|---|---|---|---|---|
| AK01 | 32 | 3 | 1.427 | 0.219 | 0.299 | 0.304 | 0.577 | 0.269 | 0.279 |
| AK03 | 31 | 3 | 2.424 | 0.226 | 0.587 | 0.597 | 0.960 | 0.616 | 0.502 |
| AK05 | 32 | 4 | 2.395 | 0.594 | 0.583 | 0.592 | 1.064 | −0.019 | 0.525 |
| AK07 | 32 | 3 | 1.683 | 0.313 | 0.406 | 0.412 | 0.734 | 0.230 | 0.370 |
| AK09 | 31 | 6 | 3.161 | 0.548 | 0.684 | 0.695 | 1.344 | 0.198 | 0.636 |
| AK10 | 32 | 4 | 1.416 | 0.250 | 0.294 | 0.299 | 0.562 | 0.150 | 0.266 |
| AK12 | 32 | 3 | 2.002 | 0.563 | 0.500 | 0.508 | 0.748 | −0.124 | 0.390 |
| AK13 | 32 | 2 | 1.032 | 0.031 | 0.031 | 0.031 | 0.080 | −0.016 | 0.030 |
| AK15 | 32 | 2 | 1.032 | 0.031 | 0.031 | 0.031 | 0.080 | −0.016 | 0.030 |
| AK23 | 32 | 3 | 1.722 | 0.406 | 0.419 | 0.426 | 0.740 | 0.031 | 0.375 |
| AK28 | 32 | 6 | 3.303 | 0.813 | 0.697 | 0.708 | 1.385 | −0.165 | 0.645 |
| Mean | 31.818 | 3.545 | 1.963 | 0.363 | 0.412 | 0.419 | 0.752 | 0.105 | 0.368 |
| SE | 0.122 | 0.413 | 0.235 | 0.074 | 0.070 | 0.071 | 0.130 | 0.067 | 0.209 |
The following parameter are reported: number of individuals (N), number of alleles (Na), number of effective alleles (Ne), observed (Ho) and expected (He) heterozygosities, unbiased expected heterozygosity (uHe), Shannon diversity index (I), inbreeding coefficient (F), Polymorphic Information Content (PIC)
The most variable loci were AK09 and AK28. These displayed the highest allelic abundance (Na = 6 for both; Table 2), together with the maximum values of He and PIC (AK09: He = 0.684, PIC = 0.636; AK28: He = 0.697, PIC = 0.645) (Table 2). Even if AK03 and AK05 showed a limited number of alleles (Na = 3 and 4, respectively), the associated PIC values are higher than the average (mean PIC = 0.368 ± 0.209; PICAK03 = 0.502, PICAK05 = 0.525) (Table 2).
Conclusions
In conclusion, we report the identification of microsatellite markers in the Ae. koreicus mosquito, the protocol for their amplification, and the preliminary genetic results on a limited number of individuals. The SSRs here implemented showed a good level of polymorphism. In addition to the novelty of designing molecular markers specific for the Korean bush mosquito, the SSRs here proposed will likely represent a useful tool for generating data and drawing hypotheses about the original introduction of Ae. koreicus in Europe, reconstructing its invasive routes and thus having more information to prevent the further spreading of the species.
Supplementary Information
Additional file 1: Figure S1 Agarose gel electrophoresis showing the amplification of all 31 microsatellite sequences identified in the genome of Aedes koreicus. For all the loci (11 validated markers and the discarded ones), four representative DNA samples of Ae. koreicus were amplified with primer pairs designed in this study. a Pictures of amplicons obtained with AK01–AK10. b Pictures of amplicons obtained with AK11–AK20. c. Pictures of amplicons obtained with AK21–AK11. M molecular marker (GeneRuler 100 bp Plus DNA Ladder, Thermo Scientific™, Waltham, MA, USA); A Adult mosquito of Ae. Koreicus, N negative control.
Additional file 2: Figure S2 Representative electropherograms of single and multiplex PCR performed to validate the efficiency of the 11 microsatellite markers identified in the genome of Aedes koreicus; electropherograms were visualised with Peak Scanner v.2 (Applied Biosystems, Waltham, MA, USA). a–k Peaks associated with a single marker (one page for each locus). l–n Multiplex A, B, and C are reported (one page for all the loci involved in the same multiplex assay). The legend displaying the dye colours, and the corresponding fluorophores/loci, together with the distribution of the microsatellite markers in the multiplex assays, are reported at the end of the document.
Acknowledgements
The authors are grateful to Dr Paolo Roberto for his help during the collection of Aedes koreicus mosquitoes in the Piedmont region. We would also like to thank the student Francesca Magna for her help during sample collections and experimental validation.
Abbreviations
- SSRs
Simple sequence repeats
- SRA
Sequence Read Archive
- BLAST
Basic Local Alignment Search Tool
- PCR
Polymerase chain reaction
- FLA
Fragment length analysis
Author contributions
LS, AN, IA, AMi, FM, AMo, SE, and PG performed sample collections and identification. LS and GN performed molecular analysis. LS, PG, and SE conducted the statistical analysis and wrote the first draft of the paper. CB contributed to the design of the study and to the manuscript revision. All authors read and approved the final manuscript.
Funding
The study has been conducted under the framework of the Project PE-13, INFACT, RESEARCH NODE2 Arthropod Vectors and Vector-Borne Diseases, which is part of the National Recovery and Resilience Plan (NRRP); this research was also partially supported by the Italian Ministry of Education, University and Research, PRIN 2017 Prot. 2017J8JR57 to CB and by the Fondazione Cariplo (2017-0798) to PG.
Availability of data and materials
All data supporting the findings of this study are available within the paper and its Supplementary Information. Microsatellite primer sequences and alleles details are provided in Table 1; agarose gel electrophoresis pictures and representative electropherograms peaks are reported in Additional File 1 and Additional File 2, respectively.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Change history
10/9/2023
Following publication of this article, the (erroneous) note ‘Open access funding provided by Università degli Studi di Milano within the CRUI-CARE Agreement’ has been removed from the Funding declaration
Contributor Information
Paolo Gabrieli, Email: paolo.gabrieli@unimi.it.
Sara Epis, Email: sara.epis@unimi.it.
References
- 1.Cebrián-Camisón S, Martínez-de la Puente J, Figuerola J. A literature review of host feeding patterns of invasive Aedes mosquitoes in Europe. Insects. 2020;11:848. doi: 10.3390/insects11120848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Giunti G, Becker N, Benelli G. Invasive mosquito vectors in Europe: from bioecology to surveillance and management. Acta Trop. 2023;239:106832. doi: 10.1016/j.actatropica.2023.106832. [DOI] [PubMed] [Google Scholar]
- 3.Derraik JGB. Exotic mosquitoes in New Zealand: a review of species intercepted, their pathways and ports of entry. Aust N Z J Public Health. 2004;28:433–444. doi: 10.1111/j.1467-842X.2004.tb00025.x. [DOI] [PubMed] [Google Scholar]
- 4.Medlock JM, Hansford KM, Schaffner F, Versteirt V, Hendrickx G, Zeller H, et al. A review of the invasive mosquitoes in Europe: ecology, public health risks, and control options. Vector Borne Zoonotic Dis. 2012;12:435–447. doi: 10.1089/vbz.2011.0814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Knox TB, Kay BH, Hall RA, Ryan PA. Enhanced vector competence of Aedes aegypti (Diptera: Culicidae) from the torres strait compared with Mainland Australia for dengue 2 and 4 viruses. J Med Entomol. 2003;40:950–956. doi: 10.1603/0022-2585-40.6.950. [DOI] [PubMed] [Google Scholar]
- 6.Fonseca DM, Widdel AK, Hutchinson M, Spichiger S-E, Kramer LD. Fine-scale spatial and temporal population genetics of Aedes japonicus, a new US mosquito, reveal multiple introductions. Mol Ecol. 2010;19:1559–1572. doi: 10.1111/j.1365-294X.2010.04576.x. [DOI] [PubMed] [Google Scholar]
- 7.Tabachnick WJ. Nature, nurture and evolution of Intra-species variation in mosquito arbovirus transmission competence. Int J Environ Res Public Health. 2013;10:249–277. doi: 10.3390/ijerph10010249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Manni M, Guglielmino CR, Scolari F, Vega-Rúa A, Failloux A-B, Somboon P, et al. Genetic evidence for a worldwide chaotic dispersion pattern of the arbovirus vector, Aedes albopictus. PLOS Negl Trop Dis. 2017;11:e0005332. doi: 10.1371/journal.pntd.0005332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vega-Rúa A, Marconcini M, Madec Y, Manni M, Carraretto D, Gomulski LM, et al. Vector competence of Aedes albopictus populations for chikungunya virus is shaped by their demographic history. Commun Biol. 2020;3:326. doi: 10.1038/s42003-020-1046-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Smitz N, De Wolf K, Deblauwe I, Kampen H, Schaffner F, De Witte J, et al. Population genetic structure of the Asian bush mosquito, Aedes japonicus (Diptera, Culicidae), in Belgium suggests multiple introductions. Parasit Vectors. 2021;14:179. doi: 10.1186/s13071-021-04676-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tanaka K, Mizusawa K, Saugstad ES. A revision of the adult and larval mosquitoes of Japan (including the Ryukyu Archipelago and the Ogasawara Islands) and Korea (Diptera: Culicidae) Contrib Am Entomol Inst. 1979;16:1–987. [Google Scholar]
- 12.Versteirt V, Pecor J, Fonseca D, Coosemans M, Bortel W. Confirmation of Aedes koreicus (Diptera: Culicidae) in Belgium and description of morphological differences between Korean and Belgian specimens validated by molecular identification. Zootaxa. 2012;3191:21–32. doi: 10.11646/zootaxa.3191.1.2. [DOI] [Google Scholar]
- 13.Aedes koreicus - current known distribution: March 2022. European Centre for Disease Prevention and Control. 2022. https://www.ecdc.europa.eu/en/publications-data/aedes-koreicus-current-known-distribution-march-2022. Accessed 10 Dec 2022.
- 14.Arnoldi I, Negri A, Soresinetti L, Brambilla M, Carraretto D, Montarsi F, et al. Assessing the distribution of invasive Asian mosquitoes in Northern Italy and modelling the potential spread of Aedes koreicus in Europe. Acta Trop. 2022;232:106536. doi: 10.1016/j.actatropica.2022.106536. [DOI] [PubMed] [Google Scholar]
- 15.Vojtíšek J, Šebesta O, Šikutová S, Kampen H, Rudolf I. First record of the invasive mosquito species Aedes koreicus (Diptera: Culicidae) in the Czech Republic. Parasitol Res. 2022;121:3701–3704. doi: 10.1007/s00436-022-07658-6. [DOI] [PubMed] [Google Scholar]
- 16.Hohmeister N, Werner D, Kampen H. The invasive Korean bush mosquito Aedes koreicus (Diptera: Culicidae) in Germany as of 2020. Parasit Vectors. 2021;14:575. doi: 10.1186/s13071-021-05077-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Capelli G, Drago A, Martini S, Montarsi F, Soppelsa M, Delai N, et al. First report in Italy of the exotic mosquito species Aedes (Finlaya) koreicus, a potential vector of arboviruses and filariae. Parasit Vectors. 2011;4:188. doi: 10.1186/1756-3305-4-188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Montarsi F, Drago A, Martini S, Calzolari M, De Filippo F, Bianchi A, et al. Current distribution of the invasive mosquito species, Aedes koreicus [Hulecoeteomyia koreica] in northern Italy. Parasit Vectors. 2015;8:614. doi: 10.1186/s13071-015-1208-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gradoni F, Bertola M, Carlin S, Accordi S, Toniolo F, Visentin P, et al. Geographical data on the occurrence and spreading of invasive Aedes mosquito species in Northeast Italy. Data Br. 2021;36:107047. doi: 10.1016/j.dib.2021.107047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ballardini M, Ferretti S, Chiaranz G, Pautasso A, Riina MV, Triglia G, et al. First report of the invasive mosquito Aedes koreicus (Diptera: Culicidae) and of its establishment in Liguria, northwest Italy. Parasit Vectors. 2019;12:334. doi: 10.1186/s13071-019-3589-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Suter T, Flacio E, Fariña BF, Engeler L, Tonolla M, Müller P. First report of the invasive mosquito species Aedes koreicus in the Swiss-Italian border region. Parasit Vectors. 2015;8:1–4. doi: 10.1186/s13071-015-1010-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Negri A, Arnoldi I, Brilli M, Bandi C, Gabrieli P, Epis S. Evidence for the spread of the alien species Aedes koreicus in the Lombardy region Italy. Parasit Vectors. 2021;14:534. doi: 10.1186/s13071-021-05031-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mosca A, Perna MF, Giovannozzi M, Roberto P. First report of two Asian invasive mosquito species, Aedes japonicus and Aedes koreicus, in Piedmont, northwest Italy. Ann Ist Super Sanità. 2022;58:162–165. doi: 10.4415/ANN_22_03_03. [DOI] [PubMed] [Google Scholar]
- 24.Kurucz K, Zeghbib S, Arnoldi D, Marini G, Manica M, Michelutti A, et al. Aedes koreicus, a vector on the rise: Pan-European genetic patterns, mitochondrial and draft genome sequencing. PLoS ONE. 2022;17:e0269880. doi: 10.1371/journal.pone.0269880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang D-X, Hewitt GM. Nuclear DNA analyses in genetic studies of populations: practice, problems and prospects. Mol Ecol. 2003;12:563–584. doi: 10.1046/j.1365-294X.2003.01773.x. [DOI] [PubMed] [Google Scholar]
- 26.Bataille A, Cunningham AA, Cruz M, Cedeño V, Goodman SJ. Adaptation, isolation by distance and human-mediated transport determine patterns of gene flow among populations of the disease vector Aedes taeniorhynchus in the Galapagos Islands. Infect Genet Evol. 2011;11:1996–2003. doi: 10.1016/j.meegid.2011.09.009. [DOI] [PubMed] [Google Scholar]
- 27.Brelsfoard CL, Dobson SL. Population genetic structure of Aedes polynesiensis in the Society Islands of French Polynesia: implications for control using a Wolbachia-based autocidal strategy. Parasit Vectors. 2012;5:80. doi: 10.1186/1756-3305-5-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Halvarsson P, Hesson JC, Lundström JO. Six polymorphic microsatellites in the flood-water mosquito Aedes sticticus. J Vector Ecol. 2013;38:404–407. doi: 10.1111/j.1948-7134.2013.12057.x. [DOI] [PubMed] [Google Scholar]
- 29.Multini LC, Wilke ABB, Suesdek L, Marrelli MT. Population genetic structure of Aedes fluviatilis (Diptera: Culicidae) PLoS ONE. 2016;11:e0162328. doi: 10.1371/journal.pone.0162328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Edillo F, Ymbong RR, Cabahug MM, Labiros D, Suycano MW, Lambrechts L, et al. Yearly variations of the genetic structure of Aedes aegypti (Linnaeus) (Diptera: Culicidae) in the Philippines (2017–2019) Infect Genet Evol. 2022;102:105296. doi: 10.1016/j.meegid.2022.105296. [DOI] [PubMed] [Google Scholar]
- 31.Widdel AK, McCuiston LJ, Crans WJ, Kramer LD, Fonseca DM. Finding needles in the haystack: single copy microsatellite loci for Aedes japonicus (Diptera: Culicidae) Am J Trop Med Hyg. 2005;73:744–748. doi: 10.4269/ajtmh.2005.73.744. [DOI] [PubMed] [Google Scholar]
- 32.Teekema S, Stroo A, Uiterwijk M, van de Vossenberg B, Jacobs F, Ibáñez-Justicia A. First finding of Aedes koreicus (Diptera: Culicidae) in the Netherlands. J Eur Mosq Control Assoc. 2022;40:3. doi: 10.52004/JEMCA2021.0005. [DOI] [Google Scholar]
- 33.Sequence Read Archive. https://www.ncbi.nlm.nih.gov/sra. Accessed: 18 May 2022.
- 34.Nucleotide Basic Local Alignment Search Tool. https://blast.ncbi.nlm.nih.gov/Blast.cgi. Accessed 18 May 2022.
- 35.Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol. 2000;132:365–386. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
- 36.Peakall R, Smouse PE. GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research—an update. Bioinformatics. 2012;28(19):2537–2539. doi: 10.1093/bioinformatics/bts460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kalinowski ST, Taper ML, Marshall TC. Revising how the computer program CERVUS accommodates genotyping error increases success in paternity assignment. Mol Ecol. 2007;16:1099–1106. doi: 10.1111/j.1365-294X.2007.03089.x. [DOI] [PubMed] [Google Scholar]
- 38.Kalinowski ST, Taper ML, Marshall TC. Corrigendum. Mol Ecol. 2010;19:1512–1512. doi: 10.1111/j.1365-294X.2010.04544.x. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Figure S1 Agarose gel electrophoresis showing the amplification of all 31 microsatellite sequences identified in the genome of Aedes koreicus. For all the loci (11 validated markers and the discarded ones), four representative DNA samples of Ae. koreicus were amplified with primer pairs designed in this study. a Pictures of amplicons obtained with AK01–AK10. b Pictures of amplicons obtained with AK11–AK20. c. Pictures of amplicons obtained with AK21–AK11. M molecular marker (GeneRuler 100 bp Plus DNA Ladder, Thermo Scientific™, Waltham, MA, USA); A Adult mosquito of Ae. Koreicus, N negative control.
Additional file 2: Figure S2 Representative electropherograms of single and multiplex PCR performed to validate the efficiency of the 11 microsatellite markers identified in the genome of Aedes koreicus; electropherograms were visualised with Peak Scanner v.2 (Applied Biosystems, Waltham, MA, USA). a–k Peaks associated with a single marker (one page for each locus). l–n Multiplex A, B, and C are reported (one page for all the loci involved in the same multiplex assay). The legend displaying the dye colours, and the corresponding fluorophores/loci, together with the distribution of the microsatellite markers in the multiplex assays, are reported at the end of the document.
Data Availability Statement
All data supporting the findings of this study are available within the paper and its Supplementary Information. Microsatellite primer sequences and alleles details are provided in Table 1; agarose gel electrophoresis pictures and representative electropherograms peaks are reported in Additional File 1 and Additional File 2, respectively.