Abstract
Aims
Congenital heart disease (CHD) is the most common genetic birth defect, which has considerable morbidity and mortality. We focused on deciphering key regulators that govern cardiac progenitors and cardiogenesis. FOXK1 is a forkhead/winged helix transcription factor known to regulate cell cycle kinetics and is restricted to mesodermal progenitors, somites, and heart. In the present study, we define an essential role for FOXK1 during cardiovascular development.
Methods and results
We used the mouse embryoid body system to differentiate control and Foxk1 KO embryonic stem cells into mesodermal, cardiac progenitor cells and mature cardiac cells. Using flow cytometry, immunohistochemistry, cardiac beating, transcriptional and chromatin immunoprecipitation quantitative polymerase chain reaction assays, bulk RNA sequencing (RNAseq) and assay for transposase-accessible chromatin using sequencing (ATACseq) analyses, FOXK1 was observed to be an important regulator of cardiogenesis. Flow cytometry analyses revealed perturbed cardiogenesis in Foxk1 KO embryoid bodies (EBs). Bulk RNAseq analysis at two developmental stages showed a significant reduction of the cardiac molecular program in Foxk1 KO EBs compared to the control EBs. ATACseq analysis during EB differentiation demonstrated that the chromatin landscape nearby known important regulators of cardiogenesis was significantly relaxed in control EBs compared to Foxk1 KO EBs. Furthermore, we demonstrated that in the absence of FOXK1, cardiac differentiation was markedly impaired by assaying for cardiac Troponin T expression and cardiac contractility. We demonstrate that FOXK1 is an important regulator of cardiogenesis by repressing the Wnt/β-catenin signalling pathway and thereby promoting differentiation.
Conclusion
These results identify FOXK1 as an essential transcriptional and epigenetic regulator of cardiovascular development. Mechanistically, FOXK1 represses Wnt signalling to promote the development of cardiac progenitor cells.
Keywords: Forkhead factors, Foxk1, Cardiovascular development, Wnt signalling
Time of primary review: 34 days
1. Introduction
Congenital heart disease is the most common genetic birth defect affecting approximately 1% of live births and having considerable morbidity and mortality.1,2 Therefore, intense interest has focused on deciphering the regulatory pathways that govern the specification and differentiation of mesodermal progenitors and to use this information to develop therapies for congenital cardiovascular diseases. The cardiovascular system consists of multiple cell lineages including: the haematopoietic, vascular, and cardiomyocyte lineages.3–5 The genesis of each of these lineages from a common germ layer requires precise and co-ordinated regulatory signals to modulate progenitors during embryonic development.3 The transcriptional regulators and signalling pathways that govern the development of these lineages are incompletely defined and warrant further investigation.
Forkhead box (FOX) proteins are a family of evolutionarily conserved transcription factors that share a DNA-binding domain known as the forkhead/winged helix domain.6 Members of this family have been shown to have essential roles during embryogenesis in lineage fate decisions, cell cycle kinetics, aging, metabolism, stem cell regulation, and chromatin remodelling (pioneer factors).6–8 Furthermore, many of these FOX factors have been shown to have important roles in cancer proliferation and tumorigenesis.9 The Foxk family consists of two members, FOXK1 and FOXK2, which have a shared structure.10 FOXK1 was discovered to be a transcription factor restricted to striated muscle (cardiac and skeletal muscles) during development in somites and the heart.11 Previous studies have characterized the role of FOXK1 as an important regulator of myogenic stem cell (satellite cell) proliferation following injury.12–18 While the role of FOXK1 has been extensively studied in skeletal muscle, the role for FOXK1 in cardiac muscle and cardiogenesis is unknown and warrants further investigation.
The Wnt signalling pathway has been shown to play essential roles in the development of the brain, limb, blood, endothelium, and heart.19 The Wnt/β-catenin signalling pathway has been shown to have positive and negative modulatory effects on the specification and the differentiation of mesodermal lineages.20–22 This biphasic role for Wnt/β-catenin signalling has been demonstrated in zebrafish, mouse embryos, and differentiating mouse embryonic stem cells. In this fashion, Wnt signalling has been shown to be procardiogenic in early pre-cardiac mesoderm and inhibitory to cardiogenesis during the later stages of cardiac differentiation.23 Moreover, β-catenin has been reported to be an upstream activator of Isl1 gene expression in the heart.24 While the Wnt/β-catenin signalling pathway has a binary role during cardiogenesis, the overexpression of the Wnt signalling cascade is associated with an expansion of the haematopoietic and endothelial lineages.22 Therefore, the activity and functional role of the Wnt signalling pathway are context dependent and, in part, modulated through protein–protein interactions. For example, bone morphogenic proteins, transforming growth factor betas, and others have been shown to interact with Wnt signalling and have a combinatorial role during development and regeneration.20
In the present study, we characterized the role of FOXK1 during mesodermal development. Using Foxk1 KO embryonic stem cell/embryoid bodies (ESC/EBs), we identify FOXK1 as an important transcriptional and epigenetic regulator of cardiogenesis. Mechanistically, our findings demonstrate that FOXK1 promotes cardiogenesis by repressing the Wnt/β-catenin signalling pathway. This study identifies a novel role for FOXK1 in the regulation and specification of mesodermal lineages during development and the interaction and regulation of distinct signalling pathways during cardiogenesis. These results identify FOXK1 as an important transcriptional and epigenetic regulator of cardiovascular development.
2. Methods
All animal studies were approved by the Institutional Animal Care and Use Committee (IACUC) at the University of Minnesota. All methods were performed in accordance with the relevant guidelines and regulations, and followed the procedures outlined in the Guide for the Care and Use of 9 Laboratory Animals (National Institutes of Health publication number. 85–23, revised 1996).
2.1. Mouse embryonic stem cell lines
iHA-Foxk1 embryonic stem cells (ESCs) (control) harbouring a tetracycline responsive element upstream of HA tagged FOXK1 was generated as previously described.25Foxk1 KO ESCs were isolated using standard techniques by harvesting blastocysts from Foxk1 heterozygous female mice bred with Foxk1 heterozygous males and sacrificed using cervical dislocation.12,26iHA-Foxk1 and Foxk1 KO ESCs were cultured in media containing 15% foetal bovine serum (FBS), 2 mM GlutaMAX, 1 × penicillin/streptomycin, 0.1 mM β-mercaptoethanol, and 1000 U/mL of leukemia inhibitory factor (LIF), at 37°C in 5% CO2 together with irradiated mouse embryonic fibroblasts as a feeder layer. We differentiated control and Foxk1 KO ESCs into EBs using mesodermal differentiation conditions as previously described.27 Briefly, ESCs were separated into single cells using 0.25% trypsin and plated for 50 min to remove fibroblast cells from the feeder layer (de-MEF (mouse embryonic fibroblasts) step). Following de-MEF, ESCs were differentiated in mesodermal media using the shaking method in media containing 15% FBS (Foundation GeminiBio), 1 × penicillin/streptomycin, 1 × GlutaMAX, 50 μg/mL Fe-saturated transferrin, 450 mM monothioglycerol, and 50 μg/mL ascorbic acid in IMDM media (Thermo). iHA-Foxk1 EBs were treated with doxycycline (0.5 μg/mL) in the differentiation media to overexpress FOXK1. D10 EBs harvested for immunohistochemistry were fixed for 10 min at 4°C in 4% paraformaldehyde, washed twice in phosphate buffered saline (PBS), and embedded in optimal cutting temperature solution before cryosectioning.
2.2. Flow cytometry analysis
Harvesting and staining of control and Foxk1 KO EBs were performed as previously described28 and analysed using a FACSAria29 machine. The antibodies used for FACS include: Flk1-APC (1:200, Cat# 560070, Lot# 8298981), platelet-derived growth factor alpha receptor-alpha (PDGFRα)-PE (1:1000, Cat# 4315814, Lot# 2049418), cardiac Troponin T (cTnT) (1:100, Cat# MS-295-P1, Lot# 295P16048), and anti-mouse AlexaFluor 488 (1:400, Cat# 715-485-151, Lot# 94650). Propidium iodide (1:1000, Cat# 1423090, Lot# 1325708) was used to gate for live cells during flow cytometry analysis.
2.3. RNA isolation and quantitative polymerase chain reaction (qPCR) analysis
Total RNA was isolated from control and Foxk1 KO EBs using the RNeasy kit (Qiagen, Cat# 74104) following the manufacturer’s protocol. Briefly, EBs were lysed in RLT-lysis buffer, followed by a column-based purification process and on-column DNA digestion. Complementary DNA (cDNA) was synthesized using the SuperScript IV VILO kit (Thermo Fisher Scientific, Cat# 11756050) following the manufacturer’s protocol. qPCR was performed using ABI Taqman probe sets. Taqman probes used in this study include VIC-labelled GAPDH:4352339E, FAM-labelled Foxk1:Mm01195488_m1, Wnt3a:Mm0437337_m1, and Wnt6:Mm00437353_m1.
2.4. Western blot
Western blots were performed as previously described.30 Briefly, embryoid body lysates were obtained from control and Foxk1 KO EBs at various time points (Days 3, 5, and 7). These were lysed using ice-cold lysis buffer for 30 min and centrifuged at 9300×g for 10 min at 4°C. Equal amounts of protein were loaded on 10% sodium dodecyl sulfate-polyacrylamide gels. Polyvinylidene fluoride membranes were blocked with 5% milk protein and incubated with a rabbit-FOXK1 antibody (1:1000, Cat# sc134550, Lot# D1911) and rabbit-GAPDH antibody (1:1000, Cat# D16H11, Lot# 7) overnight at 4°C. The membrane was subsequently incubated with a goat-anti-rabbit (1:2000, Cat#SC-2004; Lot# G247) horseradish peroxidase-conjugated secondary antibody and visualized using the Pico luminescence kit (Invitrogen) following the manufacturer’s instructions. The protein bands were visualized and imaged using the Image Lab software.
2.5. Analysis of bulk RNAseq
The sequencing reads of the bulk RNAseq data were mapped to the mouse genome (mm10) using Kallisto (0.46.2)31 with default parameters. The read counts data were normalized by DESeq2, followed by differential expression analysis.32
2.6. Generation of ATACseq libraries, sequencing, and analysis
Control and Foxk1 KO EBs were collected at different time periods during differentiation and disaggregated in 0.25% trypsin at 37°C for 3 min incubation with gentle agitation followed by inactivation with culture medium containing 10% FBS. Cells were collected by centrifugation at 500 g for 5 min at 4°C and washed once with ice-cold PBS. ATAC reaction was performed with 50 000 cells as previously described33 using the Tn5 transposase (Illumina) and libraries were created at the University of Minnesota Genome Center. Libraries were then sequenced on a NextSeq Illumina platform (2 × 50 bp) aiming for 25 million reads per sample. The sequencing reads where mapped to the mouse genome (mm10) using Bowtie2 (v2.2.4)34 and ATACseq peaks were called using MACS2 (v2.1.1).35 The ATACseq laying within the blacklisted genomic regions for functional genomics analysis were excluded.36 ChromVAR (v1.10) was used for transcription factor-based chromatin accessibility analysis. We used NucleoATAC (v0.3.4)37 to estimate the nucleosome signals at the Foxk1 motif-binding positions.
2.7. Cardiobeating assays
ESC/EBs were differentiated as described above using mesodermal conditions. At D8, EBs were transferred to a 96-well gelatin-coated flat bottom plate containing EB media at a concentration of 1 EB/well. Individual beating potential (presence or absence of beating) was assessed and quantified at D10.
2.8. Transcriptional assays
Transcriptional assays were performed as previously described.38 Briefly, HEK 293T cells were cultured in 35 mm dishes containing Dulbecco's modified Eagle's medium supplemented with 10% foetal bovine serum and penicillin/streptomycin. 100–200 k cells were seeded and transfected using the lipofectamine reagent and assayed for activity. Reporter assay was performed using the Promega Luciferase Assay System following the manufacturer's instructions. Luciferase activity was analysed with dual-luciferase reporter assay system (Promega), and normalized with the Renilla luciferase. All transfection experiments were performed in triplicate and replicated three times.
2.9. Immunohistochemistry
Cryosectioning and immunohistochemical analyses were performed as previously reported39 using the following antibodies: cTnT (1:100, Cat# MS-295-P1, Lot# 295P16048), anti-chicken AlexaFluor 488 (1:400, Cat# 703545155, Lot# 136424), anti-mouse AlexaFluor 594 (1:400, Cat# 715585150, Lot# 134514), and anti-mouse AlexaFluor 488 (1:400, Cat# 715-485-151, Lot# 94650). The total number of EBs positive for cTnT fluorescence was divided over the total number of EBs in each field of view. The percentage of cTnT+ EBs was plotted (cTnT+ EBs %) and compared between different groups.
2.10. Data analysis
No data were excluded from these studies, and all attempts at replication for standard assays (flow cytometry, qPCR, cardiogenic beating assays, and immunohistochemistry) were successful. Data collection and analysis were not performed in blind to the conditions of the experiments.
3. Results
3.1. FOXK1 regulates mesodermal progenitor cell development
While previous studies have demonstrated that FOXK1 expression during development is restricted to striated muscle (skeletal and cardiac lineages),11 the role for FOXK1 in the development of the cardiac lineage has yet to be characterized. We hypothesized that FOXK1 is an important regulator for mesodermal and cardiac progenitor cells. To test this hypothesis, we engineered Foxk1 KO mouse ESCs to examine the role of FOXK1 during mesoderm and the cardiac lineage development in vitro.
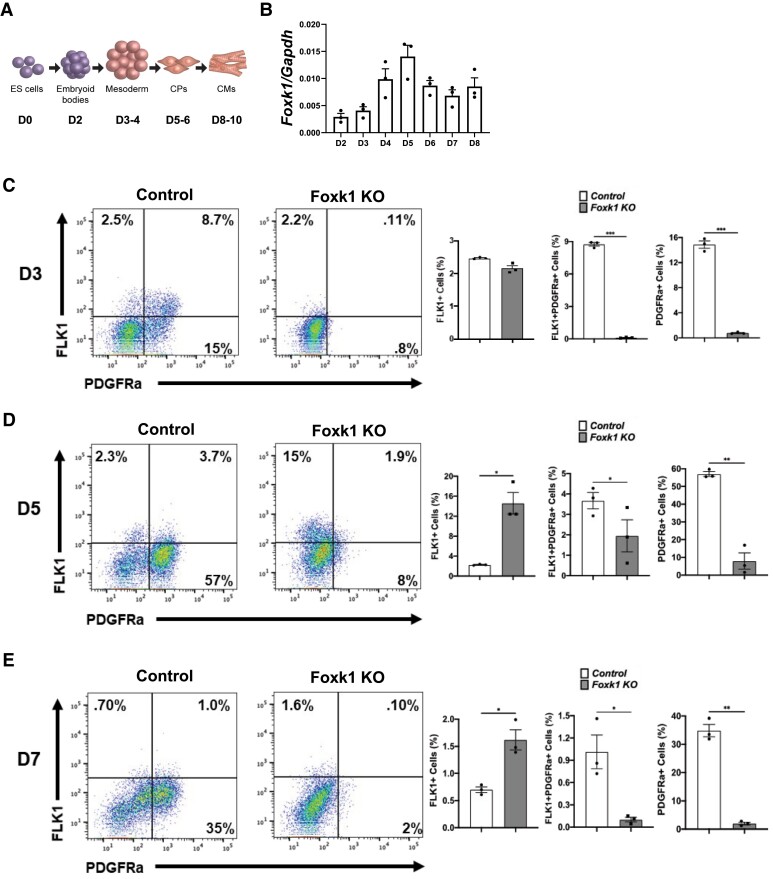
Lineage-tracing studies using the ESC/EB system support the existence of cardiovascular multipotent progenitors that can give rise to endothelial, myocardial, smooth muscle, and haematopoietic lineages.40–44 Using a previously published mesodermal and cardiac differentiation protocol for shaking ESC/EBs,30 we first differentiated control and Foxk1 KO mESCs to examine the expression of FOXK1 (Figure 1A and B and Supplemental material online, Figure S1). Both qPCR and western blot assays demonstrated that FOXK1 expression peaks at approximately Day 5 (D5) of differentiation (Figure 1B and see Supplementary material online, Figure S1), a time period where mesoderm is formed in these EBs and progenitors start differentiating towards the cardiac, haematoendothelial, and skeletal muscle lineages.45,46 To determine whether FOXK1 plays an important role in the development of mesoderm, we analysed the lineage commitment of control and Foxk1 KO EBs at Day 3 (D3) by staining for FLK1 and PDGFRα using flow cytometry (Figure 1C and see Supplementary material online, Figure S2). We observed that in the absence of FOXK1, skeletal myogenic (FLK1 − PDGFRα+) and cardiac (FLK1 + PDGFRα+), but not haematoendothelial (FLK1 + PDGFRα−) progenitors were absent on D3 compared to the control EBs (Figure 1C). To further characterize this phenotype, we examined later time periods of differentiation (Figure 1D and E). Similar to D3, flow cytometry analysis of D5 and D7 EBs demonstrated a significant reduction in cardiac and skeletal muscle progenitors in the Foxk1 KO EBs compared to their respective controls (Figure 1D and E). However, unlike D3, D5, and D7, Foxk1 KO EBs demonstrated a significant increase in blood and vascular progenitor cells (FLK1 + PDGFRα−), suggesting a cell fate change in the cardiac/skeletal muscle progenitor cells that can no longer form due to the absence of FOXK1. Based on these results, we hypothesized that FOXK1 was an important mesodermal regulator of cardiac and skeletal muscle progenitor cell development.
Figure 1.
FOXK1 regulates mesodermal progenitor cell development. (A) Schematic of embryoid body in vitro differentiation and cardiac milestones. (B) Foxk1 transcript expression during EB differentiation from Day 2 (D2) to Day 8 (D8). Note that the expression of Foxk1 peaks at Day 5 (D5). (C–E) Representative flow cytometry profile of control and Foxk1 KO EBs at D3, D5, and D7 of mesodermal differentiation protocol with quantitation of the results. Note that throughout differentiation, there is a significant defect in the ability of Foxk1 null EBs to form mesodermal progenitors, particularly cardiac and skeletal myogenic (n = 3, *P < 0.05). Statistical test: Student’s t-test. Data are presented as mean ± SEM.
3.2. FOXK1 regulates cardiac developmental transcriptional networks
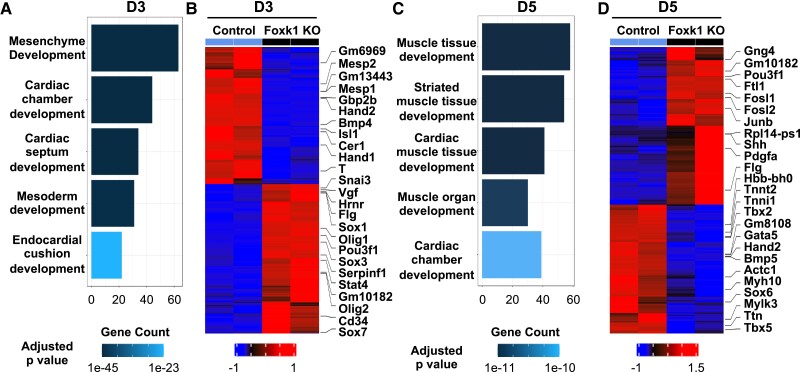
Having established a role for FOXK1 in mesodermal progenitor cell development, we isolated RNA and performed bulk RNA sequencing (RNAseq) from D3 and D5 differentiating control and Foxk1 KO EBs to define the transcriptional networks of these cells before and after mesoderm formation. We identified 644 and 951 up-regulated genes in the control D3 and D5 groups compared to the Foxk1 KO groups, respectively. Pathway analysis of D3 differentiating EBs identified mesodermal and cardiovascular developmental networks in the top five signalling pathways enriched in the control but not the Foxk1 KO EBs (Figure 2A and see Supplementary material online, Figure S3A). Among the top up-regulated genes in the control EBs, we identified important regulators of early cardiovascular development such as Mesp1, Mesp2, Isl1, and Hand2 (Figure 2B and see Supplementary material online, Figure S3B). These are known master regulators of cardiovascular development that are responsible for regulating commitment and differentiation of early progenitors towards the lineage.47 Furthermore, pathway analysis of D5 EBs highlighted cardiac developmental networks in the top five enriched pathways in the control EBs compared to the Foxk1 KO EBs, with the top genes enriched in the control group including important regulators of cardiovascular development such as Hand1, Hand2, and Tbx5 (Figure 2C and D and see Supplementary material online, Figure S3C and D). The transcriptional signature that is significantly affected in the absence of FOXK1 is known to regulate heart morphogenesis in the developing embryo. For example, Hand1 and Hand2 are important regulators of left and right ventricular morphogenesis.47 Therefore, we would hypothesize that loss of FOXK1 would significantly affect cardiovascular morphogenesis. When we examined the Foxk1 KO group, we identified 703 and 1164 up-regulated genes in the D3 and D5 Foxk1 KO groups, respectively (see Supplementary material online, Figure S3). Pathway analysis of the genes up-regulated in the KO group demonstrated that vasculature development was significantly enriched in both the D3 and D5 Foxk1 KO EBs (see Supplementary material online, Figure S3E and F). These results further supported the notion that in the absence of FOXK1, cardiac and myogenic progenitor cells were redirected to a haematoendothelial fate as we observed a significant increase FLK1+ cells in the D5 and D7 Foxk1 KO groups (Figure 1D and E). While previous studies have characterized the role of FOXK1 as a regulator of myogenic stem cell development,15,38 these RNAseq analyses identified a functional role for FOXK1 as an important regulator of other lineages within the mesodermal germ layer such as the cardiac and vascular lineages.
Figure 2.
FOXK1 regulates cardiac developmental transcriptional networks. (A, C) Gene ontology (GO) pathway analysis highlights pathways and development related terms in D3 and D5 control EBs, the x-axis represents the counts of genes in each GO term. The colour scale shows the increased significance of biological processes using the over-representation test with an adjusted P < 0.05. (B, D) The heatmap represents up-regulated and down-regulated in the control EBs vs. the Foxk1 KO EBs at D3 and D5, respectively. The heatmap colour scheme key is provided, with red representing up-regulated and blue representing down-regulated genes.
3.3. FOXK1 modulates dynamic chromatin accessibility of cardiac genes
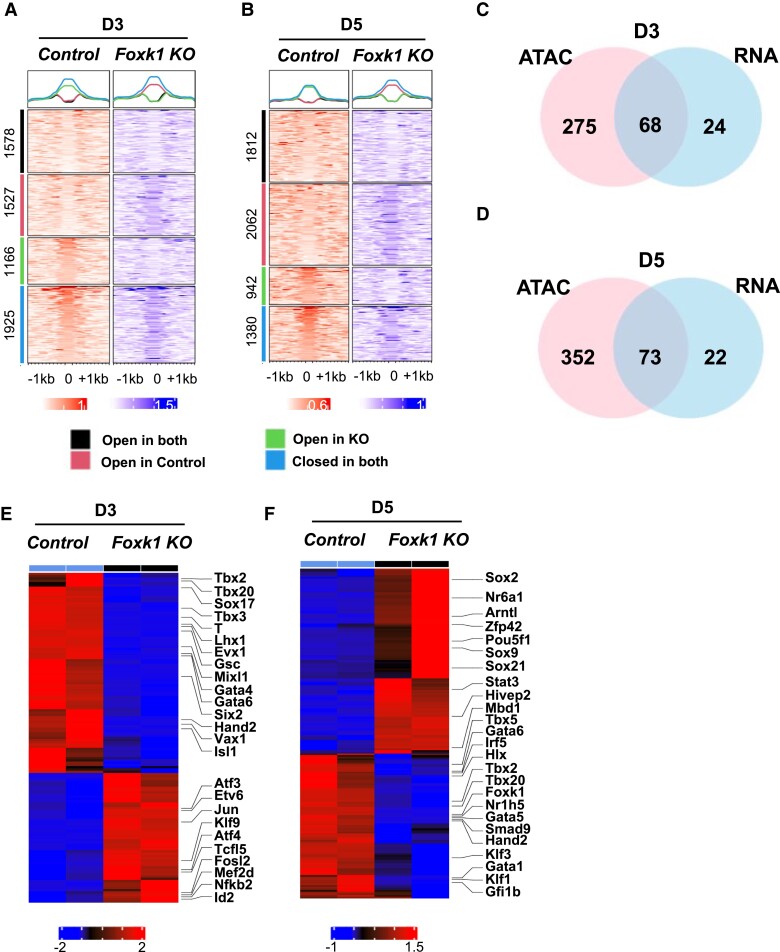
Forkhead transcription factors have been shown to have important epigenetic regulatory roles during development, reprogramming (pioneer factors), and tumorigenesis.7,8 Additionally, previous work has demonstrated that FOXK1 can interact with histone deacetylase 3 to regulate skeletal muscle regeneration and antiviral immune responses.48,49 Therefore, we hypothesized that FOXK1 regulates cardiovascular development by modulating chromatin accessibility in differentiating EBs. Using the assay for transposase-accessible chromatin using sequencing (ATACseq),33 we profiled chromatin changes that occurred during mesodermal and cardiac differentiation in the absence of FOXK1 during EB differentiation (D3 and D5 EBs). We extracted FOXK1 motif-binding positions using motifmatchr and the ATACseq peaks, and based on the nucleosome signal obtained from NucleoATAC within the centre 200 bp region, we divided the ATACseq analysis into four regions (all containing FOXK1 motifs)—nucleosome free regions (NFR) in both control and Foxk1 KO, (2) NFR in control and nucleosome occupied regions (NORs)50 in Foxk1 KO,51 NOR in control and NFR in Foxk1 KO, and NFR in both (Figure 3A and B). More than 43% of the chromatin surrounding regions containing a FOXK1 motif were significantly affected (open or closed) by the absence of FOXK1 at D3 (2693/6196) and D5 (3004/6196) (Figure 3A and B). Our analysis demonstrated reduced chromatin accessibility in 1527 regions at D3 and 2062 regions at D5 in the Foxk1 KO group compared to control (Figure 3A and B). Additionally, the absence of FOXK1 led to an increase in chromatin accessibility in 1166 regions at D3 and 942 regions at D5 in the Foxk1 KO group compared to control (Figure 3A and B). These results suggested that FOXK1 has an important role as an epigenetic regulator in promoting both chromatin relaxation and compaction during differentiation, just like other forkhead transcription factors. However, its role as a promoter of chromatin relaxation is more significant based on the number of regions affected (Figure 3A and B).
Figure 3.
FOXK1 is an epigenetic regulator of cardiac development.52 Enriched heatmap of the nucleoATAC data showing more nucleosome free regions (NFR) in control samples as compared to Foxk1 KO samples at D3 and D5 at the Foxk1-binding sites. We divided the FOXK1-binding sites into four regions, showing NFR in both, NFR in control and nucleosome occupied region50 in Foxk1 KO, NOR in control and NFR in Foxk1 KO, and NFR in both. (C–D) Venn diagram shows the overlap of increased accessibility and up-regulated genes between ATACseq and RNAseq, respectively, in the control group at D3 and D5 over the Foxk1 KO group. (E–F) The heatmap shows commonly expressed transcription factors in both the ATACseq and RNAseq datasets up-regulated and down-regulated in the control EBs over the Foxk1 KO EBs at D3 and D5. The heatmap colour scheme key is provided, with red representing up-regulated and blue representing down-regulated genes.
We performed pathway analysis of genes whose transcription start site is located within 3000 bp within the putative FOXK1-binding sites that have altered chromatin accessibility between control and Foxk1 KO EBs at both D3 and D5. We identified the top 10 pathways that are enriched in nearby regions to FOXK1 DNA-binding sites that are open in D3 control (red group in Figure 3A), open in D3 Foxk1 KO (green group in Figure 3A), open at D5 control (red group in Figure 3B) and open at D5 Foxk1 KO (green group in Figure 3B), respectively. Heart development related pathways were enriched in genes within nearby regions to FOXK1 DNA-binding sites which became closed in the absence of FOXK1 (Foxk1 KO), supporting the hypothesis that FOXK1 plays a functional role in regulating cardiac genes (see Supplementary material online, Figure S4).
Integration of the RNAseq and ATACseq datasets from D3 and D5 EBs enabled the identification of transcription factors whose expression and chromatin accessibility were significantly affected (Figure 3C–F). Analysis of this integration further identified important cardiac regulatory transcription factors whose expression and chromatin accessibility were significantly reduced due to the absence of FOXK1 (Figure 3C–F and see Supplementary material online, Figures S5–S6). Eight transcription factors (TF) were identified whose expression levels and TF-associated chromatin accessibility were both significantly up-regulated in D3 control (see Supplementary material online, Figure S6A), one up-regulated in D3 Foxk1 KO (see Supplemental material online, Figure S6C), seven up-regulated in D5 control (see Supplemental material online, Figure S6B), and one up-regulated in D5 Foxk1 KO (see Supplemental material online, Figure S6D), respectively. The expression levels and TF-associated chromatin accessibility of heart development transcription factors Smad2, Hand1, Hand2, Sox17, Foxc1, Mixl1, Rxrg, Gata4, Foxp1, Rxra, Gata5, Tbx5, and Gli2 were significantly up-regulated in control compared with Foxk1 KO, suggesting that FOXK1 may play an important role of regulating these TFs (see Supplemental material online, Figure S6E and F). These results further supported our hypothesis that FOXK1 has an important role during cardiovascular development and identified a role for FOXK1 as an epigenetic regulator of cardiovascular development by regulating chromatin dynamics.
3.4. FOXK1 regulates cardiogenesis in differentiating EBs
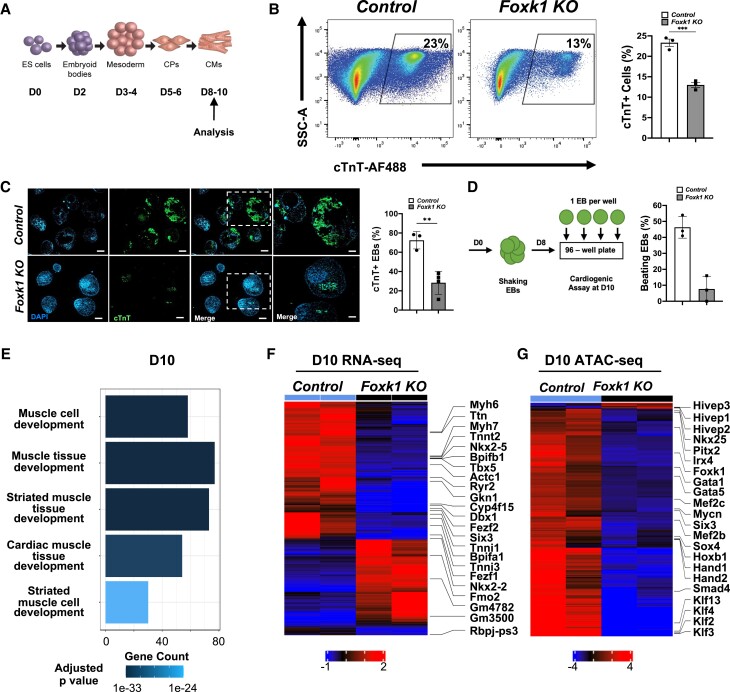
After establishing that FOXK1 regulated cardiac transcriptional and epigenetic networks in mesodermal and cardiac progenitors, we hypothesized that the absence of FOXK1 would affect the differentiation and maturation of early progenitors (Figure 4A). Therefore, we characterized D10 control and Foxk1 KO EBs for the expression of cTnT and their beating potential. Foxk1 KO EBs demonstrated a significant reduction in the number of cTnT+ and beating EBs compared to control EBs (Figure 4B–D). Bulk RNAseq of Foxk1 KO EBs demonstrated a failure to activate gene expression, which was important for cardiac muscle development compared to control EBs based on pathway analysis (Figure 4E). Among the genes affected in the Foxk1 KO group, important cardiac transcription factors (Nkx2-5 and Tbx5) and mature structural/functional cardiac genes (Myh6, Myh7, Ttn, and Ryr2) were significantly down-regulated compared to control EBs (Figure 4F), further demonstrating the important role of FOXK1 during the differentiation and maturation of cardiac progenitor cells. Similar to D3 and D5 EBs, ATACseq analysis of D10 EBs also demonstrated a significant reduction in the chromatin accessibility of transcription factors important for the regulation of later stages of cardiac development such as Mef2c, Klf2, Klf3, and Klf13 (Figure 4G). Pathway analysis of the genes up-regulated in the D10 Foxk1 KO group demonstrated an enrichment of other cellular lineages including limb, bone, cartilage, and vasculature development. These results suggested that the progenitor cells in the Foxk1 KO group were redirected to other lineages and formed terminally differentiated cells (see Supplementary material online, Figure S7). To further validate these observations, we hypothesized that the excess FLK1 + PDGFRα− progenitor cell population observed in the absence of FOXK1 (Figure 1) led to an increase in the expression of vascular progenitor cell markers that would explain the enriched vascular developmental pathways observed in the D10 Foxk1 KO group (see Supplemental material online, Figure S7). Flow cytometry analysis of D5 EBs demonstrated a significant increase in the expression of CD31 and TIE2 markers in the Foxk1 KO compared to the control group (see Supplemental material online, Figure S7). These results demonstrate that FOXK1 regulates not only the early development of cardiac progenitor cells but also their differentiation and maturation at later stages.
Figure 4.
FOXK1 regulates cardiogenesis in differentiating EBs. (A) Schematic of embryoid bodies53 during in vitro differentiation with notation of cardiac milestones. (B) Representative flow cytometry profile of control and Foxk1 KO D10 EBs with quantification of the results. Note the significant decrease in cTnT+ cells in the Foxk1 KO group compared to the control (n = 3, *P < 0.05). (C) Immunohistochemical analysis of D10 EBs demonstrates that Foxk1 KO EBs have perturbed cardiogenesis compared to the control group as assayed by cTnT staining. Quantification of the immunohistochemical results demonstrates normal cardiac differentiation in Day 10 EBs in control group that is significantly reduced in the absence of FOXK1 (n = 3, *P < 0.05). (D) Schematic of control and Foxk1 KO EBs, cardiogenic beating assay, and quantification of results. Note the significant decrease in the number of beating EBs at D10 of differentiation in the Foxk1 KO EB group compared to control (n = 3, *P < 0.05). Statistical test: Student’s t-test. Data are presented as mean ± SEM. (E) Pathway analysis highlights GO pathways and development related terms in D10 control EBs, the x-axis represents the counts of genes in each GO term. The colour scale shows the increased significance of the biological processes using the over-representation test with an adjusted P < 0.05. (F) The heatmap shows significantly (adjusted P < 0.05) differentially expressed transcripts up-regulated and down-regulated with a two-fold between control EBs vs. Foxk1 null EBs at D10. Red represents up-regulation of transcripts and blue represents down-regulation of transcripts. (G) The heatmap represents a significant (adjusted P < 1e−04) change in chromatin accessibility at D10 for control and Foxk1 KO. Red represents an increase in accessibility and blue represents reduced accessibility for the transcription factor.
3.5. FOXK1 regulates Wnt signalling in cardiac progenitor cells
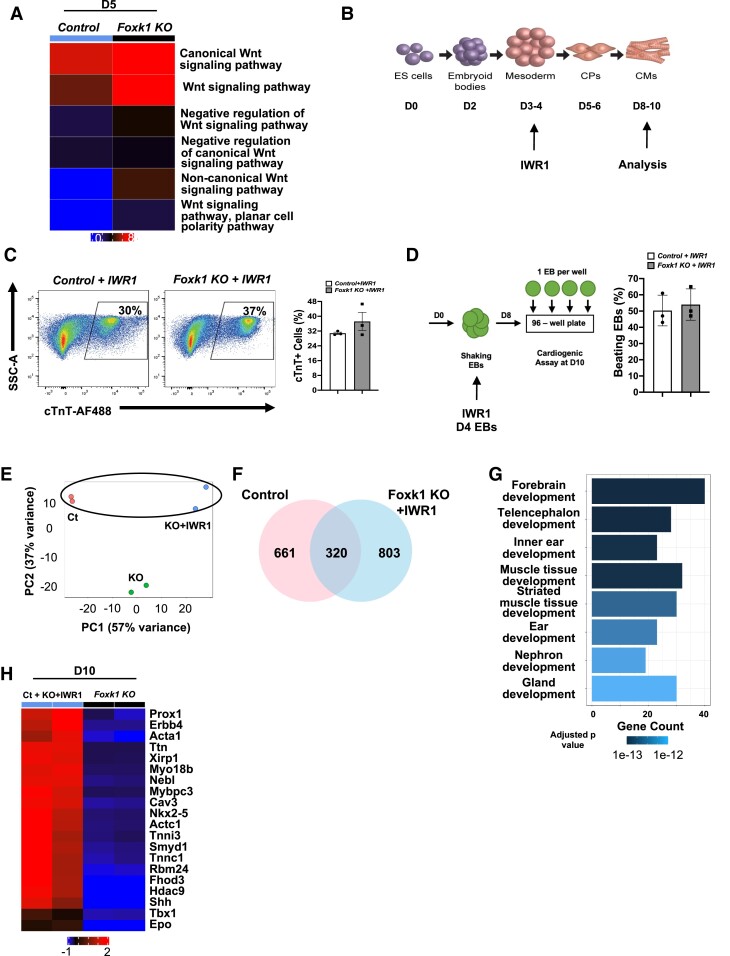
Wnt is an essential signalling pathway for cardiogenesis,23,24 and previous studies have demonstrated that forkhead/winged helix factors can modulate Wnt signalling.54,55 For example, FOXG1 has been shown to repress Wnt5a during brain development while FOXK1 has been reported to modulate Wnt signalling by translocating disheveled proteins to the nucleus.54,55 Therefore, we hypothesized that FOXK1 represses Wnt signalling in cardiac progenitor cells. To test this hypothesis, we first evaluated the temporal expression of Wnt signalling in the EB differentiation system. Our EB differentiation system recapitulated the biphasic role of Wnt signalling observed during cardiogenesis where it was expressed early (D3–D4) and repressed later (D5–D6) during development to allow for cardiogenesis to proceed (see Supplementary material online, Figure S8). These results supported the hypothesis that FOXK1 represses Wnt signalling at D5 (peak expression of Wnt signalling) of differentiation. To examine this hypothesis, we queried our D5 bulk RNA sequencing dataset and determined that in the absence of FOXK1, Wnt signalling pathways were enriched in the Foxk1 KO over the control group (Figure 5A), suggesting that FOXK1 repressed Wnt signalling at distinct stages of cardiogenesis.
Figure 5.
FOXK1 regulates Wnt signalling to promote cardiogenesis. (A) RNAseq pathway analysis of Wnt signalling in D5 EBs comparing control and Foxk1 KO groups. Wnt signalling is significantly up-regulated in the Foxk1 KO group over the control group, and expression persists (in the absence of Foxk1) during differentiation. (B) Schematic of EB in vitro differentiation with the Wnt signalling inhibitor (IWR1) and cardiac milestones noted. (C) Representative flow cytometry profile of control + IWR1 and Foxk1 KO + IWR1 D10 EBs with quantification of the results. Note that no significant differences were observed between the two groups (n = 3). (D) Schematic of the beating assay of control + IWR1 and Foxk1 KO + IWR1 D10 EBs with quantification of the results. Note that no significant differences were observed between the two groups (n = 3). Statistical test: Student’s t-test. Data presented as mean ± SEM. (E) The principal component analysis (PCA) of RNAseq of EB differentiation at D10 shows samples with similar gene expression cluster together. The second principal components (PC2) show similarities between control and Foxk1 KO + IWR1 (Ct, control; KO, Foxk1 KO; KO + IWR1, Foxk1 KO + IWR1). (F) Venn diagram shows 320 genes up-regulated in both Ct and KO + IWR1 samples when compared to Foxk1 KO samples. (G) Top 10 GO pathways and development terms significantly enriched using the genes commonly up-regulated in control and Foxk1 KO + IWR1 conditions. The x-axis represents the counts of genes in each GO term. The colour scale shows the increased significance of the biological processes using the over-representation test with an adjusted P < 0.05. (H) The heatmap shows top 20 genes from two GO terms (muscle tissue development and striated muscle tissue development) commonly up-regulated in control and Foxk1 KO + IWR1 samples. Red represents up-regulation of genes and blue represents down-regulation of genes.
If FOXK1 acts as a repressor of Wnt signalling during cardiogenesis, then we hypothesized that inhibition of the Wnt signalling pathway in the Foxk1 KO EBs would rescue the perturbation of cardiogenesis (Figure 4B–D). To this end, we added a defined Wnt signalling inhibitor (IWR1) to our differentiation conditions at D4 and characterized the EBs at D10 (Figure 5B). The addition of IWR1 to our differentiation conditions rescued both the cTnT+ cell percentage and beating potential of the Foxk1 KO EBs as no significant differences were observed between control and Foxk1 KO groups (Figure 5C and D). RNAseq analysis of D10 EBs demonstrated a significant overlap in the principal component analysis (PC2) between control56 and Foxk1 KO + IWR1 (KO + IWR1) but not the Foxk1 KO (KO) alone (no IWR1) group (Figure 5E). We further examined this overlap by identifying the commonly up-regulated genes between control and Foxk1 KO + IWR1 groups that were not up-regulated in the Foxk1 KO group to identify important regulators of cardiogenesis that were responsible for this rescue (Figure 5F). We identified 320 commonly up-regulated genes unique to these two groups (Figure 5F). Muscle tissue development as well as important regulators of cardiovascular development such as Nkx2-5, Shh, and Ttn were enriched between these two groups (Figure 5G and H).
3.6. FOXK1 is a transcriptional repressor of Wnt signalling
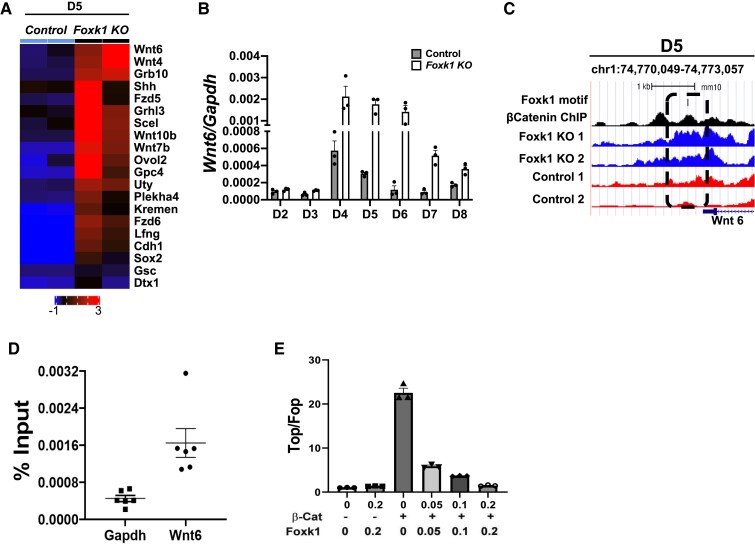
To further characterize the mechanism whereby FOXK1 represses Wnt signalling, we evaluated the role of FOXK1 as a transcriptional regulator of Wnt signalling. To this end, we first identified target genes that were dysregulated at D5 in the absence of FOXK1. Analysis of our D5 bulk RNAseq dataset identified Wnt related genes that were dysregulated during mesodermal and cardiac differentiation in the absence of FOXK1 (Figure 6A). Wnt6 was the top hit of the dysregulated genes, which we validated using qPCR to demonstrate that it was overexpressed not only at D5 but also at later stages of differentiation compared to control EBs (Figure 6A and B). WNT6 is an important regulator of cardiovascular development, and its overexpression results in a hypomorphic heart in Xenopus, suggesting that down-regulation of Wnt6 is essential during cardiogenesis.57 To determine whether FOXK1 is a transcriptional regulator of Wnt6, we examined the upstream region of the Wnt6 gene to identify any potential FOXK1 DNA-binding motifs. FOXK1 DNA-binding motifs were identified and when combined with the ATACseq datasets, we confirmed that the chromatin surrounding this DNA motif was significantly more relaxed in the absence of FOXK1 (Figure 6C). Additionally, this region had a β-catenin DNA-binding site,58 suggesting that in the absence of FOXK1, the chromatin upstream of the Wnt6 gene cannot be closed to other factors such as β-catenin that promote Wnt6 expression. To validate the capacity of FOXK1 to bind to the upstream region of the Wnt6 gene, we performed chromatin immunoprecipitation (ChIP) with a FOXK1 antibody followed by qPCR of the selected region in D5 EBs. ChIP qPCR analysis using FOXK1 demonstrated a 3.6-fold enrichment for FOXK1 in the Wnt6-binding site over the non-specific Gapdh site (Figure 6D). Because of the identified β-catenin DNA-binding site upstream of Wnt6, we also hypothesized that both FOXK1 and β-catenin could compete for binding to repress or activate Wnt signalling pathways, respectively. To this end, we used the TOP/FOP-flash reporter system to evaluate the role of FOXK1 as a transcriptional repressor of Wnt signalling in the presence of β-catenin (Figure 6E). Analysis of this luciferase reporter demonstrated that FOXK1 significantly repressed β-catenin activity in a dose dependent fashion (Figure 6E), further confirming the role of FOXK1 as a transcriptional regulator of Wnt signalling. Collectively, these data demonstrate that mechanistically, FOXK1 acts as a transcriptional and epigenetic repressor of Wnt signalling to regulate cardiovascular development.
Figure 6.
FOXK1 is a transcriptional repressor of Wnt signalling. (A) Heatmap highlighting the top 20 transcription factors associated with the Wnt signalling pathway enriched in the Foxk1 null D5 EB group from the RNAseq dataset. (B) Wnt6 transcript expression during EB differentiation from D2 to D8 in the presence and absence of FOXK1. Note that the expression of Wnt6 remains high at later stages of differentiation compared to the control group where it is down-regulated. (C) A region upstream of Wnt6 (chr1:74,770,049-74,773,057) that contains a FOXK1 DNA motif, β-catenin-binding site, and decreased chromatin accessibility in the absence of Foxk1 compared to control EBs at D5. (D) qPCR analysis using FOXK1 ChIP demonstrates a significant enrichment at the Wnt6 upstream region compared to the Gapdh control region. (E) β-Catenin transactivates the TOP-flash reporter, which is inhibited by FOXK1. Statistical test: Student’s t-test. Data presented as mean ± SEM.
4. Discussion
The heart is the first organ to form and function during development, and its proper formation is crucial for mammals to develop without any congenital anomalies or congenital heart disease.4,47,59 Several transcription factors and signalling cascades have been identified and have an important role during cardiovascular development; however, the mechanisms that govern this process are still unclear. Forkhead/winged helix transcription factors have been shown to regulate key cellular processes such as cell proliferation, differentiation, chromatin remodelling, metabolism, and others.59 We and others have shown that FOXK1 expression is restricted to striated muscle (skeletal and cardiac muscles) during development and that it is essential during skeletal muscle development and regeneration. In the present study, we have used gene disruption strategies, computational genomics, and cellular and molecular techniques to make several fundamental discoveries to decipher an important role for FOXK1 as a regulator of cardiovascular development.
First, we identified an important role for FOXK1 in the regulation of mesodermal progenitor cell development. Transcriptional analysis of these developing progenitors further demonstrated a significant decrease in the expression of cardiovascular developmental pathways and key regulatory transcription factors such as Isl1, Hand1, Hand2, and Tbx5 in the absence of FOXK1. These data identified FOXK1 as a key regulator of cardiovascular development, particularly of first and second heart field (SHF) structures by regulating the expression of these transcription factors.47 While deletion of other forkhead factors can lead to cardiac developmental defects, they appear to be due to secondary effects as their expression is not restricted to developing striated muscle, unlike FOXK1.59
Interestingly, in the absence of FOXK1, progenitor cells that could not form cardiac and skeletal muscle progenitors, instead formed vascular progenitor cells. This observation was further validated with the RNAseq analysis that showed an enrichment of vascular developmental pathways in the cells lacking FOXK1. Similar fate changes in the absence of a transcription factor during mesodermal development has been observed with the pioneer factor ETV2,60 where Etv2 KO mesodermal progenitors preferentially generated cardiac instead of vascular and blood progenitor cells.52 These data suggested that one of the mechanisms by which FOXK1 promoted cardiovascular development was by repressing other lineages such as the vascular lineage.
Our next discovery identified FOXK1 as an epigenetic regulator of cardiovascular development. More than 40% of the chromatin surrounding regions with a FOXK1 DNA-binding motif demonstrated a significant impact in the chromatin dynamics in the absence of FOXK1. More importantly, the absence of FOXK1 led to a decrease in the chromatin accessibility of the same key regulatory cardiovascular development genes identified in our RNAseq analysis (Isl1, Hand1/2, and Tbx5), along with other important regulators (Gata4 and Tbx20). Forkhead factors are known epigenetic regulators that can function by interacting with a chromatin remodeller or through their unique protein structure (resembling that of linker histones) that allows them to interact with heterochromatin and relax the chromatin landscape by displacing linker histones.7,8,49 FOXK1 has been shown to play a role as an epigenetic regulator in skeletal muscle through interactions with histone deacetylases, however, the exact mechanism whereby FOXK1 regulates the chromatin landscape near cardiac genes remains to be elucidated. The SWI/SNF complex has been recently shown to play an important role in cardiac development by assisting in the chromatin remodelling process of different important cardiac regulators.45,46,61 Whether this complex assists FOXK1 or whether FOXK1 remodels chromatin by itself like other forkhead/winged helix factors such as FOXA18 remains to be elucidated.
Another major finding from this work is that mechanistically, we have identified that FOXK1 regulates cardiovascular development by transcriptionally and epigenetically repressing Wnt signalling in cardiac progenitor cells, and we identified Wnt6 as a major downstream target of FOXK1. Absence of FOXK1 during mesodermal and cardiac differentiation led to a significant increase in the expression of Wnt6 that persisted until later stages of differentiation compared to the control group. WNT6 is a known regulator of cardiovascular development whose expression is tightly regulated as continuous overexpression of Wnt6 has been shown to be detrimental for cardiac development.57 The Wnt signalling pathway has been shown to regulate the development of SHF structures such as the outflow tract and right ventricle by regulating the expansion of Isl1 progenitors.62 Isl1 expression, along with that of other SHF regulatory genes such as Hand2, was significantly down-regulated in the bulk RNAseq data from early differentiating EBs (D3 and D5, Figure 2). Whether FOXK1 regulates Isl1 signalling and progenitor cell expansion in the SHF in vivo remains to be elucidated. Additionally, while these current studies identified FOXK1 as a repressor of Wnt signalling, others have shown that FOXK1 positively regulates Wnt signalling (in a tumorigenic setting), suggesting that the function of FOXK1 can be context dependent.55 Therefore, it will be important to identify upstream regulators of FOXK1 during cardiovascular development that might regulate this apparent context dependent expression and function.
While these studies provide the first evidence for a role of FOXK1 in cardiovascular development, future studies will focus on the role of FOXK1 and its impact on cardiac development during murine embryogenesis. While FOXK1 has already been shown to be expressed in the developing heart, the characterization of the global Foxk1 KO embryos and the conditional deletion of Foxk1 will be instrumental to dissect the precise mechanisms of action during cardiogenesis.
In summary, we have identified an important role for FOXK1 in the regulation of cardiovascular development and showed that FOXK1 is a direct transcriptional and epigenetic repressor of Wnt signalling, particularly Wnt6.
Supplementary material
Supplementary material is available at Cardiovascular Research online.
Supplementary Material
Acknowledgements
We acknowledge the FACS Core services at the LHI for the assistance with the FACS characterization and sorting experiments. We acknowledge Cynthia Faraday for the help with the design of the differentiation schematic.
Contributor Information
Javier E Sierra-Pagan, Cardiovascular Division, Department of Medicine, University of Minnesota, 401 East River ParkwayVCRC 1st Floor, Suite 131 Minneapolis, MN 55455, USA.
Nikita Dsouza, Cardiovascular Division, Department of Medicine, University of Minnesota, 401 East River ParkwayVCRC 1st Floor, Suite 131 Minneapolis, MN 55455, USA.
Satyabrata Das, Cardiovascular Division, Department of Medicine, University of Minnesota, 401 East River ParkwayVCRC 1st Floor, Suite 131 Minneapolis, MN 55455, USA.
Thijs A Larson, Cardiovascular Division, Department of Medicine, University of Minnesota, 401 East River ParkwayVCRC 1st Floor, Suite 131 Minneapolis, MN 55455, USA.
Jacob R Sorensen, Cardiovascular Division, Department of Medicine, University of Minnesota, 401 East River ParkwayVCRC 1st Floor, Suite 131 Minneapolis, MN 55455, USA.
Xiao Ma, Cardiovascular Division, Department of Medicine, University of Minnesota, 401 East River ParkwayVCRC 1st Floor, Suite 131 Minneapolis, MN 55455, USA.
Patricia Stan, Cardiovascular Division, Department of Medicine, University of Minnesota, 401 East River ParkwayVCRC 1st Floor, Suite 131 Minneapolis, MN 55455, USA.
Erik J Wanberg, Cardiovascular Division, Department of Medicine, University of Minnesota, 401 East River ParkwayVCRC 1st Floor, Suite 131 Minneapolis, MN 55455, USA.
Xiaozhong Shi, Department of Physiology, Basic Medical College, Nanchang University, Nanchang, Jiangxi 330006, China.
Mary G Garry, Cardiovascular Division, Department of Medicine, University of Minnesota, 401 East River ParkwayVCRC 1st Floor, Suite 131 Minneapolis, MN 55455, USA; Stem Cell Institute, University of Minnesota, 2001 6th Street SE Minneapolis, MN 55455, USA; Paul and Sheila Wellstone Muscular Dystrophy Center, University of Minnesota, 516 Delaware ST SE Minneapolis, MN 55455, USA.
Wuming Gong, Cardiovascular Division, Department of Medicine, University of Minnesota, 401 East River ParkwayVCRC 1st Floor, Suite 131 Minneapolis, MN 55455, USA.
Daniel J Garry, Cardiovascular Division, Department of Medicine, University of Minnesota, 401 East River ParkwayVCRC 1st Floor, Suite 131 Minneapolis, MN 55455, USA; Stem Cell Institute, University of Minnesota, 2001 6th Street SE Minneapolis, MN 55455, USA; Paul and Sheila Wellstone Muscular Dystrophy Center, University of Minnesota, 516 Delaware ST SE Minneapolis, MN 55455, USA.
Funding
Funding support was obtained from the National Institute of Health (R01HL148599 & P01HL160476).
Data availability
The bulk RNAseq and ATACseq datasets of differentiating EBs were deposited at NCBI Gene Expression Omnibus (GEO) database with the accession code GSE202563. All data will be available upon request. All unique materials used in these studies are readily available from the authors or from commercial sources.
Translational perspective.
Congenital heart disease is the most common birth defect. Deciphering the networks that govern cardiomyocyte specification, proliferation, and differentiation will provide insights regarding therapeutic interventions for cardiovascular disease. The winged helix/forkhead family of transcription factors has been shown to have critical roles in epigenetics, organogenesis, cellular proliferation, and differentiation. FOXK1 is an important transcription factor that regulates cardiovascular development through the Wnt signalling pathway. This FOXK1-Wnt pathway defines a network that may be therapeutically targeted to promote cardiogenesis.
References
- 1. Hoffman JIE. Incidence of congenital heart disease: I. Postnatal incidence. Pediatr Cardiol 1995;16:103–113. [DOI] [PubMed] [Google Scholar]
- 2. Bruneau BG. The developmental genetics of congenital heart disease. Nature 2008;451:943–948. [DOI] [PubMed] [Google Scholar]
- 3. Ferretti E, Hadjantonakis AK. Mesoderm specification and diversification: from single cells to emergent tissues. Curr Opin Cell Biol 2019;61:110–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chang CP, Bruneau BG. Epigenetics and cardiovascular development. Annu Rev Physiol 2012;74:41–68. [DOI] [PubMed] [Google Scholar]
- 5. Tam PPL, Behringer RR. Mouse gastrulation: the formation of a mammalian body plan. Mech Dev 1997;68:3–25. [DOI] [PubMed] [Google Scholar]
- 6. Golson ML, Kaestner KH. Fox transcription factors: from development to disease. Development (Cambridge, England) 2016;143:4558–4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Iwafuchi-Doi M, Donahue G, Kakumanu A, Watts JA, Mahony S, Pugh BF, Lee D, Kaestner KH, Zaret KS. The pioneer transcription factor FoxA maintains an accessible nucleosome configuration at enhancers for tissue-specific gene activation. Mol Cell 2016;62:79–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Iwafuchi-Doi M, Zaret KS. Cell fate control by pioneer transcription factors. Development (Cambridge, England) 2016; 143:1833–1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Liu Y, Ding W, Ge H, Ponnusamy M, Wang Q, Hao X, Wu W, Zhang Y, Yu W, Ao X, Wang J. FOXK transcription factors: regulation and critical role in cancer. Cancer Lett 2019;458:1–12. [DOI] [PubMed] [Google Scholar]
- 10. Ji Z, Donaldson IJ, Liu J, Hayes A, Zeef LA, Sharrocks AD. The forkhead transcription factor FOXK2 promotes AP-1-mediated transcriptional regulation. Mol Cell Biol 2012;32:385–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Garry DJ, Yang Q, Bassel-Duby R, Williams RS. Persistent expression of MNF identifies myogenic stem cells in postnatal muscles. Dev Biol 1997;188:280–294. [DOI] [PubMed] [Google Scholar]
- 12. Garry DJ, Meeson A, Elterman J, Zhao Y, Yang P, Bassel-Duby R, Williams RS. Myogenic stem cell function is impaired in mice lacking the forkhead/winged helix protein MNF. Proc Natl Acad Sci U S A 2000;97:5416–5421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hawke TJ, Garry DJ. Myogenic satellite cells: physiology to molecular biology. J Appl Physiol 2001;91:534–551. [DOI] [PubMed] [Google Scholar]
- 14. Shi X, Garry DJ. Myogenic regulatory factors transactivate the Tceal7 gene and modulate muscle differentiation. Biochem J 2010;428:213–221. [DOI] [PubMed] [Google Scholar]
- 15. Shi X, Garry DJ. Sin3 interacts with foxk1 and regulates myogenic progenitors. Mol Cell Biochem 2012;366:251–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hawke TJ, Jiang N, Garry DJ. Absence of p21CIP rescues myogenic progenitor cell proliferative and regenerative capacity in Foxk1 null mice. J Biol Chem 2003;278:4015–4020. [DOI] [PubMed] [Google Scholar]
- 17. Meeson AP, Shi X, Alexander MS, Williams RS, Allen RE, Jiang N, Adham IM, Goetsch SC, Hammer RE, Garry DJ. Sox15 and Fhl3 transcriptionally coactivate Foxk1 and regulate myogenic progenitor cells. EMBO J 2007;26:1902–1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Yang Q, Kong Y, Rothermel B, Garry DJ, Bassel-Duby R, Williams RS. The winged-helix/forkhead protein myocyte nuclear factor beta (MNF-beta) forms a co-repressor complex with mammalian sin3B. Biochem J 2000; 345(Pt 2):335–343. [PMC free article] [PubMed] [Google Scholar]
- 19. Li F, Chong ZZ, Maiese K. Winding through the WNT pathway during cellular development and demise. Histol Histopathol 2006;21:103–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Clevers H. Wnt/beta-catenin signaling in development and disease. Cell 2006;127:469–480. [DOI] [PubMed] [Google Scholar]
- 21. Davis LA, Zur Nieden NI. Mesodermal fate decisions of a stem cell: the Wnt switch. Cell Mol Life Sci 2008;65:2658–2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Murry CE, Keller G. Differentiation of embryonic stem cells to clinically relevant populations: lessons from embryonic development. Cell 2008;132:661–680. [DOI] [PubMed] [Google Scholar]
- 23. Ueno S, Weidinger G, Osugi T, Kohn AD, Golob JL, Pabon L, Reinecke H, Moon RT, Murry CE. Biphasic role for Wnt/beta-catenin signaling in cardiac specification in zebrafish and embryonic stem cells. Proc Natl Acad Sci U S A 2007;104:9685–9690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lin L, Cui L, Zhou W, Dufort D, Zhang X, Cai CL, Bu L, Yang L, Martin J, Kemler R, Rosenfeld MG, Chen J, Evans SM. Beta-catenin directly regulates Islet1 expression in cardiovascular progenitors and is required for multiple aspects of cardiogenesis. Proc Natl Acad Sci U S A 2007;104:9313–9318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Iacovino M, Bosnakovski D, Fey H, Rux D, Bajwa G, Mahen E, Mitanoska A, Xu Z, Kyba M. Inducible cassette exchange: a rapid and efficient system enabling conditional gene expression in embryonic stem and primary cells. Stem cells (Dayton, Ohio) 2011;29:1580–1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bryja V, Bonilla S, Arenas E. Derivation of mouse embryonic stem cells. Nat Protoc 2006;1:2082–2087. [DOI] [PubMed] [Google Scholar]
- 27. Koyano-Nakagawa N, Kweon J, Iacovino M, Shi X, Rasmussen TL, Borges L, Zirbes KM, Li T, Perlingeiro RC, Kyba M, Garry DJ. Etv2 is expressed in the yolk sac hematopoietic and endothelial progenitors and regulates Lmo2 gene expression. Stem cells (Dayton, Ohio) 2012;30:1611–1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Caprioli A, Koyano-Nakagawa N, Iacovino M, Shi X, Ferdous A, Harvey RP, Olson EN, Kyba M, Garry DJ. Nkx2-5 represses Gata1 gene expression and modulates the cellular fate of cardiac progenitors during embryogenesis. Circulation 2011;123:1633–1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Roth GA, Mensah GA, Johnson CO, Addolorato G, Ammirati E, Baddour LM, Barengo NC, Beaton AZ, Benjamin EJ, Benziger CP, Bonny A, Brauer M, Brodmann M, Cahill TJ, Carapetis J, Catapano AL, Chugh SS, Cooper LT, Coresh J, Criqui M, DeCleene N, Eagle KA, Emmons-Bell S, Feigin VL, Fernandez-Sola J, Fowkes G, Gakidou E, Grundy SM, He FJ, Howard G, Hu F, Inker L, Karthikeyan G, Kassebaum N, Koroshetz W, Lavie C, Lloyd-Jones D, Lu HS, Mirijello A, Temesgen AM, Mokdad A, Moran AE, Muntner P, Narula J, Neal B, Ntsekhe M, de Oliveira GM, Otto C, Owolabi M, Pratt M, Rajagopalan S, Reitsma M, Ribeiro ALP, Rigotti N, Rodgers A, Sable C, Shakil S, Sliwa-Hahnle K, Stark B, Sundstrom J, Timpel P, Tleyjeh IM, Valgimigli M, Vos T, Whelton PK, Yacoub M, Zuhlke L, Murray C, Fuster V, GBD-NHLBI-JACC global burden of cardiovascular diseases writing group . Global burden of cardiovascular diseases and risk factors, 1990-2019: update from the GBD 2019 study. J Am Coll Cardiol 2020;76:2982–3021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Singh BN, Sierra-Pagan JE, Gong W, Das S, Theisen JWM, Skie E, Garry MG, Garry DJ. ETV2 (Ets variant transcription factor 2)-Rhoj cascade regulates endothelial progenitor cell migration during embryogenesis. Arterioscler Thromb Vasc Biol 2020;40:2875–2890. [DOI] [PubMed] [Google Scholar]
- 31. Bray NL, Pimentel H, Melsted P, Pachter L. Near-optimal probabilistic RNA-seq quantification. Nat Biotechnol 2016;34:525–527. [DOI] [PubMed] [Google Scholar]
- 32. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014;15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat Methods 2013;10:1213–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods 2012;9:357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, Liu XS. Model-based analysis of ChIP-seq (MACS). Genome Biol 2008;9:R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Amemiya HM, Kundaje A, Boyle AP. The ENCODE blacklist: identification of problematic regions of the genome. Sci Rep 2019;9:9354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Schep AN, Buenrostro JD, Denny SK, Schwartz K, Sherlock G, Greenleaf WJ. Structured nucleosome fingerprints enable high-resolution mapping of chromatin architecture within regulatory regions. Genome Res 2015;25:1757–1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Shi X, Wallis AM, Gerard RD, Voelker KA, Grange RW, DePinho RA, Garry MG, Garry DJ. Foxk1 promotes cell proliferation and represses myogenic differentiation by regulating Foxo4 and Mef2. J Cell Sci 2012;125:5329–5337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Singh BN, Kawakami Y, Akiyama R, Rasmussen TL, Garry MG, Gong W, Das S, Shi X, Koyano-Nakagawa N, Garry DJ. The Etv2-miR-130a network regulates mesodermal specification. Cell Rep 2015;13:915–923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Garry DJ, Olson EN. A common progenitor at the heart of development. Cell 2006;127:1101–1104. [DOI] [PubMed] [Google Scholar]
- 41. Kattman SJ, Huber TL, Keller GM. Multipotent flk-1 + cardiovascular progenitor cells give rise to the cardiomyocyte, endothelial, and vascular smooth muscle lineages. Dev Cell 2006;11:723–732. [DOI] [PubMed] [Google Scholar]
- 42. Masino AM, Gallardo TD, Wilcox CA, Olson EN, Williams RS, Garry DJ. Transcriptional regulation of cardiac progenitor cell populations. Circ Res 2004;95:389–397. [DOI] [PubMed] [Google Scholar]
- 43. Moretti A, Caron L, Nakano A, Lam JT, Bernshausen A, Chen Y, Qyang Y, Bu L, Sasaki M, Martin-Puig S, Sun Y, Evans SM, Laugwitz KL, Chien KR. Multipotent embryonic isl1 + progenitor cells lead to cardiac, smooth muscle, and endothelial cell diversification. Cell 2006;127:1151–1165. [DOI] [PubMed] [Google Scholar]
- 44. Wu SM, Fujiwara Y, Cibulsky SM, Clapham DE, Lien CL, Schultheiss TM, Orkin SH. Developmental origin of a bipotential myocardial and smooth muscle cell precursor in the mammalian heart. Cell 2006;127:1137–1150. [DOI] [PubMed] [Google Scholar]
- 45. Alexander JM, Hota SK, He D, Thomas S, Ho L, Pennacchio LA, Bruneau BG. Brg1 modulates enhancer activation in mesoderm lineage commitment. Development (Cambridge, England) 2015;142:1418–1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Hota SK, Rao KS, Blair AP, Khalilimeybodi A, Hu KM, Thomas R, So K, Kameswaran V, Xu J, Polacco BJ, Desai RV, Chatterjee N, Hsu A, Muncie JM, Blotnick AM, Winchester SAB, Weinberger LS, Huttenhain R, Kathiriya IS, Krogan NJ, Saucerman JJ, Bruneau BG. Brahma safeguards canalization of cardiac mesoderm differentiation. Nature 2022;602:129–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Srivastava D. Making or breaking the heart: from lineage determination to morphogenesis. Cell 2006;126:1037–1048. [DOI] [PubMed] [Google Scholar]
- 48. Shi X, Seldin DC, Garry DJ. Foxk1 recruits the Sds3 complex and represses gene expression in myogenic progenitors. Biochem J 2012;446:349–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Yang L, Chen S, Zhao Q, Pan C, Peng L, Han Y, Li L, Ruan J, Xia J, Yang H, Xu F, Cheng G. Histone deacetylase 3 contributes to the antiviral innate immunity of macrophages by interacting with FOXK1 to regulate STAT1/2 transcription. Cell Rep 2022;38:110302. [DOI] [PubMed] [Google Scholar]
- 50. Chronis C, Fiziev P, Papp B, Butz S, Bonora G, Sabri S, Ernst J, Plath K. Cooperative binding of transcription factors orchestrates reprogramming. Cell 2017;168:442–459 e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM III, Hao Y, Stoeckius M, Smibert P, Satija R. Comprehensive integration of single-cell data. Cell 2019; 177:1888–1902 e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Rasmussen TL, Kweon J, Diekmann MA, Belema-Bedada F, Song Q, Bowlin K, Shi X, Ferdous A, Li T, Kyba M, Metzger JM, Koyano-Nakagawa N, Garry DJ. ER71 directs mesodermal fate decisions during embryogenesis. Development (Cambridge, England) 2011;138:4801–4812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Sukonina V, Ma H, Zhang W, Bartesaghi S, Subhash S, Heglind M, Foyn H, Betz MJ, Nilsson D, Lidell ME, Naumann J, Haufs-Brusberg S, Palmgren H, Mondal T, Beg M, Jedrychowski MP, Taskén K, Pfeifer A, Peng X-R, Kanduri C, Enerbäck S. FOXK1 and FOXK2 regulate aerobic glycolysis. Nature 2019;566:279–283. [DOI] [PubMed] [Google Scholar]
- 54. Ni Y, Liu B, Wu X, Liu J, Ba R, Zhao C. FOXG1 directly suppresses Wnt5a during the development of the hippocampus. Neurosci Bull 2021;37:298–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Wang W, Li X, Lee M, Jun S, Aziz KE, Feng L, Tran MK, Li N, McCrea PD, Park JI, Chen J. FOXKs promote Wnt/beta-catenin signaling by translocating DVL into the nucleus. Dev Cell 2015;32:707–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Baden LR, El Sahly HM, Essink B, Kotloff K, Frey S, Novak R, Diemert D, Spector SA, Rouphael N, Creech CB, McGettigan J, Khetan S, Segall N, Solis J, Brosz A, Fierro C, Schwartz H, Neuzil K, Corey L, Gilbert P, Janes H, Follmann D, Marovich M, Mascola J, Polakowski L, Ledgerwood J, Graham BS, Bennett H, Pajon R, Knightly C, Leav B, Deng W, Zhou H, Han S, Ivarsson M, Miller J, Zaks T, Group CS. Efficacy and safety of the mRNA-1273 SARS-CoV-2 vaccine. N Engl J Med 2021;384:403–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Lavery DL, Martin J, Turnbull YD, Hoppler S. Wnt6 signaling regulates heart muscle development during organogenesis. Dev Biol 2008;323:177–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Tao F, Soffers J, Hu D, Chen S, Gao X, Zhang Y, Zhao C, Smith SE, Unruh JR, Zhang D, Tsuchiya D, Venkatraman A, Zhao M, Li Z, Qian P, Parmely T, He XC, Washburn M, Florens L, Perry JM, Zeitlinger J, Workman J, Li L. Beta-catenin and associated proteins regulate lineage differentiation in ground state mouse embryonic stem cells. Stem Cell Rep 2020;15:662–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Zhu H. Forkhead box transcription factors in embryonic heart development and congenital heart disease. Life Sci 2016;144:194–201. [DOI] [PubMed] [Google Scholar]
- 60. Gong W, Das S, Sierra-Pagan JE, Skie E, Dsouza N, Larson TA, Garry MG, Luzete-Monteiro E, Zaret KS, Garry DJ. ETV2 functions as a pioneer factor to regulate and reprogram the endothelial lineage. Nat Cell Biol 2022;24:672–684. [DOI] [PubMed] [Google Scholar]
- 61. Hota SK, Bruneau BG. ATP-dependent chromatin remodeling during mammalian development. Development (Cambridge, England) 2016;143:2882–2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Tian Y, Cohen ED, Morrisey EE. The importance of Wnt signaling in cardiovascular development. Pediatr Cardiol 2010;31:342–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The bulk RNAseq and ATACseq datasets of differentiating EBs were deposited at NCBI Gene Expression Omnibus (GEO) database with the accession code GSE202563. All data will be available upon request. All unique materials used in these studies are readily available from the authors or from commercial sources.