Abstract
Nicotinamide adenine dinucleotide (NAD+) is a multifunctional molecule. Beyond redox metabolism, NAD+ has an equally important function as a substrate for post-translational modification enzymes, the largest family being the poly-ADP–ribose polymerases (PARPs, 17 family members in humans). The recent surprising discoveries of noncanonical NAD (NAD+/NADH)-binding proteins suggests that the NAD interactome is likely larger than previously thought; yet, broadly useful chemical tools for profiling and discovering NAD-binding proteins do not exist. Here, we describe the design, synthesis, and validation of clickable, photoaffinity labeling (PAL) probes, 2- and 6-ad-BAD, for interrogating the NAD interactome. We found that 2-ad-BAD efficiently labels PARPs in a UV-dependent manner. Chemical proteomics experiments with 2- and 6-ad-BAD identified known and unknown NAD+/NADH-binding proteins. Together, our study shows the utility of 2- and 6-ad-BAD as clickable PAL NAD probes.
Nicotinamide adenine dinucleotide (NAD+) wears different hats in the cell: on the one hand, it serves as a coenzyme for oxidoreductases in metabolism, and on the other, it is a substrate for signaling enzymes that mediate post-translational modifications.1 Unlike oxidoreductases, which mediate the reversible two-electron reduction of NAD+ to NADH, the enzymes that use NAD+ as a substrate cleave the nicotinamide glycosidic bond of NAD+ leading to the consumption of NAD+. The most prominent NAD+ consumers in human cells are poly-ADP–ribose polymerases (PARP-1–17) and sirtuins (SIRT1–7).2 Intriguingly, recent studies show that noncanonical NAD+ consumers exist (e.g., SARM1,3–6 DTX3L7), which have NAD+-binding sites that are quite distinct from the conserved structural motifs found in canonical NAD+ consumers. Additionally, NADH can act as an allosteric modulator of proteins (e.g., NAD-dependent isocitrate dehydrogenase).8 Hence, the NAD (NAD+/NADH) interactome is likely much more diverse than previously anticipated based solely on analysis of protein sequences.
We sought an unbiased strategy to profile NAD-binding proteins. Chemical proteomics using photoaffinity labeling (PAL) is a powerful approach for unbiased profiling of proteome-wide small-molecule–protein interactions.9 Small-molecule probes for PAL are bifunctional: they contain a photoreactive moiety as well as a “clickable” tag (e.g., alkyne). While several photoreactive groups have been used for PAL, diazirines are popular because of their compact structure and excellent photo-cross-linking properties.10 Although clickable PAL probes have been developed for nucleotides such as S-adenosyl methionine (SAH),11 and, more recently, adenosine triphosphate (ATP),12 a clickable PAL NAD probe has heretofore not been described.
We reasoned that a clickable PAL NAD probe should contain an enzymatically stable nicotinamide glycosidic bond. There are several NAD+ analogues that fit this criterion: benzamide adenine dinucleotide (BAD),13 carba-NAD+,14 and 4-thioribose NAD+ (S-NAD+).15 Importantly, these NAD+ analogues are not cleaved by NAD+ consumers.13,14 BAD, but not carba-NAD+, binds to PARP-1 and inhibits its enzymatic activity.16 Additionally, BAD inhibits NAD+/NADH-binding enzymes.13 Therefore, we focused our design efforts based on BAD.
To convert BAD into a clickable PAL probe for NAD+ consumers and other NAD-binding proteins, we sought positions on BAD that could be modified with a photoreactive group and a clickable tag without perturbing BAD’s interactions with its targets. We focused on the adenine ring of BAD, because our previous studies on orthogonal NAD+ analogues for engineered PARPs showed that substitutions on the nicotinamide ring were not tolerated by wild-type PARPs.17,18 On the adenine ring of BAD, there are three possible positions that could be modified: N-6, C-2, and C-8. We scrutinized the crystal structure of BAD bound to PARP-1 (PDB: 6bhv).16 In this structure, the C-2 position of the adenine ring of BAD is solvent-exposed, whereas the N-6 position is partially solvent-exposed (Figure 1a). In contrast, the C-8 position is buried in the NAD+-binding pocket. We therefore designed and synthesized modified BAD analogues with a “minimalist” linker19 containing a diazirine and a terminal alkyne at the C-2 or N-6 positions of the adenine ring (2-ad-BAD, 1, and 6-ad-BAD, 2) (Figure 1b and Schemes S1–6).
Figure 1.
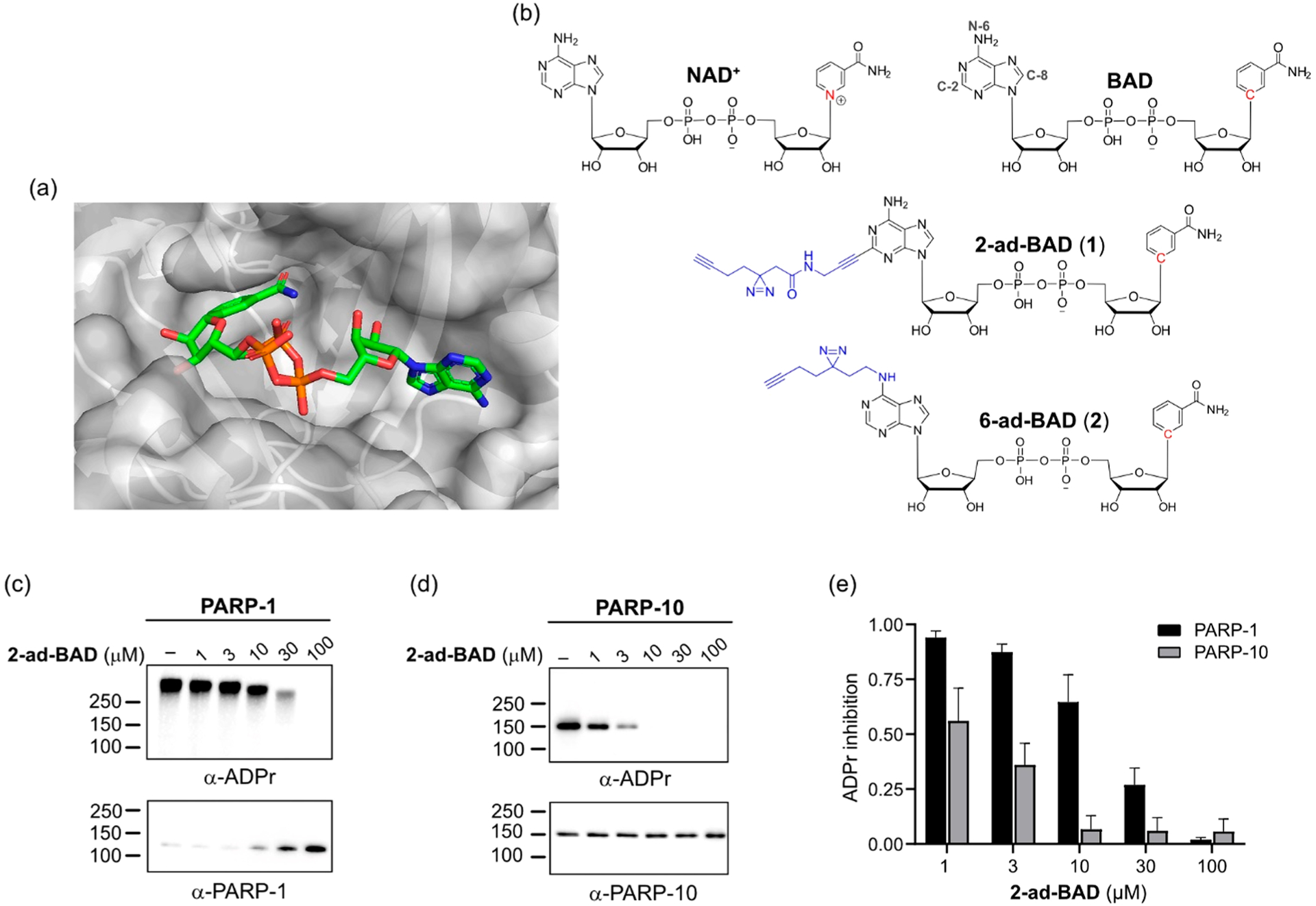
Clickable PAL NAD probe inhibits the activity of PARP-1 and PARP-10. (a) Crystal structure of BAD bound to PARP-1 (PDB: 6BHV). (b) Chemical structures of NAD+, benzamide adenine dinucleotide (BAD), 2-ad-BAD (1), and 6-ad-BAD (2). (c) Dose-dependent inhibition of PARP-1 auto-PARylation activity by 2-ad-BAD. Auto-PARylation of PARP-1 was detected using an anti-ADPr antibody, which detects both PARylation and MARylation. (d) Dose-dependent inhibition of PARP-10 auto-MARylation activity by 2-ad-BAD. Auto-MARylation of PARP-10 was detected as described in (c). (e) Quantification of results in (c) and (d). Data are mean ± SEM of three independent experiments.
We first evaluated our clickable PAL NAD probes on PARP enzymatic activity. PARPs catalyze the transfer of the ADP–ribose (ADPr) moiety of NAD+ to target proteins in a process known as ADP-ribosylation. In humans, PARPs fall into two major subgroups: (i) PARPs that catalyze poly-ADP-ribosylation (PARylation) and (ii) PARPs that catalyze mono-ADP-ribosylation (MARylation).20 In previous work, we and others found that C-2-modified NAD+ analogues are much better substrates than N-6-modified NAD+ analogues for PARP-1 and several other PARP family members.21,22 Therefore, we focused on 2-ad-BAD for PARP studies. We tested the activity of 2-ad-BAD against members from each subgroup: PARP-1 (PARylating enzyme) and PARP-10 (MARylating enzyme). Similarly to BAD (Scheme S7), 2-ad-BAD dose-dependently inhibited PARP-1 auto-PARylation and PARP-10 auto-MARylation (Figures 1c–e and S1). 2-ad-BAD was more potent against PARP-10 compared to PARP-1 (Figure 1c–e); however, the opposite was true for BAD (Figure S1). Thus, the minimalist linker for PAL at the C-2 position of 2-ad-BAD is tolerated for both PARylating and MARylating PARPs.
We next determined the ability of 2-ad-BAD to label PARP-1 in a UV-dependent manner. Previous studies showed that BAD binding to PARP-1 is DNA-dependent. In the absence of damaged DNA, the NAD+-binding pocket is sterically occluded.16 When PARP-1 binds damaged DNA, the steric block is relieved via long-range allosteric coupling between the DNA-binding domains and the catalytic domain (Figure 2a).16 We incubated PARP-1 in the presence or absence of damaged DNA prior to incubation with 2-ad-BAD, followed by UV irradiation (350 nm) for 5 min, and copper-catalyzed conjugation to a TAMRA–azide probe. Labeling of PARP-1 by 2-ad-BAD is dependent strictly on the presence of damaged DNA (Figure 2b). To determine if this is a unique feature of 2-ad-BAD compared to a PAL probe based on a PARP-1 inhibitor, we synthesized an olaparib (FDA-approved PARP-1 inhibitor) analogue containing an alkyne diazirine minimal linker (ad-olaparib) (Scheme S8). Ad-olaparib labels PARP-1 independent of PARP-1 binding to damaged DNA (Figure 2c). Thus, 2-ad-BAD, but not ad-olaparib, is an NAD+-binding site conformational reporter of PARP-1. In addition to PARP-1, we found that 2-ad-BAD labeled PARP-10; however, labeling of PARP-10 was independent of damaged DNA (Figures 2b and S2). Unlike 2-ad-BAD, 6-ad-BAD did not label PARP-1 or PARP-10 (Figure S3). Together, these results show that 2-ad-BAD is an efficient clickable PAL NAD probe for both PARylating and MARylating PARPs.
Figure 2.
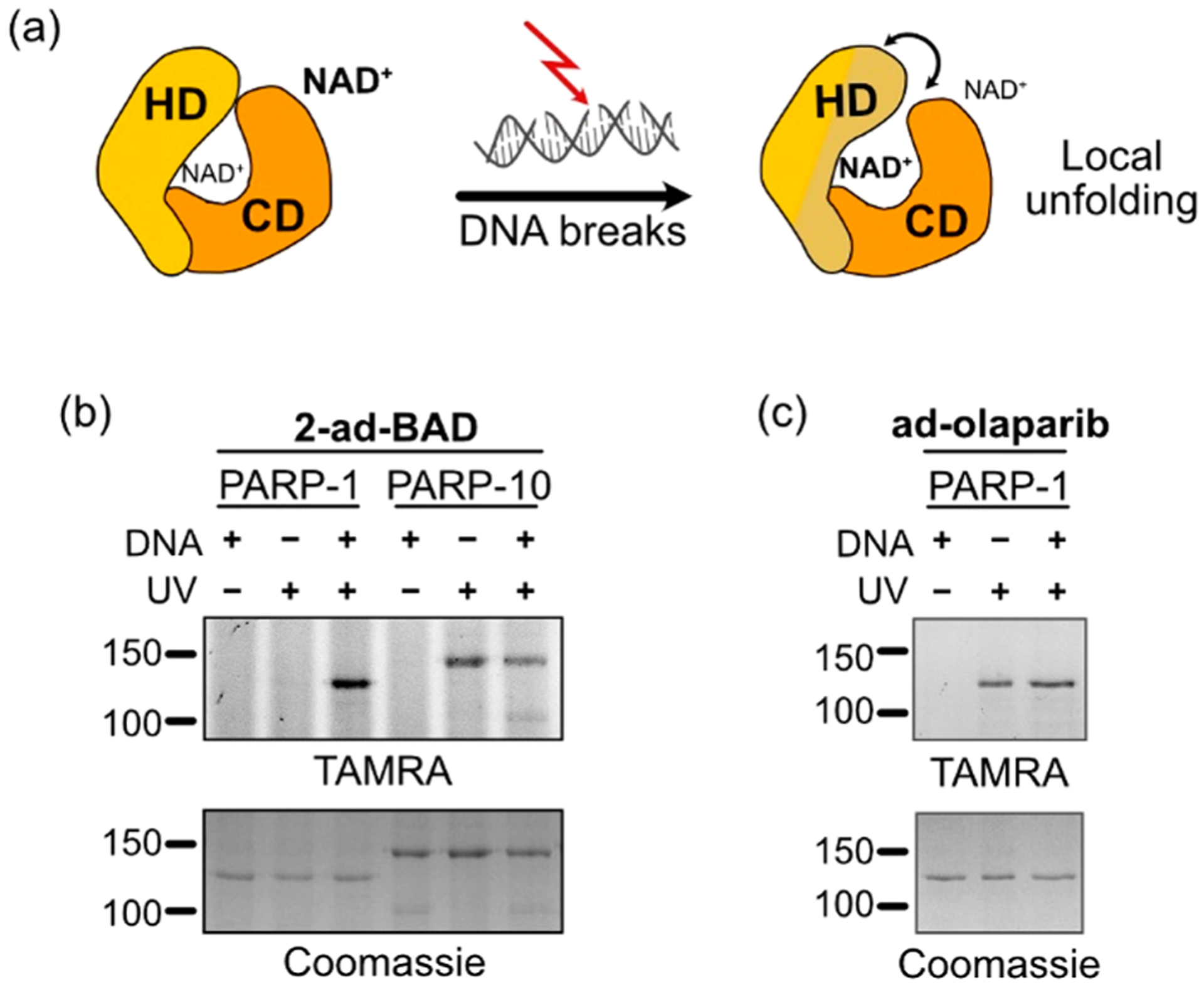
2-ad-BAD labels the NAD+ consumers PARP-1 and PARP-10. (a) Schematic of the activation of PARP-1 by damaged DNA. In the basal state, the helical domain (HD, yellow) sterically occludes the NAD+-binding site in the catalytic domain (CD, orange). Upon binding damaged DNA, the HD partially unfolds relieving the steric block on the NAD+-binding site. (b) Activated DNA is required for PARP-1, but not PARP-10, labeling by 2-ad-BAD. UV-dependent, covalent labeling of PARPs was detected by in gel fluorescence scanning (TAMRA). Coomassie was used as a loading control. (c) The labeling of PARP-1 by the clickable olaparib PAL probe, ad-olaparib, is independent of damaged DNA binding.
We next examined the labeling specificity of 2-ad-BAD. We preincubated either PARP-1 or PARP-10 with a series of adenine-containing nucleotides prior to incubation with 2-ad-BAD. NAD+ and related dinucleotides (BAD, NADH) reduced 2-ad-BAD labeling of PARP-1 and PARP-10 (Figures 3a,b and S4a). By contrast, the mononucleotides ADPr, ADP, or ATP did not block labeling of PARP-1 and PARP-10 by 2-ad-BAD (Figures 3a,b and S4a). These results further confirm the labeling specificity of 2-ad-BAD for the NAD+-binding site in PARP family members.
Figure 3.
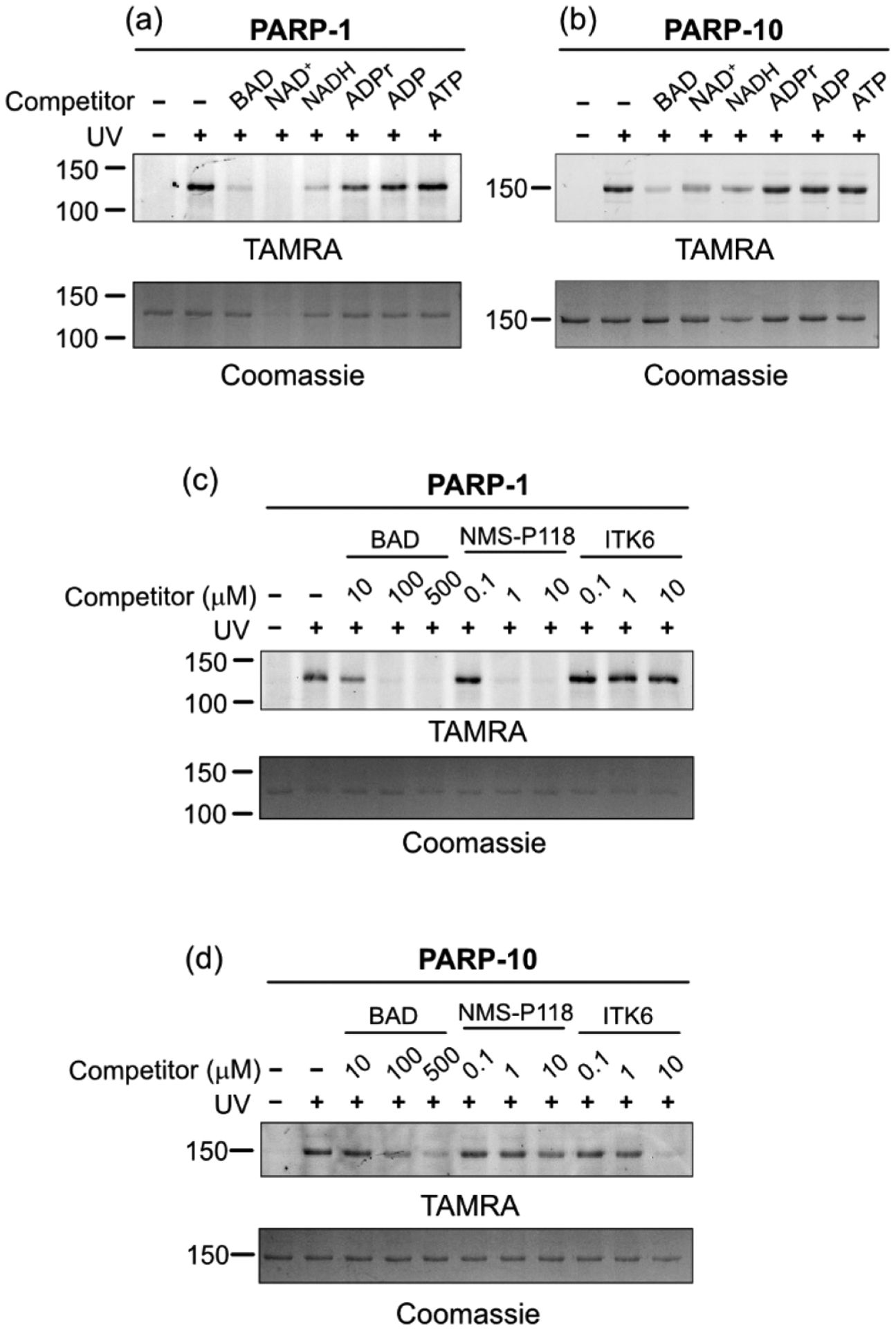
2-ad-BAD labels the NAD+-binding site and can be used for profiling PARP inhibitor selectivity. Dinucleotides, but not mono-nucleotides, block 2-ad-BAD labeling of PARP-1 (a) and PARP-10 (b). PARP inhibitor competition labeling experiments for PARP-1 (c) and PARP-10 (d).
PAL probes that target broadly enzymes belonging to particular family have found utility in competition labeling experiments, especially for examining family-wide inhibitor selectivity.23 We envisioned that 2-ad-BAD could be used for examining inhibitor selectivity across the PARP family. To test this idea, we performed competition labeling experiments with two PARP inhibitors: NMS-P118, a selective inhibitor of PARP-1,24 and ITK6, a pan-MARylating PARP inhibitor that inhibits PARP-10.25 Consistent with the effects of these compounds on PARP-1 or PARP-10 enzymatic activity (Figure S1a,b), NMS-P118 blocked the labeling of PARP-1, whereas ITK6 blocked the labeling of PARP-10 by 2-ad-BAD in dose-dependent manner (Figures 3c,d and S4b). By contrast, NMS-118 did not block the labeling of PARP-10 and ITK6 did not block the labeling of PARP-1 by 2-ad-BAD (Figures 3c,d and S4b). These results show that 2-ad-BAD can be used for profiling the selectivity of PARP inhibitors across multiple PARP family members.
We next used our clickable PAL NAD probes in chemical proteomic experiments for profiling the NAD interactome in whole cell lysates. We treated HEK 293T cell lysates with either 2-ad-BAD or 6-ad-BAD alone or in the presence of BAD (competitor), followed by UV irradiation. Protein targets of 2-ad-BAD or 6-ad-BAD were detected by click conjugation with desthiobiotin-azide. We detected the labeling of several proteins, many of which were competed by BAD (Figure 4a,b). We enriched desthiobiotinylated proteins using streptavidin agarose (Figure 4a,b) as well as proteolyzed and subjected eluted peptides to LC–MS/MS. We identified 261 and 141 proteins in 2-ad-BAD and 6-ad-BAD samples, respectively, that were at least 2-fold enriched in UV-irradiated samples in duplicate LC–MS/MS experiments (Table S1). A total of 74 proteins were competed at least 2-fold by BAD; of these, 10 were identified in both data sets (Figure 4c). Hence, 2-ad-BAD and 6-ad-BAD mostly label distinct proteins but also share some targets.
Figure 4.
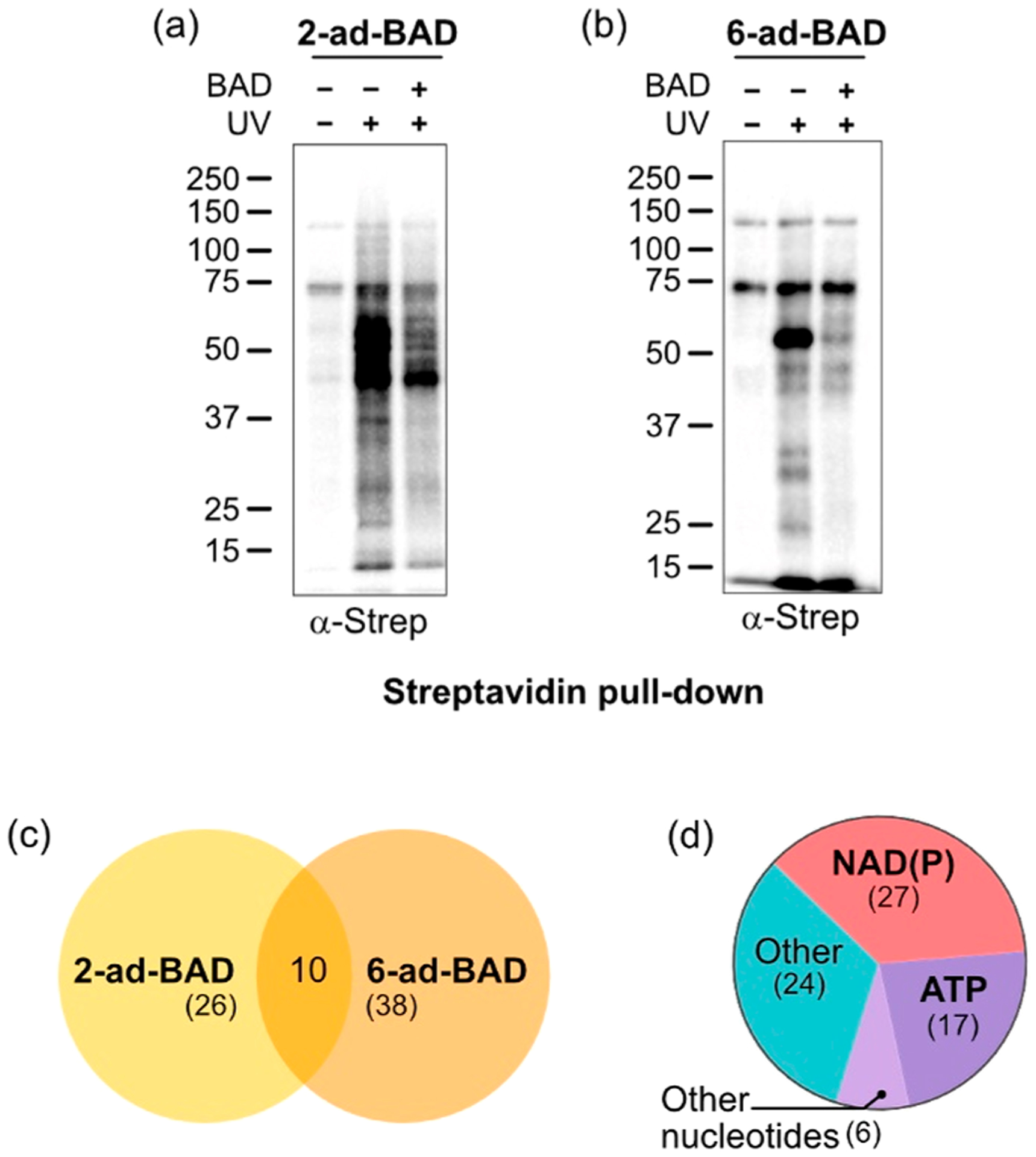
Chemical proteomics using 2- and 6-ad-BAD identifies NAD-binding proteins. (a,b) Labeling in cell lysates and enrichment of labeled proteins (containing desthiobiotin after click reaction) using streptavidin agarose. (c) Venn diagram showing targets of 2-ad-BAD and of 6-ad-BAD. (d) Pie chart showing distribution of nucleotide binding among targets.
We performed further analyses on the 74 targets that were competed by BAD. The largest group of nucleotide-binding proteins are NAD+/NADH/NADP+/NADPH binders (27), followed by ATP binders (17), AMP (2), GTP (2), ADPr (1), and FAD (1) binders (Figure 4d). Intriguingly, the other 24 targets are not known to bind NAD or related nucleotides (Figure 4d). Next, we confirmed our LC–MS/MS results using streptavidin enrichment followed by immunoblot detection. In addition to PARP-1, we confirmed the unknown NAD-binders 26S proteasome regulatory subunit 7 (Psmc2) and adenylate kinase (AK) 1 and 2 as targets of 2-ad-BAD but not 6-ad-BAD (Figure S5). We also confirmed targets of 6-ad-BAD, including the known NAD-binder C-terminal binding protein 1 (Ctbp1),26,27 a transcriptional regulator whose corepressor function is regulated by NAD+/NADH binding, as well as the unknown NAD-binder tRNA methyltransferase 10 homologue C (TRMT10C/MRPP1) (Figure S5b). Together, our results show that 2-ad-BAD and 6-ad-BAD are useful in chemical proteomics applications for identifying known—and previously unknown—NAD-binding proteins.
Lastly, we further evaluated the binding of NAD+ and related nucleotides to the 2-ad-BAD target AK1. AK1 is an enzyme that transfers the terminal phosphate group between ATP and AMP and thus is critical in cellular energy state monitoring and stress response.28 Labeling of recombinant AK1 by 2-ad-BAD is effectively competed by its known ligands, AMP and ATP, but also BAD, NADH, and to a lesser extent NAD+ and ADPr (Figure S6a). We further evaluated nucleotide binding to AK1 by thermal shift analysis using differential scanning fluorimetry (DSF). While both NAD+ and NADH caused a shift in AK1 thermal stabilization compared to vehicle control, NADH showed a thermal shift similar to ATP and ADP (Figure S6b). Together, these results suggest that NADH, and perhaps NAD+, can bind to AK1. Interestingly, squid mantle AK is less susceptible to denaturation by heat or proteolysis in the presence of NADH. NADH was also shown to act as a noncompetitive inhibitor of squid AK.29 In another study, it was found that AK1 can be eluted from blue dextran–sepharose chromatography (used historically for purifying NAD+/NADH dehydrogenases) by NADH, and its activity was competitively inhibited by both NAD+ and NADH.30
In this study, we developed new clickable PAL NAD probes that label NAD-binding proteins in a UV-dependent manner. 2-ad-BAD labels the NAD+ consumers PARP-1 and PARP-10 in a UV-dependent manner and can be used for profiling the selectivity of PARP inhibitors across multiple PARP family members. We also demonstrated that 2-ad-BAD and 6-ad-BAD can identify known and unknown NAD-binding proteins in a cellular context. A limitation of 2- and 6-ad-BAD is that they might not capture all possible NAD-binding proteins in cells due to steric constraints or lack of efficient photo-cross-linking. Nevertheless, we envision that our clickable PAL NAD probes will be useful in future chemical proteomics studies for profiling the NAD+ interactome across different tissues as well as in disease contexts.
Supplementary Material
ACKNOWLEDGMENTS
We thank Ashok Reddy and John Klimek from the OHSU Proteomics core for assistance with LC-MS/MS analysis. We thank Chadwick Smith and John Koberstein from the OHSU Vollum Institute for advice on differential scanning fluorimetry experiments. We thank Ivan R. Siordia and Daniel Sanderson for protein purification. Mass spectrometric analysis was performed by the OHSU Proteomics Shared Resource with partial support from NIH core grants P30EY010572 & P30CA069533. This work was funded by the NIH (NIH 2R01NS088629) and Fondation Leducq (16CVD04).
Footnotes
The authors declare no competing financial interest.
Supporting Information
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/jacs.1c01302.
Table S1: 2-ad-BAD and 6-ad-BAD targets as identified by LC–MS/MS analysis (XLSX)
Figures S1–S6, Schemes S1–S8, experimental procedures, and compound synthesis and characterization (PDF)
Complete contact information is available at: https://pubs.acs.org/10.1021/jacs.1c01302
Contributor Information
Justina Šileikytė, Department of Chemical Physiology and Biochemistry, Oregon Health and Science University, Portland, Oregon 97239, United States;.
Sunil Sundalam, Department of Chemical Physiology and Biochemistry, Oregon Health and Science University, Portland, Oregon 97239, United States.
Larry L. David, Department of Chemical Physiology and Biochemistry, Oregon Health and Science University, Portland, Oregon 97239, United States
Michael S. Cohen, Department of Chemical Physiology and Biochemistry, Oregon Health and Science University, Portland, Oregon 97239, United States;
REFERENCES
- (1).Cohen MS Interplay between Compartmentalized NAD+ Synthesis and Consumption: A Focus on the PARP Family. Genes Dev. 2020, 34, 254–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Liu L; Su X; Quinn WJ; Hui S; Krukenberg K; Frederick DW; Redpath P; Zhan L; Chellappa K; White E; Migaud M; Mitchison TJ; Baur JA; Rabinowitz JD Quantitative Analysis of NAD Synthesis-Breakdown Fluxes. Cell Metab. 2018, 27 (5), 1067–1080.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Essuman K; Summers DW; Sasaki Y; Mao X; DiAntonio A; Milbrandt J The SARM1 Toll/Interleukin-1 Receptor Domain Possesses Intrinsic NAD+ Cleavage Activity That Promotes Pathological Axonal Degeneration. Neuron 2017, 93 (6), 1334–1343.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Essuman K; Summers DW; Sasaki Y; Mao X; Yim AKY; DiAntonio A; Milbrandt J TIR Domain Proteins Are an Ancient Family of NAD+-Consuming Enzymes. Curr. Biol 2018, 28 (3), 421–430.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Horsefield S; Burdett H; Zhang X; Manik MK; Shi Y; Chen J; Qi T; Gilley J; Lai JS; Rank MX; Casey LW; Gu W; Ericsson DJ; Foley G; Hughes RO; Bosanac T; Von Itzstein M; Rathjen JP; Nanson JD; Boden M; Dry IB; Williams SJ; Staskawicz BJ; Coleman MP; Ve T; Dodds PN; Kobe B NAD+ Cleavage Activity by Animal and Plant TIR Domains in Cell Death Pathways. Science (Washington, DC, U. S.) 2019, 365 (6455), 793–799. [DOI] [PubMed] [Google Scholar]
- (6).Loring HS; Icso JD; Nemmara VV; Thompson PR Initial Kinetic Characterization of Sterile Alpha and Toll/Interleukin Receptor Motif-Containing Protein 1. Biochemistry 2020, 59 (8), 933–942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Chatrin C; Gabrielsen M; Buetow L; Nakasone MA; Ahmed SF; Sumpton D; Sibbet GJ; Smith BO; Huang DT Structural Insights into ADP-Ribosylation of Ubiquitin by Deltex Family E3 Ubiquitin Ligases. Sci. Adv 2020, 6 (38), No. eabc0418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Liu Y; Hu L; Ma T; Yang J; Ding J Insights into the Inhibitory Mechanisms of NADH on the Aγ Heterodimer of Human NAD-Dependent Isocitrate Dehydrogenase. Sci. Rep 2018, 8 (1), 3146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Murale DP; Hong SC; Haque MM; Lee JS Photo-Affinity Labeling (PAL) in Chemical Proteomics: A Handy Tool to Investigate Protein-Protein Interactions (PPIs). Proteome Sci. 2016, 15, 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Guo H; Li Z Developments of Bioorthogonal Handle-Containing Photo-Crosslinkers for Photoaffinity Labeling. MedChem-Comm 2017, 8, 1585–1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Cravatt BF; Wright AT; Kozarich JW Activity-Based Protein Profiling: From Enzyme Chemistry to Proteomic Chemistry. Annu. Rev. Biochem 2008, 77 (1), 383–414. [DOI] [PubMed] [Google Scholar]
- (12).Jelcic M; Wang K; Hui KL; Cai X-C; Enyedi B; Luo M; Niethammer P A Photo-Clickable ATP-Mimetic Reveals Nucleotide Interactors in the Membrane Proteome. Cell Chem. Biol 2020, 27 (8), 1073–1083.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Zatorski A; Watanabe KA; Carr SF; Goldstein BM; Pankiewicz KW Chemical Synthesis of Benzamide Adenine Dinucleotide: Inhibition of Inosine Monophosphate Dehydrogenase (Types I and II). J. Med. Chem 1996, 39 (12), 2422–2426. [DOI] [PubMed] [Google Scholar]
- (14).Slama JT; Simmons AM Carbanicotinamide Adenine Dinucleotide: Synthesis and Enzymological Properties of a Carbocyclic Analogue of Oxidized Nicotinamide Adenine Dinucleotide. Biochemistry 1988, 27 (1), 183–193. [DOI] [PubMed] [Google Scholar]
- (15).Dai Z; Zhang XN; Nasertorabi F; Cheng Q; Pei H; Louie SG; Stevens RC; Zhang Y Facile Chemoenzymatic Synthesis of a Novel Stable Mimic of NAD+. Chem. Sci 2018, 9 (44), 8337–8342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Langelier MF; Zandarashvili L; Aguiar PM; Black BE; Pascal JM NAD+ Analog Reveals PARP-1 Substrate-Blocking Mechanism and Allosteric Communication from Catalytic Center to DNA-Binding Domains. Nat. Commun 2018, 9 (1), 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Carter-O’Connell I; Jin H; Morgan RK; David LL; Cohen MS Engineering the Substrate Specificity of ADP-Ribosyltransferases for Identifying Direct Protein Targets. J. Am. Chem. Soc 2014, 136 (14), 5201–5204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Carter-O’Connell I; Jin H; Morgan RK; Zaja R; David LL; Ahel I; Cohen MS Identifying Family-Member-Specific Targets of Mono-ARTDs by Using a Chemical Genetics Approach. Cell Rep. 2016, 14 (3), 621–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Li Z; Hao P; Li L; Tan CYJ; Cheng X; Chen GYJ; Sze SK; Shen HM; Yao SQ Design and Synthesis of Minimalist Terminal Alkyne-Containing Diazirine Photo-Crosslinkers and Their Incorporation into Kinase Inhibitors for Cell- and Tissue-Based Proteome Profiling. Angew. Chem., Int. Ed 2013, 52 (33), 8551–8556. [DOI] [PubMed] [Google Scholar]
- (20).Sanderson DJ; Cohen MS Mechanisms Governing PARP Expression, Localization, and Activity in Cells. Crit. Rev. Biochem. Mol. Biol 2020, 55, 541. [DOI] [PubMed] [Google Scholar]
- (21).Wallrodt S; Buntz A; Wang Y; Zumbusch A; Marx A Bioorthogonally Functionalized NAD + Analogues for In-Cell Visualization of Poly(ADP-Ribose) Formation. Angew. Chem., Int. Ed 2016, 55 (27), 7660–7664. [DOI] [PubMed] [Google Scholar]
- (22).Rodriguez KM; Buch-Larsen SC; Kirby IT; Siordia I; Hutin D; Rasmussen M; Grant DM; David LL; Matthews J; Nielsen ML; Cohen MS Chemical Genetics and Proteome-Wide Site Mapping Reveal Cysteine MARylation by PARP-7 on Immune-Relevant Protein Targets. eLife 2021, 10, e60480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Horning BD; Suciu RM; Ghadiri DA; Ulanovskaya OA; Matthews ML; Lum KM; Backus KM; Brown SJ; Rosen H; Cravatt BF Chemical Proteomic Profiling of Human Methyltransferases. J. Am. Chem. Soc 2016, 138 (40), 13335–13343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Papeo G; Posteri H; Borghi D; Busel AA; Caprera F; Casale E; Ciomei M; Cirla A; Corti E; D’Anello M; Fasolini M; Forte B; Galvani A; Isacchi A; Khvat A; Krasavin MY; Lupi R; Orsini P; Perego R; Pesenti E; Pezzetta D; Rainoldi S; Riccardi-Sirtori F; Scolaro A; Sola F; Zuccotto F; Felder ER; Donati D; Montagnoli A Discovery of 2-[1-(4,4-Difluorocyclohexyl)Piperidin-4-Yl]-6-Fluoro-3-Oxo-2,3-Dihydro-1H-Isoindole-4-Carboxamide (NMS-P118): A Potent, Orally Available, and Highly Selective PARP-1 Inhibitor for Cancer Therapy. J. Med. Chem 2015, 58 (17), 6875–6898. [DOI] [PubMed] [Google Scholar]
- (25).Kirby IT; Kojic A; Arnold MR; Thorsell AG; Karlberg T; Vermehren-Schmaedick A; Sreenivasan R; Schultz C; Schüler H; Cohen MS A Potent and Selective PARP11 Inhibitor Suggests Coupling between Cellular Localization and Catalytic Activity. Cell Chem. Biol 2018, 25 (12), 1547–1553.e12. [DOI] [PubMed] [Google Scholar]
- (26).Fjeld CC; Birdsong WT; Goodman RH Differential Binding of NAD+ and NADH Allows the Transcriptional Corepressor Carboxyl-Terminal Binding Protein to Serve as a Metabolic Sensor. Proc. Natl. Acad. Sci. U. S. A 2003, 100 (16), 9202–9207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Madison DL; Wirz JA; Siess D; Lundblad JR Nicotinamide Adenine Dinucleotide-Induced Multimerization of the Co-Repressor CtBP1 Relies on a Switching Tryptophan. J. Biol. Chem 2013, 288 (39), 27836–27848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Klepinin A; Zhang S; Klepinina L; Rebane-Klemm E; Terzic A; Kaambre T; Dzeja P Adenylate Kinase and Metabolic Signaling in Cancer Cells. Front. Oncol 2020, 10, 660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Storey KB Purification and properties of squid mantle adenylate kinase. Role of NADH in control of the enzyme. J. Biol. Chem 1976, 251, 7810. [PubMed] [Google Scholar]
- (30).Watanabe K; Tanaka K; Kojima H; Imanishi T; Yamamoto S Interaction between Cytosolic Adenylate Kinase and Nicotinamide Adenine Dinucleotide. Jpn. J. Vet. Sci 1988, 50 (2), 509–512. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


