Abstract
Objectives
T2-weighted (w) fat sat (fs) sequences, which are important in spine MRI, require a significant amount of scan time. Generative adversarial networks (GANs) can generate synthetic T2-w fs images. We evaluated the potential of synthetic T2-w fs images by comparing them to their true counterpart regarding image and fat saturation quality, and diagnostic agreement in a heterogenous, multicenter dataset.
Methods
A GAN was used to synthesize T2-w fs from T1- and non-fs T2-w. The training dataset comprised scans of 73 patients from two scanners, and the test dataset, scans of 101 patients from 38 multicenter scanners. Apparent signal- and contrast-to-noise ratios (aSNR/aCNR) were measured in true and synthetic T2-w fs. Two neuroradiologists graded image (5-point scale) and fat saturation quality (3-point scale). To evaluate whether the T2-w fs images are indistinguishable, a Turing test was performed by eleven neuroradiologists. Six pathologies were graded on the synthetic protocol (with synthetic T2-w fs) and the original protocol (with true T2-w fs) by the two neuroradiologists.
Results
aSNR and aCNR were not significantly different between the synthetic and true T2-w fs images. Subjective image quality was graded higher for synthetic T2-w fs (p = 0.023). In the Turing test, synthetic and true T2-w fs could not be distinguished from each other. The intermethod agreement between synthetic and original protocol ranged from substantial to almost perfect agreement for the evaluated pathologies.
Discussion
The synthetic T2-w fs might replace a physical T2-w fs. Our approach validated on a challenging, multicenter dataset is highly generalizable and allows for shorter scan protocols.
Key Points
• Generative adversarial networks can be used to generate synthetic T2-weighted fat sat images from T1- and non-fat sat T2-weighted images of the spine.
• The synthetic T2-weighted fat sat images might replace a physically acquired T2-weighted fat sat showing a better image quality and excellent diagnostic agreement with the true T2-weighted fat images.
• The present approach validated on a challenging, multicenter dataset is highly generalizable and allows for significantly shorter scan protocols.
Supplementary Information
The online version contains supplementary material available at 10.1007/s00330-023-09512-4.
Keywords: Magnetic resonance imaging, Spine, Artificial intelligence
Introduction
Magnetic resonance imagining (MRI) plays an outstanding role in the assessment of spine pathologies due to its high soft tissue contrast, its non-invasiveness, the lack of radiation exposure, and the possibility for a multiparametric image acquisition [1, 2].
Routinely, sagittal T1-weighted (w) sequences (− / + contrast agent) and T2-w sequences are acquired [2]. Additionally, sagittal T2-w sequences combined with fat suppression or separation techniques have become an important part of spine imaging [2]. The removal of the contribution of the fat signal to the overall MR signal enhances contrast resolution, improves assessment of pathologies characterized by changes of the fluid concentration, reduces artifacts, and facilitates the decision of whether additional contrast agent is needed [2–14]. Particularly for the diagnosis of acute pathologies such as inflammation or acute vertebral fractures, T2-w fs images are essential [15].
However, acquiring an additional T2-w fat sat (fs) sequence requires longer scan protocols, which decreases the MR throughput [16]. Prolonged acquisition times reduce patient comfort which could contribute to motion artifacts in imaging data. Additionally, spectral fat saturation techniques are particularly prone to artifacts caused by field inhomogeneities, e.g., around metal implants [4].
Parallel to advancement of MRI acceleration techniques [17, 18], recently, virtually generated MR images offer a promising approach for scan time reduction, as the physical acquisition of particular sequences is no longer necessary. Generative adversarial networks (GANs) based on a deep-learning (DL) architecture can be used to generate such synthetic images from different MR contrasts as input [19–23]. The iterative interaction of two networks, one generating images and one learning to differentiate between synthetic and true images [24, 25], has already been used on MRI data from a variety of anatomical regions [26–28]. In the spine, GANs can generate T2-fs images from conventional T1-w and non-fs T2-w images [15, 29]. Thereby, apart from scan time acceleration, the synthetic T2-w fs images might be less prone to artifacts, as the synthetic images are based on technically stable T1-w and non-fs T2-w images as input.
To foster a widespread implementation of GAN-based T2-w fs images in research and clinical spine imaging, synthetic images need to pass a validation by radiologists’ perception and the GAN framework has to prove external validity.
Hence, our work aims to investigate the diagnostic performance of a sagittal, GAN-based T2-w fs of the spine generated from heterogenous, multicenter T1-w and T2-w images. We hypothesized that synthetic T2-w fs images represent a good alternative to true T2-w fs images consequently allowing shorter scan protocols. Therefore, synthetic T2-w fs images were compared to their true counterparts regarding (1) image quality (quantitatively, qualitatively and with a visual Turing test) and fs quality (qualitatively) and (2) diagnostic agreement (qualitatively).
Methods
Magnetic resonance imaging data
Subject population
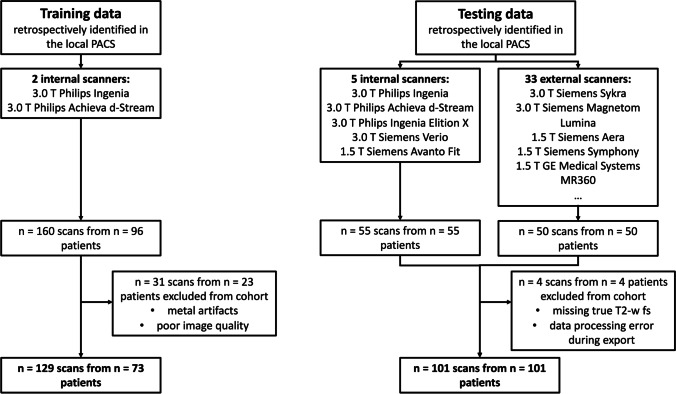
We retrospectively identified 201 patients with sagittal T1-w turbo spin echo (TSE), T2-w TSE, and T2-w TSE fs images of the spine. The study design was approved by the local ethics commission. Informed consent was waived due to the retrospective character.
Training data
Training data for the GAN was retrospectively retrieved from 160 sagittal T1-w, T2-w, and T2-w fs spine images of 96 patients. Due to metal artifacts or poor image quality, 31 scans were excluded (only in the training data) (Fig. 1). All scans originated from two in-house 3 T scanners (Ingenia and Achieva d-stream, Philips Healthcare) using a similar protocol. Sequence parameters are given in Table SM1.
Fig. 1.
Flow chart describing inclusion and exclusion criteria of training and testing data
Testing data
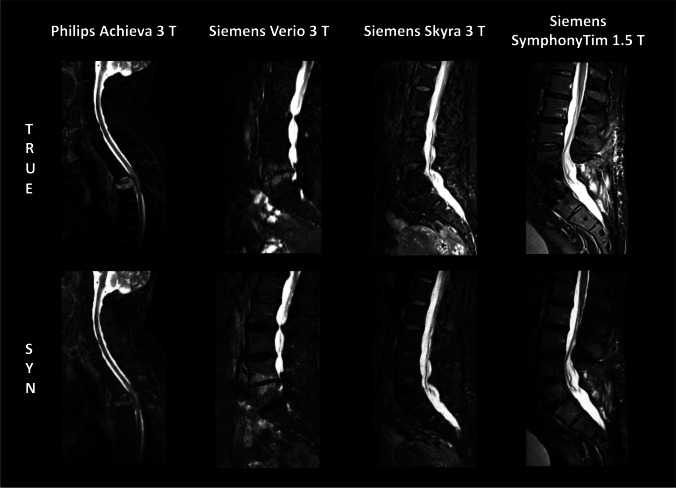
We retrospectively identified 105 MRI datasets of 105 patients consisting of sagittal T1-w, T2-w, and T2-w fs scans. Starting with date 2020/10/01 and going backward, all subsequent spine scans uploaded to the PACS were included up to a number of 105 datasets. Thereby, in-house scans (n = 55) and scans from other institutions (n = 50) being imported for clinical review were included. Four datasets were excluded due to missing true T2-w fs images or data processing errors during export (Fig. 1). Notably, artifacts, e.g., due to foreign material or poor image quality, did not represent an exclusion criterion to assess the performance of the GAN also in these challenging situations. The remaining 101 datasets originated from n = 38 scanners from three vendors (Philips Healthcare; Siemens Healthineers; GE Healthcare). Figure 2 shows images of true and synthetic T2-w fs from different scanner hardware. n = 41 datasets were acquired at 1.5 T, n = 60 datasets at 3 T. Slice thickness ranked from 2.2 to 5.5 mm; field of view (FOV) x/y/z dimensions ranked from 48/200/30 mm to 420/420/420 mm. The mean/range of sequence parameters is given in Table SM1.
Fig. 2.
Exemplary images of true and synthetic T2-w fs from different scanner hardware
In order to account for data origin bias, testing data originating from the two 3 T scanners, which were also used in the training phase (Ingenia and Achieva d-Stream, Philips Healthcare), was excluded in an additional analysis resulting in n = 66 remaining datasets. Respective results are provided in the supplementary material.
Synthesis of sagittal T2-w fs images
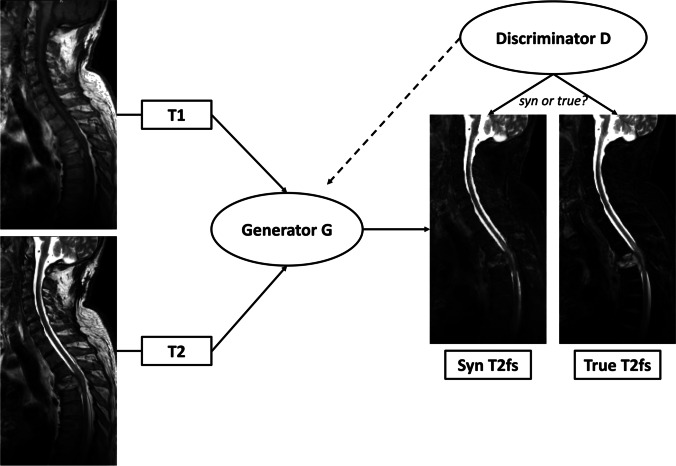
The GAN for synthesis of sagittal T2-w fs images from T1-w and non-fs T2-w images is based on the pix2pix architecture by Isola et al. [30] (details are given in SM Appendix 1). The artificial generation of one T2-w fs dataset takes on average less than 5 min depending on the computational power. Most of this time is needed for image registration; the image synthesis by the GAN takes less than 30 s. A schematic diagram with exemplary images of the GAN architecture and the training process of image synthesis is shown in Fig. 3. The GAN model and one test case can be found in the following repository: https://doi.org/10.6084/m9.figshare.16627576
Fig. 3.
Diagram of architecture and training process of the synthesis task. The Generator G uses T1- and T2-w images to generate synthetic T2-w fs images. Feedback on the similarity between synthetic T2-w fs and true T2-w fs is offered by the Discriminator D and causes modifications in network weightings until the loss of function to discriminate between both images is minimal
Evaluation of GAN performance
Objective image quality evaluation
One neuroradiologist with six years of experience in spine imaging performed apparent signal- and contrast-to-noise ratio (aSNR/aCNR) measurements comparable to the work by Pennig et al. [31] in ten representative datasets of corresponding synthetic and true T2-w fs images (including internal and external data). A region of interest (ROI) was manually drawn in the same position on synthetic and true T2-w fs images in (i) a healthy-appearing vertebral body and (ii) a region of bone marrow abnormality. Additionally, a ROI was placed in the paraspinal muscles as a reference standard for background noise, assuming relatively homogenous muscle tissue and therefore relating signal standard deviation mainly to noise. The aSNR and aCNR were calculated as follows:
| 1 |
| 2 |
where SI is the signal intensity, and SD is the standard deviation. For each dataset, aSNR and aCNR were calculated.
Subjective image and fat saturation quality evaluation
The 101 test datasets (T1-w, T2-w, synthetic T2-w fs, and true T2-w fs images) were investigated by two neuroradiologists (reader 1 with six years of experience; reader 2 with three years of experience in spine imaging). The expert readers blindly graded synthetic and true T2-w fs images regrading image quality based on a 5-point scale [16] and fat saturation quality based on a 3-point scale by assessing presence of artifacts, overall SNR, and image contrast (Table 1 (a)).
Table 1.
Grading scores for image and fat saturation quality (a) and for the six different spine pathologies (b)
| (a) | Grade | |||||
| 1 | 2 | 3 | 4 | 5 | ||
| Image quality | Poor | Marginal | Acceptable with moderate artifacts | Good with some artifacts | Excellent with minimal/no artifacts | |
| Fat suppression/separation | Weak | Medium | Good | – | – | |
| (b) Pathologies | Grade | |||||
| 0 | 1 | 2 | 3 | 4 | ||
| Bone marrow abnormalities | Absent | Focal | One-third of vertebral body | Two-thirds of vertebral body | Whole vertebral body or affection of pedicles/proc. spinosus | |
| Spondylodiscitis expansion | Absent | One-third of vertebral body | Two-thirds of vertebral body | Whole vertebral body | – | |
| Juxtadiscal Modic changes (inflammatory) | Absent | Present | – | – | – | |
| Vertebral fractures | Absent | Acute (edema present) | Chronic | – | – | |
| Cord lesions | Absent | Present | – | – | – | |
| Paravertebral tissue abnormalities | Absent | Inflammation | Hematoma | Other | – | |
To assess whether synthetic and true T2-w fs images are indistinguishable, a visual Turing test was performed. From the testing dataset 25 synthetic and 25 true T2-w fs images of the same patient, respectively, were presented randomized and blinded to eleven neuroradiologists (one to 20 years of experience in spine MRI) using a website-based graphical user interface (GUI) [32, 33]. Participants were obliged to classify the shown image as a synthetic or a true T2-w fs. Without learning whether the classification was correct or wrong, the subsequent image was presented.
Evaluation of diagnostic agreement
In each of the 101 test datasets, five consecutive vertebral segments were defined as ROI based on T1-w, T2-w, and true T2-w fs images. Thereby, throughout all datasets cervical, thoracic and lumbar spine segments were included. Subsequently, the two aforementioned expert readers assessed diagnostic agreement of the images by grading six different pathologies in the ROI: bone marrow abnormalities, spondylodiscitis expansion, Modic changes, vertebral fractures, spinal cord lesions, and paravertebral tissue abnormalities. The six named pathologies were chosen, as they are among the most common spinal pathologies. Particularly for these six pathologies, a sufficient fluid contrast is important for assessment and, therefore, the analysis of T2-w fs images is of significant diagnostic relevance. Grading scores are given in Table 1 (b). The two readers independently graded pathologies on the synthetic (T1-w, T2-w, and synthetic T2-w fs images) and the original protocol (T1-w, T2-w, and true T2-w fs images) in a randomized and blinded assessment.
Gold standard definition for accuracy
After completion of the blinded expert readings, a ground truth (GT) grading of the 101 test datasets was defined. T1-w, T2-w, and true T2-w fs images were assessed in a consensus grading of both expert readers, additionally incorporating the information of pre- or follow-up scans, other imaging modalities, and clinical information.
Statistical analysis
Statistical analysis was performed with SPSS (version 27.0, IBM SPSS Statistics for MacOS, IBM Corp.) and Microsoft Excel (2021). A p-value of 0.05 was set as threshold for statistical significance.
Significant difference between aSNR and aCNR of synthetic and true T2-w fs images from the ten representative datasets was evaluated using the Wilcoxon signed-rank test.
Image and fat saturation quality grading of synthetic and true T2-w fs was analyzed using descriptive statistics. Significant differences between image and fat saturation quality grading of synthetic and true T2-w fs were evaluated using the Wilcoxon signed-rank test.
The Turing test was analyzed using descriptive statistics. Significant difference real condition versus expert grading between true and synthetic T2-w fs images was evaluated using McNemar’s test.
To evaluate the intermethod agreement of pathology assessment based on the synthetic versus the original protocol, Cohen’s kappa (ĸ) coefficients were calculated [34]. Also, the interrater agreement for pathology grading was calculated using Cohen’s ĸ coefficients. Significant differences between Cohen’s ĸ coefficients were evaluated using the Wilcoxon signed-rank test.
For comparison with the gold standard, accuracy of grading was calculated and corresponding significance was evaluated using a McNemar’s test.
Results
Image and fat saturation quality of synthetic versus true T2-w fs
aSNR and aCNR values for synthetic and true T2-w fs images of ten representative datasets were not significantly different (p > 0.05). The detailed results are provided in Table SM2a. For a comparison of objective and subjective image quality measures, Table SM2b provides corresponding image-quality grades of both expert readers for synthetic and true T2-w fs images, respectively.
The image quality of the synthetic T2-w fs was graded higher than that of the true T2-w fs by both readers (97.0% of synthetic T2-w fs images versus 87.6% of true T2-w fs images graded at least acceptable) (Table 2 (a)). The difference in image quality grading was statistically significant (p = 0.023). Quality of fat saturation grading was not significantly different between synthetic T2-w fs and true T2-w fs, with 84.7% of synthetic T2-w fs images and 81.7% of true T2-w fs images graded as good fat saturation (p > 0.05) (Table 2 (b)).
Table 2.
Cross table image (a) and fat saturation (b) quality grading synthetic versus true T2-w fs for both readers. 1 indicates worst quality. In (a) significantly more cases favor synthetic images (bold italic, n = 67), than true T2-w fs images (italic; n = 49; p = 0.023). n = 86 cases in which image quality gradings of synthetic and true T2-w fs correspond
| (a) Image quality | Synthetic T2-w fs | |||||
| True T2-w fs | 1 (poor) | 2 | 3 | 4 | 5 (excellent) | Total |
| 1 (poor) | 0 | 0 | 0 | 1 | 1 | 2 |
| 2 | 0 | 2 | 6 | 12 | 3 | 23 |
| 3 | 0 | 2 | 18 | 18 | 9 | 47 |
| 4 | 0 | 2 | 13 | 16 | 17 | 48 |
| 5 (excellent) | 0 | 0 | 10 | 22 | 50 | 82 |
| Total | 0 | 6 | 47 | 69 | 80 | 202 |
| (b) Fat saturation quality | Synthetic T2-w fs | |||||
| True T2-w fs | 1 (weak) | 2 | 3 (good) | Total | ||
| 1 (weak) | 0 | 1 | 3 | 4 | ||
| 2 | 4 | 8 | 21 | 33 | ||
| 3 (good) | 4 | 14 | 147 | 165 | ||
| Total | 8 | 23 | 171 | 202 | ||
Analysis of image and fat saturation quality of the remaining 66 datasets, when test data originating from the two scanners, that were also used in the training phase (Ingenia and Achieva d-Stream, Philips Healthcare) was excluded is provided in Table SM3.
Visual inspection of cases with metal implants revealed a higher image quality in synthetic images. Figure 4 shows synthetic and true T2-w fs images with metal implants. The synthetic T2-w fs images were based on T1-w and T2-w sequences with specific metal artifact reduction techniques. Also, the true T2-w fs were sequences with metal artifact reduction. In both cases, the synthetic T2-w fs provided a better image quality than the true T2-w fs, offering a better SNR, higher contrast, and less artifacts surrounding the metal implants.
Fig. 4.
Representative true and synthetic T2-w fs images with metal implants (intervertebral disk cages and pedicle screws)
Based on the Turing test performed by eleven independent neuroradiologists, no significant difference real condition versus expert grading was observed between synthetic and true T2-w fs images (p > 0.05) (Table 3). 42.9% of synthetic T2-w fs images and 38.5% of true T2-w fs images were graded incorrectly as the respective counterpart.
Table 3.
Cross-table visual Turing test condition (true/synthetic) versus grading (true/synthetic). Differences were not significant (p > 0.05)
| Condition | |||
|---|---|---|---|
| Grading | True | Synthetic | Total |
| True | 169 | 118 | 287 |
| Synthetic | 106 | 157 | 263 |
| Total | 275 | 275 | 550 |
Diagnostic agreement between synthetic and original protocol
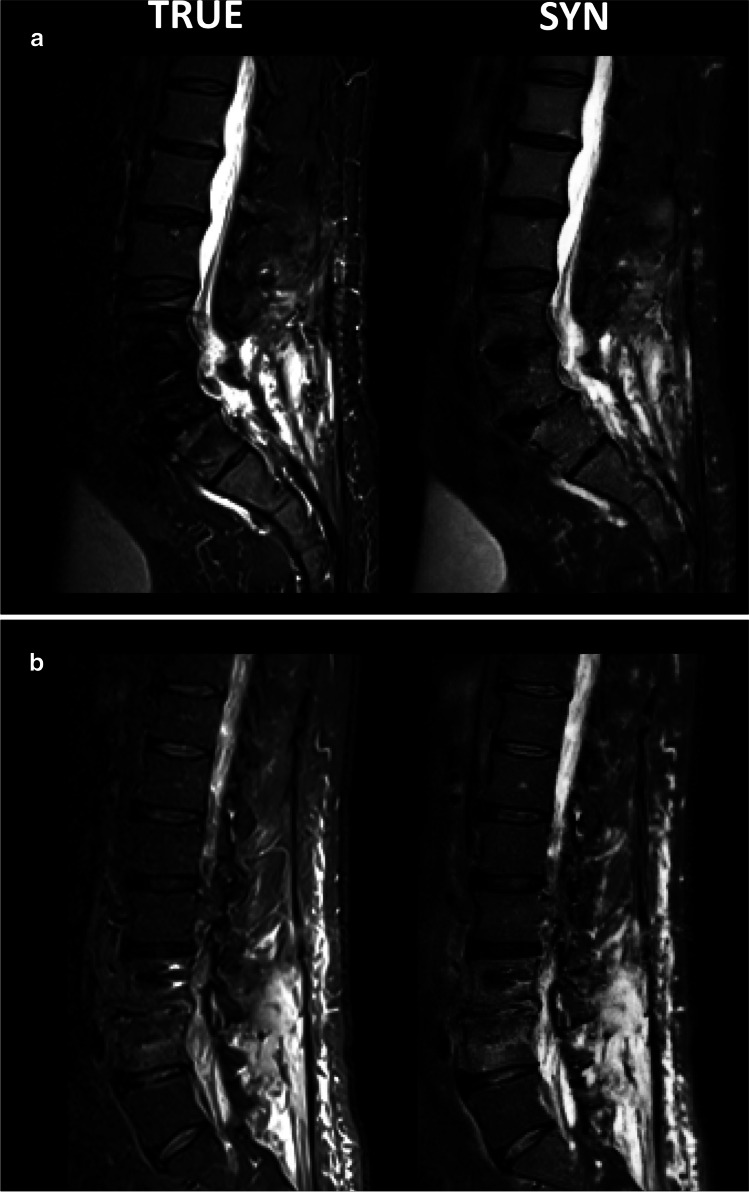
Figure 5 shows representative synthetic and true T2-w fs images with bone marrow abnormalities, vertebral fractures, and paravertebral tissue abnormalities. The original images originate from different scanner vendors and field strengths. A purely qualitative visual comparison of the two juxtaposed images shows the similar diagnostic performance of synthetic versus true T2-w fs images regarding the detection of the presented spine pathologies.
Fig. 5.
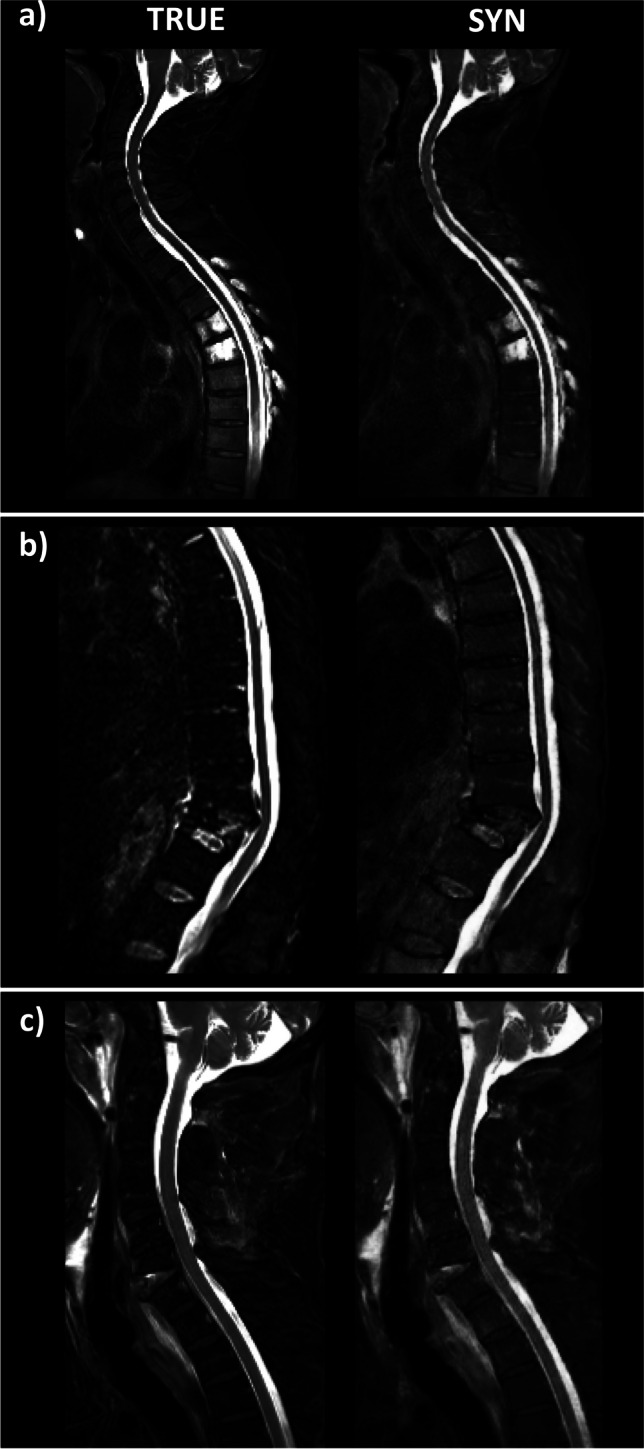
Representative true and synthetic T2-w fs images for different pathologies: a bone marrow abnormalities, b vertebral facture, and c paravertebral tissue abnormalities
Table 4 shows the intermethod agreement (Cohen’s ĸ coefficients) for grading based on the synthetic protocol compared with the original protocol for reader 1 and reader 2, respectively. For both readers, the intermethod agreement ranged from substantial to almost perfect agreement for all evaluated pathologies (bone marrow abnormalities, spondylodiscitis expansion, inflammatory Modic changes, vertebral fractures, cord lesions, and paravertebral tissue abnormalities), except for grading of spinal cord lesions by reader 1 which showed a moderate agreement. Cohen’s ĸ coefficients were significantly different between reader 1 and reader 2 (p = 0.046) (Table 4). The agreement between synthetic and original protocol by the same reader was higher than interrater agreement except for spinal cord lesions (Table 4, significance only found for reader 2, p = 0.028).
Table 4.
Intermethod agreement (Cohen’s kappa coefficient) between synthetic protocol (T1-w, T2-w, and synthetic T2-w fs) and original protocol (T1-w, T2-w, and true T2-w fs) for reader 1 and 2; interrater agreement (Cohen’s kappa coefficient) for synthetic protocol and original protocol
| Intermethod Cohen's kappa | Interrater Cohen's kappa | |||
|---|---|---|---|---|
| Pathology | Reader 1 | Reader 2 | Synthetic protocol | Original protocol |
| Bone marrow abnormalities | 0.76 | 0.91 | 0.70 | 0.81 |
| Spondylodiscitis expansion | 0.85 | 0.91 | 0.74 | 0.59 |
| Juxtadiscal Modic changes (inflammatory) | 0.75 | 0.74 | 0.66 | 0.61 |
| Vertebral fracture | 0.78 | 0.91 | 0.80 | 0.81 |
| Cord lesions | 0.56 | 0.66 | 0.59 | 0.70 |
| Paravertebral tissue abnormalities | 0.79 | 0.86 | 0.74 | 0.77 |
Resulting Cohen’s ĸ coefficients of the remaining 66 datasets, when test data originating from the two scanners, that were also used in the training phase (Ingenia and Achieva d-Stream, Philips Healthcare) was excluded, are provided in Table SM4.
No significant difference between accuracy of synthetic and original protocol was shown ranging between 82.2% for grading of bone marrow abnormalities and 95.0% for grading of spondylodiscitis expansion (p > 0.05) (Table 5).
Table 5.
Accuracy in % of grading based on the synthetic protocol and the original protocol, respectively. No significant difference was shown (p > 0.05)
| Pathology | n (ground truth) | Accuracy synthetic protocol (%) | Accuracy original protocol (%) |
|---|---|---|---|
| Bone marrow abnormalities | 61 | 82.2 | 82.7 |
| Spondylodiscitis expansion | 5 | 95.0 | 95.0 |
| Juxtadiscal Modic changes (inflammatory) | 28 | 87.1 | 85.1 |
| Vertebral fracture | 21 | 92.1 | 92.1 |
| Cord lesions | 15 | 90.0 | 93.6 |
| Paravertebral tissue abnormalities | 25 | 88.6 | 92.1 |
Scan time reduction
In the validation dataset, acquisition duration of T1-w sequence was on average 155 s; of non-fs T2-w sequences, 207 s; and of T2-w fs sequences, 207 s. Waiving the physical acquisition of T2-w fs images consequently shortens the scan protocol by around 40% in a conventional spine examination.
Discussion
Our work demonstrates the diagnostic potential of a GAN-based, sagittal T2-w fs in spine imaging. The synthetic T2-w fs images provided an overall better image quality than the true T2-w fs images, and pathology assessment on the synthetic protocol showed an excellent agreement with the original protocol. We could prove the generalizability of our approach as our assessment is based on a challenging, multicenter test dataset. Consequently, the synthetic T2-w fs might replace a physically acquired T2-w fs in the future, leading to a relevant reduction of scan time for pathology assessment in the spine.
With the introduction of DL techniques into the radiological workflow, synthetic MR contrasts based on GAN frameworks are emerging. Recently, feasibility studies demonstrated the clinical benefit of GAN-based MR images, e.g., a synthetic double inversion recovery (DIR) sequence improved lesion detection in multiple sclerosis [26]. Intrinsic MR contrasts such as T1 or T2 unlike gadolinium contrast can be synthesized without artifacts from other MR contrasts using GANs [21], potentially rendering the physical acquisition of particular MR sequences no longer necessary and thus reducing scan time.
Whereas objective image quality evaluation did not reveal significant differences between synthetic and true T2-w fs images, synthetic images showed a significantly better image quality than true T2-w fs images based on the grading by two expert readers. Our approach of virtually generating T2-w fs images with a GAN allows for an overall scan time reduction of around 40% in conventional spine examinations. This not only increases MR throughout, but might also be one reason for the significantly better image quality of synthetic T2-w fs images compared to true T2-w fs images. Due to reduced patient comfort during prolonged acquisition times and as the fs sequences are often acquired at the end, true T2-w fs images might be affected by motion artifacts. Additionally, MR fat saturation techniques are, depending on the used technique, prone to magnetic field inhomogeneities or inherently suffer from a lower SNR [4]. This is of particular concern, when regions with implanted hardware are scanned. In contrast, the T2-w fs generated by the GAN uses conventional T1-w and non-fs T2-w images as input, which are technically more stable, are less prone to artifacts, and offer higher SNR. Consequently, although it is known that artificially generated images using GANs can show particular artifacts [35], our synthetic T2-w fs images showed improved image quality.
Next to convincing image quality, synthetic images have to represent reality. Therefore, an excellent diagnostic agreement with the original protocol and high accuracy are of particular importance.
For five of the six evaluated pathologies, the expert grading based on the synthetic protocol (including the synthetic T2-w fs) showed a substantial to almost perfect agreement with the original protocol (including the true T2-w fs images). The assessment of spinal cord lesions by reader 1 merely showed a moderate agreement between the synthetic and the original protocol. Remarkably also, the interrater Cohen’s ĸ coefficient for evaluation of cord lesions based on the synthetic protocol is lower than the other interrater Cohen’s ĸ coefficients. Two aspects might explain the lower Cohen’s ĸ coefficients for grading of cord lesion: (1) The GAN was trained exclusively on T2-w Dixon fs images. However, particularly for the detection of cord lesions, T2-w short tau inversion recovery (STIR) images are recommended, whereas the Dixon fs technique is not considered ideal [12]. (2) Hyperintensities on T2-w fs images characterizing cord lesions on sagittal images are often subtle and inconclusive. Additional axial imaging can be helpful to distinguish hyperintensities on T2-w fs images from artifacts and to detect small, marginally located lesions [12]. Such sequences were not available here.
The excellent accuracy of expert grading based on the synthetic as well as on the original protocol, which showed no significant difference, underlines the good agreement of pathology assessment on synthetic images with the gold standard.
For a clinical implementation of GAN-based synthetic images, external validity is required. To the best of the authors’ knowledge, to date, the only two publications presenting GAN-based T2-w fs images in the spine employed MR images from one single vendor [15, 29]. In our work, the GAN framework has been tested on multicenter data. The 101 testing datasets consisting of T1-w and non-fs T2-w images originated from 38 different scanners, with 41 datasets from 1.5 T and 60 datasets from 3 T systems. In contrast to previous studies in brain and spine datasets with a homogeneous FOV, our study demonstrated that GANs can reliably be applied in cases with a highly variable FOV. We were able to demonstrate the generalizability of our approach, by training the network with images from two scanners only and validating it on unseen images derived from 38 different scanners of various field strengths, acquisition protocols, and manufacturers.
The present study has limitations. First, the higher image quality of synthetic compared to true T2-w fs images might lead to bias, when in the course of the grading procedure readers are learning to notice subtle intrinsic image features allowing a differentiation in few samples. In order to rule out a relevant learning bias, we additionally performed a visual Turing test. By randomly presenting synthetic and true T2-w fs images to a broad annotator group without giving feedback about mistakes [36], we could prove that synthetic and true T2-w fs images cannot be significantly distinguished from each other.
Second, the two expert readers had slightly different clinical experience, which might account for some interrater variability.
Third, in our study, only sagittal images have been assessed, although in the clinical routine potentially axial and coronal images are part of spine MRI examinations [37]. However, all current imaging protocol recommendations do not include axial fs images in their recommendations [2]. Sagittal images are often used as screening images to guide the exact ROI for (non-fs) axial imaging. As consequently, sagittal imaging plays a major role in radiological spine assessment, the present study is meant to concentrate on sagittal images.
Fourth, for the proposal of a new technique in the clinical setting, a power analysis is necessary. However, to perform a power analysis, we need some initial information of the performance and suspected diagnostic value of such a new technique that was not available prior to our work presented here. Our study is meant to preliminarily analyze the general potential of GAN-based, synthetic T2-w fs images of the spine and shows the non-inferiority of synthetic T2-w fs images compared to true T2-w fs images in a heterogenous testing datasets. Further research with a power analysis simulating the routine radiological workflow is necessary to assess the additional diagnostic value of synthetic images particularly in the clinical setting.
Conclusion
Our work underlines the potential of a GAN-based T2-w fs for scan time reduction in spine imaging. The overall better image quality and the excellent intermethod agreement render the synthetic T2-w fs a good alternative compared to the true T2-w fs. Our approach is highly generalizable as the assessment is based on a challenging, multicenter test dataset. Therefore, our GAN-based T2-w fs might replace a physically acquired T2-w fs in the future.
Supplementary Information
Below is the link to the electronic supplementary material.
Abbreviations and acronyms
- aCNR
Apparent contrast-to-noise ratio
- DIR
Double inversion recovery
- DL
Deep learning
- fs
Fat sat
- FOV
Field of view
- GAN
Generative adversarial network
- GT
Ground truth
- GUI
Graphical user interface
- ĸ
Kappa
- MRI
Magnetic resonance imaging
- ROI
Region of interest
- STIR
Short tau inversion recovery
- aSNR
apparent signal-to-noise ratio
- TSE
Turbo spin echo
- w
Weighted
Funding
Open Access funding enabled and organized by Projekt DEAL. JSK was supported by DFG (project 432290010), BMBF (German Ministry of Education and Re-search, 13GW0469D), and ERC. SS was supported by an internal faculty grant (KKF, 8700000708). This work has received research funding from the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation program (101045128—iBack-epic—ERC-2021-COG).
Declarations
Guarantor
The scientific guarantor of this publication is Jan S. Kirschke.
Conflict of interest
Jan S. Kirschke is co-founder of Bonescreen GmbH. All other authors declare no relationships with any companies, whose products or services may be related to the subject matter of the article.
Statistics and biometry
PD Dr. Alexander Hapfelmeier (Dipl.-Stat.) (Institute of General Practice and Health Services Research and Institute of Medical Informatics, Statistics and Epidemiology, Technical University of Munich, Munich, Germany) kindly provided statistical advice for this manuscript.
Informed consent
Written informed consent was not required for this study due to the retrospective character.
Ethical approval
Institutional Review Board approval was obtained (593/21 S-SR).
Methodology
• retrospective
• diagnostic or prognostic study
• performed at one institute with multicenter data
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Benedikt Wiestler and Jan S. Kirschke contributed equally to this work
References
- 1.Winegar BA, Kay MD, Taljanovic M. Magnetic resonance imaging of the spine. Pol J Radiol. 2020;85:e550–e574. doi: 10.5114/pjr.2020.99887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.ACR–ASNR–SCBT-MR–SSR practice parameter for the performance of magnetic resonance imaging (MRI) of the adult spine. https://www.acr.org/-/media/ACR/Files/Practice-Parameters/mradult-spine.pdf
- 3.Grande FD, Santini F, Herzka DA, et al. Fat-suppression techniques for 3-T MR imaging of the musculoskeletal system. Radiographics. 2014;34:217–233. doi: 10.1148/rg.341135130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Delfaut EM, Beltran J, Johnson G, Rousseau J, Marchandise X, Cotten A. Fat suppression in MR imaging: techniques and pitfalls. Radiographics. 1999;19:373–382. doi: 10.1148/radiographics.19.2.g99mr03373. [DOI] [PubMed] [Google Scholar]
- 5.Bley TA, Wieben O, François CJ, Brittain JH, Reeder SB. Fat and water magnetic resonance imaging. J Magn Reson Imaging. 2010;31:4–18. doi: 10.1002/jmri.21895. [DOI] [PubMed] [Google Scholar]
- 6.Wang B, Fintelmann FJ, Kamath RS, Kattapuram SV, Rosenthal DI. Limited magnetic resonance imaging of the lumbar spine has high sensitivity for detection of acute fractures, infection, and malignancy. Skeletal Radiol. 2016;45:1687–1693. doi: 10.1007/s00256-016-2493-5. [DOI] [PubMed] [Google Scholar]
- 7.Baker LL, Goodman SB, Perkash I, Lane B, Enzmann DR. Benign versus pathologic compression fractures of vertebral bodies: assessment with conventional spin-echo, chemical-shift, and STIR MR imaging. Radiology. 1990;174:495–502. doi: 10.1148/radiology.174.2.2296658. [DOI] [PubMed] [Google Scholar]
- 8.O'Sullivan GJ, Carty FL, Cronin CG. Imaging of bone metastasis: An update. World J Radiol. 2015;7:202–211. doi: 10.4329/wjr.v7.i8.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hong SH, Choi J-Y, Lee JW, Kim NR, Choi J-A, Kang HS. MR imaging assessment of the spine: infection or an imitation? Radiographics. 2009;29:599–612. doi: 10.1148/rg.292085137. [DOI] [PubMed] [Google Scholar]
- 10.Sollmann N, Mönch S, Riederer I, Zimmer C, Baum T, Kirschke JS. Imaging of the degenerative spine using a sagittal T2-weighted DIXON turbo spin-echo sequence. Eur J Radiol. 2020;131:109204. doi: 10.1016/j.ejrad.2020.109204. [DOI] [PubMed] [Google Scholar]
- 11.Mascalchi M, Dal Pozzo G, Bartolozzi C. Effectiveness of the short TI inversion recovery (STIR) sequence in MR imaging of intramedullary spinal lesions. Magn Reson Imaging. 1993;11:17–25. doi: 10.1016/0730-725X(93)90407-5. [DOI] [PubMed] [Google Scholar]
- 12.Wattjes MP, Ciccarelli O, Reich DS, et al. 2021 MAGNIMS-CMSC-NAIMS consensus recommendations on the use of MRI in patients with multiple sclerosis. Lancet Neurol. 2021;20:653–670. doi: 10.1016/S1474-4422(21)00095-8. [DOI] [PubMed] [Google Scholar]
- 13.Mahnken AH, Wildberger JE, Adam G, et al. Is there a need for contrast-enhanced T1-weighted MRI of the spine after inconspicuous short tau inversion recovery imaging? Eur Radiol. 2005;15:1387–1392. doi: 10.1007/s00330-005-2719-8. [DOI] [PubMed] [Google Scholar]
- 14.Özcan-Ekşi EE, Yayla A, Orhun Ö, Turgut VU, Arslan HN, Ekşi M. Is the distribution pattern of Modic changes in vertebral end-plates associated with the severity of intervertebral disc degeneration?: a cross-sectional analysis of 527 Caucasians. World Neurosurg. 2021;150:e298–e304. doi: 10.1016/j.wneu.2021.02.128. [DOI] [PubMed] [Google Scholar]
- 15.Haubold J, Demircioglu A, Theysohn JM et al (2021) Generating virtual short tau inversion recovery (STIR) images from T1- and T2-weighted images using a conditional generative adversarial network in spine imaging. Diagnostics (Basel) 11(9):1542. https://www.mdpi.com/2075-4418/11/9/1542 [DOI] [PMC free article] [PubMed]
- 16.Low RN, Austin MJ, Ma J. Fast spin-echo triple echo dixon: initial clinical experience with a novel pulse sequence for simultaneous fat-suppressed and nonfat-suppressed T2-weighted spine magnetic resonance imaging. J Magn Reson Imaging. 2011;33:390–400. doi: 10.1002/jmri.22453. [DOI] [PubMed] [Google Scholar]
- 17.Nölte I, Gerigk L, Brockmann MA, Kemmling A, Groden C. MRI of degenerative lumbar spine disease: comparison of non-accelerated and parallel imaging. Neuroradiology. 2008;50:403–409. doi: 10.1007/s00234-008-0363-0. [DOI] [PubMed] [Google Scholar]
- 18.Bratke G, Rau R, Weiss K, et al. Accelerated MRI of the lumbar spine using compressed sensing: quality and efficiency. J Magn Reson Imaging. 2019;49:e164–e175. doi: 10.1002/jmri.26526. [DOI] [PubMed] [Google Scholar]
- 19.Nie D, Trullo R, Lian J, et al. Medical image synthesis with deep convolutional adversarial networks. IEEE Trans Biomed Eng. 2018;65:2720–2730. doi: 10.1109/TBME.2018.2814538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lv J, Zhu J, Yang G. Which GAN? A comparative study of generative adversarial network-based fast MRI reconstruction. Philos Trans A Math Phys Eng Sci. 2021;379:20200203. doi: 10.1098/rsta.2020.0203. [DOI] [PubMed] [Google Scholar]
- 21.Lee D, Moon W-J, Ye JC. Assessing the importance of magnetic resonance contrasts using collaborative generative adversarial networks. Nat Mach Intelle. 2020;2:34–42. doi: 10.1038/s42256-019-0137-x. [DOI] [Google Scholar]
- 22.Qasim AB, Ezhov I, Shit S et al (2020) Red-GAN: Attacking class imbalance via conditioned generation. Yet another perspective on medical image synthesis for skin lesion dermoscopy and brain tumor MRI. http://proceedings.mlr.press/v121/qasim20a/qasim20a.pdf. Accessed 20 Sept 2022
- 23.Li H, Paetzold JC, Sekuboyina A, et al. DiamondGAN: unified multi-modal generative adversarial networks for MRI sequences synthesis. Cham: Springer International Publishing; 2019. pp. 795–803. [Google Scholar]
- 24.Radford A, Metz L, Chintala S (2015) Unsupervised representation learning with deep convolutional generative adversarial networks. arXiv. 10.48550/arXiv.1511.06434
- 25.Goodfellow I. J. P-AJ, Mirza M, Xu B, Warde-Farley D, Ozair S, Courville A, Bengio Y (2014) Generative adversarial networks. Adv Neural Inf Process Syst. https://papers.nips.cc/paper/2014/hash/5ca3e9b122f61f8f06494c97b1afccf3-Abstract.html
- 26.Finck T, Li H, Grundl L, et al. Deep-learning generated synthetic double inversion recovery images improve multiple sclerosis lesion detection. Invest Radiol. 2020;55:318–323. doi: 10.1097/RLI.0000000000000640. [DOI] [PubMed] [Google Scholar]
- 27.Kazuhiro K, Werner RA, Toriumi F, et al. Generative adversarial networks for the creation of realistic artificial brain magnetic resonance images. Tomography. 2018;4:159–163. doi: 10.18383/j.tom.2018.00042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fayad LM, Parekh VS, de Castro LR, et al. A deep learning system for synthetic knee magnetic resonance imaging: is artificial intelligence-based fat-suppressed imaging feasible? Invest Radiol. 2021;56:357–368. doi: 10.1097/RLI.0000000000000751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim S, Jang H, Hong S, et al. Fat-saturated image generation from multi-contrast MRIs using generative adversarial networks with Bloch equation-based autoencoder regularization. Med Image Anal. 2021;73:102198. doi: 10.1016/j.media.2021.102198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Isola P, Zhu J, Zhou T, Efros AA (2017) Image-to-image translation with conditional adversarial networks 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), pp 5967–5976
- 31.Pennig L, Kabbasch C, Hoyer UCI, et al. Relaxation-enhanced angiography without contrast and triggering (REACT) for fast imaging of extracranial arteries in acute ischemic stroke at 3 T. Clin Neuroradiol. 2021;31:815–826. doi: 10.1007/s00062-020-00963-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kofler F, Ezhov I, Isensee F et al (2021) Are we using appropriate segmentation metrics? Identifying correlates of human expert perception for CNN training beyond rolling the DICE coefficient. arXiv preprint arXiv:210306205. 10.48550/arXiv.2103.06205. Accessed date 20 Sept 2022
- 33.de Leeuw JR. jsPsych: A JavaScript library for creating behavioral experiments in a Web browser. Behav Res Methods. 2015;47:1–12. doi: 10.3758/s13428-014-0458-y. [DOI] [PubMed] [Google Scholar]
- 34.Jakobsson U, Westergren A. Statistical methods for assessing agreement for ordinal data. Scand J Caring Sci. 2005;19:427–431. doi: 10.1111/j.1471-6712.2005.00368.x. [DOI] [PubMed] [Google Scholar]
- 35.Odena A, Dumoulin V, Olah C. Deconvolution and checkerboard artifacts. Distill. 2016;1:e3. doi: 10.23915/distill.00003. [DOI] [Google Scholar]
- 36.Salimans T, Goodfellow I, Zaremba W, Cheung V, Radford A, Chen X. Improved techniques for training gans. Adv Neural Inf Process Syst. 2016;29:2234–2242. [Google Scholar]
- 37.Ekşi M, Özcan-Ekşi EE, Orhun Ö, Turgut VU, Pamir MN. Proposal for a new scoring system for spinal degeneration: Mo-Fi-Disc. Clin Neurol Neurosurg. 2020;198:106120. doi: 10.1016/j.clineuro.2020.106120. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.