Abstract
Background
With the growing availability of cannabis and the popularization of additional routes of cannabis use beyond smoking, including edibles, the prevalence of cannabis use in pregnancy is rapidly increasing. However, the potential effects of prenatal cannabis use on fetal developmental programming remain unknown.
Results
We designed this study to determine whether the use of edible cannabis during pregnancy is deleterious to the fetal and placental epigenome. Pregnant rhesus macaques consumed a daily edible containing either delta-9-tetrahydrocannabinol (THC) (2.5 mg/7 kg/day) or placebo. DNA methylation was measured in 5 tissues collected at cesarean delivery (placenta, lung, cerebellum, prefrontal cortex, and right ventricle of the heart) using the Illumina MethylationEPIC platform and filtering for probes previously validated in rhesus macaque. In utero exposure to THC was associated with differential methylation at 581 CpGs, with 573 (98%) identified in placenta. Loci differentially methylated with THC were enriched for candidate autism spectrum disorder (ASD) genes from the Simons Foundation Autism Research Initiative (SFARI) database in all tissues. The placenta demonstrated greatest SFARI gene enrichment, including genes differentially methylated in placentas from a prospective ASD study.
Conclusions
Overall, our findings reveal that prenatal THC exposure alters placental and fetal DNA methylation at genes involved in neurobehavioral development that may influence longer-term offspring outcomes. The data from this study add to the limited existing literature to help guide patient counseling and public health polices focused on prenatal cannabis use in the future.
Supplementary Information
The online version contains supplementary material available at 10.1186/s13148-023-01519-4.
Keywords: Delta-9-tetrahydrocannabinol (THC), Cannabis, Marijuana, DNA methylation, Prenatal substance use
Background
Epigenetic regulation and modulation of gene expression are essential for normal fetal development [1]. Chromatin remodeling through DNA methylation is one of the most studied epigenetic processes. The epigenome, and particularly DNA methylation, is responsive to exogenous exposures and can elicit gene dysregulation, which may influence cellular function and tissue development [2]. DNA methylation is a critical mediator of normal placental [3] and fetal development, including the central nervous system [4, 5]. Epigenetic responses to maternal psychoactive substance use may lead to long-term molecular alterations implicated in addiction and psychiatric disorders [6, 7]. Cannabis, commonly known as marijuana, refers to a group of plants with psychoactive and medicinal properties. Use of cannabis and cannabis-derived products in the USA has increased as a greater number of states have legalized both recreational and medicinal cannabis use, which has resulted in diminished public perception of potential physiological risks of cannabis use [8]. Cannabis use among pregnant individuals [9], especially during the critical developmental window in the first trimester to mitigate morning sickness symptoms, has doubled in the past decade [10] and half of those that use cannabis will continue to use throughout pregnancy [11–13].
Cannabis acts by targeting the endocannabinoid system to exert neuromodulatory and paracrine effects, in part via epigenetic modifications, that impact organogenesis, neurogenesis, and gliogenesis [7, 14]. The main psychoactive cannabinoid delta-9-tetrahydrocannabinol (THC) mimics endogenous cannabinoids by crossing the placenta and binding to cannabinoid receptors, CB1 and CB2, in the placenta and major fetal organs, including the brain [15–20], leading to concern for detrimental fetal and offspring outcomes [21–24].
Although limited in number and mechanistic understanding, studies suggest adverse effects of prenatal cannabis exposure that include preterm birth, stillbirth and small for gestational age infants [24–28]. Maternal cannabis use has also been associated with an increased risk for neurobehavioral morbidity in human offspring, including autism spectrum disorder (ASD), attention deficit hyperactivity disorder (ADHD), intellectual disability and learning disorders, and other neuropsychiatric disorders [29–35]. Preliminary data in humans, mice and rats indicate that prenatal cannabis exposure results in altered DNA methylation in both the placenta and the brains of exposed offspring and that these epigenetic marks may be a potential mechanistic link between maternal cannabis use and associated negative offspring outcomes [36–38]. Prenatal cannabinoid exposure has also resulted in increased anxiety-like behavior and altered genome-wide brain DNA methylation in mouse offspring [38].
Although there are limited clinical data regarding the effects of cannabis exposure during pregnancy, the rising prevalence of prenatal cannabis use [39] is of significant concern, as the potential short-term medicinal benefits for nausea and pain may be outweighed by the possibility of longer-term adverse impacts to offspring. Despite its potential adverse impact, there is a striking paucity of in vivo data on the effects of THC on fetal development in part because of the regulatory challenges, heterogeneity, and confounds in human studies. In addition, despite evidence suggesting that maternal substance use can impact the fetal epigenome [14, 37], our understanding of the impact of prenatal cannabis exposure on placental and fetal epigenetic regulation is impeded by the limitations and feasibility of tissue sampling.
To address key gaps in the evidence and overcome the limitations of previous human studies, we developed a novel non-human primate (NHP) model of chronic cannabis exposure via edible THC consumption [40, 41]. The NHP is a strong translational model that recapitulates human gene expression and regulation during development and has a similar fetal ontogeny [42, 43], placental structure [42, 43], and THC plasma disposition to humans. Use of a NHP model overcomes the limitations of previous human studies by facilitating tissue sampling [44], resulting in observations that are directly translatable to human pregnancies [45]. Edibles are the second most common mode of cannabis delivery [46] and are often recommended by dispensaries to pregnant individuals for nausea [47]. Our NHP model of THC edible consumption uniquely recapitulates typical human use, minimizes subject variability and allows precise THC dosing to elucidate direct biological consequences of chronic prenatal cannabis exposure, while methodically controlling for potential confounders.
The incidence of cannabis use is rising, and there is a critical need for investigation of the effects of in utero exposure to cannabis at the molecular level, in order to increase our understanding of the risks on fetal development, the potential underlying mechanisms, and future health outcomes. Our objective was to use our novel NHP model of maternal THC edible use to help bridge this knowledge gap by determining the impact of prenatal THC exposure on the placental and offspring epigenome.
Results
Study sample characteristics
Tissues were collected following cesarean section delivery during the third trimester at gestational day 155 (term is ~ 168 days) following chronic prenatal THC-exposure (THC, n = 5) from preconception throughout gestation or placebo-exposure (CON, n = 5). There were no significant differences in baseline characteristics between THC- and CON-exposed dams, placental, or offspring birth characteristics as shown in Table 1. DNA methylation was measured using the Infinium Methylation EPIC platform in all available tissues. Heart DNA was not available for one THC-exposed animal, and one cerebellum dataset (CON) was removed following quality control analysis due to low intensity. After quantile normalization and retaining probes previously validated in Macaca mulatta [48], we observed clear separation of datasets by tissue (Additional file 1: Figure S1).
Table 1.
Demographics and sample characteristics by treatment group
| Characteristic | Control | THC | p value |
|---|---|---|---|
| n | 5 | 5 | |
| Maternal age (years) | 10.0 ± 2.3 | 9.9 ± 2.1 | 0.945 |
| Parity | 3.6 ± 0.5 | 2.2 ± 2.2 | 0.226 |
| Maternal weight at G60 (kg) | 7.53 ± 1.02 | 7.74 ± 0.47 | 0.692 |
| Maternal weight at delivery (G155) (kg) | 8.94 ± 1.20 | 8.95 ± 0.48 | 0.990 |
| Fetal birth weight (kg) | 0.48 ± 0.08 | 0.46 ± 0.07 | 0.676 |
| Total placental weight (g) | 105.2 ± 17.6 | 105.0 ± 18.7 | 0.984 |
| Fetal sex (male:female) | 2:3 | 3:2 | –- |
Data are means ± SD. Statistical analysis performed using Welch’s t test
Prenatal delta-9-tetrahydrocannabinol exposure is associated with changes in placental and offspring tissue DNA methylation
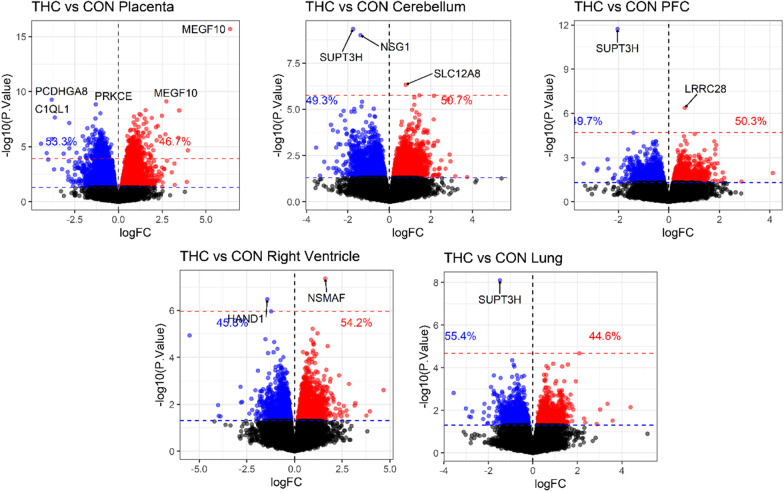
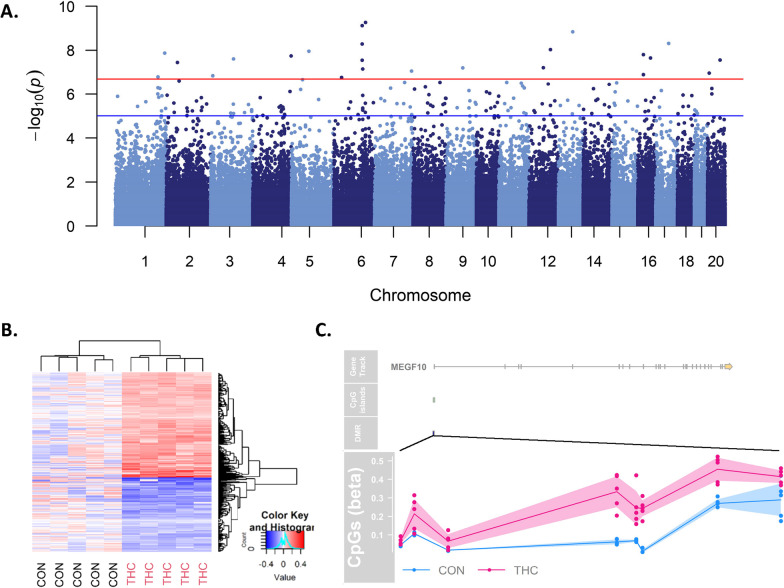
We tested for differentially methylated CpGs (DMC)s with prenatal THC exposure in each tissue using 234,836 CpG-specific linear models with factorial design of group and tissue, accounting for within-subject correlation. Genomic inflation factors from the tissue-specific contrasts ranged from lambda = 0.47–1.01 (Additional file 1: Figure S2). A total of 581 CpGs were significantly associated with prenatal THC exposure after adjusting for multiple comparisons using the Benjamini and Hochberg method to control false discovery rate (FDR) (Fig. 1) [49]. The top 5 DMCs per tissue are presented in Table 2, and the full list of FDR significant (< 0.05) DMCs is available in Additional file 2: Table S1. Out of 581 FDR DMCs, 573 (98%; 335 hypermethylated/ 238 hypomethylated) were identified in placenta and appeared to be dispersed across the genome with the exception of a spike in association located at chromosome 6 based on rhesus annotation (Mmul_10; Fig. 2A). Hierarchical clustering of placental DMCs (Fig. 2B) and a MDS plot of the most variable CpGs (Additional file 1: Figure S3) showed clear separation of THC versus CON animal DNA methylation in the placenta. The top CpG associated with THC exposure, cg23018092, was specific to placental tissue (adjusted p value = 4.85e−11; logFC = 6.38), located at chr6:124772737 in the rhesus genome. Of note, cg23018092 mapped to a CpG island overlapping the first exon of MEGF10 and there were 5 additional CpGs within this CpG island all hypermethylated with THC exposure at FDR significance (Fig. 2C).
Fig. 1.
Prenatal THC is associated with differential methylation at individual CpGs in each tissue tested. Volcano plots for each tissue show CpG-specific associations with THC exposure using limma with a factorial design of group and tissue, accounting for within-subject correlation. Each plot depicts the logFC for THC versus CON on the x-axis, − log10(p values) on the y-axis. Percentages indicate relative proportions of nominally significant hypomethylated (blue) and hypermethylated (red) CpGs. The dotted blue line indicates nominal p < 0.05, and the dotted red line indicates CpGs reaching significance after adjusting for multiple comparisons using the Benjamini and Hochberg method to control false discovery rate (FDR)
Table 2.
Top five DNA methylation loci per tissue associated with prenatal THC exposure
| Probe | Mmul_10 | Nearest gene | logFC | p value | Adj. p val | ||
|---|---|---|---|---|---|---|---|
| Chr | Pos | ||||||
| Placenta | cg23018092 | 6 | 124,772,737 | MEGF10 | 6.38 | 2.06E−16 | 4.85E−11 |
| cg27346707 | 6 | 124,772,721 | MEGF10 | 2.74 | 7.65E−10 | 5.99E−05 | |
| cg05494467 | 6 | 138,890,510 | PCDHGA8 | − 3.81 | 5.40E−10 | 5.99E−05 | |
| cg20825437 | 13 | 63,151,971 | PRKCE | − 1.30 | 1.45E−09 | 8.54E−05 | |
| cg03721362 | 6 | 124,772,671 | MEGF10 | 3.47 | 5.12E−09 | 0.0002 | |
| Cerebellum | cg00617180 | 4 | 124,660,978 | SUPT3H | − 1.75 | 4.49E−10 | 0.000105 |
| cg19948549 | 5 | 8,503,869 | NSG1 | − 1.39 | 9.65E−10 | 0.000113 | |
| cg17478992 | 2 | 151,164,369 | SLC12A8 | 0.78 | 4.69E−07 | 0.036697 | |
| cg13699963 | 16 | 69,873,069 | SDK2 | 2.14 | 1.86E−06 | 0.08718 | |
| cg11031701 | 16 | 35,959,387 | TBX2 | 1.44 | 1.74E−06 | 0.08718 | |
| Prefrontal Cortex | cg00617180 | 4 | 124,660,978 | SUPT3H | − 2.04 | 1.87E−12 | 4.38E−07 |
| cg01898225 | 7 | 78,635,212 | LRRC28 | 0.63 | 4.18E−07 | 0.049039 | |
| cg02616023 | 1 | 88,639,226 | NOS1AP | − 1.40 | 2.05E−05 | 0.999987 | |
| cg14964115 | 9 | 115,397,494 | FAM160B1 | 1.02 | 2.48E−05 | 0.999987 | |
| cg13915892 | 13 | 1,072,439 | LMAN2L | 0.60 | 3.62E−05 | 0.999987 | |
| Heart | cg22754569 | 8 | 59,305,442 | NSMAF | 1.61 | 4.36E−08 | 0.010244 |
| cg19504702 | 6 | 151,889,356 | HAND1 | − 1.43 | 3.34E−07 | 0.039244 | |
| cg00617180 | 4 | 124,660,978 | SUPT3H | − 1.23 | 1.10E−06 | 0.085922 | |
| cg03396151 | 7 | 13,532,030 | MEIS2 | 0.95 | 6.03E−06 | 0.353767 | |
| cg06933370 | 7 | 13,532,249 | MEIS2 | 1.21 | 9.42E−06 | 0.442549 | |
| Lung | cg00617180 | 4 | 124,660,978 | SUPT3H | − 1.48 | 7.89E−09 | 0.001852 |
| cg17760405 | 4 | 72,836,058 | SIM1 | 2.08 | 2.11E−05 | 0.999998 | |
| cg10762132 | 13 | 15,816,921 | SLC20A1 | − 0.94 | 4.46E−05 | 0.999998 | |
| cg23616524 | 3 | 156,111,969 | UBE2H | 0.91 | 6.44E−05 | 0.999998 | |
| cg11126485 | 14 | 12,810,784 | TMEM138 | 1.57 | 7.09E−05 | 0.999998 | |
Tested using limma with CpG-wise statistical models with factorial design of group and tissue, accounting for within subject correlation
Bold text indicates FDR adjusted p-value < 0.05
Fig. 2.
Placental methylation is significantly associated with THC exposure across the genome. A Manhattan plot of CpGs in placenta associated with prenatal THC exposure with the uncorrected log10 (p values) plotted on the y-axis and chromosomal location on the x-axis (Mmul_10). The blue line is drawn to indicate CpGs that surpassed p value < 1e−5, a threshold for suggestive association, and the red line is to highlight CpGs that passed a stringent Bonferroni threshold of p value < 2e−07. B Heatmap of placental DMCs passing FDR significance (n = 573 CpGs) clearly separate by treatment group. Values represent the log-transformed and mean centered average methylation values per CpG. C Prenatal THC is associated with methylation changes in placental MEGF10 promoter. Gviz plot [4] showing from top to bottom the gene structure, CpG island proximity, and individual CpG beta values for each animal (THC animals are shown in pink and Control in blue)
In fetal brain, we assessed DNA methylation of the cerebellum and prefrontal cortex (PFC), separately. Three CpGs reached FDR significant (< 0.05) association with THC exposure in cerebellum annotated to SUPT3H, NSG1, and SLC12A8; and 2 in PFC annotated to SUPT3H and LRRC28 (Fig. 1; Table 2). In the right heart ventricle, we identified 2 FDR significant DMCs annotated to NSMAF and HAND1 (Fig. 1; Table 2). In fetal lung, 1 CpG reached FDR significance annotated to SUPT3H. However, this CpG (cg00617180) was 1 bp upstream of a common single nucleotide polymorphism (SNP) in our rhesus macaque colony (allele frequency = 0.567) and showed a consistently large effect size across all tissues, suggesting a potentially genetic, rather than treatment effect, at this loci (Additional file 1: Figure S4A).
Given the larger number of CpGs differentially methylated with THC exposure in placental DNA, we also extracted and annotated placental differentially methylated regions (DMRs). We identified 403 significant placental THC-DMRs after FDR correction (Additional file 2: Table S2). Consistent with individual CpG analysis results, the top DMR was annotated to MEGF10 on chr5 (based on EPIC hg19 annotation; chr6 in the rhesus genome) and included 8 CpGs with a mean increase in methylation of 23% with THC exposure. The second and third most significant DMRs ranked by Fisher’s multiple comparison statistic were located on the same chromosome and mapped to ADAMTS19 and a large cluster of protocadherin genes, respectively (Additional file 1: Figure S5A; Additional file 2: Table S2).
Gene ontologies (GO) and biological pathways in genes differentially methylated with THC
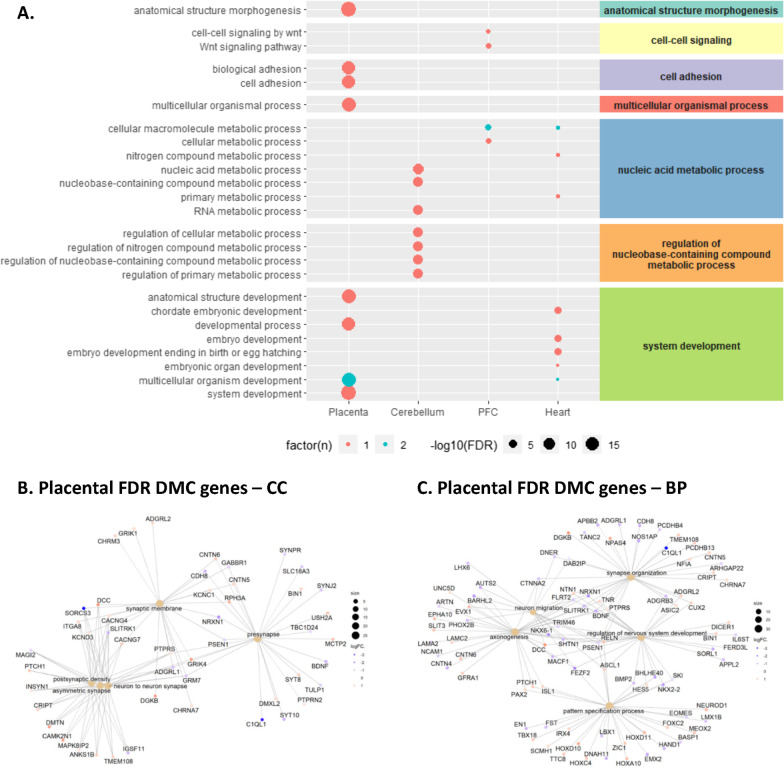
We next identified enriched Gene ontology (GO) terms and Kyoto encyclopedia of genes and genomes (KEGG) pathways within each tissue using the missMethyl package which adjusts for bias related to gene length and probe density [50]. Given our sample size, we included DMCs with a nominal p value < 0.05 in order to inform future functional studies. Top FDR significant GO biological processes per tissue are shown in Fig. 3A, and the full list of FDR significant terms and pathways is presented in Additional file 2: Table S3. In placenta, there were 549 significant GO terms after multiple testing correction, and the top enriched biological processes among placental DMCs included several terms related to development and morphogenesis including “anatomical structure morphogenesis” and “system development”, while “calcium signaling pathway” was the top KEGG term. In the brain, we observed enrichment of terms related to “nucleic acid metabolic process” and “cell–cell signaling by wnt”, while heart tissue was enriched for terms related to system development (e.g., “embryo development” and “tube development”) and also “nucleic acid metabolic process”. There were no FDR significant GO terms in lung tissue. In the placenta only, we also tested for enrichment using the more stringent set of 573 FDR significant DMCs and the clusterProfiler R package to visualize enriched cellular compartments, biological processes, and annotated genes. Placental FDR DMCs were enriched for cellular compartments and biological process terms related to neuronal synapse, nervous system development, neuron migration, and axonogenesis (Fig. 3B, C).
Fig. 3.
DMC genes are enriched for morphogenesis, nucleic acid metabolism, and nervous system development related terms. A The top 10 FDR significant GO terms in the biological processes category are presented for each tissue clustered according to larger parent categories identified using rrvgo. Color indicates the number of tissues enriched for each term and the size of circles indicates − log10 FDR p value across terms. There was no FDR significant enrichment of GO terms in lung tissue. In the placenta, the clusterProfiler R package was used to visualize enriched cellular compartments (B), biological processes (C), and connected genes using the more stringent set of 573 FDR significant DMCs
Prenatal THC changes in DNA methylation are enriched for candidate autism genes
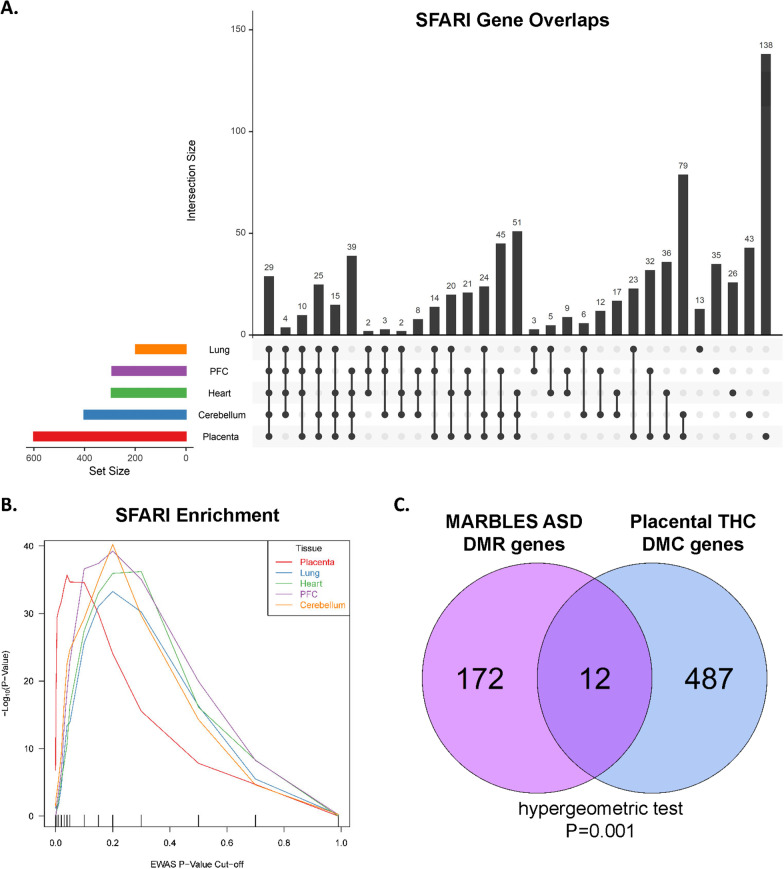
Based on prior reports suggesting an association between prenatal cannabis exposure and offspring risk of ASD [30, 51], we next tested for enrichment of candidate ASD genes among THC-exposure DMCs. From the Simons Foundation Autism Research Initiative (SFARI) database [52], there were 925 candidate ASD genes which annotated to one or more CpG included in our differential methylation analysis. We tested for enrichment at multiple p-value thresholds and performed permutation testing of random gene sets as a negative control. Twenty-nine candidate ASD genes were differentially methylated with THC in all tissues at nominal p < 0.05 (Fig. 4A) and 789 candidate ASD genes (85%) were annotated to 1 or more DMC in any tissue (Additional file 2: Table S4). In each of the 5 tissues analyzed, there was significant enrichment of SFARI genes annotated to DMCs at multiple p-value thresholds, with the greatest enrichment in placenta (Fig. 4B; Table 3). Permutation testing with random gene sets the same size as the SFARI list were not enriched among DMC genes (Additional file 1: Figure S6). Additionally, we compared our placental THC-DMC gene list to a list of differentially methylated regions (DMRs) in human placentas from pregnancies where the newborn was later diagnosed with ASD [53]. We found significant enrichment of human ASD-associated DMR genes among our FDR significant placental THC DMCs (Fig. 4C), which included 12 genes overlapping between the two datasets.
Fig. 4.
Candidate autism spectrum disorder genes are enriched among prenatal THC-DMCs for each tissue type. A UpSet plot of the overlap of SFARI genes annotated to one or more DMC for each tissue. The y-axis represents the number of intersections for each combination of sets across the x-axis. Twenty-nine SFARI genes were represented in DMC lists across all five tissues. B THC-DMC-associated genes were tested for enrichment for SFARI genes across a range of p value thresholds. The x-axis indicates p value threshold for association and the y-axis indicates the − log10 p value for enrichment at each threshold. Line colors represent unique tissues. C Venn diagram demonstrates the overlap of genes annotated to ASD DMRs in a prospective birth cohort study (MARBLES) with placental THC DMC genes. The hypergeometric test was used to test for significance of overlap
Table 3.
SFARI gene enrichment at multiple p value thresholds
| Threshold | Measure | Placenta | Lung | PFC | Heart | Cerebellum |
|---|---|---|---|---|---|---|
| p < 1e−04 | Actual | 49 | 2 | 0 | 1 | 5 |
| Expected | 21.9 | 0.4 | 0.5 | 1.0 | 1.9 | |
| OR | 2.5 | 5.7 | 0.0 | 1.0 | 2.9 | |
| p < 0.001 | Actual | 143 | 5 | 2 | 3 | 19 |
| Expected | 69.1 | 1.8 | 2.6 | 4.4 | 9.2 | |
| OR | 2.4 | 3.0 | 0.7 | 0.7 | 2.2 | |
| p < 0.005 | Actual | 285 | 13 | 23 | 28 | 56 |
| Expected | 149.3 | 8.3 | 13.2 | 20.3 | 34.0 | |
| OR | 2.4 | 1.6 | 1.8 | 1.4 | 1.8 | |
| p < 0.01 | Actual | 356 | 27 | 49 | 57 | 102 |
| Expected | 203.9 | 17.2 | 28.7 | 38.9 | 65.1 | |
| OR | 2.3 | 1.6 | 1.8 | 1.5 | 1.7 | |
| p < 0.05 | Actual | 601 | 198 | 291 | 294 | 402 |
| Expected | 417.4 | 116.5 | 167.9 | 188.0 | 257.4 | |
| OR | 2.3 | 2.0 | 2.2 | 1.9 | 2.1 |
Statistical analysis performed using a chi-squared test statistic [54]
PFC prefrontal cortex
Differential methylation signatures following prenatal THC are associated with RNA expression
We next investigated the potential regulation of placental mRNA expression by DNA methylation using expression quantitative trait methylation (eQTM) analysis. We performed crude association between levels of 631 mRNA transcripts differentially expressed (DE) following prenatal THC with methylation levels at flanking CpGs within ± 250 kb. We tested correlation between 52,474 CpG-transcript pairs, of which 3309 individual CpGs were significantly correlated to 520 unique DE genes. The top DE gene was DNAJA4 and was negatively correlated with methylation of 12 CpGs including the top eQTM CpG cg21248347 (p value = 7.05–06; Beta = − 20.67). Of note, the top differentially methylated gene is this study, MEGF10, did not meet differential expression criteria for inclusion in eQTM analysis, although we observed a trend for lower expression in the THC group relative to CON (log2FC = − 1.08; p value = 0.21). Interestingly, differential expression of several placental transcripts was associated with large numbers of unique CpGs. For example, 329 unique CpGs were significantly associated with the expression of 6 genes in a 2 megabase (Mb) block of chromosome 4 (equivalent to the 6p21.33 chromosome region of the human genome). This region included 99 CpGs associated with CFB, 61 with GPANK1, 56 with NOTCH4, 48 with SKIV2L, 46 with PPP1R18, and 19 with VWA7 (Additional file 2: Table S5). Protein–protein interaction (PPI) networks of significant placental eQTM genes identified 73 enriched GO biological processes, including terms related to tissue development and response to external stimuli (Additional file 2: Table S6). Visual inspection of network connectivity suggested SMAD3, JUN and HSP90 proteins as candidate network hubs with high confidence (Additional file 1: Figure S7). Additionally, we observed increased expression of SHANK3, which correlated with DNA methylation at several CpGs upstream of a large hypomethylated block identified in a prospective ASD cohort DMR analysis [53] (Additional file 1: Figure S8).
Discussion
Our multi-tissue study demonstrates the utility of our rhesus macaque model to study the impact of prenatal THC use on fetal development and future offspring health, and to investigate the underlying molecular mechanisms. We focus our discussion on several key findings. First, differential methylation following THC exposure was present in all 5 tissue types, with the greatest effect measured in placenta. Second, prenatal THC exposure altered placental and fetal tissue epigenetic signatures at genes involved in embryonic development that may influence longer-term offspring outcomes. Third, changes in methylation in the placenta were associated with changes in RNA expression in genes enriched for tissue development and morphogenesis-related processes. Fourth, loci differentially methylated with THC were enriched for candidate ASD genes in all tissues. Lastly, we found significant overlap at the gene level when comparing differentially methylated loci in THC-exposed placentas from this study with DMRs in human placentas from pregnancies where the newborn was later diagnosed with ASD.
A total of 573 FDR (< 0.05) DMCs and 403 DMRs were associated with THC exposure in placenta. The top DMC and DMR in placenta were annotated to MEGF10 (multiple EGF like domains 10), a gene highly expressed in brain tissue that encodes a protein critical to synaptic number and function in postnatal brain [55]. Interestingly, SNPs in the transcription regulatory region of MEGF10 have been associated with increased risk of autism in a Chinese Han cohort, and MEGF10 expression was lower in peripheral blood of autistic individuals compared to healthy controls [55]. In this study, we observed increased methylation over the transcription start site with THC exposure in placental tissue, which would suggest potential downregulation of expression. However, MEGF10 expression in our placental RNA-seq dataset was relatively low in all placental samples, in agreement with the Human Protein Atlas [56]. Therefore, although we observed lower expression in the THC group relative to CON, the difference was not statistically significant (log2FC = − 1.08; p value = 0.21). Further work is necessary to determine the functional significance of MEGF10 differential methylation in THC-exposed placentas.
We detected fewer differentially methylated CpGs with THC exposure in fetal tissues after multiple testing correction relative to placenta. This may be because placental DNA contains a higher frequency of partially methylated domains relative to blood and other tissues [57] and CpG sites with intermediate levels of methylation can be measured with greater reliability and precision than at extreme values (i.e., near β = 0 or fully unmethylated and near β = 1 or fully methylated) [58]. This may also be related to a lower relative THC exposure of fetal tissues compared with the placenta. However, the FDR DMCs identified in the fetal tissues warrant further investigation based on their biological relevance in those respective tissues. For example, in cerebellum we identified one hypomethylated CpG in the 3’UTR of neuron-specific gene family member 1 (NSG1; aka NEEP21), which is highly enriched in developing cerebellum [59]. Loss of this gene in mice is associated with increased anxiety-related behavior in certain tasks such as spending 50% less time in the open arms of the elevated plus maze [60]. In the right ventricle of the heart, we identified FDR significant hypomethylation of one CpG upstream of the heart and neural crest derivatives expressed 1 (HAND1) gene, which is essential to the formation of the right ventricle [61]. Additionally in the right ventricle, we observed hypermethylation of one CpG annotated to the neutral sphingomyelinase activation associated factor gene (NSMAF), which may play a role in activation of neutral sphingomyelinase in response to cardiac ischemia or reperfusion injury [62].
We observed several placental DMRs annotated to a large block of protocadherin (PCDH) genes and individual CpGs with decreased methylation that were negatively correlated with increased expression of PCDHB8 following THC exposure. Consistent with our observed effect of THC on placental methylation, a study of placental DNA methylation in autism cases versus control identified several differentially methylated CpGs in this region, the majority of which were hypomethylated [63]. The PCDH gene block is organized into 3 clusters containing PCDHA, PCDHB, and PCDHG genes, which are highly expressed in the developing nervous system where they play an important role in neuronal cellular diversity [64]. In mice, there is evidence that differential expression of Pcdh isoforms is regulated by differential methylation of their promoter regions [65], and in humans dysregulated PCDH methylation has been observed in several neurological and psychiatric disorders [66].
Our findings are also consistent with a recent study that used whole genome bisulfite sequencing to identify novel differentially methylated regions in placenta samples from a prospective ASD study. The study by Zhu et al. [67] included functional characterization of a novel transcript within a hypomethylated block at 22q13.33, renamed NHIP (neuronal hypoxia inducible, placenta associated). Within this region, we observed increased methylation and expression of SHANK3, a gene involved in glutamatergic synapse formation with known mutations, epigenetic dysregulation, and structural variations observed in patients with ASD and other neurological syndromes [68–71].
We recently reported that these THC-exposed pregnancies demonstrated findings suggestive of placental insufficiency including decreased amniotic fluid volume, placental perfusion, and fetal oxygen availability [72]. The placenta plays a vital role in overall fetal health, development, and growth through the supply of oxygen and nutrients, gas and waste exchange, and endocrine signaling [73, 74]. Impaired placental function can result in adverse offspring outcomes including growth restriction, stillbirth, and miscarriage [75]. The placenta also plays an important role in producing neurotransmitters that may directly affect fetal brain development, and there is growing evidence linking placental dysfunction with adverse neurobehavioral outcomes in offspring [76–78]. This intimate relationship between placental function and fetal brain development has led to the coinage of terms such as the “placenta-brain-axis” [79] and “neuroplacentology” [80].
To better understand the potential underlying mediators of our previous findings [72], in the current study we examined correlation of DNA methylation with THC-associated gene expression [72] and examined the potential functional relevance of these eQTM loci using network analysis. Out of 631 genes differentially expressed with THC exposure, 520 were associated with methylation at one or more CpG. Notably, placental eQTM genes were enriched for GO terms related to morphogenesis, response to wounding, and response to oxygen-containing compounds. Taken together, these results suggest that prenatal THC exposure dysregulates placental gene expression and function through effects on the epigenome, and these effects are also likely to impact brain development.
Several studies have also focused on using placental ‘omics to gain mechanistic insight into the relationship between the in utero environment and birth outcomes such as fetal growth, preterm delivery, and birthweight, and infant and childhood health outcomes such as neurocognition and behavior [81, 82]. As observed in our study from prenatal cannabis exposure, dysregulation of placental DNA methylation (DNAm) in response to the in utero environment is a common finding [81]. A prior literature review that included studies of placental DNAm and/or transcriptomics in response to the prenatal environment identified 28 articles meeting their search criteria. In 16 of these studies, there was evidence of mediation or biological plausibility linking these exposures to fetal and infant health outcomes through the placental epigenome [82]. More recently, the term “placenta epigenome–brain axis” was used to describe the relationship between disrupted placental function (linked to placental gene regulation) and neurocognitive function later in life [83]. Altogether, studies from this field of research suggest that neurobehavioral disorders such as ASD, likely originate with dysregulated placental function.
Although the current evidence is not sufficient to conclude that prenatal cannabis exposure is a cause or a risk factor for development of ASD in offspring, there are a few large cohort studies that examine long-term outcomes related to prenatal cannabis exposure longitudinally. A retrospective Canadian study linked pregnancy and birth data to provincial health administrative databases to ascertain child neurodevelopmental outcomes and found an association between maternal cannabis use in pregnancy and increased incidence of ASD in the offspring [30]. However, due to limitations in data availability, this study was not able to account for key risk factors that can increase the likelihood of developing ASD. Our study provides biological plausibility for the findings of increased likelihood of developing ASD reported by this large Canadian study.
Another recent study assessed the effects of maternal cannabis use on psychosocial and physiological measures in young children along with the potential relevance of the in utero environment reflected in the placental transcriptome [35]. This study identified a relationship between maternal cannabis use and transcriptome changes in the placenta as a potential mediator of risk for anxiety-related problems in early childhood. We have now identified DNA methylation as a potential mediator of gene expression changes following prenatal THC exposure related to placental function and offspring neurobehavioral development, which may be related to this study’s observations of increased offspring anxiety-related problems in early childhood with in utero cannabis exposure.
Strengths of our study are that it utilized a translational rhesus macaque model to overcome the limitations of existing studies such as the inability to measure cannabis exposure including timing, duration, frequency, dose, type of product, and administration route. Our study used weight-based THC dosing for rigor and reproducibility, and chose THC edibles as the mode of administration to recapitulate typical human THC use in pregnancy and to examine the direct effects of THC-only on the placental and fetal epigenome without other confounders. Additionally, our study also overcomes the paucity of human epigenetic studies on prenatal cannabis exposure due to lack of feasibility in obtaining fetal tissue and ethical challenges related to research on maternal use of federally illegal drugs, such as cannabis. We were able to extend our investigation beyond the placenta to include 4 additional fetal tissues, not accessible in human pregnancies, for comparison of epigenetic signatures in response to prenatal THC exposure. All placental and fetal tissue were collected at time of cesarean section delivery from pregnancies with similar environmental exposures, including diet. This controlled tissue collection minimizes potential confounders, such as inflammation. We utilized the Illumina MethlyationEPIC platform in order to allow for direct comparison of our results with those from human studies. Future studies should consider whole genome or reduced representation bisulfite sequencing in order to identify potentially novel loci not covered by MM valid EPIC probes. Additionally, this study was limited by a smaller cohort size, which did not provide the power to examine fetal sex as a biological variable.
Conclusions
Ultimately, the long-term consequences of the altered placental and fetal epigenome that we report are unknown, but may impact offspring development and health. The findings of this study add to the limited safety data on prenatal cannabis exposure and provide potential underlying mechanisms, such as placental dysfunction and altered placental DNA methylation, for the observations of altered offspring neurobehavior associated with maternal cannabis use in pregnancy reported in the literature. Taken together, our findings lay the groundwork for ongoing postnatal studies focused on determining the impact of chronic prenatal and postnatal THC exposure on infant neurodevelopment, sensorimotor development and socioemotional behavior in the rhesus macaque. As the prevalence of prenatal cannabis use is rising along with significant increases in potency, there is an urgent need for evidence-driven recommendations on the safety of use both prenatally and postnatally. The data from this study bridge this current gap in knowledge to help guide patient counseling and public health polices focused on cannabis use.
Materials and methods
Experimental design
This study used indoor-housed sexually mature, female rhesus macaques (n = 10) maintained on a standard chow diet (TestDiet, St. Louis, Missouri) with an additional cookie with either THC (n = 5) or placebo (n = 5) daily. Cookies containing THC (THC edible) were made using research-grade THC obtained directly from the National Institute on Drug Abuse (NIDA) Drug Supply Program [40, 41]. Placebo cookies were identical to those containing THC except without the THC-vehicle. Tap water was available ad libitum. Edibles were administered prior to the animal’s daily chow to ensure consumption on an empty stomach and to confirm complete ingestion. Animals were slowly titrated up to 2.5 mg/7 kg/day of THC using published weight-based medical cannabis acclimation recommendations [72] approximately 4 months prior to undergoing time-mated breeding as previously published [40]. We previously established [40, 41] that NHP peak THC levels are within the expected reported contemporary dosing range (e.g., 5–8 ng/mL) in humans 3 h following a similar oral THC dose [84, 85]. Each THC-exposed pregnant animal (n = 5) continued to consume a daily THC edible of 2.5 mg/7 kg/day throughout pregnancy. All animals (n = 10) underwent immediate cesarean section delivery with placental collection and fetal necropsy on gestational day 155 (term is ~ 168 days) to minimize confounding variables secondary to spontaneous labor. Collected placental tissue was processed in RNAlater for RNA-sequencing. Placental tissue, in addition to fetal lung, right ventricle of the heart, prefrontal cortex, and cerebellum tissues were also collected and flash-frozen in liquid nitrogen with storage at − 80 °C for DNA studies. All protocols were approved by the Oregon National Primate Research Center (ONPRC) Institutional Animal Care and Use Committee and conformed to all guidelines for humane animal care (IP0001389).
DNA extraction and DNA methylation (DNAm) array acquisition and preprocessing
Placental and fetal tissue DNA was extracted using the AllPrep miRNA kit (Qiagen) and the QIAcube for automated nucleic acid extraction (Qiagen, USA). Epigenome-scale DNAm was measured with the Infinium MethylationEPIC BeadChip (Illumina, San Diego, California) at the Fred Hutchinson Cancer Genomics Resource (Seattle, WA). Processed BeadChips were scanned using the Illumina iScan + with ICS v3.3.28, and intensity data were extracted with Illumina GenomeStudio software (GenomeStudio v2011.1 with Methylation Analysis Module v1.9.0). All analysis was performed using R statistical software (version 4.1.1). Data normalization and QC were performed using the minfi [86] and ChAMP [87] packages. We examined density plots of beta values at each preprocessing and filtering step. Stratified quantile normalization preprocessing was performed using minfi to adjust for probe-type bias in beta distributions. Probes previously determined to be functional and valid in Macaca mulatta (referred to as MM_valid) [48] were retained prior to removing probes with failed detection p value > 0.01 in ≥ 25% of samples. For each tissue, we visually inspected the median intensity of measurements on the X and Y chromosome and confirmed that predicted sex agreed with biological sex. In addition, for each tissue, we performed hierarchical clustering and multidimensional scaling (MDS) plots to check that technical duplicates clustered together. Duplicate samples were removed prior to downstream analysis. Lastly, we removed probes mapping to non-autosomal chromosomes and probes with a colony SNP allele frequency > 0.1 at the CpG of interest [88]. We also identified common SNPs overlapping probe regions not at the CpG site, which could potentially influence probe binding or estimated DNA methylation.
Analysis of differentially methylated CpGs (DMCs) and differentially methylated regions (DMRs) between treatment groups
We performed differential methylation analysis using limma [89] with methylation for each CpG site as the response variable on the M-scale (logit2 beta). Given that our samples came from the same set of animals, we first performed within subject correlation across tissues using the dupCor function. We then fit CpG-specific models that included a factorial design for group and subject, a blocking term for subject, and the inter-animal correlation value. We used quantile–quantile plots of p values to visualize genomic inflation and computed estimated coefficients and standard errors separately for each tissue. We also calculated the mean beta values per CpG and the delta beta for each tissue and treatment group (mean THC—mean CON), for more intuitive biological interpretation. We identified DMRs using the DMRcate package, and the same statistical design used for DMC analysis and tissue-specific contrasts. DMRcate computes a kernel estimate against a null distribution. We used the default lambda setting of 1000 bp to bookend DMRs (i.e., CpGs > 1000 bp apart are split into separate DMRs) and FDR = 0.05 cutoff threshold to determine DMR significance [90].
Enrichment analysis of biological pathways and gene ontology (GO) terms
We performed enrichment analysis of DMCs with nominal p value < 0.05 for each tissue separately using the missMethyl package [91], which accounts for bias due to either (1) a greater number of probes per gene covered in the array dataset, or (2) CpGs which are annotated to multiple genes. As background for enrichment, we used the genes annotated to our filtered MM_valid probe set. For each tissue and set of nominally significant DMCs, we tested for enrichment of GO terms and KEGG pathways, separately. In the placenta only, we performed GO enrichment analysis using the ClusterProfiler package to test for and visualize enrichment of FDR significant DMCs [92].
Expression quantitative trait methylation (eQTM) analysis
We performed expression quantitative trait methylation (eQTM) analysis using the MEAL package for correlation of expression and methylation [93]. Genome-wide RNA-sequencing and EPIC DNAm was available for 10 placental and 10 lung samples. We used the biomart package to convert mmulatta ensembl IDs to hsapiens (version 75) [94]. We used the default flanking parameter to identify CpGs located within 250 kb of the transcription start site (TSS) of differentially expressed mRNA transcripts.
Enrichment analysis of candidate autism disorder genes
We used the publicly available SFARI database [52] to generate a list of genes previously implicated in autism risk. We compared the overlap of candidate autism genes with genes annotated to CpGs differentially methylated with THC (p < 0.05) for each tissue using Venn diagrams and the hypergeometric test for significant overlap. Enrichment p values < 0.05 were considered significant. As a negative control, and to ensure that the MM_valid probes were not biased for candidate ASD genes, we also performed enrichment testing at multiple p value thresholds and compared our results to enrichment p values for randomly selected gene sets of the same size [54]. We next compared our list of genes differentially methylated with THC in rhesus placenta to a published list of DMR genes in human placentas from pregnancies where the newborn was later diagnosed with ASD using the hypergeometric test for significant overlap [53].
Supplementary Information
Additional file 1. Figure S1. Density and MDS plot of quantile normalized beta values by tissue type; Figure S2. Tissue specific QQ-plots and inflation measures; Figure S3. MDS plot of most variable probes in placenta colored by treatment group; Figure S4. Boxplots of FDR significant DMCs in fetal tissues; Figure S5. Visual summary of placental methylation and expression within protocadherin gene cluster.Gviz plot showing genomic ranges, significant eQTM correlation coefficients, average difference in methylation in THC versus CON animals, and average methylation per CpG. On the “Difference THC” track, DMCs are shown in grey and DMRs are shown in red or blue indicating hyper or hypomethylation, respectively.Boxplot of PCDHB8 expression.Top CpG associated with PCDHB8 expression; Figure S6. Permutation testing with 10 random gene sets the same size as the SFARI candidate ASD gene list were not enriched among DMC genes; Figure S7. STRING protein–protein interaction plot of placental genes with significant differential expression between THC and CON animals and correlation with methylation at one or more CpG. We retained protein with high confidence interaction and color performed k-means clustering into 3 clusters, indicated by color of each bubble; Figure S8. Visual summary of placental methylation and expression within SHANK3 gene region.Gviz plot showing genomic ranges, significant eQTM correlation coefficients, average difference in methylation in THC versus CON animals, and average methylation per CpG. On the “Difference THC” track, DMCs are shown in red or blue indicating hyper or hypomethylation, respectively.Boxplot of SHANK3 expression.Top CpG associated with SHANK3 expression;
Additional file 2. Table S1. DMCs associated with prenatal THC exposure at FDR significance in one or more tissues; Table S2. DMRs associated with prenatal THC exposure at FDR significance in placenta; Table S3. MissMethyl FDR significant GO and KEGG Terms; Table S4. Summary of SFARI genes which overlap with THC DMC genes; Table S5. Significant eQTM for placental differentially expressed genes sorted by genomic coordinates using EPIC annotation; Table S6. Placental eQTM gene FDR significant GO Terms from STRINGdb
Acknowledgements
We would like to thank the veterinary and husbandry staff at ONPRC who provided excellent care for the animals used in this study, in particular Dr. Lauren Drew Martin, Dr. Heather Sidener, Travis Hodge, and Trent Crowley. Additionally, we would like to thank the OHSU Integrated Genomics Laboratory and Gene Profiling Shared Resource in addition to the Fred Hutchinson Cancer Center Genomics and Bioinformatics Shared Resources Core, Medgenome, and Novogene.
Abbreviations
- THC
Delta-9-tetrahydrocannabinol
- ASD
Autism spectrum disorder
- ADHD
Attention deficit hyperactivity disorder
- SFARI
Simons Foundation Autism Research Initiative
- NHP
Non-human primate
- FDR
False discovery rate
- DMCs
Differentially methylated CpGs
- PFC
Prefrontal cortex
- SNP
Single nucleotide polymorphism
- GO
Gene ontology
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- eQTM
Expression quantitative trait methylation
- DE
Differentially expressed
- DMRs
Differentially methylated regions
- ONPRC
Oregon National Primate Research Center
- MDS
Multidimensional scaling
- TSS
Transcription start site
Author contributions
LESK did conceptualization, investigation, data curation, formal analysis, writing—original draft. VHJR done conceptualization, investigation, data curation, writing—review and editing. RDM performed formal analysis, writing—review and editing. ELS was involved in investigation, formal analysis, writing—review and editing. done formal analysis, writing—review and editing. OJTM, JCH, CAE and DJS done writing—review and editing. AJM did data curation, formal analysis, writing—review and editing. JJT performed investigation, data curation, and resources. ERS did study design, supervision, writing—review and editing. JOL was involved in conceptualization, methodology, validation, investigation, resources, data curation, formal analysis, writing—original draft, visualization, and funding acquisition. All authors read and approved the final manuscript.
Funding
All Oregon National Primate Research Center (ONPRC) cores and units were supported by the National Institutes of Health (NIH) Grant P51 OD011092. Research reported in this publication was supported by the Reproductive Scientist Development Program, March of Dimes, and Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD), National Institute of Drug Abuse (NIDA) Drug Supply Program, March of Dimes Foundation, and Silver Family Innovation Award under Award Numbers K12 HD000849 (to J.L.), R03 HD097116 (to J.L.), and DP1 DA056493 (to J.L.). The contents of this study are solely the responsibility of the authors and do not necessarily represent the official view of NIH NICHD and NIDA.
Availability of data and materials
The raw and processed DNA methylation and RNA-seq datasets used and/or analyzed during the current study are publicly accessible through NCBI Gene Expression Omnibus (GEO) via accession series GSE223825.
Declarations
Ethics approval and consent to participate
All protocols were approved by the Oregon National Primate Research Center Institutional Animal Care and Use Committee and conformed to all guidelines for humane animal care (IP0001389).
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Gicquel C, El-Osta A, Le Bouc Y. Epigenetic regulation and fetal programming. Best Pract Res Clin Endocrinol Metab. 2008;22(1):1–16. doi: 10.1016/j.beem.2007.07.009. [DOI] [PubMed] [Google Scholar]
- 2.Martin EM, Fry RC. Environmental influences on the epigenome: exposure-associated DNA methylation in human populations. Annu Rev Public Health. 2018;39:309–333. doi: 10.1146/annurev-publhealth-040617-014629. [DOI] [PubMed] [Google Scholar]
- 3.Robinson WP, Price EM. The human placental methylome. Cold Spring Harb Perspect Med. 2015;5(5):a023044. doi: 10.1101/cshperspect.a023044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li L, Maire CL, Bilenky M, Carles A, Heravi-Moussavi A, Hong C, et al. Epigenomic programming in early fetal brain development. Epigenomics. 2020;12(12):1053–1070. doi: 10.2217/epi-2019-0319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shallie PD, Naicker T. The placenta as a window to the brain: a review on the role of placental markers in prenatal programming of neurodevelopment. Int J Dev Neurosci Off J Int Soc Dev Neurosci. 2019;73:41–49. doi: 10.1016/j.ijdevneu.2019.01.003. [DOI] [PubMed] [Google Scholar]
- 6.Smith A, Kaufman F, Sandy MS, Cardenas A. Cannabis exposure during critical windows of development: epigenetic and molecular pathways implicated in neuropsychiatric disease. Curr Environ Health Rep. 2020;7(3):325–342. doi: 10.1007/s40572-020-00275-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Szutorisz H, Hurd YL. Epigenetic effects of cannabis exposure. Biol Psychiatry. 2016;79(7):586–594. doi: 10.1016/j.biopsych.2015.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Young-Wolff KC, Foti TR, Green A, Altschuler A, Does MB, Jackson-Morris M, et al. Perceptions about cannabis following legalization among pregnant individuals with prenatal cannabis use in California. JAMA Netw Open. 2022;5(12):e2246912-e. doi: 10.1001/jamanetworkopen.2022.46912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Martin CE, Longinaker N, Mark K, Chisolm MS, Terplan M. Recent trends in treatment admissions for marijuana use during pregnancy. J Addict Med. 2015;9(2):99–104. doi: 10.1097/ADM.0000000000000095. [DOI] [PubMed] [Google Scholar]
- 10.Brown QL, Sarvet AL, Shmulewitz D, Martins SS, Wall MM, Hasin DS. Trends in marijuana use among pregnant and nonpregnant reproductive-aged women, 2002–2014. JAMA. 2017;317(2):207–209. doi: 10.1001/jama.2016.17383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Beatty JR, Svikis DS, Ondersma SJ. Prevalence and perceived financial costs of marijuana versus tobacco use among urban low-income pregnant women. J Addict Res Ther. 2012;3(4):1000135. doi: 10.4172/2155-6105.1000135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Passey ME, Sanson-Fisher RW, D'Este CA, Stirling JM. Tobacco, alcohol and cannabis use during pregnancy: clustering of risks. Drug Alcohol Depend. 2014;134:44–50. doi: 10.1016/j.drugalcdep.2013.09.008. [DOI] [PubMed] [Google Scholar]
- 13.Moore DG, Turner JD, Parrott AC, Goodwin JE, Fulton SE, Min MO, et al. During pregnancy, recreational drug-using women stop taking ecstasy (3,4-methylenedioxy-N-methylamphetamine) and reduce alcohol consumption, but continue to smoke tobacco and cannabis: initial findings from the Development and Infancy Study. J Psychopharmacol. 2010;24(9):1403–1410. doi: 10.1177/0269881109348165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wanner NM, Colwell ML, Faulk C. The epigenetic legacy of illicit drugs: developmental exposures and late-life phenotypes. Environ Epigenetics. 2019;5(4):22. doi: 10.1093/eep/dvz022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reggio PH. Endocannabinoid binding to the cannabinoid receptors: what is known and what remains unknown. Curr Med Chem. 2010;17(14):1468–1486. doi: 10.2174/092986710790980005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Volkow ND, Compton WM, Wargo EM. The risks of marijuana use during pregnancy. JAMA. 2017;317(2):129–130. doi: 10.1001/jama.2016.18612. [DOI] [PubMed] [Google Scholar]
- 17.Hutchings DE, Martin BR, Gamagaris Z, Miller N, Fico T. Plasma concentrations of delta-9-tetrahydrocannabinol in dams and fetuses following acute or multiple prenatal dosing in rats. Life Sci. 1989;44(11):697–701. doi: 10.1016/0024-3205(89)90380-9. [DOI] [PubMed] [Google Scholar]
- 18.Cristino L, Di Marzo V. Fetal cannabinoid receptors and the “dis-joint-ed” brain. EMBO J. 2014;33(7):665–667. doi: 10.1002/embj.201488086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kenney SP, Kekuda R, Prasad PD, Leibach FH, Devoe LD, Ganapathy V. Cannabinoid receptors and their role in the regulation of the serotonin transporter in human placenta. Am J Obstet Gynecol. 1999;181(2):491–497. doi: 10.1016/S0002-9378(99)70583-1. [DOI] [PubMed] [Google Scholar]
- 20.Lorenzetti V, Solowij N, Fornito A, Ian Lubman D, Yucel M. The association between regular cannabis exposure and alterations of human brain morphology: an updated review of the literature. Curr Pharm Des. 2014;20(13):2138–2167. doi: 10.2174/13816128113199990435. [DOI] [PubMed] [Google Scholar]
- 21.Fergusson D, Horwood L, Northstone K. Avon longitudinal 505 study of pregnancy and childhood. Maternal use of cannabis and pregnancy outcome 506:21–7. [DOI] [PubMed]
- 22.El Marroun H, Tiemeier H, Steegers EA, Jaddoe VW, Hofman A, Verhulst FC, et al. Intrauterine cannabis exposure affects fetal growth trajectories: the Generation R Study. J Am Acad Child Adolesc Psychiatry. 2009;48(12):1173–1181. doi: 10.1097/CHI.0b013e3181bfa8ee. [DOI] [PubMed] [Google Scholar]
- 23.Hurd Y, Wang X, Anderson V, Beck O, Minkoff H, Dow-Edwards D. Marijuana impairs growth in mid-gestation fetuses. Neurotoxicol Teratol. 2005;27(2):221–229. doi: 10.1016/j.ntt.2004.11.002. [DOI] [PubMed] [Google Scholar]
- 24.Lo JO, Shaw B, Robalino S, Ayers CK, Durbin S, Rushkin MC, et al. Cannabis use in pregnancy and neonatal outcomes: a systematic review and meta-analysis. Cannabis Cannabinoid Res. 2023. Epub 20230201. [DOI] [PMC free article] [PubMed]
- 25.Varner MW, Silver RM, Hogue CJR, Willinger M, Parker CB, Thorsten VR, et al. Association between stillbirth and illicit drug use and smoking during pregnancy. Obstet Gynecol. 2014;123(1):113. doi: 10.1097/AOG.0000000000000052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Conner SN, Bedell V, Lipsey K, Macones GA, Cahill AG, Tuuli MG. Maternal marijuana use and adverse neonatal outcomes: a systematic review and meta-analysis. Obstet Gynecol. 2016;128(4):713–723. doi: 10.1097/AOG.0000000000001649. [DOI] [PubMed] [Google Scholar]
- 27.Marchand G, Masoud AT, Govindan M, Ware K, King A, Ruther S, et al. Birth outcomes of neonates exposed to marijuana in utero: a systematic review and meta-analysis. JAMA Netw Open. 2022;5(1):e2145653. doi: 10.1001/jamanetworkopen.2021.45653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gunn JK, Rosales CB, Center KE, Nuñez A, Gibson SJ, Christ C, et al. Prenatal exposure to cannabis and maternal and child health outcomes: a systematic review and meta-analysis. BMJ Open. 2016;6(4):e009986. doi: 10.1136/bmjopen-2015-009986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nashed MG, Hardy DB, Laviolette SR. Prenatal cannabinoid exposure: emerging evidence of physiological and neuropsychiatric abnormalities. Front Psychiatry. 2020;11:624275. doi: 10.3389/fpsyt.2020.624275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Corsi DJ, Donelle J, Sucha E, Hawken S, Hsu H, El-Chaâr D, et al. Maternal cannabis use in pregnancy and child neurodevelopmental outcomes. Nat Med. 2020;26(10):1536–1540. doi: 10.1038/s41591-020-1002-5. [DOI] [PubMed] [Google Scholar]
- 31.Day NL, Goldschmidt L, Thomas CA. Prenatal marijuana exposure contributes to the prediction of marijuana use at age 14. Addiction. 2006;101(9):1313–1322. doi: 10.1111/j.1360-0443.2006.01523.x. [DOI] [PubMed] [Google Scholar]
- 32.Day NL, Leech SL, Goldschmidt L. The effects of prenatal marijuana exposure on delinquent behaviors are mediated by measures of neurocognitive functioning. Neurotoxicol Teratol. 2011;33(1):129–136. doi: 10.1016/j.ntt.2010.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Goldschmidt L, Day NL, Richardson GA. Effects of prenatal marijuana exposure on child behavior problems at age 10. Neurotoxicol Teratol. 2000;22(3):325–336. doi: 10.1016/S0892-0362(00)00066-0. [DOI] [PubMed] [Google Scholar]
- 34.Leech SL, Larkby CA, Day R, Day NL. Predictors and correlates of high levels of depression and anxiety symptoms among children at age 10. J Am Acad Child Adolesc Psychiatry. 2006;45(2):223–230. doi: 10.1097/01.chi.0000184930.18552.4d. [DOI] [PubMed] [Google Scholar]
- 35.Rompala G, Nomura Y, Hurd YL. Maternal cannabis use is associated with suppression of immune gene networks in placenta and increased anxiety phenotypes in offspring. Proc Natl Acad Sci U S A. 2021;118(47):e2106115118. doi: 10.1073/pnas.2106115118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gillies R, Lee K, Vanin S, Laviolette SR, Holloway AC, Arany E, et al. Maternal exposure to Δ9-tetrahydrocannabinol impairs female offspring glucose homeostasis and endocrine pancreatic development in the rat. Reprod Toxicol. 2020;94:84–91. doi: 10.1016/j.reprotox.2020.04.070. [DOI] [PubMed] [Google Scholar]
- 37.Knopik VS, Marceau K, Bidwell LC, Rolan E. Prenatal substance exposure and offspring development: does DNA methylation play a role? Neurotoxicol Teratol. 2019;71:50–63. doi: 10.1016/j.ntt.2018.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wanner NM, Colwell M, Drown C, Faulk C. Developmental cannabidiol exposure increases anxiety and modifies genome-wide brain DNA methylation in adult female mice. Clin Epigenetics. 2021;13(1):4. doi: 10.1186/s13148-020-00993-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Young-Wolff KC, Sarovar V, Tucker LY, Conway A, Alexeeff S, Weisner C, et al. Self-reported daily, weekly, and monthly cannabis use among women before and during pregnancy. JAMA Netw Open. 2019;2(7):e196471. doi: 10.1001/jamanetworkopen.2019.6471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ryan KS, Mahalingaiah S, Campbell LR, Roberts VHJ, Terrobias JJD, Naito CS, et al. The effects of delta-9-tetrahydrocannabinol exposure on female menstrual cyclicity and reproductive health in rhesus macaques. F&S Sci. 2021;2(3):287–294. doi: 10.1016/j.xfss.2021.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hedges JC, Hanna CB, Bash JC, Boniface ER, Burch FC, Mahalingaiah S, et al. Chronic exposure to delta-9-tetrahydrocannabinol impacts testicular volume and male reproductive health in rhesus macaques. Fertil Steril. 2022;117(4):698–707. doi: 10.1016/j.fertnstert.2021.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wolf DP, Thomson JA, Zelinski-Wooten MB, Stouffer RL. In vitro fertilization-embryo transfer in nonhuman primates: the technique and its applications. Mol Reprod Dev. 1990;27(3):261–280. doi: 10.1002/mrd.1080270313. [DOI] [PubMed] [Google Scholar]
- 43.Sibal LR, Samson KJ. Nonhuman primates: a critical role in current disease research. Ilar J. 2001;42(2):74–84. doi: 10.1093/ilar.42.2.74. [DOI] [PubMed] [Google Scholar]
- 44.Ganu RS, Harris RA, Collins K, Aagaard KM. Early origins of adult disease: approaches for investigating the programmable epigenome in humans, nonhuman primates, and rodents. Ilar J. 2012;53(3–4):306–321. doi: 10.1093/ilar.53.3-4.306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Grant KS, Petroff R, Isoherranen N, Stella N, Burbacher TM. Cannabis use during pregnancy: pharmacokinetics and effects on child development. Pharmacol Ther. 2018;182:133–151. doi: 10.1016/j.pharmthera.2017.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Steigerwald S, Wong PO, Cohen BE, Ishida JH, Vali M, Madden E, et al. Smoking, vaping, and use of edibles and other forms of marijuana among US adults. Ann Intern Med. 2018;169(12):890–892. doi: 10.7326/M18-1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dickson B, Mansfield C, Guiahi M, Allshouse AA, Borgelt LM, Sheeder J, et al. Recommendations from cannabis dispensaries about first-trimester cannabis use. Obstet Gynecol. 2018;131(6):1031. doi: 10.1097/AOG.0000000000002619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pichon F, Shen Y, Busato F, Jochems SP, Jacquelin B, Grand RL, et al. Analysis and annotation of DNA methylation in two nonhuman primate species using the Infinium Human Methylation 450K and EPIC BeadChips. Epigenomics. 2021;13(3):169–186. doi: 10.2217/epi-2020-0200. [DOI] [PubMed] [Google Scholar]
- 49.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B (Methodol) 1995;57(1):289–300. [Google Scholar]
- 50.Maksimovic J, Oshlack A, Phipson B. Gene set enrichment analysis for genome-wide DNA methylation data. Genome Biol. 2021;22(1):173. doi: 10.1186/s13059-021-02388-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sajdeya R, Brown JD, Goodin AJ. Perinatal cannabis exposures and autism spectrum disorders. Med Cannabis Cannabinoids. 2021;4(1):67–71. doi: 10.1159/000515871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Banerjee-Basu S, Packer A. SFARI Gene: an evolving database for the autism research community. The Company of Biologists Limited; 2010. [DOI] [PubMed]
- 53.Schroeder DI, Schmidt RJ, Crary-Dooley FK, Walker CK, Ozonoff S, Tancredi DJ, et al. Placental methylome analysis from a prospective autism study. Mol Autism. 2016;7:51. doi: 10.1186/s13229-016-0114-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bakulski KM, Dou JF, Feinberg JI, Aung MT, Ladd-Acosta C, Volk HE, et al. Autism-associated DNA methylation at birth from multiple tissues is enriched for autism genes in the early autism risk longitudinal investigation. Front Mol Neurosci. 2021;14:775390. doi: 10.3389/fnmol.2021.775390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wu Z, Qin J, You Y, Ma Y, Jia M, Wang L, et al. Genetic variants in the transcription regulatory region of MEGF10 are associated with autism in Chinese Han population. Sci Rep. 2017;7(1):2292. doi: 10.1038/s41598-017-02348-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Karlsson M, Zhang C, Méar L, Zhong W, Digre A, Katona B, et al. A single-cell type transcriptomics map of human tissues. Sci Adv. 2021;7(31):eabh2169. doi: 10.1126/sciadv.abh2169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schroeder DI, Blair JD, Lott P, Yu HO, Hong D, Crary F, et al. The human placenta methylome. Proc Natl Acad Sci U S A. 2013;110(15):6037–6042. doi: 10.1073/pnas.1215145110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xu Z, Taylor JA. Reliability of DNA methylation measures using Illumina methylation BeadChip. Epigenetics. 2021;16(5):495–502. doi: 10.1080/15592294.2020.1805692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Barford K, Yap CC, Dwyer ND, Winckler B. The related neuronal endosomal proteins NEEP21 (Nsg1) and P19 (Nsg2) have divergent expression profiles in vivo. J Comp Neurol. 2017;525(8):1861–1878. doi: 10.1002/cne.24168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Austin R, Chander P, Zimmerman AJ, Overby M, Digilio L, Yap CC, et al. Global loss of Neuron-specific gene 1 causes alterations in motor coordination, increased anxiety, and diurnal hyperactivity in male mice. Genes Brain Behav. 2022;21(6):e12816. doi: 10.1111/gbb.12816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li L, Wang J, Liu XY, Liu H, Shi HY, Yang XX, et al. HAND1 loss-of-function mutation contributes to congenital double outlet right ventricle. Int J Mol Med. 2017;39(3):711–718. doi: 10.3892/ijmm.2017.2865. [DOI] [PubMed] [Google Scholar]
- 62.O'Brien NW, Gellings NM, Guo M, Barlow SB, Glembotski CC, Sabbadini RA. Factor associated with neutral sphingomyelinase activation and its role in cardiac cell death. Circ Res. 2003;92(6):589–591. doi: 10.1161/01.RES.0000066290.29715.67. [DOI] [PubMed] [Google Scholar]
- 63.Bahado-Singh RO, Vishweswaraiah S, Aydas B, Radhakrishna U. Placental DNA methylation changes and the early prediction of autism in full-term newborns. PLoS ONE. 2021;16(7):e0253340. doi: 10.1371/journal.pone.0253340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chen WV, Maniatis T. Clustered protocadherins. Development. 2013;140(16):3297–3302. doi: 10.1242/dev.090621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Toyoda S, Kawaguchi M, Kobayashi T, Tarusawa E, Toyama T, Okano M, et al. Developmental epigenetic modification regulates stochastic expression of clustered protocadherin genes, generating single neuron diversity. Neuron. 2014;82(1):94–108. doi: 10.1016/j.neuron.2014.02.005. [DOI] [PubMed] [Google Scholar]
- 66.El Hajj N, Dittrich M, Haaf T. Epigenetic dysregulation of protocadherins in human disease. Semin Cell Dev Biol. 2017;69:172–182. doi: 10.1016/j.semcdb.2017.07.007. [DOI] [PubMed] [Google Scholar]
- 67.Zhu Y, Gomez JA, Laufer BI, Mordaunt CE, Mouat JS, Soto DC, et al. Placental methylome reveals a 22q13.33 brain regulatory gene locus associated with autism. Genome Biol. 2022;23(1):46. doi: 10.1186/s13059-022-02613-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Boccuto L, Lauri M, Sarasua SM, Skinner CD, Buccella D, Dwivedi A, et al. Prevalence of SHANK3 variants in patients with different subtypes of autism spectrum disorders. Eur J Hum Genet EJHG. 2013;21(3):310–316. doi: 10.1038/ejhg.2012.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Uchino S, Waga C. SHANK3 as an autism spectrum disorder-associated gene. Brain Dev. 2013;35(2):106–110. doi: 10.1016/j.braindev.2012.05.013. [DOI] [PubMed] [Google Scholar]
- 70.Loureiro LO, Howe JL, Reuter MS, Iaboni A, Calli K, Roshandel D, et al. A recurrent SHANK3 frameshift variant in Autism Spectrum Disorder. NPJ Genom Med. 2021;6(1):91. doi: 10.1038/s41525-021-00254-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhu L, Wang X, Li XL, Towers A, Cao X, Wang P, et al. Epigenetic dysregulation of SHANK3 in brain tissues from individuals with autism spectrum disorders. Hum Mol Genet. 2014;23(6):1563–1578. doi: 10.1093/hmg/ddt547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Roberts VHJ, Schabel MC, Boniface ER, D'Mello RJ, Morgan TK, Terrobias JJD, et al. Chronic prenatal delta-9-tetrahydrocannabinol exposure adversely impacts placental function and development in a rhesus macaque model. Sci Rep. 2022;12(1):20260. doi: 10.1038/s41598-022-24401-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Jansson T. Placenta plays a critical role in maternal-fetal resource allocation. Proc Natl Acad Sci U S A. 2016;113(40):11066–11068. doi: 10.1073/pnas.1613437113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Garnica AD, Chan WY. The role of the placenta in fetal nutrition and growth. J Am Coll Nutr. 1996;15(3):206–222. doi: 10.1080/07315724.1996.10718591. [DOI] [PubMed] [Google Scholar]
- 75.American College of Obstetricians and Gynecologists ACOG practice bulletin No. 204: fetal growth restriction. Obstet Gynecol. 2019;133(2):e97–e109. doi: 10.1097/AOG.0000000000003070. [DOI] [PubMed] [Google Scholar]
- 76.Andescavage N, duPlessis A, Metzler M, Bulas D, Vezina G, Jacobs M, et al. In vivo assessment of placental and brain volumes in growth-restricted fetuses with and without fetal Doppler changes using quantitative 3D MRI. J Perinatol. 2017;37(12):1278–1284. doi: 10.1038/jp.2017.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Baschat AA. Neurodevelopment following fetal growth restriction and its relationship with antepartum parameters of placental dysfunction. Ultrasound Obstet Gynecol. 2011;37(5):501–514. doi: 10.1002/uog.9008. [DOI] [PubMed] [Google Scholar]
- 78.Miller SL, Huppi PS, Mallard C. The consequences of fetal growth restriction on brain structure and neurodevelopmental outcome. J Physiol. 2016;594(4):807–823. doi: 10.1113/JP271402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Rosenfeld CS. The placenta-brain-axis. J Neurosci Res. 2021;99(1):271–283. doi: 10.1002/jnr.24603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kratimenos P, Penn AA. Placental programming of neuropsychiatric disease. Pediatr Res. 2019;86(2):157–164. doi: 10.1038/s41390-019-0405-9. [DOI] [PubMed] [Google Scholar]
- 81.Mortillo M, Marsit CJ. Select early-life environmental exposures and DNA methylation in the placenta. Curr Environ Health Rep. 2023;10(1):22–34. doi: 10.1007/s40572-022-00385-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lapehn S, Paquette AG. The placental epigenome as a molecular link between prenatal exposures and fetal health outcomes through the DOHaD hypothesis. Curr Environ Health Rep. 2022;9(3):490–501. doi: 10.1007/s40572-022-00354-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Freedman AN, Eaves LA, Rager JE, Gavino-Lopez N, Smeester L, Bangma J, et al. The placenta epigenome-brain axis: placental epigenomic and transcriptomic responses that preprogram cognitive impairment. Epigenomics. 2022;14(15):897–911. doi: 10.2217/epi-2022-0061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Karschner EL, Darwin WD, Goodwin RS, Wright S, Huestis MA. Plasma cannabinoid pharmacokinetics following controlled oral Δ9-tetrahydrocannabinol and oromucosal cannabis extract administration. Clin Chem. 2011;57(1):66–75. doi: 10.1373/clinchem.2010.152439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ohlsson A, Lindgren JE, Wahlen A, Agurell S, Hollister L, Gillespie H. Plasma delta-9-tetrahydrocannabinol concentrations and clinical effects after oral and intravenous administration and smoking. Clin Pharmacol Ther. 1980;28(3):409–416. doi: 10.1038/clpt.1980.181. [DOI] [PubMed] [Google Scholar]
- 86.Fortin JP, Triche TJ, Jr, Hansen KD. Preprocessing, normalization and integration of the Illumina HumanMethylationEPIC array with minfi. Bioinformatics. 2017;33(4):558–560. doi: 10.1093/bioinformatics/btw691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Tian Y, Morris TJ, Webster AP, Yang Z, Beck S, Feber A, et al. ChAMP: updated methylation analysis pipeline for Illumina BeadChips. Bioinformatics. 2017;33(24):3982–3984. doi: 10.1093/bioinformatics/btx513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Bimber BN, Yan MY, Peterson SM, Ferguson B. mGAP: the macaque genotype and phenotype resource, a framework for accessing and interpreting macaque variant data, and identifying new models of human disease. BMC Genom. 2019;20(1):176. doi: 10.1186/s12864-019-5559-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(7):e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Peters TJ, Buckley MJ, Statham AL, Pidsley R, Samaras K, Lord RV, et al. De novo identification of differentially methylated regions in the human genome. Epigenetics Chromatin. 2015;8:6. doi: 10.1186/1756-8935-8-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Phipson B, Maksimovic J, Oshlack A. missMethyl: an R package for analyzing data from Illumina's HumanMethylation450 platform. Bioinformatics. 2016;32(2):286–288. doi: 10.1093/bioinformatics/btv560. [DOI] [PubMed] [Google Scholar]
- 92.Yu G, Wang LG, Han Y, He QY. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16(5):284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Gonzalez CR-AaJR. MEAL: perform methylation analysis. 1.24.0 ed2021.
- 94.Durinck S, Spellman PT, Birney E, Huber W. Mapping identifiers for the integration of genomic datasets with the R/Bioconductor package biomaRt. Nat Protoc. 2009;4(8):1184–1191. doi: 10.1038/nprot.2009.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. Figure S1. Density and MDS plot of quantile normalized beta values by tissue type; Figure S2. Tissue specific QQ-plots and inflation measures; Figure S3. MDS plot of most variable probes in placenta colored by treatment group; Figure S4. Boxplots of FDR significant DMCs in fetal tissues; Figure S5. Visual summary of placental methylation and expression within protocadherin gene cluster.Gviz plot showing genomic ranges, significant eQTM correlation coefficients, average difference in methylation in THC versus CON animals, and average methylation per CpG. On the “Difference THC” track, DMCs are shown in grey and DMRs are shown in red or blue indicating hyper or hypomethylation, respectively.Boxplot of PCDHB8 expression.Top CpG associated with PCDHB8 expression; Figure S6. Permutation testing with 10 random gene sets the same size as the SFARI candidate ASD gene list were not enriched among DMC genes; Figure S7. STRING protein–protein interaction plot of placental genes with significant differential expression between THC and CON animals and correlation with methylation at one or more CpG. We retained protein with high confidence interaction and color performed k-means clustering into 3 clusters, indicated by color of each bubble; Figure S8. Visual summary of placental methylation and expression within SHANK3 gene region.Gviz plot showing genomic ranges, significant eQTM correlation coefficients, average difference in methylation in THC versus CON animals, and average methylation per CpG. On the “Difference THC” track, DMCs are shown in red or blue indicating hyper or hypomethylation, respectively.Boxplot of SHANK3 expression.Top CpG associated with SHANK3 expression;
Additional file 2. Table S1. DMCs associated with prenatal THC exposure at FDR significance in one or more tissues; Table S2. DMRs associated with prenatal THC exposure at FDR significance in placenta; Table S3. MissMethyl FDR significant GO and KEGG Terms; Table S4. Summary of SFARI genes which overlap with THC DMC genes; Table S5. Significant eQTM for placental differentially expressed genes sorted by genomic coordinates using EPIC annotation; Table S6. Placental eQTM gene FDR significant GO Terms from STRINGdb
Data Availability Statement
The raw and processed DNA methylation and RNA-seq datasets used and/or analyzed during the current study are publicly accessible through NCBI Gene Expression Omnibus (GEO) via accession series GSE223825.