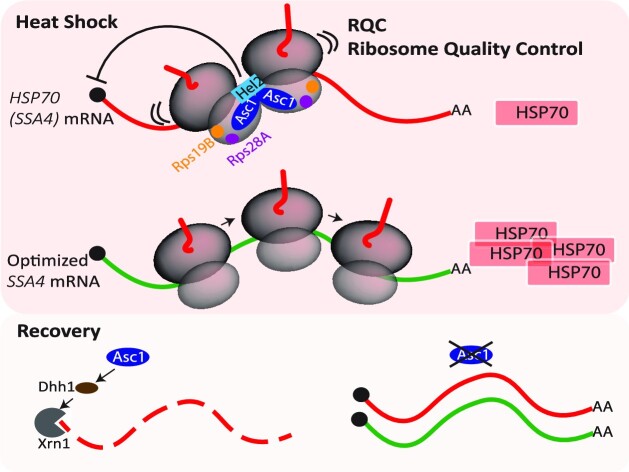
Abstract
Cells survive harsh environmental conditions by potently upregulating molecular chaperones such as heat shock proteins (HSPs), particularly the inducible members of the HSP70 family. The life cycle of HSP70 mRNA in the cytoplasm is unique—it is translated during stress when most cellular mRNA translation is repressed and rapidly degraded upon recovery. Contrary to its 5′ untranslated region's role in maximizing translation, we discovered that the HSP70 coding sequence (CDS) suppresses its translation via the ribosome quality control (RQC) mechanism. The CDS of the most inducible Saccharomyces cerevisiae HSP70 gene, SSA4, is uniquely enriched with low-frequency codons that promote ribosome stalling during heat stress. Stalled ribosomes are recognized by the RQC components Asc1p and Hel2p and two novel RQC components, the ribosomal proteins Rps28Ap and Rps19Bp. Surprisingly, RQC does not signal SSA4 mRNA degradation via No-Go-Decay. Instead, Asc1p destabilizes SSA4 mRNA during recovery from heat stress by a mechanism independent of ribosome binding and SSA4 codon optimality. Therefore, Asc1p operates in two pathways that converge to regulate the SSA4 mRNA life cycle during stress and recovery. Our research identifies Asc1p as a critical regulator of the stress response and RQC as the mechanism tuning HSP70 synthesis.
Graphical Abstract
Graphical Abstract.
Asc1 is a key regulator of the HSP70 mRNA life cycle during heat shock and recovery.
INTRODUCTION
Cells mitigate the detrimental effects of environmental stressors, such as heat, by rapidly inducing the expression of molecular chaperones known as heat shock proteins (HSPs) (1,2). Cells tailor HSP levels to the burden of misfolded proteins by tuning the heat shock response (HSR). Its activation during stress leads to the potent upregulation of HSP transcription and preferential translation. Then, cells avoid unnecessary HSP accumulation during recovery by rapidly halting transcription and increasing HSP mRNA instability (3). This fast switch from induction to attenuation of the HSR is critical for cell function.
HSPs were initially classified into families based on their molecular weights and further categorized as constitutive or inducible based on their steady-state expression levels (4). Constitutive and inducible members of the HSP70 family play a key role in preserving protein homeostasis (proteostasis), preventing protein aggregation by assisting unfolded proteins back into their functional conformations, and clearing misfolded proteins in concert with the ubiquitin-proteasome system and autophagy (5). Fast resolution of their response is also crucial, as the persistent expression of inducible HSP70 under permissive conditions causes growth defects in Drosophila (6) and promotes transformation in mammalian cells (7).
In stress, activation of heat shock factor 1 (HSF1) induces robust HSP70 transcription (1,3,8–11). Newly synthesized HSP70 mRNAs are translated despite the repression of cap-dependent translation initiation and elongation to prevent the accumulation of misfolded polypeptides (12–16). Translation initiation is further dampened by the phosphorylation of eIF2α, which inhibits GDP-GTP exchange (2,3,17–19). Co-transcriptional processing during stress favors HSP70 translation via a cap-independent pathway (20,21). This mechanism involves the translation elongation factor eEF1A1 and co-transcriptional modifications to the HSP70 5′ untranslated region (UTR) that are recognized by the translation initiation factor eIF3 (22–25). On-going translation promotes the mRNA’s stability (26). During recovery, cells resume cap-dependent translation, and de novo synthesized HSP70 binds to the transactivation domain of HSF1 to repress its own expression and attenuate the HSR (2,3,10,27). To rapidly shut down HSP70 expression, efficient degradation of its mRNA is critical, which requires its 3′ UTR (28,29). Therefore, HSP70 transcripts go from highly stable during stress to highly unstable during recovery (28,29). Although HSP70 translation is needed for its mRNA turnover, the factors tuning its fate in the cytoplasm in response to the cellular stress status remain unknown (3,30).
The regulation of HSP70 mRNA translation and stability relates to cellular changes in protein synthesis. The traditional mRNA surveillance model suggests that highly efficient translation increases mRNA stability because ribosomes protect the mRNA being translated from degradation (26,31–36). Contrary to this model, recent findings showed that high translation initiation rates destabilize mRNAs containing pro-stalling codons in the budding yeast Saccharomyces cerevisiae (37–39). Increased ribosome loading favors collisions between stalled ribosomes, which result in the formation of di-ribosomes (disomes) consisting of the leading stalled ribosome and the subsequent colliding ribosome (40,41). Colliding ribosomes signal to the ribosome quality control (RQC) mechanism to recycle stalled ribosomes and the mRNA surveillance mechanism No-Go Decay (NGD) to degrade the faulty mRNA (33,42,43).
The initiation of collision-associated RQC is mediated by Asc1p and Hel2p (the orthologs of RACK1 and ZNF598 in mammals, respectively), which stabilize the disomes (41,43–47). Asc1p is a scaffold protein located at the head of the 40S subunit near the mRNA exit channel (48). In the context of the RQC, Asc1p-Asc1p interactions between disome's 40S subunits stabilize the collision and provide an interface for recognition by the E3 ubiquitin ligase Hel2p (41,46,49). Hel2p ubiquitinates the 40S ribosomal protein Rps20p (uS10) in yeast and also Rps10p (eS10) in humans and promotes the splitting of the first stalled polyubiquitinated ribosome by the helicase Slh1p (44,47,50,51). Alternatively, Not4 ubiquitinates Rps7p (eS7), which is a substrate for Hel2p polyubiquitination (41,46,52). The E3 ubiquitin ligase, Ltn1, ubiquitinates the aberrant peptide stocked in the 60S ribosomal subunit to be degraded by the ubiquitin proteosome system (UPS) and prevent its aggregation(53,54). Disome stabilization by Asc1p and Hel2p is necessary to recruit endonucleases Cue2p or protein Syh1p that target the mRNA for degradation by NGD (42,52,55–59). To prevent the accumulation of partially synthesized peptides, ribosome collisions do not necessarily have to trigger NGD (55,60–62). Instead, they can signal to repress translation initiation. In mammalian cells, ZNF598 recruits GIGYF2 and 4EHP to inhibit translation initiation by outcompeting eIF4E binding to the cap of the mRNA on a stalled ribosome (63).
Based on these studies, we hypothesized that by resuming cap-dependent translation during recovery, cells could link an increase in HSP70 mRNA translation efficiency to its decay by NGD. To test this hypothesis, we studied the regulation of the four Stress Seventy sub-family A (Ssa) members, Ssa1-4, in S. cerevisiae (64). Ssa1p and Ssa2p are constitutively expressed, while Ssa3p and Ssa4p are inducible. The codon sequences of SSA4 and SSA3 mRNAs are biased toward low-frequency codons, which promote ribosome stalling and regulate their expression via the RQC and NGD. Accordingly, we found that the RQC factors Asc1p and Hel2p regulate the SSA4 mRNA life cycle, but in unexpected ways. Firstly, the RQC mechanism downregulates Ssa4p synthesis during heat shock, which prevents its overproduction during stress. This regulation depends on the low codon optimality of the SSA4 mRNA CDS and involves the ribosomal proteins Rps28Ap and Rps19Bp, which emerge as new RQC components. Secondly, the RQC mechanism does not lead to NGD nor the degradation of SSA4 mRNA during stress or recovery. Instead, Asc1p destabilizes SSA4 mRNAs during recovery independently of its ribosome binding. This result points to two distinct functions for Asc1p that converge to control the fate of SSA4 mRNA in the cytoplasm. Thus, we have identified Asc1p as a novel critical regulator of the yeast HSR.
MATERIALS AND METHODS
Yeast culture
All yeast strains are derived from the parental strain BY4741, and their genotypes are summarized in Supplementary Table S1. They were grown in yeast extract peptone dextrose (YPD) medium or the conditional medium appropriate for their genotype at 25°C with constant shaking at 250 rpm. Knock-in and deleted strains were created by homologous recombination of the parental strain after the transformation of a PCR fragment amplified from a plasmid carrying selection-specific markers. Gene deletions and knock-ins were verified by PCR analyses of genomic DNA extracted from individual colonies, as previously described (65). The primers and plasmids used are listed in Supplementary Tables S2 and S3, respectively.
Heat shock and recovery
For northern and western blot experiments, cells in the logarithmic growth phase (optical density at 600 nm (OD600): 0.4–0.6) were heat shocked in a 42°C water bath with constant shaking at 150 rpm until the indicated time points. Immediately after heat shock, the heated medium was replaced with the same volume of room temperature (RT) medium. The culture flasks were then placed in a 25°C shaker incubator and rotated at 250 rpm until cultures were collected for downstream sample processing. For spot assays, cells at OD600 1.0–1.5 were diluted in water to OD600 0.5 and then serially diluted at a 1:5 ratio in water five times. Five μl of serially diluted cells were plated on YPD-agar, then either incubated at 25°C or at 42°C for 16 h and then at 25°C. The preconditioning was performed by heat shock at 37°C for 1 h followed by 5 h of recovery at 25°C before moving the plates to 42°C. The plates were then checked for colonies every 24 h and images were acquired. For growth curves, cells at OD600 ∼0.5 were heat shocked in a 42°C water bath with constant shaking at 150 rpm. Absorbance at 600nm was measured from the culture collected after heat shock and followed by 30, 60, 90 and 120 min of recovery.
Protein extraction, western blotting, and polysome profiling
Unstressed, heat-shocked, and recovered yeast (5 ml) were collected and centrifuged at 3000 × g for 5 min. Cell pellets were first washed with 2 M LiOAc at RT and then with 0.4 M NaOH on ice. Cells were lysed with sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) buffer (60 mM Tris–HCl pH 6.8, 10% glycerol, 2% SDS, 5% 2-mercaptoethanol and 0.0025% bromophenol blue). The lysates were heated for 10 min at 95°C, resolved by 10% SDS-PAGE, and transferred to nitrocellulose membranes. Ponceau S staining was used to confirm equal protein loading. Membranes were blocked with 5% skim milk in 1× phosphate-buffered saline with 0.05% Tween 20 (PBST) for 1 h at RT and then incubated with specific antibodies (eIF2α, phospho eIF2α (Ser51) (Cat# 9722, 9721S, Cell Signaling Technology, Danvers, MA, USA)), HA (Cat# 901501, BioLegend), tubulin (Developmental Studies Hybridoma Bank), and β-actin (Cat# A2228, EMD Millipore Corp) overnight at 4°C. Followed by three washes in PBST, membranes were incubated with horseradish peroxidase-conjugated goat-anti-mouse (Cat# 1706516, Bio-Rad) or goat-anti-rabbit antibody (Cat# 1706515, Bio-Rad) for 2 h at RT. Three washes with PBST were performed, followed by Clarity Western ECL treatment and imaging on a ChemiDoc Gel Imaging System (Bio-Rad). The intensity of the target protein signal was quantified using ImageJ version 2.1.0 (National Institutes of Health, Bethesda, MD, USA) and normalized to that of the loading control (β-actin or tubulin).
For protein isolation from polysomes, polysomes were prepared from heat shocked yeast extracts as previously described (66). Protein was extracted by adding 3 volumes of 100% cold ethanol to the monosome or polysome fractions. RNA–protein complexes were precipitated overnight at –20°C and centrifugated for 30 min at 13 000 rpm at 4°C. The pellets were washed once again with 70% ethanol, allowed to air-dry, and dissolved in the above-mentioned SDS-PAGE loading buffer.
RNA extraction, northern blotting, and RT-QPCR
Unstressed, heat-shocked, and recovered yeast (5 ml) were collected and centrifuged at 3000 × g for 5 min at 4°C. For the protocol below, all centrifugations were performed at 12 000 × g. The pellets were resuspended in 0.5 mL of RNA extraction lysis buffer (10 mM Tris–HCl pH 8.5, 5 mM EDTA, 2% SDS, 2% stock 2-mercaptoethanol), and transferred to 1.5 ml tubes. Cells were lysed by incubating the tubes in a heat block at 83°C for 20 min. After centrifugation for 5 min, the supernatant was transferred to a fresh tube containing 0.55 ml of pH 8 phenol. After vortexing for 30 s and centrifugation for 5 min, the top layer was transferred to a new tube labeled N. RNA extraction lysis buffer (0.25 ml) was added to the previous tube, which was vortexed briefly. An equal volume (0.25 ml) of chloroform was added, the tube was vortexed and centrifuged, and the top layer was transferred to tube N. Another 0.55 ml of pH 8 phenol was added to tube N, which was vortexed and spun as above, and the top layer was transferred to a new tube containing 0.55 ml of acid phenol–chloroform, pH 4.5 (Cat# AM9720, Thermofisher Scientific, St. Austin, TX, USA). The tubes were vortexed briefly, spun and 0.45 ml of the top layer was transferred to a new set of tubes containing 0.2 ml of 0.6 M sodium acetate, pH 4.5. The contents were mixed by flicking followed by a quick spin. Once again, Acid Phenol-Chloroform, pH 4.5, 0.6 ml was added to the tubes accompanied by a vortex and spin. Approximately 0.35 ml of the top layer was once again transferred to new tubes containing 1.1 ml of 100% ethanol and 0.03 ml of 5 M ammonium acetate. After mixing, the samples were placed at −80°C overnight. The next day, the samples were spun at 4°C for 15 min and the supernatant was discarded. The pellet was washed twice with 80% ethanol and allowed to air dry, then dissolved in 0.04 ml of RNase-free water and the RNA was quantified. Equal amounts (1000–2000 ng) of RNA were aliquoted into fresh tubes and dried in a SpeedVac for 45 min at 45°C. Samples were resuspended in 5 μl of RNase-free water and mixed with 7 μl of homemade RNA loading dye. The RNA samples were run in a 1% denaturing gel in 1 × MESA buffer. Transfer to zeta probe nylon membranes was set up using capillary electrophoresis overnight. The membrane was UV-crosslinked at 1200 mJ, stained for total RNA, prehybridized, hybridized, exposed to a phosphorscreen, and developed using a phosphorimager. Northern blotting and radiolabeling of probes were performed as in (67). Genomic BY4741 was used as a template to PCR amplify probes that target SSA1, SSA2, SSA3, SSA4, SSA4-Opt 3′ UTR (MS2V6) using primers listed in Supplementary Table S2.
For reverse transcription, 1 μg of total RNA was treated with 1 unit of DNaseI (Promega) for 30 min and 100 ng were reverse transcribed using 4 μl of iScript RT supermix (Biorad) in a 20 μl reaction following manufacturer instruction. For qPCR, cDNAs were diluted two-fold in water. PCR was performed in 5 μl reactions consisting of 1 μl of cDNA, 2.5 μl PowerUp SYBR Green master mix (ThermoFisher) and 0.25 μl of 1 μM of each primer. Standard curves were generated using a log titration of BY4741 WT or SSA4 optimized genomic DNA (50–0.05 ng). Data was collected using Viaa7 PCR system with 45 cycles. The standard curve was used to calculate RNA amounts.
mRNA half-life calculations
Northern blots were quantified using ImageJ and normalized to corresponding methylene blue staining. Considering the intensity of the heat shock sample (timepoint: 0 min) to be 100% induction, the relative intensity was calculated for recovery samples (timepoints: 15, 30, 60, 90 min). A polynomial curve was plotted for time vs the percentage of mRNA decayed. The polynomial equation was obtained for the curve and solved for X, given that Y is 50 using what-if analysis in Microsoft Excel.
Preferred codon percentage determination
A list of preferred codons in S. cerevisiae was procured from (68) and the CDSs of SSA mRNAs were obtained from the Saccharomyces Genome Database (https://www.yeastgenome.org/). A Python script was developed to count the occurrence of each preferred codon and divide it by the total number of codons to calculate the percentages of preferred codons (https://github.com/LR-MVU/YEAST-SSA.git) and (https://doi.org/10.5281/zenodo.7847569).
Single-molecule in situ hybridization (smFISH) and imaging analysis
The smFISH procedure was performed as previously described (69). Briefly, yeast strains were grown in 25 mL YPD at 25°C to early log phase and heat shocked at 42°C. At the indicated time points, they were fixed in 4% paraformaldehyde, permeabilized in spheroplast buffer containing lyticase (as described in (69), (Cat# L2524, Sigma Aldrich, Oakville, ON, Canada)), and seeded onto poly-L-lysine-coated coverslips. After ethanol incubation, rehydration with 2× saline sodium citrate buffer, and prehybridization, the cells were hybridized with Stellaris smFISH probes to detect MS2V6 sequence in the tagged SSA4 or SSA2 mRNAs (LGC Biosearch Technologies) as previously described in (69). The coverslips were washed, dried, and mounted in Prolong Gold Antifade Mounting Medium (Invitrogen, Bulington, ON, Canada), then imaged using a wide-field inverted Nikon Ti-2 wide-field microscope equipped with a Spectra X LED light engine (Lumencor), and an Orca-Fusion sCMOS camera (Hamamatsu) controlled by NIS-Elements Imaging Software. For yeast cells, a 100 × 1.49 NA oil immersion objective lens (Nikon) was used with an xy pixel size of 67.5 nm and a z-step of 200 nm. Outlines were created using the CellProfiler pipeline, and single mRNAs were quantified using FISH-quant (70).
Ribosome profiling analysis
Ribosome profiling and RNA sequencing (RNA-Seq) data in yeast under heat shock conditions performed by Mühlhofer et al. (71) were downloaded from the Gene Expression Omnibus (accession numbers: Riboseq data: SRR9265440 and SRR9265438; RNA-seq data: SRR9265437 and SRR9265428). Raw sequencing reads were processed first by trimming adapters using Cutadapt 3.4 and discarding the low-quality reads. The reads were next mapped to rRNAs and aligned reads were discarded. The remaining reads were then mapped to the yeast transcriptome and the resulting SAM files were further processed to SQLite files using a Python script from the Trips-Viz webserver (https://trips.ucc.ie/) for compatibility with PausePred (72), the built-in function of Trips-Viz (73). Default settings were used to detect ribosome stall sites on SSA4 mRNAs, which were visualized using Trips-Viz.
Protein immunoprecipitation and mass spectrometry
FLAG-tagged Asc1 (Asc1p-3 × FLAG) yeast cultures were grown in 400 ml YPD at 25°C to early log phase and heat shocked for 1 h at 42°C. The cells were pelleted by centrifugation at 3750 rpm for 3 min at 4°C. The pellets were washed with 10 ml water, transferred to Eppendorf tubes, snap-frozen with liquid nitrogen, thawed on ice, and resuspended in 2 ml of lysis buffer (100 mM HEPES pH 8.0, 20 mM magnesium acetate, 10% glycerol, 10 mM EGTA, 0.1 mM EDTA) containing Complete X protease inhibitor and phosphatase inhibitors (1 M NaF, 0.2M NaAPi, and Na2vO4). Glass beads (0.5 mm) were added, and tubes were vortexed 20 times with 30 s on-off cycles in a cold room. Lysates were separated from the beads by a quick centrifugation at 4°C and transferred to new tubes. About 100 μl of M2-anti-FLAG beads (Cat# M8823, Millipore sigma, Oakville, ON, Canada) were washed three times with 500 μl of lysis buffer, then incubated with the lysates for 2 h on a nutator in a cold room. The beads were magnetized, and the flowthrough was collected. The beads were then washed twice with 1 ml of lysis buffer and each wash was collected. Samples of the beads, lysate, flow-through, and washes were boiled in SDS-PAGE buffer, resolved by SDS-PAGE, and western blotted for FLAG. Immunoprecipitation samples were sent to Proteomics Services at the McGill University Health Centre Research Institute for mass spectrometry. For each sample, proteins on the beads were loaded onto a single stacking gel band to remove lipids, detergents, and salts. The single gel band containing all proteins was reduced with DTT, alkylated with iodoacetic acid, and digested with trypsin. Extracted peptides were re-solubilized in 0.1% aqueous formic acid and loaded onto a Thermo Acclaim Pepmap (Thermo, 75 uM ID × 2 cm C18 3 uM beads) precolumn and then onto an Acclaim Pepmap Easyspray (Thermo, 75 uM × 15 cm with 2 uM C18 beads) analytical column separation using a Dionex Ultimate 3000 uHPLC at 250 nl/min with a gradient of 2–35% organic (0.1% formic acid in acetonitrile) over 3 h. Peptides were analyzed using a Thermo Orbitrap Fusion mass spectrometer operating at 120 000 resolution (FWHM in MS1) with HCD sequencing (15000 resolution) at top speed for all peptides with a charge of 2+ or greater. The raw data were converted into *.mgf format (Mascot generic format) for searching using the Mascot 2.6.2 search engine (Matrix Science) against yeast protein sequences (Uniprot 2022). The database search results were loaded onto Scaffold Q + Scaffold_5.0 (Proteome Sciences) for statistical treatment and data visualization. We only consider proteins as enriched or depleted upon HS if they exhibit a fold change >1.5 and have a P-value ≤0.05 (Supplementary Table S4).
For co-immunoprecipitation, Flag-tagged Asc1, Flag-tagged Asc1/TAP-tagged RPS28, and Flag-tagged Asc1/HA-tagged RPS19 strains were grown in 25 ml of YPD at 25°C to early log phase and heat shocked for 1 h at 42°C. The cells were pelleted by centrifugation at 3750 rpm for 3 min at 4°C. The pellets were washed with 10 ml water, transferred to Eppendorf tubes, snap-frozen with liquid nitrogen, thawed on ice, and resuspended in 400 μl of lysis buffer (100 mM HEPES pH 8.0, 20 mM magnesium acetate, 10% glycerol, 10 mM EGTA, 0.1 mM EDTA) containing Complete X protease inhibitor. Glass beads (0.5 mm) were added, and tubes were vortexed 20 times with 30 s on-off cycles in a cold room. Lysates were separated from the beads by a quick centrifugation at 4°C and transferred to new tubes. About 10 μl of M2-anti-FLAG beads were washed three times with 200 μl of lysis buffer, then incubated with the lysates for 2 h on a nutator in a cold room. The beads were magnetized and washed thrice with 500 μl of lysis buffer and each wash was collected. Samples of the beads and lysate were boiled in SDS-PAGE buffer, resolved by SDS-PAGE, and immunoblotted for FLAG (Anit-FLAG_M2 antibody, F1804, Sigma, Saint Louis, MO, USA), HA (HA antibody, Cat#901501, Biolegend, San Diego, CA, USA.) or TAP (Peroxidase Anti-Peroxidase Soluble Complex antibody (PAP) antibody, Cat#P1291 Sigma, Saint Louis, MO, USA).
RESULTS
The RQC factors asc1p and hel2p repress ssa4p expression during heat shock
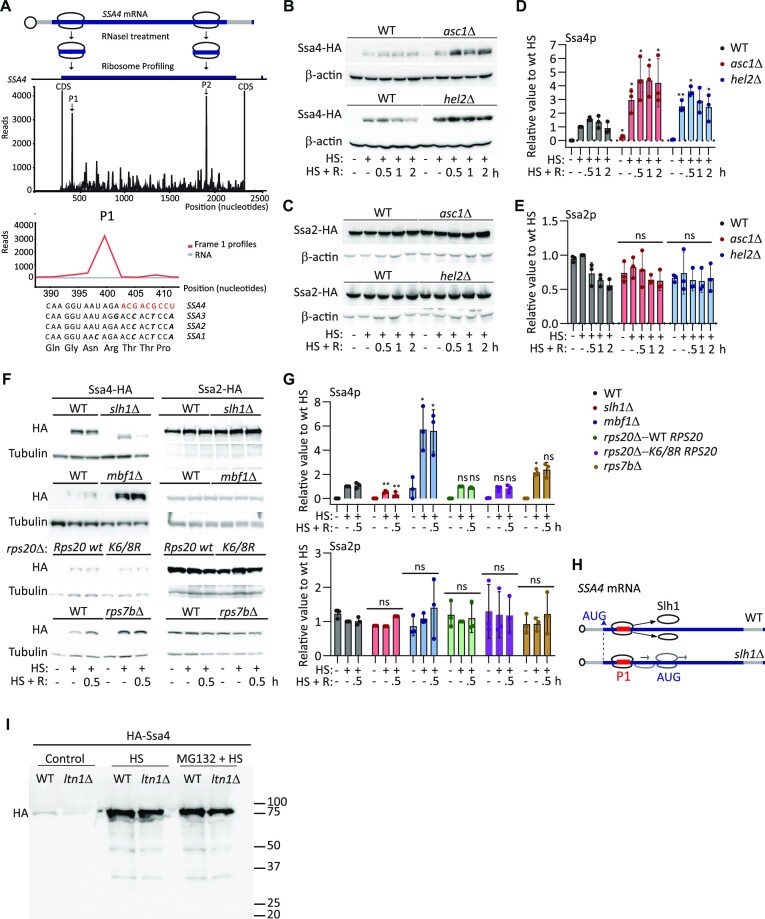
In the SSA subfamily of yeast HSP70 genes, the CDSs of inducible SSA3 and SSA4 are biased towards low-frequency codons (50% optimal codons) compared to the constitutive SSA1 and SSA2 CDSs (75% optimal codons; Supplementary Figure S1A) (68). SSA3 mRNA translation initiation is regulated by an upstream open reading frame (uORF), making SSA4 the most inducible member of this subfamily (74). The presence of nonoptimal codons in highly translated mRNAs leads to slow decoding and ribosome stalling, favoring collisions (33). Therefore, we analyzed previously published ribosome profiling data (71) to identify the presence of stalled ribosomes over the SSA4 mRNA CDS under heat shock (30 min at 42°C) (Figure 1A and Supplementary Figure S1B). PausePred analysis identified stalled ribosomes (i.e. peaks with 20-fold more ribosome occupancy than the following mRNA position) (72,73) at position 400 (P1) on the SSA4 mRNA in both experimental replicates and at position 1800 (P2) in one replicate. Although the SSA4 mRNA codon sequence protected by the ribosome is identical to that of two other SSA mRNAs, the SSA4 sequence contains three low-frequency codons following the stalled ribosome (Figure 1A). The presence of stalled ribosomes on SSA4 transcripts and its enrichment in low-frequency codons led us to investigate the roles of the RQC mechanism and NGD in regulating SSA4 mRNA translation and decay, respectively (75).
Figure 1.
Deletion of Asc1p or Hel2p increases SSA4 mRNA translation during heat shock. (A) Ribosome profiling analysis of S. cerevisiae SSA4 mRNA after 30 min of heat shock at 42°C. Top: Schematic of SSA4 mRNA with ribosomes stalled at two positions. Middle: A single transcript plot of the ribosome sequencing analysis aligned with RNA-Seq data (65). P1 and P2 indicate ribosome stall sites. Bottom: The SSA4 nucleotide sequence protected by the ribosome at P1 and the corresponding nucleotide and amino acid sequences in SSA1, SSA2 and SSA3. (B, C) Western blots of 3×-HA-tagged Ssa4p (B) and Ssa2p (C) in the WT, asc1Δ and hel2Δ strains under basal conditions (25°C), after 30 min of heat shock at 42°C (HS), and at the indicated recovery (R) time points. β-actin was used as a loading control. (D, E) Quantification of Ssa4p (D) and Ssa2p (E) expression. HA band intensities were first normalized to their corresponding β-actin band and are expressed relative to the normalized expression of heat-shocked WT yeast. Bars indicate the mean and standard deviation (SD) of three independent experiments, each represented by a dot. *P < 0.05, **P < 0.001, ***P < 0.0001 (by unpaired t-test). (F) Western blots of 3×-HA-tagged Ssa4p and Ssa2p in the WT, slh1Δ, mbf1Δ and WT Rps20 versus Lys 6/8 Arg Rps20 and rps7BΔ strains under basal conditions (25°C), after 30 min of heat shock at 42°C (HS), and after 30 min recovery (R). Tubulin was used as a loading control. (G) Quantification of Ssa4p and Ssa2p expression. HA band intensities were first normalized to their corresponding Tubulin band and are expressed relative to the normalized expression of heat-shocked WT yeast. Bars indicate the mean and standard deviation (SD) of three independent experiments, each represented by a dot. *P < 0.05, **P < 0.001 (by unpaired t-test). (H) Scheme of the SSA4 ORF where stalled ribosomes are disassembled by Slh1p in WT cells. In the absence of Slh1p, stalled ribosomes can be disassembled at a lower frequency and the 40S subunit will continue scanning the mRNA and initiate translation in the next AUG. (I) Western blots of 3×-HA-N-terminal tagged Ssa4p under basal conditions (25°C) and after 30 min of heat shock at 42°C (HS) previously exposed or not to 0.25 M of MG132 for 6 h. Right side numbers indicate the ladder molecular weight.
The RQC factors Asc1p and Hel2p stabilize ribosome collisions and repress the translation of the affected mRNA (40,42). To investigate their roles in regulating Ssa4p synthesis during heat shock and subsequent recovery, we deleted ASC1 or HEL2 genes from haploid BY4741 wild-type (WT) S. cerevisiae. Given the high similarity between the four SSAs, we inserted 3 × Hemagglutinin (HA) epitopes in the C-terminus and 12 × MS2V6 RNA stem-loops in the 3’UTR of each of the endogenous SSA genes to distinguish their proteins and mRNAs by western blotting and smFISH, respectively. Compared to the WT strain, the asc1Δ and hel2Δ strains had significantly higher expression of Ssa4p during heat shock; however, no changes in Ssa1p, Ssa2p, Ssa3p, nor the non-heat shock protein Doa1p were observed between the basal (25°C), heat shock, and recovery conditions (Figure 1B–E, Supplementary Figure S1C and D). Increased Ssa4p expression in heat-shocked asc1Δ and hel2Δ yeast persisted during recovery, but the difference in expression compared to the WT strain did not increase further. To exclude Ssa4p dilution by cell division during recovery, we measured yeast duplication after 30 min of heat shock (Supplementary Figure S1E). None of the strains duplicated for the two hours that followed the heat shock, suggesting that SSA4 mRNA is not translated during recovery. This indicates that the enrichment in low-frequency codons in the SSA4 mRNA CDS downregulates Ssa4p expression via the RQC only during heat shock.
We modified other factors involved in RQC to confirm the regulation of Ssa4p synthesis by this mechanism. We deleted the RNA helicase Slh1p that splits the leading stalled ribosomes (50,51) and the Multi-protein Bridging Factor 1 (Mbf1p) that prevents the leading ribosome from +1 frameshifting (76–79). Compared to WT, the slh1Δ strain had a significantly lower expression of Ssa4p during heat shock and recovery (Figure 1F and G). Interestingly, the band corresponding to Ssa4-HA was slightly smaller (by ∼5 kDa). This result was unexpected since SLH1 deletion has the same effect as HEL2 deletion in stalling mRNA reporters studied in yeast and mammalian cells (51,46,80). To explain both the lower expression and smaller Ssa4p band, we propose that in the absence of Slh1p, the leading stalled ribosome cannot be efficiently split and recycled and remains stabilized with the collided ribosomes by Asc1p and Hel2p. As a result, they could form a roadblock at P1 that prevents them from finishing translation, decreasing Ssa4p full-length expression. Interestingly, SSA4 has a downstream AUG in-frame with the start codon at nucleotide 177 in the CDS, where translation could reinitiate and produce a polypeptide that is 53 amino acids shorter than the full length (Figure 1H). Thus, we suggest that from the few 80S ribosomes stalled in P1 that can disassemble without Slh1p, the 40S subunit remains bound to the mRNA and scans the CDS until the next AUG (nucleotide 177), where a new 80S is assembled reinitiating translation (similar to what happened in uORF containing mRNAs) (81,82) (Figure 1H). This result fits with ribosomes stalled at P1, as detected by ribosome profiling (Figure 1A). Similarly to the asc1Δ and hel2Δ strains, the mbf1Δ strain expressed significantly more Ssa4p than WT. These results further support the regulation of SSA4 mRNA translation by colliding ribosomes and RQC (Figure 1F and G).
Two 40S ribosomal proteins, Rps7Bp and Rps20p, are involved in RQC. Monoubiquitination of Rps7Bp by Not4p followed by its polyubiquitination by Hel2p is required to resolve stalled ribosomes. Besides Rps7p ubiquitylation, a more canonical way to resolve stalling ribosomes implies ubiquitylation of Rps20p at Lys 6 and 8 by Hel2p (41,46). To evaluate the role of these proteins in Ssa4p synthesis, we expressed the Rps20p (Lys 6/8 Arg) mutant or deleted Rps7Bp (Figure 1F and G). Compared to WT, strains expressing the Rps20p (Lys 6/8 Arg) mutant or deletion of RPS7B had the same or only a two-fold increase Ssa4p expression (Figure 1F and G). Thus, during heat shock, an alternative mechanism probably mediates the ubiquitination of a different 40S ribosomal protein. The essential protein Rps3p is a good candidate because it regulates ribosome-associated quality control during the mammalian unfolded protein response in a Hel2- and Asc1-dependent manner (83).
The truncated Ssa4 nascent peptide should be excised from the 60S ribosomal subunit for degradation. Peptides are ubiquitinated by the E3 ligase Ltn1p and degraded by the UPS (53,54). We HA-tagged Ssa4p in the N-terminus to identify an expected Ssa4 truncated peptide of ∼3.5 kDa. Since small proteins are challenging to detect by western blotting, we aimed to find the larger polyubiquitinated form by inhibiting its UPS-mediated degradation with MG132. Under these conditions, we expected the polyubiquitinated 3xHA-Ssa4p to accumulate in WT cells and be absent in ltn1Δ yeast. However, we only identified the full-length 3xHA-Ssa4p and two smaller aberrant products that were the same in WT and ltn1Δ cells under mock- or MG132-treated conditions (Figure 1I). This result suggested that an alternative mechanism degraded the aberrant Ssa4 peptide. Since it only has one Lys in its sequence, it might not be a subject for Ltn1-directed ubiquitination, and it could be degraded instead by autophagy or proteases in the cytosol (84,85).
Asc1p promotes SSA4 mRNA degradation during recovery from heat shock
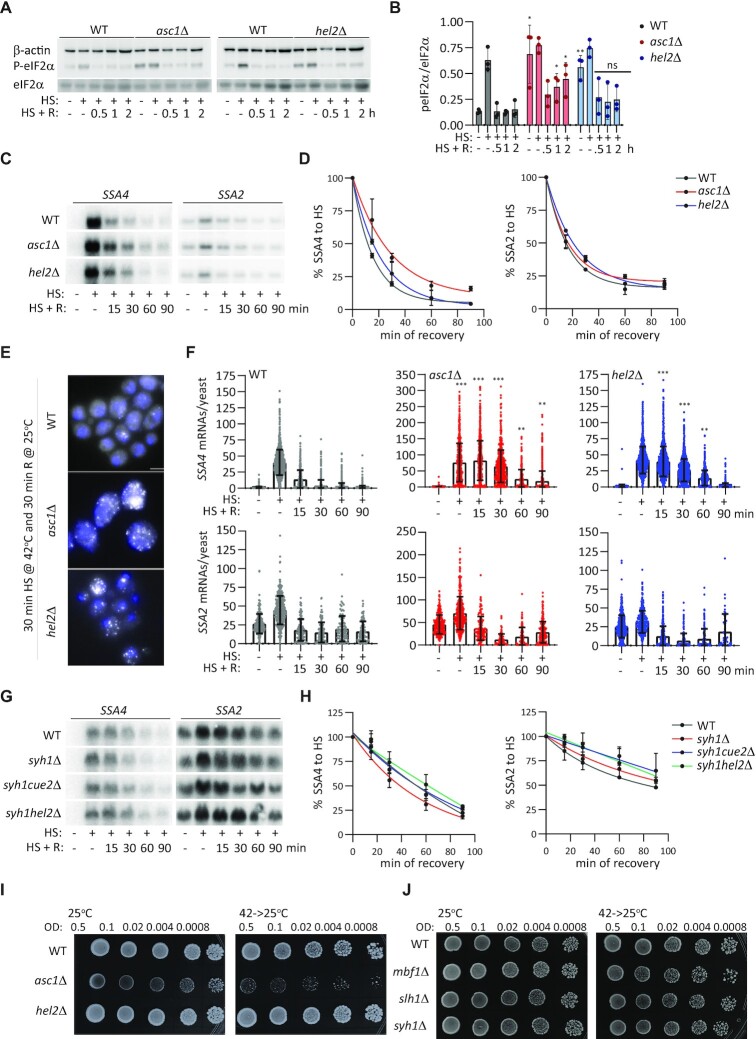
We pondered two non-exclusive options to explain the regulation of Ssa4p synthesis during heat shock but not during recovery: Asc1p and Hel2p could only repress SSA4 mRNA translation during heat shock, and/or rapid SSA4 mRNA degradation at the permissive temperature could prevent its translation during recovery. To investigate the first option, we considered that heat shock triggers the phosphorylation of eIF2α, favoring the translation of inducible HSP mRNAs, and that in asc1Δ and hel2Δ cells, Gcn2p recognizes the ribosome collision and phosphorylates eIF2α even under basal conditions to activate the integrated stress response (ISR) (61,86). Accordingly, asc1Δ and hel2Δ strains had higher P-eIF2α levels than the WT strain under basal conditions, and all strains displayed P-eIF2α during heat shock. Interestingly, P-eIF2α decreased to WT basal levels within 15 min of recovery, even in the asc1Δ and hel2Δ cells, which could hinder SSA4 mRNA translation during recovery and thus decrease the regulatory effects of Asc1p and Hel2p (Figures 2A, B, and 1B). These results point to specific repression of SSA4 mRNA translation by Asc1p and Hel2p during heat shock.
Figure 2.
SSA4 mRNA is stabilized in asc1Δ and hel2Δ cells during recovery from heat shock. (A) Western blots to detect eIF2α phosphorylation. P-eIF2α, total eIF2α and β-actin were quantified in the WT, asc1Δ, and hel2Δ strains under basal, heat shock, and recovery conditions. (B) Quantification of eIF2α phosphorylation. P-eIF2α band intensities were divided by their corresponding total eIF2α intensities. Bars indicate the mean and SD of three independent experiments, each represented by a dot. *P < 0.05, **P < 0.001 by unpaired t-test. (C) Northern blots to detect the expression of SSA4 and SSA2 mRNAs in the WT, asc1Δ and hel2Δ strains under basal conditions (25°C), after 30 min of heat shock at 42°C (H), and at the indicated recovery time points (R). (D) Quantification of the half-lives of SSA mRNAs during recovery. Band intensities were normalized to the methylene blue staining and are expressed relative to the heat shock band for each strain (considered to be 100% induction) to obtain decay curves and calculate half-lives (t1/2 of SSA4 mRNA: WT 16’, asc1Δ 25′, and hel2Δ 19’; t1/2 of SSA2 mRNA: WT 19’, asc1Δ 22’ and hel2Δ 27’). Datapoints on the curves indicate the mean and SD of two independent experiments. (E) Representative smFISH images of SSA4-MS2V6 mRNA after 30 min recovery in the WT, asc1Δ, and hel2Δ yeast strains. Scale bar: 5 μm. (F) Quantification of SSA4 or SSA2 mRNAs per yeast cell in each strain were detected by smFISH. Bars indicate the mean and SD of three experiments; dots represent individual cells (n= 600–1200). The unpaired t-test was used to compare each time point to the basal condition. (G) Northern blots to detect the expression of SSA4 and SSA2 mRNAs in the WT, syh1Δ, syh1/cue2Δ and syh1/hel2Δ strains under basal conditions (25°C), after 30 min of heat shock at 42°C (H), and at the indicated recovery time points (R). (H) Decay plot of SSA mRNAs during recovery. Band intensities were normalized to the methylene blue staining and are expressed relative to the heat shock band for each strain (considered to be 100% induction) to obtain decay curves and calculate half-lives (I, J). Spot assays of the WT, asc1Δ, and hel2Δ strains and WT, mbf1Δ, slh1Δ and syh1Δ strains grown at 25ºC and recovering at 25ºC after 16 h at 42ºC. OD, optical density at 600 nm.
To consider the role of mRNA stability in Ssa4p expression during recovery, we quantified SSA mRNA levels at the basal temperature, during heat shock, and after recovery for 15, 30, 60 and 90 min. Ribosome collisions can lead to endonucleolytic cleavage and the rapid degradation of problematic mRNAs by NGD (42,52,56,58,59). Thus, the stabilization of ribosome collisions by Asc1p and Hel2p could promote rapid SSA4 mRNA decay during recovery. We detected full-length SSA4, SSA3, SSA2 and SSA1 mRNAs by northern blot in the untagged strains using antisense probes against their 3′ UTRs, which contain the most distinct nucleotide sequences between them (Figure 2C, D and Supplementary Figure S2A and S2B). As expected, all SSA mRNAs were highly induced upon heat shock and rapidly returned to basal levels during recovery in the WT strain. The asc1Δ and hel2Δ strains only prolonged SSA4 mRNA expression during recovery without affecting the decay of the rest of the SSA mRNAs. We plotted the intensities of the SSA4 and SSA2 mRNA bands to calculate their half-lives by non-linear regression, which best fitted the curves connecting mRNA intensities during heat shock and recovery conditions. The effect was specific to SSA4 mRNA, with 2.5- and 1.35-fold increases in its half-life upon deletion of ASC1 and HEL2, respectively (Figure 2D).
We confirmed this prolonged SSA4 mRNA stability and the lack of effects on SSA2 mRNA stability during recovery by smFISH in the SSA4- and SSA2-3 × HA-12MSV6 strains, respectively (Figure 2E and F), using a fluorescent probe against the MS2V6 sequence, and quantified them with FISH-quant (70). The average number of single SSA4 mRNAs per cell was doubled in the asc1Δ strain compared to the WT and hel2Δ strains, probably because they are larger cells and produce more mRNAs to compensate for their volume, as SSA2 mRNA was also more abundant. In the WT strain, most SSA4 mRNAs were cleared within 30 min of recovery, while some hel2Δ cells and the majority of asc1Δ cells retained significantly higher numbers of SSA4 mRNAs until after 90 min of recovery (P < 0.05, by unpaired t-test). Since only 5–10% of cells in all strains show staining for transcription sites during recovery, this result indicated a prolonged SSA4 mRNA half-live. All strains showed similar patterns of SSA2 mRNA induction and decay during heat shock and recovery (Figure 2F). In the asc1Δ and hel2Δ strains, smFISH analyses revealed a delay in peak SSA4 mRNA expression from heat shock to after 15 min of recovery. Since we could not quantify the contributions of nascent transcripts to the total mRNA pools by smFISH, we attribute this discrepancy with the northern blot results to turnover of the cytoplasmic mRNA population during the first 15 min of recovery. SSA4 mRNA decay may occur faster than the export of nascent transcripts to the cytoplasm in WT cells but not in the asc1Δ and hel2Δ strains, shifting the timing of peak expression.
To analyze the role of colliding ribosomes and RQC in SSA4 mRNA degradation by NGD, we quantified SSA4 mRNA levels in yeast in which the main players of the two NGD pathways were deleted. The predominant pathway involves Syh1p, and the secondary comprises Hel2p and the endonucleolytic cleavage by Cue2p (59). We detected full-length SSA4 and SSA2 mRNAs by northern blot in the untagged strains using antisense probes against their 3′ UTRs (Figure 2G). Compared to WT, syh1Δ, syh1Δ /cue2Δ and syh1Δ /hel2Δ had the same SSA4 and SSA2 mRNA expression during heat shock and recovery and similar half-lives during recovery (Figure 2H). These results indicate that the degradation of SSA4 mRNA is independent of NGD factors. We suggest that the prolonged SSA4 mRNA half-lives detected in asc1Δ and hel2Δ cells might be due to elongating ribosomes protecting the mRNA from degradation or an NGD-independent role of these proteins in destabilizing SSA4 mRNA during recovery.
Of all strains, asc1Δ has the highest Ssa4p expression and the longest SSA4 mRNA half-live; thus, it should be better equipped to survive heat shock than the WT and strains deleted of other RQC and NGD factors. However, as previously reported (48), the asc1Δ strain grew the slowest and was the most sensitive to heat shock, suggesting that its growth phenotype is independent of the expression of the cytoprotective Ssa4p (Figure 2I). Overall, our results indicate that SSA4 mRNA degradation during recovery is independent of NGD and asc1Δ cells prolonged SSA4 mRNA stability longer than hel2Δ cells, indicating that Asc1p plays an additional role in the SSA4 mRNA decay during recovery from heat shock. Since prolonged SSA4 mRNA stability during recovery did not further increase Ssa4p levels, Asc1p and Hel2p might regulate SSA4 mRNA translation during heat shock and stability during recovery by independent mechanisms.
Optimizing the SSA4 coding sequence escapes the RQC, but its mRNA is still stabilized in asc1Δ cells
To gain insight into the mechanisms by which Asc1p and Hel2p regulate SSA4 mRNA translation and decay, we optimized the SSA4 CDS to bypass ribosome stalling. We synonymized the CDS with a computational pipeline that considers the codon context of mRNA translate under specific conditions (87). Interestingly, the optimal SSA4 CDS acquired the codons used in SSA3, SSA2, and SSA1 for conserved amino acids, further supporting a role for SSA4’s specific enrichment in low-frequency codons (Supplementary Figure S3). To generate an SSA4 codon-optimized (Opt) strain, we substituted the endogenous SSA4 CDS with the SSA4-Opt sequence, conserving the 5′- and 3′-UTR sequences. We tagged SSA4-Opt with 3 × HA and 12MSV6 to study its effects on SSA4 mRNA translation in WT cells (Opt-WT) and the roles of Asc1p and Hel2p in its translational regulation.
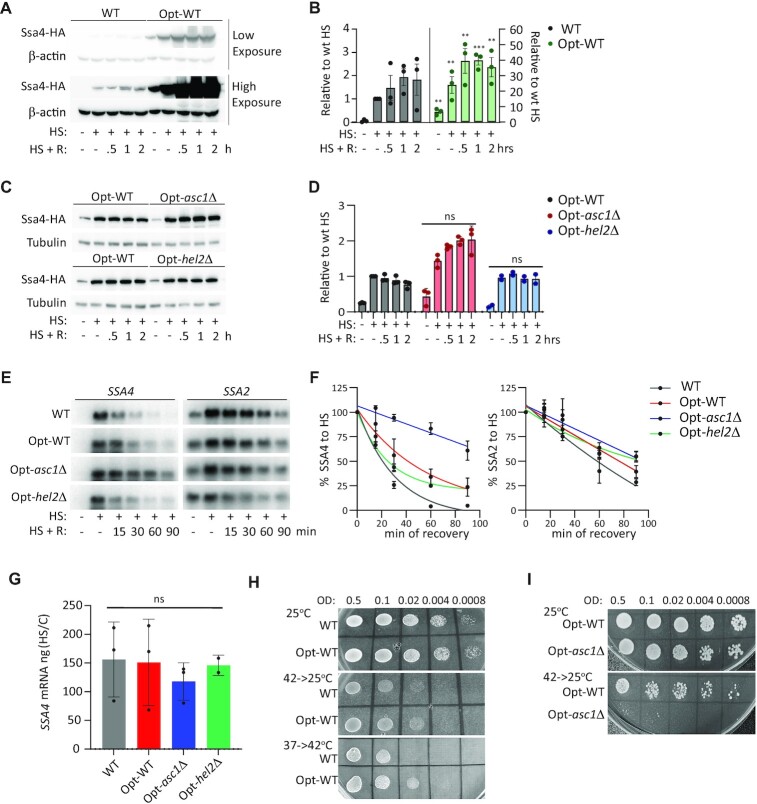
Opt-WT cells expressed Ssa4p even in basal conditions, indicating that the low-frequency codons in the WT CDS prevent the spurious accumulation of Ssa4p in the absence of stress. In addition, codon optimization dramatically increased Ssa4p upregulation during heat shock and recovery by 20- and 40-fold, respectively (Figure 3A and B). Therefore, the WT SSA4 CDS suppresses Ssa4p synthesis during heat shock. Remarkably, Ssa4p induction upon heat shock was similar in the Opt-WT and Opt-hel2Δ strains, further demonstrating that Hel2p's repression of Ssa4p synthesis depends on these low-frequency codons (Figure 3C and D). Likewise, SSA4 codon optimization decreased Ssa4p induction during heat shock from 6-fold in the WT-asc1Δ strain to less than 2-fold in the Opt-asc1Δ strain (Figures 1B, D, 3C, and D). We concluded that the WT SSA4 CDS is necessary for Asc1p and Hel2p to repress its translation during heat shock.
Figure 3.
A codon-optimized SSA4 mRNA escapes the RQC mechanism, but it is still destabilized by Asc1p. (A) Expression of the 3 × HA-Ssa4p in the WT and Opt strains under basal conditions (25°C), after 30 min of heat shock at 42°C (HS), and at the indicated recovery time points (R). β-Actin was used as a loading control. (B) Quantification of 3 × HA-Ssa4p expression. Band intensities were normalized to their corresponding β-actin band and are expressed relative to the normalized expression of SSA4-WT yeast under heat shock. Bars indicate the mean and SD of three independent experiments, each represented by a dot. *P< 0.05, **P< 0.001 and ***P< 0.0001 (by unpaired t-test). (C) Expression of 3 × HA-Ssa4p in the WT, Opt-WT, Opt-asc1Δ and Opt-hel2Δ strains under basal conditions (25°C), after 30 min of heat shock at 42°C, and at the indicated recovery time points. Tubulin was used as a loading control. (D) Quantification of Ssa4p expression. Band intensities were normalized to their corresponding tubulin band and are expressed relative to the normalized expression of WT yeast under heat shock. Bars indicate the mean and SD of three independent experiments, each represented by a dot. *P< 0.05, **P< 0.001, ***P< 0.0001, ns, not significant (by unpaired t-test). (E) Northern blot detection of SSA4 and SSA2 mRNAs in the WT, Opt-WT, Opt-asc1Δ and Opt-hel2Δ strains under basal conditions, after 30 min of heat shock at 42°C, and at the indicated recovery time points. (F) Quantification of the half-lives of SSA4 and SSA2 mRNAs during recovery. Band intensities were normalized to the methylene blue staining and are expressed relative to the heat shock band for each strain (considered to be 100% induction) to obtain decay curves and calculate half-lives (t1/2 of OPT mRNA: in WT 22’, Opt-WT 40’, Opt-asc1Δ 100’ and Opt-hel2Δ 27’;t1/2 of SSA2 mRNA: in WT 57’, Opt-WT 77’, Opt-asc1Δ 92’ and Opt-hel2Δ 118’). (G) Quantification of the fold induction of SSA4 mRNA at 30 min of heat shock at 42°C. Induction was calculated by dividing the amount of cDNA in ng in heat shock to control. Bars indicate the mean and SD of three independent experiments, each represented by a dot (by unpaired t-test); ns, not significant. (H and I) Spot assays of SSA4-WT and -Opt (H) and Opt-WT, Opt-asc1Δ and Opt-hel2Δ (I) strains grown at 25ºC, recovering at 25ºC after 16 h at 42ºC, and preconditioned by mild stress (37ºC for 1 h, 6 h at 25ºC, and then heat shock at 42ºC). OD, optical density at 600 nm.
We next investigated whether the SSA4-Opt CDS stabilizes SSA4 mRNA and prevents Asc1p and Hel2p from destabilizing it during recovery. We compared the expression and stability of SSA4-Opt mRNA (and SSA2 mRNA as a control) in the Opt-WT, Opt-asc1Δ and Opt-hel2Δ strains after 30 min heat shock at 42°C followed by 15, 30, 60 and 90 min of recovery at 25°C by northern blot. SSA4-Opt mRNA was 1.5 times more stable than SSA4-WT mRNA during recovery and completely abolished the effect of Hel2p on SSA4 mRNA stability. Therefore, increasing the translational efficiency of Opt-SSA4 mRNA slightly increased its mRNA stability. However, the rapid degradation of SSA4-Opt mRNA further supports that its main degradation pathways are independent of its codon optimality and NGD. Remarkably, the SSA4-Opt mRNA half-life was 2.5 times longer in the Opt-asc1Δ strain compared to Opt-WT, while the SSA2 mRNA half-life was unaffected (Figures 3E and 4E). Optimizing SSA4 CDS did not change the upregulation of SSA4 mRNA induction at 30 min of heat shock. Likewise, the Opt-asc1Δ and Opt-hel2Δ strains had similar SSA4 mRNA induction as the Opt-WT (Figure 3G). This result supports a role for Asc1p in promoting SSA4 mRNA decay during recovery that it is independent of the SSA4 CDS.
Figure 4.
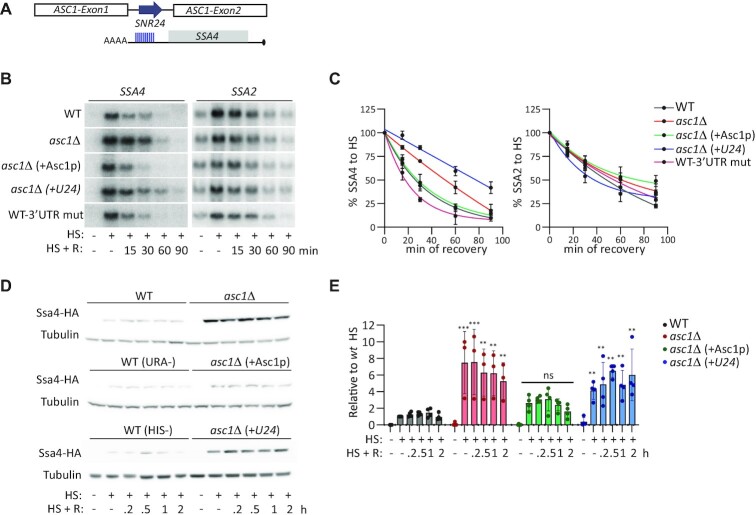
Asc1p, not SNR24, regulates SSA4 mRNA translation and stability. (A) Schematic of the ASC1 locus. It contains two exons and an intron, which encodes the small nucleolar RNA SNR24 (U24) upon splicing. The 10-nucleotide region in the 3′ UTR of SSA4 mRNA that is complementary to U24 is indicated by blue lines. (B) Northern blots to detect the expression of SSA4 and SSA2 mRNAs in the WT, asc1Δ, asc1Δ expressing the CDS of Asc1p (asc1Δ +Asc1p), and asc1Δ+U24 strains, and the WT strain with five 3′ UTR mutations in the U24 complementarity region (WT-3′ UTR mut) under basal conditions, after 30 min of heat shock at 42°C (HS), and at the indicated recovery time points (R). (C) Quantification of the half-lives of SSA4 and SSA2 mRNAs during recovery. Band intensities were normalized to the methylene blue staining and are expressed relative to the heat shock band for each strain (considered to be 100% induction) to obtain decay curves and calculate half-lives (t1/2 of SSA4 mRNA: in WT 28’, asc1Δ 51’, asc1Δ +Asc1p 21’, asc1Δ +U24 78’, WT-3′ UTR mut 20’;t1/2 of SSA2 mRNA: WT 48’, asc1Δ 60’, asc1Δ +Asc1 68’, asc1Δ +U24 36’, WT-3′ UTR mut 55′). (D) Expression of 3 × HA-Ssa4p in the indicated strains under basal conditions (25°C), after 30 min of heat shock at 42°C, and at the indicated recovery time points. Tubulin was used as a loading control. (E) Quantification of Ssa4p expression. Band intensities were normalized to their corresponding tubulin band and are expressed relative to the normalized expression of WT yeast under heat shock. Bars indicate the mean and SD of three independent experiments, each represented by a dot. **P< 0.001, ***P< 0.0001, ns, not significant (by unpaired t-test).
Given the existing notion that heat shock maximizes Ssa4p production to cope with protein misfolding, learning that the SSA4 CDS attenuates its own translation via the RQC was unexpected. Thus, we investigated if Ssa4p overexpression is toxic. The WT and Opt-WT strains grew similarly at the permissive temperature and during recovery from stress, and Ssa4p overexpression enhanced the survival of Opt-WT cells to heat shock upon preconditioning (Figure 3H). However, Ssa4p overexpression did not overcome the increased vulnerability of asc1Δ cells to heat shock (Figure 3I). Thus, this strain's inability to survive heat shock is independent of Asc1p's role in regulating Ssa4p expression. Overall, our results support two separate functions of Asc1p in controlling the life cycle of SSA4 mRNA in the cytoplasm: translational repression during heat shock, a role which relies in the RQC mechanism (shared with Hel2p, Slh1p and Mbf1p) and depends on the WT SSA4 CDS, and mRNA decay during recovery, which is independent of the CDS and SSA4 mRNA translation efficiency.
Asc1p regulates SSA4 mRNA, not its intronic U24 small nucleolar (sno) RNA
The ASC1 locus contains an intron that encodes the snoRNA SNR24, known as U24 (88,89). U24 is a C/D box snoRNA that guides 2'-O-methylation of the 25S ribosomal RNA (rRNA). This role requires at least 10 nucleotides (nts) of perfect complementarity (90–93). Sequence analysis of the SSA4 3′ UTR revealed 10 nts of perfect complementarity with the U24 sequence (TGAAGTAGCA; Figure 4A). Since the 3′ UTR is required to destabilize SSA4 mRNA during recovery (29), we sought to determine whether the deletion of U24 stabilizes SSA4 mRNA in the asc1Δ strain, rather than Asc1p. Thus, we restored either the expression of Asc1p or U24 by inserting centromeric plasmids into the asc1Δ strain (as described in (88)). We also mutated the 3′ UTR of the endogenous SSA4 mRNA to prevent U24 binding (TGTTCATGCA; WT-3′ UTR mut) to determine whether this sequence destabilizes SSA4 mRNA during recovery. Northern blot analysis of SSA4 mRNA levels in basal, heat shock, and recovery conditions showed that Asc1p expression destabilized SSA4 mRNA during recovery, while exogenous U24 expression in the Asc1Δ strain increased SSA4 mRNA stability by 3.4 times. Accordingly, mutating the 3′ UTR U24 binding sequence did not affect SSA4 mRNA stability during recovery (Figure 4B and C). To validate our results, we investigated the roles of Asc1p and U24 in destabilizing the SSA4-Opt mRNA. As expected, Asc1p expression was sufficient to destabilize SSA4-Opt mRNA in the Opt-asc1Δ strain and revert its half-life to that observed in the Opt-WT strain. In contrast, U24 did not change the SSA4-Opt mRNA half-life and neither Asc1p nor U24 changed SSA2 mRNA stability in the Opt-asc1Δ (Figure 4B, C, and Supplementary Figure S4B and C). These results indicate that Asc1p regulates SSA4 mRNA stability during recovery.
Although it is well known that Asc1p and Hel2p act together to regulate the translation of faulty mRNAs and trigger the RQC mechanism (47), we investigated whether U24 also regulates SSA4 mRNA translation. We quantified Ssa4p expression in asc1Δ cells expressing either Asc1p or U24. While Asc1p expression restored Ssa4p induction to the WT level, yeast expressing U24 without Asc1p failed to rescue the high Ssa4p expression of the asc1Δ strain (Figure 4D and E). Restoring either Asc1p or U24 in the Opt-Asc1Δ strain did not change Ssa4p synthesis during heat shock and recovery. Altogether, these experiments indicate that U24 does not regulate the SSA4 mRNA life cycle, strongly supporting two independent roles for Asc1p in deciding the fate of cytoplasmic SSA4 mRNA. First, Asc1p regulates SSA4 mRNA translation during heat shock in response to its low-frequency codons and second, it regulates SSA4 mRNA stability during recovery independently of its CDS or translation efficiency.
Asc1p binding to ribosomes is required for the RQC mechanism to regulate SSA4 mRNA translation but not to destabilize it during recovery
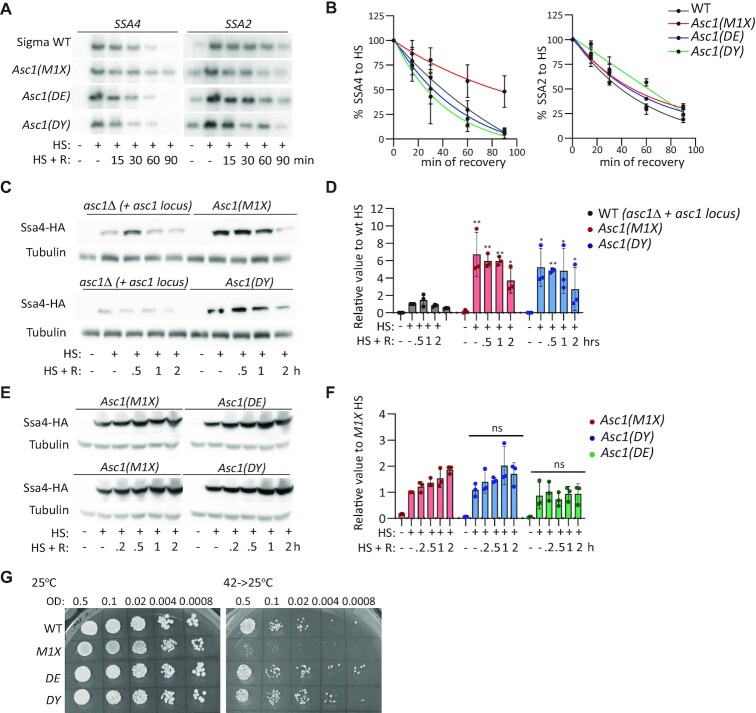
Asc1p is a multifunctional protein with roles in and out of the ribosome (89). We examined whether Asc1p binding to the ribosome is needed to regulate SSA4 mRNA translation during heat shock and destabilize SSA4 mRNA during recovery. We obtained three ASC1 mutants, M1X, DE and DY, in the yeast sigma background described by Thompson et al (89). In the M1X mutant the start codon was substituted by a stop codon in the ASC1 CDS that prevents Asc1p expression but maintains ASC1 and U24 RNA levels. The DE mutant holds two substitutions, R38D and K40E, in the N-terminus that decrease Asc1p's binding to ribosomes. The DY mutant, D109Y, has a lower ribosome binding capacity than the DE mutant and defects in NGD (89). Of these mutants, only M1X prolonged the half-life of SSA4 mRNA during recovery, and none of the strains changed SSA2 mRNA stability (Figure 5A and B). This result confirmed our previous findings in the BY4741 background, showing that Asc1p, but not U24, destabilizes SSA4 mRNA during recovery from heat shock. They also show that SSA4 mRNA decay is not mediated by NGD, because the DY mutant behaved like the WT strain. Finally, since the SSA4 mRNA half-lives in the DE (24 min) and D109Y (27 min) strains were similar to the WT sigma strain (30 min), Asc1p's ability to regulate SSA4 mRNA stability is unrelated to ribosome binding. These results were consistent with Asc1p destabilizing both SSA4-WT and SSA4-Opt mRNAs, and an additional role for Asc1p independent of the SSA4 CDS (Figures 2C–F, 3E and F).
Figure 5.
Roles of ASC1 gene, Asc1p-null, and ribosome binding mutants on SSA4 mRNA stability and translation and in heat shock survival. (A) Northern blots to detect the expression of SSA4 and SSA2 mRNAs in the sigma WT, Asc1_M1X, Asc1(DE) and Asc1(DY) strains under basal conditions (25°C), after 30 min of heat shock at 42°C (HR), and at the indicated recovery time points (R). (B) Quantification of the half-lives of SSA4 and SSA2 mRNAs during recovery. Band intensities were normalized to the methylene blue staining and are expressed relative to the heat shock band for each strain (considered to be 100% induction) to obtain decay curves and calculate half-lives (t1/2 of SSA4 mRNA: in sigma WT 30’, Asc1_M1 × 84’, Asc1 DE 24’and Asc1 DY 27’;t1/2 of SSA2 mRNA: in sigma WT 39’, Asc1_M1 × 47’, Asc1 DE 43′ and Asc1 DY 53′). (C) Expression of 3 × HA-Ssa4p in asc1Δ BY4741 strains expressing the full ASC1 locus or ASC1 M1X or DY mutants under basal conditions (25°C), after 30 min of heat shock at 42°C, and at the indicated recovery time points. Tubulin was used as a loading control. (D) Quantification of Ssa4p expression. Band intensities were normalized to their corresponding tubulin band and are expressed relative to the normalized expression of WT yeast under heat shock. Bars indicate the mean and SD of three independent experiments, each represented by a dot. *P< 0.05, **P< 0.001 (by unpaired t-test). (E) Expression of 3 × HA-Ssa4p in sigma strains expressing Asc1p mutants under basal conditions (25°C), after 30 min of heat shock at 42°C, and at the indicated recovery time points. Tubulin was used as a loading control. (F) Quantification of Ssa4p expression. Band intensities were normalized to their corresponding tubulin band and are expressed relative to the normalized expression of WT yeast under heat shock. Bars indicate the mean and SD of three independent experiments, each represented by a dot (by unpaired t-test); ns, not significant. (G) Spot assays of sigma WT, M1X, DE and DY strains plated on YPD under control (25ºC, left) and recovery (42ºC for 16 h then incubated at 25ºC, right) conditions. OD, optical density at 600 nm.
We next investigated whether Asc1p needs to bind the ribosome to repress Ssa4p synthesis. We used centromeric plasmids to express the WT, M1X, or DY ASC1 genes in the asc1Δ BY4741 strain, which had the SSA4 locus tagged with 3 × HA-12MS2V6. The M1X strain expressed ∼7 times more Ssa4p than the WT strain upon heat shock, as we previously observed with the asc1Δ. Interestingly, Ssa4p induction in the DY strain resembled that of the M1X strain upon heat shock, implying that low binding of Asc1p to the ribosome is not sufficient to repress SSA4 mRNA translation (Figure 5C and D). We confirmed these results in the original sigma strains by tagging the SSA4 locus with 3 × HA-12MS2V6 in the M1X, DE, and DY strains. All three strains showed similar Ssa4p induction upon heat shock and recovery (Figure 5E and F). Therefore, Asc1p binding to the ribosome is needed for its translational control of SSA4 mRNA during heat shock, further confirming the involvement of the RQC mechanism in regulating Ssa4p synthesis.
We also investigated whether Asc1p must bind to ribosomes to promote heat shock survival. The expression of the low ribosome-binding DE and DY mutants enabled asc1Δ yeast to survive heat shock. Thus, Asc1p's prosurvival role in heat shock is independent of ribosomal binding and the regulation of SSA4 mRNA stability and translation (Figure 5G). Collectively, these results suggest that Asc1p repression of SSA4 mRNA translation requires its binding to the ribosome, and Asc1p-mediated destabilization of SSA4 mRNA is independent of ribosome binding. Thus, Asc1p probably uses two independent mechanisms, in and out of the ribosome, to regulate SSA4 mRNA translation and decay.
Heat shock enhances asc1p binding to rps28bp and rps19ap, suppressing SSA4 mRNA translation
To identify the molecular partners sustaining the translational regulation and decay roles of Asc1p in SSA4 mRNA fate in the cytoplasm, we FLAG-tagged endogenous Asc1p, immunoprecipitated it from yeast growing at 25°C or after 60 min of heat shock at 42°C, and performed liquid chromatography coupled to mass spectrometry (Figures 6A, Supplementary Figure 5A, and Supplementary Table S4). Heat shock did not induce any posttranslational modifications of Asc1p, but significantly changed its interactome as detected in three independent replicas. Two main cluster of proteins were preferentially bound by Asc1p upon stress, stress-related proteins such as chaperones (proteins labeled in green circles), suggesting that Asc1p might partially unfold during heat shock, and 40S ribosomal proteins (labeled in red circles), suggesting that heat changes the interaction between ribosomal proteins or their composition (Figure 6A). The four ribosomal proteins with a significantly tighter interaction with Asc1p during heat shock were Rps2, Rps7B, Rps19B (and paralog Rps19A) and Rps28A.
Figure 6.
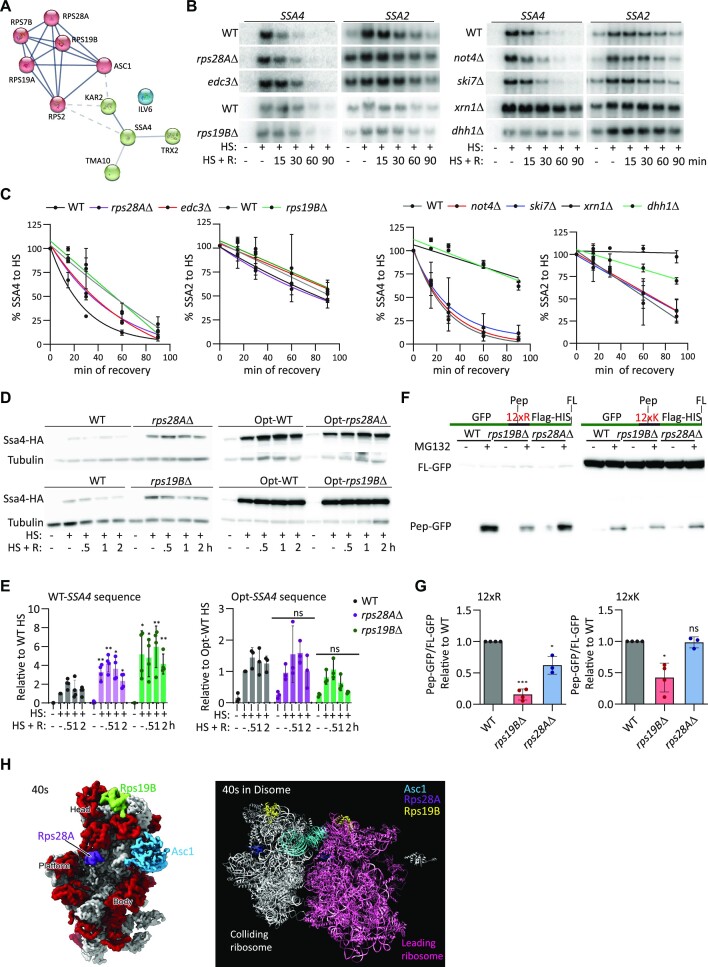
The ribosomal proteins Rps28Ap and Rps19Bp interact with Asc1p during heat shock to repress Ssa4p synthesis. (A) Asc1p interaction network showing proteins significantly enriched (P< 0.05) >1.5-fold after 30 min of heat shock compared to basal conditions in three replicas ((plotted with STRING), (blue lines = known interactions), (dotted lines = edges between clusters)). (B) Northern blots to detect the expression of SSA4 and SSA2 mRNAs in WT, rps28AΔ and edc3Δ and WT vs rps19BΔ (left) and WT, not4Δ, ski7Δ, xrn1Δ and dhh1Δ (right) under basal conditions (25°C), after 30 min of heat shock at 42°C, and at the indicated recovery time points. (C) Quantification of the half-lives of SSA4 and SSA2 mRNAs during recovery. Band intensities were normalized to the methylene blue staining and are expressed relative to the heat shock band for each strain (considered to be 100% induction) to obtain decay curves and calculate half-lives (t1/2 of SSA4 mRNA: in WT 32’, rps28AΔ 34’, edc3Δ 35′, WT 25′and rps19BΔ 27’;t1/2 of SSA2 mRNA: in WT 70’, rps28AΔ 78, edc3Δ 92’, and WT 45’, rps19BΔ 48’). (D) Expression of 3 × HA-Ssa4p expression in WT, rps28AΔ and rps19BΔ yeast and Opt-WT, Opt-rps19BΔ, and Opt-rps28AΔ strains under basal conditions (25°C), after 30 min of heat shock at 42°C, and at the indicated recovery time points. Tubulin was used as a loading control. (E) Quantification of Ssa4p expression. Band intensities were normalized to their corresponding tubulin band and are expressed relative to the normalized expression of WT yeast under heat shock. Bars indicate the mean and SD of three independent experiments, each represented by a dot. *P< 0.05, **P< 0.001, ns, not significant (by unpaired t-test). (F) Ectopic expression of GFP-R12-HIS or GFP-K12-HIS in WT, rps19BΔ and rps28AΔ strains treated with or without 0.25 M of MG132 treatment for 6 hours. G. Quantification of ratio of Pep-GFP (fragment) to Full length (FL)-GFP relative to WT expression in WT, rps19BΔ and rps28AΔ strains under MG132 exposure. Bars indicate the mean and SD of three independent experiments, each represented by a dot. *P< 0.05, **P< 0.001 (by unpaired t-test). (H) Structure of the S. cerevisiae 40S subunit as a monomer (left, surface representation, ribosomal proteins in red and ribosomal RNA in gray) and as a disome (right, ribbon representation) with Asc1p, Rps28Ap and Rps19Bp indicated.
Next, we investigated the contribution of these ribosomal proteins to Asc1p activity on SSA4 expression. We previously demonstrated that RPS7B deletion did not affect Ssa4p synthesis (Figure 1E and F). Since Rps2p is essential, we validated the interaction of Asc1p with Rps28Ap and Rps19Bp by immunoprecipitation followed by western blotting (Supplementary Figure S5). RPS28A or RPS19B genes were deleted to evaluate their role in SSA4 mRNA stability and translation (Figure 6B–E). The ribosomal protein Rps28Ap was of particular interest because it functions outside the ribosome to degrade YRA1 pre-mRNA and RPS28B mRNA by interacting with enhancer of mRNA decapping protein 3 (Edc3p) (94). Therefore, we also deleted EDC3 to analyze their roles in SSA4 mRNA decay and translation. SSA4 and SSA2 mRNAs had similar half-lives in the rps19BΔ and WT strains (Figure 6B and C). SSA2 mRNA was also unaffected in the rps28AΔ and edc3Δ cells, while the half-life of SSA4 mRNA was prolonged by 1.5 times (Figure 6B and C). This increase in SSA4 mRNA stability was attenuated compared to ASC1 deletion, which increased it by 2.5 times (Figure 2B). We concluded that Rps28Ap, Edc3p and Rps19Bp do not function with Asc1p outside the ribosome to destabilize SSA4 mRNA during recovery.
To define the mechanism by which SSA4 mRNA is degraded during recovery, we investigated the role of well-known decapping and deadenylation factors. We found that SSA4 mRNA degradation during recovery depends on the DEAD-box helicase and mRNA decapping enzyme Dhh1p and the exoribonuclease Xrn1p and is independent of deadenylase CCR4-Not complex component Not4p and the exosome component Ski7p (Figure 6B and C). These results suggest a role for Asc1p in activating factors that decap the SSA4 mRNA and favor its 5′ to 3′ degradation by the exonuclease Xrn1p independently of the Ccr4–Not complex, which has been implicated in the degradation of mRNAs with pro-stalling codons (55). Not4Δ yeast had similar Ssa4p induction that WT cells, further supporting that ubiquitination of Rps7Bp is not important for the regulation of SSA4 mRNA translation by ribosomal collisions and RQC (Figure 1F, G, Supplementary Figure S5B and C). It is important to note that the prolonged half-life of SSA4 mRNA in the xrn1Δ and dhh1Δ strains did not further increase Ssa4p during recovery, indicating that SSA4 mRNA translation is suppressed during recovery (Supplementary Figure S5B and C).
Given the increased interaction of Rps28Ap and Rps19Bp with Asc1p during heat shock, we next investigated their roles in regulating SSA4 mRNA translation. Remarkably, rps28aΔ and rps19bΔ cells induced significantly more Ssa4p than WT cells during heat shock, supporting a role for Rps28Ap and Rps19Bp in the regulation of SSA4 mRNA translation, probably through the RQC mechanism (Figure 6D and E). This induction is comparable to that exhibited by asc1Δ cells (Figures 1B, D, 4D and E). To determine if this novel role for Rps28Ap and Rps19Bp as translational regulators of SSA4 expressiondepends on its binding to the ribosome and the presence of low-frequency codons, we deleted RPS28A or RPS19B in the SSA4-Opt strain. In this case, the absence of either Rps28Ap or Rps19Bp did not affect Ssa4p expression during heat shock, pointing to Rps28Ap and Rps19Bp as new ribosomal components of the RQC mechanism, at least under heat shock conditions (Figure 6D and E). To rule out the possibility that these factors play an indirect role in RQC by recruiting Asc1p to the polysomes during heat shock, we analyze the distribution of Asc1p in monosome and polysomes obtained from heat shocked WT, rps28AΔ and rps19BΔ cells (Supplementary Figure S5D). Heat shocked yeast had a wide 80S peak and flat polysomes because heat shock represses global translation (95). We pooled together the polysome fractions to enrich their proteins and detected Asc1-3xFlag in the monosome and polysome by western-blotting. In all strains, Asc1p is almost exclusively localized with polysomes (Supplementary Figure S5D). Therefore, Rps28Ap and Rps19Bp directly affect RQC and SSA4 mRNA translation regulation by ribosome collisions during heat shock.
Since we proposed a novel role of Rps28Ap and Rps19Bp in quality controls induced by ribosome collisions, we clarified their broader RQC activity using two standard stalling reporters: GFP-R12-FlagHIS3 and GFP-K12-FlagHIS3 (96). We detected truncated GFP peptides (Pep-GFP) in MG132-treated WT yeast that were more prominent in the R12 than the K12 construct (Figure 6E and F). Compared to the WT, the fraction of truncated GFP peptides was significantly lower in the rps19BΔ strain. We observed the deletion of RPS19B to have a higher effect on R12 than K12 stretches. Deletion of RPS28A only reduced the formation of truncated peptides in the R12 construct, but the effect of Rps28A was much lower than the effect of Rps19Ap. These results indicate that Rps19Bp is a bona fide RQC factor. However, Rps28Ap might only affect certain stalling mRNAs under certain conditions. We identified the positions of Asc1p, Rps28Ap and Rps19Bp in both the 40S subunit ((4V7R) PDB data bank) and the published structure of the yeast disome (41); however, direct interactions between these three ribosomal proteins were not detected (Figure 6H). Since these structures were obtained from yeast growing under permissive temperatures, we speculate that the increased interactions between these ribosomal proteins upon heat shock could be due to either temperature-mediated changes in the 40S ribosome structure and/or the positions or amounts of additional factors linking them.
DISCUSSION
Cells rapidly adapt to survive harsh environmental conditions through the potent upregulation of HSPs. Regulatory elements controlling this quick and transient activation have been identified in the HSP70 promoter, which contains heat shock elements that direct transcription, and the 5′ and 3′ UTRs of HSP70 mRNA, which regulate translation and mRNA stability, respectively (3). Our work demonstrates that the CDS of SSA4, the most induced HSP70 gene in yeast, also regulates its expression. Surprisingly, its enrichment in low-frequency codons dampens Ssa4p synthesis during heat shock by activating the RQC mechanism, which feeds back to repress its own translation. Hence, our data argue that not all stress-induced gene expression pathways act to increase HSP70 expression during stress. Recently, a mechanism to attenuate HSP70 synthesis was found in mammalian cells undergoing heat shock. In this case, the regulation was independent of the HSP70 mRNA sequence and relied on the heat-induced non-coding RNA Heat, which reduces HSP70 transcriptional induction (97). In addition to the role of the SSA4 CDS during heat shock, we discovered that it also prevents the spurious accumulation of Ssa4p under optimal growth conditions. This observation suggests that the SSA4 CDS lessens the synthesis of Ssa4p at permissive temperatures and provides an extra checkpoint to tailor HSP70 levels to the burden of misfolded proteins both with and without stress.
As the four Ssa proteins have more than 80% amino acid identity, the use of nonoptimal codons provides the means to specifically regulate translation through ribosome decoding kinetics. In our study, we fully reverted the SSA4 CDS to the optimal codons used by SSA1 and SSA2 (87), revealing that for SSA4, codon low-optimality causes ribosome stalling instead of stretches of polybasic amino acids (which are shared by all SSA genes) (33,42,98,99). RQC and NGD components have primarily been studied under permissive conditions either by inserting a stretch of polybasic or rare amino acids in an endogenous or reporter gene or by deleting a stop codon so the polyA tail is translated into a stretch of basic arginine residues (60,96,100,101). Hence, we have revealed SSA4 as one of the few known endogenous mRNAs whose translation is controlled by ribosome stalling and the RQC mechanism (102).
Interestingly, RQC regulation of Ssa4p synthesis is restricted to heat shock, as indicated by experiments done in yeast deleted of ASC1 or HEL2. First, asc1Δ and hel2Δ cells did not augment the translation of SSA4-WT mRNA during recovery (Figure 1B and D). Secondly, expressing SSA4-Opt in the ASC1 and HEL2 deletion strains did not further augment the spurious accumulation of Ssa4p under non-stress conditions (Figure 3C and D). These results imply a mechanism boosting SSA4 mRNA translation under heat shock, which probably depends on eIF2α phosphorylation, as asc1Δ and hel2Δ cells only exhibited basal eIF2α phosphorylation at early recovery time points. Identifying these factors will also help define the pathway used by the ribosomes stalled in SSA4 mRNAs to repress translation. In mammalian cells, the RQC mechanism signals to inhibit translation initiation via ZNF598’s recruitment of GIGYF2 and 4EHP. 4EHP outcompetes eIF4E binding to the cap of the mRNA holding the stalled ribosomes (63). Yeast does not have a 4EHP orthologue, but SSA4 mRNA translation is unaffected by the deletion of SYH1, which has a GYF domain, suggesting an alternative mechanism in yeast (Supplementary Figure S2C and D).
Besides Asc1p and Hel2p, we found the canonical downstream RQC factors, Slh1p and Mbf1p, to regulate Ssa4p expression indicating that ribosomes stalled in SSA4 mRNA are disassembled by Slh1p and prevented from frameshifting by Mbf1 (76–79). However, not all RQC steps described for stalling mRNAs applied to SSA4. In the case of SLH1 deletion, the SSA4 mRNA translation is down- instead of up-regulated, as previously described for stalling mRNA reporters (51,46,80). In slh1Δ cells, reporters have an increased readthrough over the stalling sequence despite higher stalling picks in the mRNA (51). We propose that most of the leading 80S ribosomes stalled in SSA4 mRNA do not disassemble. However, if they do, collided ribosomes will not be able to continue translation due to their stabilization in collisions by Asc1p and Hel2p, forming a roadblock (61). Since we observe a smaller Ssa4-HAp, we propose that a few 80S disassemble, allowing the 40S to scan the downstream CDS and reinitiate translation at an appropriate AUG. Future experiment will determine the factors mediating ribosome splitting in the absence of Slh1p and whether they are specific of heat shock or certain stalling sequences.
Likewise, future experiments will identify the ubiquitinated ribosomal proteins acting in the SSA4 ribosome quality controls as neither Rps20p nor Rps7 does so. It might be possible that a stress dependent-E3 ligase cooperates with Hel2 to solve ribosome collisions induced under non-permissive conditions (83,103). The analysis of Asc1 interactome upon heat stress helped us to define the ribosomal proteins Rps28Ap and Rps19Bp as new RQC components. Beyond SSA4 mRNA, the Rps19B regulates well-known stalling RNA at permissive conditions. However, Rps28A had a lower than Rps19B or no effect in the formation of truncated peptides produced from stalled ribosomes. Thus, their tighter interaction with Asc1 upon heat stress might point to changes in the conformation of components of disomes that are either mRNA or temperature specific.
In yeast, the RQC mechanism and NGD are intimately connected (42,56). Further, recent work has shown that stalled ribosomes signal to the CCR4–NOT complex via Not5p to deadenylate the mRNA and trigger its decay (55). However, NGD did not trigger SSA4 mRNA decay, as shown by its high stability during heat shock and the discrete increase in the half-life of the optimized over the WT SSA4 mRNA during recovery. None of the factors involved in NGD pathways, Syh1p and Cue2p, destabilized SSA4 mRNA, further suggesting a decay mechanism independent of the stalled ribosomes (42,52,55–59). Therefore, it was unexpected to discover Asc1p's involvement in destabilizing SSA4 mRNA during recovery, independently of the SSA4 CDS or ribosome binding affinity. Asc1p is a multifunctional protein with diverse roles in and out of the ribosome (89). It is possible that the mechanism used by Asc1p to destabilize SSA4 mRNA is independent of direct interactions with regulatory factors. Instead, Asc1p's capacity to regulate the assembly of processing bodies might facilitate the release of decay enzymes that degrade SSA4 mRNA (104). Since condensate formation is critical for cells to survive stress (74,105), it is tempting to speculate that this role of Asc1p explains the inability of asc1Δ cells to recover from heat stress while strains expressing Asc1p mutants DE and DY survive to heat stress.
Previously Asc1 was identified as a factor communicating the stress signal from RQC to HSF1 and regulating the heat shock response independently of its control by HSP70/HSP40 (44,106). Our work establishes a new role for the RQC mechanism in regulating the expression of the inducible HSP Ssa4p. Interestingly, the RQC factor Asc1p also mediated SSA4 mRNA decay during recovery and regulated heat shock survival independently of Ssa4p expression. Overall, Asc1p emerges as a critical prosurvival factor that operates in independent pathways, on and off the ribosome, to regulate the fate of SSA4 mRNA in the cytoplasm, the strength, and duration of the heat shock response, and whether the cell survives heat stress. If these roles are conserved in mammalian cells, its ortholog RACK1 could be targeted therapeutically to recover proteostasis under pathological conditions like cancer and neurodegeneration.
DATA AVAILABILITY
Proteomics data is accessible through ProteomeXchange via the PRIDE Archive (Project accession: PXD037545), and the program to calculate codon optimality is available at https://github.com/LR-MVU/YEAST-SSA.git and https://doi.org/10.5281/zenodo.7847569.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank Drs Wendy Gilbert (Yale School of Medicine), Roy Parker (University of Boulder, Colorado), Daniel Klionsky (University of Michigan), Arlen Johnson (The University of Texas at Austin), Rachel Green (John Hopkins University), and Toshifumi Inada (The University of Tokyo) for providing yeast strains and plasmids, Delina Efrem, Kevin Munoz Portocarrero, Jessamine Mattson, Ryan Huang, Suleima Jacob-Tomas, Sonya Madan and Maanasa Koripalli from Vera's lab for technical help, and Dr Joaquin Ortega (McGill University) for help localizing proteins in the structures of the 40S monosome and disomes (Figure 6H). We also thank Dr Rafael Cuesta Sanchez for technical help and critical reading and High-Fidelity Science Communications for manuscript editing.
Notes
Present address: Irene Garcia de-Andres, Technische Universität Dresden, Germany.
Present address: Aitana Garcia-Bueno Rico, Department of Experimental and Health Sciences, Universidad Pompeu Fabra, Barcelona 08003, Spain.
Contributor Information
Lokha R Alagar Boopathy, Department of Biochemistry. McGill University, Montreal, Quebec, H3G 1Y6, Canada.
Emma Beadle, Department of Biochemistry. McGill University, Montreal, Quebec, H3G 1Y6, Canada.
Alan RuoChen Xiao, Department of Biochemistry. McGill University, Montreal, Quebec, H3G 1Y6, Canada.
Aitana Garcia-Bueno Rico, Department of Biochemistry. McGill University, Montreal, Quebec, H3G 1Y6, Canada.
Celia Alecki, Department of Biochemistry. McGill University, Montreal, Quebec, H3G 1Y6, Canada.
Irene Garcia de-Andres, Department of Biochemistry. McGill University, Montreal, Quebec, H3G 1Y6, Canada.
Kyla Edelmeier, Department of Biochemistry. McGill University, Montreal, Quebec, H3G 1Y6, Canada.
Luca Lazzari, Department of Biochemistry. McGill University, Montreal, Quebec, H3G 1Y6, Canada.
Mehdi Amiri, Department of Biochemistry. McGill University, Montreal, Quebec, H3G 1Y6, Canada.
Maria Vera, Department of Biochemistry. McGill University, Montreal, Quebec, H3G 1Y6, Canada.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Natural Sciences and Engineering Research Council of Canada [RGPIN-2019-04767]; Fonds de recherche du Québec (FRQS) [NC-2999446]; Lokha R. Alagar Boopathy is funded by the FRQS scholarship program [315076]; Centre de Recherche en Biologie Structurale at McGill University; Dr Celia Alecki is funded by an FRQS postdoctoral fellowship [300232]. Funding for open access charge: NSERC; FRQNT; FRQS.
Conflict of interest statement. None declared.
REFERENCES
- 1. Morimoto R.I. Cells in stress: transcriptional activation of heat shock genes. Science. 1993; 259:1409–1410. [DOI] [PubMed] [Google Scholar]
- 2. Silver J.T., Noble E.G.. Regulation of survival gene hsp70. Cell Stress Chaperones. 2012; 17:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Boopathy L.R.A., Jacob-Tomas S., Alecki C., Vera M.. Mechanisms tailoring the expression of heat shock proteins to proteostasis challenges. J. Biol. Chem. 2022; 298:101796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kampinga H.H., Hageman J., Vos M.J., Kubota H., Tanguay R.M., Bruford E.A., Cheetham M.E., Chen B., Hightower L.E.. Guidelines for the nomenclature of the human heat shock proteins. Cell Stress Chaperones. 2009; 14:105–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Rosenzweig R., Nillegoda N.B., Mayer M.P., Bukau B.. The Hsp70 chaperone network. Nat. Rev. Mol. Cell Biol. 2019; 20:665–680. [DOI] [PubMed] [Google Scholar]
- 6. Feder J.H., Rossi J.M., Solomon J., Solomon N., Lindquist S.. The consequences of expressing hsp70 in Drosophila cells at normal temperatures. Genes Dev. 1992; 6:1402–1413. [DOI] [PubMed] [Google Scholar]
- 7. Kumar S., Stokes J., Singh U.P., Gunn K.S., Acharya A., Manne U., Mishra M.. Targeting Hsp70: a possible therapy for cancer. Cancer Lett. 2016; 374:156–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Anckar J., Sistonen L.. Regulation of HSF1 function in the heat stress response: implications in aging and disease. Annu. Rev. Biochem. 2011; 80:1089–1115. [DOI] [PubMed] [Google Scholar]
- 9. Baler R., Dahl G., Voellmy R.. Activation of human heat shock genes is accompanied by oligomerization, modification, and rapid translocation of heat shock transcription factor HSF1. Mol. Cell. Biol. 1993; 13:2486–2496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dayalan Naidu S., Dinkova-Kostova A.T.. Regulation of the mammalian heat shock factor 1. FEBS J. 2017; 284:1606–1627. [DOI] [PubMed] [Google Scholar]
- 11. Hentze N., Le Breton L., Wiesner J., Kempf G., Mayer M.P.. Molecular mechanism of thermosensory function of human heat shock transcription factor Hsf1. Elife. 2016; 5:e11576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Alain T., Morita M., Fonseca B.D., Yanagiya A., Siddiqui N., Bhat M., Zammit D., Marcus V., Metrakos P., Voyer L.-A.et al.. eIF4E/4E-BP ratio predicts the efficacy of mTOR targeted therapies. Cancer Res. 2012; 72:6468–6476. [DOI] [PubMed] [Google Scholar]
- 13. Andrei M.A., Ingelfinger D., Heintzmann R., Achsel T., Rivera-Pomar R., Lührmann R.. A role for eIF4E and eIF4E-transporter in targeting mRNPs to mammalian processing bodies. RNA. 2005; 11:717–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Vries R.G.J., Flynn A., Patel J.C., Wang X., Denton R.M., Proud C.G.. Heat shock increases the association of binding protein-1 with initiation factor 4E *. J. Biol. Chem. 1997; 272:32779–32784. [DOI] [PubMed] [Google Scholar]
- 15. Liu B., Han Y., Qian S.-B.. Cotranslational response to proteotoxic stress by elongation pausing of ribosomes. Mol. Cell. 2013; 49:453–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Shalgi R., Hurt J.A., Krykbaeva I., Taipale M., Lindquist S., Burge C.B.. Widespread regulation of translation by elongation pausing in heat shock. Mol. Cell. 2013; 49:439–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kimball S.R., Fabian J.R., Pavitt G.D., Hinnebusch A.G., Jefferson L.S.. Regulation of guanine nucleotide exchange through phosphorylation of eukaryotic initiation factor eIF2α: role of the α- and δ-subunits of eIF2B *. J. Biol. Chem. 1998; 273:12841–12845. [DOI] [PubMed] [Google Scholar]
- 18. Liu B., Qian S.-B.. Translational reprogramming in stress response. Wiley Interdiscip. Rev. RNA. 2014; 5:301–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yoshizawa F., Kimball S.R., Jefferson L.S.. Modulation of translation initiation in rat skeletal muscle and liver in response to food intake. Biochem. Biophys. Res. Commun. 1997; 240:825–831. [DOI] [PubMed] [Google Scholar]
- 20. Guzikowski A.R., Chen Y.S., Zid B.M.. Stress-induced mRNP granules: form and function of processing bodies and stress granules. Wiley Interdiscip. Rev. RNA. 2019; 10:e1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zid B.M., O’Shea E.K.. Promoter sequences direct cytoplasmic localization and translation of mRNAs during starvation in yeast. Nature. 2014; 514:117–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Coots R.A., Liu X.-M., Mao Y., Dong L., Zhou J., Wan J., Zhang X., Qian S.-B.. m6A facilitates eIF4F-independent mRNA translation. Mol. Cell. 2017; 68:504–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Meyer K.D., Patil D.P., Zhou J., Zinoviev A., Skabkin M.A., Elemento O., Pestova T.V., Qian S.-B., Jaffrey S.R.. 5′ UTR m6A promotes Cap-independent translation. Cell. 2015; 163:999–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Vera M., Pani B., Griffiths L.A., Muchardt C., Abbott C.M., Singer R.H., Nudler E.. The translation elongation factor eEF1A1 couples transcription to translation during heat shock response. Elife. 2014; 3:e03164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zhou J., Wan J., Gao X., Zhang X., Jaffrey S.R., Qian S.-B.. Dynamic m6A mRNA methylation directs translational control of heat shock response. Nature. 2015; 526:591–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Parker R. RNA Degradation in Saccharomyces cerevisae. Genetics. 2012; 191:671–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Krakowiak J., Zheng X., Patel N., Feder Z.A., Anandhakumar J., Valerius K., Gross D.S., Khalil A.S., Pincus D. Hsf1 and Hsp70 constitute a two-component feedback loop that regulates the yeast heat shock response. Elife. 2018; 7:e31668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bönisch C., Temme C., Moritz B., Wahle E.. Degradation of hsp70 and other mRNAs in Drosophila via the 5′–3′ pathway and its regulation by heat shock. J. Biol. Chem. 2007; 282:21818–21828. [DOI] [PubMed] [Google Scholar]
- 29. Petersen R.B., Lindquist S.. Regulation of HSP70 synthesis by messenger RNA degradation. Cell Regul. 1989; 1:135–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Didomenico B.J., Bugaisky G.E., Lindquist S.. The heat shock response is self-regulated at both the transcriptional and posttranscriptional levels. Cell. 1982; 31:593–603. [DOI] [PubMed] [Google Scholar]
- 31. Chan L.Y., Mugler C.F., Heinrich S., Vallotton P., Weis K.. Non-invasive measurement of mRNA decay reveals translation initiation as the major determinant of mRNA stability. Elife. 2018; 7:e32536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Heck A.M., Wilusz J.. The interplay between the RNA decay and translation machinery in eukaryotes. Cold Spring Harb. Perspect. Biol. 2018; 10:a032839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Park H., Subramaniam A.R.. Inverted translational control of eukaryotic gene expression by ribosome collisions. PLoS Biol. 2019; 17:e3000396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Roy B., Jacobson A.. The intimate relationships of mRNA decay and translation. Trends Genet. 2013; 29:691–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Schwartz D.C., Parker R.. Mutations in translation initiation factors lead to increased rates of deadenylation and decapping of mRNAs in Saccharomyces cerevisiae. Mol. Cell. Biol. 1999; 19:5247–5256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Schwartz D.C., Parker R.. mRNA decapping in yeast requires dissociation of the Cap binding protein, eukaryotic translation initiation factor 4E. Mol. Cell. Biol. 2000; 20:7933–7942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Presnyak V., Alhusaini N., Chen Y.-H., Martin S., Morris N., Kline N., Olson S., Weinberg D., Baker K.E., Graveley B.R.et al.. Codon optimality is a major determinant of mRNA stability. Cell. 2015; 160:1111–1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Radhakrishnan A., Chen Y.-H., Martin S., Alhusaini N., Green R., Coller J.. The DEAD-Box protein Dhh1p couples mRNA decay and translation by monitoring codon optimality. Cell. 2016; 167:122–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Webster M.W., Chen Y.-H., Stowell J.A.W., Alhusaini N., Sweet T., Graveley B.R., Coller J., Passmore L.A.. mRNA deadenylation is coupled to translation rates by the differential activities of Ccr4-Not nucleases. Mol. Cell. 2018; 70:1089–1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ikeuchi K., Izawa T., Inada T.. Recent progress on the molecular mechanism of quality controls induced by ribosome stalling. Front. Genet. 2019; 9:743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ikeuchi K., Tesina P., Matsuo Y., Sugiyama T., Cheng J., Saeki Y., Tanaka K., Becker T., Beckmann R., Inada T.. Collided ribosomes form a unique structural interface to induce Hel2-driven quality control pathways. EMBO J. 2019; 38:e100276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Doma M.K., Parker R.. Endonucleolytic cleavage of eukaryotic mRNAs with stalls in translation elongation. Nature. 2006; 440:561–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Simms C.L., Yan L.L., Zaher H.S.. Ribosome collision is critical for quality control during No-Go decay. Mol. Cell. 2017; 68:361–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Brandman O., Stewart-Ornstein J., Wong D., Larson A., Williams C.C., Li G.-W., Zhou S., King D., Shen P.S., Weibezahn J.et al.. A ribosome-bound quality control complex triggers degradation of nascent peptides and signals translation stress. Cell. 2012; 151:1042–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Juszkiewicz S., Hegde R.S.. Initiation of quality control during Poly(A) translation requires site-specific ribosome ubiquitination. Mol. Cell. 2017; 65:743–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Matsuo Y., Ikeuchi K., Saeki Y., Iwasaki S., Schmidt C., Udagawa T., Sato F., Tsuchiya H., Becker T., Tanaka K.et al.. Ubiquitination of stalled ribosome triggers ribosome-associated quality control. Nat. Commun. 2017; 8:159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Sitron C.S., Brandman O.. Detection and degradation of stalled Nascent chains via ribosome-associated quality control. Annu. Rev. Biochem. 2020; 89:417–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Gerbasi V.R., Weaver C.M., Hill S., Friedman D.B., Link A.J.. Yeast Asc1p and mammalian RACK1 are functionally orthologous core 40S ribosomal proteins that repress gene expression. Mol. Cell. Biol. 2004; 24:8276–8287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Winz M.-L., Peil L., Turowski T.W., Rappsilber J., Tollervey D.. Molecular interactions between Hel2 and RNA supporting ribosome-associated quality control. Nat. Commun. 2019; 10:563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Best K., Ikeuchi K., Kater L., Best D., Musial J., Matsuo Y., Berninghausen O., Becker T., Inada T., Beckmann R.. Structural basis for clearing of ribosome collisions by the RQT complex. Nat. Commun. 2023; 14:921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Sitron C.S., Park J.H., Brandman O.. Asc1, Hel2, and Slh1 couple translation arrest to nascent chain degradation. RNA. 2017; 23:798–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Tomomatsu S., Watanabe A., Tesina P., Hashimoto S., Ikeuchi K., Li S., Matsuo Y., Beckmann R., Inada T.. Two modes of Cue2-mediated mRNA cleavage with distinct substrate recognition initiate no-go decay. Nucleic Acids Res. 2023; 51:253–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Joazeiro C.A.P. Mechanisms and functions of ribosome-associated protein quality control. Nat. Rev. Mol. Cell Biol. 2019; 20:368–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Doamekpor S.K., Lee J.-W., Hepowit N.L., Wu C., Charenton C., Leonard M., Bengtson M.H., Rajashankar K.R., Sachs M.S., Lima C.D.et al.. Structure and function of the yeast listerin (Ltn1) conserved N-terminal domain in binding to stalled 60S ribosomal subunits. Proc. Natl. Acad. Sci. U.S.A. 2016; 113:E4151–E4160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Buschauer R., Matsuo Y., Sugiyama T., Chen Y.-H., Alhusaini N., Sweet T., Ikeuchi K., Cheng J., Matsuki Y., Nobuta R.et al.. The Ccr4-Not complex monitors the translating ribosome for codon optimality. Science. 2020; 368:eaay6912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. D’Orazio K.N., Wu C.C.-C., Sinha N., Loll-Krippleber R., Brown G.W., Green R.. The endonuclease Cue2 cleaves mRNAs at stalled ribosomes during No Go Decay. Elife. 2019; 8:e49117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Shoemaker C.J., Green R.. Translation drives mRNA quality control. Nat. Struct. Mol. Biol. 2012; 19:594–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Navickas A., Chamois S., Saint-Fort R., Henri J., Torchet C., Benard L.. No-Go Decay mRNA cleavage in the ribosome exit tunnel produces 5′-OH ends phosphorylated by Trl1. Nat. Commun. 2020; 11:122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Veltri A.J., D’Orazio K.N., Lessen L.N., Loll-Krippleber R., Brown G.W., Green R.. Distinct elongation stalls during translation are linked with distinct pathways for mRNA degradation. Elife. 2022; 11:e76038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Goldman D.H., Livingston N.M., Movsik J., Wu B., Green R.. Live-cell imaging reveals kinetic determinants of quality control triggered by ribosome stalling. Mol. Cell. 2021; 81:1830–1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Meydan S., Guydosh N.R.. Disome and trisome profiling reveal genome-wide targets of ribosome quality control. Mol. Cell. 2020; 79:588–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Meydan S., Guydosh N.R.. A cellular handbook for collided ribosomes: surveillance pathways and collision types. Curr. Genet. 2021; 67:19–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Hickey K.L., Dickson K., Cogan J.Z., Replogle J.M., Schoof M., D’Orazio K.N., Sinha N.K., Hussmann J.A., Jost M., Frost A.et al.. GIGYF2 and 4EHP inhibit translation initiation of defective messenger RNAs to assist ribosome-associated quality control. Mol. Cell. 2020; 79:950–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Andersson R., Eisele-Bürger A.M., Hanzén S., Vielfort K., Öling D., Eisele F., Johansson G., Gustafsson T., Kvint K., Nyström T.. Differential role of cytosolic Hsp70s in longevity assurance and protein quality control. PLoS Genet. 2021; 17:e1008951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Hocine S., Vera M., Zenklusen D., Singer R.H.. Promoter-autonomous functioning in a controlled environment using single molecule FISH. Sci. Rep. 2015; 5:9934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Coudert L., Adjibade P., Mazroui R.. Analysis of translation initiation during stress conditions by polysome profiling. J. Vis. Exp. 2014; 87:e51164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Amador-Cañizares Y., Bernier A., Wilson J.A., Sagan S.M.. miR-122 does not impact recognition of the HCV genome by innate sensors of RNA but rather protects the 5′ end from the cellular pyrophosphatases, DOM3Z and DUSP11. Nucleic Acids Res. 2018; 46:5139–5158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Bennetzen J.L., Hall B.D.. Codon selection in yeast. J. Biol. Chem. 1982; 257:3026–3031. [PubMed] [Google Scholar]
- 69. Tutucci E., Vera M., Biswas J., Garcia J., Parker R., Singer R.H.. An improved MS2 system for accurate reporting of the mRNA life cycle. Nat. Methods. 2018; 15:81–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Mueller F., Senecal A., Tantale K., Marie-Nelly H., Ly N., Collin O., Basyuk E., Bertrand E., Darzacq X., Zimmer C.. FISH-quant: automatic counting of transcripts in 3D FISH images. Nat. Methods. 2013; 10:277–278. [DOI] [PubMed] [Google Scholar]
- 71. Mühlhofer M., Berchtold E., Stratil C.G., Csaba G., Kunold E., Bach N.C., Sieber S.A., Haslbeck M., Zimmer R., Buchner J.. The heat shock response in yeast maintains protein homeostasis by chaperoning and replenishing proteins. Cell Rep. 2019; 29:4593–4607. [DOI] [PubMed] [Google Scholar]
- 72. Kumari R., Michel A.M., Baranov P.V.. PausePred and Rfeet: webtools for inferring ribosome pauses and visualizing footprint density from ribosome profiling data. RNA. 2018; 24:1297–1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Kiniry S.J., Judge C.E., Michel A.M., Baranov P.V.. Trips-Viz: an environment for the analysis of public and user-generated ribosome profiling data. Nucleic Acids Res. 2021; 49:W662–W670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Iserman C., Desroches Altamirano C., Jegers C., Friedrich U., Zarin T., Fritsch A.W., Mittasch M., Domingues A., Hersemann L., Jahnel M.et al.. Condensation of Ded1p promotes a translational switch from housekeeping to stress protein production. Cell. 2020; 181:818–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Brandman O., Hegde R.S.. Ribosome-associated protein quality control. Nat. Struct. Mol. Biol. 2016; 23:7–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Juszkiewicz S., Slodkowicz G., Lin Z., Freire-Pritchett P., Peak-Chew S.-Y., Hegde R.S.. Ribosome collisions trigger cis-acting feedback inhibition of translation initiation. Elife. 2020; 9:e60038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Houston L., Platten E.M., Connelly S.M., Wang J., Grayhack E.J.. Frameshifting at collided ribosomes is modulated by elongation factor eEF3 and by integrated stress response regulators Gcn1 and Gcn20. RNA. 2022; 28:320–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Hendrick J.L., Wilson P.G., Edelman I.I., Sandbaken M.G., Ursic D., Culbertson M.R.. Yeast frameshift suppressor Mutations in the genes coding for transcription factor Mbf1p and ribosomal protein S3: evidence for autoregulation of S3 synthesis. Genetics. 2001; 157:1141–1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Wang J., Zhou J., Yang Q., Grayhack E.J.. Multi-protein bridging factor 1(Mbf1), Rps3 and Asc1 prevent stalled ribosomes from frameshifting. Elife. 2018; 7:e39637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Juszkiewicz S., Speldewinde S.H., Wan L., Svejstrup J.Q., Hegde R.S.. The ASC-1 complex disassembles collided ribosomes. Mol. Cell. 2020; 79:603–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Zhang H., Wang Y., Lu J.. Function and evolution of upstream ORFs in eukaryotes. Trends Biochem. Sci. 2019; 44:782–794. [DOI] [PubMed] [Google Scholar]
- 82. Munzarová V., Pánek J., Gunišová S., Dányi I., Szamecz B., Valášek L.S.. Translation reinitiation relies on the interaction between eIF3a/TIF32 and progressively folded cis-acting mRNA elements preceding short uORFs. PLoS Genet. 2011; 7:e1002137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Sundaramoorthy E., Leonard M., Mak R., Liao J., Fulzele A., Bennett E.J.. ZNF598 and RACK1 regulate mammalian ribosome-associated quality control function by mediating regulatory 40S ribosomal ubiquitylation. Mol. Cell. 2017; 65:751–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Sitron C.S., Brandman O.. CAT tails drive degradation of stalled polypeptides on and off the ribosome. Nat. Struct. Mol. Biol. 2019; 26:450–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Thrun A., Garzia A., Kigoshi-Tansho Y., Patil P.R., Umbaugh C.S., Dallinger T., Liu J., Kreger S., Patrizi A., Cox G.A.et al.. Convergence of mammalian RQC and C-end rule proteolytic pathways via alanine tailing. Mol. Cell. 2021; 81:2112–2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Wu C.C.-C., Peterson A., Zinshteyn B., Regot S., Green R.. Ribosome collisions trigger general stress responses to regulate cell fate. Cell. 2020; 182:404–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Lanza A.M., Curran K.A., Rey L.G., Alper H.S.. A condition-specific codon optimization approach for improved heterologous gene expression in Saccharomyces cerevisiae. BMC Syst. Biol. 2014; 8:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Li Z., Lee I., Moradi E., Hung N.-J., Johnson A.W., Marcotte E.M.. Rational extension of the ribosome biogenesis pathway using network-guided genetics. PLoS Biol. 2009; 7:e1000213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Thompson M.K., Rojas-Duran M.F., Gangaramani P., Gilbert W.V.. The ribosomal protein Asc1/RACK1 is required for efficient translation of short mRNAs. Elife. 2016; 5:e11154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Galardi S., Fatica A., Bachi A., Scaloni A., Presutti C., Bozzoni I.. Purified box C/D snoRNPs are able to reproduce site-specific 2′-O-methylation of target RNA in vitro. Mol. Cell. Biol. 2002; 22:6663–6668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Kiss-László Z., Henry Y., Bachellerie J.P., Caizergues-Ferrer M., Kiss T.. Site-specific ribose methylation of preribosomal RNA: a novel function for small nucleolar RNAs. Cell. 1996; 85:1077–1088. [DOI] [PubMed] [Google Scholar]
- 92. Qu L.H., Henry Y., Nicoloso M., Michot B., Azum M.C., Renalier M.H., Caizergues-Ferrer M., Bachellerie J.P.. U24, a novel intron-encoded small nucleolar RNA with two 12 nt long, phylogenetically conserved complementarities to 28S rRNA. Nucleic Acids Res. 1995; 23:2669–2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Reichow S.L., Hamma T., Ferré-D’Amaré A.R., Varani G.. The structure and function of small nucleolar ribonucleoproteins. Nucleic Acids Res. 2007; 35:1452–1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. He F., Li C., Roy B., Jacobson A.. Yeast Edc3 targets RPS28B mRNA for decapping by binding to a 3′ untranslated region decay-inducing regulatory element. Mol. Cell. Biol. 2014; 34:1438–1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Kramer S., Queiroz R., Ellis L., Webb H., Hoheisel J.D., Clayton C., Carrington M.. Heat shock causes a decrease in polysomes and the appearance of stress granules in trypanosomes independently of eIF2α phosphorylation at Thr169. J. Cell Sci. 2008; 121:3002–3014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Dimitrova L.N., Kuroha K., Tatematsu T., Inada T.. Nascent peptide-dependent translation arrest leads to Not4p-mediated protein degradation by the proteasome*. J. Biol. Chem. 2009; 284:10343–10352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Ji Q., Zong X., Mao Y., Qian S.-B.. A heat shock–responsive lncRNA Heat acts as a HSF1-directed transcriptional brake via m6A modification. Proc. Natl. Acad. Sci. U.S.A. 2021; 118:e2102175118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Chen L., Muhlrad D., Hauryliuk V., Cheng Z., Lim M.K., Shyp V., Parker R., Song H.. Structure of the Dom34-Hbs1 complex and implications for no-go decay. Nat. Struct. Mol. Biol. 2010; 17:1233–1240. [DOI] [PubMed] [Google Scholar]
- 99. Letzring D.P., Dean K.M., Grayhack E.J.. Control of translation efficiency in yeast by codon–anticodon interactions. RNA. 2010; 16:2516–2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Ito-Harashima S., Kuroha K., Tatematsu T., Inada T.. Translation of the poly(A) tail plays crucial roles in nonstop mRNA surveillance via translation repression and protein destabilization by proteasome in yeast. Genes Dev. 2007; 21:519–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Wilson M.A., Meaux S., van Hoof A.. A genomic screen in yeast reveals novel aspects of nonstop mRNA metabolism. Genetics. 2007; 177:773–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Matsuo Y., Uchihashi T., Inada T.. Decoding of the ubiquitin code for clearance of colliding ribosomes by the RQT complex. Nat. Commun. 2023; 14:79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Sugiyama T., Li S., Kato M., Ikeuchi K., Ichimura A., Matsuo Y., Inada T.. Sequential ubiquitination of ribosomal protein uS3 triggers the degradation of non-functional 18S rRNA. Cell Rep. 2019; 26:3400–3415. [DOI] [PubMed] [Google Scholar]
- 104. Tkach J.M., Yimit A., Lee A.Y., Riffle M., Costanzo M., Jaschob D., Hendry J.A., Ou J., Moffat J., Boone C.et al.. Dissecting DNA damage response pathways by analysing protein localization and abundance changes during DNA replication stress. Nat. Cell Biol. 2012; 14:966–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Franzmann T.M., Alberti S.. Protein phase separation as a stress survival strategy. Cold Spring Harb. Perspect. Biol. 2019; 11:a034058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Alford B.D., Tassoni-Tsuchida E., Khan D., Work J.J., Valiant G., Brandman O.. ReporterSeq reveals genome-wide dynamic modulators of the heat shock response across diverse stressors. Elife. 2021; 10:e57376. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Proteomics data is accessible through ProteomeXchange via the PRIDE Archive (Project accession: PXD037545), and the program to calculate codon optimality is available at https://github.com/LR-MVU/YEAST-SSA.git and https://doi.org/10.5281/zenodo.7847569.