Abstract
Capicua transcriptional repressor (CIC)-rearranged sarcoma represents a distinct pathologic entity, which constitutes the second most prevalent category of undifferentiated round cell sarcomas (URCS) after Ewing sarcoma. The two most common translocations are t(4;19) and t(10;19), resulting in CIC fusions with either DUX4 and DUX4L paralog, respectively, however, other rare variant fusions have also been reported. In this study we expand the molecular spectrum of CIC-gene partners, reporting on 5 cases of URCS showing CIC fusions with AXL, CITED1, SYK, and LEUTX by targeted RNA or DNA sequencing. There were four females and one male with a wide age range (12-70; median, 36 years). Four cases occurred in the deep soft tissues (lower extremity, 3; neck, 1) and one case in the central nervous system (midbrain/thalamus). All cases showed similar histologic findings within the spectrum of URCS. By Immunohistochemistry, ETV4 showed variable positivity in 4/4 cases, ERG was positive in 3/4, and WT1 in 1/4 cases. CD31 was positive in 2/3, including one co-expressing ERG. Unsupervised clustering of methylation profiles by t-SNE performed in 4 cases showed that all clustered tightly together and along the CIC sarcoma methylation class. RNAseq data showed consistent upregulation of ETV1 and ETV4 mRNA in all cases examined, at similar levels to CIC::DUX4 URCS. Our study expands on the molecular diversity of CIC-rearranged URCS to include novel and rare partners, providing morphologic, immunohistochemical, gene expression, and methylation evidence supporting their classification within the family of tumors harboring the more common DUX4/DUX4L partner genes.
Keywords: CIC, AXL, CITED1, SYK, LEUTX, undifferentiated round cell sarcoma
INTRODUCTION
Capicua transcriptional repressor (CIC)-rearranged sarcomas constitute the second most common category of undifferentiated round cell sarcomas (URCS), being a distinct pathologic entity with a worse prognosis than Ewing sarcoma and less responsive to chemotherapy (1). The most common fusions result from either t(4;19) or t(10;19) translocations, where CIC is fused to either DUX4 or DUX4L paralog, respectively (2, 3), although other rare gene partners (FOXO4, NUTM1, and NUTM2A) have also been reported (1, 4-7).
CIC protein is a high-mobility group box transcription repressor that binds to target promoters and enhancers. Its default inhibitory activity relies on suppressing transcription in the absence of signaling, whereas conversely its phosphorylation prevents its nuclear import and attenuates its DNA binding activity (8-11). The CIC-fusion oncoprotein is a potent transcriptional activator that binds and upregulates the promoters of downstream targets such as polyoma enhancer activator 3 (PEA3) subfamily of genes (including ETV1/4/5) both at the mRNA and protein levels (12-14). This gene expression signature is highly pathognomonic and quite specific for diagnostic purposes among URCS; the latter being especially important as both RNA sequencing (RNAseq) and fluorescence in situ hybridization (FISH) have low sensitivity in this setting and may produce false negative results (15, 16). Consequently, ETV4 immunohistochemistry and RNA in situ hybridization of ETV1/4/5 have been reported to be useful ancillary studies in supporting the diagnosis (17, 18). Gene expression profiling has also shown upregulation of WT1, which can be exploited immunohistochemically with similar sensitivity albeit inferior specificity to ETV4 (13, 17). By unsupervised hierarchical clustering of RNA sequencing, CIC-rearranged sarcomas appear to cluster in a distinct group separate from other small round blue cell tumors (15).
Herein, we report five cases of CIC-rearranged URCS with novel and very rare gene partners expanding on their molecular diversity. We provide morphologic, immunohistochemical, and molecular support that they are similar to their more common CIC::DUX4 counterparts.
MATERIALS AND METHODS
Study cohort
After approval from the Institutional Review Board, CIC-rearranged URCS were identified from the Memorial Sloan Kettering Cancer Center (MSKCC) Department of Pathology files and the personal consultation files of the senior author (C.R.A). Thus 5 cases with novel and rare partners were identified and included in this study. Clinical data, including age, sex, anatomic site, treatment, and follow-up were retrieved from pathology reports and clinical records. Hematoxylin and eosin-stained slides and immunohistochemical stains from all specimens were re-reviewed by two of the authors (K.L and C.R.A).
Immunohistochemistry
The relevant antibodies and dilutions used in this study can be seen in Supplementary Table 1.
Fluorescence In Situ Hybridization (FISH)
FISH analysis was performed on interphase nuclei from paraffin-embedded 4mm sections using bacterial artificial chromosome (BAC clone), flanking CIC in 19q13 (1, 3). Two hundred tumor nuclei were evaluated using a Zeiss fluorescence microscope (Zeiss Axioplan, Oberkochen, Germany), controlled by Isis 5 software (Metasystems). ). A cutoff of >20% nuclei showing a break-apart signal was considered to be positive for rearrangement. Nuclei with an incomplete set of signals were omitted from the score
Targeted RNA sequencing (MSK-Fusion)
Detailed descriptions of MSK-Fusion, an amplicon-based targeted RNA NGS assay using the Archer™ FusionPlex™ standard protocol, were described previously (19). Briefly, RNA was extracted from tumor formalin-fixed paraffin-embedded material followed by cDNA synthesis. cDNA libraries were made using the Archer™FusionPlex™ standard protocol. Fusion unidirectional GSPs have been designed to target specific exons in 123 genes known to be involved in chromosomal rearrangements based on current literature. The final targeted amplicons were ready for 2 x 150 bp sequencing on an Illumina MiSeq sequencer. FASTQ files were automatically generated using the MiSeq reporter software (Version 2.6.2.3) and analyzed using the Archer™ analysis software (Version 5.0.4). Each fusion call was supported by a minimum of 5 unique reads and a minimum of 3 reads with unique start sites. Archer™ FusionPlex™ was performed in 4 cases.
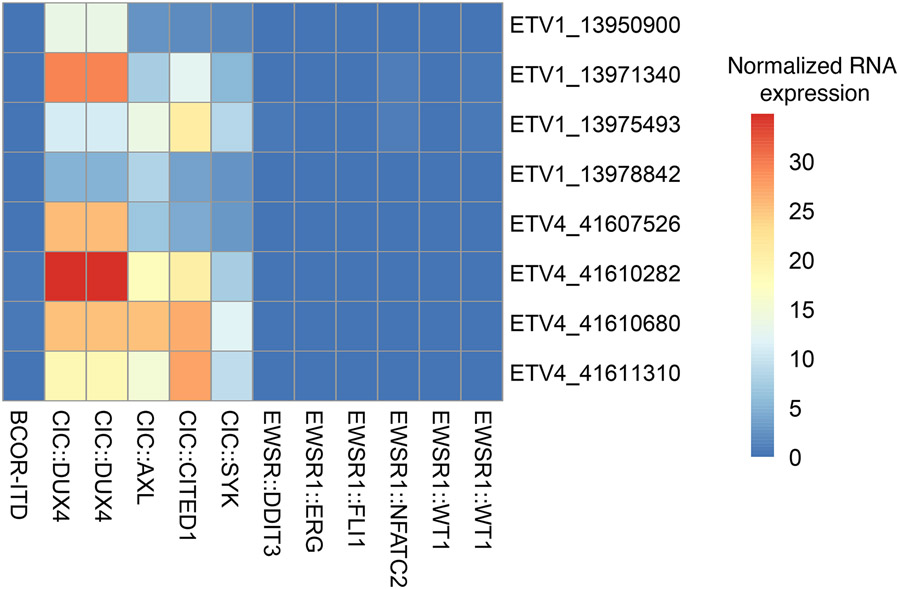
RNA expression values for Archer probes targeting ETV1 and ETV4 for CIC-rearranged samples (n=5) and control clinical samples consisting of various small blue round cell tumors (n=7)(Ewing sarcoma, BCOR-rearranged sarcoma, desmoplastic small round cell tumor, myxoid/round cell liposarcoma, EWSR1::NFATC2 sarcoma) were downloaded from Archer data portal. The expression levels were arbitrary values normalized to average expression of Archer’s internal control. Heatmap was generated by the pHeatmap package version 1.0.12 using R version 4.1.0.
Targeted DNA sequencing (MSK-IMPACT Solid)
Detailed descriptions of MSK-IMPACT workflow and data analysis, a hybridization capture-based targeted DNA Next Generation Sequencing (NGS) assay for solid tumors have been described previously in detail (20). MSK-IMPACT Solid was performed in all 5 cases.
Methylation Profiling and Clustering Analysis
Four CIC-rearranged tumors with novel and rare partners were tested by methylation profiling. Details on methylation profiling have been published previously (21). Briefly, genomic DNA was extracted from formalin-fixed paraffin-embedded (FFPE) tissue sections for each of the samples. Next, 250 ng of genomic DNA was subjected to bisulfite conversion and processed on the Illumina (San Diego, CA) methylation EPIC/850k platform according to manufacturer’s instructions.
We used several external, publicly available data sets to enhance the analysis. First, we obtained raw IDAT files for 148 samples from the Heidelberg sarcoma methylation classifier reference cohort [Gene Expression Omnibus (GEO) study accession number GSE140668](22): 36 alveolar rhabdomyosarcoma (ARMS), 30 embryonal rhabdomyosarcoma (ERMS), 26 desmoplastic small round cell tumor (DSRCT), 37 Ewing sarcoma (ES), 11 CIC-rearranged round cell sarcoma, and 8 BCOR-altered round cell sarcoma.
IDAT processing and data analysis on all 152 samples was performed using R version 4.1.0 and the “minfi” package version 1.38.0 (23). Normalization was performed using the preprocess Illumina function and probes with a detection P value > 0.01 were filtered as were single nucleotide polymorphisms (SNP)-related probes, and probes on sex chromosomes. Methylation levels were measured using beta values for all cases (24).
For unsupervised clustering, dimensionality reduction was performed by the T-distributed stochastic neighborhood embedding (t-SNE) method. After normalizing the input data matrix (centering the mean of each column to zero), the top 10,000 most variable CpGs by variance were analyzed using the “Rtsne” package version 0.15 with the following non-default parameters: perplexity = 10, max_iter = 5000, and theta = 0.
RESULTS
Clinical and histopathologic findings (Fig. 1-4 & Table 1)
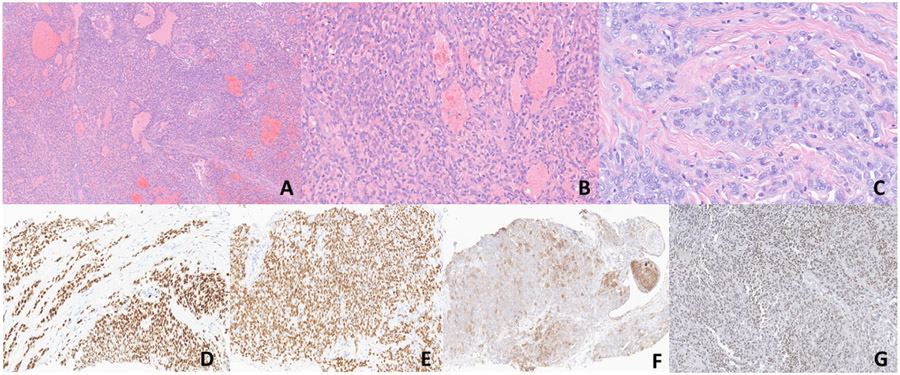
Fig. 1.
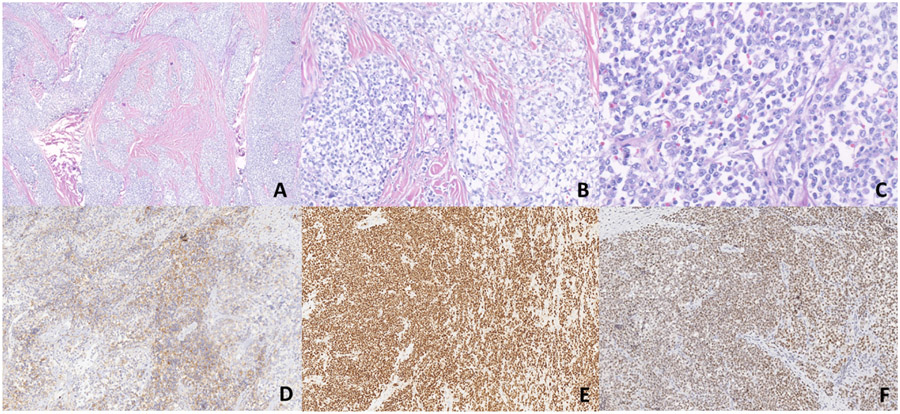
A-G. Morphologic and immunohistochemical findings of Case 1 (CIC::AXL). (A-C). Sections show sheets of round to spindle cells without any obvious line of differentiation. Pseudovascular spaces lined by neoplastic cells are evident. On higher magnification, the cells exhibit high nuclear to cytoplasmic ratio with vesicular chromatin pattern and inconspicuous nucleoli (x50, x200, x400). D-E) ERG and WT1 show diffuse strong nuclear positivity respectively. F) CD99 shows patchy membranous expression. G) ETV4 shows diffuse weak to moderate nuclear positivity.
Fig. 4.
A-F. Morphologic and immunohistochemical findings of Case 5 (CIC::SYK). (A). Sections show large areas of necrosis with perivascular accentuation of viable tumor (x50). B-C) Medium and high-power magnification show a proliferation of round to vaguely spindle cells with no obvious line of differentiation morphologically. The cells exhibit vesicular chromatin pattern with inconspicuous nucleoli (x100, x400). D) Diffuse nuclear expression of ERG. Note internal positive control of background endothelial cells with no expression in the surrounding pericytes. E) Diffuse membranous and cytoplasmic expression of CD31. F) ETV4 shows focal weak immunoreactivity.
Table 1.
Clinical and Pathologic Features
| Case # |
Age /Sex |
Location | Tumor size (cm) |
Pertinent IHC | Chemotherapy/radiation treatment |
Surgical treatment, margins |
Histologic treatment response |
Follow-up (months) |
|---|---|---|---|---|---|---|---|---|
| 1 | 29/F | Left lower neck | 5.1 | ERG +, WT-1+, CD99 + (patchy), ETV4 +*, CD31− | Neoadjuvant VAC/IE# | Resection, negative margins | No treatment response^, | NED (2) |
| 2 | 36/F | Right foot | 4.2 | ERG+, WT-1 −, CD99 + (patchy), ETV 4+**, CD31− | Neoadjuvant VAC/IE, adjuvant temozolomide &irinotecan | Resection, negative margins | 30-40% in the form of fibrosis & necrosis | AWD [lung & brains metastases] (19) |
| 3 | 12/F | Left buttock | 3.0 | WT-1 −, CD99 (paranuclear dot-like), CD31− | Adjuvant radiation to primary site (55.8 Gy), ifosfamide with doxorubicin or etoposide for lung metastases, whole lung radiation (15 Gy) | Primary excision, positive margins | N / A | NED (108) |
| 4 | 42/F | Midbrain/thalamus | N/A | ERG−, WT1−, CD31+ (strong), ETV4 +/−*** | Neoadjuvant brain radiation therapy and temozolomide | No surgical treatment | N/A | NED (51) |
| 5 | 70/M | Lower extremity | 11.5 | ERG+, CD31 + (strong), ETV4 +/− **** | Doxorubicin, paclitaxel plus nivolumab for local recurrence and lung metastasis, gemcitabine & docetaxel post local recurrence resection | Resection of local recurrence, negative margins | 30% necrosis | NED (12) |
y- years, N/A- not available, CNS- central nervous system, IHC- immunohistochemistry, NED- No evidence of disease
diffuse weak to focally moderate +
diffuse moderate +
focal moderate + (<5% tumor volume),
focal weak + (<5% tumor volume)
Vincristine, doxorubicin, cyclophosphamide alternating with ifosfamide & etoposide
No treatment response histologically but tumor reduced in size from 5.1cm to 3.2 cm in greatest dimension
The cohort comprises four females and one male, all except one patient occurring in adults, with an age range of 12 to 70 years (median: 36y, mean: 37.8y). Four tumors involved the deep soft tissues (neck, foot, buttock, lower extremity) and one occurred in the central nervous system (midbrain/thalamic). The tumors’ size ranged from 3 to 11.5 cm (median 4.6 cm, mean: 5.9 cm) in greatest dimension. All five patients presented with primary tumors at diagnosis. All tumors showed similar histologic findings within the spectrum of undifferentiated round cell sarcomas (URCS). They were composed of solid sheets and nests of round to ovoid to epithelioid cells with high nuclear to cytoplasmic ratio, amphophilic to clear cytoplasm, vesicular chromatin pattern, and mild to moderately conspicuous nucleoli. One tumor showed a minor spindle cell component and a focal pseudoalveolar pattern. In one case a more prominent myxoid matrix was evident, in which the cells were arranged in a reticular or trabecular fashion. In the CNS tumor, the cells were variably discohesive with a pattern of growth ranging from nests to sheet-like to papillary. In some cells apparent fibrillary and/or refractile cytoplasmic inclusions were seen, whereas occasionally multinucleation was also noted. Two tumors were multilobulated with interlobular fibrous bands. The mitotic figures ranged from 8 to 35 per 1mm2. Two patients had resections of their primary tumors post-neoadjuvant therapy, whereas one patient had a primary resection without neoadjuvant therapy. One patient presented with a recurrence near the stump of a below-the-knee amputation 6 months before. A subsequent above-the-knee amputation was performed post-neoadjuvant therapy. Necrosis was present in the pre-treatment specimens of cases 2 and 5; in their post-treatment resections, 30-40% necrosis and fibrosis and 30% necrosis respectively were evident. In case 1 there was no histologic treatment effect post-neoadjuvant resection, however, the tumor was reduced in size from 5.1cm to 3.2cm in greatest dimension. The surgical margin was positive in 1/4 of cases (case 3, primary resection without neoadjuvant treatment).
Immunohistochemical findings (Fig. 1-4 & Table 1)
ETV4 was evaluated in four tumors and showed diffuse positivity with weak to moderate intensity in two (CIC::AXL and CIC::CITED1) and focal (<5% of tumor volume) weak to moderate expression in the other two (CIC::LEUTX and CIC::SYK). ERG was positive in 3/4 cases evaluated, whereas WT1 showed diffuse nuclear positivity in one tumor and it was essentially negative in the other three cases examined. CD31 was positive in 2/3; it was co-expressed with ERG in one case, whereas in the other case, ERG was negative. CD99 showed non-specific positivity in 3/3 tumors evaluated (patchy cytoplasmic in two and paranuclear dot-like pattern in one). CAM5.2 showed rare positive cells in 1/2, whereas EMA and SMA were focally and weakly positive in 1/3 and 1/2 cases respectively. Vimentin and TLE1 (patchy) were positive in 1/1 cases. Nuclear expression of SMARCB1 (BAF-47, INI1) was retained in 3/3, and SMARCA4 (BRG1) was retained in one case tested. The following immunohistochemical stains were negative in the cases evaluated: S100, desmin, myogenin, MyoD1, CKMNF116, CD34, CD45, ALK, Pan-Trk, SS18-SSX, PanCK, CK7, CK20, EMA, BCOR, SATB2, TdT, HMB45, MelanA, CD163, p63, calponin, GFAP, AFP, inhibin, CD10, TTF-1, PAX-8, OCT-4, PR, chromogranin, synaptophysin, and neurofilament protein.
Fluorescence In Situ Hybridization (Table 2)
Table 2.
FISH, RNA and DNA sequencing results
| Case# | RNA sequencing (Archer FusionPlex) |
DNA sequencing (MSK-IMPACT) | FISH* |
|---|---|---|---|
| 1 | In-frame CIC Exon 20:: AXL Exon 2 with a 2bp insertion | CIC::AXL: c.4663:CIC_c.151:AXLdup | Loss of 5 portion of CIC locus Indicative of fusion |
| 2 | In-frame CIC Exon20::CITED1 Exon 3 | CIC::CITED1: t(19;X)(q13.2;q13.1)** | Negative for EWSR1, FUS & SS18 rearrangements |
| 3 | Not done | CIC::LEUTX: c.4797:CIC_c.354:LEUTXdup | Negative for EWSR1 rearrangements |
| 4 | In-frame CIC Exon 20:: LEUTX Exon 3 | CIC::LEUTX: c.4769:CIC_c.321:LEUTXdup | None |
| 5 | Negative | CIC breakpoint in exon 20: t(9;19)(q22.2;q13.2). *** | None |
Fluorescence in situ hybridization
The same fusion was detected in the primary tumor and brain metastasis
The nearest gene on chr9 where the breakpoint occurred is SYK.
In case 1 with CIC::AXL fusion, the initial FISH showed loss of the 5’ portion of CIC locus in 93% of cells, indicative of a CIC fusion. In two cases FISH was negative for EWSR1 rearrangement, whereas in another case FISH was negative for FUS and SS18 gene rearrangements.
Targeted RNA sequencing results (Archer™ FusionPlex™) [Fig. 5 & Table 2]
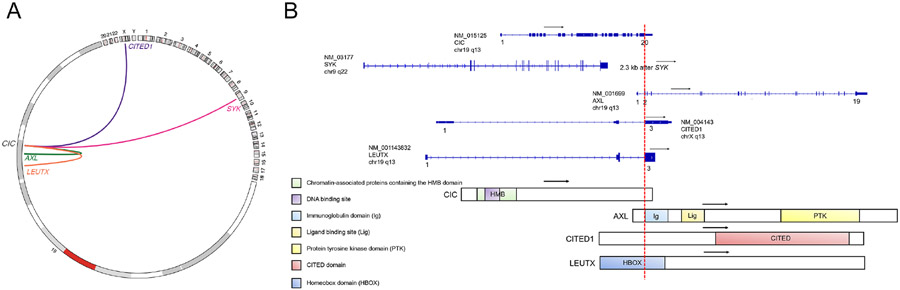
Fig. 5.
A: Circos plot depicting CIC fusions represented by links between cytobands (hg19 genome). Plot generated using R package “circlicize” version 0.4.13 (Gu 2014). B: Schematics of CIC fusion transcripts annotated by NCBI RefSeq accession numbers and the predicted chimeric protein domains. Numbers, black arrows, and red dotted lines represent exons, directions of transcript, and fusion breakpoint, respectively.
Archer™ FusionPlex™ was performed in 4 tumors and detected in-frame fusions between genes CIC Exon 20 (NM_015125) and AXL Exon 2 (NM_001699) [case 1], CIC Exon 20 (NM_015125) and CITED1 Exon 3 (NM_004143) [case 2] and CIC Exon 20 (NM_015125) and LEUTX Exon 3 (NM_001143832) [case 4]. Archer™ FusionPlex™ was negative for gene fusions in case 5, whereas it was not done in case 3.
Targeted DNA sequencing results (MSK-IMPACT Solid) [Fig. 5 & Table 2]
In case 3 a CIC (NM_015125)-LEUTX (NM_001143832) fusion: c.4797:CIC c.354:LEUTXdup was detected, resulting in the fusion of CIC exons 1-20 to a part of LEUTX exon 3. In three cases (cases 1,2 & 4) MSK IMPACT was able to confirm the gene fusions detected at the RNA level by Archer™ FusionPlex™. In case 2 MSK IMPACT also detected the same fusion in the brain metastasis. In case 5, in which Archer™ FusionPlex™ was negative, MSK-IMPACT detected a CIC (NM_015125) rearrangement with a breakpoint in exon 20: t(9;19)(q22.2;q13.2) (chr9:g.93665094::chr19:g.42799213); the breakpoint on chr9 being 2.3 kilobases downstream of the nearest coding gene (SYK).
In case 1 the CIC::AXL rearrangement was a duplication that resulted in a fusion of CIC exon 1-20 to AXL exons 2-20, which includes the kinase domain of AXL. The somatic alteration FAT1 (NM_005245) exon 10 p.L1625 *(c.4874T>G) was also detected, whereas the copy number profile was suggestive of broad copy number gain on chromosome 12 and broad copy number loss of chromosome arm 16q23.24 and 19p13.
Additional copy number changes in the remaining cases were broad copy number gains on chromosome 8 and broad copy number losses of chromosome arms 9q, 11q22-23, and 17q11-17q21. The estimated tumor mutation burden (TMB) of the entire cohort ranged from 0.8 to 1.6 mutations per megabase (mt/Mb). All tumors were microsatellite stable (MSIsensor score ranged from 0.12 to 0.76)(25). The somatic alterations and copy number changes of all cases can be found in the supplementary Table 2.
Unsupervised t-SNE clustering of methylation profiles and sarcoma methylation classifier (Fig. 6)
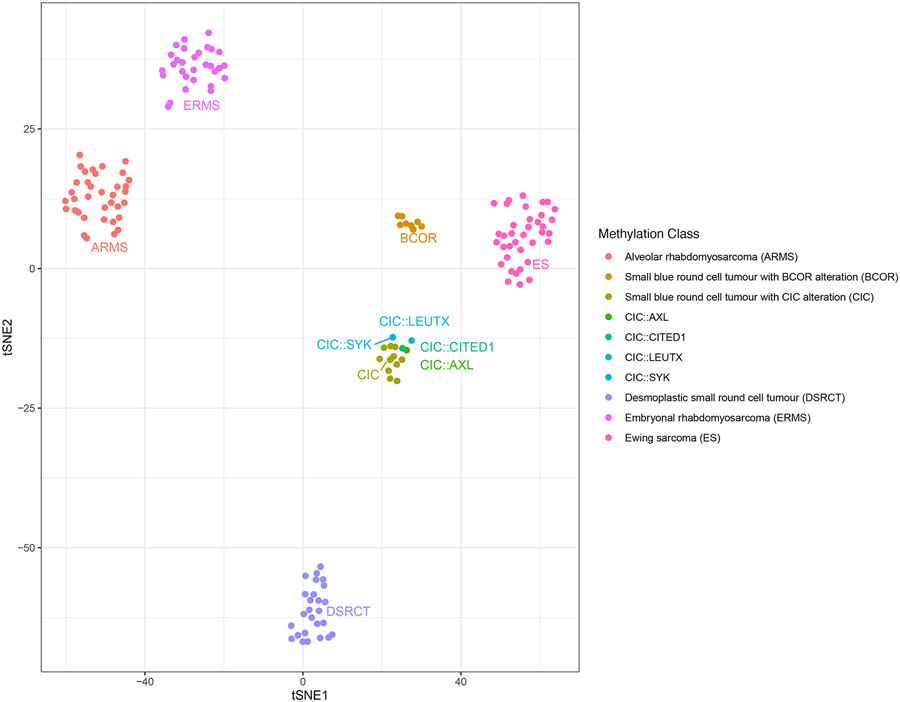
Fig. 6.
Unsupervised clustering of methylation profiles by t-SNE showed that the four cases (AXL, LEUTX [case 4], CITED1 and SYK) clustered tightly together with the CIC sarcoma methylation class and were distinct from other small round blue cell tumors (ARMS, ERMS, DSRCT, Ewing sarcoma, BCOR-rearranged sarcomas)
Unsupervised clustering of methylation profiles by t-SNE showed that the four cases (AXL, LEUTX [case 4], CITED1, and SYK) clustered tightly together with the CIC sarcoma methylation class as described by Koelsche et al (22), and were distinct from other small round blue cell tumors (ARMS, ERMS, DSRCT, Ewing sarcoma, BCOR-rearranged sarcomas).
Upregulation of ETV1/4 mRNA (Fig. 7)
Fig. 7.
Heatmap depicting RNA expression values for Archer probes targeting ETV1 and ETV4 for CIC-rearranged samples and control clinical samples consisting of various small blue round cell tumors (Ewing sarcoma, BCOR-rearranged sarcoma, desmoplastic small round cell tumor, myxoid/round cell liposarcoma, EWSR1::NFATC2 sarcoma). Row names denote gene symbol and chromosomal start coordinate of probe targets. Column names denote underlying gene fusions of the corresponding sample. Expression levels were normalized to average expression of each probe of Archer’s internal control.
Targeted RNAseq data showed that the CIC::AXL, CIC::CITED1 and CIC::SYK cases exhibited consistent upregulation of ETV1 and ETV4 mRNA at various levels; this was similar to CIC::DUX4 URCS and in contrast to other round cell/undifferentiated sarcoma controls with various fusions which expressed near-zero levels.
Treatment and follow-up (Table 1)
Two patients received neoadjuvant chemotherapy with VAC/IE (Vincristine, doxorubicin, cyclophosphamide/ifosfamide, etoposide) [cases 1 &2)]. One patient (case 2) received temozolomide and irinotecan, palliative brain radiation, and gemcitabine with docetaxel for metastatic disease. One patient (case 3) received ifosfamide with doxorubicin or etoposide for lung metastases with near completion of distant disease followed by localized adjuvant radiation to the primary site (55.8 Gy) and whole lung radiation (15Gy). One patient (case 4) received neoadjuvant brain radiation with subsequent temozolomide, and one patient (case 5) received doxorubicin followed by paclitaxel and nivolumab as part of a clinical trial for local recurrence and possible metastases.
The follow-up ranged from 2 to 108 months (median: 19m, mean: 38.4m). Overall, one patient developed lung and brain metastases, one patient solely lung metastases, and one patient local recurrence at the stump of a previous below-the-knee amputation and also lung metastases. Three patients were alive with no evidence of disease, one patient was alive with localized disease and one patient was alive with distant disease.
DISCUSSION
Our study reports on five new cases of CIC-rearranged undifferentiated round cell sarcomas (URCS) with novel and very rare fusion partners (AXL, CITED1, LEUTX, and SYK). In contrast to CIC::DUX4 sarcomas in which there is a slight male dominance, in this cohort, there was a female predominance (M/F: 1:4), whereas the mean age at diagnosis (37.8y) was slightly older. In all cases, the morphologic features were those of an undifferentiated sarcoma within the spectrum that has been described in CIC-rearranged sarcomas(1). Moreover, similar to cases harboring the canonical fusion, the immunohistochemical profile of this study group also showed expression of ETV4, ERG, and WT1 with variable positivity. In all our cases the breakpoints were within exon 20 of CIC similar to most reported cases harboring CIC::DUX4, CIC::FOXO4, and CIC::LEUTX fusions, with retention of the high mobility group (HMG) box DNA-binding domain (3, 11, 26). NUTM1, albeit still very rare, appears to be the second most common partner in CIC-rearranged undifferentiated sarcomas with otherwise typical morphology (27). In contrast to our cohort, these tumors have a predisposition to axial location with frequent bone involvement(7, 27, 28). Although initially thought to be predominant in childhood it appears that they have a broader age distribution closer to that of CIC::DUX4 undifferentiated sarcomas. Breakpoints within exons 16 to 20 of CIC have been documented with retention of the high mobility group (HMG) box DNA binding domain as well. In case 2 a broad copy number gain of chromosome 8 was detected which is a common recurrent abnormality in CIC::DUX4 sarcomas and has also been reported in undifferentiated sarcomas with CIC-NUTM1 fusion(27, 29). In the three cases with post-neoadjuvant therapy resection, the treatment response was poor (up to 40%, grade I response) similar to other CIC-rearranged sarcomas (1). However, the patient with primary CNS tumor (case 4) showed post-treatment no evidence of disease radiologically and in cases 3 and 5 the lung metastases were not detectable after radiation and chemotherapy. In contrast to patients with CIC::DUX4 or CIC::NUTM1 sarcomas who have a poor 5-year survival, all patients in our cohort were alive, albeit with a limited median follow-up of 19 months. Nevertheless, the number of subjects in this study is admittedly very low to draw meaningful statistical conclusions.
AXL is a member of the TAM family of tyrosine kinase receptors (Tyro-3, AXL, MER) (30). Ligand binding to the extracellular domain usually leads to its activation by dimerization and transautophosphorylation of tyrosine residues(30). In adult tissues, it is expressed ubiquitously mediating a diverse array of cellular functions; in neoplasia, it has been implicated in the development and progression of many malignancies by playing a role in cancer cell proliferation, migration, and invasion, angiogenesis, and metastasis(30). In case 1, exon 20 of CIC was fused with exon 2 of AXL, expected to encode for the entire protein kinase domain of AXL presumably leading to activation of CIC by phosphorylation.
CITED1 (CREB-binding protein/P300 Interacting Transactivator with glutamic acid [E]/aspartic acid [D]-rich Carboxy-Terminal Domain 1) is located on chromosome Xq13.1 and belongs to the CITED family of nuclear proteins (CITED1-4), which co-regulate transcriptional nuclear proteins via the transactivation domain (31). In mesenchymal neoplasia, CITED1 has been described in SRF-fused perivascular tumors (SRF::CITED1), whereas CITED2 has been reported in spindle cell rhabdomyosarcoma (VGLL2::CITED2) and PRDM10-rearranged soft tissue tumors (PRDM10::CITED2) (31-33). Only one case of undifferentiated round cell sarcoma with an in-frame CIC::CITED1 has been published in the literature (34). The breakpoints were between exon 21 of CIC and exons 4 and 5 of CITED1. The patient received chemotherapy, ponatinib, and pazopanib and eventually deceased.
LEUTX gene is a paired homeodox gene (PRD) located at chromosome 19q13, which belongs to the same class as DUX4 (35). It plays an important role in embryonal development and its expression is mostly suppressed postnatally (36). Fusions of LEUTX have been described in a case of malignant epithelioid peripheral nerve sheath tumor (BRD4::LEUTX) and a case of therapy-related acute myeloid leukemia (KAT6A::LEUTX)(37, 38). To this date, CIC::LEUTX has been described in six cases [2 angiosarcomas (soft tissue and CNS), 1 intraspinal extramedullary subdural undifferentiated round cell sarcoma, 1 anaplastic ganglioglioma, 1 anaplastic astrocytoma, and 1 CNS embryonal tumor](11, 36, 39-41). One of our cases represents the first extra-CNS case of URCS with this fusion. In both of our cases the breakpoints were between CIC exon 20 and LEUTX exon 3 which are identical with the 4 out of 6 reported CIC::LEUTX cases in which information regarding breakpoints is known. Intriguingly, 2 out of 6 CIC::LEUTX reported (one CNS angiosarcoma and one embryonal tumor) harbored TSC2 somatic mutations similar to our CNS case (39, 41) [supplementary table 2]; it is unknown if the other cases also harbored TSC2 mutations as they were not reported. In the CNS embryonal tumor, the detected somatic TSC2 variant c.G2714A was evaluated as pathogenic mutation by public database ClinVar. The patient received chemotherapy and radiotherapy along with mTOR inhibitor (everolimus) with significant clinical response (39). Nevertheless, in our cohort the detected TSC2 mutation in case 4 (c.3889G>A p.A1297T, see supplementary table 2) is predicted to be benign or likely neutral in ClinVar.
SYK gene (Spleen Tyrosinase Kinase), located on 9q22, encodes for a member of the family of nonreceptor type Tyr protein kinases. It is primarily expressed in neutrophils, myeloid derived suppressor cells, macrophages, mast cells, and B-lymphocytes but can also be expressed in non-immune cells (42). Its function in solid tumors is not entirely clear with a possible tumor suppressive function (43). In Ewing sarcoma an oncogenic signaling axis involving SYK, c-MYC, and MALAT1 has been described and may potentially serve as a therapeutic target (44). Small-molecule inhibitors of SYK have shown antineoplastic properties and are currently explored in clinical trials for malignant and autoimmune diseases(45)
As described in other CIC-rearranged sarcomas, in 3 of the CIC-rearranged sarcomas with AXL, CITED1, and SYK partners, there was ETV1 and ETV4 mRNA overexpression noted in keeping with strong transcriptional activation by CIC HMG box binding to the promoter of PEA genes (11, 12, 46). By IHC, all examined cases showed ETV4 positivity albeit with variable expression, while WT1 was only detected in 1/4 cases. However, there were two cases in which ETV4 immunohistochemical expression was only focal (<5% of tumor volume) and in one of them WT1 was also negative (WT1 not performed in the other). These findings may suggest a lower sensitivity for ETV4 and WT1 immunoexpression in tumors with rare CIC-partners compared to CIC::DUX4 sarcomas, which have been reported as 90% and 95%, respectively (17).
One case of our cohort (case 5 with CIC::SYK) showed co-expression of CD31 and ERG and had been initially diagnosed as epithelioid angiosarcoma. In 2017 Huang et al reported three cases of epithelioid angiosarcomas with diffuse expression of CD31/ERG and CIC-rearrangements (11). They occurred in the kidney and soft tissues of adult females and histologically they were composed of solid round to epithelioid cells with no vasoformative features. CIC::LEUTX fusion was detected in one case in which the fusion partner was available, whereas concomitant CIC mutations were found in two cases. Very recently Kojima et al reported 9 cases of CIC-rearranged sarcomas with co-expression of CD31 and ERG; four cases showed diffuse or strong expression of CD31, whereas none showed combined uniform diffuse and strong expression of both markers (47). Their clinicopathologic characteristics did not differ from the remaining 21 cases of their cohort. Three cases were initially diagnosed and treated as angiosarcoma with no clinical response. In contrast to conventional epithelioid angiosarcomas, these CIC-rearranged ‘angiosarcomas’ did not exhibit vasoformative features and expressed ETV4 and/or WT1 immunohistochemically. Four cases examined by next generation sequencing did not harbor CIC missense mutations; by DNA methylation profiling a CD31-positive CIC-rearranged sarcoma clustered with CD31-negative CIC-rearranged sarcomas and distant from angiosarcomas. Unsupervised clustering of methylation profiles by t-SNE showed that the CD31+/ERG+ case in our cohort clustered tightly with the CIC sarcoma methylation class. These findings suggest that CIC-rearranged sarcomas with CD31/ERG co-expression likely belong to the CIC-rearranged URCS family rather than bone-fide angiosarcomas. Moreover, two of the previously reported ‘epithelioid angiosarcomas’ with CIC rearrangements showed overlapping gene signature with CIC::DUX4 undifferentiated sarcomas by unsupervised hierarchical clustering, with significant overexpression of ETV1/4/5 and WT1 genes compared to a control group of conventional angiosarcomas (11).
CIC-rearranged sarcomas have a distinct methylation profile (22). Unsupervised clustering of methylation profiles by t-SNE showed that the four evaluated cases of our cohort clustered tightly together and along the CIC sarcoma methylation class. Intriguingly, rare cases of undifferentiated sarcomas (four in the CNS and one of unknown origin) with ATXN1::DUX4, ATXN1:NUTM2A, and ATXN1L-NUTM2A fusions showed similar morphologic and immunohistochemical profile, ETV1/4/5 upregulation and matched with the CIC-rearranged sarcomas by DNA methylation profile (48-50). In less well-documented reports, one case with ATXN1::NUTM1 and one with CIC frame-shift deletion along with a cell line with an in-frame CIC-deletion also clustered with CIC-rearranged sarcomas (46, 48, 51, 52). The ATXN1/ATXN1L forms a transcriptional repressor complex with CIC, which represses its target genes by binding to DNA (53). It is hypothesized that the ATXN1/ATXN1L fusions alter their protein structure and destabilize the protein complex with CIC, which leads to downstream gene overexpression and oncogenesis (50). This very recent data suggest that the categorization as ‘CIC-rearranged undifferentiated sarcoma’ may be too restrictive to capture otherwise identical tumors and perhaps the term “undifferentiated sarcoma with upregulation of ETV1/4/5” may actually be more inclusive.
In conclusion, we report five cases of CIC-rearranged sarcomas with novel and rare partners. They were within the morphologic spectrum of undifferentiated round cell sarcomas, showed upregulation of ETV1 and ETV4, and were classified under CIC-rearranged sarcomas by unsupervised methylation clustering. This report expands on the molecular diversity of this rare neoplasm.
Supplementary Material
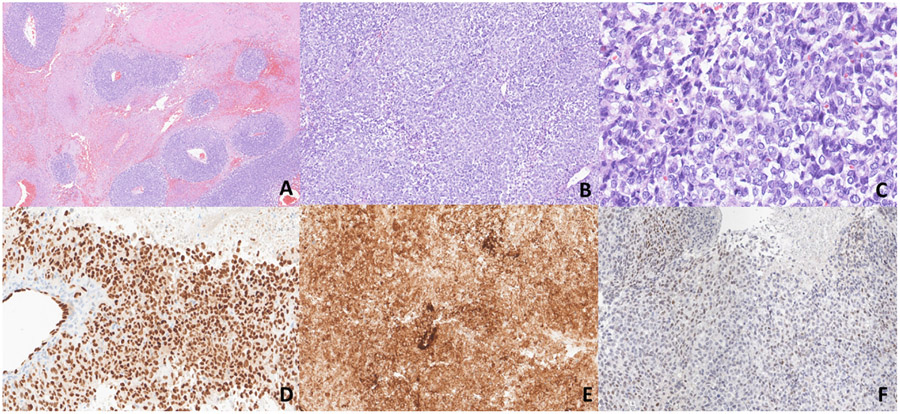
Fig. 2.
A-F. Morphologic and immunohistochemical findings of case 2 (CIC::CITED1). A-C) Low-power magnification shows a multinodular growth pattern in a collagenous background. Medium and high-power magnification show epithelioid cells with minimal eosinophilic to clear cytoplasm and vesicular chromatin pattern with mildly conspicuous nucleoli (x50, x100, x400). D) Immunohistochemistry for CD99 shows patchy positivity. E-F) Diffuse positivity for ERG and ETV4 respectively.
Fig. 3.
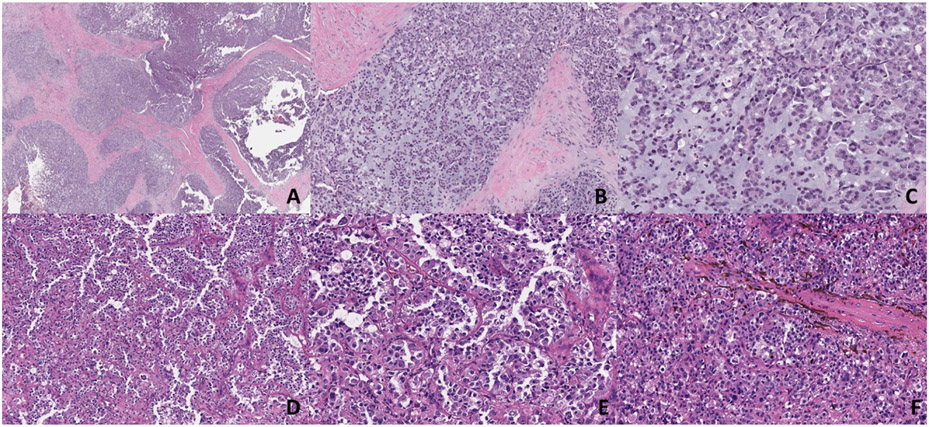
A-F. Morphologic findings of cases 3 and 4 (CIC::LEUTX). A-C) Low-power magnification shows a multinodular growth pattern, whereas medium and high-power magnification show epithelioid cells with eosinophilic cytoplasm in a myxoid background (x50, x100, x200). (D-F) This primary CNS tumor shows dyshesive epithelioid cells in a pseudoalveolar pattern, whereas in other areas there is more solid growth. Cells show ample eosinophilic cytoplasm with nuclear pleomorphism (x100, x200).
FUNDING
Supported by: P50 CA217694 (C.R.A, W.T, S.S), P30 CA008748 (C.R.A), Cycle for Survival (C.R.A), Kristin Ann Carr Foundation (C.R.A)
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
ETHICAL APPROVAL AND CONSENT TO PARTICIPATE
Ethical approval was obtained for this study from the Institutional Review Board (IRB) of Memorial Sloan Kettering Cancer Canter.
DATA AVAILABILITY
All data used in this study are available and can be accessed upon reasonable request
REFERENCES
- 1.Antonescu CR, Owosho AA, Zhang L. et al. Sarcomas With CIC-rearrangements Are a Distinct Pathologic Entity With Aggressive Outcome: A Clinicopathologic and Molecular Study of 115 Cases. Am J Surg Pathol. 2017;41(7):941–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yoshimoto M, Graham C, Chilton-MacNeill S. et al. Detailed cytogenetic and array analysis of pediatric primitive sarcomas reveals a recurrent CIC-DUX4 fusion gene event. Cancer Genet Cytogenet. 2009;195(1):1–11. [DOI] [PubMed] [Google Scholar]
- 3.Italiano A, Sung YS, Zhang L. et al. High prevalence of CIC fusion with double-homeobox (DUX4) transcription factors in EWSR1-negative undifferentiated small blue round cell sarcomas. Genes Chromosomes Cancer. 2012;51(3):207–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yoshida A, Goto K, Kodaira M. et al. CIC-rearranged Sarcomas: A Study of 20 Cases and Comparisons With Ewing Sarcomas. Am J Surg Pathol. 2016;40(3):313–23. [DOI] [PubMed] [Google Scholar]
- 5.Sugita S, Arai Y, Aoyama T. et al. NUTM2A-CIC fusion small round cell sarcoma: a genetically distinct variant of CIC-rearranged sarcoma. Hum Pathol. 2017;65:225–30. [DOI] [PubMed] [Google Scholar]
- 6.Sugita S, Arai Y, Tonooka A. et al. A novel CIC-FOXO4 gene fusion in undifferentiated small round cell sarcoma: a genetically distinct variant of Ewing-like sarcoma. Am J Surg Pathol. 2014;38(11):1571–6. [DOI] [PubMed] [Google Scholar]
- 7.Watson S, Perrin V, Guillemot D. et al. Transcriptomic definition of molecular subgroups of small round cell sarcomas. J Pathol. 2018;245(1):29–40. [DOI] [PubMed] [Google Scholar]
- 8.Dissanayake K, Toth R, Blakey J. et al. ERK/p90(RSK)/14-3-3 signalling has an impact on expression of PEA3 Ets transcription factors via the transcriptional repressor capicua. Biochem J. 2011;433(3):515–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Astigarraga S, Grossman R, Diaz-Delfin J, Caelles C, Paroush Z, Jimenez G. A MAPK docking site is critical for downregulation of Capicua by Torso and EGFR RTK signaling. EMBO J. 2007;26(3):668–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jimenez G, Guichet A, Ephrussi A, Casanova J. Relief of gene repression by torso RTK signaling: role of capicua in Drosophila terminal and dorsoventral patterning. Genes Dev. 2000;14(2):224–31. [PMC free article] [PubMed] [Google Scholar]
- 11.Huang SC, Zhang L, Sung YS. et al. Recurrent CIC Gene Abnormalities in Angiosarcomas: A Molecular Study of 120 Cases With Concurrent Investigation of PLCG1, KDR, MYC, and FLT4 Gene Alterations. Am J Surg Pathol. 2016;40(5):645–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kawamura-Saito M, Yamazaki Y, Kaneko K. et al. Fusion between CIC and DUX4 up-regulates PEA3 family genes in Ewing-like sarcomas with t(4;19)(q35;q13) translocation. Hum Mol Genet. 2006;15(13):2125–37. [DOI] [PubMed] [Google Scholar]
- 13.Specht K, Sung YS, Zhang L, Richter GH, Fletcher CD, Antonescu CR. Distinct transcriptional signature and immunoprofile of CIC-DUX4 fusion-positive round cell tumors compared to EWSR1-rearranged Ewing sarcomas: further evidence toward distinct pathologic entities. Genes Chromosomes Cancer. 2014;53(7):622–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Oh S, Shin S, Janknecht R. ETV1, 4 and 5: an oncogenic subfamily of ETS transcription factors. Biochim Biophys Acta. 2012;1826(1):1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kao YC, Sung YS, Chen CL. et al. ETV transcriptional upregulation is more reliable than RNA sequencing algorithms and FISH in diagnosing round cell sarcomas with CIC gene rearrangements. Genes Chromosomes Cancer. 2017;56(6):501–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yoshida A, Arai Y, Kobayashi E. et al. CIC break-apart fluorescence in-situ hybridization misses a subset of CIC-DUX4 sarcomas: a clinicopathological and molecular study. Histopathology. 2017;71(3):461–9. [DOI] [PubMed] [Google Scholar]
- 17.Hung YP, Fletcher CD, Hornick JL. Evaluation of ETV4 and WT1 expression in CIC-rearranged sarcomas and histologic mimics. Mod Pathol. 2016;29(11):1324–34. [DOI] [PubMed] [Google Scholar]
- 18.Smith SC, Palanisamy N, Martin E. et al. The utility of ETV1, ETV4 and ETV5 RNA in-situ hybridization in the diagnosis of CIC-DUX sarcomas. Histopathology. 2017;70(4):657–63. [DOI] [PubMed] [Google Scholar]
- 19.Zhu G, Benayed R, Ho C. et al. Diagnosis of known sarcoma fusions and novel fusion partners by targeted RNA sequencing with identification of a recurrent ACTB-FOSB fusion in pseudomyogenic hemangioendothelioma. Mod Pathol. 2019;32(5):609–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zehir A, Benayed R, Shah RH. et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat Med. 2017;23(6):703–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Benhamida JK, Hechtman JF, Nafa K. et al. Reliable Clinical MLH1 Promoter Hypermethylation Assessment Using a High-Throughput Genome-Wide Methylation Array Platform. J Mol Diagn. 2020;22(3):368–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Koelsche C, Schrimpf D, Stichel D. et al. Sarcoma classification by DNA methylation profiling. Nat Commun. 2021;12(1):498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Aryee MJ, Jaffe AE, Corrada-Bravo H. et al. Minfi: a flexible and comprehensive Bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics. 2014;30(10):1363–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Du P, Zhang X, Huang CC. et al. Comparison of Beta-value and M-value methods for quantifying methylation levels by microarray analysis. BMC Bioinformatics. 2010;11:587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Niu B, Ye K, Zhang Q. et al. MSIsensor: microsatellite instability detection using paired tumor-normal sequence data. Bioinformatics. 2014;30(7):1015–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Solomon DA, Brohl AS, Khan J, Miettinen M. Clinicopathologic features of a second patient with Ewing-like sarcoma harboring CIC-FOXO4 gene fusion. Am J Surg Pathol. 2014;38(12):1724–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Le Loarer F, Pissaloux D, Watson S. et al. Clinicopathologic Features of CIC-NUTM1 Sarcomas, a New Molecular Variant of the Family of CIC-Fused Sarcomas. Am J Surg Pathol. 2019;43(2):268–76. [DOI] [PubMed] [Google Scholar]
- 28.Yang S, Liu L, Yan Y. et al. CIC-NUTM1 Sarcomas Affecting the Spine. Arch Pathol Lab Med. 2022;146(6):735–41. [DOI] [PubMed] [Google Scholar]
- 29.Smith SC, Buehler D, Choi EY. et al. CIC-DUX sarcomas demonstrate frequent MYC amplification and ETS-family transcription factor expression. Mod Pathol. 2015;28(1):57–68. [DOI] [PubMed] [Google Scholar]
- 30.Linger RM, Keating AK, Earp HS, Graham DK. TAM receptor tyrosine kinases: biologic functions, signaling, and potential therapeutic targeting in human cancer. Adv Cancer Res. 2008;100:35–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Karanian M, Kelsey A, Paindavoine S. et al. SRF Fusions Other Than With RELA Expand the Molecular Definition of SRF-fused Perivascular Tumors. Am J Surg Pathol. 2020;44(12):1725–35. [DOI] [PubMed] [Google Scholar]
- 32.Alaggio R, Zhang L, Sung YS. et al. A Molecular Study of Pediatric Spindle and Sclerosing Rhabdomyosarcoma: Identification of Novel and Recurrent VGLL2-related Fusions in Infantile Cases. Am J Surg Pathol. 2016;40(2):224–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Puls F, Carter JM, Pillay N. et al. Overlapping morphological, immunohistochemical and genetic features of superficial CD34-positive fibroblastic tumor and PRDM10-rearranged soft tissue tumor. Mod Pathol. 2022;35(6):767–76. [DOI] [PubMed] [Google Scholar]
- 34.Kumar-Sinha C, Anderson B, Heider A. et al. Clinical Sequencing of High-Grade Undifferentiated Sarcomas: A Case Series and Report of an Aggressive Primary Cardiac Tumor With Multiple Oncogenic Drivers. JCO Precis Oncol. 2020;4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Holland PW, Booth HA, Bruford EA. Classification and nomenclature of all human homeobox genes. BMC Biol. 2007;5:47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Song K, Huang Y, Xia CD, Zhu HQ, Wang J. A case of CIC-rearranged sarcoma with CIC-LEUTX gene fusion in spinal cord. Neuropathology. 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Barresi S, Giovannoni I, Rossi S. et al. A novel BRD4-LEUTX fusion in a pediatric sarcoma with epithelioid morphology and diffuse S100 expression. Genes Chromosomes Cancer. 2021;60(9):647–52. [DOI] [PubMed] [Google Scholar]
- 38.Chinen Y, Taki T, Tsutsumi Y. et al. The leucine twenty homeobox (LEUTX) gene, which lacks a histone acetyltransferase domain, is fused to KAT6A in therapy-related acute myeloid leukemia with t(8;19)(p11;q13). Genes Chromosomes Cancer. 2014;53(4):299–308. [DOI] [PubMed] [Google Scholar]
- 39.Hu W, Wang J, Yuan L. et al. Case Report: A Unique Case of Pediatric Central Nervous System Embryonal Tumor Harboring the CIC-LEUTX Fusion, Germline NBN Variant and Somatic TSC2 Mutation: Expanding the Spectrum of CIC-Rearranged Neoplasia. Front Oncol. 2020;10:598970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lake JA, Donson AM, Prince E. et al. Targeted fusion analysis can aid in the classification and treatment of pediatric glioma, ependymoma, and glioneuronal tumors. Pediatr Blood Cancer. 2020;67(1):e28028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Noch E NB, Chan J, Wolden S, Tap W, Antonescu C, Khakoo Y. A 43 year-old woman with primary central nervous system angiosarcoma with CIC-LEUTX gene rearrangement Neurology 2019;92:P3.6–017. [Google Scholar]
- 42.Aguirre-Ducler A, Gianino N, Villarroel-Espindola F. et al. Tumor cell SYK expression modulates the tumor immune microenvironment composition in human cancer via TNF-alpha dependent signaling. J Immunother Cancer. 2022;10(7). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bailet O, Fenouille N, Abbe P. et al. Spleen tyrosine kinase functions as a tumor suppressor in melanoma cells by inducing senescence-like growth arrest. Cancer Res. 2009;69(7):2748–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sun H, Lin DC, Cao Q. et al. Identification of a Novel SYK/c-MYC/MALAT1 Signaling Pathway and Its Potential Therapeutic Value in Ewing Sarcoma. Clin Cancer Res. 2017;23(15):4376–87. [DOI] [PubMed] [Google Scholar]
- 45.Sharman J, Hawkins M, Kolibaba K. et al. An open-label phase 2 trial of entospletinib (GS-9973), a selective spleen tyrosine kinase inhibitor, in chronic lymphocytic leukemia. Blood. 2015;125(15):2336–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sturm D, Orr BA, Toprak UH. et al. New Brain Tumor Entities Emerge from Molecular Classification of CNS-PNETs. Cell. 2016;164(5):1060–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kojima N, Arai Y, Satomi K. et al. Co-expression of ERG and CD31 in a subset of CIC-rearranged sarcoma: a potential diagnostic pitfall. Mod Pathol. 2022. [DOI] [PubMed] [Google Scholar]
- 48.Pratt D, Kumar-Sinha C, Cieslik M. et al. A novel ATXN1-DUX4 fusion expands the spectrum of 'CIC-rearranged sarcoma' of the CNS to include non-CIC alterations. Acta Neuropathol. 2021;141(4):619–22. [DOI] [PubMed] [Google Scholar]
- 49.Satomi K, Ohno M, Kubo T. et al. Central nervous system sarcoma with ATXN1::DUX4 fusion expands the concept of CIC-rearranged sarcoma. Genes Chromosomes Cancer. 2022. [DOI] [PubMed] [Google Scholar]
- 50.Xu F, Viaene AN, Ruiz J. et al. Novel ATXN1/ATXN1L::NUTM2A fusions identified in aggressive infant sarcomas with gene expression and methylation patterns similar to CIC-rearranged sarcoma. Acta Neuropathol Commun. 2022;10(1):102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Siegfried A, Masliah-Planchon J, Roux FE. et al. Brain tumor with an ATXN1-NUTM1 fusion gene expands the histologic spectrum of NUTM1-rearranged neoplasia. Acta Neuropathol Commun. 2019;7(1):220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fults D, Pedone CA, Morse HG, Rose JW, McKay RD. Establishment and characterization of a human primitive neuroectodermal tumor cell line from the cerebral hemisphere. J Neuropathol Exp Neurol. 1992;51(3):272–80. [DOI] [PubMed] [Google Scholar]
- 53.Lu HC, Tan Q, Rousseaux MW. et al. Disruption of the ATXN1-CIC complex causes a spectrum of neurobehavioral phenotypes in mice and humans. Nat Genet. 2017;49(4):527–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data used in this study are available and can be accessed upon reasonable request