Abstract
Background:
Alternaria alternata and house dust mite exposure evokes IL-33 secretion from the airway epithelium, which functions as an alarmin to stimulate type 2 immunity. Extracellular DNA (eDNA) is also an alarmin that intensifies inflammation in cystic fibrosis, chronic obstructive pulmonary disease, and asthma.
Objective:
We investigated the mechanisms underlying allergen-evoked DNA mobilization and release from the airway epithelium and determined the role of eDNA in type 2 immunity.
Methods:
Human bronchial epithelial (hBE) cells were used to characterize allergen-induced DNA mobilization and extracellular release using comet assays to measure DNA fragmentation, Qubit double-stranded DNA assays to measure DNA release, and DNA sequencing to determine eDNA composition. Mice were used to investigate the role of eDNA in type 2 immunity.
Results:
Alternaria extract rapidly induces mitochondrial and nuclear DNA release from human bronchial epithelial cells, whereas house dust mite extract induces mitochondrial DNA release. Caspase-3 is responsible for nuclear DNA fragmentation and becomes activated after cleavage by furin. Analysis of secreted nuclear DNA showed disproportionally higher amounts of promotor and exon sequences and lower intron and intergenic regions compared to predictions of random DNA fragmentation. In mice, Alternaria-induced type 2 immune responses were blocked by pretreatment with a DNA scavenger. In caspase-3–deficient mice, Alternaria-induced DNA release was suppressed. Furthermore, intranasal administration of mouse genomic DNA with Alternaria amplified secretion of IL-5 and IL-13 into bronchoalveolar lavage fluid while DNA alone had no effect.
Conclusion:
These findings highlight a novel, allergen-induced mechanism of rapid DNA release that amplifies type 2 immunity in airways.
Keywords: Asthma, allergic inflammation, furin, caspase-3, Alternaria, house dust mite
Graphical Abstract
Asthma is often exacerbated by exposure to respiratory viruses or environmental allergens.1-4 However, the underlying mechanisms responsible for initiation or amplification of type 2 immune responses and exacerbation of the disease are poorly understood.1 Recently a significant correlation was discovered between DNA release and exacerbation of type 2 inflammation in humans after rhinovirus infection.5 Furthermore, in mice, rhinovirus exposure induced DNA release from neutrophils and potentially other cell types including epithelial cells, and when the DNA was degraded with DNase exacerbation of the type 2 inflammatory response was blocked.5 Similarly, in patients with treatment-resistant severe asthma, airway inflammation was associated with extracellular DNA (eDNA) within bronchial biopsy samples or sputum specimens, which correlated with decreases in lung function and asthma control.5,6 Thus, eDNA appears to promote type 2 airway inflammation and represents a potential target for reducing virus- and allergen-induced exacerbations of asthma.
Recognition of pathogen-derived or “foreign” DNA is one of the most fundamental mechanisms of host defense,7 and host-derived, or “self,” DNA can elicit strong inflammatory responses.8 Our mechanistic understanding of the contribution of self-DNA in diseases has increased significantly in the past 10 years. Indeed, sensing of self-derived mitochondrial DNA (mtDNA) or nuclear DNA (nDNA) has been implicated in sterile inflammation in various organs, including pancreas, liver, heart, and brain.8 Furthermore, immunologic activities of aluminum-based adjuvants (aluminum salt or alum), which are used widely in human vaccination and mouse models of type 2 immunity, are attributed to self-DNA derived from dying host cells9 or activated CD4+ T cells.10
Multiple cell death pathways, including necroptosis, pyroptosis, and ETosis, have all been proposed as sources of self-DNA by immune and inflammatory cells that contribute to airway inflammation.11 “ETosis” is a term that describes the formation of extracellular traps (ETs) composed of double-stranded genomic DNA or oxidized mtDNA organized as weblike structures associated with antimicrobial molecules, which serve to immobilize and kill microorganisms.12-15 Under conditions of microbial and fungal infection, ET formation has been shown to represent a unique cell death process exhibited by various immune cells (neutrophils, eosinophils, macrophages, and mast cells) and plays an important role in vertebrate innate immunity.16-18 Interestingly, ETs are also produced by root cap cells of plants in response to a variety of microbial and fungal pathogens.19 Perhaps the most extensively characterized form of ETosis in vertebrates occurs in neutrophils, which involves extrusion of DNA and intracellular constituents after nuclear and plasma membrane disruption.12,20 This process, known as suicide NETosis (after neutrophil ET), represents one form of DNA release from neutrophils. A second form, called vital NETosis, was discovered several years ago and involves vesicular release of DNA, which occurs much more rapidly (5-60 minutes) than suicide NETosis.21 Remarkably, neutrophils undergoing vital NETosis remain viable and are capable of continuing neutrophil functions.22-24 Notably, DNA-depleted neutrophil cytoplasts remain capable of tracking, imprisoning, and killing microorganisms including Staphylococcus25 and Candida albicans.26 However, no information is available regarding whether structural cells, such as airway epithelial cells, are capable of releasing nDNA while maintaining viability.
In the present study, we report that airway epithelial cells engage in a distinct form of vital DNA release in response to allergen exposure. Unlike vital NETosis where eDNA forms weblike structures to entrap microorganisms, epithelial cells release mtDNA and nDNA fragments (~1000-5000 bp) that activate signaling pathways that amplify type 2 immunity.
METHODS
Materials
Alternaria alternata and house dust mite (HDM; Dermatophagoides farina) extracts were obtained from Greer Laboratories (Lenoir, NC). Hydrogen peroxide solution, l-glutathione (GSH), N-acetyl-l-cysteine (NAC), PAMAM dendrimer, and mouse genomic DNA were purchased from Sigma-Aldrich (St Louis, Mo). AZ 10417808 (AQZ-1), PD 150606, and decanoyl-RVKR-CMK were procured from Tocris Bioscience (Bristol, United Kingdom). Hanks balanced salt solution (HBSS), acetoxymethyl ester form of fura-2 (fura-2 AM), SYBR Gold, YoYo-1, SYTOX Green, 4′,6-diamidino-2-phenylindole (DAPI), and EDTA were obtained from Thermo Fisher Scientific (Waltham, Mass). Etoposide was acquired from Abcam (Cambridge, United Kingdom). Naphthofluroscein, caspase-3, and calpain small interfering RNA (siRNA), control siRNA (fluorescein conjugate), siRNA transfection reagent, and media were purchased from Santa Cruz Biotechnology (Santa Cruz, Calif). Fluorescently labeled antibodies to CD3 (145-2C11), CD25 (PC51), CD44 (IM7), CD16/CD32 (2.4G2), CD14 (rmC5-3), CD45R/B220 (RA3-6B2), and IgG2a isotype control were purchased from BD Biosciences (San Jose, Calif).
Cell culture
Primary human bronchial epithelial (hBE) cells were purchased from Lonza (Basel, Switzerland) and grown in bronchial epithelial cell growth medium with growth factor supplements (PromoCell, Heidelberg, Germany) at 37°C in a humidified atmosphere of 5% CO2 in air. Immortalized hBE cells were produced after transfection of genes encoding cyclin-dependent kinase-4 and human telomerase reverse transcriptase.27,28 16HBE14o− hBE cells (Sigma) were grown in minimum essential medium with 10% fetal bovine serum on 24 mm, 0.4 xμm pore size transwell polyester membranes (Corning Life Sciences, Lowell, Mass). hBE cells were grown on 2-well chamber slides (Laboratory-Tek, VWR International, Chicago, Ill) for DNA dye experiments, caspase-3 assays, and Ca2+ imaging experiments, or on 12-well plates or for DNA release measurements and comet assays.
In vivo experiments
Six- to 13-week-old female wild-type (WT) BALB/c mice and caspase-3 knockout mice on a BALB/c background were purchased from The Jackson Laboratory (Bar Harbor, Maine). IL-13eGFP mice were the kind gift of Andrew McKenzie (MRC Laboratory of Molecular Biology, Cambridge, United Kingdom).29 Animals were maintained under specific-pathogen–free conditions, and all experiments were performed with the approval of, and following the regulatory guidelines and standards set by, the Institutional Animal Care and Use Committee of Mayo Clinic.
Mice were treated intranasally with 25 μg (submaximal) or 50 μg of Alternaria extract with or without DNA scavenger PAMAM-G3, DNase, or mouse genomic DNA under isoflurane anesthesia. After 1 or 4.5 hours after administration, mice were humanely killed with pentobarbital and the tracheas cannulated in order to perform bronchoalveolar lavage (BAL) using PBS (0.5 mL). BAL fluid samples were stored at −20°C until use in DNA or cytokine ELISA assays.
Alternatively, to examine the roles of eDNA in development of type 2 adaptive immunity, naive WT BALB/c mice were treated intranasally with 10 μg ovalbumin (OVA) plus 50 μg Alternaria extract with or without PAMAM-G3 (100 μg) in 50 μL PBS once on day 0. On days 21, 22, and 23, the mice were challenged intranasally with 10 μg OVA. Twenty-four hours after the last OVA challenge, mice were humanely killed, and BAL fluid, lungs, and plasma were collected. BAL cell numbers were counted, and differentials were determined in Wright-Giemsa–stained cytospin preparations. More than 200 cells were counted using conventional morphologic criteria. The lungs were homogenized in 0.5 mL PBS and centrifuged at 10,000 × g at 40°C for 15 minutes. Cytokine levels in the supernatants of BAL fluids or lung homogenates were measured using ELISA, as described below. The plasma levels of OVA-specific IgE, IgG1 IgG2a, and IgG2b were measured using sandwich ELISA as described previously.30
RNA interference and CRISPR/Cas9 experiments
hBE cells were transfected with caspase-3 siRNAs using siRNA transfection reagent according to the manufacturer’s protocol. Briefly, 0.5 to 1 μg siRNA and control (fluorescein conjugate) siRNA as a positive control in transfection reagent and medium was transfected into the cells after growing them in antibiotic-free growth medium to obtain approximate 50% confluence while incubating them at 37°C in a CO2 incubator (~6 hours).
hBE cells were transfected with furin guide RNAs (Integrated DNA Technologies, Coralville, Iowa) using a Neon Transfection System (Thermo Fisher) following the manufacturer’s guidelines. Cells (1 × 106) in R buffer (Neon kit) were mixed with 4 μg of furin guide RNAs and 4 μg of Cas9 nuclease, then transfected using the Neon Transfection System (1600 V, 20 ms, 3 pulses). Next, cells were incubated in antibiotic-free media for 24 hours, followed by replacement with fresh growth media containing antibiotics for 72 to 96 hours before experiments. The level of knockdown after transfection was confirmed by Western blot analysis.
Detection of eDNA with fluorescent DNA dyes
hBE cells growing on 2-well chamber slides were incubated with fluorescent DNA dyes YoYo-1 (1.0 μmol), SYTOX Green (10 μmol), or DAPI (0.1 μg/mL) in HBSS solution containing 10 mmol HEPES buffer for 15 minutes before exposure to 100 μg/mL Alternaria in HBSS solution for 30 minutes. Fluorescent dye bound to eDNA was visualized with a Nikon Diaphot fluorescence microscope at excitation/emission wavelengths of 358 nm/460 nm (DAPI), 491 nm/509 nm (YoYo-1), and 504 nm/523 nm (SYTOX Green) with a 40× fluorescence objective (Nikon, Tokyo, Japan).
DNA release measurements
hBE cells were treated with 25 to 200 μg/mL Alternaria or HDM extract in HBSS solution for 0.5 hours or at various time points between 0.5 and 12 hours; then extracellular fluid samples were collected to measure eDNA content. HBSS or BAL fluid samples were analyzed with a Qubit double-stranded (ds) DNA high sensitivity assay kit, and DNA concentrations were determined with a Qubit Fluorometer (Thermo Fisher). As a control, the HBSS or BAL sample was treated with DNase 1 (2 IU for 30 minutes at 37°C) to verify DNase sensitivity of the fluorescence signal associated with the DNA concentration in the sample.
Comet assay
DNA fragmentation was examined with a CometAssay Kit (Trevigen, Gaithersburg, Md) in accordance with a previously published protocol.31 Briefly, Alternaria-exposed cells were trypsinized and fixed onto a comet slide with molten LM Agarose (1:10 vol/vol ratio). The cells were then lysed in 40°C lysis solution and the DNA denatured in an alkaline electrophoresis solution (200 nmol NaOH, 1 mmol EDTA) before electrophoresis using the CometAssay ES unit. After washing and drying the slides, DNA was stained with SYBR Gold in TE buffer and then visualized with a Nikon fluorescence microscope with 10× fluorescence objective at excitation/emission wavelengths of 496 nm/522 nm. DNA fragmentation was quantitatively analyzed by CometScore 2.0 software and expressed as percentage DNA in the head or tail.
Caspase-3 activity assay
In situ measurement of Alternaria-induced caspase-3 activation involved the use of Incucyte Caspase-3 dye (Sartorius, Göttingen, Germany) loaded into hBE cells. Cells were incubated with 5 μmol Incucyte dye reagent for 30 minutes with or without caspase-3 or furin inhibitors or after silencing of caspase-3 or knockdown of furin, then exposed to 100 μg/mL Alternaria in HBSS. A Nikon fluorescence microscope with 40× fluorescence objective was used for live-cell imaging at excitation/emission wavelengths of 500 nm/530 nm.
Quantitative PCR
Total eDNA from hBE cells exposed to Alternaria or HDM was isolated using the Quick-DNA Miniprep Plus Kit (Zymo Research, Irvine, Calif). One microgram of total DNA (quantified by the Qubit Fluorometric assay), TaqMan probes for MT-CO1, MT-ND1, or GAPDH (Hs02596864_g1, Hs02596873_s1, Hs02786624_g1), and TaqMan Fast Advanced Master Mix (Thermo Fisher) were used to perform quantitative reverse transcriptase PCR on a Thermo Fisher Step-One-Plus Real-Time PCR System. The threshold cycle (Ct) was used to determine the relative expression level of specific genes.
DNA sequencing and analysis
hBE cells were exposed to Alternaria (100 μg/mL) for 4 hours before collecting extracellular fluid in 3 separate experiments. DNA extraction was accomplished using Quick-DNA Universal Kit (Zymo) and stored in elution buffer (10 mmol Tris-HCl, pH 8.5, 0.1 mmol EDTA) at a concentration of ~4 ng/μL. DNA samples were submitted to the University of Minnesota Genomics Center for sequencing (Illumina MiSeq, 2 × 300 PE, 16 million reads; Illumina, San Diego, Calif). Sample quality control was determined with Fastqc/0.11.732 compiled together with Multiqc. The percentage of duplicate reads was low, and the libraries that were generated possessed good diversity of DNA fragments that were not overamplified during library preparation. Raw paired-end reads were trimmed of Illumina sequencing adapters and low-quality sequences using Trimmomatic v0.33 with the following settings: IL-LUMINACLIP:all_illumina_adapters.fa:2:30:10:2:true LEADING:3 TRAILING:3 SLIDINGWINDOW:4:15 MINLEN:50.33 Reads were aligned to the human ENSEMBL (GRCh38) genome with star/2.7.1a,34 and PCR-duplicated reads were removed using Picard (v2.18.16) MarkDuplicates (broadinstitute.github.io/picard). Genomic analysis was performed by Genrich, a peak caller that analyzes alignment files for significant enrichment, using the default settings (github.com/jsh58/Genrich). Peaks were associated with human ENSEMBL genes from the EnsDb.Hsapiens.v86 R package (R Project; www.r-project.org) using ChIPpeakAnno.35 eDNA peak densities were visualized across the human hg38 chromosomes using karyoploteR.36 Sequence data is available through the Gene Expression Omnibus site at: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE198721.
Connecting gene-associated eDNA peaks with gene expression
Human airway epithelial cell total RNA sequencing data was downloaded from Gene Expression Omnibus (www.ncbi.nlm.nih.gov/geo) study GSE78672 to identify expressed genes. Transcripts per million (TPM)-normalized gene (hg38) expression tables (ENCFF592BLV and ENCFF788JHJ) served as replicates. Gene expression was averaged between replicates and sorted. The top 20,000 expressed genes were then divided into gene expression quantiles: 5045 highly expressed genes (expression > 194 TPM), 5046 mid-to-high expressed genes (expression between 2.68 and 194 TPM), 5046 mid-to-low expressed genes (expression between 0.41 and 2.68 TPM), and low expressed genes (expression between 0.02 and 0.41 TPM). eDNA peaks that were in common between the 3 replicates were associated with the 20,000 top genes. A total of 4157 eDNA peaks were found within the gene promoter or within the body of these highly expressed genes. The percentage of eDNA peaks associated with genes from each expression quantile was plotted. To create a random set of peaks, 4157 eDNA peaks were randomly assigned to genes, and the percentages of the eDNA peaks associated with genes of each expression quantile were plotted.
Western blot analysis
To determine the level of protein knockdown, cells were lysed with radioimmunoprecipitation assay buffer, and total protein was quantified with the Qubit Protein Assay Kit (Thermo Fisher). Protein samples (25 μg) along with Chameleon Duo–prestained protein ladder (LI-COR, Lincoln, Neb) were separated by electrophoresis on Novex 10-20% tricine protein gels in tricine SDS running buffer (125 V, 1 hour) and transferred to a nitrocellulose membrane in Tris-glycine transfer buffer (Thermo Fisher) (25 V, 1.5 hours). After blocking nonspecific binding with Odyssey blocking buffer (LI-COR) for 1 hour, the membrane was incubated overnight with primary antibodies (Santa Cruz) for caspase-3 (sc-7272), furin (sc-133142), and β-actin (sc-69879), followed by incubation with IRDye 680RD secondary antibodies (LI-COR) for 1 hour. Proteins were visualized with an Odyssey CLx imager (LI-COR); fluorescence intensity of the protein bands was measured by densitometry by ImageJ software (imagej.nih.gov/ij). Protein expression was normalized to β-actin.
Immunocytochemistry
After exposure to Alternaria extract for 0.5 hours, hBE cells grown on chamber slides were fixed in 4% paraformaldehyde for 20 minutes and permeabilized with 0.3% Triton-X for 15 minutes. After a 1-hour blocking step using 3% bovine serum albumin, cells were initially incubated with anti-H2B antibodies (ab1790, Abcam) overnight, then with Alexa Fluor 488 chicken anti-rabbit IgG secondary antibody (Thermo Fisher) for 1 hour. DAPI diluted in PBS was used to label nuclei. An Olympus FV1000 confocal microscope (Olympus, Tokyo, Japan) with a 40× fluorescence objective was used to record images of H2B and DAPI. Western blot analysis established that the primary anybody used for immunocytochemistry detected a protein with a molecular weight corresponding to the size of H2B.
Cytokine release measurements
Measurements of IL-33, IL-17, IFN-γ, IL-5, IL-6, and IL-13 concentrations in BAL fluid after exposure to Alternaria or IL-33 were performed with Quantikine ELISA kits (R&D Systems, Minneapolis, Minn), following the manufacturer’s instructions. Protein concentrations in the BAL fluid were quantitated with a BCA Protein Assay Kit (Bio-Rad, Hercules, Calif).
Fluorescence-activated cell sorting analyses
IL-13 production by lung group 2 innate lymphoid cells (ILC2s) was examined by fluorescence-activated cell sorting (FACS). IL-13eGFP mice (BALB/c background) were exposed to 50 μg per dose of Alternaria extract with or without the DNA scavenger PAMAM-G3. After 4.5 hours, lungs were collected, and single-cell suspensions were obtained by digestion with a cocktail of collagenases (Roche Diagnostics, Indianapolis, Ind) at 0.2 Wünsch units per lung for 60 minutes. Cells were stained with phycoerythrin-conjugated antibodies to CD3, CD14, CD16/CD32, B220, PerCP Cy5.5-conjugated anti-CD44, and APC-conjugated anti-CD25, then analyzed by FACS with a BD FACSAria device. IL-13eGFP expression by lung ILC2s was examined by gating on the lineage-negative (Lin−) CD25+CD44high cells as described previously.37 Data were acquired by Fortessa (BD Biosciences Immunocytometry Systems, Franklin Lakes, NJ) and analyzed by FlowJo v10.6.2 software (Treestar, Ashland, Ore).
Intracellular [Ca2+] measurements
Intracellular Ca2+ concentration ([Ca2+]i) was measured in hBE cells grown on chamber slides for 48 hours, then incubated with 1 μmol fura-2 AM for 1 hour. After washing cells twice and bathing them in HBSS solution, the chamber side was mounted onto the stage of an inverted Nikon fluorescence microscope with a 20× fluorescence objective, and fluorescence ratios were measured at excitation/emission wavelengths of 340-380 nm/510 nm. MetaMorph software was used for image acquisition and data analysis. Relative changes in [Ca2+]i were determined and expressed as the fluorescence ratio when the cells were excited at 340 nm and 380 nm (F340/F380).
Statistical analysis
Data are presented as means ± standard errors of the mean. Statistical analyses and curve fitting were performed by GraphPad Prism v9.0 software (GraphPad Software, La Jolla, Calif). Specific statistical tests are indicated in the figures.
RESULTS
Alternaria extract exposure induces DNA release into extracellular media
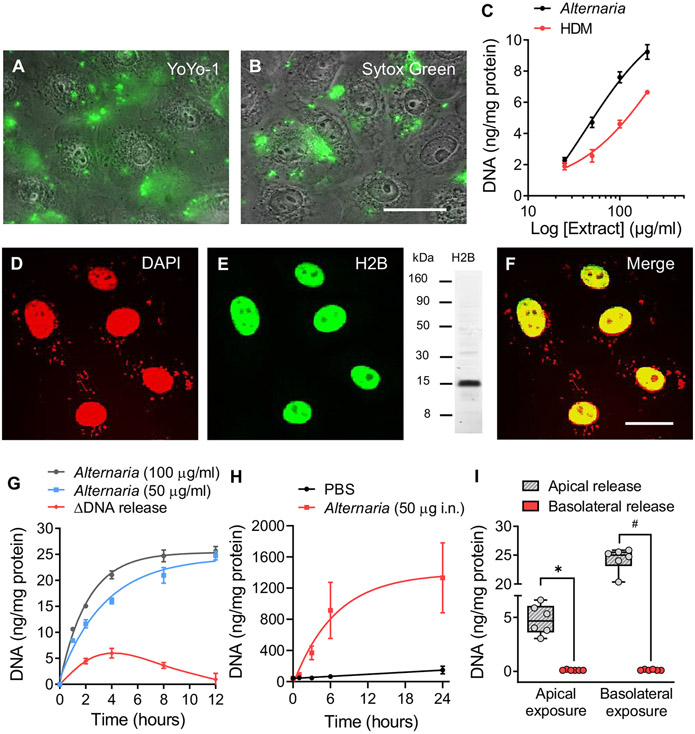
When confluent monolayers of hBE cells were bathed in saline solution containing a membrane-impermeable fluorescent DNA dye (YoYo-1, 1.0 μmol) and stimulated with Alternaria (100 μg/mL) for 30 minutes, fluorescence associated with YoYo-1 binding to eDNA was detected on the apical surface of the cells. Unbound YoYo-1 fluorescence is undetectable in solution; however, fluorescence intensity dramatically increased after binding to dsDNA.38 The merged fluorescence/phase contrast image of hBE cells in Fig 1, A, also shows that the plasma membrane and nuclear membrane remained intact throughout Alternaria exposure; otherwise, nuclei would have displayed YoYo-1 fluorescence. Similarly, SYTOX Green (10 μmol), another membrane-impermeable DNA dye (Fig 1, B), and extranuclear labeling with DAPI (see Fig E1, A, in the Online Repository at www.jacionline.org) revealed the presence of eDNA after treatment with Alternaria (100 μg/mL) for 30 minutes. As a negative control, we stimulated YoYo-1 (apical, 1 μmol)-treated hBE cell monolayers with heat-treated Alternaria extract for 10 minutes at 100°C and detected no fluorescence on the apical surface, indicating that heating blocked the effect of Alternaria on DNA release (Fig E1, B and C). Fig 1, C, compares concentration response results for DNA release induced by Alternaria and HDM extracts using the Qubit dsDNA High Sensitivity Assay Kit. Both Alternaria and HDM extracts evoked concentration-dependent increases in eDNA after 30 minutes of exposure, with half maximal effective concentration values of 42 and 274 μg/mL, respectively. In Fig 1, D-F, Alternaria-stimulated DNA release from the nucleus appears as punctate staining with DAPI. In contrast, the nucleus retained histone 2B, indicating that the nuclear membrane remained intact.
FIG 1.
Alternaria stimulates DNA release. Fluorescence/phase contrast images (original magnification 40×, scale bar = 10 μm, n = 3) showing eDNA with (A) YoYo-1 (1 μmol) and (B) SYTOX Green (10 μmol) after Alternaria (30 minutes). (C) Concentration effects of Alternaria and HDM on eDNA (Alternaria, half maximal effective concentration [EC50] = 42 μg/mL; r2 = 0.89 and HDM, EC50 = 274 μg/mL; r2 = 0.90). (D) DAPI-labeled nuclei 30 minutes after Alternaria (100 μg/mL) treatment (n = 3). (E) Histone 2B (H2B) localization 30 minutes after Alternaria (100 μg/mL) exposure. Western blot showing H2B labeling with antibodies used for immunocytochemistry. (F) Colocalization (yellow) of DNA and H2B in the nucleus (original magnification 60×, scale bar = 5 μm). (G) DNA release at 2 Alternaria concentrations (50 and 100 μg/mL), n = 6 each. ΔDNA release (red) represents differences in DNA accumulation between time intervals after Alternaria (100 μg/mL) exposure. (H) eDNA in BAL fluid of mice (n = 5) treated intranasally with Alternaria (50 μg). (I) Apical and basolateral treatment of 16HBE14o− monolayers exposed to 100 μg/mL Alternaria for 30 minutes (n = 6 each, #P < .0004, *P < .0001; unpaired, 2-tailed ttests with Welch correction).
DNA release is transient and directed across the apical membrane
Time course measurements demonstrated that Alternaria exposure induced transient DNA release (Fig 1, G). The data were fit using a first-order exponential function with time constants (τ) of 3.5 hours (50 μg/mL; n = 6) and 2.1 hours (100 μg/mL; n = 6), with the change in DNA release (ΔDNA at 100 μg/mL) over time indicated. The maximum level of release occurred at 4 hours and ended after 12 hours of continuous Alternaria exposure. The kinetics of DNA release into BAL fluid of mice exposed to Alternaria (50 μg intranasally) is reported in Fig 1, H. Similar to the results with hBE cells, DNA release followed an exponential time course (τ = 6.7 hours; n = 5) that reached a plateau after 12 hours. Sidedness of DNA release occurred in confluent 16HBE14o− cell monolayers grown on membrane filters (Fig 1, I). Alternaria (100 μg/mL) exposure on the apical or basolateral side of the monolayer resulted in DNA release into the apical bathing solution, but not the basolateral solution. Interestingly, basolateral exposure produced a 5-fold increase in the magnitude of apical DNA release compared to apically stimulated monolayers.
Extracellular DNA composition
Approximately equal amounts of mtDNA and nDNA were released into the extracellular media after Alternaria exposure. HDM, however, induced mtDNA release, but unlike Alternaria, nDNA fragmentation was not observed. To determine whether mtDNA was a component of eDNA, PCR analysis was used to probe for 2 mitochondrial genes, cytochrome c oxidase (MT-CO1) and NADH dehydrogenase (MT-ND1). The Ct values for MT-CO1 and MT-ND1 after Alternaria (100 μg/mL) exposure (30 minutes) were 21.8 ± 0.1 (n = 4) and 22.9 ± 0.1 (n = 4), respectively, and 17.1 ± 0.1 (n = 4) and 17.4 ± 0.2 (n = 4), respectively, after HDM (200 μg/mL) exposure, suggesting that HDM-induced eDNA is largely derived from mitochondria. eDNA samples also contained GAPDH (Alternaria: Ct = 26.6 ± 0.1; n = 4; HDM: Ct = 26.9 ± 0.1; n = 4), but at a much lower level than expressed in cells (Ct = 14.5 ± 0.1, n = 4).
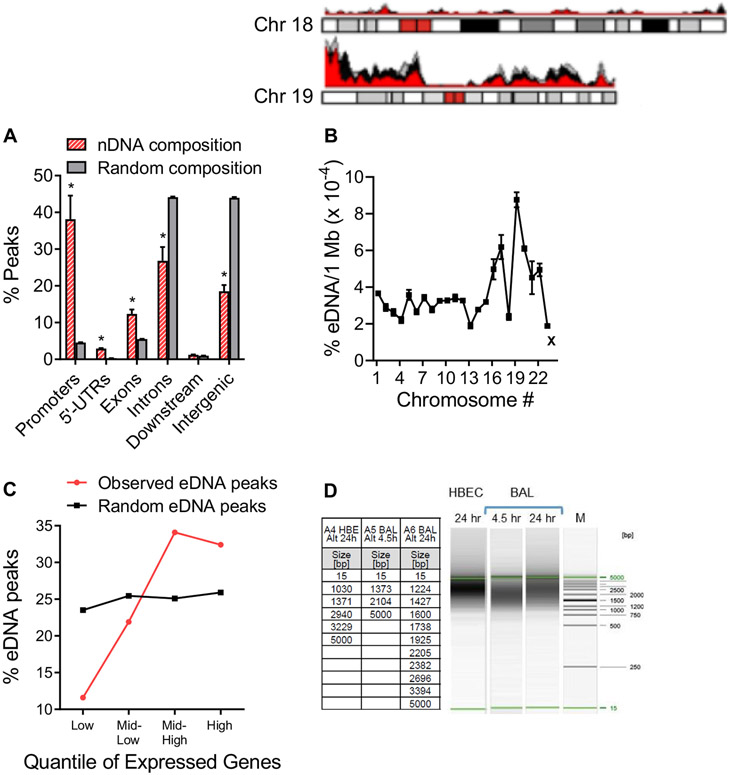
nDNA released by hBE cells in response to Alternaria (100 μg/mL) was isolated from the apical fluid, and Illumina libraries were prepared for sequencing (MiSeq, 300 bp paired-end run). Sequenced eDNA fragments from 3 biological replicates were aligned to the human genome, and regions of genomic enrichment, or peaks, were called within each. Fig 2, A, shows the genomic distribution of 8782 shared eDNA peaks across 6 major regions of the human genome compared to the expected distribution based on cumulative sequence lengths of those regions. The composition of eDNA was significantly different in 5 of the 6 genomic regions, indicating that nDNA was not randomly released as a consequence of cell death. Similarly, Fig 2, B, shows a karyotype analysis that compares the normalized fraction (eDNA/1 Mb) of each chromosome present in eDNA. We noted a greater representation of DNA from smaller chromosomes (eg, chromosome 19) compared to the larger ones and chromosome 18. We also noted consistency between experiments, indicating that this pattern of nDNA representation is reproducible and not to be expected from random DNA fragmentation and release. Fig 2, C, shows the correlation of gene-associated eDNA peaks (defined as peaks within the gene body or at a gene promoter) with highly expressed genes in hBE cells determined from RNA sequencing analysis with ribosomal RNA depletion (RNA sequencing data available at www.encodeproject.org/experiments/ENCSR822SUG). The top 20,000 expressed genes were placed into 4 categories (expression quantiles) on the basis of their level of expression (low, mid-low, mid-high, and high). The percentage eDNA peaks for each quantile was determined and compared to what would be predicted if the distribution was random. The results show a positive correlation between eDNA peaks and increasing levels of gene expression, whereas no correlation was observed for the random distribution of eDNA peaks. To determine the size of DNA fragments released in response to allergen exposure, apical extracellular fluid from hBE cells cultured with Alternaria and BAL fluids from mice exposed to Alternaria were analyzed using high-resolution capillary electrophoresis. DNA was extracted from extracellular media and mouse BAL fluid collected at 4.5 and 24 hours after Alternaria (100 μg/mL or 50 μg intranasally) exposure (Fig 2, D). The DNA fragment sizes observed in apical fluid samples from hBE cells and BAL samples were comparable and ranged from 1000 to 5000 bp. (Note that 15 bp and 5000 bp are size markers serving as internal standards.) The typical DNA laddering pattern for apoptosis (fragment sizes spaced at ~180-200 bp intervals) was not observed in any of these samples.
FIG 2.
Alternaria (50 μg/mL)-induced nDNA release is not random. (A) eDNA peaks from genome regions common to all replicates (red bars, n = 3; Promoters *P = .0343, 5′-UTRs *P = .0028, Exons *P = .0302, Introns *P = .0434, Intergenic *P = .0039; unpaired, 2-tailed t tests with Welch correction) compared to the percentage expected if DNA mobilization and secretion was random (gray bars, n = 3). (B) Karyotype analysis showing percentage of eDNA associated with each chromosome normalized to 1 Mb of eDNA (n = 3). Regions of chromosomes 18 and 19 detected in each of 3 eDNA samples are indicated as overlapping red, gray, and black peaks. “X” refers to the X chromosome. (C) Positive correlation between 4157 gene-associated eDNA peaks and the level of expressed genes in hBE cells determined from RNA sequencing analysis. Plot shows quantiles for the top 20,000 expressed genes. (D) DNA gel showing the range of DNA sizes in extracellular fluid of hBE cultures exposed to Alternaria (100 μg/mL, 24 hours) and BAL samples from mice treated with 50 μg (intranasally [i.n.]) Alternaria (4.5 and 24 hours). Lane M shows the DNA size markers; the table gives the size fragments detected from each gel.
DNA fragmentation is repaired within 24 hours
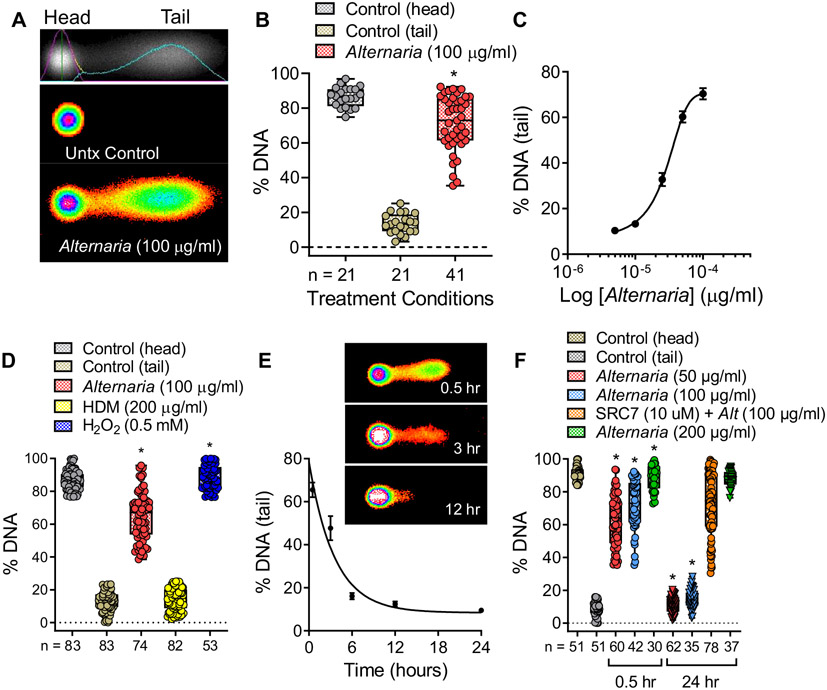
To assess the effects of Alternaria and HDM on nDNA cleavage, comet assays were performed on cultured hBE cells. The assay involves treatment of cells with Alternaria for 30 minutes, embedding them in an agarose gel, then disrupting the plasma and nuclear membranes with an alkaline buffer solution. Next, DNA fragments were separated from nonfragmented DNA by electrophoresis and fluorescently labeled with SYBER Gold. In untreated control cells, nDNA remained within the circular nucleoid disk after electrophoresis (Fig 3, A). However, Alternaria exposure induced tail formation, indicating the presence of DNA fragments migrating away from the nucleoid disk. Fig 3, B and C, shows the percentage of nDNA present in the tail and that the extent of DNA fragmentation depends on Alternaria concentration. In contrast to the effects of Alternaria (100 μg/mL) or H2O2 (0.5 mmol, used as a positive control), HDM exposure up to 200 μg/mL did not induce DNA fragmentation after 30 minutes (Fig 3, D). Furthermore, longer Alternaria exposures resulted in decreases in percentages of DNA present within comet tails, indicating that DNA repair occurs over time in the presence of Alternaria extract (Fig 3, E). The kinetics of the decrease in percentage of DNA in the tail exhibited a first-order exponential decay, reaching a level that was not significantly different from untreated control cells at 12 hours (Fig 3, E). The extent of DNA repair after Alternaria exposure was concentration dependent (Fig 3, F). DNA fragmentation induced by Alternaria up to 100 μg/mL ceased by 24 hours, whereas that induced by 200 μg/mL Alternaria continued. As a positive control, cells were pretreated with SCR7 pyrazine, an inhibitor of nonhomologous end joining repair,39 along with 100 μg/mL Alternaria and no reduction in percentage of DNA within comet tails was observed after 24 hours. This result showed that inhibiting nonhomologous end joining blocks the DNA repair that occurs within 24 hours after Alternaria exposure (Fig 3, F). In addition, treatment with the apoptosis-inducing agent etoposide (100 μmol) or DNA oxidation with H2O2 (0.5 mmol) did not exhibit complete repair by 24 hours (see Fig E2 in the Online Repository at www.jacionline.org). Moreover, hBE cells maintained their normal morphology at 24 hours after exposure to 100 μg/mL Alternaria (see Fig E3 in the Online Repository).
FIG 3.
Alternaria induces nDNA fragmentation followed by DNA repair. (A) Alternaria (100 μg/mL) exposure (30 minutes) produced DNA fragmentation indicated by tail formation in comet assays. (B) Percentage DNA in comet tails after Alternaria treatment (30 minutes, *P < .0001 compared to control). (C) DNA fragmentation increases with increasing Alternaria concentrations (half maximal effective concentration [EC50] = 26.3 ± 4 μg/mL, n = 6; data were fit with a 3 parameter logistic function). (D) In contrast to Alternaria (100 μg/mL) or H2O2 (0.5 mmol), HDM (200 μg/mL) does not induce DNA fragmentation (*P < .0001 compared to control). (E) Representative images of comets at various times after Alternaria (50 μg/mL) exposure. The percentage of DNA in comet tails decreased with time, indicating DNA repair after Alternaria (50 μg/mL) exposure (n = 20-61). (F) Effects of increasing Alternaria (50 [n = 60], 100 [n = 42], and 200 [n = 30] μg/mL) concentration on percentage of DNA in the tail at 0.5 hours and 24 hours after Alternaria exposure (*P < .0001 compared to 50 and 100 μg/mL Alternaria, respectively). Statistical analysis of data in (B), (D), and (F) was performed using a Brown-Forsythe and Welch ANOVA followed by Dunnett T3 posttest.
Oxidative stress–induced increases in [Ca2+]i stimulate DNA release
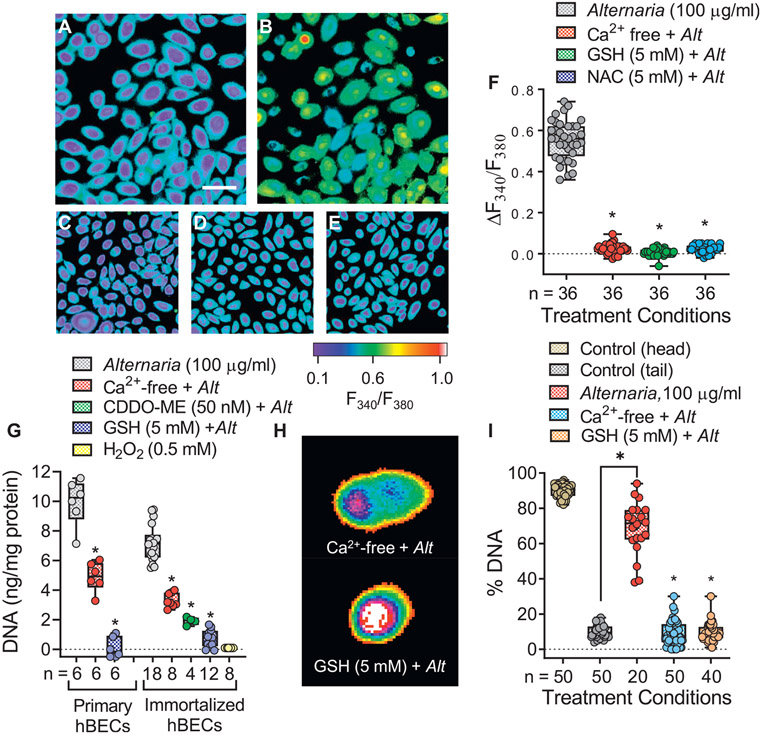
Previous studies have shown that treatment with Alternaria induces reactive oxygen species (ROS) production in hBE cells, resulting in oxidative stress.40 In the present study, Alternaria exposure stimulated Ca2+ uptake in fura-2 AM–loaded hBE cells, which was blocked in the absence of extracellular Ca2+ (Ca2+-free conditions) and when oxidative stress was prevented by pretreating cells with the ROS scavengers 5 mmol GSH or 5 mmol N-acetyl-cysteine (NAC) (Fig 4, A-E). Measurements of fluorescence ratios from fura-2 AM imaging experiments are shown in Fig 4, F. Alternaria-induced DNA release was suppressed by ~50% in both primary and immortalized hBE cells under Ca2+-free conditions and blocked when oxidative stress was prevented in cells pretreated with GSH (5 mmol) or 50 nM CDDO-ME, an Nrf-2 activator that induces antioxidant enzyme expression41 (Fig 4, G). Unlike Alternaria, DNA fragmentation after H2O2 (0.5 mmol) treatment did not produce DNA release (Fig 4, G). Comet assays revealed that inhibiting Alternaria-dependent Ca2+ uptake (Ca2+-free conditions) or preventing oxidative stress by pretreating cells with GSH (5 mmol) blocked DNA fragmentation (Fig 4, H and I).
FIG 4.
Alternaria-induced oxidative stress and increases in [Ca2+], are required for DNA fragmentation and release. (A) Image (original magnification 20×, scale bar = 10 μm) of fura-2 AM–labeled hBE cells before Alternaria exposure. (B) Increases in [Ca2+], after Alternaria (100 μg/mL) treatment (10 minutes). (C-E) Ca2+-free conditions (C) or pretreatment with ROS scavengers. Application of (D) 5 mmol GSH or (E) 5 mmol NAC block Alternaria (100 μg/mL) induced increases in [Ca2+]i. (F) Fura-2 AM fluorescence ratios (340 nm/380 nm) after Alternaria (100 μg/mL) and when hBE cells were maintained under Ca2+-free conditions or pretreated with 5 mmol GSH or NAC (*P < .0001; ANOVA followed by Dunnett posttest). (G) Ca2+-free and 5 mmol GSH inhibits Alternaria (100 μg/mL)-induced DNA release from primary and immortalized hBE cells (*P < .0001; ANOVA followed by Dunnett posttest). CDDO-ME (50 nmol) inhibited DNA release in immortalized hBE cells (ANOVA followed by Tukey posttest), but 0.5 mmol H2O2 alone did not induce DNA release. (H) Images of comets after Alternaria (100 μg/mL) exposure under Ca2+-free conditions and in the presence of 5 mmol GSH. (I) DNA fragmentation was blocked under Ca2+-free conditions and after pretreatment with 5 mmol GSH (*P < .0001; ANOVA followed by Dunnett posttest).
Furin and caspase-3 are involved in nDNA mobilization and release
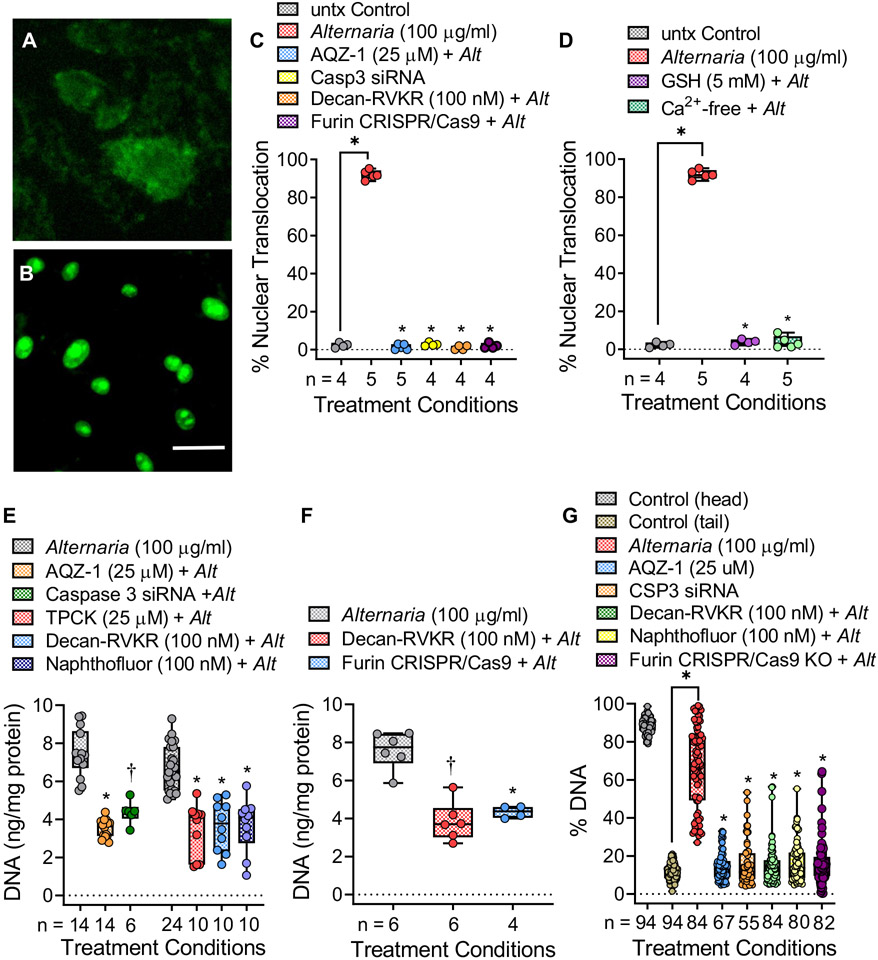
Alternaria-dependent activation of caspase-3 was examined in live cells using a membrane-permeable fluorogenic substrate, which localizes within the nucleus after cleavage by caspase-3 (Fig 5, A and B). The small molecule caspase-3 inhibitor AQZ-1 (AZ10417808)42 and siRNAs targeting caspase-3 (see Fig E4, A, in the Online Repository at www.jacionline.org) completely blocked nuclear translocation of the fluorescent probe, indicating inhibition of caspase-3 catalytic activity (Fig 5, C). Similarly, pretreatment with the furin inhibitor decanoyl-RVKR-CMK43 or CRISPR/Cas9-mediated gene knockdown of furin expression (Fig E4, B and C) also blocked caspase-3 activity. Moreover, Ca2+-free conditions and ROS scavenging with GSH inhibited caspase-3 activation (Fig 5, D). Fig 5, E, shows that DNA release was inhibited by AQZ-1 and by caspase-3 siRNAs as well as by 3 known inhibitors of furin, including TPCK, decanoyl-RVKR-CMK, and naphthofluorescein. Fig 5, F, and Fig E4, B, show that CRISPR/Cas9-mediated knockdown of furin expression inhibited DNA release to the same extent as the furin inhibitor decanoyl-RVKR-CMK. In addition, inhibition of caspase-3 or furin activity/expression blocked Alternaria-induced DNA fragmentation (Fig 5, G). Finally, the amino acid sequence of human pro–caspase-3 (see Fig E5 in the Online Repository) shows the location of a putative furin cleavage site near the C-terminal end of the sequence.
FIG 5.
Alternaria-induced DNA fragmentation and release involves furin-mediated activation of caspase-3. (A and B) hBE cells labeled with fluorescent caspase-3 substrate before (A) and after (B) 100 μg/mL Alternaria (original magnification 40×, scale bar = 10 μm). (C) Nuclear fluorescence detected after Alternaria (100 μg/mL) alone and after caspase-3 inhibitor AQZ-1 (25 μmol), caspase-3 siRNA, decanoyl-RVKR-CMK (100 nmol), and furin knockdown (KD) (*P < .0001). (D) Nuclear fluorescence detected after Alternaria (100 μg/mL) alone and after 5 mmol GSH or Ca2+-free conditions (*P < .0001). (E) AQZ-1 (25 μmol) or caspase-3 siRNAs inhibited ~50% of Alternaria-induced DNA release (AQZ-1: *P < .0004; CSP3 siRNA; †P < .01). Furin inhibition with TPCK (25 μmol), decanoyl-RVKR-CMK (100 nmol), or naphthofluorescein (100 nmol) blocked ~50% of Alternaria-induced DNA release (*P < .0001). (F) Furin KD blocked Alternaria-induced DNA release, similar to decanoyl-RVKR-CMK (*P < .0005, †P < .0001). (G) Comet assay showing inhibition of Alternaria-evoked DNA fragmentation after AQZ-1 (25 μmol) and caspase-3 siRNAs (*P < .0001). DNA fragmentation was blocked by decanoyl-RVKR-CMK (100 nmol), naphthofluorescein (100 nmol), and furin KD (*P < .0001). Statistics (Fig 3, C-G) involved comparisons with Alternaria alone using Brown-Forsythe and Welch ANOVA followed by Dunnett T3 posttest.
Alternaria-evoked DNA release amplifies type 2 immune responses
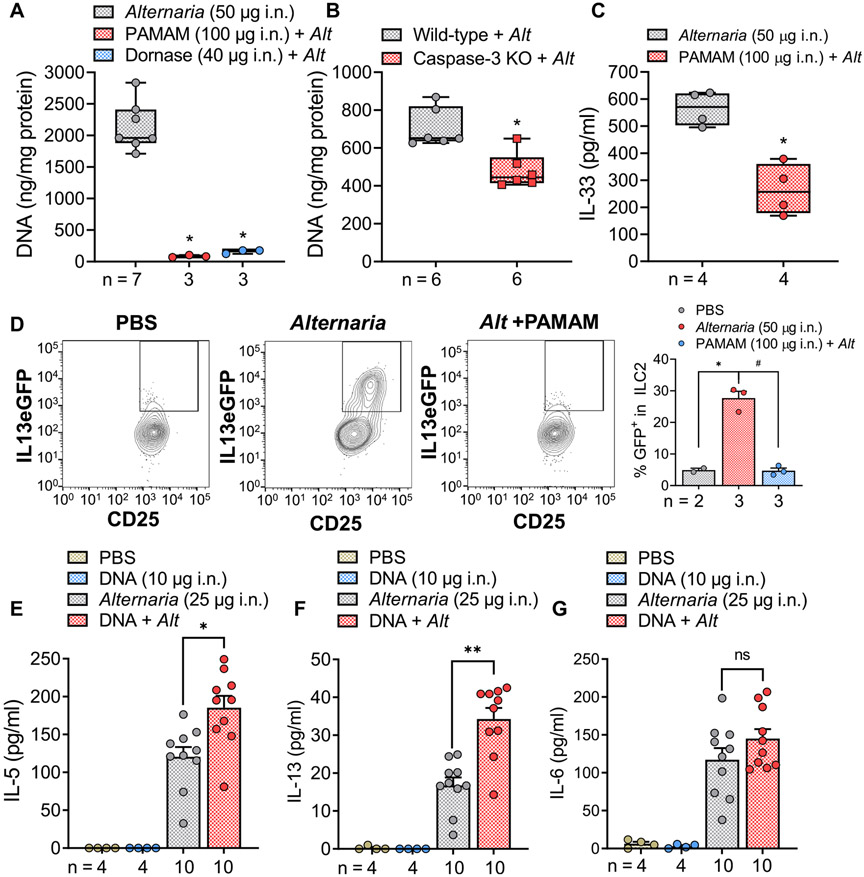
We examined the roles of eDNA in a mouse model of Alternaria-induced innate type 2 immunity. The amount of eDNA in the BAL fluid of mice after treatment with Alternaria for 4.5 hours was dramatically reduced when the DNA scavenger compound PAMAM-G3; ([NH2−(CH2)2NH2]:(G=3); dendri PAMAM (NH2)32), a cationic polymer (mw = 6909), was coadministered with Alternaria (Fig 6, A). A similar result was obtained when DNA was degraded by coadministering DNase with Alternaria. Moreover, caspase-3–deficient mice treated with Alternaria exhibited 32% lower levels of DNA in the BAL fluid after 4.5 hours compared to Alternaria-treated WT mice (Fig 6, B). Fig 6, C, shows that pretreatment with PAMAM-G3 partially inhibited the secretion of IL-33 into BAL fluids. Furthermore, when heterozygous IL-13eGFP mice were exposed to Alternaria (50 μg per dose intranasally) for 4.5 hours, FACS analysis revealed increased green fluorescent protein fluorescence associated with Il13 transcription in ILC2s (Fig 6, D). Alternaria-induced expression of IL-13eGFP was significantly inhibited by administration of PAMAM-G3, indicating that eDNA contributes to Alternaria-induced type 2 immunity. Fig 6, E and F, demonstrates the amplifying effect of mouse genomic DNA (10 μg) coadministered intranasally with a submaximal dose of Alternaria (25 μg) on IL-5 and IL-13 secretion into BAL fluid after 4.5 hours. DNA alone had no effect, but in the presence of Alternaria, significant increases in both cytokines were observed. In contrast, DNA coadministration did not amplify the effect of Alternaria on IL-6 secretion into the BAL (Fig 6, G). Furthermore, Alternaria did not induce IFN-γ or IL-17 secretion, and DNA coadministration did not have any additional effect, suggesting that DNA amplification was selective for IL-5 and IL-13 (see Fig E6, A and B, in the Online Repository at www.jacionline.org).
FIG 6.
DNA stimulates innate type 2 immunity in mice. (A) DNA scavenger PAMAM-G3 (100 μg i.n.) or DNase (40 μg i.n.) reduced (*P < .0001; ANOVA followed by Tukey posttest) DNA content in BAL fluids after Afternaria (50 μg i.n.). (B) Alternaria increases in BAL [DNA] from caspase-3 knockout (KO) mice was lower than in BAL samples from Alternaria-stimulated WT mice (*P = .0024; unpaired t test). (C) Alternaria-stimulated (50 μg i.n.) increases in BAL [IL-33] were reduced (*P = .0019; unpaired, t test) during coadministration of PAMAM-G3 (100 μg i.n.). (D) FACS analysis of lung ILC2 cells in IL-13 eGFP mice exposed to Alternaria (50 μg i.n.) showed increases (*P = .0207) in IL-13 that were blocked after pretreatment with PAMAM-G3 (#P = .0055; compared to Alternaria alone; Brown-Forsythe and Welch ANOVA followed by Dunnett T3 posttest). (E and F) Submaximal (i.n.) Alternaria (25 μg) and mouse genomic DNA (10 μg) amplifies IL-5 and IL-13 secretion into BAL fluid (*P = .0057 for IL-5 and **P < .0001 for IL-13, ANOVA followed by Tukey posttest), while DNA alone had no effect. (G) Alternaria-induced increases in BAL [IL-6] were not amplified by 10 μg of mouse genomic DNA (ANOVA followed by Tukey posttest). i.n., Intranasally.
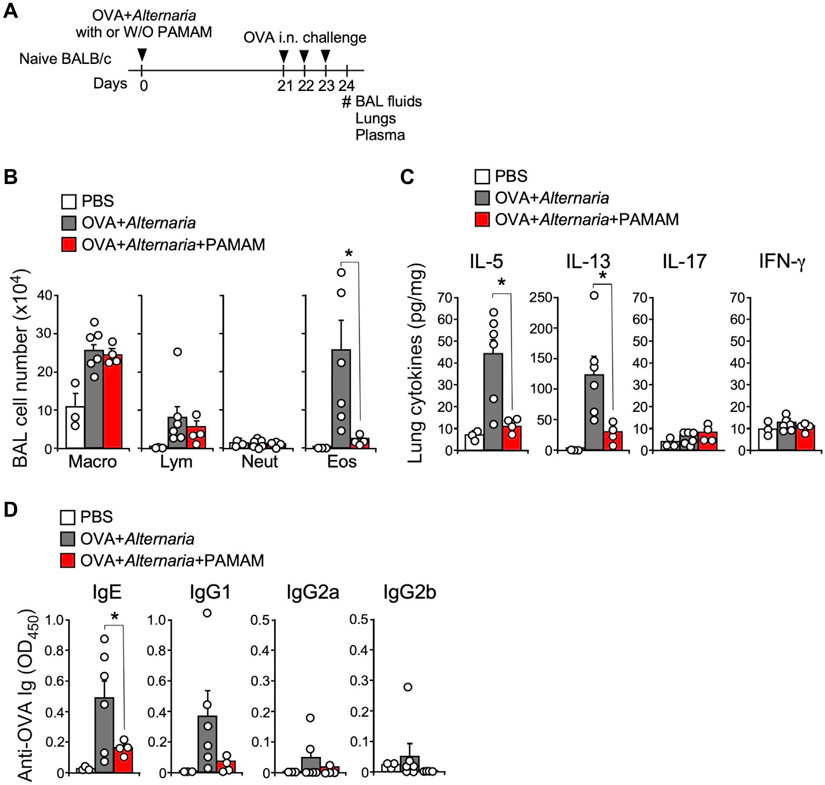
The role of eDNA in Alternaria-induced adaptive type 2 immunity is presented in Fig 7. Mice were initially treated intranasally with OVA (10 μg) and Alternaria (50 μg) at day 0 with or without PAMAM-G3 (100 μg), then challenged intranasally with OVA (10 μg) at days 21, 22, and 23 before BAL fluid, lung, and plasma samples were collected on day 24 (Fig 7, A). Analysis of immune cell counts in the BAL showed that the OVA-induced increase in eosinophils is nearly abolished when mice were exposed to PAMAM-G3 (Fig 7, B). Similarly, OVA-induced increases in lung IL-5 and IL-13 were reduced in the presence of PAMAM-G3, but no effect was observed on the levels of IL-17 and IFN-γ (Fig 7, C). Finally, measurements of OVA-induced IgE showed significant inhibition when PAMAM-G3 was administered. However, no significant OVA induction or PAMAM-G3 inhibition of IgG1, IgG2a, or IgG2b was detected. These findings support the conclusion that Alternaria-induced DNA release plays an important role in amplifying type 2 immunity in mice.
FIG 7.
eDNA amplifies Alternaria-induced adaptive type 2 immunity. (A) Experimental protocol was to examine the effect of DNA scavenging on Alternaria-induced type 2 immune responses; naive mice were exposed i.n. to OVA 1 Alternaria extract with or without PAMAM-G3 and then challenged i.n. with OVA alone. (B) Alternaria-stimulated increases in BAL eosinophil (Eos) counts were significantly reduced in the presence of 100 μg PAMAM-G3 (*P < .05; ANOVA followed by Tukey posttest). (C) OVA-challenge increases in IL-5 and IL-13 were significantly reduced (*P < .05) by pretreatment with PAMAM-G3 (100 μg). (D) Increases in IgE after OVA challenge were significantly inhibited (*P < .05) by pretreatment with PAMAM-G3 (100 μg). i.n., Intranasally.
DISCUSSION
The present study describes a novel response of the airway epithelium to Alternaria and HDM extracts. The results show that Alternaria-induced oxidative stress produces apical but not basolateral release of both nDNA and mtDNA fragments from hBE cells into the extracellular media within minutes after exposure. Kinetic measurements further demonstrated that DNA release terminated after 12 hours, and unlike cells undergoing ETosis, it was not associated with nuclear or plasma membrane disruption.44 Sequence analysis revealed that 5 of 6 genomic regions present within eDNA did not correspond with their predicted distribution within the genome, indicating that generation and release of nDNA fragments was selective. Karyotype analysis showed that DNAs associated with smaller chromosomes, especially chromosome 19, were consistently more highly represented within eDNA compared to larger chromosomes, again indicating a selective pattern of production and release of nDNA fragments. The previously reported sequence of chromosome 19 revealed that it has the highest gene density compared to all other human chromosomes—approximately twice that of the genome-wide average.45 Moreover, sequences from the genes that are most highly expressed in hBE cells showed greater representation within eDNA compared to genes with a low level of expression. These observations suggest that Alternaria-stimulated endonuclease activity targeted regions of the DNA associated with the most highly expressed genes, presumably because of greater access to sites where active transcription was taking place.
The results of the present study show that although Alternaria and HDM extracts elicit rapid release of DNA, HDM appears to induce secretion of mtDNA, not nDNA, as observed in response to Alternaria exposure for 30 minutes. This is likely because short-term HDM exposure did not elicit nDNA cleavage (Fig 3, D), which is necessary for generation of short nDNA fragments observed after Alternaria exposure. However, a previous study showed that treatment of BEAS-2B cells with 200 μg/mL HDM for 6 hours caused double-stranded breaks (DSBs) in nDNA leading to DNA damage and apoptosis, suggesting that induction of DSBs serves as an underlying driver of asthma pathophysiology.46 The DSBs and cytotoxicity associated with HDM exposure were attributed to the generation of reactive oxygen and nitrogen species. In the present study, we demonstrated that caspase-3–mediated DNA fragmentation was dependent on Alternaria-induced increases in [Ca2+]i and activation of furin (Fig 5, C and D), resulting in pro–caspase-3 activation by a noncanonical mechanism. This effect depended on a persistent increase in [Ca2+]i, which occurs after Alternaria exposure, but not when cells are treated with HDM.31 Persistent elevations in [Ca2+]i may also be important in supporting nDNA release across the plasma membrane, but not mtDNA secretion evoked by HDM exposure.
Inhibition of DNA fragmentation and DNA release by pretreatment with GSH and the Nrf-2-activating compound CDDO-ME suggested a role for ROS in the epithelial response to Alternaria. Interestingly, ROS production induced by H2O2 also caused DNA fragmentation, but not DNA release (Fig 4, G). This result suggests that oxidative stress alone was not sufficient to trigger DNA release, and that Alternaria must act by other mechanisms to facilitate trafficking and delivery of DNA fragments into the extracellular media. Importantly, Alternaria-induced DNA fragmentation and release was transient and ceased within 24 hours, whereas proapoptotic agents, such as etoposide, induced continuous DNA fragmentation over the same time period. It is also notable that the nature of Alternaria-induced nDNA release from the airway epithelium is distinct from nDNA ejection that occurs after immune cell ETosis. The long strands of nDNA and associated antimicrobial molecules that are ejected during vital and suicide NETosis or after catapult release from eosinophils form weblike structures that serve to entrap and kill microorganisms.12,14,17,21,23 Hence, ETosis does not involve extensive nDNA fragmentation before ejection from the nucleus. In contrast, the much smaller fragments of nDNA released by airway epithelial cells exposed to Alternaria require prior activation of caspase-3 and DNA fragmentation factor before DNA fragments exit the cell. Furthermore, fragments of nDNA would not be effective at entrapment of microorganisms, and thus it is unlikely that they would play a significant role in antimicrobial defense. However, evidence from the present study indicates that these smaller nDNA fragments appear to have a signaling role, serving to amplify type 2 immune responses that underlie allergic inflammation.
Inhibition of DNA cleavage and extracellular release occurred when caspase-3 activity was blocked by (1) treatment with a selective nonpeptide inhibitor, (2) transfection with siRNAs to reduce caspase-3 protein expression, and (3) removal of extracellular Ca2+. Previous studies showed that caspase-3 activation leads to nuclear translocation and cleavage of DNA fragmentation factor to generate an active endonuclease that catalyzes DNA cleavage by creating DSBs in nDNA.47,48 Furthermore, prior studies showed that sustained increases in [Ca2+]i initiated cleavage and activation of pro–caspase-3 independent of apoptosis.49,50 Ca2+-dependent cleavage of pro–caspase-3 produced a catalytically active protein of ~25 kDa; however, the protease responsible for noncanonical, Ca2+-dependent caspase-3 activation remained unidentified.50 In the present study, we demonstrate that selective inhibitors of furin, along with knockdown of furin expression using CRISPR/Cas9 gene editing, blocked Alternaria-evoked activation of caspase-3 and inhibited both nDNA fragmentation and nDNA release. Importantly, the amino acid sequence of human pro–caspase-3 contains a consensus furin cleavage site (R-X-X-RKY ↓ V) located 34 amino acids upstream of the C terminus, and cleavage at this site produces a protein product containing an intact catalytic site with predicted molecular mass of ~25 kDa.51 These findings indicate that furin functions as a caspase-3 activator and serves as a critical enzyme necessary for noncanonical activation of caspase-3 by Alternaria and potentially other allergens. Interestingly, a furin cleavage site consisting of polybasic residues was also identified in the spike protein of severe acute respiratory syndrome coronavirus 2, the coronavirus responsible for the recent coronavirus disease 2019 global pandemic. Cleavage of the spike protein by furin was shown to be important in facilitating infection and pathogenesis of the virus.52
Previous studies have shown that alarmins, such as ATP, uric acid, and IL-33, initiate type 2 immunity.53 The present study demonstrated that Alternaria-induced DNA mobilization and release amplified type 2 immunity in mice. Intranasal administration of Alternaria produced time-dependent increases in DNA within the BAL fluid that reached a maximum between 12 and 24 hours, similar to the kinetics of DNA release from hBE cells. Furthermore, analysis of the DNA showed that the size fragments in the BAL fluid were within the same range as observed in the extracellular media from Alternaria-stimulated hBE cells (Fig 2, D). Treatment of the BAL fluid with a DNA scavenger (PAMAM-G3) or with DNase abolished DNA detection within the BAL samples. Furthermore, Alternaria-stimulated DNA release into BAL fluid was partially inhibited in caspase-3–deficient mice, which was similar to results from hBE cells where caspase-3 expression was reduced by RNA interference or when its activity was inhibited with a selective, nonpeptide inhibitor. FACS analysis of IL-13eGFP mice showed that Alternaria exposure induced expression of IL-13 in ILC2s and pretreatment with a DNA scavenger blocked Alternaria-induced IL-13 expression, indicating a role for eDNA in amplifying innate type 2 immunity. Similarly, IL-33 release into the BAL fluid was inhibited after pretreatment with a DNA scavenger. The potentiating effects of eDNA on innate type 2 cytokine production were also demonstrated by coadministration of mouse genomic DNA with submaximal doses of Alternaria. Treatment with DNA alone had no effect on IL-5 or IL-13 secretion into the BAL fluid; however, in the presence of Alternaria, a significant increase in the secretion of these cytokines was observed. These findings imply that self-DNA released after allergen exposure enhances the secretion of IL-33 and promotes type 2 cytokine production by ILC2s in vivo. Further evidence supporting a role for eDNA in type 2 immunity was obtained from OVA sensitization studies in mice, which showed that pretreatment with PAMAM-G3 inhibited the adjuvant activity of Alternaria to promote adaptive type 2 immune responses to the innocuous OVA antigen (Fig 7). Moreover, DNA scavenging also inhibited OVA-stimulated increases in eosinophil counts within BAL fluid as well as the increase in OVA-specific IgE levels detected in plasma. Thus, DNA scavengers may have some therapeutic benefit in reducing allergic inflammation induced by environmental allergens that stimulate DNA release.
The molecular components of Alternaria and HDM extracts responsible for DNA mobilization and release are presently unknown. It is possible that Toll-like receptor (TLR) ligands, such as Alt a 1, may play a role. In an earlier study, Alt a 1, perhaps the most clinically significant allergen secreted by Alternaria, was shown to induce immune responses in human airway epithelial cells that involved binding to TLR2 and TLR4 and their subsequent interactions with adaptor proteins, including MyD88 and TIRAP.54 Likewise, Alternaria-evoked IL-33 and IL-1α expression, TH2 cytokine responses, and IgE antibody production were suppressed in MyD88-deficient mice, although TH2 responses and lung inflammation remained unaltered in TLR4 knockout mice.55 Heat treatment of Alternaria extract inhibited IgE production and reduced TH2 mediator release in C57BL/6 mice,55 and the results of the present study showed that heat treatment abolishes DNA release (Fig E1). However, Alt a 1 is known to be heat stable,56,57 suggesting that it may not promote DNA release. Alternaria extract does contain several other allergens,58 many of them enzymes, including enolase (Alt a 6), mannitol dehydrogenase (Alt a 8), alcohol dehydrogenase (Alt a 10), and glutathione-S-transferase (Alt a 13), which are sensitive to heat inactivation. The extract also contains heat-labile protease activity that also contributes to innate immune responses of airway epithelial cells.59-62 Similarly, several allergens present in HDM extract (such as Der f 1, Der p 1, Der f 3, Der f 6) also exhibit cysteine or serine protease activity,63,64 indicating a common set of enzyme activities in both extracts that might be important in DNA release. Finally, it is also possible that molecular components other than enzymes and proteins within Alternaria and HDM extracts may stimulate DNA release, so further experiments will be needed in order to identify the specific molecules involved.
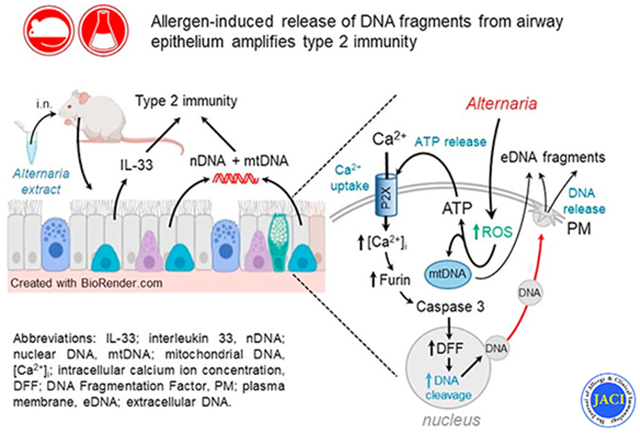
Our proposed model for Alternaria and HDM-induced DNA release from the airway epithelium is summarized in the graphical abstract. Previous studies have demonstrated that Alternaria exposure induces ATP secretion by 2 mechanisms, one consisting of conductive ATP efflux mediated by VDAC-1 (aka voltage-dependent anion channel 1) located in the apical membrane, and a second pathway that involves vesicular ATP release.28,65 Extracellular ATP stimulates P2X7 receptors, leading to Ca2+ uptake and a sustained rise in [Ca2+]i that initially activates furin, which cleaves procaspase-3, resulting in stimulation of caspase-3 activity. Caspase-3 then enters the nucleus, where it activates DNA fragmentation factor to produce DNA fragmentation. DNA exit from the cell does not involve disruption of the nuclear envelope or the plasma membrane and may occur by a mechanism similar to vital NETosis.23,24 This process involves vesicle-mediated DNA transport from the nucleus to the plasma membrane where DNA exocytosis occurs. It is important to emphasize that the DNA release described here is distinct from DNA ejection by ETosis, where long filaments of eDNA play a role in antimicrobial defense.20,44 Instead, the small nDNA and mtDNA fragments released in response to Alternaria appear to function as alarmins, promoting type 2 immune responses that include release of IL-33 and type 2 cytokines, which facilitate type 2 immunity and allergic airway inflammation.
Supplementary Material
Key messages.
hBE cells and mouse airways rapidly release eDNA after Alternaria and HDM exposure.
Alternaria induces noncanonical, furin-mediated activation of caspase-3, which results in DNA mobilization and extracellular release of small (1000-5000 bp) DNA fragments.
In mice, eDNA amplifies IL-5 and IL-13 secretion and type 2 immunity.
Acknowledgments
We thank Kelly O’Grady (Bio-Techne Corporation) for designing the guide RNAs for the furin CRISPR/Cas9 experiments. We also thank Milena Saqui Salces and Nevin Young (University of Minnesota) for their reviews and helpful comments.
Supported by grants from the National Institutes of Health (AI128729) and by the Minnesota Agricultural Experiment Station (AES 0016097).
Abbreviations used
- BAL
Bronchoalveolar lavage
- DAPI
4′,6-Diamidino-2-phenylindole
- DSB
Double-stranded break
- dsDNA
Double-stranded DNA
- eDNA
Extracellular DNA
- ET
Extracellular trap
- FACS
Fluorescence-activated cell sorting
- GSH
l-Glutathione
- hBE
Human bronchial epithelial
- HBSS
Hanks balanced salt solution
- HDM
House dust mite
- ILC2
Group 2 innate lymphoid cell
- mtDNA
Mitochondrial DNA
- NAC
N-Acetyl-l-cysteine
- nDNA
Nuclear DNA
- NET
Neutrophil ET
- OVA
Ovalbumin
- ROS
Reactive oxygen species
- siRNA
small interfering RNA
- TLR
Toll-like receptor
- TPM
Transcripts per million
- WT
Wild type
Footnotes
Disclosure of potential conflict of interest: The authors declare that they have no relevant conflicts of interest.
REFERENCES
- 1.Gern JE. Virus/allergen interaction in asthma exacerbation. Ann Am Thorac Soc 2015;12:S137–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Grayson MH, Feldman S, Prince BT, Patel PJ, Matsui EC, Apter AJ. Advances in asthma in 2017: mechanisms, biologics, and genetics. J Allergy Clin Immunol 2018;142:1423–36. [DOI] [PubMed] [Google Scholar]
- 3.López-Rodríguez JC, Benedé S, Barderas R, Villalba M, Batanero E. Airway epithelium plays a leading role in the complex framework underlying respiratory allergy. J Investig Allergol Clin Immunol 2017;27:346–55. [DOI] [PubMed] [Google Scholar]
- 4.Steinke JW, Borish L. Immune responses in rhinovirus-induced asthma exacerbations. Curr Allergy Asthma Rep 2016;16:78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Toussaint M, Jackson DJ, Swieboda D, Guedán A, Tsourouktsoglou TD, Ching YM, et al. Host DNA released by NETosis promotes rhinovirus-induced type-2 allergic asthma exacerbation. Nat Med 2017;23:681–91. Erratum in: Nat Med 2017;23:1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lachowicz-Scroggins ME, Dunican EM, Charbit AR, Raymond W, Looney MR, Peters MC, et al. Extracellular DNA, neutrophil extracellular traps, and inflammasome activation in severe asthma. Am J Respir Crit Care Med 2019;199:1076–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Briard B, Place DE, Kanneganti TD. DNA sensing in the innate immune response. Physiology 2020;35:112–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ablasser A, Chen ZJ. CGAS in action: expanding roles in immunity and inflammation. Science 2019;363:1–9. [DOI] [PubMed] [Google Scholar]
- 9.Marichal T, Ohata K, Bedoret D, Mesnil C, Sabatel C, Kobiyama K, et al. DNA released from dying host cells mediates aluminum adjuvant activity. Nat Med 2011;17:996–1002. [DOI] [PubMed] [Google Scholar]
- 10.Costanza M, Poliani PL, Portararo P, Cappetti B, Musio S, Pagani F, et al. DNA threads released by activated CD4+ T lymphocytes provide autocrine costimulation. Proc Natl Acad Sci U S A 2019;116:8985–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tait SWG, Ichim G, Green DR. Die another way—non-apoptotic mechanisms of cell death. J Cell Sci 2014;127:2135–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sollberger G, Tilley DO, Zychlinsky A. Neutrophil extracellular traps: the biology of chromatin externalization. Dev Cell 2018;44:542–53. [DOI] [PubMed] [Google Scholar]
- 13.Lood C, Blanco LP, Purmalek MM, Carmona-Rivera C, De Ravin SS, Smith CK, et al. Neutrophil extracellular traps enriched in oxidized mitochondrial DNA are interferogenic and contribute to lupus-like disease. Nat Med 2016;22:146–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Simon D, Simon HU, Yousefi S. Extracellular DNA traps in allergic, infectious, and autoimmune diseases. Allergy Eur J Allergy Clin Immunol 2013;68:409–16. [DOI] [PubMed] [Google Scholar]
- 15.Yousefi S, Stojkov D, Germic N, Simon D, Wang X, Benarafa C, et al. Untangling “NETosis” from NETs. Eur J Immunol 2019;49:221–7. [DOI] [PubMed] [Google Scholar]
- 16.Pertiwi KR, de Boer OJ, Mackaaij C, Pabittei DR, de Winter RJ, Li X, et al. Extracellular traps derived from macrophages, mast cells, eosinophils and neutrophils are generated in a time-dependent manner during atherothrombosis. J Pathol 2018;247:505–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yousefi S, Gold JA, Andina N, Lee JJ, Kelly AM, Kozlowski E, et al. Catapult-like release of mitochondrial DNA by eosinophils contributes to antibacterial defense. Nat Med 2008;14:949–53. [DOI] [PubMed] [Google Scholar]
- 18.Yousefi S, Morshed M, Amini P, Stojkov D, Simon D, Von Gunten S, et al. Basophils exhibit antibacterial activity through extracellular trap formation. Allergy Eur J Allergy Clin Immunol 2015;70:1184–8. [DOI] [PubMed] [Google Scholar]
- 19.Driouich A, Smith C, Ropitaux M, Chambard M, Boulogne I, Bernard S, et al. Root extracellular traps versus neutrophil extracellular traps in host defence—a case of functional convergence? Biol Rev 2019;94:1685–700. [DOI] [PubMed] [Google Scholar]
- 20.Dąbrowska D, Jabłońska E, Garley M, Ratajczak-Wrona W, Iwaniuk A. New aspects of the biology of neutrophil extracellular traps. Scand J Immunol 2016;84:317–22. [DOI] [PubMed] [Google Scholar]
- 21.Pilsczek FH, Salina D, Poon KKH, Fahey C, Yipp BG, Sibley CD, et al. A novel mechanism of rapid nuclear neutrophil extracellular trap formation in response to Staphylococcus aureus. J Immunol 2010;185:7413–25. [DOI] [PubMed] [Google Scholar]
- 22.Zhao W, Fogg DK, Kaplan MJ. A novel image-based quantitative method for the characterization of NETosis. J Immunol Methods 2015;423:104–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yipp BG, Kubes P. NETosis: how vital is it? Blood 2013;122:2784–94. [DOI] [PubMed] [Google Scholar]
- 24.Zhou E, Silva LMR, Conejeros I, Velásquez ZD, Hirz M, Gärtner U, et al. Besnoitia besnoiti bradyzoite stages induce suicidal- and rapid vital-NETosis. Parasitology 2020;147:401–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yipp BG, Petri B, Salina D, Jenne CN, Scott BNV, Zbytnuik LD, et al. Infection-induced NETosis is a dynamic process involving neutrophil multitasking in vivo. Nat Med 2012;18:1386–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Byrd AS, O’Brien XM, Johnson CM, Lavigne LM, Reichner JS. An extracellular matrix–based mechanism of rapid neutrophil extracellular trap formation in response to Candida albicans. J Immunol 2013;190:4136–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ramirez RD, Sheridan S, Girard L, Sato M, Kim Y, Pollack J, et al. Immortalization of human bronchial epithelial cells in the absence of viral oncoproteins. Cancer Res 2004;64:9027–34. [DOI] [PubMed] [Google Scholar]
- 28.Srisomboon Y, Squillace DL, Maniak PJ, Kita H, O’Grady SM. Fungal allergen-induced IL-33 secretion involves cholesterol-dependent, VDAC-1–mediated ATP release from the airway epithelium. J Physiol 2020;598:1829–45. [DOI] [PubMed] [Google Scholar]
- 29.Neill DR, Wong SH, Bellosi A, Flynn RJ, Daly M, Langford TKA, et al. Nuocytes represent a new innate effector leukocyte that mediates type-2 immunity. Nature 2010;464:1367–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kobayashi T, Iijima K, Radhakrishnan S, Mehta V, Vassallo R, Lawrence CB, et al. Asthma-related environmental fungus, Alternaria, activates dendritic cells and produces potent Th2 adjuvant activity. J Immunol 2009;182:2502–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Srisomboon Y, Ohkura N, Iijima K, Kobayashi T, Maniak PJ, Kita H, et al. Airway exposure to polyethyleneimine nanoparticles induces type 2 immunity by a mechanism involving oxidative stress and ATP release. Int J Mol Sci 2021;22:9071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Andrews S. A quality control tool for high throughput sequence data. Available at: https://www.bioinformatics.babraham.ac.uk/projects. [Google Scholar]
- 33.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 2014;30:2114–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 2013;29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhu LJ, Gazin C, Lawson ND, Pagès H, Lin SM, Lapointe DS, et al. ChIPpeakAnno: a Bioconductor package to annotate ChIP-seq and ChIP-chip data. BMC Bioinformatics 2010;11:237–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gel B, Serra E. KaryoploteR: an R/Bioconductor package to plot customizable genomes displaying arbitrary data. Bioinformatics 2017;33:3088–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tei R, Iijima K, Matsumoto K, Kobayashi T, Lama J, Jacobsen EA, et al. TLR3-driven IFN-β antagonizes STAT5-activating cytokines and suppresses innate type 2 response in the lung. J Allergy Clin Immunol 2021;149:1044–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Licari G, Brevet PF, Vauthey E. Fluorescent DNA probes at liquid/liquid interfaces studied by surface second harmonic generation. Phys Chem Chem Phys 2016;18:2981–92. [DOI] [PubMed] [Google Scholar]
- 39.Yang Z, Chen S, Xue S, Li X, Hu J, Sun Z, et al. Injection of an SV40 transcriptional terminator causes embryonic lethality: a possible zebrafish model for screening nonhomologous end-joining inhibitors. Onco Targets Ther 2018;11:4945–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Uchida M, Anderson EL, Squillace DL, Patil N, Maniak PJ, Iijima K, et al. Oxidative stress serves as a key checkpoint for IL-33 release by airway epithelium. Allergy Eur J Allergy Clin Immunol 2017;72:1521–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Robledinos-Antón N, Fernández-Ginés R, Manda G, Cuadrado A. Activators and inhibitors of NRF2: a review of their potential for clinical development. Oxid Med Cell Longev 2019;2019:1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Scott CW, Sobotka-briner C, Wilkins DE, Jacobs RT, Folmer JJ, Frazee WJ, et al. Novel small molecule inhibitors of caspase-3 block cellular and biochemical features of apoptosis. J Pharmacol Exp Ther 2003;304:433–40. [DOI] [PubMed] [Google Scholar]
- 43.Cheng YW, Chao TL, Li CL, Chiu MF, Kao HC, Wang SH, et al. Furin inhibitors block SARS-CoV-2 spike protein cleavage to suppress virus production and cytopathic effects. Cell Rep 2020;33:108254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wartha F, Henriques-Normark B. ETosis: a novel cell death pathway. Sci Signal 2008;1:23–6. [DOI] [PubMed] [Google Scholar]
- 45.Dunham A, Matthews LH, Burton J, Ashurst JL, Howe KL, Ashcroft KJ, et al. The DNA sequence and analysis of human chromosome 19. Nature 2004;428:522–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chan TK, Loh XY, Peh HY, Tan WNFSD, Tan WNFSD, Li N, et al. House dust mite–induced asthma causes oxidative damage and DNA double-strand breaks in the lungs. J Allergy Clin Immunol 2016;138:84–96. [DOI] [PubMed] [Google Scholar]
- 47.Liu X, Zou H, Slaughter C, Wang X. DFF, a heterodimeric protein that functions downstream of caspase-3 to trigger DNA fragmentation during apoptosis. Cell 1997;89:175–84. [DOI] [PubMed] [Google Scholar]
- 48.Widlak P, Garrard WT. Discovery, regulation, and action of the major apoptotic nucleases DFF40/CAD and endonuclease G. J Cell Biochem 2005;94:1078–87. [DOI] [PubMed] [Google Scholar]
- 49.Juin P, Pelletier M, Oliver L, Tremblais K, Grégoire M, Meflah K, et al. Induction of a caspase-3–like activity by calcium in normal cytosolic extracts triggers nuclear apoptosis in a cell-free system. J Biol Chem 1998;273:17559–64. [DOI] [PubMed] [Google Scholar]
- 50.Pelletier M, Oliver L, Meflah K, Vallette FM. Caspase-3 can be pseudo-activated by a Ca2+-dependent proteolysis at a non-canonical site. FEBS Lett 2005;579:2364–8. [DOI] [PubMed] [Google Scholar]
- 51.Duckert P, Brunak S, Blom N. Prediction of proprotein convertase cleavage sites. Protein Eng Des Sel 2004;17:107–12. [DOI] [PubMed] [Google Scholar]
- 52.Johnson BA, Xie X, Bailey AL, Kalveram B, Lokugamage KG, Muruato A, et al. Loss of furin cleavage site attenuates SARS-CoV-2 pathogenesis. Nature 2021;591:293–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hammad H, Lambrecht BN. Barrier epithelial cells and the control of type 2 immunity. Immunity 2015;43:29–40. [DOI] [PubMed] [Google Scholar]
- 54.Hayes T, Rumore A, Howard B, He X, Luo M, Wuenschmann S, et al. Innate immunity induced by the major allergen Alt a 1 from the fungus Alternaria is dependent upon Toll-like receptors 2/4 in human lung epithelial cells. Front Immunol 2018;9:30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Denis O, Vincent M, Havaux X, De Prins S, Treutens G, Huygen K. Induction of the specific allergic immune response is independent of proteases from the fungus Alternaria alternata. Eur J Immunol 2013;43:907–17. [DOI] [PubMed] [Google Scholar]
- 56.Garrido-Arandia M, Silva-Navas J, Ramírez-Castillejo C, Cubells-Baeza N, Gómez-Casado C, Barber D, et al. Characterisation of a flavonoid ligand of the fungal protein Alt a 1. Sci Rep 2016;6:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chruszcz M, Chapman MD, Osinski T, Solberg R, Demas M, Porebski PJ, et al. Alternaria alternata allergen Alt a 1: a unique β-barrel protein dimer found exclusively in fungi. J Allergy Clin Immunol 2012;130:241–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kustrzeba-Wojcicka I, Siwak E, Terlecki G, Wolanczyk-Medrala A, Medrala W. Alternaria alternata and its allergens: a comprehensive review. Clin Rev Allerg Immunol 2014;47:354–65. [DOI] [PubMed] [Google Scholar]
- 59.Chiu LL, Perng DW, Yu CH, Su SN, Chow LP. Mold allergen, Pen c 13, induces IL-8 expression in human airway epithelial cells by activating protease-activated receptor 1 and 2. J Immunol 2007;178:5237–44. [DOI] [PubMed] [Google Scholar]
- 60.Matsuwaki Y, Wada K, White T, Moriyama H, Kita H. Alternaria fungus induces the production of GM-CSF, interleukin-6 and interleukin-8 and calcium signaling in human airway epithelium through protease-activated receptor 2. Int Arch Allergy Immunol 2012;158:19–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bartemes KR, Kita H. Innate and adaptive immune responses to fungi in the airway. J Allergy Clin Immunol 2018;142:353–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kouzaki H, O’Grady SMM, Lawrence CBB, Kita H. Proteases induce production of thymic stromal lymphopoietin by airway epithelial cells through protease-activated receptor-2. J Immunol 2009;183:1427–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Thomas WR. Hierarchy and molecular properties of house dust mite allergens. Allergol Int 2015;64:304–11. [DOI] [PubMed] [Google Scholar]
- 64.Cao H, Liu Z. Clinical significance of dust mite allergens. Mol Biol Rep 2020;47:6239–46. [DOI] [PubMed] [Google Scholar]
- 65.O’Grady SM, Patil N, Melkamu T, Maniak PJ, Lancto C, Kita H. ATP release and Ca2+ signalling by human bronchial epithelial cells following Alternaria aeroallergen exposure. J Physiol 2013;591:4595–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.