Abstract
The Escherichia coli replication origin oriC contains the initiator ATP-DnaA-Oligomerization Region (DOR) and its flanking duplex unwinding element (DUE). In the Left-DOR subregion, ATP-DnaA forms a pentamer by binding to R1, R5M and three other DnaA boxes. The DNA-bending protein IHF binds sequence-specifically to the interspace between R1 and R5M boxes, promoting DUE unwinding, which is sustained predominantly by binding of R1/R5M-bound DnaAs to the single-stranded DUE (ssDUE). The present study describes DUE unwinding mechanisms promoted by DnaA and IHF-structural homolog HU, a ubiquitous protein in eubacterial species that binds DNA sequence-non-specifically, preferring bent DNA. Similar to IHF, HU promoted DUE unwinding dependent on ssDUE binding of R1/R5M-bound DnaAs. Unlike IHF, HU strictly required R1/R5M-bound DnaAs and interactions between the two DnaAs. Notably, HU site-specifically bound the R1-R5M interspace in a manner stimulated by ATP-DnaA and ssDUE. These findings suggest a model that interactions between the two DnaAs trigger DNA bending within the R1/R5M-interspace and initial DUE unwinding, which promotes site-specific HU binding that stabilizes the overall complex and DUE unwinding. Moreover, HU site-specifically bound the replication origin of the ancestral bacterium Thermotoga maritima depending on the cognate ATP-DnaA. The ssDUE recruitment mechanism could be evolutionarily conserved in eubacteria.
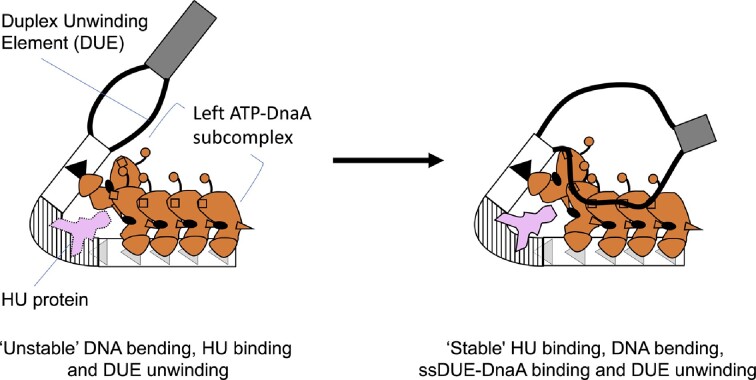
Graphical Abstract
Graphical Abstract.
The left-oriC subregion responsible for DUE unwinding is shown. ATP-DnaA constructs a specific pentamer, causing initial unstable DNA bending and DUE unwinding, which are stabilized by bent DNA-preferential binding of HU protein and specific ssDUE-DnaA binding.
INTRODUCTION
The replication of bacterial chromosomal DNA requires the formation of highly ordered nucleoprotein complexes at oriC, the chromosomal DNA origin of replication (1–4). In Escherichia coli, the ATP-bound DnaA (ATP-DnaA) initiator protein forms a complex with oriC to promote local unwinding of duplex DNA within oriC. This complex formation is aided by IHF, a nucleoid-associated protein with sequence-specific binding activity. DnaB replicative DNA helicases are recruited to oriC-bound DnaA complexes and loaded onto the resulting single-stranded (ss) DNA via specific interactions with the DnaC helicase loader. This results in expansion of the unwound region, which allows loading of DnaG primase and DNA polymerase to initiate DNA synthesis (5–10).
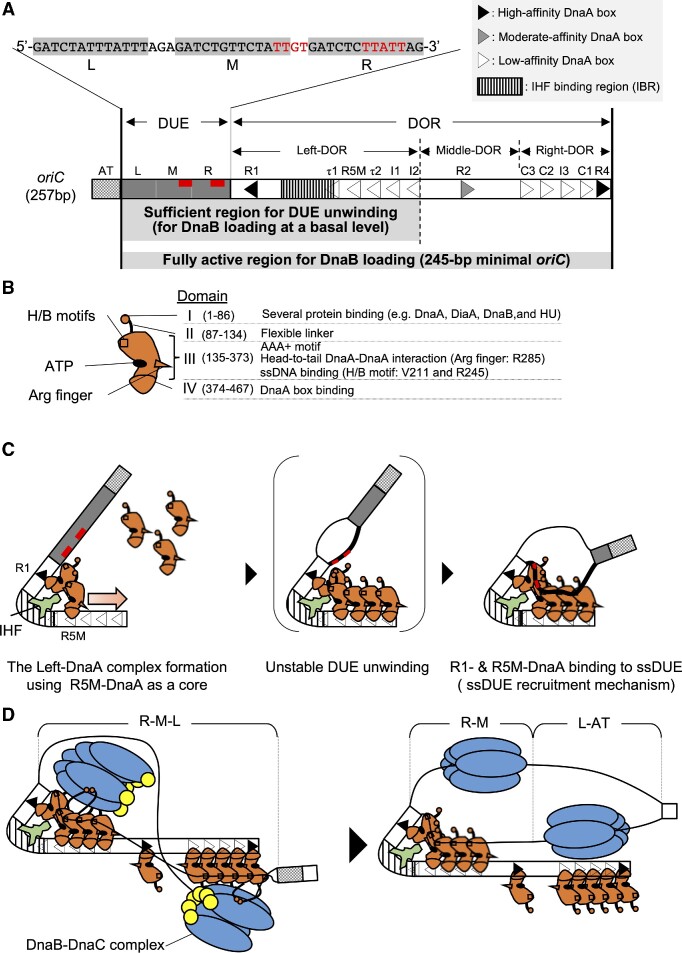
The 245-bp minimal oriC of E. coli is composed of a Duplex-Unwinding Element (DUE) and a DnaA-Oligomerization Region (DOR) (Figure 1A) (1,3,4). The DUE contains three AT-rich 13-mer repeats (L, M, and R) and M/R-DUE includes two T-rich sequence motifs, TT[G/A]T(T) (11,12). Binding of DOR-bound ATP-DnaA complexes to the single-stranded TT[G/A]T(T) motifs stabilizes the unwound state of DUE (Figure 1A–C) (12) (also see below). L-DUE is less stably unwound than M/R-DUE (13,14). M/R-DUE is essential and L-DUE is stimulatory for loading of a pair of DnaB helicases onto the unwound regions (14). Upon loading of this pair of DnaB helicases, the stable unwound region is expanded to the L-DUE and the AT-cluster region immediately upstream of L-DUE (Figure 1D) (14,15). The AT-cluster region assists in DnaB helicase loading when formation of DnaA complexes is incomplete (14).
Figure 1.
Schematic structures of oriC and DnaA and the ssDUE recruitment mechanism. (A) Overall structure of oriC (257 bp). Minimal oriC (245 bp) includes the duplex-unwinding element (DUE; gray bar) and the DnaA-Oligomerization Region (DOR; open bar). DUE is composed of three AT-rich 13-mer repeats termed L, M and R. The sequence of the DUE-upper (T-rich) strand is shown over the structure. DnaA binding motifs TT[A/G]T(T) are indicated by red characters or red bars. The AT-cluster (a dotted square) flanking DUE outside the minimal oriC is a supplementary unwinding region. The DOR contains 12 DnaA boxes (filled, gray and open arrowheads representing high-, moderate- and low-affinity sites respectively) and an IHF binding region (IBR; vertical striped box). The DOR is subdivided into Left, Middle, and Right subregions. Major functions of the subregions are described briefly below the structure. The Middle-DOR bearing DnaA box R2 assists in DnaA assembly at Left/Right-DORs. (B) Domains of DnaA. Domains I-IV are shown schematically, with amino acid residue numbers shown in each bracket. H/B motifs (V211 and R245, squares) and Arg finger (R285, triangle) are indicated. The major functions of each domain are described on the right side of the structure. (C) DUE unwinding mechanism by which IHF promotes ssDUE recruitment. DnaA and IHF are indicated by red and pale green diagrams, respectively. For simplicity, only DUE and Left-DOR bearing IHF and ATP-DnaA are shown. ATP-DnaA levels increase during the cell cycle, resulting in oligomerization on low-affinity DnaA boxes using the R5M-DnaA protomer as a core, and resulting in the construction of DnaA pentamers, including R1-DnaA (panels 1 and 2). DUE is unwound unstably by thermal motion and torsional stress (panel 2). The M/R-region bearing TT[A/G]T(T) sequences (red line) of the DUE-upper strand (black bold line) binds to R1-DnaA and R5M-DnaA via IHF-induced DNA bending (panel 3). The resulting ssDUE–ATP-DnaA–DOR ternary complex stabilizes the unwinding of DUE. (D) Model for DnaB loading. In addition to M/R-DUE, L-DUE is moderately unwound and interacts with the Right-DnaA subcomplex. Each Left-DnaA and Right-DnaA subcomplex binds to a DnaB hexamer (blue ovals) complexed with DnaC (yellow circles) (left panel). Upon DnaB loading, the stable unwound region expands to the AT-rich cluster (right panel).
The DOR carries twelve DnaA-binding sequences (DnaA boxes) termed R1-2, R4, R5M, I1-3, τ1-2 and C1-3 (Figure 1A) (3,16–18). R1 and R4 are high-affinity DnaA boxes, and R2 is a moderate-affinity DnaA box. Each DnaA box is composed of typical 9-mer consensus sequences (TT[A/T]TNCACA) to which ATP-DnaA or ADP-DnaA can bind. The other nine boxes (R5M, I1-3, τ1-2 and C1-3) are low-affinity DnaA boxes with degenerated consensus sequences. ATP-DnaA rather than ADP-DnaA preferentially binds to the low-affinity DnaA boxes cooperatively via specific DnaA-DnaA interactions (18–23) (see below).
The DOR is subdivided into three subregions: the Left-, Middle-, and Right-DOR (Figure 1A) (23). The Left-DOR contains the left-directional array of DnaA boxes (R1, τ1, R5M, τ2, I1 and I2) and an IHF binding region (IBR) that overlaps with DnaA box τ1 (21,24,25). The Right-DOR consists of the right-directional DnaA boxes (R4, C1, I3, C2 and C3). ATP-DnaA binding to Left- and Right-DOR results in the formation of the Left- and Right-DnaA subcomplexes, respectively. The Middle-DOR has only one DnaA box (R2). In the presence of IHF, the Left-DnaA subcomplex is sufficient for M/R-DUE unwinding (Figure 1C) (14,21,24). The Right-DnaA subcomplex binds with moderate affinity to T-rich ssDNA (upper strand) spanning M/L-DUE, stimulating the unwinding of DUE, which is essential for efficient loading of DnaB helicases (Figure 1D) (14). The Left- and Right-DnaA subcomplexes each recruits a DnaB-DnaC complex through direct DnaA-DnaB binding, leading to DnaB loading onto ssDUE (14,21). The Left-DnaA subcomplex alone binds a single DnaB–DnaC complex and can sustain a moderate level of the DnaB loading.
DnaA consists of four functional domains I-IV, which play specific roles in DnaA assembly and DnaB loading (Figure 1BC) (3,26,27). In E. coli DnaA, domain I interacts with multiple proteins, including the DnaA-assembly stimulator DiaA as well as DnaB, and HU protein, a structural homolog of IHF, does with domains I-II (8,28–31). In addition, weak domain I-domain I interactions support interactions of R2-bound DnaA with DnaA molecules bound to Left- and Right-DOR (20,23,32). Domain II is a flexible linker (29,33). Domain III contains AAA+ (ATPase Associated with various cellular activities) motifs involved in tight ATP/ADP binding, ATP hydrolysis, and domain III-domain III interactions (2,3,18,19,34–36). The arginine finger motif Arg285 within this domain interacts with ATP bound to the flanking DnaA protomer, sustaining the formation of the head-to-tail ATP-DnaA oligomerization on Left- and Right-DOR (Figure 1BC) (18). AID motifs (Arg 227 and Leu 290) residing near Arg285 may assist in specific domain III-domain III interactions, stimulating the formation of functional Left-DnaA subcomplexes (19). Furthermore, the H/B motifs (Val211 and Arg245) of domain III play an essential role in capturing the T-rich strand of ssDUE by binding to TT[G/A]T(T) sites during DUE unwinding processes (Figure 1A–C) (12,24,37). Domain IV binds specifically to the DnaA box (38).
Moreover, in E. coli oriC, IHF binds to its consensus recognition sequences in the IBR located between DnaA boxes R1 and R5M, enabling stable DUE unwinding in the presence of ATP-DnaA (Figure 1C). IHF is a heterodimer consisting of highly homologous α and β subunits encoded by ihfA and ihfB, respectively (39–41). IHF tightly binds to 30–35 bp DNA regions by recognizing the 13-mer core sequence (WATCAAnnnnTTA), resulting in a sharp bend in DNA, up to 160° (Figure 1A and C) (42–44). DnaA box τ1 partly overlaps with IBR, and IHF predominantly binds to this region (24).
The mechanism underlying IHF-promoted unwinding of DUE has been elucidated in E. coli. ATP-DnaA molecules assemble on the low-affinity DnaA boxes R5M-I2 using R5M-bound DnaA as a DnaA assembly core (Figure 1C, left) (1,3,22–24). DNA bending by IHF at IBR stimulates the interaction of R1-bound DnaA with R5M-bound DnaA in a head-to-tail manner, resulting in the formation of the Left-DnaA subcomplex. DNA superhelicity and heat energy cause the initial unstable unwinding of the DUE (Figure 1C, middle). DNA bending by IHF at IBR also brings the resulting ssDUE close to the Left-DnaA subcomplex, allowing formation of the ssDUE-Left-DnaA subcomplex and stabilizing the unwound state, a prerequisite for DnaB loading. R1- and R5M-bound DnaA protomers are thought to be important in the ssDUE-DnaA binding process, with H/B motifs of the protomers binding to TT[G/A]T(T) sequences in the ssDUE M-R region (Figure 1C, right). Collectively, these processes are referred to as the ssDUE recruitment mechanism (21,24). Recent intensive studies support that principles of this mechanism are conserved in oriCs from other bacteria such as Thermotoga maritima and Helicobacter pylori in addition to the Vibrio cholerae chromosome 2 (Chr2) (12,45,46).
Despite the importance of IHF in the ssDUE recruitment mechanism in E. coli oriC, IHF is present only in proteobacteria, nitrospirae, and nitrospinae (Supplementary Figure S1) (41,47). By contrast, both DnaA and HU, a structural homolog of IHF and a major nucleoid-associated protein, are ubiquitously conserved throughout eubacterial species (39,41) (Supplementary Figure S1). Unlike IHF, HU binds to DNA in a sequence-independent manner, although it preferentially binds to bent DNA: The dissociation constants Kd values for bent DNA and for linear DNA are reported to be 5–20 nM and 1.8–760 μM, respectively (39,41,48). Despite this difference, HU can replace IHF in E. coli oriC DUE unwinding in vitro (13,25,49). Moreover, HU can support the in vivo initiation of chromosomal replication in E. coli. IHF-deficient E. coli cells can survive despite disturbed regulation of replication initiation (25); however, cells in which both IHF and HU are disrupted grow at a very slow rate or can die (50). Compared with wild-type cells, IHF-deficient cells have lower ratios of oriC to DnaA (51). Moreover, the origin unwinding of T. maritima, one of the most evolutionarily ancient organisms, can be reconstituted in vitro using T. maritima DnaA (TmaDnaA) and E. coli HU, which is homologous to that of T. maritima HU (52). These characteristics suggest that HU may be a general DUE-unwinding stimulator in eubacteria.
The precise mechanisms by which E. coli HU promotes DUE unwinding remain unclear. In vitro analyses using E. coli oriC and proteins, based on sequence-non-specific DNA binding of HU, have suggested that HU modulates overall DNA topology to promote oriC unwinding (53,54). Alternatively, it has been suggested that E. coli HU may be included in oriC-DnaA complexes through unknown mechanisms (25,49,53,54). Even if so, the DnaA assembly mechanism on E. coli oriC in the presence of HU is not completely identical to that in the presence of IHF, in that DnaA binding to the low-affinity site I3 is inefficient in the presence of HU, but not in the presence of IHF (25). As such initiation mechanism using HU has remained as a long-standing mystery. The high structural homology between IHF and HU suggests that the ssDUE recruitment mechanism can be involved in HU-promoted unwinding of DUE in E. coli. The requirement for ssDNA binding by H/B motifs of TmaDnaA during HU-dependent T. maritima oriC (Tma-oriC) unwinding in vitro also suggests that HU can be involved in initiation even in other eubacterial species in a principally similar manner (12). However, the HU-promoting mechanisms as well as the structure of putative oriC-HU-DnaA complexes have not yet been determined even in E. coli.
The present study was designed to uncover the molecular mechanisms of HU-promoted DUE unwinding (the HU system). In addition to E. coli oriC, Tma-oriC was also analyzed to have insights into evolutional conservation. First, for E. coli oriC, the minimal region required for unwinding and the specific roles of key DnaA protomers in the HU system were determined. Subsequently, the HU binding sites in E. coli in the presence of the cognate ATP-DnaA were identified. The specificity of binding of ssDUE to DnaA was also analyzed in the HU system in vitro. All results were consistent with those of in vivo analyses and suggested that the ssDUE recruitment mechanism was the primary mechanism, even in ATP-DnaA-HU-oriC complexes. Unlike during IHF-promoted DUE unwinding (the IHF system), R1/R5M-bound DnaAs as well as interactions between the two were essential for HU-associated DUE unwinding. Taken these together with results of footprint experiments, the present findings suggest that R1-bound DnaA binds to R5M-bound DnaA, triggering DNA bending between the two sites and unstable DUE unwinding, which is stabilized by binding of ssDUE to Left-DnaA subcomplex and preferential binding of HU to the bent region. Next, we analyzed Tma-oriC and results suggested that HU binds to a specific region within interspace of two TmaDnaA binding sites, depending on TmaDnaA binding to these sites. These are fundamentally consistent with the proposed mechanism based on analyzes of E. coli oriC. Because DnaA and HU are highly conserved in eubacteria, this ssDUE recruitment mechanism is conceivable to be an evolutionarily conserved feature of origin unwinding of eubacteria.
MATERIALS AND METHODS
Proteins, DNA, plasmids and strains
DnaA and its derivatives used in this study were prepared as described (12,22,24). HU was prepared as previously reported (55), except for additional purification on a heparin column. Plasmids and primers used in this study are listed in Supplementary Tables S1 and S2, respectively. All plasmids used here are described previously (21–24,52). To construct ssMR-Left DOR, the upper strand was extended using the 5′ overhang primer M28-R1f and the 119-mer ssDNA of the bottom strand (Left half DORr), the 3′ end of which was modified with amino-modifier-C7-CPG that prevents extension of the bottom strand. Similarly, dsMR-Left DOR was constructed by primer extension with the Left half DORr oligo without the 3′ end modification.
All E. coli strains used in this study are listed in Supplementary Table S3. To eliminate a kanamycin-resistant cassette (kan) from oriC mutant strains, FLP recombinase encoded on pCP20 was used (24). Elimination of kan was verified by checking sensitivity to 50 μg/ml kanamycin in LB agar plates. NY20-frt strain and oriC mutant strains (SYM25-frt, SYM5-frt, SYM6-frt, SYM7-frt, SYM9-frt, NY24-frt, SYM-24-frt) were constructed by eliminating kan from NY20 strain and strains SYM25, SYM5, SYM6, SYM7, SYM9, NY24, SYM-24 (Supplementary Table S3). ΔqueG::frt-kan, ΔihfA::frt-kan and ΔihfB::spec derived from strains SR08, KMG-5 and KX95, respectively, were introduced by P1 phage transduction into the oriC mutant strains without the kanamycin resistant gene. Details for these constructions are described in Supplementary Data.
Buffers
Buffer P contained 60 mM HEPES–KOH (pH 7.6), 0.1 mM zinc acetate, 8 mM magnesium acetate, 30% [v/v] glycerol, and 0.32 mg/ml bovine serum albumin (BSA). Buffer N contained 50 mM HEPES–KOH (pH 7.6), 2.5 mM magnesium acetate, 0.3 mM EDTA, 7 mM dithiothreitol (DTT), 0.007% [v/v] Triton X-100, and 20% [v/v] glycerol. Buffer F’ contained 25 mM HEPES–KOH (pH 7.6), 5 mM calcium acetate, 2.8 mM magnesium acetate, 4 mM DTT, 10% [v/v] glycerol, 0.2% [v/v] Triton X-100, and 0.5 mg/ml BSA. Buffer X contained 30 mM Tris–HCl (pH 8.0), 3 mM magnesium chloride, 0.2 mM DTT, and 0.1 mM EDTA.
DUE unwinding assay
DUE unwinding assays were performed essentially as described (22,24). Briefly, EcoDnaA, ChiDnaA, and their derivatives were preincubated with 3 μM ATP for 10 min at 4°C, resulting in their ATP forms. M13oriCMS9 oriC plasmid or its derivatives (1.3 nM) were incubated with 43 nM HU and the indicated amounts of the ATP form of EcoDnaA or its derivatives at 38°C for 3 min in 20 μl buffer P containing 5 mM ATP and 125 mM potassium chloride, followed by further incubation with 1.5 units of P1 nuclease (Wako) for 200 s. The reactions were halted by the addition of 20 μl of stop buffer (1% SDS and 25 mM EDTA), and the DNA was purified by phenol-chloroform extraction and ethanol precipitation. One-half of each purified DNA was digested with EcoRI, which yielded 3.8 and 3.9 kb fragments for DUE unwinding of M13oriCMS9 (24). The resultant DNA fragments were analyzed by 1% agarose gel electrophoresis and subjected to Gelstar (Lonza) staining. Gel images were taken using a FAS V transilluminator, and products derived from unwound plasmids were quantified using ImageJ software.
DNase I footprint assay
The assay was essentially performed as described previously (18,24), with minor modifications, including the design of the oriC fragment. DnaA and HU were incubated for 10 min at 30°C in 10 μl buffer F’ containing 2.4 nM of end-labeled ssMR-Left DOR (102-bp double strand DNA with a 28-mer single-stranded region) or dsMR-Left DOR (130-bp double strand DNA), 20 mM ammonium sulfate, 50 mM sodium chloride, 1 μg/ml poly (dI-dC), 1 μg/ml poly (dA-dT) and 3 mM ATP, followed by the addition of 10 mU of DNase I and further incubation at 30°C for 4 min. Purified DNA was analyzed by 6% sequencing gel electrophoresis and a Typhoon FLA9500 image analyzer (GE Healthcase).
For analyzing τ1-I2 DOR, DnaA and HU were incubated for 10 min at 30°C in 10 μl buffer F’ containing 2.4 nM of end-labeled wild-type τ1-I2 DOR or its τ1non mutant derivative, 20 mM ammonium sulfate, 50 mM sodium chloride, 0.7 μg/ml poly (dI-dC), 0.7 μg/ml poly (dA-dT), 3 mM ATP or ADP, followed by the addition of 15 mU of DNase I and further incubation at 30°C for 4 min. Purified DNA was analyzed similarly.
Dimethyl sulfate (DMS) footprint assay
This assay was performed basically accordingly to a previously described method (25). To analyze HU binding to oriC, supercoiled oriC plasmid M13oriCMS9 (2.1 nM) was incubated with the indicated amounts of HU and DnaA in buffer X containing 100 mM potassium chloride and 5 mM ATP or ADP at 38°C for 5 min, followed by the addition of 0.25% DMS and further incubation for 5 min. DMS was quenched by the addition of 650 mM 2-mercaptoethanol, and the DNA was purified by phenol-chloroform extraction and ethanol precipitation. The purified DNA was incubated at 90°C for 30 min with 1 M piperidine, followed by ethanol precipitation. One sixth portion of the resultant DNA fragments were analyzed by primer extension using 0.4 units of Vent (exo-) DNA polymerase and 0.1 pmol 32P-labeled ori2 for the upper strand or 0.1 pmol 32P-labeled KWSmaIoriCFwd for the lower strand. Extension products were analyzed by electrophoresis on 6% sequencing gels, followed by exposure of the dried gels to imaging plates. For densitometry, the imaging plates were scanned with Typhoon FLA 9500 (GE Healthcare) and images were quantified using ImageJ.
To analyze Tma-oriC, 6 nM pOZ14 was incubated with the indicated amounts of HU and TmaDnaA in buffer X at 48°C for 5 min, followed by the procedure used in the analysis of M13oriCMS9.
Flow cytometry analysis
Flow cytometry analysis was performed essentially as described (56). Briefly, cells were grown at 30°C in LB medium including 100 μg/ml ampicillin until the absorbance of the culture (A600) reached 0.1. Portions of the cultures were diluted thousand folds into 5 ml LB medium, which were incubated at 30°C until the absorbance of the culture (A600) reached 0.1. The remaining portions were further incubated to deduce the doubling time (Td) by measuring A600 every 20 min. At A600 of 0.1, the aliquots of the cultures were fixed in 70% ethanol to analyze cell mass using Multisizer 3 Coulter counter (Beckman Coulter). The remaining cultures were further incubated for 4 h with 0.3 mg/ml rifampicin and 0.01 mg/ml cephalexin for run-out replication of the chromosomal DNA. The resultant cells were fixed in 70% ethanol. After DNA staining with SYTOX Green (Life Technologies), cellular DNA contents were analyzed on a FACS Calibur flow cytometer (BD Bioscience). We deduced the number of the origins/cell (ori/cell) from histograms of the flowcytometry analysis. The values of ori/cell were divided by the mean cell mass deduced from the cell mass analysis, resulting in the values of ori/mass.
Immunoblot analysis
The assay was essentially performed as described previously (24,57). Briefly, exponentially growing cells were prepared as described above for flow cytometry analysis. When A600 of the culture reached 0.1, cells were harvested by centrifugation and dissolved in SDS sample buffer to adjust the calculated A600 of 50. Proteins in the portions (10 μl) were separated using 10% SDS-PAGE and were transferred to polyvinylidene difluoride membranes (Millipore) using a semi-dry blotting method (Bio-Rad). After blocking at 30°C for 30 min in 3% gelatin-TBS (3% gelatin, 20 mM Tris–HCl, 500 mM NaCl, pH 7.5), the membrane was incubated overnight at 4°C in 1% gelatin-TTBS (20 mM Tris–HCl, 500 mM NaCl, 0.05% Tween-20, pH 7.5) containing anti-DnaA rabbit antiserum (1:3000). After washing, the membrane was incubated at 30°C for 1 h in 1% gelatin-TTBS containing goat anti-rabbit IgG antibody conjugated to alkaline phosphatase (Bio-Rad), followed by development using AP Conjugate substrate Kit (Bio-Rad).
When colonies were directly analyzed, colony suspension of fresh transformants grown at 30°C for 24 h on LB agar plates including 100 μg/ml of ampicillin were used, instead of liquid culture.
RESULTS
Determination of the minimal DOR for HU-promoted DUE unwinding
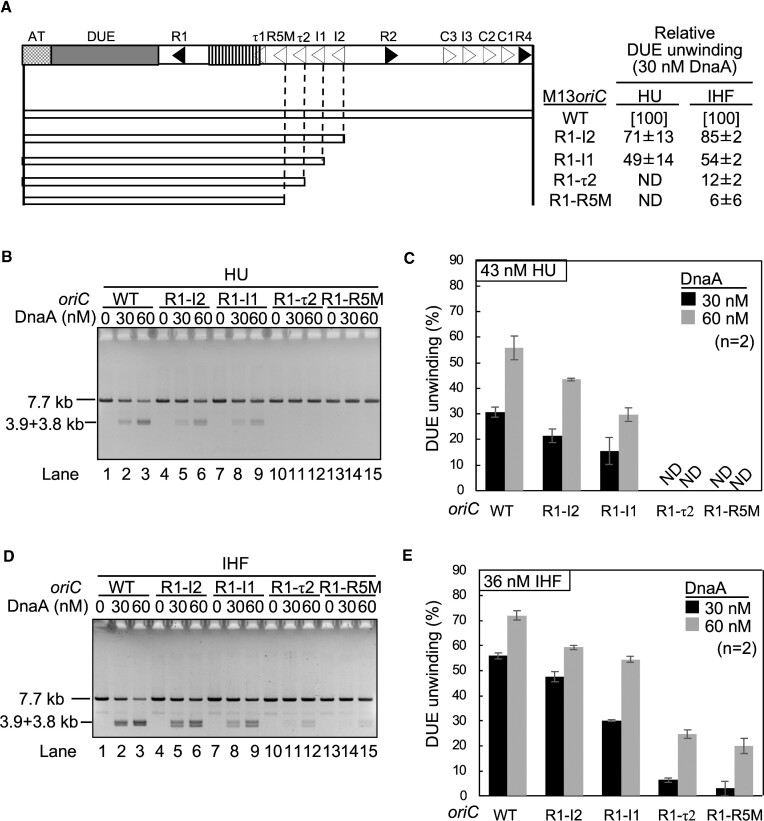
In the presence of IHF, the Left-DOR was sufficient for DUE unwinding by ATP-DnaA (21,24). To investigate the specificity of DOR in the HU system, minimal DOR for DUE unwinding in the presence of HU or IHF was assessed using deletion derivatives of the supercoiled oriC plasmid (M13oriCMS9) (Figure 2A). In this assay, the unwound DUE was digested by the single strand-specific nuclease P1, followed by digestion with EcoRI, which yields specific DNA fragments indicative of DUE-specific digestion by P1 nuclease (24). The unwinding activity of M13oriCMS9 R1-I2 was slightly less than that of the wild-type oriC in the HU system (Figure 2B and C). M13oriCMS9 R1-τ2 and M13oriC R1-R5M exhibited little DUE unwinding activity in the presence of HU (Figure 2B, lanes 10–15; Figure 2C), but both had moderate activities in the presence of IHF at a higher DnaA level (Figure 2D, lanes 10–15; Figure 2E). These results suggested that the Left-DOR was generally sufficient for DUE unwinding, as shown for the IHF system and that Left-DOR-DnaA complexes formed with HU are relatively more labile than those with IHF, as basically consistent with previous reports (21,24). In particular, the R1-I1 region was found to be the HU-specific minimum Left-DOR region (Figure 2B, lanes 10–15; Figure 2C; Figure 2D, lanes 10–15; Figure 2E). Thus, DnaA molecules bound to R1, τ2 and I1 are likely crucial for HU-promoted DUE unwinding.
Figure 2.
Determination of the minimal oriC for DUE unwinding in the HU system. DUE unwinding activities of wild-type oriC plasmid M13oriCMS9 (WT) and its deletion derivatives were analyzed by P1 nuclease assay in the presence of HU or IHF. The oriC regions (open bars) included in each plasmid are shown below the wild-type oriC structure depicted as in Figure 1. (A) The relative unwinding activities of the mutant oriCs relative to that of wild-type oriC are shown in panel A as ‘DUE unwinding’ using the data obtained with 30 nM ATP-DnaA (see below). The supercoiled form of oriC plasmids (1.3 nM) were incubated with the indicated amounts of ATP-DnaA in the presence of HU (43 nM) (B and C) or IHF (36 nM) (D and E), followed by co-incubation with P1 nuclease. The DNA products were extracted, further digested with EcoRI, and analyzed using 1% agarose gel electrophoresis. Representative gel images of two independent experiments were shown (B and D). Band intensities of each lane in the gel image were analyzed by densitometric scanning. The percentages of the P1 nuclease-digested oriC DNA molecules per input DNA molecules are shown as ‘DUE unwinding (%)’ (C and E). Means and standard deviations (SDs) are also shown (n = 2). ND, not detected.
Analysis of individual DnaA boxes in HU-promoted DUE unwinding
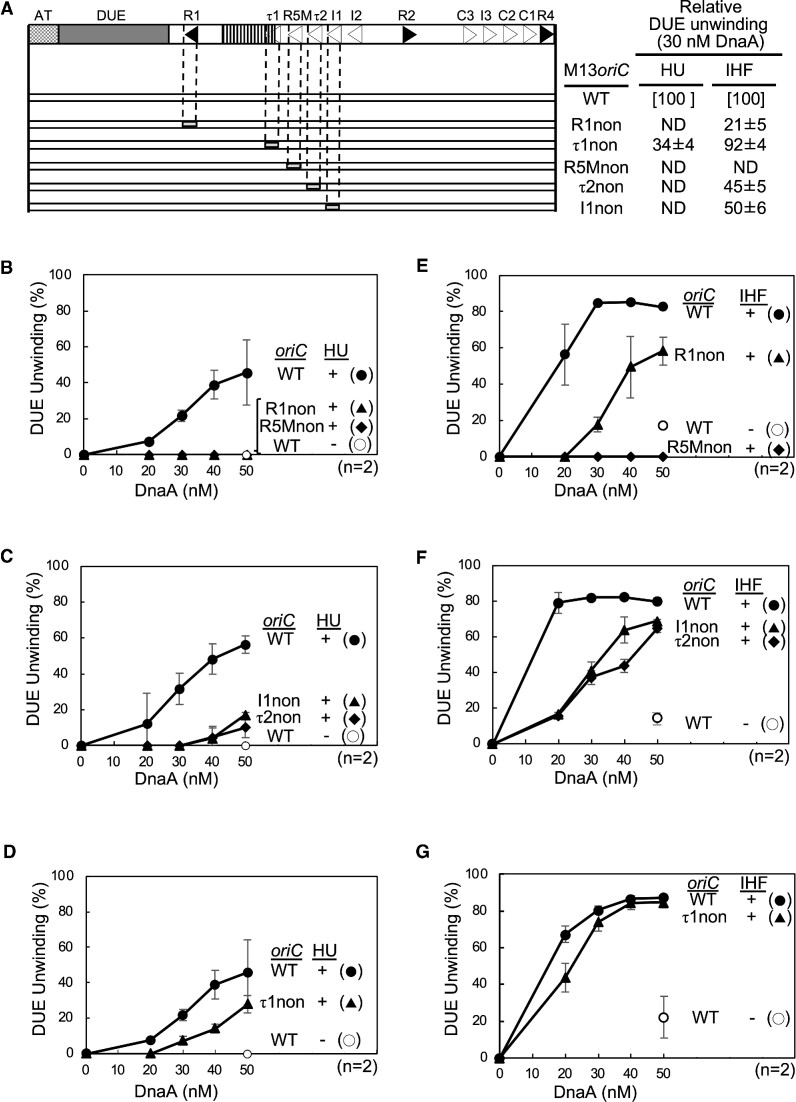
Requirements for individual DnaA boxes were analyzed using full-length oriC with a DnaA box mutation in the Left-DOR. Because DnaA box I2 was non-essential for DUE unwinding (Figure 2), DnaA boxes R1, τ1, R5M, τ2 and I1 were individually substituted with a sequence (nonsense box) lacking specific affinity for DnaA (Figure 3A) (16,24). Specific unwinding of these mutant oriC plasmids was assessed by the P1 nuclease assay in the presence of HU or IHF (Figure 3B–G). The R5M box was found to be essential for DUE unwinding in both the HU and IHF systems (Figure 3B and E; Supplementary Figure S2A and D), in agreement with previous results showing that R5M box-bound DnaA (R5M-DnaA) is essential as a DnaA assembly core in low-affinity DnaA box cluster spanning boxes R5M to I2 (24). DnaA boxes τ2 and I1 were more important for DUE unwinding in the HU than in the IHF system (Figure 3C and F; Supplementary Figure S2B and E). These strict requirements in the HU system were consistent with the results of deletion analysis.
Figure 3.
Determination of the key DnaA boxes required for the HU system. DUE unwinding activities of wild-type oriC plasmid M13oriCMS9 (WT) and its substitution derivatives in the presence of HU or IHF as determined by P1 nuclease assays. The wild-type oriC structure is shown as in Figure 2; below it, the oriC derivatives (open bars) with nonsense boxes (light-gray) substituted with DnaA box (R1non, τ1non, R5Mnon, τ2non, or I1non) are shown (A). Unwinding activities of mutant oriC DUE relative to that of wild-type oriC DUE shown in panel A as ‘DUE unwinding’ using the data obtained with 30 nM ATP-DnaA shown in panels B–G (B–D for the HU system and E–G for the IHF system). Means and standard deviations (SDs) are also shown (n = 2). Representative gel images are shown in Supplementary Figure S2. ND, not detected.
DnaA box R1 was essential in the HU system, whereas it was moderately important in the IHF system (Figure 3B and E; Supplementary Figure S2A and D). In addition, DnaA box τ1 moderately assisted in DUE unwinding activity in the HU system (Figure 3D and G; Supplementary Figure S2C and F). Given that even in the absence of IHF and HU, R1-bound DnaA (R1-DnaA) interacts with ATP-DnaA molecules bound to the R5M-I2 region (21,24), these findings suggest that R1-DnaA plays an essential role in construction of ATP-DnaA complexes competent to DUE unwinding specifically in the HU system and that τ1 box-bound DnaA (τ1-DnaA) plays a subordinate role in these processes in the HU system.
Specific roles for R1- and R5M-DnaA in the HU system
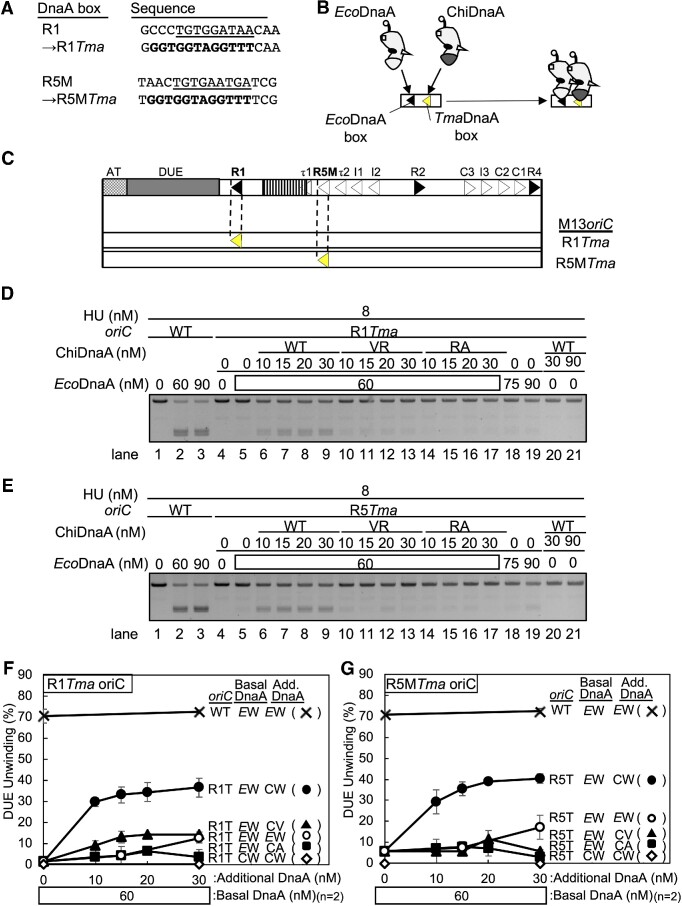
In DUE unwinding processes, the R1- and R5M-DnaA protomers directly bind to ssDUE to stabilize the unwound form of DUE in the ATP-DnaA-IHF-oriC complex (Figure 1C) (24). If DUE unwinding by HU and DnaA depends on a ssDUE recruitment mechanism, R1- and R5M-DnaA protomers should similarly interact with ssDUE directly even in the HU system. Thus, the specific activities of R1- and R5M-DnaA protomers in the HU system were analyzed using a chimeric DnaA-oriC system. The chimeric DnaA (ChiDnaA) consisted of E. coli DnaA (EcoDnaA) domains I-III connected to TmaDnaA domain IV. The chimeric oriC had a unique 12-mer TmaDnaA box (consensus sequence, AAACCTACCACC) instead of E. coli-type 9-mer DnaA boxes (consensus sequence, TTATnCACA) (Figure 4A). Unlike EcoDnaA, ChiDnaA binds specifically to the TmaDnaA box but little to the E. coli DnaA box (Figure 4B) (14,22–24,58). In addition to ChiDnaA, the oriCR1Tma plasmid bearing a TmaDnaA box substitution at the R1 box site (M13oriCMS9 R1Tma) and the oriCR5MTma plasmid bearing the identical substitution at the R5M box site (M13oriC MS9 R5MTma) were used (Figure 4C). Both chimeric oriC plasmids unwound only when both EcoDnaA and ChiDnaA (ChiDnaA WT) were co-incubated with HU (Figure 4D, lanes 4–9, and 18–21; Figure 4E, lanes 4–9, and 18–21; Figure 4F and G). However, unlike ChiDnaA WT, ChiDnaA bearing V211A and R245A substitutions in the ssDNA binding H/B motifs (ChiDnaA VR) barely or only slightly stimulated the unwinding of DUE of the both oriC plasmids (Figure 4D, lanes 10–17; Figure 4E, lanes 10–17; Figure 4F and G). Slight residual activities of the ChiDnaA VR were possibly due to EcoDnaA non-specific binding to a TmaDnaA box at the R1 or R5M box site in a cooperatively stimulated manner. These results support the importance of ssDUE binding by R1- and R5M-DnaA protomers in the HU system and the idea that ssDUE recruitment mechanism operates in the HU system.
Figure 4.
Roles of H/B motifs and the arginine fingers of R1-DnaA and R5M-DnaA in HU promotion of DUE unwinding. (A) Sequences of R1 and R5M boxes substituted with TmaDnaA box (R1Tma and R5Tma, respectively). The R1 and R5M sequences are underlined and the consensus TmaDnaA box is indicated in bold letters. (B) The principle of the chimeric DnaA assay. ChiDnaA has domains I–III (amino acids 1–374) of E. coli DnaA (white) and domain IV (amino acids 342–440) of T. maritima DnaA (gray). ChiDnaA, but not EcoDnaA, specifically binds to TmaDnaA box (yellow arrowhead). DnaA is illustrated as in Figure 1C. (C) Schematic depiction of chimeric oriCs, as in Figure 1A. The positions of R1Tma and R5Tma are indicated (yellow arrowhead). (D–G) DUE unwinding activities analyzed by P1 nuclease assay. The supercoiled form of the oriC plasmid (M13oriCMS9) or its derivatives (M13oriCMS9 R1Tma and M13oriCMS9 R5MTma) (1.3 nM) were incubated with ATP-EcoDnaA (50 nM) and HU (8 nM) in the presence of the indicated amounts of ChiDnaA (WT) or its mutants ChiDnaA V211A/R245A (VR) or R285A (RA), followed by P1 nuclease assay. Gel analysis was performed as described for Figure 3B–E. The images shown are representative of two independent experiments (panels D and E). Percentages of P1 nuclease-digested oriC DNA molecules per input DNA molecules are shown as ‘DUE unwinding (%)’ (panels F and G). Means and standard deviations (SDs) are also shown (n = 2). Abbreviations in panels D and E: EW, wild-type E. coli DnaA; CW, wild-type ChiDnaA; CV, ChiDnaA V211A/R245A; CA, ChiDnaA R285A.
In the IHF system, R1-DnaA interacts with neighboring R5M-bound ATP-DnaA through the R1-DnaA Arg finger, stimulating ssDUE recruitment (Figure 1C) (22). The importance of the Arg finger of R1-DnaA in the HU system was investigated using the chimeric DnaA-oriC system. Unlike ChiDnaA WT, ChiDnaA R285A (ChiDnaA RA) did not substantially stimulate DUE unwinding of M13oriCMS9 R1Tma in the presence of EcoDnaA (Figure 4D, lanes 14–17; Figure 4F). These results support the importance of specific interaction between R1- and R5M-DnaA protomers in the HU-dependent DUE unwinding system.
The role of the R5M-DnaA Arg finger was similarly analyzed. Unlike ChiDnaA WT, ChiDnaA RA did not substantially stimulate DUE unwinding of M13oriCMS9 R5MTma in the HU system (Figure 4E, lanes 14–17; Figure 4G), which is consistent with the results showing that R5M-DnaA acts as the assembly core for cooperative DnaA binding in the R5M-I2 low-affinity DnaA box cluster (24).
Specific importance of R1, R5M and τ1 boxes in cells lacking IHF and bearing HU
The importance of R1 and R5M boxes was assessed in live cells lacking IHF and bearing HU. The synthetic effects of disruptions of the ihfA gene encoding the IHF α subunit, and of each DnaA box of the Left-DOR were analyzed. A single disruption in ihfA or ihfB encoding the IHF β subunit, was sufficient to inactivate the functional activity of IHF, as flow cytometry showed that inhibition levels for replication initiation were similar among cells with single or double disruptions (Supplementary Figure S3A: also see below). In these experiments, exponentially growing cells were further incubated in the presence of rifampicin and cefalexin to inhibit the initiation of replication and cell division, allowing run-out replication of the whole chromosome. Flow cytometry analysis of these cells shows the number of oriC copies per cell at the time of drug addition (7,14,24).
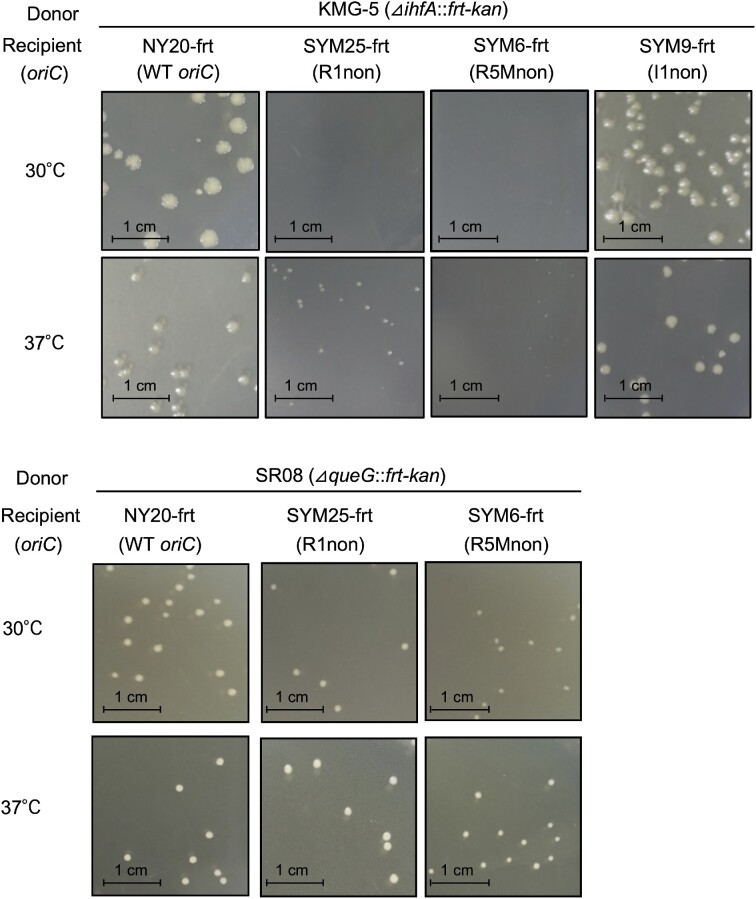
IHF-intact cells with individual substitutions at R1, R5M, and I1 with the nonsense box (i.e. R1non, R5Mnon and I1non) grow at a rate similar to cells with intact oriC, whereas initiation of chromosomal replication is inhibited moderately by R1non or R5Mnon and more mildly by I1non (24). However, we found that the R1 and R5M boxes were essential for maintaining the growth rates of IHF-deficient cells. When cells were incubated at 30°C for 16 h, the transduction frequency of ΔihfA::kan cells with R1non or R5Mnon, but not I1non, was markedly lower than that of wild-type cells (Table 1, Figure 5 and Supplementary Figure S3BC). When incubated at 37°C for 12 h, ΔihfA::kan transductants with R1non or R5Mnon formed only small colonies (Table 2 and Figure 5). Disruption of a non-essential gene (ΔqueG::kan) was used as a control: ΔqueG::kan cells bearing R1non or R5Mnon grew at 30°C and 37°C similarly to those of wild-type cells (Table 1; Figure 5).
Table 1.
Transduction frequency of ΔihfA::kan and ΔqueG::kan at 30°C
| Donor | Recipient (relevant genotype) | Average number of transductants | Transduction frequency (×10−7) | Relative frequency |
|---|---|---|---|---|
| ΔihfA::kan | NY20-frt (WT oriC) | 180 ± 91 | 2.5 ± 1.2 | [1] |
| SYM25-frt (R1non) | <1 | <0.014 | <0.006 | |
| SYM6-frt (R5Mnon) | <1 | <0.014 | <0.006 | |
| SYM9-frt (I1non) | 154 ± 70 | 2.1 ± 1.0 | 0.90 | |
| ΔqueG::kan | NY20-frt (WT oriC) | 145 ± 47 | 2.0 ± 0.6 | [1] |
| SYM25-frt (R1non) | 92 ± 50 | 1.2 ± 0.7 | 0.60 | |
| SYM6-frt (R5Mnon) | 39 ± 3 | 0.53 ± 0.0 | 0.30 |
NY20-frt (wild-type oriC; WT), SYM25-frt (R1non), SYM6-frt (R5Mnon), or SYM9-frt (I1non) was crossed with P1 phage lysate prepared from the strain KMG-5 (ΔihfA::frt-kan) or SR08 (ΔqueG::frt-kan). Transductants were incubated at 30°C for 16 h on LB agar plates containing 50 μg/ml kanamycin, and the numbers of colonies were counted. Transduction frequency was calculated by dividing the number of colonies by the number of infection events (plaque-forming units). For relative frequency, the efficiency of NY20-frt was defined as 1. Results represent the mean of two independent experiments.
Figure 5.
Requirement of DnaA boxes R1 and R5M in rapid growth of IHF-deficient cells. Colony formation of oriC mutants lacking the ihfA gene. NY20-frt (wild-type oriC; WT) and its DnaA box mutant derivatives SYM25-frt (R1non), SYM6-frt (R5Mnon), and SYM9-frt (I1non), were crossed with P1 phage lysates prepared from the strain KMG-5 (ΔihfA::frt-kan) or control SR08 (ΔqueG::frt-kan). Transductants were incubated on LB agar plates at 30°C for 16 h or 37°C for 12 h. Transduction frequencies are shown in Tables 1 and 2.
Table 2.
Transduction frequency of ΔihfA::kan at 37°C
| Recipient (relevant genotype) | Average number of transductants | Transduction frequency (×10−7) | Relative frequency |
|---|---|---|---|
| NY20-frt (WT oriC) | 167 ± 59 | 2.3 ± 0.8 | [1] |
| SYM25-frt (R1non) | 120 ± 9* | 1.6 ± 0.1 | 0.70 |
| SYM6-frt (R5Mnon) | 96 ± 2* | 1.3 ± 0 | 0.60 |
| SYM9-frt (I1non) | 182 ± 40 | 2.5 ± 0.6 | 1.1 |
To confirm the synergistic effects of mutations in IHF and mutations in DnaA boxes R1 and R5M, colony formation was analyzed in a series of oriC mutant cells with spec-substitution mutations in the ihfB gene. Consistent with cells bearing ΔihfA::kan, the growth of ΔihfB::spec cells bearing R1non or R5Mnon at 30°C was markedly lower than that of wild-type oriC cells bearing ΔihfB::spec (Supplementary Figure S3BC), indicating that the R1 and R5M boxes sustain the growth of cells lacking IHF and bearing HU. The elevated importance of the R1 and R5M boxes in cells lacking IHF is in agreement with the in vitro results described above.
The importance of τ1, τ2 and I1 boxes was tested using ΔihfA::kan cells bearing the nonsense box substitution to each (i.e., τ1non, τ2non, and I1non). The growth of ΔihfA::kan cells bearing these substitutions on LB agar plates at 30°C was similar to that of IHF-intact cells bearing the same substitutions (Supplementary Figure S4A). However, flow cytometry analysis showed that the DNA replication initiation frequency was lower in ΔihfA::kan cells bearing τ1non or τ2non than in ΔihfA::kan cells without those mutations, as shown by a decrease in the eight chromosomes peak and an increase in the two to four chromosomes peaks (Supplementary Figure S4B). In IHF-intact cells, initiation was not substantially inhibited by τ1non or by τ2non (Supplementary Figure S4B), in agreement with previous findings (24). I1non markedly inhibited initiation even in IHF-intact cells, which was exacerbated in ΔihfA::kan cells (Supplementary Figure S4B). These differences caused by the DnaA box sites could be related to the residual levels in stability of DnaA complexes formed only by the intact DnaA boxes.
DnaA protomers bound to τ2 and I1 assist in stabilizing the Left-DnaA subcomplex (24), basically consistent with the results of inhibited initiation in ΔihfA::kan cells bearing τ2non or I1non. These suggest an idea that the compromised initiation in ΔihfA::kan cells bearing τ1non was due to a decrease in the stability of the Left-DnaA subcomplex. To assess such a role for τ1, we performed DNase I footprint experiments using the oriC τ1-I2 fragments with wild-type τ1 box or τ1non. ATP-DnaA binding to R5M box was moderately enhanced depending on DnaA binding to τ1 box (Supplementary Figure S4C and D), consistent with the idea that τ1 moderately contributes to stability of the Left-DnaA subcomplex in the system lacking IHF. The moderate effect might come from relatively unstable binding of ATP-DnaA binding to τ1 because of the distance to the flanking R5M box (four bp) being greater than the other box-to-box spaces (two bp each) in this region (24). Slight protection of the τ1non region was detected in the presence of high DnaA concentrations, which might be caused by non-specific unstable interaction of a DnaA molecule bound to R5M-bound DnaA via domain I–domain I interaction. The τ1non fragment showed altered digestion patterns within the τ1 box-corresponding region, which is due to changes of the nucleotide sequence.
Specific importance in vivo of Arg finger and ssDNA binding motifs of R1/R5M-DnaAs in HU-dependent cells
The Arg finger motif of R1-DnaA has been shown to play an important role in the initiation of chromosome replication in cells bearing IHF and HU (22). Similarly, the ssDNA-binding H/B motifs (V211 and R245) of R1- and R5M-DnaA were shown to be important in the in vivo initiation of replication (24). The in vivo importance of these motifs was analyzed using cells with chimeric oriC and ChiDnaA under ihfA-null conditions. To this end, ΔihfA::kan cells were modified by substitution of a TmaDnaA box for boxes at the R1 (R1Tma) and R5M (R5MTma) positions. ΔihfA::kan cells bearing R1Tma showed marked inhibition of colony formation at 30°C, although slight growth was observed (Supplementary Figure S3B). Because the growth of these cells with R1non was even lower (Supplementary Figure S3B), the residual growth of the R1Tma cells may result from the slight binding of EcoDnaA to the TmaDnaA box under in vivo conditions. Similarly, ΔihfA::kan cells with R5MTma also showed marked inhibition of colony formation but with slight residual growth (Supplementary Figure S3C).
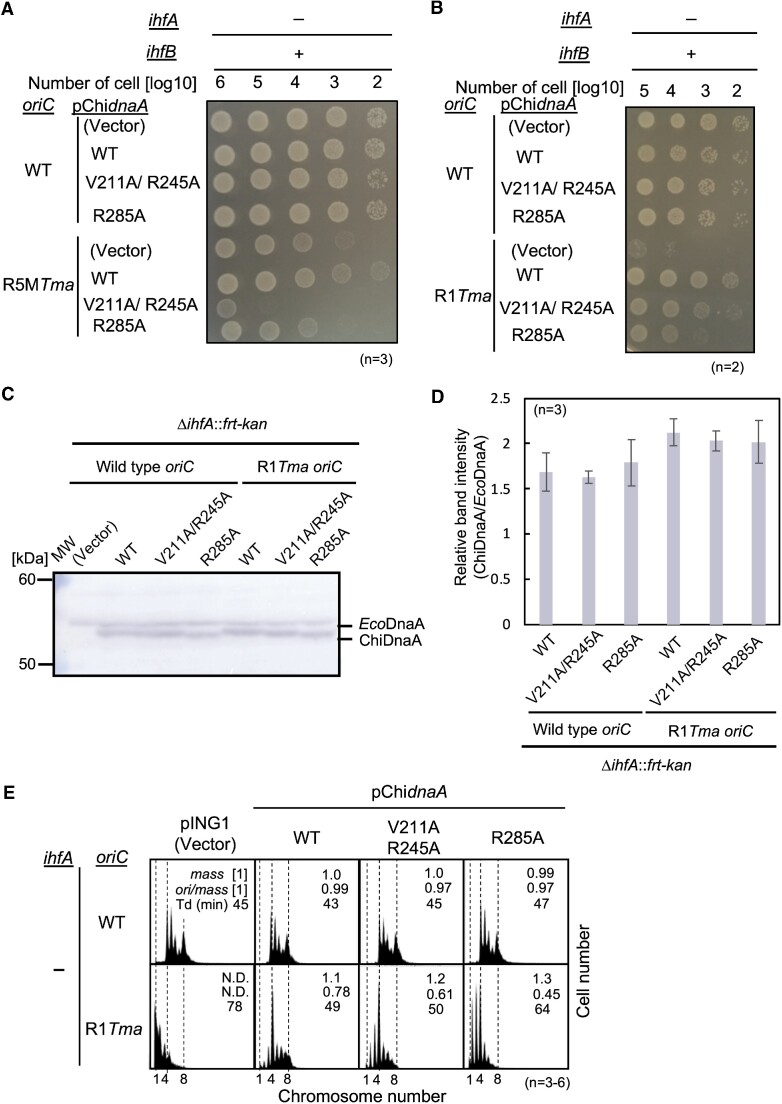
To test whether the Arg finger motif and the H/B motifs of R5M-DnaA are crucial for colony formation in cells lacking IHF and bearing HU, a series of pING1 vector-based plasmids encoding ChiDnaA WT (pChidnaA) or ChiDnaA mutants, pChidnaA V211A/R245A (pChidnaA VR) and pChidnaA R285A (pChidnaA RA), were introduced into ΔihfA::kan cells with R5MTma. Although none of these plasmids significantly affected colony formation by ΔihfA::kan cells bearing wild-type oriC, pChidnaA WT rescued the colony formation deficiency of ΔihfA::kan cells bearing R5MTma (Figure 6A). Consistent with previous studies (22,24), these findings indicate that even leaky expression can supply sufficient ChiDnaA to support the growth of R5MTma cells. Notably, the rescuing activities of pChidnaA RA and pChidnaA VR were markedly lower than that of pChidnaA WT. These results suggest that R5M-DnaA requires both the Arg finger motif and the H/B motifs to sustain functional initiation complexes even in cells with HU but without IHF.
Figure 6.
Roles of R1- and R5M-DnaA protomers in IHF-deficient cells. (A and B) Colony forming abilities. ΔihfA cells with wild-type oriC or chimeric R5Tma oriC (A) or R1Tma oriC (B) were transformed with pING1 plasmid (Vector) or its derivatives expressing ChidnaA (WT), ChidnaA V211A/R245A (V211A/R245A), or ChidnaA R285A (R285A). The transformants were grown overnight at 30°C and 10-fold serial dilutions of the cultures (∼109 cells/ml) were incubated on LB agar medium for 14 h at 30°C. + wild-type; – deletion. (C and D) ΔihfA cells with wild-type oriC (NY20-dihfA) or R1Tma oriC (NY24-dihfA) bearing pING1 (Vector), pChidnaA WT, pChidnaA V211A/R245A or pChidnaA R285A were grown in LB agar plates including ampicillin for 24 h at 30°C and were harvested for immunoblot analysis. Purified EcoDnaA and ChiDnaA were also analyzed as size markers. MW, molecular weight markers. A representative gel image is shown in panel C. Band intensities of each lane in the gel image were analyzed by densitometric scanning. The relative band intensities of ChiDnaA to EcoDnaA are shown as ‘Relative intensity (ChiDnaA/EcoDnaA)’ (D). Means and standard deviations (SDs) are also shown (n = 3). (E) Flow cytometry analyses of ΔihfA cells with or without oriC R1Tma (NY20-dihfA and NY24-dihfA) bearing pING1 vector or its pChidnaA-derivatives, as described above. Cells were grown at 30°C in LB medium containing ampicillin, followed by further incubation with rifampicin and cephalexin for run-out replication. DNA contents were quantified by flow cytometry. Cell sizes (mass) at the time of drug addition were measured by a Coulter counter. The mean mass, ori/mass ratio, and doubling time of each strain are indicated at the top right of each panel. Experiments were repeated three to six times.
Similarly, ΔihfA::kan cells bearing R1Tma and the above plasmids were analyzed. The results confirmed the importance of the Arg finger motif in R1-DnaA, as well as the moderate importance of the H/B motifs of R1-DnaA at 30°C and 25°C (Figure 6B and Supplementary Figure S5A). The ΔihfA::kan cells expressed moderately similar levels of WT or mutant ChiDnaA from corresponding plasmids without the inducer arabinose: in particular, cellular levels of ChiDnaA V211A/R245A and ChiDnaA R285A in ΔihfA R1Tma cells were very similar (Figure 6C and D).
The function of H/B motifs of R1-DnaA was investigated in more detail by flow cytometry to determine the number of oriC copies in each growing cell. Most of the ΔihfA::kan cells with wild-type oriC bearing each plasmid had more than four chromosomes (Figure 6E, upper row). Consistent with previous findings (25,56), ΔihfA::kan cells initiated chromosome replication asynchronously, likely due to changes in oriC function as well as DnaA regulation by the loss of function of DARS2 and datA, which are IHF-dependent regulators for DnaA activity (see Discussion) (56,59). By contrast, ΔihfA::kan cells with R1Tma bearing the vector pING1 grew at a slower rate, with most cells harboring one to three chromosomes (Figure 6E; first histogram of the lower row). Introduction of pChidnaA WT restored cell growth rates and enhanced the initiation of chromosomal replication, with most cells having four to eight chromosomes (Figure 6E, second histogram of the lower row). In contrast, introduction of pChidnaA RA only moderately restored the growth rates of cells and cells bearing pChidnaA RA or pChidnaA VR had increased percentages of cells containing fewer than four chromosomes compared to cells bearing pChidnaA WT, with most cells bearing pChidnaA RA having two to four chromosomes, fewer than those bearing pChidnaA VR (Figure 6E, third and fourth histograms of the lower row). These findings indicate that in vivo initiation activity of R1-ChiDnaA bearing V211A/R245A is moderately lower than that of R1-ChiDnaA WT, and that of R1-ChiDnaA R285A is much lower, consistent with the results of colony formation of mutant cells (Figure 6B and Supplementary Figure S5A). Cellular levels of ChiDnaA V211A/R245A and ChiDnaA R285A in ΔihfA R1Tma cells were very similar even under these growth conditions (Supplementary Figure S5B and S5C). Taken together, these results suggest that the Arg finger of R1-DnaA plays a very important role in HU-promoted initiation of replication, whereas the H/B motifs play a stimulatory role.
HU binds to IBR in a DnaA complex-dependent manner.
Based on the findings showing that R1-bound DnaA is more crucial in the HU system than in the IHF system and that interaction of R1/R5M-bound DnaAs is important in the both systems, HU-promoted initiation mechanisms were further analyzed by footprint experiments. HU prefers to bind to a bent region of DNA (41,60) and IBR resides between the R1 and R5M boxes. These and present findings suggest that, in the absence of IHF, R1- and R5M-DnaA molecules transiently interact with each other, stimulating loop formation of the intervening region, and that HU preferentially binds to the bent region, stabilizing the overall complex. The resultant structure of the overall complex would mimic the configuration of ATP-DnaA–IHF–Left-DOR complexes, thereby adopting the ssDUE recruitment mechanism (Figure 1C).
This hypothesis was initially tested in DMS footprint experiments using supercoiled M13oriCMS9. DMS modifies guanine residues, which are detected by primer extension assays (Figure 7 and Supplementary Figure S6). If present, G residues at the fourth position of a DnaA box become more sensitive, whereas those at the second position become less sensitive, to DMS in a manner depending on stable binding of ATP/ADP-DnaA (Supplementary Figure S6A) (25). Primers for the lower and upper strands were used accordingly to the different orientations of DnaA box sequences (Figures 1A and 7A, Supplementary Figure S6B–D). In good agreement, addition of ATP-DnaA or ADP-DnaA altered intensities of the bands corresponding to DnaA box R2 (Figure 7A and Supplementary Figure S6B). Intensities of the bands corresponding to the cluster of low-affinity DnaA boxes R5M, τ2, I1 and I2 were changed in an ATP-DnaA-dependent manner (Supplementary Figure S6C and D).
Figure 7.
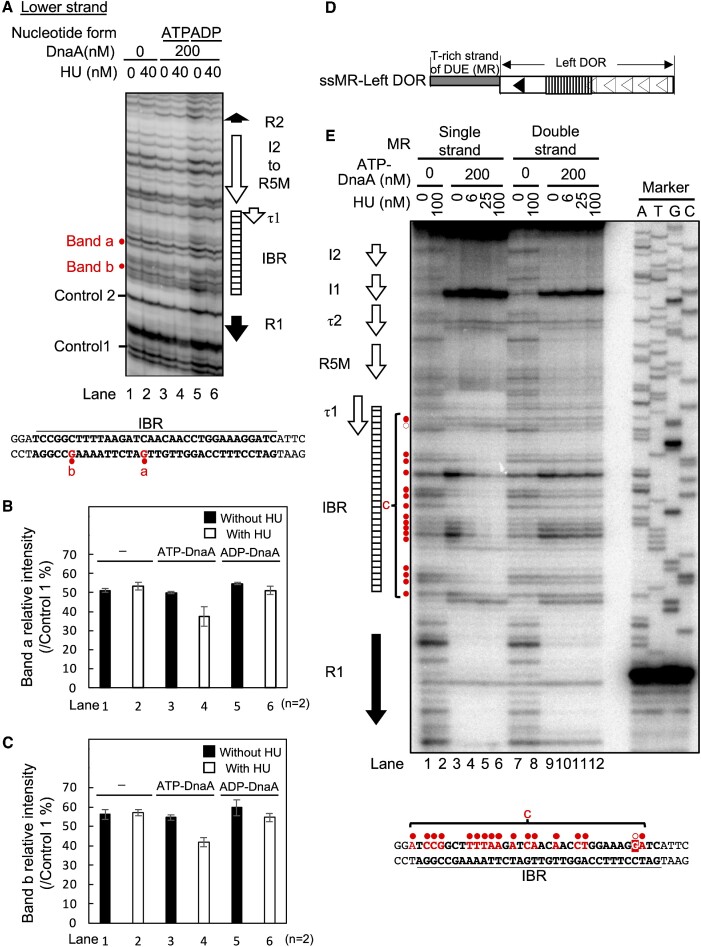
Specific HU binding to oriC in the presence of ATP-DnaA. (A–C) DMS footprint using oriC plasmid. The supercoiled form of the oriC plasmid (M13oriCMS9) was incubated at 38°C for 5 min with the indicated amount of HU in the presence or absence of ATP-DnaA or ADP-DnaA (200 nM), followed by further incubation with 0.25% DMS for 5 min. DMS-treated plasmids were cleaved with 1 M piperidine, and the resultant DNA fragments were analyzed by primer extension assays using the 32P-labeled primer KWSmaIoriCFwd for the lower strand modifications. The primer extension products were resolved on 6% sequencing gels. The positions of the DnaA boxes and IBR are shown alongside the gel image. The intensities of the bands corresponding to specific guanine residues within IBR (a and b) were reduced in a manner dependent on both ATP-DnaA and HU. Those guanine residues are highlighted in red on the IBR sequence below the gel image (A). Intensities of bands a and b relative to the control 1 band quantified as ‘Relative band intensity of a’ (B) and ‘Relative band intensity of b’ (C). Results are the means and standard deviations (SD) of two independent experiments. Histograms of the footprint patterns are shown in Supplementary Figure S6E. (D, E) HU binding to the ssDUE overhang-oriC DNA fragment. The ssMR-Left DOR consisted of the Left-DOR with a 5′-overhang of the upper (T-rich) strand of M/R-DUE (D). DNaseI footprint using the ssMR-Left DOR in the presence of ATP-DnaA and HU (E). 32P-labeled ssMR-left DOR and dsMR-left DOR were incubated at 30°C for 10 min with the indicated amount of HU in the presence or absence of ATP-DnaA (200 nM), followed by further incubation for 4 min with DNase I (10 mU). DNase I-digested products were analyzed on 6% sequencing gels. The positions of DnaA boxes and IBR are shown on the left side of the gel image. Region c includes specific sites which caused HU-dependent protection (closed red circles) or enhancement (an open red circle) in the presence of ATP-DnaA. The IBR sequence is shown below the gel image. Region c and the sites marked by the red circles on the left side of the gel image, are indicated by red characters.
When HU alone was incubated with the oriC plasmid, the DMS modification was unaffected, consistent with the non-specific and dynamic DNA interaction of HU (Figure 7A, lanes 1 and 2). By contrast, when HU and ATP-DnaA were incubated together, the intensities of bands within IBR were moderately reduced (bands a and b in Figure 7A, lanes 3 and 4; and Figure 7B and C). Little change was observed for the two bands with mixtures of HU and ADP-DnaA (Figure 7A, lanes 5 and 6; and Figure 7B and C; Supplementary Figure S6E). In the case of ADP-DnaA, intensities of two bands at the R1-proximal terminus and the middle of IBR were moderately reduced by HU, which might come from interaction of HU with IBR via interaction with R1-bound DnaA. These results would be in agreement with the hypothesis, that the construction of ATP-DnaA-specific Left-DOR complexes is a prerequisite for specific HU-IBR interactions.
To further consolidate the possible structures of ATP-DnaA–HU–Left-DOR complexes, DNase I footprint assays were performed using the Left-DOR fragment with the T-rich ssDUE overhang (ssMR-Left DOR in Figure 7D). Based on the ssDUE recruitment mechanism (Figure 1C), an ATP-DnaA pentamer is likely constructed via DNA bending in IBR, forming an unstable complex, which could be stabilized by preferential binding of HU to the bent IBR and by ssDUE binding to the ATP-DnaA pentamer. When ssMR-Left DOR and ATP-DnaA were mixed, regions of DnaA boxes were protected (Figure 7E, lanes 1 and 3). The DNase I susceptibility of IBR was significantly stimulated depending on ATP-DnaA, suggesting structural changes of IBR (region c in Figure 7E, lanes 3–6). Moreover, the upper region of the R1 box was protected (Figure 7E, lanes 3–6), which may have resulted from wrapping of this DNA region by the DnaA complex, consistent with the ssDUE recruitment mechanism (23). Notably, when HU was co-incubated with ATP-DnaA, most bands in region c within IBR were protected in an HU dose-dependent manner (Figure 7E, lanes 3–6). In this region, the intensities of several specific bands detected in the presence of 100 nM HU and in the absence of DnaA, were markedly reduced even with 6 and 25 nM HU in the presence of DnaA. DnaA box-like sequence was not present around these sites (Figure 7E. lanes 1–6). These results are consistent with the enhanced binding of HU to bent DNA: the dissociation constants Kd values for bent and linear DNA are reported to be 5–20 nM and 1.8–760 μM, respectively (48). Thus, these results suggest that HU directly and specifically binds to the bent IBR within the ATP-DnaA–Left-DOR complex. In addition, intensities of bands within DnaA box τ1/IBR-overlapping region are slightly changed depending on HU (Figure 7E). Interaction of HU with this region might exclude a DnaA molecule from τ1 box (see Discussion).
In addition, we performed similar experiments using Left-DOR fragment with double strand DUE, but not ssDUE (dsMR-Left DOR) (Figure 7E and Supplementary Figure S6H). When ATP-DnaA was co-incubated, regions for DnaA boxes were protected like those of ssMR-Left DOR. However, unlike the case of ssMR-Left DOR, the band intensities in region c within IBR in the presence of DnaA were only slightly changed by the addition of HU, implying labile interaction of HU with this region. These results thus show contribution of ssDUE to the site-specific HU binding, consistent with the importance of the ssDUE binding of DnaA (Figure 4). Interaction of ssDUE with Left DOR-DnaA complex is inferred to indirectly enhance the HU binding through stabilization of DNA bending (see Discussion). This is also consistent with the features with supercoiled oriC plasmid that can promote DUE unwinding (Figure 7A).
Furthermore, we performed similar experiments using ssMR-Left DOR, HU and ATP-DnaA with deletion of N-terminal domains I-II (ATP-DnaA III-IV). Weak interaction between HU and DnaA domains I-II is previously suggested (49). DNase I footprint patterns with ATP-DnaA III-IV in the presence and absence of HU were basically similar to those with ATP-DnaA WT, although HU-dependent protection of IBR was slightly less intense with ATP-DnaA III-V than with ATP-DnaA WT (Supplementary Figure S6I). These results suggest that interaction of HU and DnaA is not essential for, but is slightly stimulatory for specific binding of HU to IBR. Also, these results are consistent with previous reports indicating that DnaA lacking domains I-II is largely active in in vitro DUE unwinding in the presence of HU and that DnaA-HU interaction stimulates stability of DnaA complexes on oriC (28,49).
HU binds to the interspace between TmaDnaA boxes 1 and 2 on Tma-oriC.
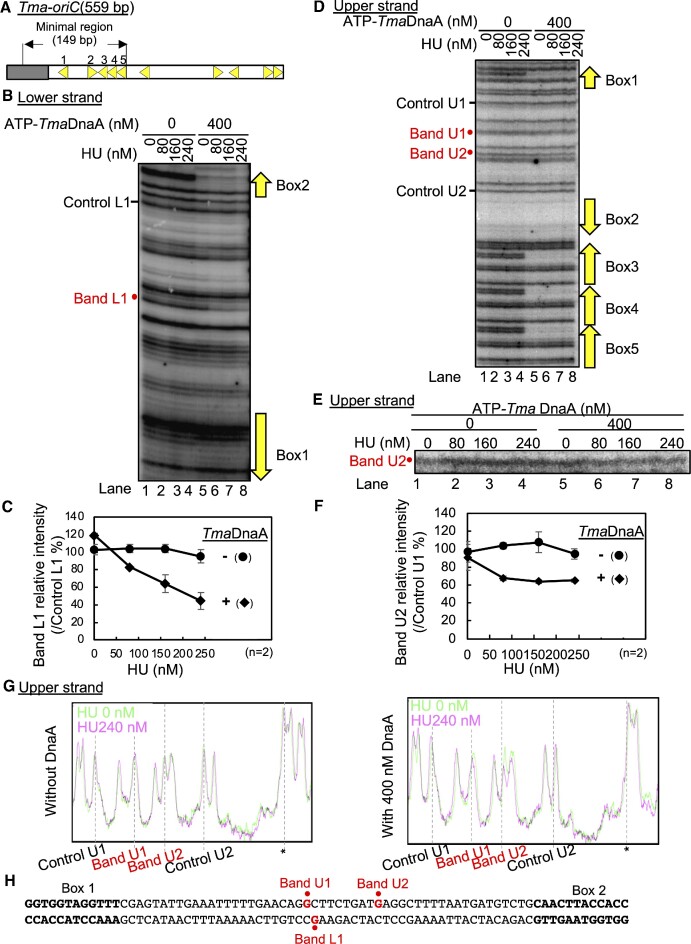
T. maritima is an organism that is evolutionarily distant from E. coli and conserves the cognate HU, but not IHF (Supplementary Figure S1). Our previous study suggests that the ssDUE recruitment mechanism operates even in the origin of this bacterial species (12,21). To gain insight into the evolutionary conservation of HU function in oriC, DnaA-HU-oriC complexes from T. maritima were analyzed by DMS footprint experiments. The 149-bp minimal Tma-oriC includes TmaDnaA boxes 1 to 5 and an AT-rich DUE, which is unwound in vitro by ATP-DnaA of T. maritima (ATP-TmaDnaA) in the presence of E. coli HU, which is homologous to T. maritima HU (Figure 8A) (52). ATP-TmaDnaA complexes constructed on the minimal Tma-oriC are suggested to bind to the cognate ssDUE via the H/B motifs of TmaDnaA (21).
Figure 8.
HU binding to Tma-oriC in the presence of TmaDnaA. (A) Overall structure of Tma-oriC. DUE and DOR of Tma-oriC are indicated by gray and open squares, respectively. TmaDOR carries ten TmaDnaA boxes (yellow arrowheads). The minimal region for DUE unwinding and the names of the TmaDnaA box in this region are shown above the structure. (B–H) DMS footprint using Tma-oriC plasmid. The supercoiled form of the Tma-oriC plasmid pOZ14 (6 nM) was incubated at 48°C for 5 min with the indicated amount of HU in the presence or absence of ATP-TmaDnaA (400 nM), followed by further incubation with 0.25% DMS for 5 min and cleavage by 1 M piperidine. The resultant DNA fragments were analyzed by primer extension assay using the 32P-labeled primer TMA28 for lower strand modification (B and C) and the 32P-labeled primer 306 for upper strand modification (D-G). The primer extension products were resolved on 6% sequencing gels (B and D). Enlarged image of Band U2 is shown in panel E. The relative intensities of Bands L1 and U2 to each control band were quantified as ‘Band L1 relative intensity’ (C) and ‘Band U2 relative intensity’ (F). Quantification of band U1 is shown in Supplementary Figure S7B and C. Results are the means and standard deviations (SD) of two independent experiments. In addition, the histogram of footprint profiles with and without HU are shown in panel G. These histograms were calibrated by a peak indicated by asterisk and overlayed. The sequence from TmaDnaA box1 to box2 is shown in panel H. Specific sites (bands 1 and 2) protected in a manner dependent on both HU and TmaDnaA are indicated in red.
A DMS footprint assay using a supercoiled plasmid containing the minimal Tma-oriC showed that ATP-TmaDnaA bound to TmaDnaA boxes 1 to 5 (Figure 8B for box 2 and Figure 8D boxes 1 and 3–5), consistent with our previous results of DNaseI footprint assays without HU (21). The band annotation for the oriC position is shown in Supplementary Figure S7. Notably, HU protected the specific position located between TmaDnaA boxes 1 and 2 depending on ATP-TmaDnaA (Band L1 in Figures 8B and C, and Supplementary Figure S7D and E). Consistent results were shown by analysis of the complementary strand although changes of band intensity were small (Band U1 and U2 in Figures 8D–G and Supplementary Figure S7A–C and F–I). In contrast, modifications in the box 3–5 region were substantially unaffected by HU (Figure 8B and D). Also, HU alone did not substantially change the footprint profiles (Figure 8B, lanes 1–4, and 8E, lanes 1–4). These results are fully consistent with the idea that HU binds to the interspace between TmaDnaA boxes 1 and 2 only when ATP-TmaDnaA forms a complex with the minimal Tma- oriC.
To determine important TmaDnaA protomers for HU binding, we similarly analyzed Tma-oriC plasmid with substitution of a TmaDnaA box with a sequence defective in specific TmaDnaA binding (Supplementary Figure S7D–I). TmaDnaA binding was detected in the intact boxes, but not in the substituted sites. HU-dependent protection in the interspace between TmaDnaA boxes 1 and 2 was reduced in Tma-oriCs with substitution of TmaDnaA box 1 or box 2, but not substantially in Tma-oriC WT or Tma-oriCs with substitution of TmaDnaA box 3, box 4 or box 5 (Supplementary Figure S7D-I). These results suggest that DnaA protomers bound to box 1 and box 2 are important for HU binding to the interspace. Slight protection in this region remained even in the presence of TmaDnaA box 1 or box 2 substitution might be caused by residual activity of TmaDnaA complex constructed on boxes 2–5 for binding to ssDUE of Tma-oriC (12). These results are consistent with proposed model for specific HU binding in E. coli oriC (see Discussion).
DISCUSSION
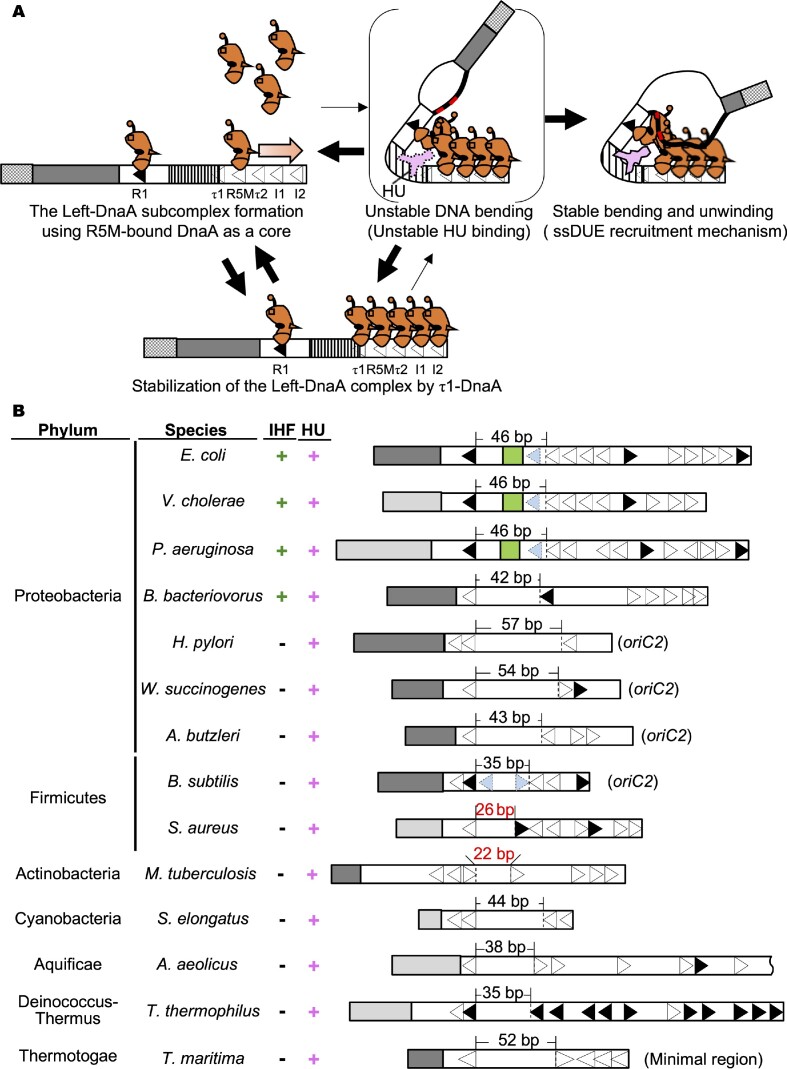
Unlike IHF, HU homologs are evolutionarily widely conserved in eubacterial species. Although both HU and IHF have been reported to stimulate origin unwinding in E. coli, the HU-specific DUE unwinding mechanism has remained as a long-standing mystery. In this study, HU-DnaA-oriC complexes were investigated intensively using in vitro reconstituted systems. Similar to the IHF-promoted unwinding of DUE, the HU-promoted mechanism showed a specific requirement for the Left-DnaA subcomplex, especially DnaA protomers bound to the R1 and R5M boxes. Moreover, the protomer specific analysis using ChiDnaA suggested that R1- and R5M-DnaA protomers directly interacted with ssDUE to promote stable DUE unwinding. These observations were supported by in vivo experiments. Further analysis revealed that HU bound to the interspace between the R1 and R5M boxes (the R1-R5M interspace), depending on ATP-DnaA-specific complex formation, which was effectively stimulated by ssDUE. These mechanisms are consistent with the hypothesis, that ATP-DnaA binding to oriC and interaction between R1- and R5M-DnaA protomers trigger DNA bending in the R1-R5M interspace, stimulating labile interaction of HU to the bending site and initial DUE unwinding, which promotes ssDUE binding to ATP-DnaA subcomplex, resulting in stable DNA bending of the R1-R5M interspace, stable HU binding and stable DUE unwinding (Figure 9A). The initial DUE unwinding would be so labile as not to be detected by P1 nuclease assay. These characteristics of the HU-DnaA-oriC complexes were consistent with those of the IHF-DnaA-oriC complexes and with the finding that the ssDUE recruitment mechanism operates even in the HU system. In addition, even in the HU system, the advantage of the ssDUE recruitment mechanism in DnaB loading would not be changed. A similar mechanism of site-specific HU binding was suggested also in Tma-oriC, supporting the hypothesis that the HU-promoted ssDUE recruitment mechanism is evolutionarily conserved in eubacteria.
Figure 9.
Model of HU-promoted ssDUE recruitment mechanism. (A) Model of HU-promoted ssDUE recruitment mechanism. The left oriC subregion including DUE and Left-DOR and DnaA are shown as in Figure 1. HU is shown in the pink diagram. An increase in ATP-DnaA level during the cell cycle results in its oligomerization on the low-affinity DnaA boxes using the R5M-DnaA protomer as a core (leftmost panel). If DnaA temporarily binds also to the τ1 box, τ1-DnaA moderately stabilizes the R5M-I2 DnaA complex (lower panel). IBR is unstably bent by unstable interaction between R1-DnaA and R5M-DnaA, forming a transient structure with unstable DUE unwinding and unstable HU binding (upper middle panel). The M/R-region bearing TT[A/G]T(T) sequences (red line) of the DUE-upper strand (black bold line) binds to R1-DnaA and R5M-DnaA, stimulating stability of the bent structure of IBR. HU binds specifically to the bent region and stabilizes the overall structure (upper right panel). The resulting ssDUE–ATP-DnaA–DOR–HU complex stabilizes the unwound state of DUE. (B) Eubacterial oriC structures. Experimentally determined DUEs (dark gray) or predicted DUEs (light gray) are shown. DORs are shown by open bars. DnaA box positions are indicated by arrowheads (Black; full consensus for EcoDnaA box, Open; DnaA box with degenerated sequence). For bacteria with bipartite origins, oriC1 and oriC2, which bears the insertion of the dnaA gene (H. pylori, W. succinogenes, A. butzleri, and B. subtilis), only DUE-proximal oriC2s are shown. For A. aeolicus oriC, only DUE-proximal region is shown because of its length. For T. maritima oriC, the minimal region for DUE unwinding is shown. DnaA box τ1 in E. coli and DnaA boxes corresponding to the E. coli τ1 in V. cholerae and P. aeruginosa oriCs are indicated by the pale blue arrowheads. The pale blue arrowheads in B. subtilis oriC2 show DnaA boxes non-essential for initiation. The length of the interspace between DnaA boxes is indicated above the structure (black characters, comparative length for E. coli IBR; red characters, shorter length for E. coli IBR). The presence of IHF and HU, the name of the species, and the phylum are also indicated on the left side of each structure. Abbreviations: E. coli, Escherichia coli K12; V. cholerae, Vibrio cholerae O1biovar eltor str. N16961; P. aeruginosa, Pseudomonas aeruginosa PAO1; B. bacteriovorus, Bdellovibrio bacteriovorus HD100; H. pylori, Helicobacter pylori 26 695; W. succinogenes, Wolinellasuccinogenes DSM 1740; A. butzleri, Arcobacter butzleri RM4018; B. subtilis, Bacillus subtilis QB928; S. aureus, Staphylococcus aureus RF112; M. tuberculosis, Mycobacterium tuberculosis H37Rv; S. elongatus, Synechococcus elongatus PCC 7942; A. aeolicus, Aquifex aeolicus VF5; T. thermophilus, Thermus thermophilus HB8; T. maritima, Thermotoga maritima MSB8.
Some of the mechanisms in the HU system differed from those in the IHF system. First, DMS footprint data revealed that the site-specific HU binding to the R1-R5M interspace depended on ATP-DnaA. By contrast, binding of IHF is sequence-specific and does not depend on ATP-DnaA. Second, in vivo analyses showed more stringent requirements for R1 and R5M boxes in initiating chromosomal replication and in colony formation by IHF-deficient cells. By contrast, colony formation is observed in IHF-wild-type cells bearing R1non or R5Mnon on the chromosome, although initiation of replication is markedly reduced in these cells (24,61,62). The importance of R1- and R5M-DnaA in HU-promoted DUE unwinding is consistent with the essentiality of R1 and R5M boxes in in vitro DUE unwinding in a reconstituted system as well as the idea that DnaA binding-dependent oriC structural changes are a prerequisite for HU binding specificities. In addition, R5M-DnaA, which functions as the core for ATP-DnaA assembly, was found to bind to ssDUE even in the HU system. R1-DnaA was also found to bind to ssDUE in the HU system.
The τ1 box was found to be more important for in vivo initiation of replication in IHF-deficient cells than in IHF-wild-type cells (Supplementary Figure S4). In IHF-wild-type cells, chromosomal replication initiates without substantial inhibition, even when oriC bears τ1non mutations. The in vitro unwinding of oriC in the IHF system is largely sustained even with the τ1non mutation, and DNase I footprint experiments show that IHF outcompeted DnaA for binding to the τ1 box (24). The present study showed that the in vitro unwinding of oriC in the HU system was only moderate in the presence of τ1non (Figure 3), consistent with in vivo observations (Supplementary Figure S4). These findings in addition to results of DNase I footprint experiments suggest that DnaA temporarily binds to the τ1 box, stabilizing the DnaA assembly on the cluster of low-affinity DnaA boxes in the Left-DOR during complex formation. DnaA-DnaA interactions can lead to the formation of a bent DNA in the interspace, such that the interaction of HU with the bent DNA region, would lead to extrusion of τ1 box-bound DnaA (τ1-DnaA), stabilizing HU-DNA binding (Figure 9A).
HU has been hypothesized to alter the overall superhelicity of oriC DNA to stimulate DUE unwinding (53,54). However, the effective amounts per oriC in replication initiation are similar for HU and IHF regardless of sequence-non-specific binding nature of HU (13,54). Moreover, the present study demonstrated similar requirements for the R1-I1 region in DUE unwinding by the both HU and IHF systems. These results suggest that basically similar mechanisms underly DUE unwinding in the two systems rather than the specific possibility that the overall superhelical modulation caused by sequence-non-specific DNA binding of HU stimulates DUE unwinding, although this possibility can not be completely excluded. In addition, although HU has been reported to directly but weakly interact with DnaA domains I-II (49), this interaction is likely non-essential based on the reported feature that DUE unwinding activity is largely sustained, even by domains I-II-deleted DnaA in the HU system (28). Consistently, in this study we showed that stimulation of HU binding to the R1-R5M interspace is largely sustained by DnaA lacking domains I-II (DnaA III-IV) although the stimulation is slightly less than that by WT DnaA (Supplementary Figure S6I). These results are fully consistent with the proposed DUE unwinding mechanism in the HU system. The WT DnaA-dependent slight stimulation might come from enhancement in stability of DnaA complex by HU-DnaA interaction or that in local concentration of HU molecules in the close vicinity of DnaA complexes (49). In live cells, HU-DnaA interaction could enhance recruitment of HU molecules to the vicinity of DnaA-oriC complexes.
Taken together, these findings suggest that E. coli uses two systems to unwind DUE, which are principally similar. IHF can bind to the R1-R5M interspace sequence-specifically prior to ATP-DnaA complex formation within oriC whereas HU binds to it depending on the formation of ATP-DnaA–Left-DOR complexes. Notably, the site-specific binding of IHF to oriC is important to ensure timely initiation during cell cycle in E. coli (63). E. coli may have acquired the IHF system to develop a sophisticated system for regulating the cell cycle-coordinated replication initiation. IHF plays various roles in DnaA regulation, not only by increasing ATP-DnaA levels, depending on DnaA reactivating sequence DARS2, but also by reducing ATP-DnaA levels through datA-dependent ATP-DnaA hydrolysis (56,58,59). DARS2 and datA as well as oriC are chromosomal loci and IHF alternates its binding targets through the cell cycle to initiate DNA replication in a timely manner.
The specific role for HU in oriC could be conserved during eubacterial evolution. In addition to E. coli, site-specific, ATP-TmaDnaA-dependent binding of HU was observed in Tma-oriC. Similar to the mechanisms underlying E. coli oriC dynamics, the mechanisms underlying these Tma-oriC dynamics can be explained by the preferential binding of HU to bent DNA and the specific configuration of the ATP-DnaA complex, consistent with the ssDUE recruitment mechanism. Because T. maritima is one of the most evolutionarily ancestral organisms, the ssDUE recruitment mechanism could be highly conserved throughout the eubacteria. Consistently, the origins of the Vibrio cholerae chromosome 2 (Chr2) and of the chromosome of H. pylori, an ϵ proteobacterium in addition to the origin of RK2 plasmid have been reported to use the same principles of the ssDUE recruitment mechanism (45,46,64). Although HU is not essential in the in vitro unwinding of the H. pylori ori2 DUE (45), various in vitro conditions for unwinding as well as in vivo significance of HU in initiation remain to analyzed. Roles for HU in the origins of V. cholerae Chr2, RK2 plasmid and other systems also remain to be elucidated. In Bacillus subtilis (B. subtilis), Mycobacterium smegmatis and Borrelia burgdorferi, HU is suggested to be involved in the initiation of chromosome replication (65–67).
Some of the oriC structures in eubacterial origins are inferred to be consistent with the HU-promoted ssDUE recruitment mechanism (Figure 9B). A relatively wider interspace between two DnaA boxes within the origin likely plays a key role in specific HU binding. The length for the HU binding region in E. coli oriC is comparable to the 35 bp IBR. Although HU has been reported to occupy a 9-bp sequence (68), the bending of the structure by two nearby DnaA molecules would likely result in the need for a longer sequence: physicochemical study suggests that an HU mode complexed with sharply bent DNA is taken with 34-bp DNA, but not 10-bp DNA (69). Moreover, HU was found to bind to both the interspace between the leftward R1 and R5M boxes in oriC and between the leftward box 1 and rightward box 2 of Tma-oriC, which suggests that the relative orientations of DnaA boxes are less important for HU binding. Based on these features, a review of various eubacterial oriCs suggests potential HU-interactive regions (Figure 9B) (24,52,70–78). An exception may be the case for B. subtilis oriC: a candidate region overlaps DnaA boxes 3 and 4 (Figure 9B). However, like τ1 box of E. coli, these boxes are non-essential for initiation, and have degenerated from the consensus sequence (75,76). Also, S. aureus and M. tuberculosis oriCs have relatively shorter interspaces, of 26- and 22-bp, respectively: DnaA boxes within or flanking these regions could have a feature like DnaA box τ1 or even shorter regions could be sufficient for the cognate HU homolog (Figure 9B). Further detailed investigations are clearly required.
DATA AVAILABILITY
The data supporting the findings of this study are available from the corresponding author upon reasonable request.
Supplementary Material
Notes
Present address: Hironori Kawakami, Laboratory for Systems Immunology, Faculty of Pharmaceutical Sciences, Sanyo‐Onoda City University, Sanyo-Onoda, Yamaguchi 756-0884, Japan.
Contributor Information
Ryusei Yoshida, Department of Molecular Biology, Graduate School of Pharmaceutical Sciences, Kyushu University, 3-1-1 Maidashi, Higashi-ku, Fukuoka 812-8582, Japan.
Shogo Ozaki, Department of Molecular Biology, Graduate School of Pharmaceutical Sciences, Kyushu University, 3-1-1 Maidashi, Higashi-ku, Fukuoka 812-8582, Japan.
Hironori Kawakami, Department of Molecular Biology, Graduate School of Pharmaceutical Sciences, Kyushu University, 3-1-1 Maidashi, Higashi-ku, Fukuoka 812-8582, Japan.
Tsutomu Katayama, Department of Molecular Biology, Graduate School of Pharmaceutical Sciences, Kyushu University, 3-1-1 Maidashi, Higashi-ku, Fukuoka 812-8582, Japan.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Japan Society for the Promotion of Science (JSPS KAKENHI) [JP20H03212, JP17H03656, JP21K19233, JP23H02438]; JSPS pre-doctoral fellowship [JP22J11077 to R.Y.]. Funding for open access charge: Japan Society for the Promotion of Science.
Conflict of interest statement. None declared.
REFERENCES
- 1. Grimwade J.E., Leonard A.C.. Blocking, bending, and binding: regulation of initiation of chromosome replication during the Escherichia coli cell cycle by transcriptional modulators that interact with origin DNA. Front. Microbiol. 2021; 12:732270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Costa A., Hood I.V., Berger J.M.. Mechanisms for initiating cellular DNA replication. Annu. Rev. Biochem. 2013; 82:25–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Katayama T., Kasho K., Kawakami H.. The DnaA cycle in Escherichia coli: activation, function and inactivation of the initiator protein. Front. Microbiol. 2017; 8:2496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wolanski M., Donczew R., Zawilak-Pawlik A., Zakrzewska-Czerwinska J.. oriC-encoded instructions for the initiation of bacterial chromosome replication. Front. Microbiol. 2015; 6:735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. O’Donnell M., Langston L., Stillman B.. Principles and concepts of DNA replication in bacteria, archaea, and eukarya. Cold Spring Harb. Perspect. Biol. 2013; 5:a010108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Arias-Palomo E., Puri N., O’Shea Murray V.L., Yan Q., Berger J.M. Physical basis for the loading of a bacterial replicative helicase onto DNA. Mol. Cell. 2019; 74:173–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sakiyama Y., Nishimura M., Hayashi C., Akama Y., Ozaki S., Katayama T.. The DnaA AAA+ domain His136 residue directs DnaB replicative helicase to the unwound region of the replication origin, oriC. Front. Microbiol. 2018; 9:2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hayashi C., Miyazaki E., Ozaki S., Abe Y., Katayama T.. DnaB helicase is recruited to the replication initiation complex via binding of DnaA domain I to the lateral surface of the DnaB N-terminal domain. J. Biol. Chem. 2020; 295:1131–1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Felczak M.M., Chodavarapu S., Kaguni J.M.. DnaC, the indispensable companion of DnaB helicase, controls the accessibility of DnaB helicase by primase. J. Biol. Chem. 2017; 292:20871–20882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bell S.P., Kaguni J.M.. Helicase loading at chromosomal origins of replication. Cold Spring Harb. Perspect. Biol. 2013; 5:a010124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bramhill D., Kornberg A.. Duplex opening by dnaA protein at novel sequences in initiation of replication at the origin of the E. coli chromosome. Cell. 1988; 52:743–755. [DOI] [PubMed] [Google Scholar]
- 12. Ozaki S., Kawakami H., Nakamura K., Fujikawa N., Kagawa W., Park S.Y., Yokoyama S., Kurumizaka H., Katayama T.. A common mechanism for the ATP-DnaA-dependent formation of open complexes at the replication origin. J. Biol. Chem. 2008; 283:8351–8362. [DOI] [PubMed] [Google Scholar]
- 13. Hwangs D.S., Kornbergs A.. Opening of the replication origin of Escherichia coli by DnaA protein with protein HU or IHF. J. Biol. Chem. 1992; 267:23083–23086. [PubMed] [Google Scholar]
- 14. Sakiyama Y., Nagata M., Yoshida R., Kasho K., Ozaki S., Katayama T.. Concerted actions of DnaA complexes with DNA-unwinding sequences within and flanking replication origin oriC promote DnaB helicase loading. J. Biol. Chem. 2022; 298:102051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Fang L., Davey M.J., O’Donnell M. Replisome assembly at oriC, the replication origin of E. coli, reveals an explanation for initiation sites outside an origin. Mol. Cell. 1999; 4:541–553. [DOI] [PubMed] [Google Scholar]
- 16. Schaper S., Messer W.. Interaction of the initiator protein DnaA of Escherichia coli with its DNA target. J. Biol. Chem. 1995; 270:17622–17626. [DOI] [PubMed] [Google Scholar]
- 17. Grimwade J.E., Ryan V.T., Leonard A.C.. IHF redistributes bound initiator protein, DnaA, on supercoiled oriC of Escherichia coli. Mol. Microbiol. 2000; 35:835–844. [DOI] [PubMed] [Google Scholar]
- 18. Kawakami H., Keyamura K., Katayama T.. Formation of an ATP-DnaA-specific initiation complex requires DnaA arginine 285, a conserved motif in the AAA+ protein family. J. Biol. Chem. 2005; 280:27420–27430. [DOI] [PubMed] [Google Scholar]
- 19. Ozaki S., Noguchi Y., Hayashi Y., Miyazaki E., Katayama T.. Differentiation of the DnaA-oriC subcomplex for DNA unwinding in a replication initiation complex. J. Biol. Chem. 2012; 287:37458–37471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Rozgaja T.A., Grimwade J.E., Iqbal M., Czerwonka C., Vora M., Leonard A.C.. Two oppositely oriented arrays of low-affinity recognition sites in oriC guide progressive binding of DnaA during Escherichia coli pre-RC assembly. Mol. Microbiol. 2011; 82:475–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ozaki S., Katayama T.. Highly organized DnaA-oriC complexes recruit the single-stranded DNA for replication initiation. Nucleic Acids Res. 2012; 40:1648–1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Noguchi Y., Sakiyama Y., Kawakami H., Katayama T.. The Arg fingers of key DnaA protomers are oriented inward within the replication origin oriC and stimulate DnaA subcomplexes in the initiation complex. J. Biol. Chem. 2015; 290:20295–20312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Shimizu M., Noguchi Y., Sakiyama Y., Kawakami H., Katayama T., Takada S.. Near-atomic structural model for bacterial DNA replication initiation complex and its functional insights. Proc. Natl. Acad. Sci. U.S.A. 2016; 113:E8021–E8030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sakiyama Y., Kasho K., Noguchi Y., Kawakami H., Katayama T.. Regulatory dynamics in the ternary DnaA complex for initiation of chromosomal replication in Escherichia coli. Nucleic Acids Res. 2017; 45:12354–12373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ryan V.T., Grimwade J.E., Nievera C.J., Leonard A.C.. IHF and HU stimulate assembly of pre-replication complexes at Escherichia coli oriC by two different mechanisms. Mol. Microbiol. 2002; 46:113–124. [DOI] [PubMed] [Google Scholar]
- 26. Ozaki S., Katayama T.. DnaA structure, function, and dynamics in the initiation at the chromosomal origin. Plasmid. 2009; 62:71–82. [DOI] [PubMed] [Google Scholar]
- 27. Chodavarapu S., Kaguni J.M.. Replication initiation in bacteria. Enzymes. 2016; 39:1–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sutton M.D., Carr K.M., Vicente M., Kaguni J.M.. Escherichia coli DnaA protein. The N-terminal domain and loading of DnaB helicase at the E. coli chromosomal origin. J. Biol. Chem. 1998; 273:34255–34262. [DOI] [PubMed] [Google Scholar]
- 29. Abe Y., Jo T., Matsuda Y., Matsunaga C., Katayama T., Ueda T.. Structure and function of DnaA N-terminal domains: specific sites and mechanisms in inter-DnaA interaction and in DnaB helicase loading on oriC. J. Biol. Chem. 2007; 282:17816–17827. [DOI] [PubMed] [Google Scholar]
- 30. Keyamura K., Abe Y., Higashi M., Ueda T., Katayama T.. DiaA dynamics are coupled with changes in initial origin complexes leading to helicase loading. J. Biol. Chem. 2009; 284:25038–25050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Keyamura K., Fujikawa N., Ishida T., Ozaki S., Su’etsugu M., Fujimitsu K., Kagawa W., Yokoyama S., Kurumizaka H., Katayama T.. The interaction of DiaA and DnaA regulates the replication cycle in E. coli by directly promoting ATP-DnaA-specific initiation complexes. Genes Dev. 2007; 21:2083–2099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Felczak M.M., Simmonst L.A., Kaguni J.M.. An essential tryptophan of Escherichia coli DnaA protein functions in oligomerization at the E. coli replication origin. J. Biol. Chem. 2005; 280:24627–24633. [DOI] [PubMed] [Google Scholar]
- 33. Nozaki S., Ogawa T.. Determination of the minimum domain II size of Escherichia coli DnaA protein essential for cell viability. Microbiology. 2008; 154:3379–3384. [DOI] [PubMed] [Google Scholar]
- 34. Kaguni J.M. Replication initiation at the Escherichia coli chromosomal origin. Curr. Opin. Chem. Biol. 2011; 15:606–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Erzberger J.P., Mott M.L., Berger J.M.. Structural basis for ATP-dependent DnaA assembly and replication-origin remodeling. Nat. Struct. Mol. Biol. 2006; 13:676–683. [DOI] [PubMed] [Google Scholar]
- 36. Felczak M.M., Kaguni J.M.. The box VII motif of Escherichia coli DnaA protein is required for DnaA oligomerization at the E. coli replication origin. J. Biol. Chem. 2004; 279:51156–51162. [DOI] [PubMed] [Google Scholar]
- 37. Duderstadt K.E., Chuang K., Berger J.M.. DNA stretching by bacterial initiators promotes replication origin opening. Nature. 2011; 478:209–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Fujikawa N., Kurumizaka H., Nureki O., Terada T., Shirouzu M., Katayama T., Yokoyama S.. Structural basis of replication origin recognition by the DnaA protein. Nucleic Acids Res. 2003; 31:2077–2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Swinger K.K., Rice P.A.. IHF and HU: flexible architects of bent DNA. Curr. Opin. Struct. Biol. 2004; 14:28–35. [DOI] [PubMed] [Google Scholar]
- 40. Dillon S.C., Dorman C.J.. Bacterial nucleoid-associated proteins, nucleoid structure and gene expression. Nat. Rev. Microbiol. 2010; 8:185–195. [DOI] [PubMed] [Google Scholar]
- 41. Kamashev D., Agapova Y., Rastorguev S., Talyzina A.A., Boyko K.M., Korzhenevskiy D.A., Vlaskina A., Vasilov R., Timofeev V.I., Rakitina T.V.. Comparison of histone-like HU protein DNA-binding properties and HU/IHF protein sequence alignment. PLoS One. 2017; 12:e0188037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hales L.M., Gumport R.I., Gardner J.F. Determining the DNA sequence elements required for binding integration host factor to two different target sites. J. Bacteriol. 1994; 176:2999–3006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Rice P.A., Yang S.W., Mizuuchi K., Nash H.A.. Crystal structure of an IHF-DNA complex: a protein-induced DNA U-turn. Cell. 1996; 87:1295–1306. [DOI] [PubMed] [Google Scholar]
- 44. Kasho K., Oshima T., Chumsakul O., Nakamura K., Fukamachi K., Katayama T.. Whole-genome analysis reveals that the nucleoid protein IHF predominantly Binds to the replication origin oriC specifically at the time of initiation. Front. Microbiol. 2021; 12:697712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Jaworski P., Zyla-uklejewicz D., Nowaczyk-cieszewska M., Donczew R., Mielke T., Weigel C., Zawilak-Pawlik A.. Putative cooperative ATP–DnaA binding to double-stranded DnaA box and single-stranded DnaA-trio motif upon Helicobacter pylori replication initiation complex assembly. Int. J. Mol. Sci. 2021; 22:6643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Chatterjee S., Jha J.K., Ciaccia P., Venkova T., Chattoraj D.K.. Interactions of replication initiator RctB with single-and double-stranded DNA in origin opening of Vibrio cholerae chromosome 2. Nucleic Acids Res. 2020; 48:11016–11029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Letunic I., Bork P.. Interactive tree of life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021; 49:W293–W296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Bonnefoy E., Takahashi M., Yaniv J.R.. DNA-binding parameters of the HU protein of Escherichia coli to cruciform DNA. J. Mol. Biol. 1994; 242:116–129. [DOI] [PubMed] [Google Scholar]
- 49. Chodavarapu S., Felczak M.M., Yaniv J.R., Kaguni J.M.. Escherichia coli DnaA interacts with HU in initiation at the E. coli replication origin. Mol. Microbiol. 2008; 67:781–792. [DOI] [PubMed] [Google Scholar]
- 50. Kano Y., Imamoto F.. Requirement of integration host factor (IHF) for growth of Escherichia coli deficient in HU protein. Gene. 1990; 89:133–137. [DOI] [PubMed] [Google Scholar]
- 51. Von Freiesleben U., Rasmussen K.V., Atlung T., Hansen F.G.. Rifampicin-resistant initiation of chromosome replication from oriC in ihf mutants. Mol. Microbiol. 2000; 37:1087–1093. [DOI] [PubMed] [Google Scholar]
- 52. Ozaki S., Fujimitsu K., Kurumizaka H., Katayama T.. The DnaA homolog of the hyperthermophilic eubacterium Thermotoga maritima an open complex with a minimal 149-bp origin region in an ATP-dependent manner. Genes Cells. 2006; 11:425–438. [DOI] [PubMed] [Google Scholar]
- 53. Baker T.A., Kornberg A.. Transcriptional activation of initiation of replication from the E. coli chromosomal origin: an RNA-DNA hybrid near oriC. Cell. 1988; 55:113–123. [DOI] [PubMed] [Google Scholar]
- 54. Skarstad K., Baker T.A., Kornberg A.. Strand separation required for initiation of replication at the chromosomal origin of E. coli is facilitated by a distant RNA-DNA hybrid. EMBO J. 1990; 9:2341–2348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Dixon N.E., Kornberg A.. Protein HU in the enzymatic replication of the chromosomal region of Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 1984; 81:424–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Kasho K., Fujimitsu K., Matoba T., Oshima T., Katayama T.. Timely binding of IHF and Fis to DARS2 regulates ATP-DnaA production and replication initiation. Nucleic Acids Res. 2014; 42:13134–13149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Su’etsugu M., Kawakami H., Kurokawa K., Kubota T., Takata M., Katayama T.. DNA replication-coupled inactivation of DnaA protein in vitro: a role for DnaA arginine-334 of the AAA+ Box VIII motif in ATP hydrolysis. Mol. Microbiol. 2001; 40:376–386. [DOI] [PubMed] [Google Scholar]
- 58. Sugiyama R., Kasho K., Miyoshi K., Ozaki S., Kagawa W., Kurumizaka H., Katayama T.. A novel mode of DnaA-DnaA interaction promotes ADP dissociation for reactivation of replication initiation activity. Nucleic Acids Res. 2019; 47:11209–11224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Kasho K., Katayama T.. DnaA binding locus datA promotes DnaA-ATP hydrolysis to enable cell cycle-coordinated replication initiation. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:936–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Kamashev D., Balandina A., Rouviere-Yaniv J.. The binding motif recognized by HU on both nicked and cruciform DNA. EMBO J. 1999; 18:5434–5444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Langer U., Richter S., Roth A., Weigel C., Messer W.. A comprehensive set of DnaA-box mutations in the replication origin, oriC, of Escherichia coli. Mol. Microbiol. 1996; 21:301–311. [DOI] [PubMed] [Google Scholar]
- 62. Kaur G., Vora M.P., Czerwonka C.A., Rozgaja T.A., Grimwade J.E., Leonard A.C.. Building the bacterial orisome: high-affinity DnaA recognition plays a role in setting the conformation of oriC DNA. Mol. Microbiol. 2014; 91:1148–1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Weigel C., Messer W., Preiss S., Welzeck M., Morigen. The sequence requirements for a functional Escherichia coli replication origin are different for the chromosome and a minichromosome. Mol. Microbiol. 2001; 40:498–507. [DOI] [PubMed] [Google Scholar]
- 64. Wegrzyn K., Fuentes-Perez M.E., Bury K., Rajewska M., Moreno-Herrero F., Konieczny I.. Sequence-specific interactions of Rep proteins with ssDNA in the AT-rich region of the plasmid replication origin. Nucleic Acids Res. 2014; 42:7807–7818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Karaboja X., Wang X.. HBsu is required for the initiation of DNA replication in Bacillus subtilis. J. Bacteriol. 2022; 204:e0011922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Holówka J., Trojanowski D., Janczak M., Jakimowicz D., Zakrzewska-Czerwińska J.. The origin of chromosomal replication is asymmetrically positioned on the mycobacterial nucleoid, and the timing of its firing depends on HupB. J. Bacteriol. 2018; 200:e00044-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Kobryn K., Naigamwalla D.Z., Chaconas G.. Site-specific DNA binding and bending by the Borrelia burgdorferi Hbb protein. Mol. Microbiol. 2000; 37:145–155. [DOI] [PubMed] [Google Scholar]
- 68. Bonnefoy E., Rouviere-Yaniv J.. HU and IHF, two homologous histone-like proteins of Escherichia coli, form different protein - DNA complexes with short DNA fragment. EMBO J. 1991; 10:687–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Koh J., Shkel I., Saecker R.M., Record M.T.. Nonspecific DNA binding and bending by HUαβ: interfaces of the three binding modes characterized by salt-dependent thermodynamics. J. Mol. Biol. 2011; 410:241–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Donczew R., Mielke T., Jaworski P., Zakrzewska-Czerwińska J., Zawilak-Pawlik A.. Assembly of Helicobacter pylori initiation complex is determined by sequence-specific and topology-sensitive DnaA-oriC interactions. J. Mol. Biol. 2014; 426:2769–2782. [DOI] [PubMed] [Google Scholar]
- 71. Kumar S., Farhana A., Hasnain S.E.. In-vitro helix opening of M. tuberculosis oriC by DnaA occurs at precise location and is inhibited by IciA like protein. PLoS One. 2009; 4:e4139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Watanabe S., Ohbayashi R., Shiwa Y., Noda A., Kanesaki Y., Chibazakura T., Yoshikawa H.. Light-dependent and asynchronous replication of cyanobacterial multi-copy chromosomes. Mol. Microbiol. 2012; 83:856–865. [DOI] [PubMed] [Google Scholar]
- 73. Schaper S., Nardmann J., Lüder G., Lurz R., Speck C., Messer W.. Identification of the chromosomal replication origin from Thermus thermophilus and its interaction with the replication initiator DnaA. J. Mol. Biol. 2000; 299:655–665. [DOI] [PubMed] [Google Scholar]
- 74. Yee T.W., Smith D.W.. Pseudomonas chromosomal replication origins: a bacterial class distinct from Escherichia coli-type origins. Proc. Natl. Acad. Sci. U.S.A. 1990; 87:1278–1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Richardson T.T., Harran O., Murray H.. The bacterial DnaA-Trio replication origin element specifies single-stranded DNA initiator binding. Nature. 2016; 534:412–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Richardson T.T., Stevens D., Pelliciari S., Harran O., Sperlea T., Murray H.. Identification of a basal system for unwinding a bacterial chromosome origin. EMBO J. 2019; 38:e101649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Jaworski P., Donczew R., Mielke T., Thiel M., Oldziej S., Weigel C., Zawilak-Pawlik A.. Unique and universal features of epsilonproteobacterial origins of chromosome replication and DnaA-DnaA box interactions. Front. Microbiol. 2016; 7:1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Makowski Ł., Donczew R., Weigel C., Zawilak-Pawlik A., Zakrzewska-Czerwinska J.. Initiation of chromosomal replication in predatory bacterium Bdellovibrio bacteriovorus. Front. Microbiol. 2016; 7:1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data supporting the findings of this study are available from the corresponding author upon reasonable request.