Abstract
The current targeting drug delivery mainly relies on cancer cell surface receptors. However, in many cases, binding affinities between protein receptors and homing ligands is relatively low and the expression level between cancer and normal cells is not significant. Distinct from conventional targeting strategies, we have developed a general cancer targeting platform by building artificial receptor on cancer cell surface via a chemical remodeling of cell surface glycans. A new tetrazine (Tz) functionalized chemical receptor has been designed and efficiently installed on cancer cell surface as “overexpressed” biomarker through a metabolic glycan engineering. Different from the reported bioconjugation for drug targeting, the tetrazine labeled cancer cells not only locally activate TCO-caged prodrugs but also release active drugs via the unique bioorthogonal Tz-TCO click-release reaction. The studies have demonstrated that the new drug targeting strategy enables local activation of prodrug, which ultimately leads to effective and safe cancer therapy.
Key words: Artificial receptor, Click and release, Local activation, Protein degradation
Graphical abstract
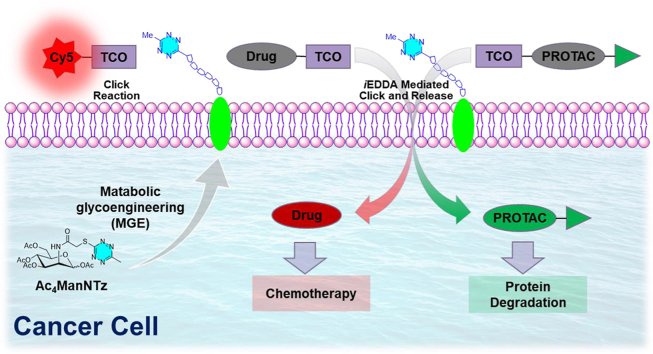
A new tetrazine (Tz) functionalized chemical receptor has been designed and efficiently installed on cancer cell surface as “overexpressed” biomarker for targeted imaging and prodrug activation.
1. Introduction
Targeted molecular imaging and therapy relying on effective identification and accumulation of cell-surface receptors have great potential to improve cancer diagnosis and treatment1, 2, 3. The current strategy mainly relies on tumor cell surface receptors for targeting4. The specific recognition of cell-surface receptors enables the targeted delivery of therapeutic agents to the site of interest and minimizes the side effects on normal tissue. However, protein receptors are rarely tumor cell specific and in many cases the expression level between cancer and normal cells is not significant2,3. Moreover, binding affinities between protein receptors and homing ligands such as folate, peptides, sugars and aptamers is relatively low5. Additionally, the therapeutic resistance mutation of protein receptors makes the targeting approach difficult and vulnerable in cancer treatment6, 7, 8. So far, no small molecule-based targeting strategies for cancer therapy have made it to the clinic.
Chemically modified prodrug with reduced systemic toxicity can be activated locally at the site of disease to perform therapeutic function on demand, which is a valuable strategy to develop pharmaceutical agents to circumvent the systemic and high dosage related undesired adverse effects9. The conventional mechanism of prodrug activation in biological system is inherent from the differences between disease and normal tissues2, 3, 4, 5. For instance, antibody–drug conjugates (ADCs) and antibody-based pre-targeting strategies have been used in delivery and local activation of cytotoxic agents10. However, these systems are limited by endogenous overexpression of cell surface receptors and encounter non-specific activation via metabolic hydrolysis of the linker between the antibody and drug11 as well as the insufficient penetration ascribed to the large size of antibodies12.
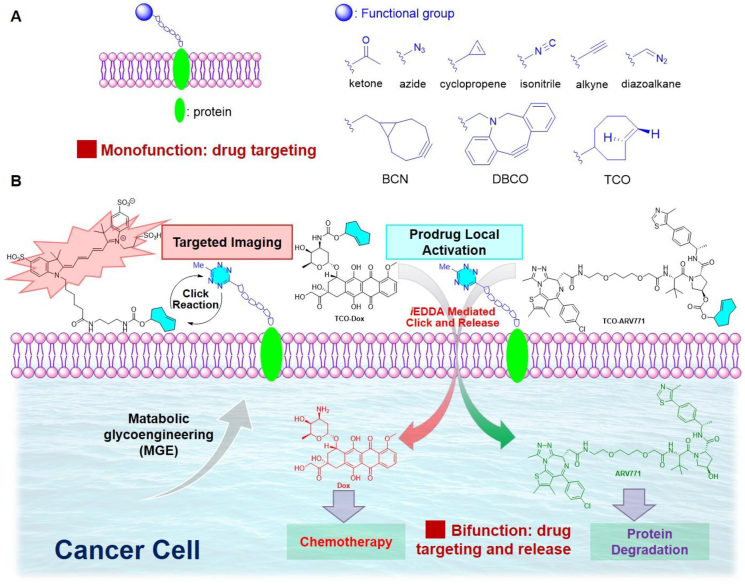
Chemical remodeling of cell surface glycans by metabolism of the corresponding unnatural sugars is a viable tool to incorporate artificial chemical receptors onto the cell surface for noninvasive cell imaging and targeted therapeutics delivery13, 14, 15, 16. The installed cell surface chemical receptors are abiotic and can be targeted high specifically through bioorthogonal chemistry14. Additionally, the small size of chemical receptors avoids the receptor saturation and immunogenicity as a comparison to conventional protein-based targeting strategies. Therefore, metabolic glycan labelling has been explored for noninvasive cell imaging and for targeted therapy. A variety of functional groups, particularly clickable moieties for click reactions such as ketone, azide, diazoalakne, isonitrile, alkyne, and strained cyclopropane, BCN, DBCO, and trans-cyclooctyne (Fig. 1A)14, have been metabolically engineered onto cell-surface glycoproteins. However, these functionalities do not have release capacity for drug delivery. With the fast reaction kinetics and bioorthogonality of tetrazine (Tz) and trans-cyclooctene (TCO) involved inverse-electron-demand Diels–Alder (iEDDA) reaction17, 18, 19, 20, 21, 22, 23, 24, the potential and effectiveness of targeting and drug delivery can be achieved at the low concentration in complex biological context. This unique mechanism is particularly appealing for developing prodrug release system25, 26, 27, 28, 29, 30. We envisioned that chemical remodeling of cell surface glycans with incorporation of a Tz moiety could generate an unprecedented Tz labeled cancer cell surface, which could serve as a handle for drug activation and delivery through the unique click-release capacity.
Figure 1.
(A) Glycometabolic engineering of functionalized glycoproteins on cell surface for selective labelling and drug targeting (known). (B) Building tetrazine-functionalized chemical receptors for biorthogonal targeting, activation, and delivery of drugs to cancer cells (this work).
Herein, we report the design and metabolic engineering of a tetrazine mannosamine onto the cancer cell surface for the first time as an endogenous “overexpressed” biomarker for targeted anti-cancer drug delivery (Fig. 1B). The metabolic glycoengineering of tetrazine chemical receptors enables specific cell surface labelling in various cancer cells efficiently. Furthermore, the tetrazine labeled on cancer cells can induce click-release reaction to locally activate TCO-caged prodrugs by iEDDA reaction-mediated uncaging process. The effectiveness of click-release strategy was validated by TCO-caged doxorubicin (Dox) prodrug. The selectivity of TCO-Dox prodrug for tetrazine labelled cancer cells is enhanced by 10 folds as a comparison to the unlabeled cells. Furthermore, this strategy was further demonstrated in click-release of caged TCO-ARV771 PROTAC (PROteolysis TArgeting Chimeras) for selective degradation of bromodomain-containing protein (BRD4).
2. Results and discussion
2.1. Design and synthesis of Ac4ManNTz and analogues
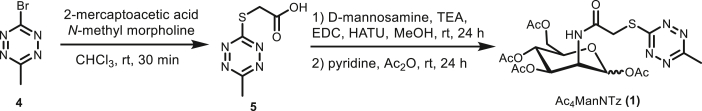
Previous studies have shown that the engineering efficiency of the Roseman-Warren biosynthetic pathway is highly sensitive to the N-acyl structure of unnatural ManNAc analogues31,32. Structures with long or branched N-acyl chains are poor substrates for the biosynthetic enzymes, while those containing short, linear side chains are better tolerated31. We therefore designed peracetylated Ac4ManNTz (1) and Ac4ManNPhTz (2) containing respective linear and rigid linkers and Ac4ManNPh (3) with a phenyl structure to replace Tz as control. We then synthesized these compounds (Scheme 1). Briefly, the key 3-bromo-6-methyl-tetrazine (4) was synthesized from hydrazinecarbothiohydrazide by following literature procedure (Supporting Information Fig. S1)33,34. Nucleophilic aromatic substitution reaction of 4 with 2-mercaptoacetic acid gave tetrazine thiol acetic acid 5. Amidation of the acid with d-mannosamine hydrochloride in the presence of EDC·HCl, and HATU as coupling reagents was achieved and followed by acylation with acetic anhydride to provide the target Ac4ManNTz (1). Ac4ManNPhTz (2) and Ac4ManNPh (3) were synthesized by the reaction of d-mannosamine hydrochloride with the corresponding tetrazine carboxylic acid (Supporting Information Figs. S1–S3).
Scheme 1.
Synthesis of Ac4ManNTz (1).
2.2. Engineering and optimization of unnatural ManNAc analogues on cell-surface
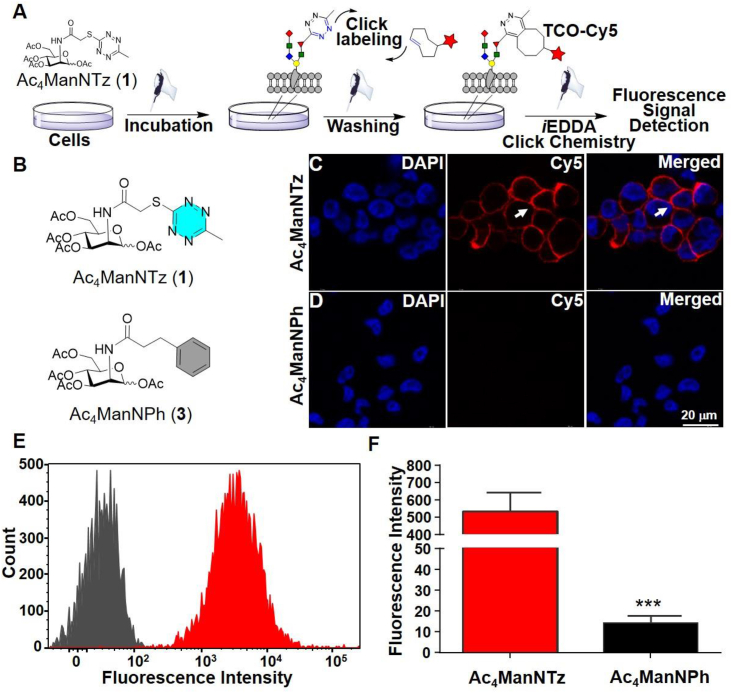
Before performing the metabolic engineering of Ac4ManNTz (1), Ac4ManNPhTz (2) and Ac4ManNPh (3) in cells, we tested their cell viability using MTT assay. We found that these compounds could be tolerated over 100 μmol/L for Ac4ManNTz (1), and Ac4ManNPh (3) in HeLa and A549 cells. For Ac4ManNPhTz (2), it shows mild toxicity under increased concentration (Supporting Information Fig. S4) but is still well compatible based on low concentration been used. It is expected that they are enzymatically deacetylated in the cytosol and then metabolically converted to the corresponding N-Tz, -PhTz and -Ph acetyl sialic acids, which are subsequently incorporated into sialoglycoconjugates31,32. Once presented on the cell surface, the Tz-labeled sialylated glycans can be visualized with a TCO-fluorescent tag, which can be used to determine their labeling efficiency. To evaluate the metabolic glycoengineering efficiency of these new tetrazine precursors, MDA-MB-231 cells were cultured in presence of Ac4ManNTz (1), and Ac4ManNPh (3) respectively (Fig. 2A and B). Subsequently, the cells were labeled with TCO-biotin conjugate followed by incubation with streptavidin-Cy5 for fluorescence imaging analysis. Confocal microscopy clearly showed strong labeling (red) of cell surface (arrow) in group of cells preincubated with Ac4ManNTz (1) (Fig. 2C). Minimal signal is seen with the cells cultured in presence of Ac4ManNPh (3), which lacks the functional tetrazine group (Fig. 2D). This indicates that the unnatural Tz derived sugar was efficiently installed on the cell surface. The metabolic engineering of tetrazine group on cell surface was also confirmed by flow cytometry analyses of MDA-MB-231 cells following the same treatment consistently with much stronger fluorescence intensity in group of Ac4ManNTz (1) than the control group (Fig. 2E and F). Moreover, similar metabolically labelling and specific cell membrane fluorescence imaging were observed in other cancer cells such as A549 and HeLa cells (Supporting Information Figs. S5 and S6). These results collectively demonstrated that Ac4ManNTz (1) was able to metabolically label cancer cells with tetrazine groups on cell surface with high efficiency.
Figure 2.
Labeling of metabolically engineered cell-surface glycoconjugates. (A) Working flowchart for engineering and detection. MDA-MB-231 cells were grown with 25 μmol/L Ac4ManNTz (1) for three days, washed with PBS and labeled with TCO-Biotin (250 μmol/L) for 1 h at 37 °C, followed by incubation with Streptavidin-Cy5 (20 μg/mL) for 20 min at rt. (B) The chemical structure of Ac4ManNTz (1) and Ac4ManNPh (3). (C, D) Confocal images showing the MDA-MB-231 cells pretreated by the Ac4ManNTz (1) could be specifically labeled on the cell surface (arrow) while the control group pretreated by the Ac4ManNPh (3) displayed barely visible fluorescent signal. Scale bar: 20 μm. (E) Flow cytometry of cells pretreated with Ac4ManNTz (1) and Ac4ManNPh (3). (F) Mean Cy5 fluorescence intensity of MDA-MB-231 cells extracted from (C, D). Data were presented as mean ± SEM (n = 3) and analyzed by Student's t-test (two-tailed), ∗∗∗P ≤ 0.001.
The reaction between tetrazine and TCO was one of the fast click reactions23,24. However, labeling of tetrazine in biological system need to be tested in low concentration and the reaction interface was limited to two-dimensional cell surface rather than the dispersed in solution. Therefore, we sought to establish the time and concentration profile necessary to maximize the observed fluorescence signal in the cell-based experiments. The cell surface tetrazine reacted with TCO-biotin, followed by immunostaining with streptavidin-AF647, which could be detected by flow cytometry. The increase of fluorescence signal was the Ac4ManNTz concentration dependent (10.0–50.0 μmol/L, Supporting Information Fig. S7). Ac4ManNTz (1) at 50 μmol/L was chosen for the following screening since it gave sufficient fluorescence signals in flow cytometry. Ac4ManNTz (1) treated and untreated cells were incubated with TCO-biotin at different concentrations (0.1–1 mmol/L) and various periods of time (5 min–1 h) followed by immunostaining with streptavidin-AF647 (Supporting Information Figs. S8 and S9). A maximal fluorescence signal for cells treated with Ac4ManNTz (1) was observed at 0.25 mmol/L of TCO-biotin. The reaction was completed in 30 min and the fluorescence signal from the tetrazine-bearing cells reached the plateau with over 80-fold greater than the background, which is more sufficient and faster compared to the Staudinger ligation reported previously.35
2.3. Identification of tetrazine labeled cell lysates
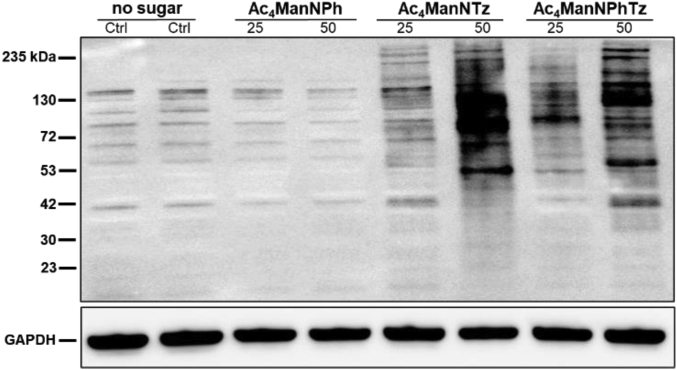
To identify tetrazine-labelled glycoconjugates on MDA-MB-231 cell, we analyzed cell lysates by Western blot. MDA-MB-231 cells were cultured with 25 μmol/L of Ac4ManNTz (1), Ac4ManNPhTz (2), Ac4ManNPh (3) or DMSO for 72 h and then lysed with RIPA buffer. The bands observed in lanes 5–8 correspond to the tetrazine tagged sialic acid post translational modified on the protein, which were labeled via bioorthogonal iEDDA chemistry. Lanes 1 and 2 (negative control), as well as lanes 3 and 4, corresponding to sugar precursor, Ac4ManNPh (3), lacking tetrazine, are silent in the following HRP-streptavidin probing (Fig. 3). Significant glycoprotein labeling was observed in lysates from cells treated with Ac4ManNTz (1) as comparison to Ac4ManNPhTz (2), which shows the high efficiency of metabolic glycoengineering of these two tetrazine sugar precursors in cells.
Figure 3.
Western blot analysis of lysates from MDA-MB-231 cells treated with 25 or 50 μmol/L of Ac4ManNTz (1), Ac4ManNPhTz (2), Ac4ManNPh (3) or DMSO (no tetrazine-sugar). MDA-MB-231 cells were incubated with biotin-TCO probe (0.25 mmol/L) for 30 min. The cell lysates were harvested and probed with anti-biotin antibody conjugated to horseradish peroxidase (HRP) (upper panel), then stripped and re-probed with an anti-GAPDH antibody as a protein loading control (lower panel).
2.4. Metabolic engineering unnatural Ac4ManNTz (1) on various cell lines
Having demonstrated Ac4ManNTz (1), which can be efficiently incorporated into MDA-MB-231 cell surface, we then examined the scope and efficiency of Ac4ManNTz (1) metabolism in variable human cancer cell lines including A549 (human lung cancer cell lines), HCT116 (human colorectal cancer cell lines), MDA-MB-231 (human breast cancer cell lines), and HeLa (human ovarian cancer cell lines) (Supporting Information Fig. S10). In these cell lines, the increased fluorescence signals were observed with the initial concentration of Ac4ManNTz (1), indicating that the efficient labeling was achieved. However, the sensitivity was cell type dependent. The metabolization of Ac4ManNTz (1) in these cell lines is variable. A549 and HCT116 cells displayed similar fluorescence enhancement when treated with 50 μmol/L of Ac4ManNTz (1). While HeLa cells displayed lower fluorescence increasing at comparable concentration of Ac4ManNTz (1). The MDA-MB-231 cells displayed highest fluorescence among these cell lines, and thus greater numbers of tetrazine on cell surface.
2.5. Concurrent labeling of the cell surface by utilization of two separate and compatible bioorthogonal chemistry
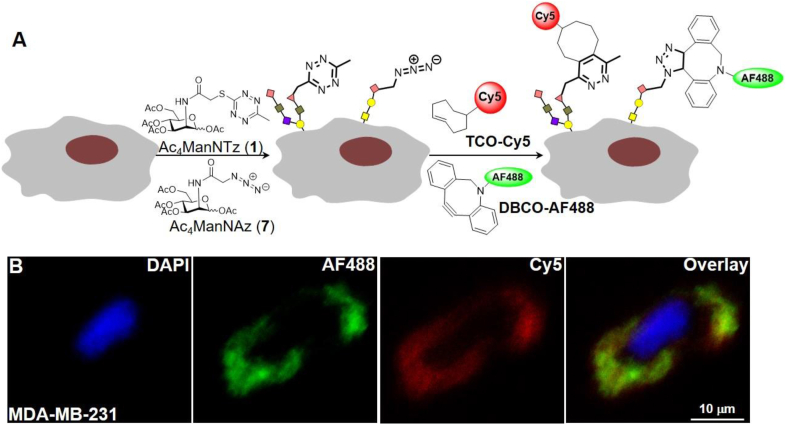
We evaluated the compatibility of tetrazine-TCO iEDDA reaction with other strain promoted click chemistry, such as azide-dibenzocyclooctyne cycloaddition36. MDA-MB-231 cells were grown in presence of one or both of the two unnatural sialic acid precursors, Ac4ManNTz (1) and Ac4ManNAz (7) (Fig. 4A). Followed by simultaneously labeling with cyclooctyne (DBCO) bearing green fluorophore FITC as well as biotinylated trans-cyclooctyne and streptavidin bearing red fluorophore Cy5, confocal microscopy revealed multiplexed fluorescence labeling on cell surface, shown in Fig. 4B. The flow cytometry was carried out in parallel to confirm the findings of expression of chemical reporters, azido and tetrazine, on cell surface by multiplexed labeling (Supporting Information Fig. S11). Only cells that were initially incubated with Ac4ManNAz (7) displayed green fluorescent signals (FITC), and only cells incubated with Ac4ManNTz (1) displayed red fluorescent signals (AF647). The cells treated with both Ac4ManNTz (1) and Ac4ManNAz (7) exhibited fluorescent signals in both emission channels. In absence of the chemical reporters, only minimal background labeling was observed for both ligation reactions. These results illustrate the value and biocompatibility of tetrazine-TCO involved iEDDA ligation for dual labeling.
Figure 4.
Simultaneous labeling of two cell surface chemical reporters. Distinct metabolic targets can be simultaneously imaged using tetrazine and azido reporters. (A) MDA-MB-231 cells were cultured in the presence of both Ac4ManNTz (25 μmol/L) and Ac4ManNAz (25 μmol/L) for 72 h, followed by concurrent treatment with DBCO-FITC (25 μmol/L, 30 min at 37 °C) to covalently tag cell surface azides and TCO-Biotin (0.25 mmol/L, 30 min at 37 °C), streptavidin-AF647 to tag tetrazines, respectively. Cells were then mounted to cover slides and counter stained with DAPI and imaged via confocal microscopy. (B) Representative images are shown. Red: Cy5, green: FITC, blue: DAPI. Scale bar: 10 μm.
2.6. Click and release study
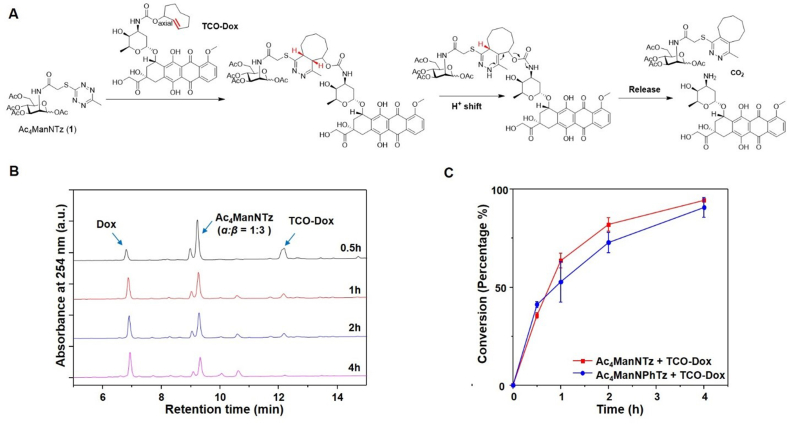
To demonstrate the targeted activation and delivery of cytotoxic agents to tetrazine functionalized cancer cells, we designed a TCO caged doxorubicin (TCO-Dox), a clinically used chemotherapy as the model drug37. We first tested this drug release system with tetrazine mannosamines, Ac4ManNTz (1) or Ac4ManNPhTz (2), and a caged doxorubicin at the allylic position of TCO with a carbamate linker, TCO-Dox in chemical system. It is expected that the tetrazine group on Ac4ManNTz (1) reacts with TCO-Dox forming a 4,5-dihydropyridazine bicyclic intermediate, following a H+-shift and tautomerization to a 1,4-dihydropyridazine. The 1,4-dihydropyridazine undergoes a conversion to form exocyclic double bond with the elimination of CO2 and release of doxorubicin from the carbamate linked TCO at allylic position (Fig. 5A). Ultra-performance liquid chromatography (UPLC) was employed to monitor the release of doxorubicin from the TCO-caged prodrug, by incubating Ac4ManNTz (1) with TCO-Dox, 20 μmol/L in 5% DMSO in PBS buffer at 37 °C. We monitored prodrug consumption and the release of parent drug doxorubicin upon reaction with tetrazine. As shown in Fig. 5B, the prodrug itself did not liberate doxorubicin for at least 24 h in PBS buffer, supporting the bioorthogonality of the click release system. Notably, more than 90% doxorubicin was released within 4 h from TCO-Dox treatment with Ac4ManNTz (1) or Ac4ManNPhTz (2) at concentration of 40 μmol/L (Fig. 5C).
Figure 5.
HPLC analysis of click release reaction between Ac4ManNTz and TCO-Dox. (A) Mechanism of the iEDDA reaction mediated release. Ac4ManNTz react with TCO-Dox through [4 + 2] cycloaddition to form dihydropyridazine which subsequently undergoes rearrangement and elimination that ultimately liberate the Dox. (B) Representative curves from high-pressure liquid chromatography (HPLC) analysis show cumulative release of Dox after mixing Ac4ManNTz (40 μmol/L) with TCO-Dox (20 μmol/L) in PBS buffer for 30 min, 1, 2, 4 h. (C) The quantification of released Dox as a function of time. Data are averages ± SEM, n = 3.
2.7. Assessment of the click and release system
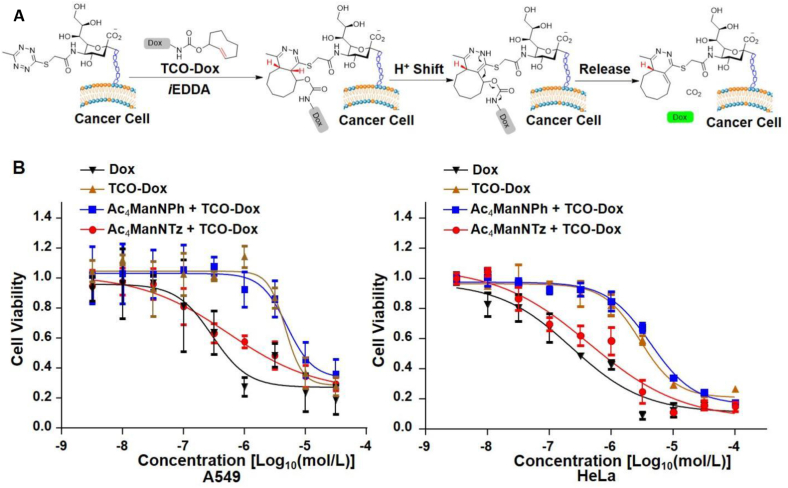
Have demonstrated the release of Dox from its TCO-prodrug in chemical system, we next evaluated the activation and release in cancer cells. We first metabolically inserted tetrazines group on A549 (human lung cancer cells) and HeLa (human cervical cancer cells) cancer cells by incubation with 50 μmol/L of Ac4ManNTz (1) and washed out free Ac4ManNTz (1) in the culture medium. The prodrug TCO-Dox was then added, and the liberation of active doxorubicin was initiated by iEDDA-mediated click release reaction to exhibit inhibitory bioactivities (Fig. 6A). Exposure of A549 cells to doxorubicin led to a significant decrease of the proliferation ability (278 nmol/L), while TCO-Dox gives attenuated bioactivity on proliferation assays (IC50 = 4.76 μmol/L) by CCK-8 proliferation assay, which shows the caged amine group effectively block inhibitory activity (Fig. 6B and Table 1). The cells pretreated with Ac4ManNTz (1) and metabolized with tetrazine gave IC50 of 548 nmol/L, while the control group without tetrazine labeling show no enhancement on inhibitory activity. A similar trend was observed with HeLa cells (Fig. 6C and Table 1). The inhibitory effect was validated on HeLa cells with IC50 of 229 nmol/L for doxorubicin and 2.93 μmol/L for TCO-Dox prodrug by CCK-8 proliferation assay (Fig. 6C). Strong inhibiting activity was observed on HeLa cells with IC50 of 439 nmol/L with Ac4ManNTz (1) treated cells, but much low effect was seen for control group with IC50 of 4.28 μmol/L (Fig. 6C and Table 1).
Figure 6.
Prodrug activation on cell surface. (A) Cancer cells metabolic glycan labeling with sialic acid tetrazine by pretreated with Ac4ManNTz (1) and induced local activation of TCO-Dox at cell surface. (B) Comparison of the 48 h cytotoxicity of TCO-Dox, Dox and activated Dox, and non-activated Dox against A549 cells and HeLa cells.
Table 1.
Calculated IC50 values for Dox, TCO-Dox and TCO-Dox activated with non-activated against each cell line.
| Drug | Doxa | TCO-Dox | TCO-Doxb Activated |
TCO-Doxc Non-activated |
|---|---|---|---|---|
| A549 | 0.278 | 4.76 | 0.548 | 5.04 |
| HeLa | 0.229 | 2.93 | 0.439 | 4.28 |
IC50 values are determined the 48 h cytotoxicity of each drug. The units of IC50 are in μmol/L.
Activated TCO-Dox indicates that each cell line was pre-incubated with 50 μmol/L Ac4ManNTz for 3 days, washing and incubated with TCO-Dox at different concentration for 48 h.
Nonactivated TCO-Dox indicates the pre-incubation with 50 μmol/L Ac4ManNPh.
2.8. PROTACs activation and delivery via the click and release system
The PROTACs have been emerged as a new promising therapeutic approach by recruiting the ubiquitin proteasome system (UPS) to degrade the targeted protein of interest (POI)38,39. PTOTACs, which do not rely on occupancy-driven pharmacology, offers unrivaled power over traditional small-molecule inhibitors in that they can degrade their protein targets in a sub-stoichiometric, catalytic fashion40. However, potential toxicity from systemic degradation of proteins in healthy cells and undesirable ligase-mediated off-target effects limit their application as both chemical tools and therapeutics41,42. Therefore, an urgent need exists for developing strategies to improve tumor-targeting ability of PROTACs.
Several targeted therapy strategies, capable of increasing the enrichment of drugs in tumor tissues and reducing side effects, have been developed for the delivery of PROTACs43. In this context, light-controllable PROTACs enable achieving spatiotemporal regulation of the activity of the PROTAC molecules44. Nonetheless, these methods are limited cancer types with light accessibility and in some cases, UV light activation is used and often induces undesired tissue damage. The tumor microenvironment-activation45, and antibody–drug conjugates (ADCs)46, folate47, and aptamer48 as vectors for targeting tumor cell surface receptors have been developed for selective delivery of PROTACs to cancer cells. However, the use of antibodies suffers from drawbacks including high production cost, receptor saturation, poor solid tumor penetration and severe immunogenicity49. In addition, each mAb developed is effective for only certain types of cancer because the targeted protein receptors vary from cancer to cancer. Tumor microenvironment-activation and folate and aptamer-caged PROTACs have been shown to exhibit improved degradation selectivity between cancer cells and normal cells. However, as discussed above, tumor microenvironment and protein receptors are rarely tumor cell specific and in many cases the expression level between cancer and normal cells is not significant2,3 and binding affinities between protein receptors and homing ligands such as folate and aptamers is relatively low.5
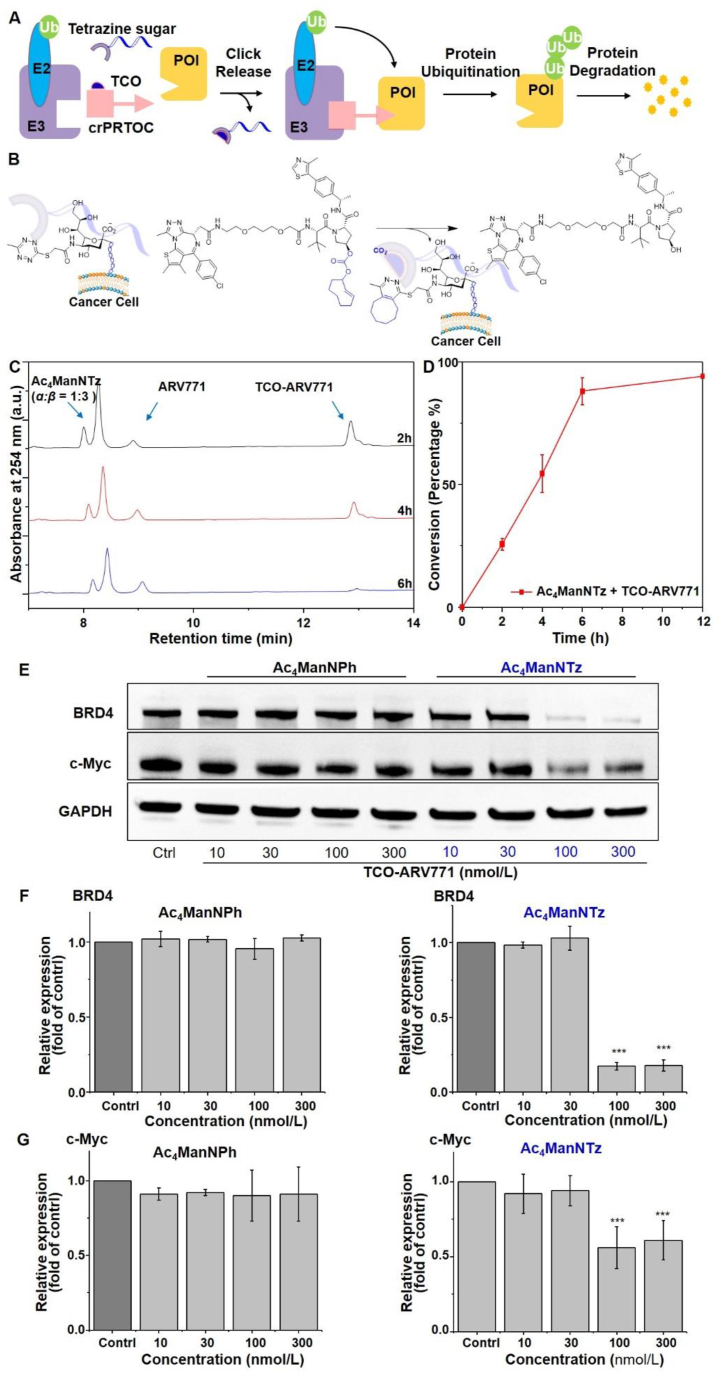
We propose a bioorthogonal click and release of PROTACs from their prodrugs, capable of conditionally activating the caged PROTACs in tumor site. To test the concept, we designed a TCO caged ARV771, TCO-ARV771. ARV771 is a potent degrader of BET protein by recruiting a VHL (von Hippel-Lindau) E3 ubiquitin ligase42. High dose of ARV771 has been reported to be toxic, due to its effects on completely depleting bromodomain family members that play critical role in modulating enhancer and transcription activity of many genes 41. This issue limits the further application of ARV771 in the clinic.
Caging the hydroxyl group, which is critical for the recruitment of VHL E3 ubiquitin ligase50,51 in the VHL ligand, abolishes the degradation activity of ARV77152. Therefore, we designed a TCO caged ARV771, TCO-ARV771, by incorporating bulky TCO moiety via a carbonate linkage onto the hydroxyl group of a well-studied VHL-based bromodomain (BRD) degrader, ARV771 (Fig. 7A)42. It is expected that caging the hydroxyl group in ARV771 by TCO will diminish the degradation function. However, the degradation activity can be regained by the removal of the TCO group via bioorthoginal click-release reaction with the cancer cell surface tetrazine group. The tetrazine moiety aids targeted enrichment of the PROTAC into cancer cells to achieve high selectivity.
Figure 7.
Click and release PROteolysis-Targeting Chimeras (crPROTACs) for BRD4 degradation. (A) Schematic description of click and release of PROTACs by iEDDA medicated uncaging mechanism. (B) In vitro activation of TCO-ARV771 by tetrazine functionalized cancer cell surface. (C) UPLC spectra of release of ARV771 by incubating Ac4ManNTz (1) and TCO-ARV771 for 2, 4, and 6 h. (D) Release profile of reaction of Ac4ManNTz (1) (40 μmol/L) with TCO-ARV771 (20 μmol/L). A 20 μL aliquot sample was taken from the reaction mixture for UPLC analysis. Data are averages ± SEM, n = 3. (E) Western blot analysis of BRD4 degradation and c-Myc expression. (F) Quantification of BRD4 protein expression. (G) Quantification of c-Myc protein expression. Data are averages ± SEM, n = 3.
TCO-ARV771 was synthesized through coupling of TCO-Ns with ARV771 in the presence of TEA in DMF (see Supporting Information). We first tested the release of ARV771 by incubation of Ac4ManNTz (1) (40 μmol/L) with the synthesized TCO-ARV771 prodrug (20 μmol/L) in PBS buffer and the event was monitored by UPLC. The appearance of ARV771 was observed and >80% of active ARV771 was released within 6 h (Fig. 7B–D). It is noted that the uncaging reaction of TCO-ARV771 was found to be inferior to the TCO-Doxorubicin, probably owing to the steric hindrance.
2.9. Assessment of the degradation of PROTACs in vitro
We then demonstrated the degradation activity of ARV771 via click-release reaction of TCO-ARV771 with cancer cell surface tetrazine. In the studies, MDA-MB-231 cells was used and was pretreated with Ac4ManNTz (1) and Ac4ManNPh (3) as control. The metabolic glycoengineering process was lasted for 3 days and then free Ac4ManNTz (1) or Ac4ManNPh (3) was washed out. The Tz and Ph engineered cells were then treated with TCO-ARV771 prodrug with concentrations ranging from 10 to 300 nmol/L of BRD4 degradation activity was analyzed by Western blot. As shown, no degradation of BRD4 with Ac4ManNPh (3) engineered cells was observed (Fig. 7E and G). The degradation with tetrazine (Tz) functionalized cells was seen in a concentration dependent manner. Significant degradation (ca. 80%) occurred at 100 nmol/L TCO-ARV771 (Fig. 7E and G). The degradation activity is compatible with ARV771. Furthermore, the suppression of c-Myc, a downstream effector of BET proteins, was observed by depletion rate about 50% at 100 nmol/L (Fig. 7E and G). Taken together, these results indicated that TCO-ARV771 is specifically enriched and can selectively degrade the POI in cancer cells versus non-Tz functionalized cancerous cells.
3. Conclusions
In this study, we have developed a new strategy for achieving targeted cancer therapy. Different from conventional targeting strategies, we have built unique tetrazine functionalized chemical receptors on cancer cells via efficient metabolic glycoengineering. The strategy serves as a general approach to engineering the tetrazine functionality on variable cancer cell lines, such as MDA-MB-231, HCT116, A549 and HeLa cells efficiently. The chemical receptors can be specifically targeted by TCO-caged prodrugs via rapid and bioorthogonal Tz-TCO click chemistry, enabling highly selective cancer cell imaging including multiplexed imaging. Also different from reported click chemistry-based drug targeting, the artificial receptors have an additional drug release function. We have demonstrated that the tetrazine labeled cancer cells can locally activate TCO-caged prodrugs doxorubicin and ARV771 to release active drugs with significantly improved selectivity. The works offer a promising method for anticancer therapy delivery. Further efforts are being pursued to enable selective engineering tetrazine on cancer cells, which could be used for in vivo drug targeting and delivery.
4. Experimental
4.1. Materials
Common materials or chemical reagents were purchased from commercial sources such as Sigma–Aldrich, Ambeed, and Fluka and used without further purification. AlexaFlour@647-labeled streptavidin and AlexaFlour@488-DBCO were purchased from Thermo Fisher Scientific. TCO-PEG3-Biotin was purchased from Thermo Fisher Scientific. The solvents were used by dry solvents system. All reactions were monitored by TLC or LC–MS from Shimadzu. Purification was conducted on preparative flash column chromatography and preparative reversed-phase high performance liquid chromatography (RP-HPLC) with solvent systems specified. Nuclear magnetic resonance (NMR) spectra were recorded on automated Bruker AVIII-400 instruments. High-resolution mass spectra (HRMS) were recorded on a Bruker microTOF II instrument in positive ion mode using an Agilent G1969 API-TOF with an electrospray ionization (ESI) source. Ultraperformance liquid chromatography (UPLC) spectra for compounds were acquired using a Shimadzu LabSolutions system.
4.2. Cell culture
All cells were cultured at 37 °C, 5% CO2 and were supplemented with 10% fetal bovine serum (GIBCO, catalog No. 10437) and 1% penicillin/streptomycin (Thermo Fisher, catalog No. 30-002-Cl). MDA-MB-231 breast cancer cells were cultured in Roswell Park Memorial Institute (RPMI) 1640 media (GIBCO, catalog No. 11875085). HCT-116 cells were cultured with McCoy's 5 A media (GIBCO, catalog No.16600082). A549 and HeLa cells were cultured with DMEM media (GIBCO, catalog No. 11995). Cells were passaged by aspirating the media, followed by treatment with trypsin (0.25%, Millipore-Sigma, catalog No. SM2003C) and incubation at 37 °C until cells began to detach form the plate (approximately 5 min). Cell aggregates were dispersed by pipetting and passing the cells to culture media. The hemocytometer was used to count cell number.
4.3. Western blotting
After treatment with indicated compounds, cells were washed twice with ice-cold PBS and extract protein using RIPA buffer with EDTA supplemented with Halt™ protease inhibitor cocktail (Thermo Scientific, catalog No. 87786) on ice for 30 min. The lysates were harvested in 1.5 mL Eppendorf tubes and centrifuged at 12,000 rpm at 4 °C for 20 min. The supernatant was collected, and the protein concentration was quantified using the Pierce BCA protein assay kit (Thermo Scientific, catalog No. 23227). Aliquots of proteins (15 μL) was loaded and running by electrophoresis on NuPAGE™ 4–12% Bis-Tris Mini Protein Gel (Thermo Fisher, catalog. No. NP0322BOX) followed by electrophoretic transfer onto PVDF membranes (Bio-Rad, catalog. No. 1620177). The membranes were blocked with 5% non-fat dry milk (Bio-Rad, catalog. No. 1706404), and subsequently incubated with primary rabbit anti-BRD4 monoclonal antibody and mouse anti-Myc monoclonal antibody from Cell Signaling Inc at a predetermined optimal concentration overnight at 4 °C. Mouse anti-GAPDH monoclonal antibody was used for loading control. After washing with TBST thrice (10 min each), the membranes were incubated with the secondary horse radish peroxidase (HRP)-linked antibody for 1 h at room temperature. Following washing thrice with TBST, the membranes were incubated with chemiluminescent HRP substrate. The blotting membranes were recorded using iBright™ Imaging System (Thermo Fisher). Information on all antibodies used in Western blot analysis is listed In Supporting Information Table S1. The Immunoblots were quantified using the ImageJ version 1.52a software.
4.4. Flow cytometry
Flow cytometry analysis was performed on BD FACS Canto II using 488 nm and 647 nm laser. At least 104 viable cells were analyzed from each sample. Cancer cells were seeded at a density of 105 cells/well in 12-well plates and grown in the presence of Ac4ManNTz (1) for 3 days in RPMI1640 media. Cells were then washed with PBS twice and incubate with TCO-PEG3-Biotin solution in media for 30 min at 37 °C. Afterwards cells were washed with PBS twice and incubated with AlexaFlour@647-Streptavidin (20 μg/mL) for 20 min in PBS buffer (0.1% FBS in PBS) dark at room temperature. The cells were then washed with PBS twice and treated with trypsin (100 μL) for 5 min at 37 °C. Subsequently media (300 μL) was added, cells were gently suspended and centrifuged at 1200 rpm for 5 min at 4 °C. The supernatant was carefully removed and wash the pellet twice with PBS, then re-suspended in PBS solution at 4 °C. The cell suspensions were placed in FACS tubes and kept on ice prior to analysis. The cellular debris and doublet cells were gated out by forward and side scatter. Each sample was run in triplicate at each experiment. Median fluorescence intensity was calculated, and the data were analyzed using FlowJo 10.4.2 software.
4.5. Confocal microscopy
Fluorescence images of cells were collected using a Leica SP5-II Confocal Microscope equipped with a 40, 1.3 NA oil immersion objective lens and CMOS cameras. Cells were seeded onto coverslip in a 12-well plate at a density of ∼5 × 104 cells per well and allowed to attach for 24 h Ac4ManNTz (50 μmol/L) was added, and the cells were incubated at 37 °C for 72 h. After washing with PBS, cells were incubated with TCO-Biotin (0.25 mmol/L) for 30 min. The cells were washed twice with PBS and incubated with Streptavidin-Cy5 (20 μg/mL) for 20 min. After washing, the cells were fixed with 4% paraformaldehyde solution, followed by staining of cell nuclei with DAPI. The coverslips were mounted onto microscope slides and imaged under a confocal laser scanning microscope. Image analysis was performed in ImageJ/Fiji.
4.6. Cell proliferation
Cell viability was determined using a cell counting kit (CCK-8, Dojindo Molecular Technologies, Inc.). Briefly, cells were harvested from culture dishes and seeded in 96-well plates with ∼5 × 103 cells/well at 37 °C in a humidified 5% CO2 environment and allowed to adhere overnight. Cells were exposed to medium containing different concentrations of test compounds or vehicle (n = 3). After incubation for an additional 24 h, 10% of CCK-8 in 100 μL culture medium was added to each well and incubated for 2 h. The absorbance was measured with a microplate spectrophotometer at abs = 450 nm. The experiments were performed in triplicate. The values were normalized by the controls and calculated by the Logit method.
4.7. Analysis of tetrazine labeled glycoproteins
A549 cells were seeded at a density of ∼106 cells each in 6 well-plate and allowed to attach for 12 h. Then 25 or 50 μmol/L of Ac4ManNTz (1), Ac4ManNPhTz (2), Ac4ManNPh (3) or control (DMSO) were added and incubated for 3 days. After washing with PBS, cells were incubated with TCO-PEG3-Biotin (0.25 mmol/L) at 37 °C for 30 min. Then washing with PBS, cells were harvested by incubating with lysis buffer. The supernatant protein was obtained by centrifugation at 12,000 rpm, 4 °C for 20 min. Aliquots of proteins (15 μL) were loaded and running for Western Blot. After blocked with 5% non-fat dry milk, the membrane was incubated with anti-α-biotin-HRP (1:5000, Thermo Scientific, catalog No. ENN100) in blocking buffer for 1 h. Then washed with PBST thrice (10 min each) and developed by ECL Western blotting Substrate.
4.8. Dual labeling with Ac4ManNTz and Ac4ManNAz
MDA-MB-231 cells were seeded at a density of ∼105 cells in 1-well plate and cultured with Ac4ManNTz (25 μmol/L) and Ac4ManNAz (25 μmol/L) for 3 days in RPMI 1640 medium. The cells were washed with PBS twice and incubated with DBCO-FITC488, 10 μmol/L in medium for 30 min. Cells were then washed with PBS twice and incubated with TCO-PEG3-Biotin (0.25 mmol/L) for 30 min at 37 °C. After washing twice with PBS, cells were incubated with Steptavidin-Cy5, 20 μg/mL in medium for 20 min. After washing, the cells were fixed with 4% paraformaldehyde solution, followed by staining of cell nuclei with DAPI. The coverslips were mounted onto microscope slides and imaged under a confocal microscope.
Acknowledgments
We gratefully acknowledge financial support from NIH 5R01GM130772, R. Ken Coit College of Pharmacy and Arizona Center for Drug Discovery at the University of Arizona, USA.
Footnotes
This work is dedicated to the memory of Professor Hualiang Jiang.
Peer review under the responsibility of Chinese Pharmaceutical Association and Institute of Materia Medica, Chinese Academy of Medical Sciences.
Supporting informaion
Supporting data to this article can be found online at https://doi.org/10.1016/j.apsb.2022.12.018.
Author contributions
Jing Chen and Peng Ji contributed equally to this work. Jing Chen carried out and designed the experiments and wrote the manuscript draft. Peng Ji and Giri Gnawali conceived the study, designed, and analysed the experiments, and edited the manuscript. Feng Gao, Mengyang Chang, and Hang Xu supported performing experiments, analysing data, and edited the manuscript. Wei Wang planned, designed, supervised, and directed the project, wrote, and edited the manuscript, acquisition of funding. All authors have given approval to the final version of the manuscript.
Conflicts of interest
The authors declare no conflicts of interest.
Appendix ASupplementary data
The following is the Supplementary data to this article.
References
- 1.Willmann J.K., van Bruggen N., Dinkelborg L.M., Gambhir S.S. Molecular imaging in drug development. Nat Rev Drug Discov. 2008;7:591–607. doi: 10.1038/nrd2290. [DOI] [PubMed] [Google Scholar]
- 2.Zhao Z., Ukidve A., Kim J., Mitragotri S. Targeting strategies for Tissue-specific drug delivery. Cell. 2020;181:151–167. doi: 10.1016/j.cell.2020.02.001. [DOI] [PubMed] [Google Scholar]
- 3.Manzari M.T., Shamay Y., Kiguchi H., Rosen N., Scaltriti M., Heller D.A. Targeted drug delivery strategies for precision medicines. Nat Rev Mater. 2021;6:351–370. doi: 10.1038/s41578-020-00269-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Misra A.R., Imran Vhora I. Receptor targeted drug delivery: current perspective and challenges. Ther Deliv. 2014;5:1007–1024. doi: 10.4155/tde.14.63. [DOI] [PubMed] [Google Scholar]
- 5.Srinivasarao M., Low P.S. Ligand-targeted drug delivery. Chem Rev. 2017;117:12133–12164. doi: 10.1021/acs.chemrev.7b00013. [DOI] [PubMed] [Google Scholar]
- 6.Mombaert P., Mizoguchi E., Grusby M.J., Glimcher L.H., Bhan A.K., Tonegawa S. Spontaneous development of inflammatory bowel disease in T cell receptor mutant mice. Cell. 1993;75:275–282. doi: 10.1016/0092-8674(93)80069-q. [DOI] [PubMed] [Google Scholar]
- 7.Puffenberger E.G., Hosoda K., Washington S.S., Nakao K., deWit D., Yanagisawa M., et al. A missense mutation of the endothelin-B receptor gene in multigenic hirschsprung's disease. Cell. 1994;79:1257–1266. doi: 10.1016/0092-8674(94)90016-7. [DOI] [PubMed] [Google Scholar]
- 8.Liaw D., Marsh D.J., Li J., Dahia P.L., Wang S.I., Zheng Z., et al. Germline mutations of the PTEN gene in Cowden disease, an inherited breast and thyroid cancer syndrome. Nat Genet. 1997;16:64–67. doi: 10.1038/ng0597-64. [DOI] [PubMed] [Google Scholar]
- 9.Rautio J., Meanwell N.A., Di L., Hageman M.J. The expanding role of prodrugs in contemporary drug design and development. Nat Rev Drug Discov. 2018;17:559–587. doi: 10.1038/nrd.2018.46. [DOI] [PubMed] [Google Scholar]
- 10.Drago J.Z., Modi S., Chandarlapaty S. Unlocking the potential of antibody–drug conjugates for cancer therapy. Nat Rev Clin Oncol. 2021;18:327–344. doi: 10.1038/s41571-021-00470-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu A.M., Senter P.D. Arming antibodies: prospects and challenges for immunoconjugates. Nat Biotechnol. 2005;23:1137–1146. doi: 10.1038/nbt1141. [DOI] [PubMed] [Google Scholar]
- 12.Dreher M.R., Liu W., Michelich C.R., Dewhirst M.W., Yuan F., Chilkoti A.J. Tumor vascular permeability, accumulation, and penetration of macromolecular drug carriers. J Natl Cancer Inst. 2006;98:335. doi: 10.1093/jnci/djj070. [DOI] [PubMed] [Google Scholar]
- 13.Dube D.H., Bertozzi C.R. Metabolic oligosaccharide engineering as a tool for glycobiology. Curr Opin Chem Biol. 2003;7:616–625. doi: 10.1016/j.cbpa.2003.08.006. [DOI] [PubMed] [Google Scholar]
- 14.Wang H., Mooney D.J. Metabolic glycan labelling for cancer-targeted therapy. Nat Chem. 2020;12:1102–1114. doi: 10.1038/s41557-020-00587-w. [DOI] [PubMed] [Google Scholar]
- 15.Mahal L.K., Yarema K.L., Bertozzi C.R. Engineering chemical reactivity on cell surfaces through oligosaccharide biosynthesis. Science. 1997;276:1125–1128. doi: 10.1126/science.276.5315.1125. [DOI] [PubMed] [Google Scholar]
- 16.Prescher J.A., Dube D.H., Bertozzi C.R. Chemical remodeling of cell surfaces in living animals. Nature. 2004;430:873–877. doi: 10.1038/nature02791. [DOI] [PubMed] [Google Scholar]
- 17.Wang H., Wang R., Cai K., He H., Liu Y., Yen J., et al. Selective in vivo metabolic cell-labeling-mediated cancer targeting. Nat Chem Biol. 2017;13:415–424. doi: 10.1038/nchembio.2297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li J., Chen P.R. Development and application of bond cleavage reactions in bioorthogonal chemistry. Nat Chem Biol. 2016;12:129–137. doi: 10.1038/nchembio.2024. [DOI] [PubMed] [Google Scholar]
- 19.Selvaraj R., Fox J.M. trans-Cyclooctene–a stable, voracious dienophile for bioorthogonal labeling. Curr Opin Chem Biol. 2013;17:753–760. doi: 10.1016/j.cbpa.2013.07.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ji X., Pan Z., Yu B., De La Cruz L.K., Zheng Y., Ke B., et al. Click and release: bioorthogonal approaches to ‘‘on-demand’’ activation of prodrugs. Chem Soc Rev. 2019;48:1077–1094. doi: 10.1039/c8cs00395e. [DOI] [PubMed] [Google Scholar]
- 21.Knall A.C., Slugovc C. Inverse electron demand Diels–Alder (iEDDA)-initiated conjugation: a (high) potential click chemistry scheme. Chem Soc Rev. 2013;42:5131–5142. doi: 10.1039/c3cs60049a. [DOI] [PubMed] [Google Scholar]
- 22.Devaraj N.K. The future of bioorthogonal chemistry. ACS Cent Sci. 2018;4:952–959. doi: 10.1021/acscentsci.8b00251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Blackman M.L., Royzen M., Fox J.M. Tetrazine ligation: fast bioconjugation based on inverse-electron-demand Diels–Alder reactivity. J Am Chem Soc. 2008;130:13518–13519. doi: 10.1021/ja8053805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Taylor M.T., Blackman M.L., Dmitrenko O., Fox J.M. Design and synthesis of highly reactive dienophiles for the tetrazine-trans-cyclooctene ligation. J Am Chem Soc. 2011;133:9646–9649. doi: 10.1021/ja201844c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fan X., Ge Y., Lin F., Yang Y., Zhang G., Ngai W., et al. Optimized tetrazine derivatives for rapid bioorthogonal decaging in living cells. Angew Chem Int Ed. 2016;55:14046–14050. doi: 10.1002/anie.201608009. [DOI] [PubMed] [Google Scholar]
- 26.Khan I., Seebald L.M., Robertson N.M., Yigit M.V., Royzen M. Controlled in-cell activation of RNA therapeutics using bond-cleaving bio-orthogonal chemistry. Chem Sci. 2017;8:5705–5712. doi: 10.1039/c7sc01380a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Versteegen R.M., ten Hoeve W., Rossin R., de Geus M.A.R., Janssen H.M., Robillard M.S. Click-to-release from trans-Cyclooctenes: mechanistic insights and expansion of scope from established carbamate to remarkable ether cleavage. Angew Chem Int Ed. 2018;57:10494–10499. doi: 10.1002/anie.201800402. [DOI] [PubMed] [Google Scholar]
- 28.Rossin R., Versteegen R.M., Wu J., Khasanov A., Wessels H.J., Steenbergen E.J., et al. Chemically triggered drug release from an antibody–drug conjugate leads to potent antitumour activity in mice. Nat Commun. 2018;9:1484. doi: 10.1038/s41467-018-03880-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zheng Y., Ji X., Yu B., Ji K., Gallo D., Csizmadia E., et al. Enrichment-triggered prodrug activation demonstrated through mitochondria-targeted delivery of doxorubicin and carbon monoxide. Nat Chem. 2018;10:787–794. doi: 10.1038/s41557-018-0055-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van Onzen A.H.A.M., Versteegen R.M., Hoeben F.J.M., Filot I.A.W., Rossin R., Zhu T., et al. Bioorthogonal tetrazine carbamate cleavage by highly reactive trans-cyclooctene. J Am Chem Soc. 2020;142:10955–10963. doi: 10.1021/jacs.0c00531. [DOI] [PubMed] [Google Scholar]
- 31.Keppler O.T., Horstkorte R., Pawlita M., Schmidt C., Reutter W. Biochemical engineering of the N-acyl side chain of sialic acid: biological implications. Glycobiology. 2001;11:11R–18R. doi: 10.1093/glycob/11.2.11r. [DOI] [PubMed] [Google Scholar]
- 32.Jacobs C.L., Goon S., Yarema K.J., Hinderlich S., Hang H.C., Chai D.H., et al. Substrate specificity of the sialic acid biosynthetic pathway. Biochemistry. 2001;40:12864–12874. doi: 10.1021/bi010862s. [DOI] [PubMed] [Google Scholar]
- 33.Fields S.C., Parker M.H., Erickson W.R. A simple route to unsymmetrically substituted 1,2,4,5-tetrazines. J Org Chem. 1994;59:8284–8287. [Google Scholar]
- 34.Yang J., Karver M.R., Li W., Sahu S., Devaraj N.K. Metal-catalyzed one-pot synthesis of tetrazines directly from aliphatic nitriles and hydrazine. Angew Chem Int Ed. 2012;51:5222–5225. doi: 10.1002/anie.201201117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Saxon E., Bertozzi C.R. Cell surface engineering by a modified Staudinger reaction. Science. 2000;287:2007–2010. doi: 10.1126/science.287.5460.2007. [DOI] [PubMed] [Google Scholar]
- 36.Chang P.V., Preschera J.A., Slettena E.M., Baskina J.M., Millera I.A., Agarda N.J., et al. Copper-free click chemistry in living animals. Proc Natl Acad Sci U S A. 2010;107:1821–1826. doi: 10.1073/pnas.0911116107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang S., Liu X., Bawa-Khalfe T., Lu L.S., Lyu Y.L., Liu L.F., et al. Identification of the molecular basis of doxorubicin-induced cardiotoxicity. Nat Med. 2012;18:1639–1642. doi: 10.1038/nm.2919. [DOI] [PubMed] [Google Scholar]
- 38.Lai A.C., Crews C.M. Induced protein degradation: an emerging drug discovery paradigm. Nat Rev Drug Discov. 2017;16:101–114. doi: 10.1038/nrd.2016.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sun X., Gao H., Yang Y., Ming M., Wu Y., Song Y., et al. PROTACs: great opportunities for academia and industry. Signal Transduct Targeted Ther. 2019;4:64. doi: 10.1038/s41392-019-0101-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Burslem G.M., Smith B.E., Lai A.C., Jaime-Figueroa S., McQuaid D.C., Bondeson D.P., et al. The advantages of targeted protein degradation over inhibition: an RTK case study. Cell Chem Biol. 2018;25:67–77. doi: 10.1016/j.chembiol.2017.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bondeson D.P., Smith B.E., Burslem G.M., Buhimschi A.D., Hines J., Jaime-Figueroa S., et al. Lessons in PROTAC design from selective degradation with a promiscuous warhead. Cell Chem Biol. 2018;25:78–87. doi: 10.1016/j.chembiol.2017.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Raina K., Lu J., Qian Y., Altieri M., Gordon D., Rossi A.M.K., et al. PROTAC-induced BET protein degradation as a therapy for castration-resistant prostate cancer. Proc Natl Acad Sci U S A. 2016;113:7124–7129. doi: 10.1073/pnas.1521738113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhong Y., Chi F., Wu H., Liu Y., Xie Z., Huang W., et al. Emerging targeted protein degradation tools for innovative drug discovery: from classical PROTACs to the novel and beyond. Eur J Med Chem. 2022;231 doi: 10.1016/j.ejmech.2022.114142. [DOI] [PubMed] [Google Scholar]
- 44.Teichmann E., Hecht S. Shining a light on proteolysis targeting chimeras. ACS Cent Sci. 2019;5:1645–1647. doi: 10.1021/acscentsci.9b00955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang C., He S., Zeng Z., Cheng P., Pu K. Smart nano-PROTACs reprogram tumor microenvironment for activatable photo-metabolic cancer immunotherapy. Angew Chem Int Ed. 2022;6 doi: 10.1002/anie.202114957. [DOI] [PubMed] [Google Scholar]
- 46.Cotton A.D., Nguyen D.P., Gramespacher J.A., Seiple I.B., Wells J.A. Development of antibody-based PROTACs for the degradation of the cell-surface immune checkpoint protein PD-L1. J Am Chem Soc. 2021;143:593–598. doi: 10.1021/jacs.0c10008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu J., Chen H., Liu Y., Shen Y., Meng F., Kaniskan H.U., et al. Cancer selective target degradation by folate-caged PROTACs. J Am Chem Soc. 2021;143:7380–7387. doi: 10.1021/jacs.1c00451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.He S., Gao F., Ma J., Ma H., Dong G., Sheng C. Aptamer-PROTAC conjugates (APCs) for tumor-specific targeting in breast cancer. Angew Chem Int Ed. 2021;60:23299–23305. doi: 10.1002/anie.202107347. [DOI] [PubMed] [Google Scholar]
- 49.Beck A., Goetsch L., Dumontet C., Corvaia N. Strategies and challenges for the next generation of antibody–drug conjugates. Nat Rev Drug Discov. 2017;16:315–337. doi: 10.1038/nrd.2016.268. [DOI] [PubMed] [Google Scholar]
- 50.Min J.H., Yang H., Ivan M., Gertler F., Kaelin W.G., Jr., Pavletich N.P. Structure of an HIF-1alpha -pVHL complex: hydroxyproline recognition in signaling. Science. 2002;296:1886–1889. doi: 10.1126/science.1073440. [DOI] [PubMed] [Google Scholar]
- 51.Hon W.C., Wilson M.I., Harlos K., Claridge T.D., Schofield C.J., Pugh C.W., et al. Structural basis for the recognition of hydroxyproline in HIF-1 alpha by pVHL. Nature. 2002;417:975–978. doi: 10.1038/nature00767. [DOI] [PubMed] [Google Scholar]
- 52.Buckle D.L., Van Molle I., Gareiss P.C., Tae H.S., Michel J., Noblin D.J., et al. Targeting the von Hippel-Lindau E3 ubiquitin ligase using small molecules to disrupt the VHL/HIF-1alpha interaction. J Am Chem Soc. 2012;134:4465–4468. doi: 10.1021/ja209924v. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.